Abstract
Background and Purpose
Ischemic stroke has a strong familial component to risk. The Siblings with Ischemic Stroke Study (SWISS) is a genome-wide family-based analysis that included use of imputed genotypes. SWISS was conducted to examine associations between SNPs and risk of stroke and stroke subtypes within pairs.
Methods
SWISS enrolled 312 probands with ischemic stroke across 70 US and Canadian centers. Affected siblings were ascertained by centers and confirmed by central record review; unaffected siblings were ascertained by telephone contact. Ischemic stroke was subtyped using TOAST criteria. Genotyping was performed using an Illumina 610 quad array (probands) and an Illumina linkage V array (affected siblings). SNPs were imputed using 1000 Genomes Project data and MACH software. Family-based association analyses were conducted using the sibling-transmission disequilibrium test.
Results
For all pairs, the correlation of age at stroke within pairs of affected siblings was r = 0.83 (95%CI, 0.78 to 0.86; P < 2.2×10−16). The correlation did not differ substantially by subtype. The concordance of stroke subtypes among affected pairs was 33.8% (kappa = 0.13; P = 5.06×10−4) and did not differ by age at stroke in the proband. Although no SNP achieved genome-wide significance for risk of ischemic stroke, there was clustering of the most associated SNPs on chromosomes 3p (NOS1) and 6p.
Conclusions
Stroke subtype and age at stroke in affected sibling pairs exhibit significant clustering. No individual SNP reached genome-wide significance. However, two promising candidate loci were identified, including one that contains NOS1, though these risk loci warrant further examination in larger sample collections.
Ischemic stroke has long been recognized to cluster in families. This clustering has been attributed to both genetic factors and common environmental exposures. Gene mutations have been identified that lead to rare ischemic stroke syndromes like CADASIL and MELAS. However, search for genetic loci associated with ischemic stroke risk has yielded meager results thus far despite several candidate gene [1] and large-scale genome-wide association studies. [2] [3] The Siblings with Ischemic Stroke Study (SWISS) provides a different and promising approach to discover novel risk factors for ischemic stroke through the study of unrelated families of affected and unaffected siblings. Here we present the results of the family-based genome-wide scan of SWISS after achieving the original recruitment goal.
Methods
Study Subjects
Probands were recruited at 70 US medical centers and Canadian medical centers. Probands were adult (>18 years old) men and women presenting to a participating center with a study neurologist-confirmed ischemic stroke. Stroke was defined as rapidly developing signs of a focal or global disturbance of cerebral function, with symptoms lasting at least 24 hours or leading to death with no apparent cause other than vascular origin (WHO definition). [4] Stroke was defined as ischemic if CT or MR imaging of the brain was performed within 7 days of onset of stroke symptoms and identified the symptomatic cerebral infarct or failed to identify an alternative cause of symptoms. Probands were required to have reported at least one living full sibling with a history of stroke. No probands were enrolled with iatrogenic vasospastic, or vasculitic stroke or if the stroke occurred in the setting of a mechanical heart valve or in the setting of untreated or actively treated bacterial endocarditis. Probands were also excluded if they were known to have CADASIL, Fabry disease, homocysteinuria, MELAS, or sickle cell anemia. Study neurologists at each center assigned to the qualifying ischemic stroke of each proband a TOAST subtype diagnosis.[5]
Stroke-affected siblings of the proband (concordant siblings) were recruited using proband-initiated contact.[6] Telephone interviews were performed to obtain demographic and clinical information and to gain permission for obtaining medical records pertaining to treatment for stroke. Medical records were compiled and adjudicated by a central committee (JFM; TGB), to verify the diagnosis of ischemic stroke and to assign a TOAST subtype diagnosis. Assignment of TOAST subtype diagnoses to SWISS concordant siblings has moderate inter-rater reliability.[7] Unaffected siblings were ascertained by telephone contact and interview.
Genotyping Considerations
The establishment of lymphoblastoid cell lines, quality control of genomic DNA, acquisition of genetic data, and genotyping quality control metrics were performed using standard procedures. Please see http://stroke.ahajournals.org for these details.
