Abstract
Extra-intestinal pathogenic E. coli (ExPEC), including avian pathogenic E. coli (APEC), pose a considerable threat to both human and animal health, with illness causing substantial economic loss. APEC strain χ7122 (O78∶K80∶H9), containing three large plasmids [pChi7122-1 (IncFIB/FIIA-FIC), pChi7122-2 (IncFII), and pChi7122-3 (IncI2)]; and a small plasmid pChi7122-4 (ColE2-like), has been used for many years as a model strain to study the molecular mechanisms of ExPEC pathogenicity and zoonotic potential. We previously sequenced and characterized the plasmid pChi7122-1 and determined its importance in systemic APEC infection; however the roles of the other pChi7122 plasmids were still ambiguous. Herein we present the sequence of the remaining pChi7122 plasmids, confirming that pChi7122-2 and pChi7122-3 encode an ABC iron transport system (eitABCD) and a putative type IV fimbriae respectively, whereas pChi7122-4 is a cryptic plasmid. New features were also identified, including a gene cluster on pChi7122-2 that is not present in other E. coli strains but is found in Salmonella serovars and is predicted to encode the sugars catabolic pathways. In vitro evaluation of the APEC χ7122 derivative strains with the three large plasmids, either individually or in combinations, provided new insights into the role of plasmids in biofilm formation, bile and acid tolerance, and the interaction of E. coli strains with 3-D cultures of intestinal epithelial cells. In this study, we show that the nature and combinations of plasmids, as well as the background of the host strains, have an effect on these phenomena. Our data reveal new insights into the role of extra-chromosomal sequences in fitness and diversity of ExPEC in their phenotypes.
Introduction
Escherichia coli are versatile bacteria; with the majority being non-pathogenic and considered as commensals. A subset of these bacteria has acquired specific virulence attributes that confer an ability to survive in different niches and cause a broad spectrum of intestinal and extra-intestinal diseases [1], [2]. One of the important aspects of the fitness of E. coli is thought to be its ability to survive and persist in a variety of environments, including varied anatomical niches, food, soils, poultry litter, and acidic conditions. Extra-intestinal pathogenic E. coli (ExPEC) cause infections outside of their normal intestinal habitat in both mammals and birds, resulting in a considerable economic and public health burden [3], [4]. Major infections associated with ExPEC in humans include urinary tract infections (UTI), newborn meningitis (NBM) and septicemia [4]. In birds, a subgroup of ExPEC, named Avian Pathogenic E. coli (APEC), causes a complex of systemic infections, mainly respiratory, often leading to death [4]. The genetic relationship between APEC and other ExPEC of human and animal origin [4] emphasizes the potential zoonotic risk of avian-derived E. coli strains. In poultry, isolates associated with fecal matter, environmental contamination and chicken meat products possess virulence gene profiles similar to those causing human outbreaks [5], [6], which suggests that retail chicken may be an important reservoir for E. coli causing ExPEC infections in humans.
ExPEC exhibit a high degree of antigenic and genetic diversity, which complicates their diagnosis and the design of cross-protective vaccines [7]. ExPEC are defined by a limited number of O-antigens, with specific O antigens being associated with certain clinical syndromes. For example, E. coli from a small number of O serogroups (O4, O6, O14, O22, O75, and O83) cause 75% of urinary tract infections [8] and a limited number of serotypes, principally O1, O2, O78, O8, and O35, are commonly implicated in avian colibacillosis [9], suggesting that not all O polysaccharides have identical virulence properties [10], [11]. The possession of multiple large plasmids is often a defining feature of ExPEC, especially APEC, in which the virulence is partly plasmid-mediated [12], [13], [14], [15], [16], [17], [18], [19].
Although many studies have been dedicated to understanding the pathogenesis of ExPEC, little is known about the mechanisms of their persistence. Since a correlation between the ecology of bacteria and their virulence exists, understanding the mechanisms of fitness and survival of these bacteria in extreme and changing conditions would not only improve our understanding of their persistence, but also will contribute to better design strategies for their prevention and treatments.
Previously, the model APEC strain χ7122 (O78∶K80∶H9), containing three large plasmids pChi7122-1, pChi7122-2, and pChi7122-3, previously named pAPEC-1, pAPEC-2, and pAPEC-3 respectively, and a cryptic plasmid pChi7122-4 (Table 1), has been used to undestand the role of large plasmids in the virulence of ExPEC [12]. Specifically, we determined that both the nature of plasmids and their combinations have an effect on the virulence and the genetic diversity of ExPEC. Although we have clearly determined that pChi7122-1 has a major role in systemic infection of APEC in chickens, the role of the remaining plasmids remained unclear.
Table 1. Strains and plasmids used in this study.
| Strain/plasmid | Relevant characteristicsa | Reference |
| Strains | ||
| χ7122 background | ||
| χ7122 | APEC O78∶K80∶H9, gyrA Nalr, Strr, Sxtr | [79] |
| χ7145 | χ7122 (χ289:hisG-zee), rfb deleted by replacement with E. coli K-12 region at 45 min | [10], [80] |
| χ7167 | χ7179 rfb + (O111) prototroph by P1χ2963 lysate χ6206 Strain H30, O26∶H11, SLT-1 | [80] |
| χ7193 | χ7179 rfb + (O1), prototroph by P1χ7112 lysate | [10], [80] |
| χ7367 | pChi7122-3, Nalr | [12] |
| χ7368 | ΔpChi7122-1, Δ pChi7122-2, ΔpChi7122-3, Nalr | [12] |
| χ7394 | pChi7122-1, Nalr | [12] |
| χ7392 | pChi7122-2, Nalr, Strr, Sxtr | [12] |
| χ7561 | pAPEC-1, pAPEC-2, Nalr, Strr, Sxtr | [12] |
| χ7562 | pChi7122-1, pChi7122-3, Nalr | [12] |
| χ7274 | pChi7122-2, pChi7122-3, Nalr, Strr, Sxtr | [90] |
| E. coli K-12 background | ||
| χ6092 | E. coli K-12, Lac− F− Tcr | [19] |
| χ7346 | χ6092 pChi7122-1, Tcr | [19] |
| χ7347 | χ6092 pChi7122-2, Tcr, Strr, Sxtr | [12] |
| χ7348 | χ6092 pChi7122-3, Tcr | [12] |
| Plasmids | ||
| pChi7122-1 | 103,275 pb plasmid of APEC χ7122 | [19], [90] |
| pChi7122-2 | 82,676 pb plasmid of APEC χ7122 | [19] |
| pChi7122-3 | 56,676 pb plasmid of APEC χ7122 | [19] |
| pChi7122-4 | 4,300 pb plasmid of APEC χ7122 | This study |
aNalr, nalidixic acid resistant; Tcr, tetracycline resistant; Strr, streptomycin resistant; Sxtr, Trimethoprim/sulfamethoxazole resistant.
Since pChi7122-2 and pChi7122-3 do not encode for common ExPEC virulence factors [12], and their roles are considered as minor in systemic infection in chickens [12], we hypothesized that these plasmids could be important in persistence of this bacterial strain in different stressful conditions encountered before and during infections. Therefore, this study aimed to (1) fully sequence and analyze the DNA of plasmids pChi7122-2, pChi7122-3, and pChi7122-4 of APEC strain χ7122; and (2) evaluate the contribution of these plasmids, as well the plasmid pChi7122-1, either individually or in combination, in the bacterial interaction with a model human intestinal epithelial cell line, bile and acid resistance, biofilm formation, and growth in iron-restricted medium and in the presence of different carbon sources. Moreover, since the plasmids can be carried by strains with different backgrounds, we aimed to determine the effect of different host strain backgrounds on plasmid-associated phenotypes. This study presents for the first time the sequence of three plasmids of APEC strain χ7122 and provides new insights into the genetic and phenotypic mechanisms that ExPEC may use for their persistence and survival in stressful conditions.
Results and Discussion
Genome sequencing has made major contributions to our knowledge of virulence and the evolution of pathogenic bacteria. So far, virulence plasmids associated with ExPEC are ColV, ColBM and Vir plasmids [20]; many of which are already fully sequenced and have been determined as belonging mainly to the IncFIB/FIIA backbone. Although PCR characterization of UPEC and APEC plasmids has revealed the presence of plasmids from other Inc groups [21], studies on their role in ExPEC has been limited. Previously, we examined the role of the three large plasmids of APEC χ7122 in pathogenesis in chickens [12] and sequenced the plasmid pChi7122-1 [19]. We were able to assign roles for pChi7122-1 in the virulence in systemic infection of bacteria; however the roles of pChi7122-2 and pChi7122-3 in APEC χ7122 were equivocal.
General sequence features of plasmids pChi7122-2, pChi7122-3, and pChi7122-4
In this study, we present the whole DNA sequences of plasmids pChi7122-2 (FR851303), pChi7122-3 (FR851304) and pChi7122-4 (FR851305). The general sequence features of the three plasmids are listed in the Table 2.
