ABSTRACT
Epstein-Barr virus (EBV), a member of the herpesvirus family, is the causative agent of common human infections and specific malignancies. EBV entry into target cells, including B cells and epithelial cells, requires the interaction of multiple virus-encoded glycoproteins. Glycoproteins H and L (gH/gL) cooperate with glycoprotein B (gB) to mediate fusion of the viral envelope with target cell membranes. Both the gH/gL complex and gB are required for fusion, whereas glycoprotein 42 (gp42) acts as a tropism switch and is required for B cell infection and inhibits epithelial cell infection. Our previous studies identified a prominent KGD motif located on the surface of gH/gL. In the current study, we found that this motif serves as a bifunctional domain on the surface of gH/gL that directs EBV fusion of B cells and epithelial cells. Mutation of the KGD motif to AAA decreased fusion with both epithelial and B cells and reduced the binding of gH/gL to epithelial cells and to gp42. We also demonstrate that deletion of amino acids 62 to 66 of gp42 selectively reduces binding to wild-type gH/gL, but not the KGD mutant, suggesting that the KGD motif of gH/gL interacts with the N-terminal amino acids 62 to 66 of gp42.
IMPORTANCE
Epithelial and B cells are the major targets of Epstein-Barr virus (EBV) infection in the human host. EBV utilizes different glycoprotein complexes to enter these cell types. For B cell fusion, EBV uses complexes containing gp42, gH/gL, and gB, whereas just gH/gL and gB are required for epithelial cell fusion. In the current study, a bifunctional domain consisting of a prominent KGD motif on the surface of the gH/gL structure was identified; this domain affects interactions with gp42 or epithelial receptors, ultimately dictating with which cell type virus-induced fusion can occur. These studies will lead to a better understanding of the mechanism of EBV-induced membrane fusion and herpesvirus-induced membrane fusion in general.
Introduction
Epstein-Barr virus (EBV) is one of eight known human herpesviruses and is a member of the Gammaherpesviridae subfamily (1). EBV has a high prevalence in humans, with more than 90% of the population latently infected with the virus (1). Typically, infection during childhood is asymptomatic, but infection in adolescents can result in the development of infectious mononucleosis. Virions are transmitted by saliva resulting in the infection of epithelial cells of the oral pharynx. Transmission by sexual, transfusion, and transplantation routes has also been reported (2, 3). Following transmission, the virus infects B cells and establishes latency in memory B cells where it persists indefinitely (4, 5). EBV is associated with a variety of hematopoietic cancers, such as Burkitt’s lymphoma and Hodgkin’s lymphoma. EBV is also associated with lymphoproliferative disorders in patients with immune dysfunction such as HIV/AIDS or in patients undergoing immune suppression for organ transplantation (2, 6). Entry into target cells is an essential step for EBV to cause disease. EBV entry is a complex process requiring the cooperation of multiple glycoproteins and cell surface receptors and ultimately resulting in fusion of the virion envelope either by direct fusion with the plasma membrane or following endocytosis (7, 8).
B cells and epithelial cells are two major target cells of EBV (1). However, different glycoproteins are involved in B cell and epithelial cell entry and fusion. Glycoprotein B (gB), the glycoprotein H and L (gH/gL) complex, and glycoprotein (gp42) are required for EBV fusion of B cells, while only gB and the gH/gL complex are required for EBV fusion of epithelial cells (7, 9). Cleaved secreted gp42 can trigger viral fusion with B cells in the absence of membrane-bound gp42, while virus lacking gp42 can bind to B cells but is not able to infect them (10). The initial attachment to B cells is mediated by the interaction of gp350/220 with complement receptor 2 (CR2) which is also designated CD21 (11, 12). Previous studies have shown that gp42 and gH/gL form a stable complex (13, 14). Interestingly, soluble gp42 can inhibit viral fusion with epithelial cells, which suggests that the site on gH/gL that gp42 binds is an important site for epithelial cell entry (9). Further support of a bifunctional domain on gH/gL that is required for gp42 binding to gH/gL and gH/gL-mediated epithelial cell entry results from the identification of N-terminal gp42 peptides that can block both B cell and epithelial cell entry and fusion (15).
The function of gp42 in B cell entry and fusion has been extensively studied using site-specific mutations based on the crystal structures of gp42 unbound and bound to the B cell receptor human leukocyte antigen class II (HLA class II) (16, 17). Two regions of gp42 displayed different conformations in the two structures. One is a hydrophobic pocket that is widened in response to HLA class II binding. The other is the N-terminal region, which projects outward from the C-type lectin domain in a path that is distinct from that observed in the HLA-bound structure. These observed conformational changes of gp42 following interaction with HLA class II have led to the model that a conformational change in gp42 triggers fusion required for B cell infection (7, 16).
All human herpesviruses have the gH/gL complex that is essential for membrane fusion and infection (7). gL is a chaperone protein that is essential for the correct folding and transport of gH to the cell surface (18, 19). The gH/gL complex is indispensable for epithelial cell fusion (9, 19). EBV virions lacking gH cannot attach to epithelial cells, and soluble gH/gL can bind epithelial cells (20, 21). This indicates that there is a receptor for gH/gL on epithelial cells and that interaction with the receptor is required for epithelial cell fusion. Recently, the putative epithelial cell receptor of gH/gL was identified as the integrins αvβ6 and αvβ8. Soluble forms of integrins αvβ6 and αvβ8 coprecipitate with soluble gH/gL and induce fusion with cells expressing gH/gL and gB (22). The crystal structure of the gH/gL complex was recently resolved and indicated that the gH/gL complex is composed of four domains, forming a flat elongated shape with individual domains arranged in tandem along the length of the molecule (23). Interestingly, an integrin binding motif, KGD, is found prominently on the surface of the gH/gL structure in an exposed loop in domain II (D-II) (23).
Previous studies have shown that EBV glycoprotein-mediated membrane fusion with epithelial cells can be blocked by saturating amounts of either gp42 or short gp42-derived peptides (15). However, how gp42 blocks epithelial cell fusion is still unclear. The prevailing hypothesis is that the binding of gp42 to gH/gL masks the region of gH/gL that binds to receptors on epithelial cells. With the identification of integrins as epithelial cell receptors for EBV gH/gL and identification of a KGD motif located on the surface of the gH/gL crystal structure, we hypothesize that soluble gp42 binding to the KGD motif of gH/gL prevents the integrin-dependent fusion with epithelial cells. To test this hypothesis. we constructed a gH/gL mutant in which the KGD motif was mutated to triple alanine (gH-AAA). This mutant was tested for its fusion function in epithelial and B cells and its association with gp42. After mutation of the KGD motif, fusion mediated by gH/gL was decreased in both B cells and epithelial cells with a concomitant decrease in binding to gp42 and epithelial cells. By examining the association of gH/gL and gH-AAA/gL with gp42 deletion mutants, we found that the gH/gL KGD motif interacts with amino acids 62 to 66 located within the gp42 N terminus.