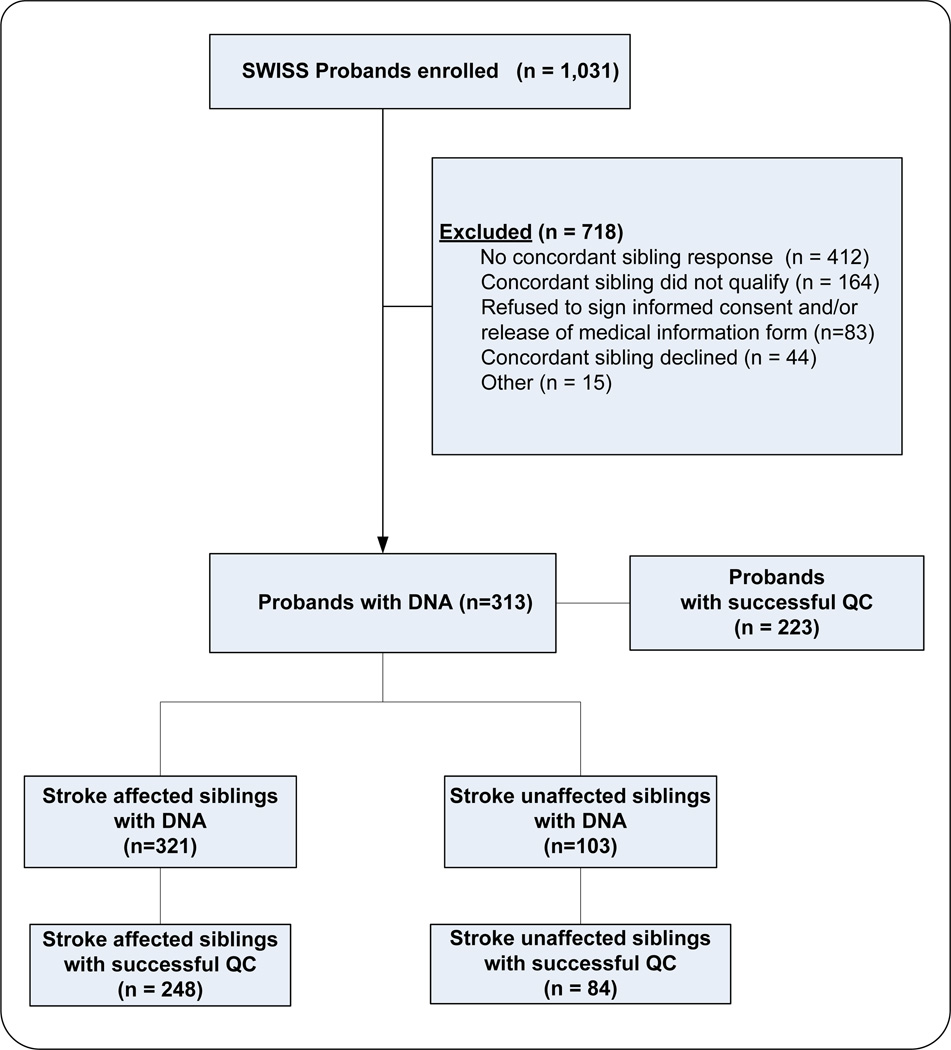
Consensus SNPs that passed QC in both phases (genome-wide association and family-based phases) were merged for all available sibships (2239 SNPs were imputed in the probands). Using all 5612 SNPs in the merged dataset, reported relationships were verified using pi_hat estimates. Sibships were confirmed if pairwise pi_hats values were between 0.35 and 0.65; samples were removed from a sibship if estimated pi_hat was not in this range. This dataset of the combined genotyping phases represents the final dataset for all subsequently described analyses. The flow of patients in the study is shown in Figure 1.
Figure 1.
Flow of participants in the study.
Genetic Data Analysis
All family-based analyses were conducted using PLINK 1.07 software. [8] The dFam utility within PLINK implements a siblings-based transmission-disequilibrium test and was used to conduct these analyses. The dFam option is a powerful test for sibling-only datasets, incorporating data across sibships as well as using data from estimated parental genotypes to calculate expected allele frequencies for comparison to observed allele frequencies. The association test is based on the Cochran-Mantel-Haenszel test. Bonferroni correction for the number of tested SNPs corresponds to a minimum p-value for genome-wide significance of P < 8.91×10−6.
Additional Statistical Analyses
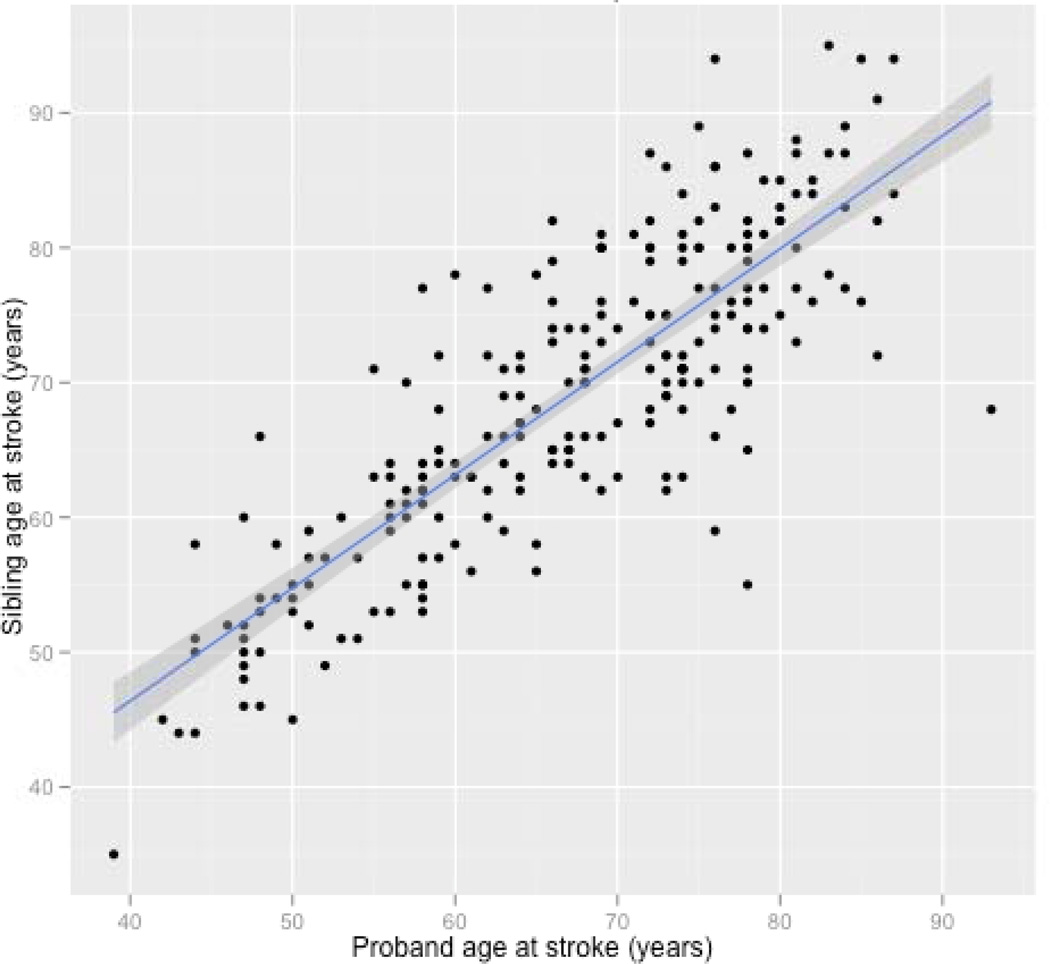
Frequencies of stroke risk factors (hypertension, hyperlipidemia and diabetes) between affected and unaffected participants were compared using chi-squared tests. The correlation between affected sibling age at stroke was estimated using the Pearson test of correlation. These analyses were conducted across all TOAST subtypes as well as following stratification by concordant and discordant subtypes among affected sibling pairs. Linear regression was used to determine the confidence intervals and linear fit of the age association as show in Figure 2 Kappa statistics were calculated to quantify concordance of phenotypes of interest within sibling pairs for all ages and stroke subtypes as well as models stratified by age (< 65 year proband as defining age strata) and stroke subtype. All analyses that did not include genetic data were conducted using scripts written in R (R Development Core Team (2008)). [9]
Figure 2.
