Abstract
Background
Targeted Induced Loci Lesions IN Genomes (TILLING) is increasingly being used to generate and identify mutations in target genes of crop genomes. TILLING populations of several thousand lines have been generated in a number of crop species including Brassica rapa. Genetic analysis of mutants identified by TILLING requires an efficient, high-throughput and cost effective genotyping method to track the mutations through numerous generations. High resolution melt (HRM) analysis has been used in a number of systems to identify single nucleotide polymorphisms (SNPs) and insertion/deletions (IN/DELs) enabling the genotyping of different types of samples. HRM is ideally suited to high-throughput genotyping of multiple TILLING mutants in complex crop genomes. To date it has been used to identify mutants and genotype single mutations. The aim of this study was to determine if HRM can facilitate downstream analysis of multiple mutant lines identified by TILLING in order to characterise allelic series of EMS induced mutations in target genes across a number of generations in complex crop genomes.
Results
We demonstrate that HRM can be used to genotype allelic series of mutations in two genes, BraA.CAX1a and BraA.MET1.a in Brassica rapa. We analysed 12 mutations in BraA.CAX1.a and five in BraA.MET1.a over two generations including a back-cross to the wild-type. Using a commercially available HRM kit and the Lightscanner™ system we were able to detect mutations in heterozygous and homozygous states for both genes.
Conclusions
Using HRM genotyping on TILLING derived mutants, it is possible to generate an allelic series of mutations within multiple target genes rapidly. Lines suitable for phenotypic analysis can be isolated approximately 8-9 months (3 generations) from receiving M3 seed of Brassica rapa from the RevGenUK TILLING service.
Keywords: TILLING, HRM, Brassica, genotyping
Background
The identification and analysis of gene mutations in plants is fundamental to the investigation of gene function. One approach, Targeted Induced Loci Lesions IN Genomes (TILLING) was originally developed in Arabidopsis [1] and has subsequently been successful in a range of crop plants [2,3]. In this reverse genetic approach, an ethyl methane sulfonate (EMS) mutagenised population is screened for SNPs within target genes [4]. EMS mutagenesis generates multiple alleles within each gene, including nonsense, missense, splicing and cis-regulatory mutants, in comparison to T-DNA and transposon mutagenesis that generate only knockout mutants [5,6]. Analysis of an allelic series can provide information on important domains or amino acids within the protein of interest. A number of TILLING populations have been developed for a variety of crops of different genome size, including rice [2], wheat [7,8], Brassica rapa [3], Brassica napus [9], Lotus japonicus [10], Medicago trunculata [11], Arachis hypogaea [12], and Solanum lycopersicum [13]. These populations have subsequently been used to isolate mutations in a variety of genes, including those involved in starch metabolism in Lotus japonicas [14], peanut allergens [12] and a gene encoding a fatty acid elongase [9].
High resolution melt (HRM) analysis is a technique that measures the disassociation of double-stranded DNA at high temperature resolution, and permits the analysis of genetic variations (SNPs, mutations, methylation) in PCR amplicons [15-17]. This technique allows genotyping and mutation scanning without the need for costly labeled probes, as it uses high fidelity heteroduplex-detecting double-stranded DNA binding dyes, such as EvaGreen which exhibit equal binding affinities for GC-rich and AT-rich regions and no sequence preference [18,19]. The heteroduplex products are detected by the presence of a second low-temperature melting transition [20]. This enables the reaction to be performed in a single tube, making it cost-effective and suitable for high-throughput screening [17]. HRM has been used extensively in the genotyping of human tissue samples for the identification of genes associated with diseases [21,22] as well as identification of clinically important fungal species [23]. HRM has been used for quantitative detection of adulteration with related species of pants used for medicinal purposes [24]. HRM has been used previously to identify mutations in TILLING populations of tomato [13], wheat [8] and Medaka [25]. In the tomato study, HRM was used to identify mutations using DNA pools from an EMS mutagenised population of plants. The wheat studies demonstrated that HRM can be used to isolate novel mutations in starch branching enzyme IIa (SBEIIa) genes from an EMS population [8] and in mutation scans in mixed PCR amplicons containing three homeologous gene fragments [19]. It was also demonstrated that homozygous and heterozygous individuals of a single mutation could be genotyped using HRM [19].
The genus Brassica includes the closest crop relatives of Arabidopsis thaliana, such as B. rapa (A-genome, 2n = 2x = 20, ~550 Mbp), which includes vegetable crops (e.g. turnip, Chinese cabbage) and oil-seed crops, B. oleracea (C-genome, 2n = 2x = 18, > 600 Mbp) which includes vegetable crops (cauliflower, broccoli, cabbage) and the amphidiploid B. napus (AC-genome, 2n = 4x = 38, ~1100 Mbp), which includes oil-seed crops (canola, oilseed rape) and swede. As with many crop plants Brassica genomes are complex arising from a series of duplication events that has resulted in most genes being present in multiple paralogous and homeologous copies [26,27]. A TILLING population has recently been generated in B. rapa to enable gene function to be analysed [3] and has previously been used to identify mutations in the B.rapa orthologue of the INDEHISCENT gene [28]. The aim of this study is to determine if HRM analysis can be used to genotype TILLING mutants in the crop species B. rapa and thereby to improve the efficiency of functional genomics approaches in this species.