Table 2. Summary of general characteristics of the three sequenced plasmids of APEC χ7122.
| Plasmids | Size (bp) | Inc group | GC% | N° ORFs | Starting codons | Gene function | virulence factors-encoded | ATB |
| pChi7122-2 | 82,676 | IncFII | 52.8 | 115 | ATG (78.26%) GTG (14.78%) TTG (6.08%) | 18.26% (known and putative virulence genes) 41.74% (involved in plasmid functions) 4.35% (ISs) 19.13% (CHP) 16.52% (HP) | EitABCD | Strr, Sxtr |
| pChi7122-3 | 56,676 | IncI2 | 42.7 | 86 | ATG (86.04%) GTG (6.8%) TTG (5.81%) | 26.74% (known and putative virulence genes) 31.39% (involved in plasmid functions) 2.32% (ISs) 8.14% (CHP) 31.39% (HP) | Type IV Pil fimbriae | - |
| pChi7122-4 | 4,300 | ColE2-like | 49.3 | 3 | ATG (100%) | 66.66% (involved in plasmid functions) 33.34% (CHP) | None | - |
ATB, antibiotic; Strr, streptomycin resistant; Sxtr, Trimethoprim/sulfamethoxazole resistant; -, absent.
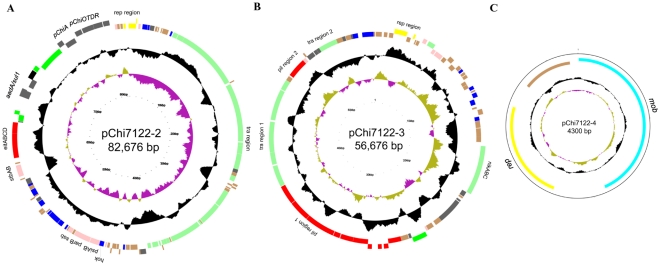
Plasmids pChi7122-2, pChi7122-3 and pChi7122-4 consist of 82,676 bp, 56,676 bp and 4,300 bp respectively (Fig. 1, Table 2) and are predicted to encode 115, 86, and 3 coding sequences (CDS) respectively (Table 2, Table S1 and S2); these CDSs include the complete sequences for the iron acquisition system eitABCD on pChi7122-2 and type IV fimbriae on pChi7122-3, which have been previously shown to be present on these plasmids by PCR [12]. Analysis of pChi7122-4 revealed 3 CDSs that were predicted to encode plasmid replication and maintenance functions only (Table 2, Fig. 1); consequently we excluded this plasmid from all further experimental analysis.
Figure 1. Circular representation of pChi7122-2 (A), pChi7122-3 (B), and pChi7122-4 (C).
The different rings represent (from outer to inner) CDS which are color coded by functional group (ring 1 and 2), deviation from average G+C content (ring 3), and GC skew [(G−C)/(G+C); ring 4]. Colors represent the following: red, virulence-associated; green, plasmid transfer; yellow, plasmid replication; grey other functions; brown, hypothetical protein.
We assessed the presence of eitA gene of pChi7122-2 and two genes of pChi7122-3 (pilS and pilV) by PCR among 225 pathogenic E. coli strains from different origins, including 100 human E. coli strains isolated from the main clinical extra-intestinal sources (50 UTI and 50 non-UTI), 80 APEC, and 45 human enteric pathogenic E. coli. PCR results show that eitA was present in 10% of non-UTI human isolates and 5% of APEC strains, but was absent in other groups. The genes pilS and pilV of pChi7122-3 were detected in 8.75% of the APEC group and in 10% of human UTI isolates, respectively. The low prevalence of pChi7122-2 (eitA) and pChi7122-3 (pilS and pilV) genes among other ExPEC of human and avian origin, as determined by PCR, could indicate the recent acquisition of these genes by these E. coli strains, enabling them to inhabit new niches.
Our past work has determined that plasmids pChi7122-2 and pChi7122-3 are self-conjugative [12]. Herein, their sequences analysis has revealed the presence of genes required for their transfer (Fig. 1, Table S1 and S2). The transfer region of pChi7122-2 is about 34 kb consisting of 24 tra and 9 trb genes (Table S1) and is identical to the one of the virulence plasmid pAA (FN554767.1; 99% identity with 92% coverage), whereas the tra region of pChi7122-3 has the same organization as its equivalent in E. coli conjugative plasmid IncI2 R721 (AP002527.1); it contains 11 tra/trb genes, grouped in two clusters separated by two pil genes (Fig. 1, Table S2). pChi7122-3 also harbors genes nikB, nikC, and nikA for relaxome formation involved in plasmid transfer [22], [23] (Fig. 1, Table S2).
Among the three plasmids, only pChi7122-2 carries antibiotic resistance genes (MM2-101, MM2-102 and MM2-103) (Table S1). These genes encode for a dihydropteroate synthase (sul1) [24], a GCN5-related N-acetyl transferase [25] and a streptomycin 3′-adenylytransferase (SP-R) (aadA) [26], respectively. The phenotypic expression of streptomycin and sulfonamide (trimethoprim/sulfamethoxazole) resistance in strains containing pChi7122-2 has been determined by disk diffusion tests. Although streptomycin has only limited current usage in clinical medicine, it remains important for therapy of, and growth promotion in, animals and bacterial disease control in plants [27]. It was suggested that sulphonamide resistance genes can be transferred from commensal bacteria via integrons, transposons or plasmids, into more virulent bacteria in the intestine [28].
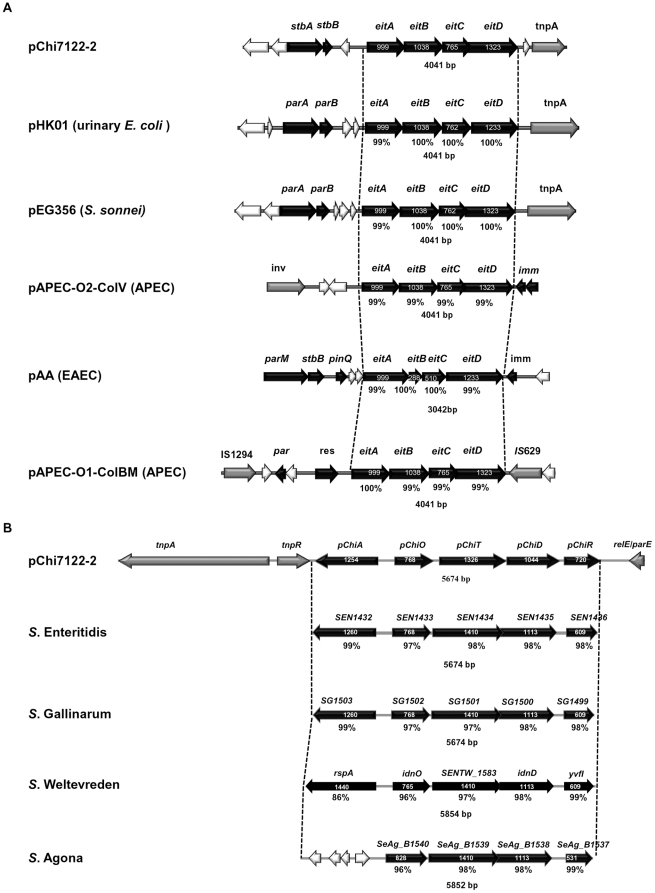
Comparative analysis of the pChi7122 plasmids with those in the public databases using BLASTn showed that pChi7122-2 shares high homology with plasmids from Shigella sonnei (pEG356), an urinary E. coli isolate (pHK01), EAEC (pAA), and K. pneumoniae (pKF3-70), respectively (with 100% identity and 70% coverage). The pChi7122-3 genome has shown homology with only one plasmid, the E. coli plasmid R721 (with 99% identity and 90% coverage), which includes the type IV fimbriae pil operon [29], [30] and shufflon [31], [32]. Mauve alignment of pChi7122-2 and pChi7122-3 with their respective homologous plasmids confirmed these homologies (Fig. S1).
The plasmid pChi7122-2 and its homologous plasmids have a 4 kb region in common, which encodes for the ABC iron uptake locus eitABCD (Fig. 2A), previously described in two other APEC plasmids, pAPEC-O2-ColV [17] and pAPEC-O1-ColBM [15]. A DNA comparison of the regions of eitABCD of the six plasmids has shown that with the exception of pAPEC-O2-ColV, this region is located downstream of the par region of plasmids and is flanked by the transposon tnpA gene in pChi7122-2, pHK01, and pEG356 respectively and by an insertion sequence IS629 in pAPEC-O1-ColBM (Fig. 2A) which could explain the dissemination of eitABCD among genomes of these bacteria. We were unable to detect the iron-uptake phenotype expression of eitABCD genes using CAS agar medium [12], even though it was efficient in revealing those of pChi7122-1 and chromosomally-encoded systems. Therefore in this study, we extended the analysis by testing the growth of strains, with and without the three plasmids, in iron-limited medium alone or supplemented with either FeSO4, heme or hemoglobin. Our results show that only pChi7122-1 increased the growth of strains in iron-sequestered environments (Fig. S2). The ability to acquire iron from heme and hemoglobin could be related to the autotransporter Tsh encoded by pChi7122-1 [33], which has previously been reported to bind to red blood cells [34]. Future studies are needed to determine conditions of expression of eitABCD, such as under in vivo conditions.