Correlation between proband and sibling age at stroke. Correlation coefficient = 0.83. P-value < 0.0001. Pairs are points, Blue line = linear model, grey shading = 95% CI.
Results
A total of 312 affected sibling pairs (312 probands) were enrolled at 70 centers across the US and Canada. After quality control filtering, the final study population consisted of 223 probands, 248 stroke-affected siblings, and 84 stroke-unaffected siblings (total sample size, 555). Ischemic stroke-affected individuals had expected high rates of conventional atherosclerotic risk factors (Table 1). Stroke-affected individuals (probands and affected-siblings) were significantly more likely to have hypertension (P < 0.0001), hyperlipidemia (P= 0.002) and diabetes (P = 0.008) than stroke-unaffected individuals. Stroke-affected siblings were somewhat older than the probands. This difference of 2 years (P = 0.057) is expected as an older sibling of the proband would be more likely to have a stroke than a younger sibling.
Table 1.
Characteristics of the Study Population after passing genomic quality controls.
| Probands | Affected Siblings |
Unaffected Siblings |
Total | |
|---|---|---|---|---|
| N total (%) | 223 (40) | 248 (45) | 84 (15) | 555 (100) |
| Age (mean, SD) | 66.99, 11.39 | 69.01, 11.55 | 66.00, 11.12 | 67.74, 11.46 |
| % Female | 48.43 | 45.97 | 58.33 | 48.83 |
| TOAST Criteria N (%) | ||||
| Cardioembolic | 25 (11) | 23 (9) | n.a. | 48/471 (10) |
| Large Vessel | 58 (26) | 47 (19) | n.a. | 105/471 (22) |
| Small Vessel | 66 (30) | 76 (31) | n.a. | 142/471 (30) |
| Other | 13 (6) | 10 (4) | n.a. | 23/471 (5) |
| Undetermined | 61 (27) | 92 (37) | n.a. | 153/471 (33) |
| Hypertension N (%) | ||||
| Yes | 155 (70) | 175 (71) | 38 (45) | 368 (66) |
| No | 68 (30) | 72 (29) | 46 (55) | 186 (34) |
| Unknown | 0 | 1 (<1) | 0 | 1 (<1) |
| Atrial Fibrillation N (%) | ||||
| Yes | 24 (11) | 57 (23) | 12 (14) | 93 (17) |
| No | 197 (88) | 188 (76) | 72 (86) | 457 (82) |
| Unknown | 2 (1) | 3 (1) | 0 | 5 (1) |
| Hyperlipidemia N (%) | ||||
| Yes | 140 (63) | 162 (65) | 39 (46) | 341 (61) |
| No | 83 (37) | 84 (34) | 45 (54) | 212 (38) |
| Unknown | 0 | 2 (1) | 0 | 2 (1) |
| Diabetes N (%) | ||||
| Yes | 50 (22) | 61 (25) | 9 (11) | 120 (22) |
| No | 173 (78) | 185 (75) | 75 (89) | 433 (78) |
| Unknown | 0 | 2 (<1) | 0 | 2 (<1) |
| Smoking N (%) | ||||
| Current | 44 (20) | 45 (18) | 14 (17) | 103 (19) |
| Never | 104 (47) | 91 (37) | 37 (44) | 232 (42) |
| Former | 74 (33) | 109 (44) | 33 (39) | 216 (39) |
| Unknown | 1 (<1) | 3 (1) | 0 | 4 (<1) |
n.a.=Not Applicable
Sibling age at time of stroke strongly correlated with proband age at time of stroke, despite the sibling being older. As shown in Figure 2 for all the sibling pairs, the correlation coefficient was r = 0.83 (95%CI, 0.78 to 0.86; P < 2.2×10−16). For affected sibling pairs who have the same stroke subtype, the correlation coefficient was not different from all pairs, r = 0.83 (95%CI, 0.75 to 0.89; P < 2.2×10−16). This was the same for sibling pairs in which the affected siblings had different stroke subtypes, r = 0.83 (95%CI, 0.77 to 0.87; P < 2.2×10−16). Over 50% of the variance in age at stroke onset in siblings could be predicted by the age of proband at time of stroke. As shown in Table 2, there was significant concordance with affected siblings for TOAST subtype (kappa = 0.13; P = 5.06×10−4); this relationship remained significant for sibling pairs where the proband was less than 65 years old at time of stroke and for sibling pairs in which the proband was 65 years or older.
Table 2.