Results and Discussion
Gene selection and TILLING
In order to demonstrate that HRM is suitable for efficient genotyping of mutant lines identified using TILLING, two target genes were chosen, BraA.CAX1.a and BraA.MET1.a. In A. thaliana, CAX1 has been shown to encode a calcium-proton antiporter [29] and MET1 has been shown to encode a DNA methyltransferase [30]. The CAX1 sequence from A. thaliana [Genbank: NM_129373.3] was used to identify the homologous sequence from B. rapa and from this sequence, 1.5 kb including the transcriptional start was used as the target for TILLING. To isolate BraA.MET1.a from B. rapa R-o-18, primers were designed based on the B. rapa Chiifu-401 sequence [Genbank:AB251937] [31]. These produced a 2110 bp DNA fragment covering 136 bp upstream of the translational start site together with the first intron and the first 407 bp of the second exon. Both sequences were submitted to the RevGenUK TILLING service, which resulted in 20 and 17 mutations being identified in BraA.CAX1.a and BraA.MET1.a respectively (Table 1). These mutations included silent, missense, nonsense and non-coding mutations.
Table 1.
Nucleotide changes in BraA.cax1.a and BraA.met1.a TILLING mutants.
| Linea | Mutation Changeb | Position (base-pair)c | Position (amino-acid)d | Type of mutatione |
|---|---|---|---|---|
| BraA.cax1.a-1 | G to A | 881 | - | Non-coding |
| BraA.cax1.a-2 | C to T | 860 | - | Non-coding |
| BraA.cax1.a-3 | G to A | 545 | 2 | Non-coding |
| BraA.cax1.a-4 | G to A | 771 | 77 | Missense |
| BraA.cax1.a-5 | C to T | 494 | - | Non-coding |
| BraA.cax1.a-6 | G to A | 517 | - | Non-coding |
| BraA.cax1.a-7 | G to A | 673 | 44 | Missense |
| BraA.cax1.a-8 | G to A | 599 | 19 | Missense |
| BraA.cax1.a-9 | G to A | 645 | 35 | Missense |
| BraA.cax1.a-10 | C to T | 736 | 65 | Missense |
| BraA.cax1.a-11 | G to A | 795 | 85 | Missense |
| BraA.cax1.a-12 | C to T | 708 | 56 | Missense |
| BraA.cax1.a-13 | C to T | 689 | 49 | Silent |
| BraA.cax1.a-14 | C to T | 661 | 40 | Missense |
| BraA.cax1.a-15 | C to T | 658 | 39 | Missense |
| BraA.cax1.a-16 | C to T | 421 | - | Non-coding |
| BraA.cax1.a-17 | G to A | 926 | - | Non-coding |
| BraA.cax1.a-18 | C to T | 818 | 92 | Silent |
| BraA.cax1.a-19 | C to T | 732 | 64 | Missense |
| BraA.cax1.a-20 | C to T | 690 | 50 | Nonsense |
| BraA.met1.a-1 | C to T | 3516 | 248 | Silent |
| BraA.met1.a-2 | G to A | 3334 | 188 | Missense |
| BraA.met1.a-3 | G to A | 3334 | 188 | Missense |
| BraA.met1.a-4 | G to A | 3084 | 104 | Silent |
| BraA.met1.a-5 | C to T | 3081 | 103 | Silent |
| BraA.met1.a-6 | G to A | 3252 | 243 | Nonsense |
| BraA.met1.a-7 | G to A | 3500 | 243 | Missense |
| BraA.met1.a-8 | G to A | 3012 | 80 | Silent |
| BraA.met1.a-9 | G to A | 3495 | 241 | Silent |
| BraA.met1.a-10 | C to T | 3335 | 188 | Missense |
| BraA.met1.a-11 | C to T | 3069 | 99 | Silent |
| BraA.met1.a-12 | G to A | 3330 | 186 | Silent |
| BraA.met1.a-13 | C to T | 3324 | 184 | Silent |
| BraA.met1.a-14 | G to A | 3213 | 147 | Nonsense |
| BraA.met1.a-15 | C to T | 3265 | 165 | Silent |
| BraA.met1.a-16 | G to A | 3330 | 186 | Silent |
| BraA.met1.a-17 | G to A | 3258 | 162 | Silent |
a - TILLING line, b - Nucleotide change in mutant line, c - Position of nucleotide change in base-pairs, d - Position of amino-acid change in peptide, e - Type of mutation caused by nucleotide change. Non-coding - nucleotide change not within coding region, missense - nucleotide change results in an amino-acid change, nonsense - nucleotide change results in the introduction of a premature stop codon, silent - no change in amino-acid.