RESULTS
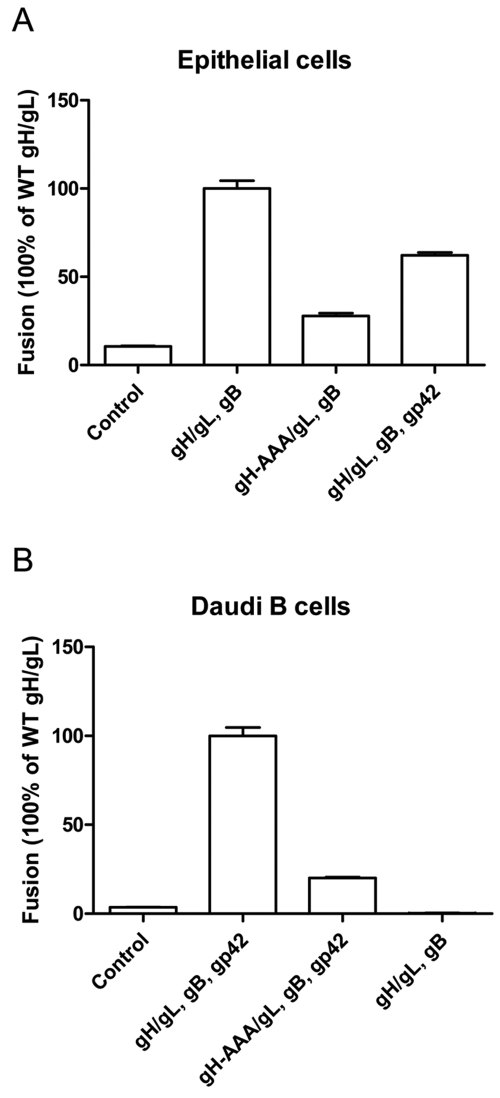
The proposed integrin binding motif KGD of EBV gH/gL is important for epithelial and B cell fusion.
To investigate whether the prominent KGD motif present on the gH/gL structure represents a bifunctional binding domain that interacts with gp42 and integrins, a triple alanine mutation (AAA) in the KGD motif (amino acids 188 to 190) of gH (gH-AAA) was constructed. We first investigated the fusion function of the gH-AAA mutant in epithelial cells. Effector CHO-K1 cells were transfected with a luciferase reporter construct under the control of a T7 promoter (T7-luciferase) and the glycoproteins essential for epithelial cell-cell fusion, gB and gH/gL or gH-AAA/gL as indicated in Fig. 1. The effector cells were then overlaid with human embryonic kidney 293 (HEK 293) target cells stably expressing T7 polymerase, and fusion was monitored by luciferase expression. HEK 293 cells were used as epithelial target cells since they fuse with effector cells in the absence of gp42 (24). We found that the gH-AAA/gL mutant had decreased fusion efficiency with HEK 293 cells (Fig. 1A), which is compatible with the putative role of the KGD motif of gH/gL interacting with the integrins αvβ6 and αvβ8 required for epithelial cell entry (22, 25). Along with these experiments, we confirmed the inhibitory effect of gp42 on epithelial cell-cell fusion (15, 26). In the presence of gp42, fusion with epithelial cells decreased about 40% (Fig. 1A).
FIG 1 .
The KGD motif of gH is required for EBV fusion with epithelial and Daudi B cells. CHO-K1 cells were transiently transfected with the T7 luciferase plasmid alone (control) or with the T7 luciferase plasmid, EBV gB, gL, together with gH or gH-AAA mutant with or without gp42, as indicated. (A and B) Transfected CHO-K1 cells were overlaid with epithelial cells expressing T7 polymerase (A) or Daudi B cells expressing T7 polymerase (B). Luciferase activity was normalized to cells with wild-type (WT) gH/gL levels, which was set at 100%. The data are means plus standard deviations (SD) (error bars) from three independent experiments.
Daudi is a human B cell lymphoma cell line for which fusion with effector cells is dependent on gp42 (27, 28). For Daudi B cell fusion, gp42 was transfected in addition to gH/gL and gB into the CHO-K1 cells. Interestingly, the gH-AAA mutation also decreased fusion activity with Daudi B cells by about 80% (Fig. 1B), suggesting a role for this motif in binding gp42 required for B cell fusion. CHO-K1 cells without gp42 (Fig. 1B, rightmost bar) show no fusion activity, which indicates the requirement of gp42 for Daudi B cell fusion.
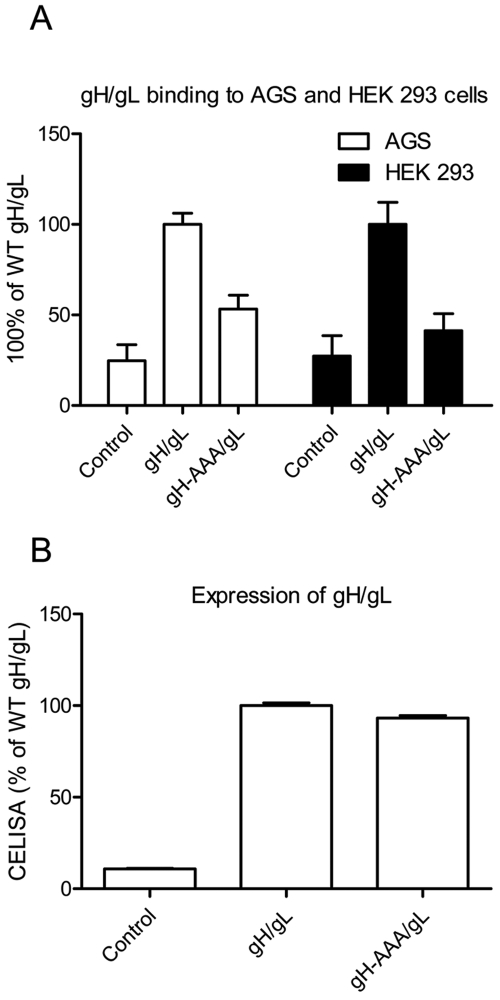
Mutation of the gH/gL KGD motif results in reduced binding of gH-AAA/gL to epithelial cells and gp42.