Concordance for ischemic stroke subtype within affected sibling pairs.
| All ages | Proband < 65 years | Proband ≥ 65 years | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Proband stroke type |
Total pairs |
% Expected Concordant |
% Observed Concordant |
Total pairs |
% Expected Concordant |
% Observed Concordant |
Total pairs |
% Expected Concordant |
% Observed Concordant |
| All Strokes | 233 | -- | 34.8 | 92 | -- | 33.7 | 141 | -- | 35.5 |
| Cardioembolic | 29 | 1.5 | 24.1 | 13 | 2.0 | 30.8 | 16 | 1.3 | 18.8 |
| Large Vessel | 58 | 6.2 | 31.0 | 26 | 8.0 | 38.5 | 32 | 5.2 | 25.0 |
| Small Vessel | 68 | 8.5 | 36.8 | 22 | 5.7 | 45.4 | 46 | 10.6 | 32.6 |
| Other | 13 | 0.3 | 7.7 | 7 | 0.6 | 14.3 | 6 | 0.2 | 0 |
| Unknown | 65 | 7.8 | 46.2 | 24 | 6.8 | 25.0 | 41 | 8.5 | 58.5 |
| Kappa Statistic | -- | -- | 0.13 | -- | -- | 0.14 | -- | -- | 0.12 |
| Kappa SE | -- | -- | 0.04 | -- | -- | 0.06 | -- | -- | 0.05 |
| Kappa Z | -- | -- | 3.48 | -- | -- | 2.46 | -- | -- | 2.45 |
| Kappa P-value | -- | -- | 5.06 × 10−4 | -- | -- | 1.38 × 10−2 | -- | -- | 1.44 × 10−2 |
Note: Kappa statistics calculated for all pairs by age strata
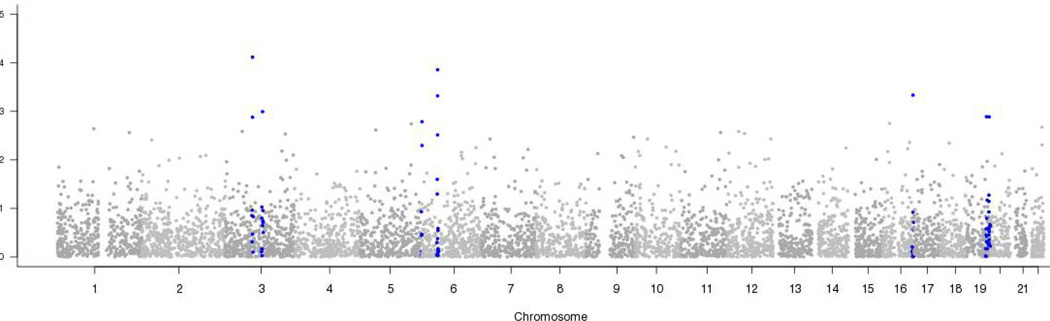
Results of the genome-wide analyses are shown in Figure 3. Although no SNP association with ischemic stroke achieved genome-wide level of significance, the ten most associated SNPs exhibited clustering in several genomic regions. These ten most significantly associated SNPs, their locations, frequencies and effect estimates are shown in Table 3. The ten SNPs represent 8 genomic loci, with minor allele frequencies ranging from 0.38–0.48 (common alleles). The effects for each are small, with odds ratios ranging from 0.96 to 1.04. The location of the most significantly associated SNPs (as well as others within 2.5 Mb) is shown in Figure 3 (indicated by blue shading). There are clusters of associated SNPs on chromosomes 3p and 6p. The SNPs on chromosome 3p lie in a strong candidate gene, NOS1.
Figure 3.
Summary of transmission-disequilibrium test results. All SNPs within 2.5 megabases of SNPs shown in table 1 are highlighted in blue.
Table 3.
Top 10 most significant SNPs representing 8 genomic loci.
| Minor | Odds Ratio | Chi-Squared | ||||||
|---|---|---|---|---|---|---|---|---|
| SNP | CHR | MB | cM | Allele | MAF | for Minor Allele | Statistic | P-value |
| rs1383407 | 3 | 78.991 | 107.18 | C | 0.4488 | 0.961 | 15.65 | 7.63 × 10−5 |
| rs328049 | 3 | 79.068 | 107.21 | A | 0.4766 | 0.968 | 10.31 | 1.32 × 10−3 |
| rs986692 | 3 | 107.766 | 116.1 | T | 0.4120 | 1.040 | 10.80 | 1.01 × 10−3 |
| rs1053110 | 5 | 180.421 | 205.94 | T | 0.4704 | 1.034 | 9.92 | 1.64 × 10−3 |
| rs1293457 | 6 | 44.866 | 68.46 | T | 0.4372 | 1.036 | 12.20 | 4.79 × 10−4 |
| rs3778507 | 6 | 45.005 | 68.69 | A | 0.4075 | 1.043 | 14.53 | 1.38 × 10−4 |
| rs179209 | 16 | 19.215 | 41.4 | A | 0.4749 | 0.967 | 9.773 | 1.77 × 10−3 |
| rs750740 | 16 | 87.335 | 129.03 | T | 0.4695 | 1.037 | 12.26 | 4.63 × 10−4 |
| rs897783 | 19 | 56.723 | 88.79 | A | 0.3779 | 1.036 | 10.36 | 1.29 × 10−3 |
| rs976192 | 20 | 1.444 | 5.81 | C | 0.3797 | 1.041 | 10.35 | 1.30 × 10−3 |
SNP denotes single nucleotide polymorphism; CHR, chromosome; MB, megabase; cM, centiMorgan; MAF, minor allele frequency.