Figure 2. Comparison of physical and genetic maps for eitABCD and sugar pathway regions.
The eitABCD region from pChi7122-2 was compared to its equivalent in pHK01 (HM355591.1), pEG356 (FN594520.1), pAPEC-O2-ColV (NC_007675.1), pAA (FN554767.1), and pAPEC-O1-ColBM (NC_009837.1) (A); and sugars pathways genome region pChiA pChiOTDR in pChi7122-2 to its equivalent found in the genomes of S. Enteritidis (AM933172.1), S. Gallinarum (AM933173.1), S. Weltevreden (FR775220.1) and S. Agona (CP001138.1) respectively (B).
New putative sugar utilization pathways identified in pChi7122-2
An important aspect of pathogenesis is the ability of bacteria to adapt their metabolism to the available nutrients by coordinating their metabolism with their life cycle [35]. Recent reports have shown that in the intestine, both commensal and enterohemorrhagic E. coli (EHEC) require multiple carbon metabolic pathways [36], [37].
In this study, DNA sequence analysis of pChi7122-2 has revealed the presence of two systems of sugar utilization pathways. This system, with two divergent operons, consists of a gene for a starvation-sensing protein (pChiA) located in the opposite orientation to four successive genes pChiOTDR (Fig. 2B, 3). These genes have no significant homology with DNA sequences of other E. coli available on public databases, as determined by MegaBLASTn search analysis, but share 94% homology (with 100% coverage) with the chromosomal DNA sequence of genomes of Salmonella Enteritidis (AM933172.1), Gallinarum (AM933173.1), Weltevreden (FR775220.1) and Agona (CP001138.1), respectively (Fig. 2B). The sequence analysis of this region in these Salmonella serovars has determined that, with the exception of S. Agona, in which the pChiA-equivalent gene is truncated, the organization of the pChiOTDR homologous genes in the genome of the four pathogens is the same (Fig. 2B). The identities of the proteins translated by these genes were between 86%–99% (Fig. 2B, Table S3, and Fig. S3).
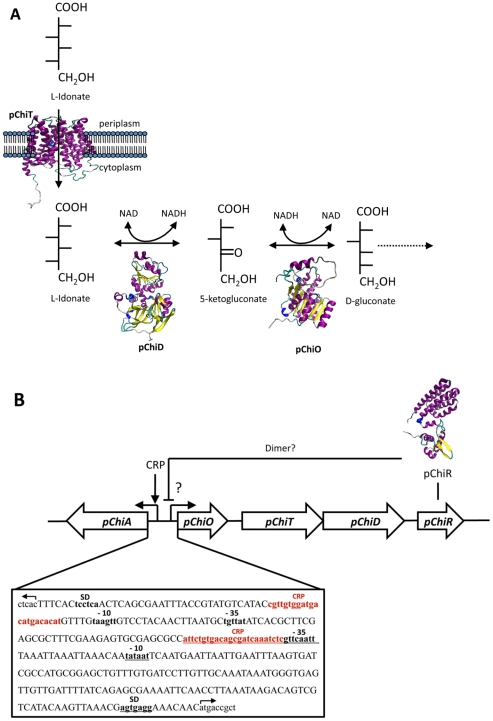
Figure 3. Plasmid pChi7122-2 putative sugar utilization pathway.
Illustration of the proposed biochemical pathway for putative sugar utilization encoded by the operon pChiOTDR (A), the predicted 3D model and function of each enzyme are presented; and diagrammatic representation of the genetic organization of the putative sugar utilization operon pChiA pChiOTDR (B). The two promoter regions identified are indicated with arrows. Bioinformatic analysis indicated that pChiR may act as a transcriptional regulator of pChiOTDR genes. The promoter region contains independent CRP binding boxes indicated in red. The promoter elements for pChiOTDR (Ppchi) located in the positive DNA strand are underlined. The promoter elements for pChiA (PpChiA) in the negative strand are in bold and red without underlining.
The putative functions and the predicted 3-D structures of the pChiA and pChiOTDR gene products, determined by Blast-PSI and HHpred [38], show that pChiA encodes for a bifunctional dehydratase that utilizes both D-mannonate and D-altronate as substrates [39] and pChiOTDR encode for a gluconate 5-dehydrogenase, pChiO; an exonate sugar transport, pChiT; an L-idonate 5 dehydrogenase, pChiD; and a regulator protein GntR-like, pChiR, respectively (Table S3, Fig. 3A). Two promoter regions, PpChiA and PpChi, with independent cAMP receptor protein (CRP) binding boxes [40], [41], were detected in the promoter region of pChiA and pChiO (Fig. 3B). Bioinformatic analysis indicated that pChiR is a putative transcriptional regulator from GntR family [42]. In the absence of glucose, the preferred carbon source for E. coli, the CRP would activate the pChi7122-2 sugars pathways [40], [41]; whereas pChiR would have an opposite effect. It is known that colonic mucus contains several sugar acids that represent an important source of nutrients and that genes involved in the catabolism of N-acetylglucosamine, sialic acid, glucosamine, gluconate, arabinose, and fucose are expressed in both commensal E. coli and EHEC [36]. It has also been reported that UPEC bacteria grown in urine express enzymes for catabolism of sialic acid, gluconate, xylose, and arabinose [43] and genes involved in the transport of gluconate and related hexonates are up-regulated in S. Typhimurium in macrophages [44], suggesting that the new pChi7122-2 sugar pathways could also be important either in the pathogenesis of APEC, as well as in Salmonella serovars Enteritidis, Gallinarum, Weltevreden and Agona or in their persistence in different hosts.
Compared to the chromosomal E. coli K-12 L-idonic acid pathway encoded by the gnTII genes, idnK idnDOTR [45], the genes of the operon pChiOTDR of pChi7122-2 have no significant homologies at the DNA level and share some sequence identity at the protein level (Table 3); moreover, the position of the gene of L-idonate 5 dehydrogenase is different in the two distinct gene clusters. Intriguingly, the gluconate kinase gene, idnK, of GnTII pathway [45] is absent in the pChi7122-2 pathway and is substituted by the gene of the starvation sensing protein, rspA-like [46] pChiA which is essential for survival of bacteria in limited nutrient conditions. The gene encoding the regulatory protein GntR in the GnTII pathways, exhibits no significant homology at both DNA and protein levels with its counterpart in pChi7122-2 (pChiR) (Table 3). In this study, although we have shown that strains have better growth in media with glucoronic acid than with other sugars tested (Fig. S4), there were no significant differences between strains with and without the plasmid pChi7122-2. The functionality of the sugar utilization pathway genes located on pChi7122-2 would be more apparent in gntII-operon-deleted strains [45], or by evaluation of their expression under in vivo conditions, such as using the selective capture of transcribed sequences (SCOTS) method [47]. Future studies will be conducted to determine the conditions of their expression and their eventual role in both APEC and Salmonella serovars.
Table 3. Comparison of the pChi7122-2-encoded sugar pathway operon with GntII L-idonic pathway of E. coli K-12.
| L-idonic acid-like catabolism pathway of pChi7122-2 rspA pChiOTDR | L-idonic acid catabolism pathway GntII of E. coli K-12 idnK idnDOTR | |||||
| GI-Numbers | Gene symbols | Gene product | DNA homology | % AA identity/%positive | Expect | Gene (Accession no.) |
| MM2_107 | pChiA | Starvation sensing protein | NSH | - | - | |
| MM2_108 | pChiO | Gluconate 5-dehydrogenase | NSH | 48%/64% | 1e-70 | idnO (AAC77203.1) |
| MM2_109 | pChiT | The major facilitator superfamily protein | NSH | 23%/46% | 0.033 | idnT (AA77222.1) |
| MM2_110 | pChiD | L-idonate 5-dehydrogenase | NSH | 46%/69% | 1e-97 | idnD (NP_418688.1) |
| MM2_111 | pChiR | Regulatory protein GntR | NSH | None | None | idnR (NP_418685.1) |
AA, amino acid; -, absent; NSH, no significant homology.
Diversity of plasmids-associated fitness phenotypes and the effect of host strain background on their expression
The genomic diversity among ExPEC isolates has been described and multiple factors have been linked to their virulence [48], [49]. However, a systematic analysis of ExPEC phenotypic diversity has not been done previously. In this study, the large plasmids-associated phenotypes related to fitness of ExPEC bacteria as well the effect of host strain backgrounds were investigated.
Intestines are suspected to be a primary reservoir of ExPEC strains causing diseases in both humans [50] and chickens [5]. To determine if large plasmids would increase the fitness of their carriers in the gastrointestinal (GI) tract environment, we assessed the ability of strains to colonize intestine cells and resist both acid and bile, attributes that allow enteric bacteria to live and persist in the intestine of their host [51].