To determine why the KGD motif was important for both epithelial cell and B cell fusion, we compared the binding of wild-type (wt) gH/gL and gH-AAA/gL to AGS cells and HEK 293 cells. Both AGS cells and HEK 293 cells have been widely used in EBV epithelial cell-cell fusion and entry assays (22, 26). We transfected Chinese hamster ovary CHO-K1 cells with control plasmid, wt gH/gL, or gH-AAA/gL, lysed the cells by freeze-thawing, and collected the supernatants containing solubilized gH/gL. The supernatants were overlaid on AGS or HEK 293 cells, and gH/gL binding to the cells was detected by cell enzyme-linked immunosorbent assay (CELISA). The mutant gH-AAA/gL had decreased binding to AGS and HEK 293 cells compared with the wt gH/gL (Fig. 2A). To exclude the possibility that the gH-AAA/gL mutant was expressed at lower levels than wt gH/gL, we verified the cell surface expression of wt or mutant gH/gL prior to preparing the cell lysates and found the expression levels comparable (Fig. 2B). These data indicate that the gH-AAA/gL has reduced binding to epithelial cells compared to wt gH/gL and that this decreased cell surface binding correlates with the decreased epithelial cell fusion activity.
FIG 2 .
gH binding with AGS and HEK 293 cells was decreased in the gH-AAA/gL mutant. CHO-K1 cells were transiently transfected with control, gH/gL, or gH AAA/gL. (A) Soluble gH/gL and gH-AAA-gL were prepared as described in Materials and Methods and used to overlay with AGS cells or HEK 293 cells in 96 wells in triplicate. After incubation for 4 hours at 4°C, binding was examined by CELISA. (B) In parallel, the transfected cells were examined for gH/gL and gH-AAA/gL expression and were found to be similar. Twenty-four hours posttransfection, 4 × 104 cells were seeded into 96-well plates in triplicate. CELISA was performed with anti-gH/gL antibody (Ab) (E1D1). The data are means plus SD from three independent experiments.
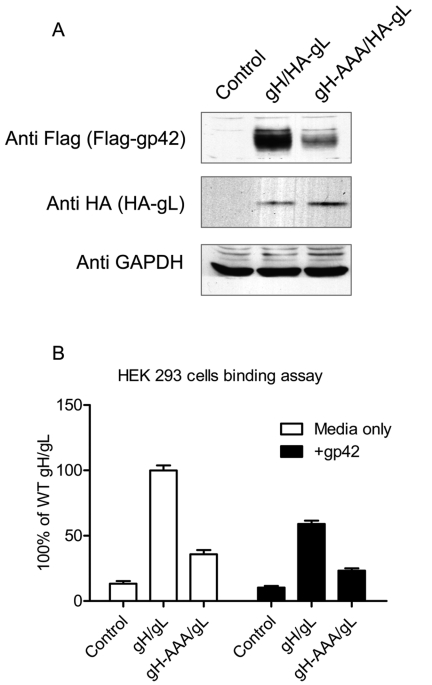
Previous studies have shown that the association of gp42 with gH/gL is crucial for B cell fusion and the essential binding site on gp42 for gH/gL is contained within the gp42 amino terminus (14, 15). Since the gH-AAA/gL mutant also exhibited decreased fusion activity for B cells (Fig. 1B), we examined the binding of a Flag-tagged soluble form of gp42 (29) to wt gH/gL and gH-AAA/gL expressed on the cell surfaces of CHO-K1 cells. Supernatants containing soluble Flag-tagged gp42 were applied to cells transfected with gH/gL, the cells were extensively washed, and gp42 binding was determined by Western blotting. Despite observing equal amounts of gL and controlling for equivalent total protein loading by monitoring glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Fig. 3A, middle and bottom panels), we found that cells expressing the gH-AAA/gL mutant had significantly decreased gp42 binding (Fig. 3A, top panel), indicating that the KGD motif is also important for the binding of gp42 with gH/gL.
FIG 3 .
The KGD motif is involved in interaction of gH/gL with gp42. (A) The gH-AAA/gL mutant has decreased association with gp42. CHO-K1 cells seeded in six-well plates were transfected with control, gH/HA-gL, or mutant gH-AAA/HA-gL. Twenty-four hours posttransfection, the cells were washed twice with ice-cold PBS and incubated with media containing Flag-tagged gp42 (isolated 24 h after transfection) for 1 h at 4°C. The cells were then washed with ice-cold PBS four times and lysed with 200 µl of 1× SDS lysis buffer. Proteins were separated on 10% SDS-polyacrylamide gels. Western blot analyses were performed using the polyclonal anti-Flag antibody (F7425) at 1:1,000 to detect the Flag-gp42 that bound to the transfected cells. Anti-GAPDH and anti-HA (gL-HA) were used as loading controls. The data are representative of three independent experiments. (B) Soluble gp42 can decrease the binding of gH/gL with epithelial cells. CHO-K1 cells were transiently transfected with control, gH/gL, or gH-AAA/gL. Twenty-four hours posttransfection, 2 × 106 cells were collected, and soluble gH/gL or gH-AAA/gL was prepared. Soluble gp42 was added as described in Materials and Methods. Binding of gH/L or gH-AAA/gL to HEK 293 cells was measured following overlay of HEK 293 cells with soluble gH/gL or gH-AAA/gL with or without soluble gp42 for 4 h at 4°C followed by detection of gH binding by CELISA with gH/gL antibody E1D1. The data are means plus SD from three independent experiments.
To confirm that the binding site on gH/gL is also the binding site for gp42, we examined whether soluble gp42 blocks the binding of gH/gL or gH-AAA/gL to epithelial cells. Supernatants containing solubilized gH/gL or gH-AAA/gL were preincubated with media only or media containing soluble gp42 for 1 h at 4°C and then overlaid onto HEK 293 cells. CELISA was then used to detect gH/gL or gH-AAA/gL binding to the HEK 293 cells. The results in Fig. 3B demonstrate that soluble gp42 decreased gH/gL binding to epithelial cells nearly 50% compared with media only. Soluble gp42 also further decreased the binding of gH-AAA/gL to epithelial cells approximately 40% of gH-AAA/gL levels and approximately 25% of wild-type gH/gL levels. These observations indicate that the gH/gL KGD motif constitutes an important functional domain of gH/gL that directs binding to gp42.