Discussion
Our genome-wide scan for risk factors for ischemic stroke was performed in the largest collection of affected sibling pairs to date and showed potential loci of interest including a locus on NOS1. We are not aware of other genetic studies showing NOS1 variants to be associated with human ischemic stroke risk. However, knock-out mouse models show that NOS1 gene deficiency causes worsening neointimal formation and constrictive vascular remodeling. [10] Genetic variants theoretically could impart elevated ischemic stroke risk by a vascular mechanism, by a thrombotic mechanism or even by a parenchymal mechanism where brain tissue is sensitized to focal ischemia. In the case of NOS1, it seems that variants would be more likely to impart risk by a vascular mechanism than by a parenchymal mechanism, since NOS1 knockout mice have smaller infarcts than control wild-type mice in an MCA occlusion model. [11] It is important not to speculate beyond the strength of our observations, as no locus achieved genome-wide statistical significance.
Genome-wide studies have had mixed results in ischemic stroke. When SWISS was initiated, the human genome had only been sequenced in draft form. [12] SWISS was predicated on the hypothesis that ischemic stroke obeyed the common disease, common variant hypothesis, which states that the genetic influences on many common disease are attributable to a limited number of allelic variants present in > 1–5% of the population. [13] It has since become less clear that the hypothesis holds for ischemic stroke. No single locus has been identified in two genome-wide association studies at a genome-wide level of significance. [14] Our study supports the idea that no single locus substantially contributes to ischemic stroke risk from the perspective of common variants contributing to disease risk, although future sequencing-based studies of rare variants may meet with substantially more success.
SWISS was designed to treat all types of ischemic stroke as a single phenotype. The phenotypic heterogeneity of ischemic stroke has long been appreciated, but categorizing subtypes of ischemic stroke historically has been done with little consistency in genetic research.[15] Despite this methodological limitation, genetic risk factors have been identified that appear to be specific for certain ischemic stroke subtypes. For example, the chromosome 9p21 locus appears to impart risk for so-called large vessel atherosclerotic stroke. [16, 17] The atrial fibrillation locus 4q25 appears to impart risk for cardioembolic stroke. [18, 19] Collaborating with investigators from Sweden, we previously assessed whether ischemic stroke subtypes clustered among affected sibships, showing low aggregation rates. [20] We continue to see low aggregation rates, but the relationship is significant. In addition to having larger numbers, the current analysis is restricted to those affected sibling pairs confirmed to be full siblings through genomic analysis. It is not known whether more complex systems of classifying stroke also show a tendency toward aggregation within families.[21]
Age at onset of stroke may be a quantitative phenotype more tractable to genomic analysis. In an interim analysis, we had observed a significant association of proband age at stroke onset and sibling age at stroke onset. [22] As with the subtype aggregation reanalysis, the current analysis involves a larger sample size and is restricted to those affected sibling pairs confirmed to be full siblings through genomic analysis. As a phenotype, age at stroke onset has the limitation that it does not necessarily reflect the burden of ischemic disease at any given moment in the lifespan of a patient. Some cerebral infarcts are asymptomatic [23] while other cerebral infarcts may be symptomatic but undiagnosed. [24]
In summary, we have described here an affected relative-based genetic analysis of ischemic stroke. This work provides preliminary evidence for the involvement of several loci in risk for this disease, and these loci certainly warrant follow-up. This work also suggests that any individual risk variants involved in ischemic stroke are likely to have a low population-attributable risk. Attributable risk could be low if the risk conferred is relatively low; it could also be low if there is extensive allelic and/or genetic heterogeneity in stroke, with no single locus being a common, high risk conferring locus. Clearly we can hope that the future application of now- and next-generation technologies in large and extremely well characterized cohorts will enable identifying genetic risks for ischemic stroke.
Supplementary Material
Acknowledgements
This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD. (http://biowulf.nih.gov).
This work used samples and clinical data from the NINDS Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds), human subjects protocol numbers 2003-081 and 2004-147.
Sources of Funding
SWISS is funded by the National Institute for Neurological Disorders and Stroke (NINDS) through grant number R01 NS39987 (J.F.M. PI), IRB 1082-99 (Mayo Clinic). The inclusion of BLSA samples was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services; project number Z01 AG000015-50, human subjects protocol number 2003-078. Dr. Hardy is supported by an MRC Returning scientist award. The Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services; project number Z01 AG000954-06.