Genotyping of TILLING lines using HRM
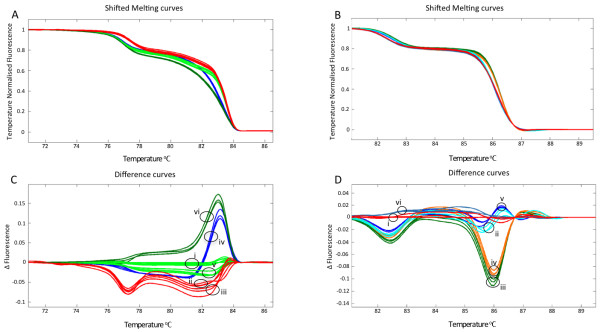
From the lines identified using TILLING, twelve (BraA.cax1.a-1, -4, -6, -7, -8, -10, -11, -12, -14, -16, -18, -20) and five (BraA.met1.a-3, -6, -7, -10 and -14) lines carrying mutations in BraA.CAX1 and BraA.MET1.a respectively were chosen to be characterised based on the nature and position of the mutation. In order to genotype the lines, a HRM assay was developed for each gene. To increase the specificity of the HRM reaction a nested PCR approach was used for the BraA.cax1.a lines, first using the TILLING primers to amplify genomic DNA, prior to using the HRM primers with this product. In total, three primer pairs were designed to genotype all 12 mutations within the BraA.CAX1.a gene (amplifying fragments of 200, 203 and 260 bp; Table 2) and one primer pair was designed to genotype all the mutations within the BraA.MET1.a gene (amplifying a 399 bp fragment; Table 2). Individual M3 plants of the TILLING lines and wild-type R-o-18 plants were then genotyped using the HRM assay using a single primer pair per plate and three technical replicates per plant, allowing multiple alleles to be genotyped in a single scanning run. Analysis of the difference plots for the fluorescent signals detected individual plants that were wild-type, homozygous or heterozygous for the mutation in alleles of both genes (Figure 1). Three distinct groups can readily be observed in the difference plots. To confirm these results and assign zygosity groups, the PCR products from selected lines were sequenced. Sequencing the HRM PCR products identified by HRM as heterozygous from BraA.cax1.a-11 and BraA.met1.a-6, showed a double peak at the position of the mutation (Figure 2), confirming that these were indeed heterozygous for the mutations. In addition, sequencing of PCR products from BraA.cax1.a-1 and BraA.met1.a-6 identified as homozygous by HRM, confirmed that they were homozygous for the mutation (Figure 2). These results demonstrate that HRM can be used both for rapid genotyping of TILLING mutants and for distinguishing wild-type, heterozygous and homozygous plants. The results also demonstrate that multiple alleles can be genotyped using a single primer pair on one PCR plate, allowing multiple lines and individual plants to be genotyped simultaneously.
Table 2.
Details of primers used for gene isolation and HRM analysis.
| Primera | Geneb | Stagec | Mutation Targetedd | Sequence 5' to 3'e |
|---|---|---|---|---|
| BrCAX1 TILLING_Forward | BraA.CAX1.a | TILLING | AGAGATTTCCTAGCCATGTG | |
| BrCAX1 TILLING_Reverse | BraA.CAX1.a | TILLING | CGACCCCTAATTGTTTTATGTG | |
| BrCAX1 HRM1_Forward | BraA.CAX1.a | HRM | 6,16 | TCCTCGAAGTTGCCTCTGAT |
| BrCAX1 HRM1_Reverse | BraA.CAX1.a | HRM | 6,16 | GCTGCTGACCATTGTTCCTG |
| BrCAX1 HRM3_Forward | BraA.CAX1.a | HRM | 7,8,12,14,20 | GCAGGAACAATGGTCAGCAG |
| BrCAX1 HRM3_Reverse | BraA.CAX1.a | HRM | 7,8,12,14,20 | CGAGAATGACTTCTTGGAGATT |
| BrCAX1 HRM6_Forward | BraA.CAX1.a | HRM | 4,10,11,18 | TCCAGAAGGTTCCATACAAAGG |
| BrCAX1 HRM6_Reverse | BraA.CAX1.a | HRM | 4,10,11,18 | CGCTGTTCTGTTTAGTAATGTGTTG |
| BrMet1aF4 | BraA.MET1.a | Gene isolation | ACTTCACAGCATCTCCTCAGGGC | |
| BrMet1aF5 | BraA.MET1.a | Gene isolation | CTTGACAATGGTGCTGTTATTCAG | |
| BrMet1aF6 | BraA.MET1.a | Gene isolation | GTCTGGTTTGATGGCAGAGGACG | |
| BrMet1aR4 | BraA.MET1.a | Gene isolation | CGCACTTCGCAGCTTCAGAAAC | |
| BrMet1aR5 | BraA.MET1.a | Gene isolation | AAATAGAGATTGTTGAACACCCCC | |
| BrMet1aR6 | BraA.MET1.a | Gene isolation | TTGGATACCACAGGTGCCATGC | |
| BrMet1aHRMF | BraA.MET1.a | HRM | 3,6,7,10,14 | TGCGATGATAAGGAGAAAGG |
| BrMet1aHRMR | BraA.MET1.a | HRM | 3,6,7,10,14 | TTTGCCGTCTCATCCAAAC |
a- Primer name, b-gene primer designed to, c-stage of analysis primer used, d-line primers used to genotype, e- sequence of primer
Figure 1.
HRM analysis for Brassica rapa R-o-18 wild-type and individual plants from M3 BraA.cax1.a-11 and BraA.met1.a-6 TILLING mutants. A: Normalised melt curves for fluorescent signals from DNA strand dissociation of triplicated technical replicates of B. rapa wild-type (light green lines) and individual plants of M3 TILLING mutant BraA.cax1.a-11, plant BraA.cax1.a-11A (red), plant BraA.cax1.a-11B (red), plant BraA.cax1.a-11C (blue), plant BraA.cax1.a-11D (light green), plant BraA.cax1.a-11E (dark green). B: Normalised melt curves for fluorescent signals from DNA strand dissociation of triplicated technical replicates of B. rapa wild type (red lines) and individual plants M3 TILLING mutant BraA.met1.a-6, plant BraA.met1.a-6-1 (light blue), plant BraA.met1.a-6-2 (green), plant BraA.met1.a-6-3 (orange), plant BraA.met1.a-6-4 (purple), plant BraA.met1.a-14-1 (blue). C: Difference plot of genotypes' fluorescence normalised to wild type samples (light green lines, i) and individual plants of M3 TILLING mutant BraA.cax1.a-11, plant BraA.cax1.a-11A (red, ii), plant BraA.cax1.a-11B (red, iii), plant BraA.cax1.a-11C (blue, iv), plant BraA.cax1.a-11D (light green, v), plant BraA.cax1.a-11E (dark green, vi). D: Difference plot of genotypes' fluorescence normalised to wild type (red, i) and individual plants of the M3 TILLING mutant BraA.met1.a-2, plant BraA.met1.a-6-1 (light blue, ii), plant BraA.met1.a-6-2 (green, iii), plant BraA.met1.a-6-3 (orange, iv), plant BraA.met1.a-6-4 (purple, v), plant BraA.met1.a-14-1 (blue, vi).