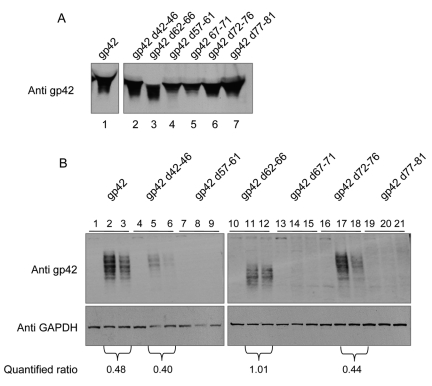
gp42 residues contained within amino acids 62 to 66 rely on the gH KGD motif to bind gH/gL.
In previous studies, we mapped the amino-terminal residues of gp42 that bind gH/gL (14, 15). From these studies, two contiguous regions of the N terminus of gp42 from approximately amino acids 36 to 61 and 67 to 81 were found to directly bind to gH/gL. A short stretch of amino acids (amino acids 62 to 66), which separated these two regions, was found to contribute to the high-affinity binding of gp42 to gH/gL. In order to further delineate the region of gp42 that binds the gH/gL KGD motif, we chose a group of mutants from previously constructed 5-amino-acid deletion mutants within the gp42 amino terminus. We were most interested in testing mutants that still bound gH/gL (mutant in which amino acids 42 to 46 were deleted [d42-46], d62-66, and d72-76) to determine whether there was any change in the binding of the gp42 mutants to the gH-AAA/gL mutant. We also chose mutants that we had previously shown did not bind gH/gL (d57-61, d67-71, and d77-81) as controls (14, 15). The mutants were first expressed in CHO-K1 cells, supernatants containing soluble gp42 were collected, and expression of each mutant and wild-type gp42 was found to be similar by SDS gel electrophoresis (Fig. 4A). The medium supernatants were then used in a monolayer cell binding assay to determine binding of each mutant to control transfected cells, wt gH/gL-transfected cells, or gH-AAA/gL mutant transfected cells (Fig. 4B). In all cases, there was no binding of wt gp42 or the gp42 mutants to control transfected cells (Fig. 4B, lanes 1, 4, 7, 10, 13, 16, and 19). In addition, we found that d57-61, d67-71, and d77-81 did not associate with wt gH/gL or mutant gH-AAA/gL (Fig. 4B, lanes 8, 9, 14 and 15, and 20 and 21) consistent with the previous CELISA studies with soluble gH/gL (15). Interestingly, the d42-46 and d72-76 mutants still associated with wt gH/gL, but less gp42 was present in the gH-AAA/gL mutant compared to wt gp42, suggesting that these two gp42 mutants bind with lower affinity to the gH-AAA/gL mutant (Fig. 4B, lanes 5 and 6 and lanes 17 and 18). The most interesting gp42 deletion mutant was the gp42 d62-66 mutant (Fig. 4B, lanes 11 and 12). As described above, previous studies had suggested that this region constitutes a linker separating the two domains of gp42 that are important for high-affinity binding to gH/gL. This mutant had a nearly identical binding profile for wt gH/gL and the gH-AAA/gL mutant in contrast to wt gp42 and the gp42 mutants d42-46 and d72-76 in which a reduction of gp42 binding was observed for gH-AAA/gL compared with gH/gL. The quantified ratio of the associated gp42 d62-66 mutant with gH-AAA/gL compared with gH/gL was approximately 1, and the quantified ratio for associated gp42, gp42 d42-46, or gp42 d72-76 with gH-AAA/gL compared with gH/gL was less than 0.5 (Fig. 4B). The d62-66 mutant selectively reduces binding to wild-type gH/gL to comparable levels of the gH-AAA/gL mutant. This suggests that the linker deletion mutant in gp42 and the AAA mutation in gH/gL have similar effects in reducing the gp42 interaction with gH/gL and that these two mutations are not additive. These sites in gH/gL and gp42 may either interact directly or propagate their binding effects through a common, nearby site on gH/gL. These results indicate that the KGD motif relies on amino acids 62 to 66 of gp42 for binding to gH/gL. Compatible with observed binding, this mutant was reduced in fusion function with B cells (approximately 50% of wt gp42) and exhibited reduced binding to gH/gL in immunoprecipitation and in a cell-based binding assay (15).
FIG 4 .
Amino acids 62 to 66 of gp42 are important for gp42 interaction with the KGD motif of gH/gL. CHO-K1 cells seeded in six-well plates were transfected with control (lanes 1, 4, 7, 10, 13, 16, and 19 in panel B), gH/gL (lanes 2, 5, 8, 11, 14, 17, and 20 in panel B), or gH-AAA/gL (lanes 3, 6, 9, 12, 15, 18, and 21 in panel B). Twenty-four hours after transfection, the cells were washed twice with ice-cold PBS and incubated with media containing gp42 or gp42 deletion mutants as indicated in panel A, lanes 1 to 7 (isolated after 24 h transfection) for 1 h at 4°C. (B) The cells were then washed with ice-cold PBS four times and lysed with 200 µl of 1× SDS lysis buffer. 2× SDS lysis buffer was added to the media containing gp42 or gp42 deletion mutants. Samples were separated on 10% SDS-polyacrylamide gels. Western blot analyses were performed using the polyclonal anti-gp42 antibody (pb1112) at 1:1,000 dilution. The gp42 d62-66 (amino acids 62 to 66 deleted) mutant is deleted for two potential N-linked glycosylation sites, which results in a change in gp42 migration as previously described (28). GAPDH was used as the loading control. The data are representative of three independent experiments. Associated gp42 or mutant gp42 with gH/gL or gH-AAA/gL was quantified and normalized against GAPDH loading control using Quantity One software (version 4.3.1; Bio-Rad). The quantified data were expressed as the average of three independent experiments as indicated underneath the blot (ratio of the amount of associated gp42 with gH-AAA/gL to the amount of associated gp42 with gH/gL).
The gp42 d62-66 mutant is impaired in blocking epithelial cell fusion.