APEC strain χ7122 associates with and invades into intestinal epithelial cells without affecting the distribution of the tight junction protein ZO-1
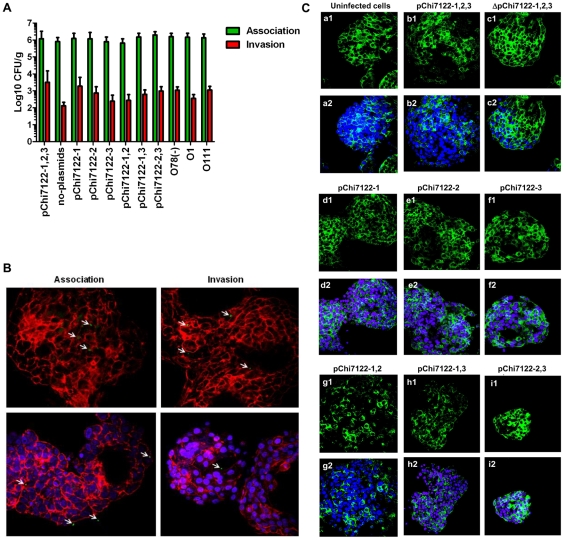
Some APEC strains are genetically similar to human ExPEC, especially to uropathogenic E. coli (UPEC) [52], and could cause human diseases [53]. Herein, we investigated the ability of APEC-derivative strains to associate with, and invade into, human cells of the kind that may be targeted by human ExPEC bacteria during their commensal life cycle in the intestine. The intestine is suspected to be a reservoir of ExPEC that cause infections in humans [50]. Since APEC strains are now considered as potential food-borne pathogens that could be transmitted to humans via poultry products [4], [6], [28], we aimed to investigate the interaction of APEC-derivative strains with 3-D organotypic models of human intestinal epithelial cells. The 3-D model of intestinal epithelium used in this study has been shown previously to mimic the in vivo parental tissue more closely than monolayer cultures with regard to morphology and function [54]. The highly differentiated character of the 3-D intestinal cells is reflected in the presence of distinct apical and basolateral polarity, increased expression and better organization of tight junctions, extracellular matrix, and brush border proteins, highly localized expression of mucins, and multiple epithelial cell types relevant to those found in vivo [55]. Our data showed that APEC-derivative strains were able to associate with, and invade into, human intestinal epithelial cells, and large plasmids did not have significant effect on these characteristics (Fig. 4). Although tight junctions efficiently restrict most microbes from penetrating into deeper tissues and contain the microbiota, some pathogens have developed specific strategies to alter or disrupt these structures as part of their pathogenesis, resulting in either pathogen penetration, or other consequences such as diarrhea. In this study, evaluation of different APEC-derivative strains for their interaction with 3-D human intestinal epithelial cells, showed that although these strains attached and invaded into these cells, they did not disturb their tight junctions, based on immunofluorescence evaluation (Fig. 4). These data suggest that invasion of the intestine and dissemination would not occur through intercellular transportation of the bacteria, which could potentially disseminate through transcellular transportation, a mechanism used by meningitis-causing bacteria, including E. coli K1 to invade brain microvascular endothelial cells (BMECs) [56]. These bacteria could live as commensals in the intestines from where they shed and cause diseases in different hosts or other sites of the same host.
Figure 4. Comparative of association and invasion of strains with 3-D INT-407 cells.
Mean of Log CFU/ml and standard deviation of bacteria association and invasion with cells (A) Representative confocal laser scanning micrographs showing association, and invasion of 3-D INT-407 cells with χ7122 and derivatives (B). Bacteria are marked in green, while the blue and red colors represent cell nuclei labeled with DAPI and F-actin cytoskeleton labeled with phalloidin, respectively; and ZO-1 staining of non-infected and infected 3-D INT-407 with plasmid-derivative strains (C). Non-infected 3-D INT-407 aggregates (a) or 3-D INT-407 aggregates infected for 2 h with wild type (χ7122) (b), ΔpChi7122-1,2,3 (χ7368) (c), pChi7122-1 (χ7394) (d), pChi7122-2 (χ7392) (e), pChi7122-3 (χ7367) (f), pChi7122-1,2 (χ7561) (g), pChi7122-1,3 (χ7562) (h), and pChi7122-2,3 (χ7274) (I). The ZO-1 antigen is marked in green, while the blue color represents cell nuclei labeled with DAPI. Images are presented with (indicated as “2”) and without (indicated as “1”) DAPI labeling for clarity purposes. Images are based on 400× magnifications. Arrows indicate the bacteria stained in green; Abbreviations used are: pChi7122-1,2,3 = pChi7122-1, pChi7122-2, and pChi7122-3; pChi7122-1,2 = pChi7122-1 and pChi7122-2; pChi7122-1,3 = pChi7122-1 and pChi7122-3; pChi7122-2-3 = pChi7122-2 and pChi7122-3.
Role of plasmids in bile and acid resistance
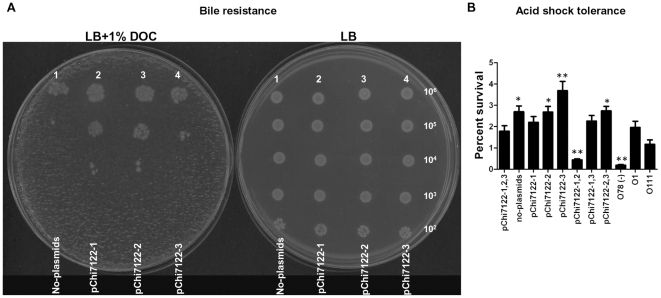
Mechanisms associated with bile resistance in bacteria are LPS synthesis, expression of efflux pump genes and regulatory genes such as marAB and phoPQ [51]. In this study, we have shown that all wild-type derived strains tested were resistant to deoxycholate (DOC), one of the most abundant bile salts in humans (data not shown); whereas the group of strains derived from E. coli K-12 behaved differently (Fig. 5A). Although, E. coli K-12 was sensitive to the bile, its plasmid derivative strains χ7346 (pChi7122-1) and χ7347 (pChi7122-2) had increased survival in LB agar media with 1% (w/v) DOC as compared to their parent χ6092. The strain χ7348 (pChi7122-3) was as sensitive to bile as its parent strain χ6092 (Fig. 5A). According to our results APEC χ7122 strain better tolerates the presence of bile salts in the media then E. coli K-12 which was sensitive to the detergent (Fig. 5, data of wild-type not shown). The mechanism of resistance of APEC could be both LPS and plasmid related. In fact, the detection of plasmid-associated resistance in E. coli K-12 background but not in the wild-type background strains, could be related to the presence of other factors, including the LPS in these strains that has masked the effect of plasmids on this phenomenon; this statement is supported by the resistance of the rough mutant which is usually hypersensitive to bile [57]. The mechanism of resistance encoded by the plasmid pChi7122-1 could be associated with proteins such as OmpT that was previously associated with bile resistance in Vibrio cholerae [58] and ABC transport proteins that are known to play a role in the protection of cells from toxic compounds [59]. Since such factors are not located on pChi7122-2, other factors predicted to be encoded by this plasmid, such as TA modules could be involved in bile tolerance of bacteria; as TA systems are now known to play an important role in bacterial stress physiology [60], [61], [62]. To our knowledge, this is the first time that plasmids have been shown to be associated with the bile resistance of E. coli.
Figure 5. Bile and acid tolerance of strains.
Bile sensitivity assay for E. coli K-12 and derivatives, no-plasmids (χ6092), pChi7122-1 (χ7346), pChi7122-2 (χ7347), and pChi7122-3 (χ7348). Five-microliters of serial ten-fold dilutions of each strain were spotted on both LB and LB +1% (w/v) DOC agar plates. The approximate numbers of bacteria present in each dilution are indicated on the right side of the plate (A). Percent acid survival of wild-type derivative strains in acid shock for 18 hours (B). Abbreviations used are: pChi7122-1,2,3 = pChi7122-1, pChi7122-2, and pChi7122-3; pChi7122-1,2 = pChi7122-1 and pChi7122-2; pChi7122-1,3 = pChi7122-1 and pChi7122-3; pChi7122-2-3 = pChi7122-2 and pChi7122-3.
Acid resistance is important for bacterial survival in acidic stomach or in foods with low pH [63]. Our results have shown that plasmids do not have any effect on the growth of the wild-type derived strains when grown in acidic medium for a short period (12 hours), as the strains with and without plasmids grew similarly (data not shown). However, at longer incubation times (18 hours), strains behaved differently (Fig. 5B). Similar to the study by Lim et al. [64] on the plasmid pO157 in E. coli O157, we have shown that in the absence of its three plasmids, the APEC strain survived better in acidic conditions than in their presence when incubated for 18 h. Moreover, our study showed that although the plasmid pChi7122-1, either alone or in combination with pChi7122-2 or pChi7122-3, decreased the acid tolerance of bacteria, the presence of pChi7122-3 had the opposite effect (Fig. 5B). Since pChi7122-1 and pO157 play a major role in the virulence of APEC [12] and E. coli O157 [64] respectively, these findings could indicate that the presence of plasmids exert a cost to bacterial fitness when exposed for a long period (>18 hours) to acidic conditions, whereas bacteria containing other plasmids such as pChi7122-3 in χ7122 would have better survivability in these conditions. Elucidation of the mechanism of acid tolerance associated with pChi7122-3 is needed to fully understanding the persistence of E. coli in acidic conditions.