Figure 2.
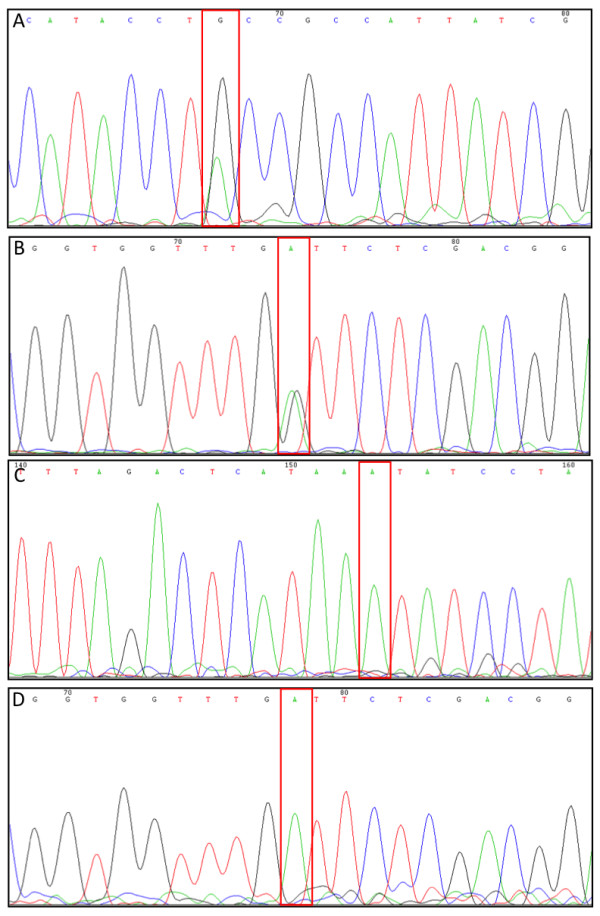
Sequencing chromatograms of M3 generation of B. rapa R-o-18 mutants. A: BraA.cax1.a-11, gene sequence from 788 bp to 807 bp shown. B: BraA.met1.a-6, gene sequence from 3246 bp to 3265 bp shown. C: BraA.cax1.a-1, gene sequence from 868 bp to 888 bp shown. D: BraA.met1.a-6, gene sequence from 3246 bp to 3265 bp shown. Rectangular box indicates zygosity state at the site of EMS induced transitional mutation.
Backcross analysis
To reduce the mutation load, reciprocal crosses of the TILLING lines and wild-type R-o-18 plants were performed with both homozygous and heterozygous mutant plants. A heterozygous plant from the BraA.met1.a-6 line was crossed to a wild-type plant and ten F1 plants were genotyped. The results from the genotyping showed the expected segregation of the mutant allele, with 5 plants having a heterozygous mutation and 5 plants being wild-type for the mutation. The genotyping results for the F1 plants from crosses of the BraA.cax1.a lines also showed the expected genotyping results (Table 3). For example, the F1 plants of a cross using a homozygous BraA.cax1.a-1 plant were all heterozygous for the mutation (n = 5). In contrast, the F1 progeny of heterozygous plants showed segregation for the mutation (Table 3). These results demonstrate that HRM can be used to follow mutations and zygosity through multiple generations and crossing events, making it an ideal method for genotyping of TILLING mutants.
Table 3.
HRM genotyping results of F1 progeny of BraA.cax1.a and BraA.met1.a lines crossed to wild-type.
| Linea | Plantb | Number of heterozygous plantsc | Number of wild-type plantsd |
|---|---|---|---|
| BraA.cax1.a-1 | E | 5 | 0 |
| BraA.cax1.a-4 | C | 2 | 0 |
| BraA.cax1.a-6 | B | 4 | 2 |
| BraA.cax1.a-7 | A | 3 | 3 |
| BraA.cax1.a-8 | A | 4 | 1 |
| BraA.cax1.a-10 | A | 3 | 2 |
| BraA.cax1.a-11 | B | 3 | 3 |
| BraA.cax1.a-12 | A | 3 | 3 |
| BraA.cax1.a-14 | B | 3 | 0 |
| BraA.cax1.a-18 | D | 2 | 4 |
| BraA.cax1.a-20 | A | 2 | 2 |
| BraA.met1.a-6 | A | 5 | 5 |
a - TILLING line crossed to B. rapa R-o-18 wild-type, b - Individual M3 plant identifier' c - Number of individual F1 heterozygous plants identified by HRM' d - Number of individual F1 wild-type plants identified by HRM
Conclusions
To analyse an allelic series of mutants generated using the TILLING approach efficiently, a cost effective, high-throughput genotyping technique is required. Techniques that have been used previously include PCR-RFLP, but this relies on the mutation causing a change in a restriction site [32] and TaqMan assays, which can require expensive probes and assay optimisation [18,33]. Alternatively, PCR products covering the mutations could be sequenced. However, this is time consuming and potentially more expensive; HRM analysis of samples in triplicate costs ~30% of Sanger sequencing costs. The results presented here demonstrate that HRM can be used to genotype mutations identified by TILLING from EMS populations in B. rapa. The results also demonstrate that a single PCR primer pair could be used to identify mutations at multiple positions with the PCR fragment. This reduces the number of primer pairs needed to genotype an allelic series and hence the cost. Due to the nature of the HRM process, the DNA extraction, PCRs and melt curve analysis can all be performed in 96-well plates. This enables high-throughput genotyping of multiple alleles of a number of genes to be performed quickly and efficiently. This is particularly important in complex crop species such as B rapa, where paralogous genes may be present and accumulation of mutations may be required to generate a phenotype.
