Abstract
Bioassay (P388 lymphocytic leukemia cell line and human cancer cell lines) -guided separation of an extract prepared from the previously chemically uninvestigated Texas grasshopper Brachystola magna led to isolation of the cancer cell growth inhibitory pancratistatin (1), narciclasine (2) and ungeremine (3). Pancratistatin (1) was first isolated from the bulbs of Hymenocallis littoralis (a.k.a. Pancratium littorale Jacq) and the original crystal structure was deduced by X-ray analysis of a monomethyl ether derivative. In the present study a crystal of pancratistatin (1) was isolated from an extract of B. magna, which led to the X-ray crystal structure of this anticancer drug. Since isoquinoline derivatives 1–3 are previously known only as constituents of amaryllidaceous plants, some of the interesting implications of their rediscovery in the grasshopper B. magna that does not appear to utilize amaryllis family plants were discussed.
In 1965–66 we began the first exploratory survey of terrestrial arthropods as potential sources of new and structurally unique anticancer drugs and by 1968 had uncovered a number of promising leads.2 Over the relatively short four year period when resources were available, we isolated, e.g. antineoplastic constituents from butterfly wings,3a an Asian beetle,3b and a wasp.3c Recently, this relatively unexplored area4 for discovering medically useful new drugs has begun to be revisited on a much larger scale.5
One of the insect leads we began to pursue in 1967 was the lubber grasshopper6 Brachystola magna collected in Texas, but this foraging generalist ranger moved north to Minnesota and west to Arizona.7 Most grasshoppers are usually polyphagous and pursue plants from a broad variety of taxa. However, B magna will even consume other insects including grasshoppers alive or dead.7,8 In this regard, it’s believed that insects vary their diet to obtain, e.g. plant constituents important for their own chemical defense.7 When 2-propanol extracts of B. magna were found to provide a 40% increase in life span (at 200 mg/Kg) against the U.S. National Cancer Institute murine P388 lymphocytic leukemia, we gave increased attention to the lead. By 1970 a combination of negative events occurred, of which the most serious was the untimely passing of our entomologist colleague, D. G. Ford. This coincided with, for practical purposes, the disappearance of B. Magna from the southwest along with the necessary financial resources. Because the extract from the initial collection (less than 1 kg) of B. magna amounted to only 11 g, isolation of the antineoplastic constituent had to await the development of improved bioassays (especially with cancer cell lines), isolation and instrumental techniques. The rather surprising solution to this research objective now follows:
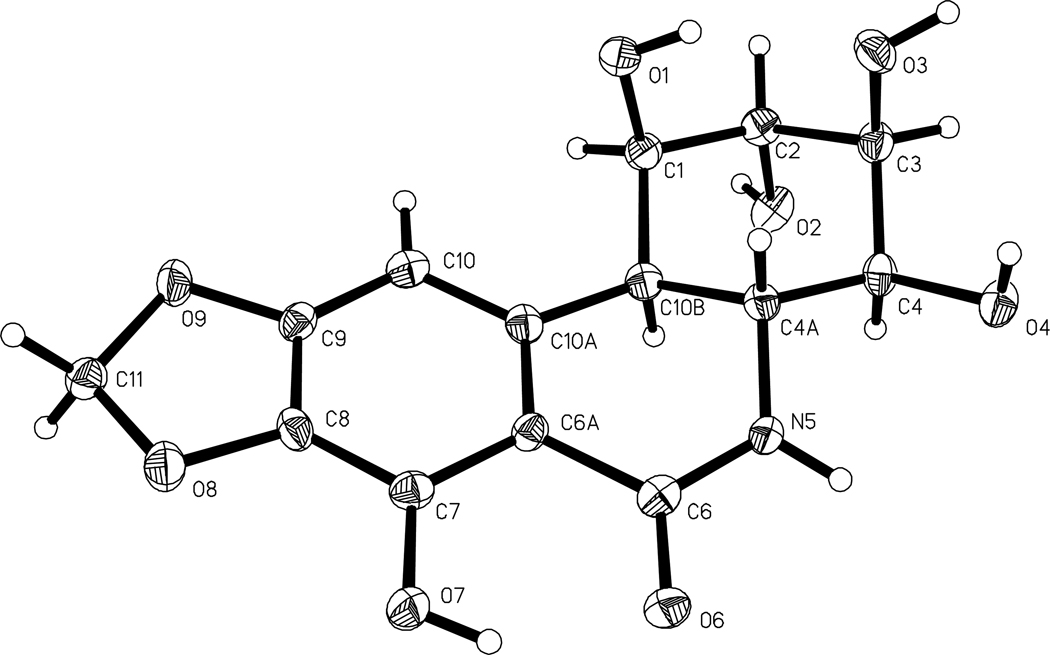