Appendix 1. SWISS Investigative Team
Executive Committee: James F. Meschia, chair; Thomas G. Brott, Robert D. Brown, Jr., Brett Kissela, John Hardy, Stephen S. Rich, Andrew Singleton, Bradford Worrall. Statistical Center: University of Virginia, Charlottesville, VA. Stephen S. Rich, Director of Center for Public Health Genomics. DNA Repository: Coriell Institute for Medical Research, Camden, New Jersey. Margaret Keller, PhD, Director. Data Management: Mayo Clinic, Rochester, Minnesota. Barry Bisbee, Data Management Specialist. Centers and Investigators (listed by proband enrollment): Neurological Associates, Inc, Richmond, Virginia (probands enrolled, 72): Principal Investigators (PI): Alan Schulman, MD; Francis McGee, MD; coordinators: Gwendolyn H. Darby; Julie Thornton. Mayo Clinic, Rochester, Minnesota (67): PI: Robert D. Brown, Jr., MD; coordinator: Colleen S. Albers. Mayo Clinic, Jacksonville, Florida (64): PI: Thomas G. Brott, MD; coordinators: Alexa Richie; Dale Gamble; Sothear Luke. University of Virginia Health System, Charlottesville, Virginia (59): PI: Bradford Worrall, MD, MSc; coordinators: Colleen Harman; Martha Davis; Kay Maupin. University of Cincinnati, Cincinnati, Ohio (51): PI: Brett Kissela, MD; coordinator: Kathleen Alwell. Shands Jacksonville, affiliated with the University of Florida, Jacksonville, Florida (50): PI: Scott Silliman, MD; coordinator: Rhonda Calhoun. Mercy Ruan Neurology Clinic, Des Moines, Iowa (46): PI: Michael Jacoby, MD; coordinator: Cheryl Inman. Washington University School of Medicine, St. Louis, Missouri (33): PI: Jin-Moo Lee, MD, PhD; coordinators: Julie Rickmann; Jill Newgent. University of Maryland, Baltimore, Maryland (27): PI: John Cole, MD; coordinators: Mark Dobbins; Mary J. Sparks; Nancy Zappala. University of Pennsylvania Medical Center, Philadelphia, Pennsylvania (23): PI: Scott Kasner, MD; Co-Investigators: Brett Cucchiara, MD; Steven Messe, MD; Michael McGarvey, MD; coordinators: Mary Liz DeSanto; Jean Luciano. Lankenau Institute for Medical Research, Bryn Mawr, Pennsylvania (22): PI: Gary H. Friday, MD; coordinators: Joan Brown; Angela Gordon. Kaleida Stroke Center, Millard Fillmore Gates Hospital, Buffalo, New York (22): PI: Richard Ferguson, MD; coordinator: Kathy Parkes. Centre Hospitalier Affilie Universitaire de Quebec, Quebec City, Province of Quebec, Canada (21): PI: Ariane Mackey MD; Co-Investigator: Steve Verrault, MD; coordinator: Annette Hache. MetroHealth Medical Center, Cleveland, Ohio (21): PI: Joseph Hanna, MD; coordinator: Dana Cook. Mayo Clinic, Scottsdale, Arizona (20): PI: Bart Demaerschalk, MD; coordinator: Erica L. Boyd. Ohio State University, Columbus, Ohio (19): PI: Andrew Slivka, MD; coordinator: Peggy Notestine. Mercy General Hospital, Sacramento, California (19): PI: Paul Akins; coordinators: Deidre Wentworth; Laura Newman. Luther-Midelfort Hospital, Eau Claire, Wisconsin (18): PI: Felix Chukwudelunzu, MD; coordinator: Karen Snobl. OSF Stroke Center, Peoria, Illinois (17): PI: Arun Talkad, MD; Co-Investigators: David Wang, DO; Maureen Matthews, APN, CNP, CNRN; coordinator: Mary Buttice. Rochester General Hospital, Rochester, New York (17): PIs: W. Scott Burgin, MD; Joshua Hollander, MD; coordinator: Cheryl Weber. University of Iowa Hospital, Iowa City, Iowa (15): PI: Patricia Davis, MD; coordinator: Jeri Sieren. University of South Alabama, Mobile, Alabama (15): PI: J. Ivan Lopez, MD; coordinator: Mel Parnell. Hôspital Charles Le Moyne, Greenfield Park, Province of Quebec, Canada (14): PI: Leo Berger, MD; coordinators: Martine Mainville; Louise-Marie Béliveau. Marshfield Clinic, Marshfield, Wisconsin (14): PI: Percy Karanjia, MD; Co-Investigator: Kenneth Madden, MD, PhD; coordinators: Kathy Mancl; Rob Bohl. University of California, Davis School of Medicine, Sacramento, California (13): PI: Piero Verro, MD; coordinators: John Bautista; Roxana Hupcey. Mercy Hospital, Sioux City, Iowa (13): PI: Jennifer Pary, MD; coordinators: Deb Motz; Darla Bauer. University of Texas Southwestern Medical Center at Dallas, Dallas, Texas (12): PI: Mark Johnson, MD; Co-Investigator: Jessica Lee, MD; coordinator: April Blair. Maine Medical Center, Scarborough, Maine (12): PI: John Belden; coordinator: Diane Diconzo-Fanning. Cleveland Clinic Florida, Weston, Florida (11): PI: Virgilio Salanga, MD; coordinator: Kilaun Thompson. Wake Forest University School of Medicine, Winston-Salem, North Carolina (11): PI: David Lefkowitz, MD; Co-Investigators: Patrick Reynolds, MD; Charles Tegeler, MD; coordinator: Emily Smith. University of Utah Stroke Center, Salt Lake City, Utah (10). PI: Elaine J. Skalabrin, MD; coordinator: Jill Austin. Helen Hayes Hospital, West Haverstraw, New York (10): PIs: Jason Greenberg, MD; Laura Lennihan, MD; coordinator: Laura Tenteromano. Stroke Prevention and Atherosclerosis Research Centre, London, Ontario Canada (10): PI: J. David Spence, MD; coordinator: Rose Freitas. Indiana University School of Medicine, Indianapolis, Indiana (9): PI: Linda Williams, MD; coordinator: Flossy Lincoln. Scripps Clinic, La Jolla, California (9): PI: Mary Kalafut, MD; coordinators: Crystal Sanchez; Gail Moore. Thomas Jefferson University Hospital, Philadelphia, Pennsylvania (8): PI: Rodney Bell, MD; coordinator: Prema Krishna. University of Kentucky, Lexington, Kentucky (7): PI: L. Creed Pettigrew, MD; coordinator: Deborah Taylor. University of Wisconsin, Madison, Wisconsin (7): PI: Robert J. Dempsey, MD; coordinator: Pam Winne. East Bay Region Associates in Neurology, Berkeley, California (7): PI: Brian Richardson, MD; coordinator: Lauren McCormick. Emory University, Atlanta, Georgia (7): PI: Barney J. Stern, MD; coordinator: Betty Jo Shipp. Inova Fairfax Hospital, Falls Church, Virginia (7): PI: Paul Nyquist, MD; coordinator: Barbara Farmer. Massachusetts General Hospital, Boston, Massachusetts (6): PI: Karen Furie, MD; coordinator: Ashley Birch. University of California Los Angeles Stroke Center, Los Angeles, California (6): PI: Jeffrey Saver, MD; coordinator: Hannah Smith. Florida Neurovascular Institute, Tampa, Florida (5): PI: Erfan Albakri, MD; coordinator: Valerie Walters. Seton Brain and Spine Institute, Seton Family of Hospitals, Austin, Texas (5): PI: Darryl Camp, MD; coordinators: Lisa Houy; Michele Ajgaonkar. Morton Plant Hospital - Neuroscience Institute, Clearwater, Florida (5): PI: Ajay Arora, MD; coordinators: Jo Simpson; Teresa Jones; Victoria Lindeman. Yale University School of Medicine, New Haven, Connecticut (5): PI: Lawrence Brass, MD; coordinators: Renee Dobos; Janet Halliday. The Stroke Center at Hartford Hospital, Hartford, Connecticut (4): PI: Isaac Silverman, MD; coordinators: Martha Ahlquist; Jennifer Blum. Southtowns Neurology, West Seneca, New York (4): PI: Peterkin Lee Kwen, MD; coordinator: Sarfaraz Semy. Vanderbilt Stroke Center, Nashville, Tennessee (4): PI: Anne E. O'Duffy, MD; coordinator: Diane Brown. University of North Carolina Chapel Hill, Chapel Hill, North Carolina (4): PI: Souvik Sen, MD; Co-Investigator: Natalie Aucutt-Walter, MD; coordinators: Roxanne Poole; Douglas Parker; Omid Akhavan. Elkhart Clinic, Elkhart, Indiana (3): PI: Thomas Vidic, MD; coordinator: Randall Gibson, PhD. USCD Stroke Center, San Diego, California (3): PI: Patrick Lyden, MD; coordinator: Nancy Kelly. Rush Presbyterian St. Luke’s Medical Center, Chicago, Illinois (3): PI: Sean Ruland, DO; coordinator: Karen Whited. University of Illinois at Chicago, Chicago, Illinois (3): PI: Cathy Helgason, MD; coordinator: Joan Martellotto. Johns Hopkins Bayview Medical Center, Baltimore, Maryland (3): PI: Rafael H. Llinas, MD; coordinator: Janice Alt. Field Neurosciences Institute, Saginaw, Michigan (3): PI: Faith Abbott, DO; coordinator: Richard Herm. Medical University of South Carolina, Charleston, South Carolina (3): PI: Timothy Carter, MD; coordinator: Feng Liu. Royal University Hospital, Saskatoon, Province of Saskatchewan, Canada (3): PI: Ali Rajput, MD; coordinator: Theresa Shirley. Chattanooga Center for Neurologic Research, Chattanooga, Tennessee (3): PI: Thomas Devlin, MD; coordinators: Katrina Baron; Tammi Owens. Lancaster General Hospital, Lancaster, Pennsylvania (2): PI: Venkatachalam Mangeshkumar, MD; coordinator: Holly Snyder. St. Mary’s Medical Center, Huntington, West Virginia (2): PI: Carl McComas, MD; coordinators: Jennifer Edwards; Cheryl Kane. Albert Einstein Medical Center, Philadelphia, Pennsylvania (2): PI: Jonathan Dissin, MD; coordinator: Rohini Bhole. Froedtert Memorial Lutheran, Milwaukee, Wisconsin (1): PI: Michel Torbey, MD; coordinator: Erin Brandenburg. Sparks Neurology Center, Fort Smith, Arkansas (1): PI: Margaret Tremwel, MD; coordinators: Cheryl Hyde; Mary Rambin. Syracuse VAMC, Syracuse, New York (1): PI: Antonio Culebras, MD; coordinator: Therese Dean. Rhode Island Hospital, Providence, Rhode Island (1): PI: Janed Wilterdink, MD; coordinator: Carol Cirillo. Charles R. Drew University of Medicine and Science, Los Angeles, California (1): PI: Lowell Nelson, MD; coordinator: Marcia Montenegro. Saint Joseph Mercy Oakland, Pontiac, Michigan (1): PI: Lionel Glass, MD; coordinator: Susan Oblak. Legacy Health System, Tualatin, Oregon (0): PI: Paul Ash, MD; coordinator: Taryn Lust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest/Disclosure
The funding source had no involvement in study design beyond peer review of the grant. The funding source had no involvement in collection, analysis, or interpretation of data; in writing of the report; or in the decision to submit the article for publication.