Methods
Plant material
Seed of the highly inbred, homozygous, self-compatible, rapid cycling B. rapa ssp. trilocularis line R-o-18 was used as wild-type control. Seeds for lines (M3 generation) containing mutations in target genes were obtained from the R-o-18 TILLING population through the RevGenUK service [3,34].
Plant growth conditions
Plants were grown under glasshouse conditions with a 16 h photoperiod at 22.3 ± 4°C and 13.3 ± 2°C mean day and night temperatures respectively. Seeds were sown directly into pots (height 11.3 cm; diameter 13 cm) containing Levington M3 high nutrient compost (Monro Group, Chichester, UK) and irrigated twice daily. Following three weeks of vegetative growth, Vitafeed® 2-1-4 nutrient solutions (N-P-K: 16-8-32 + micronutrients; Vitax Ltd., Coalville, Leicestershire, UK) were applied to plants weekly at a rate of 3 g l-1 via overhead irrigation. Plants initiated flowering after approximately 6 weeks of growth and seeds were collected approximately 4 weeks thereafter. Perforated bread bags (150 mm × 700 mm, WR Wright & Sons Ltd, Liverpool, UK) were used to enclose inflorescences to prevent cross-pollination from neighboring plants.
In silico data mining to identify gene sequences
Basic Local Alignment Search Tool (BLAST) [35] software was employed to identify sequences from all Brassica spp. that were orthologous to the A. thaliana CAX1 gene [Genbank: NM_129373.3]. All A. thaliana sequences of interest were retrieved from GenBank [36]. For the BLASTn algorithm [37] the entire GenBank database (March 2009) was interrogated to identify all Brassica sequences sharing > 80% sequence identity at an e-value of < 1e-30 with the A. thaliana query sequence. The Brassica Genome Sequencing Project (BrGSP) database [38] was interrogated (March 2009) using the WuBLAST [39] algorithm for all Brassica BAC-end sequences, fully sequenced BACs, ESTs and genome survey sequence which shared > 80% sequence identity with this query sequence. A. thaliana genes sharing high sequence similarities to the query gene i.e. members of the same gene family including AtCAX2 [Genbank:NM_001036119.1] and AtCAX3 [Genbank:NM_115045.3] were used to interrogate both databases. After in silico comparative sequence analyses, Brassica sequences were assigned to the member of the A. thaliana gene family with which they shared the highest sequence identity, thus removing false positives. All selected sequences which were orthologous to AtCAX1 were formatted into FASTA and entered into a contig assembly program (ContigExpress, VectorNTI 11, Invitrogen, Paisley, UK) using default settings to form longer contiguous Brassica sequences and identify possible locus specific paralogues. These nascent Brassica gene sequences were aligned to the A. thaliana gene of interest, flanked at either end with 1 Mbp of sequence, using AlignX software at default settings. The genomic structure of these contigs was elucidated and annotated in the Vector map software (VectorNTI 11). Primers (Table 2) were designed to amplify ~ 1.2 kbp fragments of BraA.CAX1.a beginning upstream from the deduced transcriptional start site to a region within the gene using Primer 3 (Version 0.4.0) [40] with the parameters "Max Self Complementarity" and "Max 3' Self Complementarity" adjusted to 2, to avoid hairpin loops and potential dimerisation. Primers to amplify the BraA.MET1.a gene (Table 2) from B. rapa R-o-18 were designed based on the sequence described by Fujimoto et al. [[31], Genbank:AB251937].
Amplification of locus specific sequences from B. rapa R-o-18
DNA was extracted from leaves of wild-type B. rapa R-o-18 using the GenElute® Plant Genomic DNA Miniprep kit (Sigma-Aldrich, Gillingham, UK) according to the manufacturer's instructions and subjected to PCR amplification using primers (Table 2) designed to Brassica contig sequence. Amplified fragments were ligated in plasmid pCR8®/GW/TOPO® (Invitrogen, Paisley, UK) and subsequently sequenced.