The original Texas collection of Brachystola magna provided a 2-propanol extract that upon solvent partitioning between 9:1→1:1 CH3OH-H2O and hexane→CH2CL2 and the H2O phase between 3:2 CH3OH-H2O and 1-butanol, gave CH2Cl2 and 1-butanol fractions that were found to significantly inhibit the P388 leukemia cell line (ED50 3.5 µg/mL and 1.9 µg/mL , respectively) and five human cancer cell lines. Bioassay-guided (P388 leukemia cell line) separation of the 1-butanol fraction (2.88 g) by a series of gel permeation and partition separations on Sephadex LH-20 columns, followed by final purification employing HPLC and recrystallization led to pancratistatin (1, 4.1 × 10−2 % yield, P388 ED50 0.048 µg/mL),9,10 narciclasine (2, 1.4 × 10−2 % yield, P388 ED50 0.018 µg/mL)11 and ungeremine (3, 2.3 × 10−2 % yield, P388 ED50 1.2 µg/mL).12 Pancratistatin (1) was recrystallized from CH3OH/H2O and small, colorless crystals were obtained. The molecular formula was assigned as C14H15NO8 on the basis of high-resolution APCI+ mass spectroscopy and the structure was confirmed by an X-ray crystal structure determination. While the X-ray crystal structure of 1 had been solved by our group in 1994, the results had not previously been published. Consequently, when the initial cell parameters of the current data collection were obtained and compared with the 1994 collection, we were surprised to discover that the anticancer compound obtained from the grasshopper extract was identical to the pancratistatin obtained from plant sources.10,13 Figure 1 shows the stereochemical drawing of 1 obtained from the current X-ray analysis.
Figure 1.
The X-ray crystal structure of pancratistatin (1) with the atomic thermal ellipsoids being displayed at 50% probabilities.
Narciclasine (2) was obtained as a colorless amorphous semisolid, and its structure was easily assigned on the basis of high field 2D-NMR analyses and high-resolution FAB mass spectroscopy that provided molecular formula C14H13NO7. Ungeremine (3) was isolated as a yellow powder. The molecular formula was assigned as C16H12NO3 on the basis of high-resolution APCI+ mass spectroscopy and combined with results of 2D-NMR analyses confirmed its structure. The isolation of isoquinoline derivatives 1–3 from B. magna represents, to our knowledge, the first results from a chemical constituent study of this grasshopper.
Presumably, if the serious constraints of the 1970 period had not occurred, we might have discovered the important anticancer/antiviral pancratistatin (1), now in preclinical development, some 11–12 years sooner than our first isolation of this promising drug from the spider lilly Hymenocallis littoralis (a.k.a. Pancratia littoralis).9 Furthermore, the amount of pancratistatin (1) contained in B. magna would suggest a defensive strategy and potential new plant source of isocarbostyril 1 as amaryllidaceous species have not been recorded among the plants selected by this grasshopper as preferred foods.7,8 Interestingly, the southern grasshopper mouse, Onychomy tossidus, views B. magna as quite palatable.14 Clearly, such biological/chemical relationships will be useful to further investigate.
The cancer cell growth inhibitory properties of B. magna constituents 1–3 were evaluated using the murine P388 lymphocytic leukemia cell line and a selection of human cancer cell lines. (Table 1). As we have consistently found, pancratistatin (1) and narciclasine (2) both exhibit strong inhibition against growth of the P388 lymphocytic leukemia and the mini-panel of human tumor cell lines.11a,14 Table 1 also includes the first cancer cell line data for ungeremine (3).