Our study also confirmed the importance of the full expression of O78-antigen LPS for the acid tolerance of E. coli [65], and demonstrated that the nature of LPS had a minor effect on this stress response (Fig. 5B).
Large plasmids increase biofilm formation at host temperatures
Bacterial biofilm formation is a major concern in both medical and industrial systems. Biofilm formation is associated with many medically-important pathogenic bacteria, as an estimated 65-80% of all human infections are thought to be biofilm-related [66]. However, elucidating the mechanisms of biofilm formation necessary for establishing strategies for their prevention and treatments is becoming a matter of urgency.
ExPEC cells are found in biofilm-like communities in both gastrointestinal [67] and urinary tracts [68] indicating the importance of biofilms in the persistence of these bacteria. ExPEC bacteria have to adapt to extreme temperature changes. In this study, our strategy using three large plasmids, either individually or in combination in both an APEC wild-type and an E. coli K-12 background, and different O-LPS at different temperatures, has revealed new insights into biofilm formation of ExPEC. Altogether, our data distinguished four groups of factor-driven biofilms, including plasmidless-, plasmid-, O-LPS-, and rough LPS-mediated biofilms in E. coli which differ in their expression conditions.
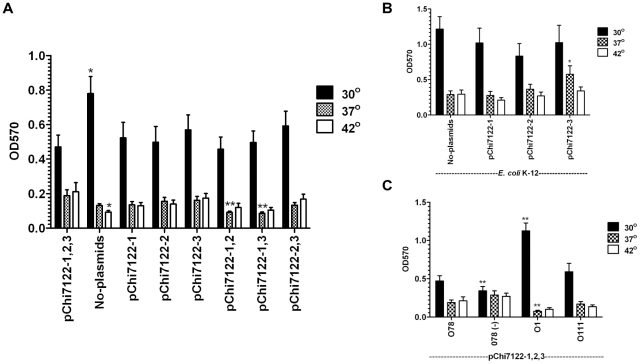
In general, the different strains tested formed more biofilms at 30°C than at 37°C or 42°C (Fig. 6A). Compared to the wild-type, the plasmidless strain produced significantly more biofilm at 30°C (P<0.05) (Fig. 6A). In the same conditions, the presence of the three plasmids, either individually or in combinations in the strains, reduced the level of biofilm formation to the level of the wild-type strain (Fig. 6A). In contrary, at host temperatures (37° and 42°) (Fig. 6A), the plasmidless strain produced less biofilm than the wild-type strain, with the data being statistically significant (P<0.05) at 42°C (Fig. 6A).
Figure 6. Effect of plasmids, O-LPS, and temperature on biofilm formation.
Biofilm formation of different strains were compared at different temperatures: χ7122 and its plasmid derivatives strains: No-plasmids (χ7368), pChi7122-1 (χ7394), pChi7122-2 (χ7392), pChi7122-3 (χ7367), pChi7122-1,2 (χ7561), pChi7122-1-3 (χ7562), and pChi7122-2,3 (χ7274) (A), E. coli K-12 (χ6092) and its derivatives: pChi7122-1 (χ7346), pChi7122-2 (χ7347), and pChi7122-3 (χ7348) (B), and χ7122 and its LPS derivative strains : rough mutant O78(-) (χ7145), smooth strains with either O1-LPS (O1) (χ7193) or O111-LPS (O111) (χ7167) (C) at either 30°C, 37°C, or 42°C.
The biofilm formed by the plasmid-cured strain, highly produced at 30°C (Fig. 6A), is probably promoted by no-plasmidic factors preferentially expressed at 30°C and at early stage of biofilm formation; among them curli required for development of biofilm and adhesion [69]. Expression of biofilm in the environment (30°C) would be beneficial for plasmidless strains; in these conditions, biofilm will allow these bacteria to be in close proximity with other bacterial species and acquire transmissible genetic elements.
It has been shown that conjugative plasmids promote bacterial biofilm formation by generating F-pili mating pairs, which is important for early biofilm formation [70], [71], [72]. In this report, we have shown that plasmid-driven biofilms are very complex and this complexity is related to the nature of the plasmids, their combinations, host strain backgrounds, and the temperature to which the strains are exposed. The presence of the three plasmids pChi7122-1, pChi7122-2, and pChi7122-3 in the wild-type strain (Fig. 6A) and pChi7122-3 in the E. coli K-12 strain (Fig. 6B), had increased biofilm formation at host temperature conditions, with data being significant at 42°C (P<0.05) (Fig. 6B). The fact that pChi7122-3-driven enhancement of bacterial biofilm was higher than those of pChi7122-2 and pChi7122-1 in both wild-type and the E. coli K-12 backgrounds could be related to not only the tra genes expression [70], [71], [72] but also to the type IV fimbriae encoded by pChi7122-3, which was previously associated with the biofilm formation in enteroaggregative E. coli [73]. Plasmid-driven biofilms could be essential in the virulence process by giving bacteria a survival advantage in different niches of the host, which could result in disease.
A controversy exists regarding the role of LPS in bacterial biofilm formation [74], [75]. In this study, we have shown that the three plasmids pChi7122-1, pChi7122-2, and pChi7122-3 in wild-type derivative strains with different O-LPS backgrounds behaved differently in their biofilm formation (Fig. 6C). In absence of O78-LPS, the rough strain produced significantly (P<0.0001) less biofilm than its smooth wild-type strain at 30°C. Even though substitution of O78-LPS with O111-LPS had little effect on biofilm formation, the substitution of O78-LPS with O1-LPS has in contrary greatly enhanced biofilm formation in these bacteria at 30°C. Since the O1-LPS-driven enhancement of bacteria biofilm occurs at 30°C condition and is repressed at host temperatures (37°C/42°C), this indicates that its role could be more important in the persistence of bacteria in the environment, and that the temperature of 30°C in early O1-LPS-associated biofilm formation is necessary. The fact that O1-LPS-driven biofilm is not highly formed at 37°C and 42°C (Fig. 6C), could be related to a change in the LPS-O1 bilayer structure at higher temperature [76], [77], [78] leading to the disturbance of the early biofilm formed. To our knowledge, this is the first report on the effect of the nature of LPS on biofilm formation.
Conclusion
A novel putative sugar utilization pathway operon that is not present in other E. coli strains but found in Salmonella serovars, an ABC iron transport system and a type IV fimbriae pil operon were located on pChi7122-2 and pChi7122-3 respectively. Multiple plasmid-encoded mechanisms, including toxin-antitoxin modules and the novel sugar pathway could be important in the fitness and persistence of APEC χ7122.
Large plasmids were involved in bile resistance (pChi7122-1 and pChi7122-2) when present in E. coli K-12 background and acid tolerance (pChi7122-3) in the wild-type background. Four different factor-driven biofilms, including plasmidless-, plasmid-, rough-LPS-, and O-LPS-mediated were demonstrated. These multiple factor-driven biofilms expressed at different temperatures could have distinct functions. Some of them could be important in the acquisition of genetic material and persistence of bacteria in the environment; others could be involved in virulence. The genotypic and phenotypic analysis of plasmid-derivative strains of an ExPEC model strain χ7122 (O78∶K80∶H9) revealed new insights into the mechanisms of fitness of ExPEC and their diversity.
Materials and Methods
Bacterial strains and growth conditions
Most of the bacterial strains used in this study, listed in Table 1, are derived from the highly virulent APEC strain χ7122 (O78∶K80∶H9) [79] and were fully described in our previous studies [10], [12], [80].
To evaluate the effect of the host strain background on plasmid-associated phenotypes, we used three derivatives of χ7122 with different LPS profiles containing the three plasmids pChi7122-1, pChi7122-2, and pChi7122-3; a rough mutant strain (O78-) of APEC χ7122, χ7145; and two derivatives of χ7145, χ7167 and χ7193, which respectively express O111 and O1 antigens rather than the native O78 antigen. We also used strains derived from an E. coli K-12, χ6092, containing either pChi7122-1, χ7346; pChi7122-2, χ7347; or pChi7122-3, χ7348 (Table 1, [10], [12], [80]).
Antibiotic susceptibility testing of strains was performed and interpreted via disk diffusion method, as recommended by the Clinical and Laboratory Standards Institute (CLSI) [81], [82].
A collection of one hundred human strains isolated from the main clinical extra-intestinal sources (50 UTI and 50 non-UTI) [19], eighty APEC strains, and forty-five enteric E. coli strains (from our collection) were used to study the distribution of pChi7122-2 and pChi7122-3-associated genes among different groups of pathogenic E. coli by PCR.
Unless otherwise stated, bacteria were routinely grown in Luria Bertani (LB) broth or on MacConkey agar supplemented with 1% lactose at 37°C. Strains were stored as stock cultures at −80°C in peptone-glycerol medium. All wild-type derivative strains grew similarly in LB and reached the OD600 ∼1.0 when incubated overnight (O/N) at 37°C standing, except the strain χ7561 (pChi7122-1, pChi7122-2) which had a slightly lower growth and its OD600 was around 0.8 when grown in the same conditions.