Plant genomic DNA extraction
Genomic DNA was extracted from the BraA.met1.a plants using the DNeasy Plant Min kit (Qiagen, Crawley, UK) following the manufacturer's instructions or using a rapid salt-extraction method [41]. For the BraA.cax1.a plants, a high throughput extraction method was developed based on a method developed by Dr. M. Poole (pers. comm.). A 0.25 cm2 diameter leaf disc from a developing or mid-expansion leaf from each plant was placed in a 96-well plate (Qiagen Collection Microtubes, Qiagen, Crawley, UK), with a single 3 mm stainless steel ball and a 600 μl aliquot of DNA extraction buffer (200 mM Tris-HCL pH 7.5, 250 mM NaCl, 25 mM EDTA and 0.5% w/v SDS) added to each well. Suspensions were shaken for 3 min in a ball mill (8000 Mixer Mill, Glen Creston, Stanmore, UK) until the majority of tissue was disrupted. Plates were subsequently centrifuged in a plate centrifuge (Centrifuge 5804, Eppendorf UK, Cambridge, UK) at 5,600 g for 20 min. After centrifugation 300 μl of the supernatant was transferred to a fresh plate (Thermofisher storage plate with rubber sealing mat, Fisher, Loughborough, UK), containing 300 μl of 100% isopropanol, mixed and incubated for 3 min at room temperature. Plates were then centrifuged at 5600 g for 20 min before discarding the supernatant. A 100 μl aliquot of 70% (v/v) ethanol was added and mixed by shaking vigorously for 5 min, centrifuged at 5,600 g for 20 min. The supernatant was discarded and dried in a vacuum manifold for 10 min. DNA was resuspended in 20 μl sterile H2O. The DNA concentration was not quantified and standardized due to the number of samples extracted simultaneously.
High Resolution Melt (HRM) analysis
Primers were designed for HRM analysis to amplify 100-250 bp regions within the BraA.CAX1.a gene and a 399 bp region of BraA.MET1.a, which were originally used to identify mutations by TILLING. Three sets of HRM primer pairs were designed for BraA.CAX1.a and one for BraA.MET1.a (Table 2), using Primer 3 (Version 0.4.0) with the parameters "Max Self Complementarity" and "Max 3' Self Complementarity" adjusted to 2, and a Tm ≥ 60°C, to avoid hairpin loops and dimerisations. Primer sequences were examined further by primer thermodynamic software (Vector NTI 11) to confirm that no secondary structures were likely to form. HRM primer amplification efficiencies and specificities, were determined in vitro by PCR amplification of wild-type B. rapa R-o-18 DNA in reaction volumes containing 10 μl of GoTaq buffer (5×), 5 μl of MgCl2 (25 mM), 1 μl of dNTPs (10 mM), 5 μl of both forward and reverse primers (10 pmol μl-1), DNA (10 ng), 0.2 μl of Taq polymerase (GoTaq) and sterile de-ionised water (SDW) to a total volume of 50 μl. PCR conditions were 95°C for 5 min and 30 cycles of 95°C for 10 sec, 55°C for 10 sec and 72°C for 30 sec.
Nested PCR approaches to amplify locus specific fragments for HRM analysis
The nested PCR approach was used for the BraA.CAX1.a lines. DNA extracted from TILLING lines were amplified with TILLING primers using Phusion™ high fidelity DNA polymerase (New England Biolabs, Hitchin, UK) to ensure sequence integrity of amplified sequences. Reaction volumes comprised of 4 μl of Phusion HF (5×), 0.4 μl of dNTPs (10 mM), 1.4 μl of forward and reverse BraA.CAX1.a TILLING primers (10 pmol μl-1), 1 μl DNA, 0.6 μl of DMSO, 0.2 μl of Phusion DNA polymerase and SDW to a total volume of 20 μl. PCR conditions involved an initial denaturation at 98°C (30 sec) followed by 30 cycles of denaturation at 98°C (5-10 sec), annealing at 58°C (15 sec) and extension at 72°C (30 sec).
HRM reactions
HRM were performed using Type-it® HRM™ PCR kit (Qiagen, Crawley, UK) following manufacturer's instructions; 1 μl of the initial nested PCR reaction for the BraA.cax1.a lines and 1 μl of genomic DNA for the BraA.met1.a lines was added as template to the HRM PCR reaction mix comprising 5 μl of 2 × HRM PCR Master Mix (including EvaGreen fluorescent dye), 1.75 μl of forward and reverse HRM primers (10 pmol μl-1) and SDW to 10 μl. Using an Eppendorf 96-well MasterCycler 5331, reaction volumes were subjected to 95°C (5 min), followed by 30 cycles of 95°C (10 sec), 55°C annealing (10 sec) and 72°C extension (30 sec). Included within each 96-well analysis plate were samples in triplicate, also triplicate wild-type DNA samples and two sets of negative controls, one without DNA added and one without HRM primer added.
Lightscanner™ analysis of HRM products
PCR plates containing HRM amplicons were placed into a Lightscanner™ (Idaho Technology, Salt Lake City, USA) and the temperature raised to 60°C for 5 mins to ensure all samples were equilibrated. The melt temperature range was set at 63.3°C-95.4°C with a ramp setting at 0.1°C and a second hold at each step. Exposure was set at "Auto", background correction to exponential, curve shift to 0.020, standards to 'Auto Group', and sensitivity at normal +2.8. HRM analysis was then performed on the dissociation of double-stranded DNA PCR products, which had been saturated with the low PCR-toxic dye, EvaGreen from the initial HRM PCR. The LightScanner™ Data Analysis software (Version 2.0, Idaho Technology) was used to analyse the data and produce normalised disassociation curves and difference plots.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SÓL developed and performed the BraA-cax1.a analysis and took the co-lead role in writing the manuscript. SA developed and performed the BraA-met1.a analysis. NG performed the BraA-cax1.a analysis and took the co-lead role in writing the manuscript. KA and JJR performed the BraA-cax1.a analysis. AS and SK developed the HRM analysis. JPH, LØ, GJK, PJW and MRB conceived and participated in development and coordination of the project. All authors contributed to the editing of the manuscript. All authors have read and approved the final manuscript.