Table 1.
Murine P388 Lymphocytic Leukemia Cell Line and Human Cancer Cell Line Inhibition Values (ED50 expressed in µg/mL) for Alkaloid 1, 2, and 3a
| Cancer cell lineb | 1 | 2 | 3 |
|---|---|---|---|
| P388 | 0.048 | 0.018 | 1.2 |
| BXPC-3 | 0.043 | 0.024 | 0.87 |
| MCF-7 | 0.035 | 0.014 | 0.48 |
| SF268 | 0.032 | 0.018 | 0.057 |
| NCI-H460 | 0.029 | 0.014 | 0.65 |
| KM20L2 | 0.048 | 0.023 | 0.50 |
| DU-145 | 0.037 | 0.012 | 0.029 |
In DMSO.
Cancer cell line type (P388, lymphocytic leukemia; BXPC-3, pancreas adenocarcinoma; MCF-7, breast adenocarcinoma; SF268, CNS glioblastoma; NCI-H460, lung large cell; KM20L2, colon adenocarcinoma; DU-145, prostate carcinoma)
Experimental Section
General Experimental Methods
All chromatographic solvents were redistilled. Sephadex LH-20 used for partition column chromatography was obtained from Pharmacia Fine Chemicals. AB Semipreparative HPLC was conducted with a Gilson model 805 HPLC coupled with a Gilson model 117 U detector. UV spectra were recorded with a Perkin-Elmer lambda 3B uv/vis spectrometer. NMR spectra were obtained with a Varian UNITY INOVA-400 and 500 spectrometers with tetramethylsilane (TMS) as an internal reference. High-resolution mass spectra were obtained using a JEOL LCMate magnetic sector instrument either in the FAB mode, with a glycerol matrix, or by APCI with a polyethylene glycol reference.
Insect Collection
The specimens of Brachystola magna, Girard (Acrididea, family Romaleidae, subfamily pomaleinae)7,8 were collected in 1967 in Texas, USA, under direction of Ford Labs, Lyford, Texas and identified by D. G. Ford (deceased May 3, 1970). A voucher specimen of B. Magna has been retained in our institute. Brachystola magna, a.k.a. lubber grasshopper, plains lubber, or homesteader6 is 3–4.5 cm long and ranges east of the U.S. Rocky Mountains from North Dakota south to Texas and Mexico and eastward with isolated populations as far as Minnesota, Iowa, Kansas and Oklahoma.7 B. magna is known for certain beneficial activities by providing some biological control of weeds in their favored grazing areas containing sunflowers and a selection of other foods. However, large populations of the insect have been very destructive to young cotton plants.15
Extraction and Initial Separation of Brachystola magna constituents
The 1967 collection of Brachystola magna (about 1 kg) preserved in 2-propanol was extracted (2 x; 10, 5 days) using 1:1 CH2Cl2-CH3OH. After each extraction, 30% by volume of H2O was added to separate a CH2Cl2 fraction. The CH2Cl2 phase was separated and solvent evaporated in vacuo, yielding a 0.92 g CH2Cl2 fraction (P388 ED50 3.5 µg/mL). The H2O phase was next partitioned between CH3OH-H2O (3:2) and 1-butanol. The 1-butanol layer was separated and solvent evaporated in vacuo, to provide a 2.28 g 1-butanol fraction (P388 ED50 1.9 µg/mL). The solvent partitioning sequence was a modification of the original procedure of Bligh and Dyer.16
Isolation Procedures
The 1-butanol fraction (2.88 g) was passed through a Sephadex LH-20 column, using CH3OH as eluent. Six P388 leukemia cell line inhibitory fractions were obtained. One fraction was recrystallized from CH3OH (3 x) to afford 2.5 mg ungeremine (3) as a yellow powder HRAPCI m/z calcd for [M + H]+ C16H12NO3, 266.0817; found 266.0810. The other five P388 cell line bioactive fractions were combined and dissolved in CH3OH (5 ml). After removing the insoluble residue, the solution components were separated by HPLC using a semipreparative Luna 5µ C8 (2) column and a gradient mobile phase (20% CH3OH in H2O for 42 min, then 100% CH3OH for 20 min at a flow rate of 2 ml/min) to afford 2.5 mg of pancratistatin (1)9,10 as colorless cubic crystals and 1.5 mg of narciclasine (2)11 as a semisolid. Compounds 1, 2, and 3 were identified by direct comparisons with known standards and with literature data.