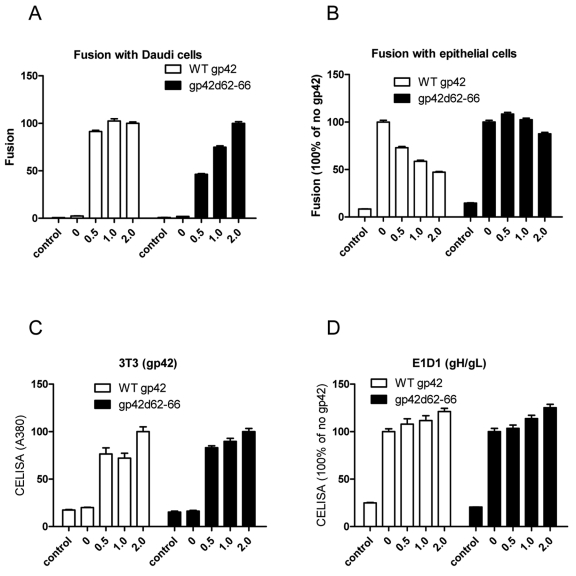
To further investigate whether amino acids 62 to 66 of gp42 may bind the gH/gL KGD motif, we examined whether this mutant was impaired in blocking HEK 293 cell fusion (Fig. 5). This might be expected if the d62-66 deletion mutant fails to bind the KGD motif of gH and makes the KGD motif of gH/gL available to bind the integrin receptor on epithelial cells required for EBV epithelial fusion. For this experiment, we monitored HEK 293 and Daudi cell fusion following transfection of increasing amounts of wt gp42 and the gp42 d62-66 mutant. As expected, for Daudi cells, there was in general an increase in cell fusion with increasing amounts of gp42 or gp42 d62-66, and the levels of fusion with the gp42 d62-66 mutant were lower than for the wt gp42, similar to what was previously published (Fig. 5A) (15). In contrast, wt gp42 but not gp42 d62-66 had an inhibitory effect on epithelial cell fusion (Fig. 5B). We also checked the cell surface expression of gp42, gp42 d62-66, and gH/gL in the transfected cells used for cell fusion shown in Fig 5A and B and as expected found a modest increase in the expression of gp42, gp42 d62-66, and gH/gL when increasing amounts of DNA were transfected (Fig. 5C and D). Thus, in contrast to wt gp42, the gp42 d62-66 mutant does not inhibit epithelial cell fusion, since it lacks a key gp42 domain important for binding the gH/gL KGD motif.
FIG 5 .
The 62-66 motif of gp42 is essential for inhibiting EBV fusion with epithelial cells. CHO-K1 cells were transiently transfected with T7 luciferase plasmid alone (control) or T7 luciferase plasmid, EBV gB, gH, and gL together with increasing amounts (in micrograms) of gp42 or gp42 d62-66. (A and B) Transfected CHO-K1 cells were overlaid with Daudi B cells (A) or epithelial cells (B), and luciferase activity was measured. (C and D) Expression of gp42, gp42 d62-66, and gH/gL. After 24 h of transfection, 4 × 104 cells were seeded into 96-well plates in triplicate. CELISA was performed with anti-gp42 Ab 3T3 (C) or with anti-gH/gL Ab (E1D1) (D). The values are shown as percentages of the value for 2.0 μg or no gp42 as shown on the y axes. The data are representative of three independent experiments.
DISCUSSION
In the current study, we investigated the role of the gH/gL KGD motif implicated in integrin and gp42 binding. The motif is located in a prominent loop on the surface of the gH/gL structure and is contained in D–II near the domain I/domain II (D-I/D-II) linker α-helix (2α-1) and maps near the D–I/D–II interface. Previous mutations in both gH and gL, including gH residue 74, which maps to a hydrophobic groove adjacent to the KGD motif, altered the activity of gH/gL, suggesting a role for this region in EBV glycoprotein entry function (23, 30). The gH/gL KGD motif has been hypothesized to be important for the interaction of gH/gL with αvβ6 and αvβ8 integrins triggering fusion of epithelial cells (22). The observation that gp42 blocks EBV entry into epithelial cells (9, 15, 31) suggests that the KGD motif and/or the area surrounding it may also be important for gp42 binding to gH/gL. It has been suggested that gp42 inhibits epithelial fusion by occluding the epithelial receptor binding domain of gH/gL (9, 13, 15). The importance of gp42 binding to gH/gL for B cell entry is evident, since previous studies have shown that mutations in gp42 that block gp42 binding to gH/gL block EBV-induced B cell membrane fusion (15). To further investigate the role of the KGD motif in membrane fusion, we mutated the KGD motif by constructing a triple alanine mutant and found that this region was important for epithelial cell fusion and also important for B cell fusion. A mutant in the KGD motif of gH/gL exhibited reduced binding to epithelial cells and bound poorly to gp42. In addition, we found by analyzing a group of gp42 N-terminal deletion mutants, that amino acids 62 to 66 contribute to the binding of gp42 to gH/gL, likely by specifically interacting with the gH/gL KGD motif.
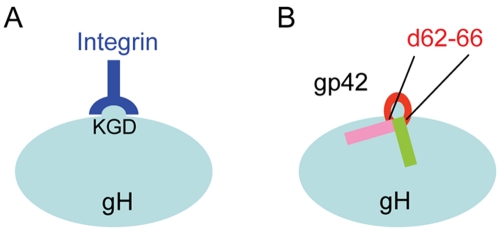
Our current studies suggest that the KGD motif of gH/gL is a bifunctional domain that mediates EBV fusion of epithelial cells and B cells through interactions with gp42 and the EBV epithelial cell receptor. The interaction of gp42 with gH/gL is complex, requiring two domains within the gp42 amino terminus (approximately amino acids 36 to 61 and 67 to 81) separated by a short linker region from amino acids 62 to 66 (14, 15). The gp42 amino terminus containing this region was disordered in the two gp42 structures representing gp42 unbound and bound to the gp42 receptor HLA class II (16, 17). Previous studies, both with deletion mutagenesis and peptides from the gp42 amino terminus, have suggested that the linker region is not important for specific interactions with gH/gL, but rather link the two gp42 gH/gL binding domains (14, 15). Interestingly, when the mutation of the KGD motif was tested by comparing the gp42 d62-66 mutant to the wt gH/gL, there was no difference in the binding of this gp42 mutant to the gH-AAA/gL mutant compared to wt gH/gL, in which a reduction in binding was observed. These results indicate that the KGD motif at least plays some part in binding amino acids 62 to 66 of gp42. The binding of this linker region to gH/gL may be mediated by specific side chain interactions or structural features at or around the KGD motif found on the gH/gL surface. In the gH/gL structure, there is a large groove located between the D-I and D-II domains that we have hypothesized may be in part the binding site for gp42. Thus, the adjacent location of the KGD motif to this groove and the observation that the KGD motif plays a bifunctional role in epithelial cell fusion and B cell fusion makes this hypothesis attractive. For the interaction of gH/gL with the αvβ6 and αvβ8 integrin epithelial receptors, the presence of the KGD motif provides a mechanism for gH/gL to specifically bind to integrins as previously shown (22, 25). RGD or KGD motifs are typical recognition sequences for integrins, while adjacent amino acids can increase affinity (32, 33). Structural and functional studies have shown that the interaction of integrins with RGD motif proteins (or peptides) depends primarily on electrostatic, specific amino acid side chain interactions, and hydrogen bonds (34). Other studies have shown that an RGD motif, found in herpes simplex virus type 1 (HSV-1) gH/gL, binds to αvβ3 integrins. Mutating the RGD motif of HSV-1 gH/gL abolished gH/gL binding to Vero cells (35, 36). Previous mutations in foot-and-mouth disease virus of this motif also block function (37), compatible with the observation of loss of function for epithelial fusion and cell binding that we observed for epithelial fusion in the gH-AAA/gL mutant.