Contributor Information
Seosamh Ó Lochlainn, Email: seosamh8@yahoo.com.
Stephen Amoah, Email: amoahscg@gmail.com.
Neil S Graham, Email: neil.graham@nottingham.ac.uk.
Khalid Alamer, Email: sbxkhha@nottingham.ac.uk.
Juan J Rios, Email: juan.rios@nottingham.ac.uk.
Smita Kurup, Email: smita.kurup@rothamsted.ac.uk.
Andrew Stoute, Email: andrew.stoute@rothamsted.ac.uk.
John P Hammond, Email: j.hammond@nottingham.ac.uk.
Lars Østergaard, Email: lars.ostergaard@bbsrc.ac.uk.
Graham J King, Email: graham.king@scu.edu.au.
Phillip J White, Email: philip.white@hutton.ac.uk.
Martin R Broadley, Email: martin.broadley@nottingham.ac.uk.
Acknowledgements
Funding for this work was provided by BBSRC (grant reference BB/G013969/1) to NG, JH, MRB and GJK and (BB/F009721/1) to GJK, SK and AS.
References
- McCallum CM, Comai L, Greene EA, Heinkoff S. Targeting Induced Lesions IN Genomes (TILLING) for plant functional genomics. Plant Physiology. 2000;123:439–442. doi: 10.1104/pp.123.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till BJ, Cooper J, Colowit P, Greene EA, Henikoff S, Comai L. Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biology. 2007;7:19. doi: 10.1186/1471-2229-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson P, Baker D, Girin T, Perez A, Amoah S, King GJ, Østergaard L. A rich TILLING resource for studying gene function in Brassica rapa. BMC Plant Biology. 2010;10:62. doi: 10.1186/1471-2229-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till BJ, Reynolds SH, Greene EA, Codomo CA, Enns LC, Johnson JE, Burtner C, Odden AR, Young K, Taylor NE, Henikoff JG, Comai L, Henikoff S. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Research. 2003;13:524–530. doi: 10.1101/gr.977903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz R, Feldman KA. T-DNA insertion mutagenesis in Arabidopsis: Going back and forth. Trends in Genetics. 1997;13:152–156. doi: 10.1016/S0168-9525(97)01094-9. [DOI] [PubMed] [Google Scholar]
- Balcells L, Swinburne J, Coupland G. Transposons as tools for the isolation of plant genes. Trends in Biotechnology. 1991;9:31–37. doi: 10.1016/0167-7799(91)90009-7. [DOI] [Google Scholar]
- Uauy C, Paraiso F, Colasuonno P, Tran RK, Tsai H, Berardi S, Comai L, Dubcovsky J. A modified TILLING approach to detect induced mutations in tetraploid and haexaploid wheat. BMC Plant Biology. 2009;9:115. doi: 10.1186/1471-2229-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botticella E, Sestili F, Hernandez-Lopez A, Phillips A, Lafiandra D. High Resolution Melting analysis for the detection of EMS induced mutations in wheat Sbella genes. BMC Plant Biology. 2011;11:156. doi: 10.1186/1471-2229-11-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Wang YJ, Tian F, King GJ, Zhang CY, Long Y, Shi L, Meng JL. A functional genomics resource for Brassica napus: development of an EMS mutagenized population and discovery of FAE1 point mutations by TILLING. New Phytologist. 2008;180:751–765. doi: 10.1111/j.1469-8137.2008.02619.x. [DOI] [PubMed] [Google Scholar]
- Perry JA, Wang TL, Welham TJ, Gardner S, Pike JM, Yoshida S, Parniske M. A TILLING reverse genetics tool and a web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiology. 2003;131:866–871. doi: 10.1104/pp.102.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Signor C, Savois V, Aubert G, Verdier J, Nicolas M, Pagny G, Moussy F, Sanchez M, Baker D, Clarke J, Thompson R. Optimizing TILLING populations for reverse genetics in Medicago trunculata. Plant Biotechnology Journal. 2009;7:430–441. doi: 10.1111/j.1467-7652.2009.00410.x. [DOI] [PubMed] [Google Scholar]
- Knoll JE, Ramos ML, Zeng Y, Holbrook CC, Chow M, Chen S, Maleki S, Bhattacharya A, Ozias-Akins P. TILLING for allergen reduction and improvement of quality traits in peanut (Arachis hypogaea L.) BMC Plant Biology. 2011;11:81. doi: 10.1186/1471-2229-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gady ALF, Hermans FWK, Van de Wal MHBJ, van Loo EN, Visser RGF, Bachem CWB. Implementation of two high through-put techniques in a novel application: detecting point mutations in large EMS mutated plant populations. Plant Methods. 2009;5:13. doi: 10.1186/1746-4811-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriet C, Welham T, Brachmann A, Pike J, Perry J, Parniske M, Sato S, Tabata S, Smith AM, Wang TL. A suite of Lotus japonicus starch mutants reveals both conserved and novel features of starch metabolism. Plant Physiology. 2010;154:643–655. doi: 10.1104/pp.110.161844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundry CN, Vandersteen JG, Reed GH, Pryor RJ, Chen J, Wittwer CT. Amplicon melting analysis with labeled primers: A closed-tube method for differentiating homozygotes and heterozygotes. Clinical Chemistry. 2003;49:396–406. doi: 10.1373/49.3.396. [DOI] [PubMed] [Google Scholar]