Crystal Structure of Pancratistatin (1)
A low temperature (−150 °C) data collection was performed on a crystal grown from D2O/methanol. 30 second frames of data were collected as −0.396 ° in ω scans on a Bruker AXS SMART 6000 CCD diffractometer using Cu radiation (1.54178 Å). Collection was conducted using the Multirun procedure in the SMART17 software, such that 96.8% of the theoretically possible reflections available were collected out to a θ limit of 68.43°. Data reduction was subsequently performed with SAINT-PLUS18 and an empirical absorbtion correction was performed with SADABS19. Structure solution and refinement were performed with SHELXTL program package. The positions for all hydrogen atoms were calculated based on atomic geometry. The final structural model obtained for pancratistatin is shown in Figure 1. Crystal data and experimental details follow: C14H15NO8, mol wt = 324.27, crystal size 0.32 × 0.13 × 0.03 mm, orthorhombic, space group P212121, no 19, a = 6.7843(4) Å, b = 9.3199(5) Å, c = 20.0409(13) Å, V = 1267.17(13) Å3, Z = 4, Dc 1.705 Mg/m3, F(000) = 680, μ(CuKα) = 1.223 mm−1, independent data, 2160 (Rint =0.0503), θ range 5.23 to 68.43°, R[I>2σ(I)] = 0.0362, Rall = 0.0560, ΔF = +0.258 and −0.248 e Å−3. Crystallographic data of 1 including atomic coordinates, bond lengths and angles, thermal parameters, and additional experimental details may be found in the Supporting Information (11 pages). This information is available free of charge via the Internet at http://pubs.acs.org. The material has also been deposited in the Cambridge Crystallographic Data Center (CCDC). Copies of can be obtained free of charge, on application to the Director, CCDC, 12 Union Road, Cambridge CB2 1EZ, UK [fax: +44-(0)1223-336033 or deposit@ccdc.cam.ac.uk].
Cancer Cell Line Bioassay Procedures
The National Cancer Institute’s standard sulforhodamine B assay was used to assess inhibition of human cancer cell growth as previously described.20 The murine P388 Lymphocytic leukemia cell line results were obtained as reported previously.21
Supplementary Material
Acknowledgment
Financial assistance was received with appreciation from Public Health Service Research Grants CA-10612-01 to CA-10612-04 from the Division of Cancer Treatment and Diagnosis, NCI, DHHS; the Arizona Disease Control Research Commission; Dr. and Mrs. Alec D. Keith; the J. W. Kieckhefer Foundation; the Margaret T. Morris Foundation; Gary L. and Diane Tooker; Caitlin Robb Foundation; Polly J. Trautman; Dr. John C. Budzinski; and the Robert B. Dalton Endowment Fund. Technical assistance was provided by Drs. J-C. Chapuis, Z. A. Cichacz, D. L. Doubek, J. L. Hartwell, C. L. Herald, F. Hogan and N. M. Melody, as well as F. Craciunescu, M. J. Dodson and L. Williams.
References and Notes
- 1.For part 552, refer to: Pettit GR, Numata A, Herald DL, Iwamoto C, Usami Y, Yamada T, Ohishi H, Cragg GM. J. Nat. Prod. doi: 10.1021/np058075+. submitted.