References
- 1.Wang X, Cheng S, Brophy VH, Erlich HA, Mannhalter C, Berger K, et al. A meta-analysis of candidate gene polymorphisms and ischemic stroke in 6 study populations: association of lymphotoxin-alpha in nonhypertensive patients. Stroke. 2009;40:683–695. doi: 10.1161/STROKEAHA.108.524587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikram MA, Seshadri S, Bis JC, Fornage M, DeStefano AL, Aulchenko YS, et al. Genomewide association studies of stroke. N Engl J Med. 2009;360:1718–1728. doi: 10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Debette S, Bis JC, Fornage M, Schmidt H, Ikram MA, Sigurdsson S, et al. Genome-wide association studies of MRI-defined brain infarcts: meta-analysis from the CHARGE Consortium. Stroke. 2010;41:210–217. doi: 10.1161/STROKEAHA.109.569194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The World Health Organization MONICA Project (monitoring trends and determinants in cardiovascular disease): a major international collaboration. WHO MONICA Project Principal Investigators. J Clin Epidemiol. 1988;41:105–114. doi: 10.1016/0895-4356(88)90084-4. [DOI] [PubMed] [Google Scholar]
- 5.Adams HP, Jr, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, 3rd, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24:35–41. doi: 10.1161/01.str.24.1.35. [DOI] [PubMed] [Google Scholar]
- 6.Worrall BB, Chen DT, Meschia JF. Ethical and methodological issues in pedigree stroke research. Stroke. 2001;32:1242–1249. doi: 10.1161/01.str.32.6.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meschia JF, Barrett KM, Chukwudelunzu F, Brown WM, Case LD, Kissela BM, et al. Interobserver Agreement in the Trial of Org 10172 in Acute Stroke Treatment Classification of Stroke Based on Retrospective Medical Record Review. Journal of Stroke and Cerebrovascular Diseases. 2006;15:266–272. doi: 10.1016/j.jstrokecerebrovasdis.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.R Development Core Team. Vienna, Austria: 2008. [Accessed on March 4, 2011]. R: A language and environment for statistical computing. R Foundation for Statistical Computing. http://www.R-project.org. [Google Scholar]
- 10.Morishita T, Tsutsui M, Shimokawa H, Horiuchi M, Tanimoto A, Suda O, et al. Vasculoprotective roles of neuronal nitric oxide synthase. FASEB J. 2002;16:1994–1996. doi: 10.1096/fj.02-0155fje. [DOI] [PubMed] [Google Scholar]
- 11.Huang Z, Huang PL, Panahian N, Dalkara T, Fishman MC, Moskowitz MA. Effects of cerebral ischemia in mice deficient in neuronal nitric oxide synthase. Science. 1994;265:1883–1885. doi: 10.1126/science.7522345. [DOI] [PubMed] [Google Scholar]
- 12.Meschia JF, Brown RD, Jr, Brott TG, Chukwudelunzu FE, Hardy J, Rich SS. The Siblings With Ischemic Stroke Study (SWISS) Protocol. BMC Med Genet. 2002;3:1. doi: 10.1186/1471-2350-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanktree MB, Dichgans M, Hegele RA. Advances in genomic analysis of stroke: what have we learned and where are we headed? Stroke. 2010;41:825–832. doi: 10.1161/STROKEAHA.109.570523. [DOI] [PubMed] [Google Scholar]
- 15.Meschia JF. Addressing the heterogeneity of the ischemic stroke phenotype in human genetics research. Stroke. 2002;33:2770–2774. doi: 10.1161/01.str.0000035261.28528.c8. [DOI] [PubMed] [Google Scholar]
- 16.Gschwendtner A, Bevan S, Cole JW, Plourde A, Matarin M, Ross-Adams, et al. Sequence variants on chromosome 9p21.3 confer risk for atherosclerotic stroke. Ann Neurol. 2009;65:531–539. doi: 10.1002/ana.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith JG, Melander O, Lovkvist H, Hedblad B, Engstrom G, Nilsson P, et al. Common genetic variants on chromosome 9p21 confers risk of ischemic stroke: a large-scale genetic association study. Circ Cardiovasc Genet. 2009;2:159–164. doi: 10.1161/CIRCGENETICS.108.835173. [DOI] [PubMed] [Google Scholar]
- 18.Gretarsdottir S, Thorleifsson G, Manolescu A, Styrkarsdottir U, Helgadottir A, Gschwendtner A, et al. Risk variants for atrial fibrillation on chromosome 4q25 associate with ischemic stroke. Ann Neurol. 2008;64:402–409. doi: 10.1002/ana.21480. [DOI] [PubMed] [Google Scholar]
- 19.Lemmens R, Buysschaert I, Geelen V, Fernandez-Cadenas I, Montaner J, Schmidt H, et al. The association of the 4q25 susceptibility variant for atrial fibrillation with stroke is limited to stroke of cardioembolic etiology. Stroke. 2010;41:1850–1857. doi: 10.1161/STROKEAHA.110.587980. [DOI] [PubMed] [Google Scholar]
- 20.Wiklund PG, Brown WM, Brott TG, Stegmayr B, Brown RD, Jr, Nilsson-Ardnor S, et al. Lack of aggregation of ischemic stroke subtypes within affected sibling pairs. Neurology. 2007;68:427–431. doi: 10.1212/01.wnl.0000252955.17126.6a. [DOI] [PubMed] [Google Scholar]
- 21.Arsava EM, Ballabio E, Benner T, Cole JW, Delgado-Martinez MP, Dichgans M, et al. The Causative Classification of Stroke system: an international reliability and optimization study. Neurology. 2010;75:1277–1284. doi: 10.1212/WNL.0b013e3181f612ce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meschia JF, Brott TG, Brown RD, Jr, Kissela BM, Hardy JA, Brown WM, et al. Correlation of proband and sibling stroke latency: the SWISS Study. Neurology. 2005;64:1061–1063. doi: 10.1212/01.WNL.0000154602.20719.E8. [DOI] [PubMed] [Google Scholar]
- 23.Vermeer SE, Longstreth WT, Jr, Koudstaal PJ. Silent brain infarcts: a systematic review. Lancet Neurol. 2007;6:611–619. doi: 10.1016/S1474-4422(07)70170-9. [DOI] [PubMed] [Google Scholar]
- 24.Howard VJ, McClure LA, Meschia JF, Pulley L, Orr SC, Friday GH. High prevalence of stroke symptoms among persons without a diagnosis of stroke or transient ischemic attack in a general population: the REasons for Geographic And Racial Differences in Stroke (REGARDS) study. Arch Intern Med. 2006;166:1952–1958. doi: 10.1001/archinte.166.18.1952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.