- Garritano S, Gemignani F, Voegele C, Nguyen-Dumont T, Le Calvez-Kelm F, De Silva D, Lesueur F, Landi S, Tavtigian SV. Determining the effectiveness of High Resolution Melting analysis for SNP genotyping and mutation scanning at the TP53 locus. BMC Genetics. 2009;10:5. doi: 10.1186/1471-2156-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino A, Mancuso T, Rossi AM. Application of high-resolution melting to large-scale, high-throughput SNP genotyping: A comparison with the TaqMan® method. Journal of Biomolecular Screening. 2010;15:623–629. doi: 10.1177/1087057110365900. [DOI] [PubMed] [Google Scholar]
- Monis PT, Giglio S, Saint CP. Comparison of SYT09 and SYBR Green I for real-time polymerase chain reaction and investigation of the effect of the dye concentration on amplification and DNA melting curve analysis. Analytical Biochemistry. 2005;340:24–34. doi: 10.1016/j.ab.2005.01.046. [DOI] [PubMed] [Google Scholar]
- Dong C, Vincent K, Sharp P. Simultaneous mutation detection of three homoeologous genes in wheat by High Resolution Melting analysis and Mutation Surveyor®. BMC Plant Biology. 2009;9:143. doi: 10.1186/1471-2229-9-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clinical Chemistry. 2003;49:853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- Poláková KM, Loptová T, Klamová H, Moravcová J. High-resolution melt curve analysis: Initial screening for mutations in BCR-ABL kinase domain. Leukemia Research. 2008;32:1236–1243. doi: 10.1016/j.leukres.2008.01.010. [DOI] [PubMed] [Google Scholar]
- Joly P, Lacan P, Garcia C, Delasaux A, Francina A. Rapid and reliable β-globin gene cluster haplotyping of sickle cell disease patients by FRET Light Cycler and HRM assays. Clinica Chimica Acta. 2011;412:1257–1261. doi: 10.1016/j.cca.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Arancia S, Sandini S, De Bernardis F, Fortini D. Rapid, simple, and low-cost identification of Candida species using high-resolution melting analysis. Diagnostic Microbiology and Infectious Disease. 2011;69:283–285. doi: 10.1016/j.diagmicrobio.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Mader E, Ruzicka J, Schmiderer C, Novak J. Quantitative high-resolution melting analysis for detecting adulterations. Analytical Biochemistry. 2011;409:153–155. doi: 10.1016/j.ab.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Kamei Y, Otozai S, Kim J, Sato A, Kuwahara Y, Tanaka M, Deguchi T, Inohara H, Tsujimura T, Todo T. High-resolution melting curve analysis for rapid detection of mutations in a Medaka TILLING library. BMC Molecular Biology. 2010;11:70. doi: 10.1186/1471-2199-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana D, Boogart T, O'Neil CM, Hynes L, Bent E, MacPherson L, Park JY, Lim YP, Bancroft I. Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. The Plant Journal. 2004;40:725–733. doi: 10.1111/j.1365-313X.2004.02244.x. [DOI] [PubMed] [Google Scholar]
- Parkin IAP, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics. 2005;171:765–781. doi: 10.1534/genetics.105.042093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girin T, Stephenson P, Goldsack CM, Kempin SA, Perez A, Pires N, Sparrow PA, Wood TA, Yanofsky MF, Ostergaard L. Brassicaceae INDEHISCENT genes specify valve margin cell fate and repress replum formation. Plant Journal. 2010;63:329–338. doi: 10.1111/j.1365-313X.2010.04244.x. [DOI] [PubMed] [Google Scholar]
- Cheng N-H, Pittman JK, Barkla BJ, Shigaki T, Hirschi KD. The Arabidopsis cax1 mutant exhibits impaired ion homeostasis, development, and hormonal responses and reveals interplay among vacuolar transporters. Plant Cell. 2003;15:347–364. doi: 10.1105/tpc.007385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES. Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Research. 1993;21:2383–2388. doi: 10.1093/nar/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto R, Sasaki T, Nishio T. Characterization of DNA methyltransferase genes in Brassica rapa. Genes & Genetic Systems. 2006;81:235–242. doi: 10.1266/ggs.81.235. [DOI] [PubMed] [Google Scholar]
- Parsons BL, Heflich RH. Genotypic selection methods for the direct analysis of point mutations. Mutation Research. 1997;387:97–121. doi: 10.1016/S1383-5742(97)00026-4. [DOI] [PubMed] [Google Scholar]
- Fitzgerald TL, Kazan K, Li Z, Morell MK, Manners JM. A high-throughput method for detection of homeologous gene deletions in hexaploid wheat. BMC Plant Biology. 2010;10:264. doi: 10.1186/1471-2229-10-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RevGenUK. http://revgenuk.jic.ac.uk
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- GenBank. http://www.ncbi.nlm.nih.gov/genbank/
- BLAST. http://blast.ncbi.nlm.nih.gov/Blast.cgi
- Brassica Genome Gateway. http://brassica.bbsrc.ac.uk
- Chao KM, Pearson WR, Miller W. Aligning two sequences within a specified diagonal band. Computer Applications in the Biosciences. 1992;8:481–487. doi: 10.1093/bioinformatics/8.5.481. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S, Misener S, editor. Humana Press, Totowa, NJ; 2000. Primer3 on the WWW for general users and for biologist programmers; pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Aljanabi SM, Martinez I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Research. 1997;25:4692–4693. doi: 10.1093/nar/25.22.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]