Plasmid sequencing and annotation
The DNA sequences of pChi7122-2, pChi7122-3 and pChi7122-4 plasmids were derived from contig sequences of the whole genomic DNA of APEC χ7122. The sequences were manipulated to the standard of an ‘Improved High-Quality Draft’ [83]. The program Artemis [84] was used to identify the plasmids and collate data. For each of the three plasmids all the sequence gaps were closed by directed polymerase chain reaction (PCR) and the products sequenced with big dye terminator chemistry on ABI3730 capillary sequencers. All the plasmids were circularized and contiguated using this method.
The DNA sequences were annotated to identify coding sequences and repeat sequences in Artemis and the vector NTI suite of programs was used to confirm the previous analysis. To identify plasmids with similar sequences, pChi7122-2 (FR851303) and pChi7122-3 (FR851304) were compared by BLASTn at NCBI. Plasmid sequences of pEG356 (FN594520.1) from Shigella sonnei, pHK01 (HM355591.1) from a urinary E. coli isolate, pAA (FN554767.1) from EAEC, and pKF3-70 (FJ494913.1) from K. pneumoniae, and R721 (AP002527.1) a trimethoprim and streptomycin resistant plasmid from an E. coli were downloaded. Plasmid sequences were aligned and then visualized using ACT and Mauve v2.3.1 [85]. Schematic plasmid drawings were constructed using DNAplotter [86].
Amino acid sequence and protein structural-based alignments were performed using the CLC Free Workbench software tool (v. 6.1 CLC bio A/S, Aarhus, Denmark) and the web-based interface for ESPript v.2.2 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) [87], respectively. The 3-D structure of the proteins pChiA, pChiT, pChiO, pChiD, and pChiR were predicted using position specific iterative (PSI)- BLAST alignment and HHpred [38].
Prevalence of pChi7122-2 (eitA) and pChi7122-3 (pilS and pilV) genes among avian and human pathogenic E. coli
One hundred human E. coli strains isolated from the main clinical extra-intestinal sources (50 UTI and 50 non-UTI) [19], eighty APEC strains, and forty-five enteric E. coli strains (from our collection) were screened by PCR [19] for the presence of the eitA, pilS, and pilV genes using the primers eitA F: 5′-AACTGCGGCTATCAGGAGAC-3′ and eitA R : 5′-CAGGTCATATCCCACAGCTT-3′; pilS F: 5′-CTTCTCTTTCTGCACACCGT-3′ and pilS R: 5′-TGTGATTGTAACGGAGCC-3′; pilV F: 5′-TCTATACAGGCGAGTATTTA-3′ and pilV R: 5′-AATTCATACCAGAATACTCA-3′. The primers were designed from the sequence of pChi7122-2 (eitA) and pChi7122-3 (pilS and pilV).
Growth comparison assays
The growth rates of strains were compared in iron-sequestered medium using LB containing 100 µM of α,α′-dipyridyl alone or supplemented with either Heme (Sigma, 600 µg ml−1), hemoglobin (Sigma, 60 µg ml−1), or FeSO4 (Sigma, 20 mM: control). The growth rates were also tested in minimal medium MM9 [12], MM9 with thiamin (1 µg ml−1) and 0.1% casamino acid, and supplemented with glucose, lactose, arabinose, mannose, galactose, glucuronic acid, or glucoronic acid (0.2%) as source of carbon at 37°C shaking (180 rpm). The OD600 was recorded every hour over 24-hour period.
Interaction of strains with 3-D INT-407 human epithelial cells
3-D cultures of human INT-407 cells (ATCC CCL6) were used as model intestinal epithelium and were prepared as previously described [54]. Approximately 106 CFU of PBS-washed bacteria, grown rotating to an OD600 1.0 in LB, were added to each well (multiplicity of infection [MOI], 10). For bacterial association assays, the 24-well plates were incubated at 37°C in 5% CO2 for 1 hour, and rinsed three times with PBS. PBS-0.1% (w/v) deoxycholic acid sodium salt was added to each well, and samples were diluted and spread on MacConkey medium plates for enumeration by viable colony counting. For invasion assays, extracellular bacteria were killed following the initial 1-h incubation period by an additional 1-h incubation in medium containing gentamicin (100 µg/ml; Sigma-Aldrich). Cells were then washed 3× with PBS and lysed. Bacterial titers in the lysates were determined by serial dilutions and plating on MacConkey agar. The results were expressed as the Log10CFU/ml.
Antibodies specific for O78-LPS (Denken Seiken) and the human tight junction protein ZO-1 (Invitrogen) were used for confocal laser scanning microscopy (CLSM) imaging. Antibodies were of porcine and mouse origins, respectively, and were used at a dilution of 1∶500 (anti-O78-LPS) and 1∶100 (anti-ZO-1). Goat anti-porcine and anti-mouse secondary antibodies labeled with Alexa Fluor 555 (Invitrogen) were used to detect the bound primary antibodies anti-O78-LPS and anti-ZO-1 respectively and were diluted 1∶500 in blocking solution (8% bovine serum albumin, 0.05% Triton-X100 in DPBS). Cell nuclei and the F-actin cytoskeleton were visualized with 4′, 6-diamidino-2-phenylindole hydrochloride (DAPI) and phalloidin (Invitrogen), respectively. The fixation and staining of 3-D aggregates was performed as described previously [88]. Optical sections of the 3-D aggregates were obtained using a Zeiss LSM 510 Duo laser scanning microscope equipped with detectors and filter sets for monitoring emissions of the selected fluorophores. Images were acquired using a Plan-Neofluar 40×/1.3 oil DIC objective and were analyzed with the Zeiss LSM software package. Axiovision 4.8 software from Carl Zeiss was used to further process collected images.
Sensitivity of strains to deoxycholate (DOC) and acid shock tolerance
To determine the ability of bacteria to survive at sub-lethal bile concentration, different strains were grown rotating to an OD600 1.0 in LB medium. Five-microliters of serial ten-fold dilutions of each strain were spotted on both LB agar and LB agar containing 1% (w/v) DOC plates and incubated overnight at 37°C.
For acid shock assays, bacterial cells were grown at 37°C in LB broth, pH 7, O/N standing. Cultures adjusted to the same OD600 of 1.0 were diluted 1∶1,000 in LB, pH 2.5, and incubated at 37°C with gentle shaking (50 rpm). Samples were cultured by direct plating on LB agar after 8 h and 18 h to determine the percent survival following acid stress. As controls, bacteria were also grown in LB, pH 7 in the same conditions to determine if the growth of bacteria was affected.
Biofilm formation assay
Biofilm formation assays were performed in 96-well polystyrene microtiter plates (Becton Dickinson, Franklin Lakes, NJ) [89]. In brief, strains were grown to stationary phase in LB at 37°C and then diluted 1∶100 in LB supplemented with 0.1% (w/v) L-glucose. Aliquots of 200 µL for each dilution were dispensed per well into a microtiter plate (four wells/strain). Each strain was tested in quadruplicate, wells containing sterile medium were used as negative controls. Plates were sealed with parafilm and cultured standing either at 30°C, 37°C or 42°C for 5 days to mimic the environmental and body temperature of humans and chickens, respectively. The media of the plates were then decanted, and the plates were washed twice with sterile PBS. Microplates were then stained with 200 µL of 1% (w/v) Crystal Violet for 30 min, followed by washing twice with PBS to remove unbound dye. After drying, dye-containing adherent cells were resolubilized with 200 µL of 30% (v/v) acetic acid solution. The absorbance was measured at 570 nm in an ELISA reader (SpectraMax M2, Molecular Devices). All tests were carried out at least three times, and the results were averaged.
Statistical analysis
Data were analyzed by one-way analysis of variance (ANOVA), followed by Bonferroni's multiple-comparison test (GraphPad Prism software, version 5.07). Differences between average values were also tested for significance by performing an unpaired, two-sided Student t test. The levels of significance (P values) are reported and values ≤0.05 were taken to be significant.
Supporting Information
Plasmids genomes comparison. Mauve pairwise nucleotide comparison of the complete pChi7122-2 DNA sequence to that of pEG356 (FN594520.1), pHK01 (HM355591.1), pAA (FN554767.1), and pKF3-70 (FJ494913.1) (A) and pChi7122-3 DNA sequence to that of R721 (AP002527.1). The colored boxes represent homologous segments completely free of genomic rearrangements. These boxes are connected by lines between genomes. Blocks below the center line indicate regions with inverse orientation. Regions outside blocks lack homology between genomes. White regions indicate the sequence specific to a genome.
(TIF)
Comparison of growth rates of bacteria in iron-restricted media. E. coli K-12 (χ6092) and its derivatives: pChi7122-1 (χ7346), pChi7122-2 (χ7347), and pChi7122-3 (χ7348) were grown in LB medium containing 2,2′-dipyridyl (- iron) or supplemented with either FeSO4 (control), Heme, or Hemoglobin at 37°C for 24 h.
(TIF)
Multiple amino acid sequence alignment. pChiD, pChiO, pChiT, and pChiA of pChi7122-2 were aligned with their homologous proteins from other bacteria. Arrows indicate β sheets; spirals α helixes and TT loops.