One of the most interesting observations in the current study is the finding that this same KGD motif plays an important role in B cell fusion and gp42 binding. The observation that gp42, in part, depends on the KGD motif for gH binding and function in fusion is not surprising, since previous studies had suggested that domains required for B cell and epithelial cell fusion and entry overlap. How the KGD motif specifically interacts with the gp42 amino acids 62 to 66 will require additional studies. Previous studies demonstrated that gp42- and gp42-derived peptides can block epithelial and B cell fusion and entry presumably by blocking the interaction of gH/gL with integrins for epithelial fusion and gp42 for B cell fusion. Similarly, our current results indicate that the addition of soluble gp42 can inhibit the binding of gH/gL to epithelial cells, indicating that the gH/gL domain for gp42 binding and epithelial cell binding overlaps. Studies with gp42 peptides have shown that deletion of the linker domain from a gp42 peptide spanning amino acids 36 to 81 does not greatly alter the affinity of the gp42 peptide for gH/gL (14). There was only a modest change (less than 2-fold) in the affinity of the linker-deleted peptide compared to the full-length peptide as determined by a fluorescence polarization (FP) assay (14). Inhibition of B cell fusion is reduced with the linker-deleted peptide in contrast to the inhibition of epithelial cell fusion which is high for both the full-length and linker-deleted peptides, indicating that the site on gH/gL required for epithelial fusion does not exactly overlap the site required for B cell fusion and presumably gp42 binding. Since our previous studies had shown that the interaction of gp42 with gH/gL is complex, requiring multiple interactions extending throughout gp42 36-81 peptide (gp42 peptide from amino acids 36 to 81), our studies using a smaller gp42 peptide may be more informative for understanding the results of our current study (14). A smaller gp42 peptide, spanning amino acids 47 to 81, binds with nearly the same affinity as the affinity of the 36-81 peptide and displays similar inhibition characteristics of B cell and epithelial cell fusion. Interestingly, when the linker is deleted from this smaller peptide, there is a greater than 70-fold reduction in affinity for gH/gL in FP and this peptide does not inhibit B cell or epithelial fusion. Further studies with two double alanine mutations (at positions 62 and 63 [62-63] and positions 64 and 65) within the linker region of the 47-81 peptide modestly altered the affinity (4-fold reduction) or modestly enhanced the affinity (2-fold) of the peptide for gH/gL, respectively (14). The peptide with enhanced affinity for gH/gL had little change in the inhibition of epithelial and B cell fusion compared to the control peptide. Interestingly, inhibition of B cell and epithelial fusion was markedly reduced for the 62-63 double alanine mutant peptide compared to the control peptide (14). These results are supportive of a role of the gp42 linker in binding to gH/gL and interfering with gH/gL-mediated induction of fusion and for binding of gp42 required for B cell fusion. Our model of the interactions between gp42 and gH/gL is shown in Fig. 6. Further studies will need to be done to clarify the interactions between gp42 and gp42 peptides with gH/gL. In particular, cocrystallization of gp42 or gp42 peptides with gH/gL would precisely map the relevant interactions and are currently ongoing. In conclusion, we identified that the conserved KGD motif in gH/gL of EBV can interact with a small region (residues 62 to 66) of gp42. The interaction is crucial for EBV fusion with B cells triggered by binding of gp42 to HLA. The KGD motif is also important for the epithelial cell fusion triggered by binding of gH/gL with epithelial cell receptors such as integrins.
FIG 6 .
Model of the bifunctional KGD motif of gH/gL. (A) In epithelial cell fusion, the KGD motif binds to integrin on epithelial cells. (B) In B cell fusion, the KGD motif binds to gp42, which is in part dependent on gp42 amino acids 62 to 66. Soluble wild-type gp42 can inhibit epithelial cell fusion, since when gp42 is bound to gH/gL, the gH/gL KGD motif is occluded, preventing binding to the epithelial receptor.
MATERIALS AND METHODS
Cell culture.
Chinese hamster ovary cells (CHO-K1) and AGS cells were grown in Ham’s F-12 medium (BioWhittaker) containing 10% FetalPlex animal serum complex (Gemini Bio-Products) and 1% penicillin-streptomycin (100 U penicillin/ml, 100 µg streptomycin/ml; BioWhittaker). The Daudi 29 cell line (for B cell fusion) and human embryonic kidney 293 (HEK 293) cells (for epithelial cell fusion) stably expressing T7 RNA polymerase (28, 30) were grown in RPMI 1640 medium with 900 µg/ml G418 (Sigma) and Dulbecco modified Eagle medium (DMEM) with zeocin, respectively, containing 10% FetalPlex animal serum complex and 1% penicillin-streptomycin.
Construction.
Mutation of the gH KGD motif (amino acids 188 to 190) to AAA in full-length gH was generated by using a QuikChange site-directed mutagenesis kit (Stratagene). The following primers were used: forward primer (5′-AAGCGAGTGACCGAGGCCGCCGCCGAGCATGTGTTGAGCCTG-3′) and reverse primer (5′-CAGGCTCAACACATGCTCGGCGGCGGCCTCGGTCACTCGCTT-3′). Sequencing was done to confirm the presence of the AAA mutation and the absence of any other mutations.
Transfection.
CHO-K1 cells at 80% confluence were transiently transfected with the mutants and other glycoproteins essential for fusion, including gB (0.8 µg), gH (0.5 µg), gL (0.5 µg), gp42 (0.8 µg), and a luciferase reporter plasmid with a T7 promoter (0.8 µg) by using Lipofectamine 2000 transfection reagent (Invitrogen) in Opti-MEM (Gibco), as previously described (38). Equal amounts of gp42 and gp42 mutants or equal amounts of gH/gL and gH-AAA mutant were used in each experiment.