- 2.Pettit GR, Traxler PM, Pase CP. Lloydia. 1973;36:202–203. [PubMed] [Google Scholar]
- 3.(a) Polonsky J, Varon Z, Jacquemin H, Pettit GR. Experientia. 1978;34:1122–1123. doi: 10.1007/BF01922904. [DOI] [PubMed] [Google Scholar]; (b) Pettit GR, Herald CL, Doubek DL, Herald DL. J. Am. Chem. Soc. 1982;104:6846–6848. [Google Scholar]; (c) Pettit GR, Blazer RM, Reierson DA. Lloydia. 1977;40:247–252. [PubMed] [Google Scholar]
- 4.Meinwald J, Eisner T. Helv. Chim. Acta. 2003;86:3633–3637. [Google Scholar]
- 5.Trowell S. The futurist. 2003:17–19. [Google Scholar]
- 6.Roisk JJ. Common Names of Insects and Related Organisms. Lanham, MD: Entomological Society of America; 1997. p. 232. [Google Scholar]
- 7.Bright KL, Bernays EA, Moran VC. J. Insect Behavior. 1994;7:779–793. [Google Scholar]
- 8.Bernays EA, Bright KL. Comp. Biochem. Physiol. 1993;104A:125–131. [Google Scholar]
- 9.Pettit GR, Gaddamidi V, Cragg GM, Herald DL, Sagawa Y. J. Chem. Soc. Chem. Commun. 1984;24:1693–1694. [Google Scholar]
- 10.Pettit GR, Gaddamidi V, Herald DL, Singh SB, Cragg GM, Schmidt JM, Boettner FE, Williams M, Sagava Y. J. Nat. Prod. 1986;49:995–1002. doi: 10.1021/np50048a005. [DOI] [PubMed] [Google Scholar]
- 11.(a) Ceriotti G. Nature. 1967;213:595–596. doi: 10.1038/213595a0. [DOI] [PubMed] [Google Scholar]; (b) Mondon A, Krohn K. Chem. Ber. 1975;37:13–16. [Google Scholar]; (c) Okmoto T, Torri Y, Isogai Y. Chem. Pharm. Bull. 1968;24:1119–1131. [Google Scholar]
- 12.Abou-Donia AH, Abib A-A, Din ASE, Evidente A, Gaber M, Scopa A. Phytochemistry. 1992;31:2139–2141. [Google Scholar]
- 13.Pettit GR, Gaddamidi V, Cragg GM. J. Nat. Prod. 1984;47:1018–1020. doi: 10.1021/np50036a020. [DOI] [PubMed] [Google Scholar]
- 14.(a) Whitman DW, Blum MS, Jones CG. Behavioural Processes. 1986;13:77–83. doi: 10.1016/0376-6357(86)90018-5. [DOI] [PubMed] [Google Scholar]; (b) Whitman DW, Blum MS, Jones CG. Ann. Entomol. Soc. Am. 1985;78:451–455. [Google Scholar]
- 15.Isely FB. Ecol. Monogr. 1938;8:551–604. [Google Scholar]
- 16.Bligh EG, Dyer WJ. Can. J. Biochem., Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 17.SMART for Windows NT v5.605. 5465 East Cheryl Parkway, Madison, WI 53711-5373, USA: BRUKER AXS Inc.; [Google Scholar]
- 18.SAINT+ for NT v6.22. 5465 East Cheryl Parkway, Madison, WI 53711-5373, USA: BRUKER AXS Inc.; [Google Scholar]
- 19. Sheldrick GM. SADABS – Bruker Area Detector ABSorption Correction Program. . See Blessing B. Acta Cryst. 1995;A51:33–38..
- 20.Monks A, Scudiero D, Skehan P, Shoemaker R, Paull K, Vistica D, Hose C, Langley J, Cronise P, Vaigro-Wolff A, Gray-Goodrich M, Campbell H, Mayo J, Boyd M. J. Natl. Cancer Inst. 1991;83:757–766. doi: 10.1093/jnci/83.11.757. [DOI] [PubMed] [Google Scholar]
- 21.Pettit GR, Meng Y, Stevenson CA, Doubek DL, Knight JC, Cichacz Z, Pettit RK, Chapuis J-C, Schmidt JM. J. Nat. Prod. 2003;66:259–262. doi: 10.1021/np020231e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.