(TIF)
Comparison of growth rates of bacteria in the presence of different carbon sources. The wild-type strain χ7122 and its derivatives: No-plasmids (χ7368), pChi7122-1 (χ7394), pChi7122-2 (χ7392), pChi7122-3 (χ7367) were tested for growth in either strict MM9 (A) or MM9 containing thiamin and casamino acid (B) without sugar, or with different sugars (glucose, lactose, arabinose, mannose, galactose, glucoronic acid, or glucoronic acid).
(TIF)
Summary of information about the coding sequences of pChi7122-2. In this table, we present details of all coding sequences found in pChi7122-2.
(DOC)
Summary of information about the coding sequences of pChi7122-3. In this table, we present details of all coding sequences found in pChi7122-3.
(DOC)
Putative functions of pChi7122-2-encoded sugar pathways genes. In this table, we present the putative functions of pChiA, pChiD, pChiT, pChiO, and pChiR genes of the sugar pathway encoded by pChi7122-2.
(DOC)
Acknowledgments
We thank Alyssa Stacy (Biodesign Institute, Arizona State University) for her technical help.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding for research was provided by the U.S. Department of Agriculture National Research Initiative (USDA NRI) CSREES grant 2007-35201-18519 to Dr. Curtiss III, and National Institutes of Health grant R21 AI090416 to Dr. Mellata. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kaper JB. Pathogenic Escherichia coli. Int J Med Microbiol. 2005;295:355–356. doi: 10.1016/j.ijmm.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 3.George DB, Manges AR. A systematic review of outbreak and non-outbreak studies of extraintestinal pathogenic Escherichia coli causing community-acquired infections. Epidemiol Infect. 2010;138:1679–1690. doi: 10.1017/S0950268810001639. [DOI] [PubMed] [Google Scholar]
- 4.Smith JL, Fratamico PM, Gunther NW. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog Dis. 2007;4:134–163. doi: 10.1089/fpd.2007.0087. [DOI] [PubMed] [Google Scholar]
- 5.Ewers C, Antao EM, Diehl I, Philipp HC, Wieler LH. Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic Escherichia coli strains with zoonotic potential. Appl Environ Microbiol. 2009;75:184–192. doi: 10.1128/AEM.01324-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson JR, Delavari P, O'Bryan TT, Smith KE, Tatini S. Contamination of retail foods, particularly turkey, from community markets (Minnesota, 1999–2000) with antimicrobial-resistant and extraintestinal pathogenic Escherichia coli. Foodborne Pathog Dis. 2005;2:38–49. doi: 10.1089/fpd.2005.2.38. [DOI] [PubMed] [Google Scholar]
- 7.Mokady D, Gophna U, Ron EZ. Virulence factors of septicemic Escherichia coli strains. Int J Med Microbiol. 2005;295:455–462. doi: 10.1016/j.ijmm.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Stenutz R, Weintraub A, Widmalm G. The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol Rev. 2006;30:382–403. doi: 10.1111/j.1574-6976.2006.00016.x. [DOI] [PubMed] [Google Scholar]
- 9.Dho-Moulin M, Fairbrother JM. Avian pathogenic Escherichia coli (APEC). Vet Res. 1999;30:299–316. [PubMed] [Google Scholar]
- 10.Mellata M, Dho-Moulin M, Dozois CM, Curtiss R, III, Brown PK, et al. Role of virulence factors in resistance of avian pathogenic Escherichia coli to serum and in pathogenicity. Infect Immun. 2003;71:536–540. doi: 10.1128/IAI.71.1.536-540.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russo TA, Sharma G, Brown CR, Campagnari AA. Loss of the O4 antigen moiety from the lipopolysaccharide of an extraintestinal isolate of Escherichia coli has only minor effects on serum sensitivity and virulence in vivo. Infect Immun. 1995;63:1263–1269. doi: 10.1128/iai.63.4.1263-1269.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellata M, Ameiss K, Mo H, Curtiss R., III Characterization of the contribution to virulence of three large plasmids of avian pathogenic Escherichia coli chi7122 (O78:K80:H9). Infect Immun. 2010;78:1528–1541. doi: 10.1128/IAI.00981-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tivendale KA, Allen JL, Browning GF. Plasmid-borne virulence-associated genes have a conserved organization in virulent strains of avian pathogenic Escherichia coli. J Clin Microbiol. 2009;47:2513–2519. doi: 10.1128/JCM.00391-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tivendale KA, Noormohammadi AH, Allen JL, Browning GF. The conserved portion of the putative virulence region contributes to virulence of avian pathogenic Escherichia coli. Microbiology. 2009;155:450–460. doi: 10.1099/mic.0.023143-0. [DOI] [PubMed] [Google Scholar]
- 15.Johnson TJ, Johnson SJ, Nolan LK. Complete DNA sequence of a ColBM plasmid from avian pathogenic Escherichia coli suggests that it evolved from closely related ColV virulence plasmids. J Bacteriol. 2006;188:5975–5983. doi: 10.1128/JB.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson TJ, Siek KE, Johnson SJ, Nolan LK. DNA sequence and comparative genomics of pAPEC-O2-R, an avian pathogenic Escherichia coli transmissible R plasmid. Antimicrob Agents Chemother. 2005;49:4681–4688. doi: 10.1128/AAC.49.11.4681-4688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson TJ, Siek KE, Johnson SJ, Nolan LK. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J Bacteriol. 2006;188:745–758. doi: 10.1128/JB.188.2.745-758.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson TJ, Wannemeuhler YM, Scaccianoce JA, Johnson SJ, Nolan LK. Complete DNA sequence, comparative genomics, and prevalence of an IncHI2 plasmid occurring among extraintestinal pathogenic Escherichia coli isolates. Antimicrob Agents Chemother. 2006;50:3929–3933. doi: 10.1128/AAC.00569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellata M, Touchman JW, Curtiss R., III Full sequence and comparative analysis of the plasmid pAPEC-1 of avian pathogenic E. coli chi7122 (O78:K80:H9). PLoS One. 2009;4:e4232. doi: 10.1371/journal.pone.0004232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson TJ, Nolan LK. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol Mol Biol Rev. 2009;73:750–774. doi: 10.1128/MMBR.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson TJ, Wannemuehler YM, Johnson SJ, Logue CM, White DG, et al. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl Environ Microbiol. 2007;73:1976–1983. doi: 10.1128/AEM.02171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida H, Furuya N, Lin YJ, Guntert P, Komano T, et al. Structural basis of the role of the NikA ribbon-helix-helix domain in initiating bacterial conjugation. J Mol Biol. 2008;384:690–701. doi: 10.1016/j.jmb.2008.09.067. [DOI] [PubMed] [Google Scholar]
- 23.Furuya N, Komano T. Specific binding of the NikA protein to one arm of 17-base-pair inverted repeat sequences within the oriT region of plasmid R64. J Bacteriol. 1995;177:46–51. doi: 10.1128/jb.177.1.46-51.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skold O. Resistance to trimethoprim and sulfonamides. Vet Res. 2001;32:261–273. doi: 10.1051/vetres:2001123. [DOI] [PubMed] [Google Scholar]
- 25.Dyda F, Klein DC, Hickman AB. GCN5-related N-acetyltransferases: a structural overview. Annu Rev Biophys Biomol Struct. 2000;29:81–103. doi: 10.1146/annurev.biophys.29.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jana S, Karan G, Deb JK. Purification of streptomycin adenylyltransferase from a recombinant Escherichia coli. Protein Expr Purif. 2005;40:86–90. doi: 10.1016/j.pep.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Sundin GW, Bender CL. Dissemination of the strA-strB streptomycin-resistance genes among commensal and pathogenic bacteria from humans, animals, and plants. Mol Ecol. 1996;5:133–143. doi: 10.1111/j.1365-294x.1996.tb00299.x. [DOI] [PubMed] [Google Scholar]
- 28.Guerra B, Junker E, Schroeter A, Malorny B, Lehmann S, et al. Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coli isolates from cattle, swine and poultry. J Antimicrob Chemother. 2003;52:489–492. doi: 10.1093/jac/dkg362. [DOI] [PubMed] [Google Scholar]
- 29.Kim SR, Komano T. The plasmid R64 thin pilus identified as a type IV pilus. J Bacteriol. 1997;179:3594–3603. doi: 10.1128/jb.179.11.3594-3603.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srimanote P, Paton AW, Paton JC. Characterization of a novel type IV pilus locus encoded on the large plasmid of locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect Immun. 2002;70:3094–3100. doi: 10.1128/IAI.70.6.3094-3100.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gyohda A, Furuya N, Ishiwa A, Zhu S, Komano T. Structure and function of the shufflon in plasmid R64. Adv Biophys. 2004;38:183–213. [PubMed] [Google Scholar]
- 32.Komano T. Shufflons: multiple inversion systems and integrons. Annu Rev Genet. 1999;33:171–191. doi: 10.1146/annurev.genet.33.1.171. [DOI] [PubMed] [Google Scholar]
- 33.Provence DL, Curtiss R., III Isolation and characterization of a gene involved in hemagglutination by an avian pathogenic Escherichia coli strain. Infect Immun. 1994;62:1369–1380. doi: 10.1128/iai.62.4.1369-1380.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kostakioti M, Stathopoulos C. Functional analysis of the Tsh autotransporter from an avian pathogenic Escherichia coli strain. Infect Immun. 2004;72:5548–5554. doi: 10.1128/IAI.72.10.5548-5554.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisenreich W, Dandekar T, Heesemann J, Goebel W. Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat Rev Microbiol. 2010;8:401–412. doi: 10.1038/nrmicro2351. [DOI] [PubMed] [Google Scholar]