Solubilized gH binding assays.
CHO-K1 cells were transfected with gH/gL or gH-AAA/gL as described above. Twenty-four hours later, 2 × 106 cells were collected, resuspended in 1 ml of media, subjected to 3 cycles of freeze-thawing in liquid nitrogen, and sonicated for 10 s. Insoluble cellular debris was removed by centrifugation at 1,500 × g for 5 min. AGS cells or HEK 293 cells were overlaid with clarified supernatants (100 µl) in a 96-well dish in triplicate. After incubation for 4 h at 4°C, gH/gL binding was examined in a cell enzyme-linked immunosorbent assay (CELISA) as described below. For gp42 inhibition assays, soluble gp42 was added to the clarified supernatants prior to overlaying with HEK 293 cells.
Fusion assay.
The virus-free cell-based fusion assay was performed as described previously (24). Briefly, effector CHO-K1 cells were transfected as described above. Twenty-four hours after transfection, the cells were detached, counted with a Beckman Coulter Z1 particle counter and mixed 1:1 with target cells (Daudi 29 B cells or HEK 293 cells, 0.25 × 106 per sample) into a 24-well plate in 1 ml Ham’s F-12 medium. Twenty-four hours later, the cells were washed twice with phosphate-buffered saline (PBS) and lysed with 100 µl of passive lysis buffer (Promega). Luciferase was quantified in duplicate by transferring 20 µl of lysed cells to a 96-well plate and adding 100 µl of luciferase assay reagent (Promega), and luminescence was measured on a Perkin-Elmer Victor plate reader.
CELISA.
The cell surface expression or cell binding of various mutants was determined as described in previous reports (38). CHO-K1 cells were transfected with various glycoproteins and split for use in fusion assays (described above) and CELISA. Twenty-four hours after transfection, 4 × 104 cells/well were transferred to a 96-well plate and incubated for another 24 h. The expression of each glycoprotein was evaluated using conformation-specific antibodies 3H3 for gp42 and E1D1 for gH/gL. After incubation with primary antibody for 30 min and fixation with 2% formaldehyde and 0.2% glutaraldehyde in PBS for 15 min, biotin-labeled secondary antibody was added at a dilution of 1:500 for 30 min. After the cells were washed, streptavidin-labeled horseradish peroxidase (1:20,000) was incubated with the fixed cells for 30 min. Peroxidase substrate was added, and the amount of cell surface staining was determined by measurement at 380 nm with Perkin-Elmer Victor plate reader.
Monolayer binding assay.
CHO-K1 cells at 80% confluence were transfected with wild-type Flag-tagged gp42 (Flag-gp42) (29) in three wells. Another three wells were transfected with gH/gL, mutant gH-AAA/gL, or control plasmid using Lipofectamine 2000, as described above. Twenty-four hours after transfection, the supernatants of the cells transfected with Flag-gp42 were collected and centrifuged to collect the soluble gp42. The cells transfected with control, gH/gL, or gH-AAA/gL were washed twice with ice-cold PBS and incubated with Flag-gp42 for 1 h at 4°C. The cells were then washed with ice-cold PBS four times and lysed with 200 µl of SDS sample buffer. Proteins were separated on 10% SDS-polyacrylamide gels after boiling for 10 min under reducing conditions. Western blot analyses were performed using the polyclonal anti-Flag antibody (F7425; Sigma) diluted 1:1,000, polyclonal anti-gp42 antibody serum (PB1112), polyclonal anti-hemagglutinin (anti-HA) (Santa Cruz) at 1:1,000, or polyclonal anti-GAPDH (Abcam) antibody at 1:1,000.
Western blotting.
Protein samples were loaded on a 10% SDS-polyacrylamide gel. After electrophoresis, proteins were transferred to nitrocellulose membranes (Schleicher & Schuell, Keene, NH). The blots were blocked with 5% nonfat dry milk in TBS/T buffer (20 mM Tris-HCl [pH 7.6], 137 mM NaCl, 0.1% Tween 20) for 2 h at room temperature (RT). The blots were washed with TBS/T buffer and incubated with primary antibodies overnight at 4°C. Anti-rabbit (Bio-Rad, Hercules, CA) peroxidase-conjugated secondary antibodies were added to the membranes at a dilution ratio of 1:3,000, and incubation was continued for 1 h at RT. The protein bands on the membrane were visualized by chemiluminescence (Pierce, Rockford, IL) and were measured by densitometry using Quantity One software (version 4.3.1; Bio-Rad).
ACKNOWLEDGMENTS
This research was supported by grants AI076183 (R.L. and T.S.J.) and AI067048 (R.L.) from the National Institute of Allergy and Infectious Diseases and by grants CA117794 (R.L. and T.S.J.) and CA133063 (R.L. and C.L.R.) from the National Cancer Institute, and by postdoctoral fellowship 12POST9380013 from American Heart Association, Midwest Affiliate.
We appreciate the help and advice from members of the Jardetzky and Longnecker laboratories, particularly Sarah Connolly, for the completion of these studies. We thank Lindsey Hutt-Fletcher for kindly providing monoclonal antibodies used in these studies.
Footnotes
Citation Chen J, Rowe CL, Jardetzky TS, and Longnecker R. 2012. The KGD motif of Epstein-Barr virus gH/gL is bifunctional, orchestrating infection of B cells and epithelial cells. mBio 3(1):e00290-11. doi:10.1128/mBio.00290-11.
REFERENCES
- 1. Rickinson AB, Kieff E. 2007. Epstein-Barrvirus, p 2657–2701 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2. Crawford DH. 2001. Biology and disease associations of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:461–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Macsween KF, Crawford DH. 2003. Epstein-Barr virus—recent advances. Lancet Infect. Dis. 3:131–140 [DOI] [PubMed] [Google Scholar]
- 4. Babcock GJ, Decker LL, Volk M, Thorley-Lawson DA. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395–404 [DOI] [PubMed] [Google Scholar]
- 5. Thorley-Lawson DA, Babcock GJ. 1999. A model for persistent infection with Epstein-Barr virus: the stealth virus of human B cells. Life Sci. 65:1433–1453 [DOI] [PubMed] [Google Scholar]
- 6. Ambinder RF. 2001. Epstein-Barr virus associated lymphoproliferations in the AIDS setting. Eur. J. Cancer 37:1209–1216 [DOI] [PubMed] [Google Scholar]
- 7. Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. 2011. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat. Rev. Microbiol. 9:369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hutt-Fletcher LM. 2007. Epstein-Barr virus entry. J. Virol. 81:7825–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang X, Kenyon WJ, Li Q, Müllberg J, Hutt-Fletcher LM. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72:5552–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang X, Hutt-Fletcher LM. 1998. Epstein-Barr virus lacking glycoprotein gp42 can bind to B cells but is not able to infect. J. Virol. 72:158–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nemerow GR, Mold C, Schwend VK, Tollefson V, Cooper NR. 1987. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J. Virol. 61:1416–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tanner J, Whang Y, Sample J, Sears A, Kieff E. 1988. Soluble gp350/220 and deletion mutant glycoproteins block Epstein-Barr virus adsorption to lymphocytes. J. Virol. 62:4452–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kirschner AN, Omerovic J, Popov B, Longnecker R, Jardetzky TS. 2006. Soluble Epstein-Barr virus glycoproteins gH, gL, and gp42 form a 1:1:1 stable complex that acts like soluble gp42 in B-cell fusion but not in epithelial cell fusion. J. Virol. 80:9444–9454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu F, Marquardt G, Kirschner AN, Longnecker R, Jardetzky TS. 2010. Mapping the N-terminal residues of Epstein-Barr virus gp42 that bind gH/gL by using fluorescence polarization and cell-based fusion assays. J. Virol. 84:10375–10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kirschner AN, Lowrey AS, Longnecker R, Jardetzky TS. 2007. Binding site interactions between Epstein-Barr virus fusion proteins gp42 and gH/gL reveal a peptide that inhibits both epithelial and B-cell membrane fusion. J. Virol. 81:9216–9229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kirschner AN, Sorem J, Longnecker R, Jardetzky TS. 2009. Structure of Epstein-Barr virus glycoprotein 42 suggests a mechanism for triggering receptor-activated virus entry. Structure 17:223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mullen MM, Haan KM, Longnecker R, Jardetzky TS. 2002. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol. Cell 9:375–385 [DOI] [PubMed] [Google Scholar]
- 18. Hutchinson L, et al. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66:2240–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Q, Buranathai C, Grose C, Hutt-Fletcher LM. 1997. Chaperone functions common to nonhomologous Epstein-Barr virus gL and varicella-zoster virus gL proteins. J. Virol. 71:1667–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Borza CM, Morgan AJ, Turk SM, Hutt-Fletcher LM. 2004. Use of gHgL for attachment of Epstein-Barr virus to epithelial cells compromises infection. J. Virol. 78:5007–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oda T, Imai S, Chiba S, Takada K. 2000. Epstein-Barr virus lacking glycoprotein gp85 cannot infect B cells and epithelial cells. Virology 276:52–58 [DOI] [PubMed] [Google Scholar]
- 22. Chesnokova LS, Nishimura SL, Hutt-Fletcher LM. 2009. Fusion of epithelial cells by Epstein-Barr virus proteins is triggered by binding of viral glycoproteins gHgL to integrins alphavbeta6 or alphavbeta8. Proc. Natl. Acad. Sci. U. S. A. 106:20464–20469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matsuura H, Kirschner AN, Longnecker R, Jardetzky TS. 2010. Crystal structure of the Epstein-Barr virus (EBV) glycoprotein H/glycoprotein L (gH/gL) complex. Proc. Natl. Acad. Sci. U. S. A. 107:22641–22646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McShane MP, Longnecker R. 2005. Analysis of fusion using a virus-free cell fusion assay. Methods Mol. Biol. 292:187–196 [DOI] [PubMed] [Google Scholar]
- 25. Hutt-Fletcher LM, Chesnokova LS. 2010. Integrins as triggers of Epstein-Barr virus fusion and epithelial cell infection. Virulence 1:395–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McShane MP, Longnecker R. 2004. Cell-surface expression of a mutated Epstein-Barr virus glycoprotein B allows fusion independent of other viral proteins. Proc. Natl. Acad. Sci. U. S. A. 101:17474–17479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haan KM, Kwok WW, Longnecker R, Speck P. 2000. Epstein-Barr virus entry utilizing HLA-DP or HLA-DQ as a coreceptor. J. Virol. 74:2451–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Silva AL, Omerovic J, Jardetzky TS, Longnecker R. 2004. Mutational analyses of Epstein-Barr virus glycoprotein 42 reveal functional domains not involved in receptor binding but required for membrane fusion. J. Virol. 78:5946–5956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rowe CL, Matsuura H, Jardetzky TS, Longnecker R. 2011. Investigation of the function of the putative self-association site of Epstein-Barr virus (EBV) glycoprotein 42 (gp42). Virology 415:122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Omerovi J, Lev L, Longnecker R. 2005. The amino terminus of Epstein-Barr virus glycoprotein gH is important for fusion with epithelial and B cells. J. Virol. 79:12408–12415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sorem J, Jardetzky TS, Longnecker R. 2009. Cleavage and secretion of Epstein-Barr virus glycoprotein 42 promote membrane fusion with B lymphocytes. J. Virol. 83:6664–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ruoslahti E. 1996. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 12:697–715 [DOI] [PubMed] [Google Scholar]
- 33. Scarborough RM, et al. 1991. Barbourin. A GPIIb-IIIa-specific integrin antagonist from the venom of Sistrurus m. Barbouri. J. Biol. Chem. 266:9359–9362 [PubMed] [Google Scholar]
- 34. Xiong JP, et al. 2002. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science 296:151–155 [DOI] [PubMed] [Google Scholar]
- 35. Gianni T, Gatta V, Campadelli-Fiume G. 2010. AlphaVbeta 3-integrin routes herpes simplex virus to an entry pathway dependent on cholesterol-rich lipid rafts and dynamin2. Proc. Natl. Acad. Sci. U. S. A. 107:22260–22265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parry C, Bell S, Minson T, Browne H. 2005. Herpes simplex virus type 1 glycoprotein H binds to alphavbeta3 integrins. J. Gen. Virol. 86:7–10 [DOI] [PubMed] [Google Scholar]
- 37. Leippert M, Beck E, Weiland F, Pfaff E. 1997. Point mutations within the betaG-betaH loop of foot-and-mouth disease virus O1K affect virus attachment to target cells. J. Virol. 71:1046–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haan KM, Lee SK, Longnecker R. 2001. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 290:106–114 [DOI] [PubMed] [Google Scholar]