- 36.Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, et al. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun. 2008;76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerlt JA, Babbitt PC, Rayment I. Divergent evolution in the enolase superfamily: the interplay of mechanism and specificity. Arch Biochem Biophys. 2005;433:59–70. doi: 10.1016/j.abb.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 40.Botsford JL, Harman JG. Cyclic AMP in prokaryotes. Microbiol Rev. 1992;56:100–122. doi: 10.1128/mr.56.1.100-122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP). J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 42.Haydon DJ, Guest JR. A new family of bacterial regulatory proteins. FEMS Microbiol Lett. 1991;63:291–295. doi: 10.1016/0378-1097(91)90101-f. [DOI] [PubMed] [Google Scholar]
- 43.Alteri CJ, Smith SN, Mobley HL. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog. 2009;5:e1000448. doi: 10.1371/journal.ppat.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- 45.Bausch C, Peekhaus N, Utz C, Blais T, Murray E, et al. Sequence analysis of the GntII (subsidiary) system for gluconate metabolism reveals a novel pathway for L-idonic acid catabolism in Escherichia coli. J Bacteriol. 1998;180:3704–3710. doi: 10.1128/jb.180.14.3704-3710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huisman GW, Kolter R. Sensing starvation: a homoserine lactone–dependent signaling pathway in Escherichia coli. Science. 1994;265:537–539. doi: 10.1126/science.7545940. [DOI] [PubMed] [Google Scholar]
- 47.Graham JE, Clark-Curtiss JE. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc Natl Acad Sci U S A. 1999;96:11554–11559. doi: 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaureguy F, Landraud L, Passet V, Diancourt L, Frapy E, et al. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genomics. 2008;9:560. doi: 10.1186/1471-2164-9-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, et al. Organized genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson JR, Russo TA. Molecular epidemiology of extraintestinal pathogenic (uropathogenic) Escherichia coli. Int J Med Microbiol. 2005;295:383–404. doi: 10.1016/j.ijmm.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 51.Gunn JS. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2000;2:907–913. doi: 10.1016/s1286-4579(00)00392-0. [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez-Siek KE, Giddings CW, Doetkott C, Johnson TJ, Fakhr MK, et al. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology. 2005;151:2097–2110. doi: 10.1099/mic.0.27499-0. [DOI] [PubMed] [Google Scholar]
- 53.Tivendale KA, Logue CM, Kariyawasam S, Jordan D, Hussein A, et al. Avian-pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infect Immun. 2010;78:3412–3419. doi: 10.1128/IAI.00347-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nickerson CA, Goodwin TJ, Terlonge J, Ott CM, Buchanan KL, et al. Three-dimensional tissue assemblies: novel models for the study of Salmonella enterica serovar Typhimurium pathogenesis. Infect Immun. 2001;69:7106–7120. doi: 10.1128/IAI.69.11.7106-7120.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barrila J, Radtke AL, Crabbe A, Sarker SF, Herbst-Kralovetz MM, et al. Organotypic 3D cell culture models: using the rotating wall vessel to study host-pathogen interactions. Nat Rev Microbiol. 2010;8:791–801. doi: 10.1038/nrmicro2423. [DOI] [PubMed] [Google Scholar]
- 56.Huang SH, Stins MF, Kim KS. Bacterial penetration across the blood-brain barrier during the development of neonatal meningitis. Microbes Infect. 2000;2:1237–1244. doi: 10.1016/s1286-4579(00)01277-6. [DOI] [PubMed] [Google Scholar]
- 57.Moller AK, Leatham MP, Conway T, Nuijten PJ, de Haan LA, et al. An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infect Immun. 2003;71:2142–2152. doi: 10.1128/IAI.71.4.2142-2152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wibbenmeyer JA, Provenzano D, Landry CF, Klose KE, Delcour AH. Vibrio cholerae OmpU and OmpT porins are differentially affected by bile. Infect Immun. 2002;70:121–126. doi: 10.1128/IAI.70.1.121-126.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annu Rev Biochem. 2004;73:241–268. doi: 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- 60.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 61.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vazquez-Laslop N, Lee H, Neyfakh AA. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J Bacteriol. 2006;188:3494–3497. doi: 10.1128/JB.188.10.3494-3497.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin J, Smith MP, Chapin KC, Baik HS, Bennett GN, et al. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lim JY, Sheng H, Seo KS, Park YH, Hovde CJ. Characterization of an Escherichia coli O157:H7 plasmid O157 deletion mutant and its survival and persistence in cattle. Appl Environ Microbiol. 2007;73:2037–2047. doi: 10.1128/AEM.02643-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barua S, Yamashino T, Hasegawa T, Yokoyama K, Torii K, et al. Involvement of surface polysaccharides in the organic acid resistance of Shiga Toxin-producing Escherichia coli O157:H7. Mol Microbiol. 2002;43:629–640. doi: 10.1046/j.1365-2958.2002.02768.x. [DOI] [PubMed] [Google Scholar]
- 66.Richards JJ, Melander C. Controlling bacterial biofilms. Chembiochem. 2009;10:2287–2294. doi: 10.1002/cbic.200900317. [DOI] [PubMed] [Google Scholar]
- 67.Macfarlane S. Microbial biofilm communities in the gastrointestinal tract. J Clin Gastroenterol. 2008;42(Suppl 3 Pt 1):S142–143. doi: 10.1097/MCG.0b013e31816207df. [DOI] [PubMed] [Google Scholar]
- 68.Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, et al. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 69.Kikuchi T, Mizunoe Y, Takade A, Naito S, Yoshida S. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol Immunol. 2005;49:875–884. doi: 10.1111/j.1348-0421.2005.tb03678.x. [DOI] [PubMed] [Google Scholar]
- 70.Ghigo JM. Natural conjugative plasmids induce bacterial biofilm development. Nature. 2001;412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- 71.Reisner A, Holler BM, Molin S, Zechner EL. Synergistic effects in mixed Escherichia coli biofilms: conjugative plasmid transfer drives biofilm expansion. J Bacteriol. 2006;188:3582–3588. doi: 10.1128/JB.188.10.3582-3588.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.May T, Tsuruta K, Okabe S. Exposure of conjugative plasmid carrying Escherichia coli biofilms to male-specific bacteriophages. ISME J. 2010 doi: 10.1038/ismej.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dudley EG, Abe C, Ghigo JM, Latour-Lambert P, Hormazabal JC, et al. An IncI1 plasmid contributes to the adherence of the atypical enteroaggregative Escherichia coli strain C1096 to cultured cells and abiotic surfaces. Infect Immun. 2006;74:2102–2114. doi: 10.1128/IAI.74.4.2102-2114.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee YW, Jeong SY, In YH, Kim KY, So JS, et al. Lack of O-polysaccharide enhances biofilm formation by Bradyrhizobium japonicum. Lett Appl Microbiol. 2010;50:452–456. doi: 10.1111/j.1472-765X.2010.02813.x. [DOI] [PubMed] [Google Scholar]
- 75.Prouty AM, Schwesinger WH, Gunn JS. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect Immun. 2002;70:2640–2649. doi: 10.1128/IAI.70.5.2640-2649.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brandenburg K. Fourier transform infrared spectroscopy characterization of the lamellar and nonlamellar structures of free lipid A and Re lipopolysaccharides from Salmonella minnesota and Escherichia coli. Biophys J. 1993;64:1215–1231. doi: 10.1016/S0006-3495(93)81488-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seydel U, Koch MH, Brandenburg K. Structural polymorphisms of rough mutant lipopolysaccharides Rd to Ra from Salmonella minnesota. J Struct Biol. 1993;110:232–243. doi: 10.1006/jsbi.1993.1026. [DOI] [PubMed] [Google Scholar]
- 78.Snyder S, Kim D, McIntosh TJ. Lipopolysaccharide bilayer structure: effect of chemotype, core mutations, divalent cations, and temperature. Biochemistry. 1999;38:10758–10767. doi: 10.1021/bi990867d. [DOI] [PubMed] [Google Scholar]
- 79.Brown PK, Curtiss R., III Unique chromosomal regions associated with virulence of an avian pathogenic Escherichia coli strain. Proc Natl Acad Sci U S A. 1996;93:11149–11154. doi: 10.1073/pnas.93.20.11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mellata M, Dho-Moulin M, Dozois CM, Curtiss R, III, Lehoux B, et al. Role of avian pathogenic Escherichia coli virulence factors in bacterial interaction with chicken heterophils and macrophages. Infect Immun. 2003;71:494–503. doi: 10.1128/IAI.71.1.494-503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.European Committee on Antimicrobial Susceptibility Testing (EU-CAST) Breakpoint tables for interpretation of MICs and zone diameters. 2008. Available: http://www.eucast.org/eucast_disk_diffusion_test/(updated 27 April, 2010)
- 82.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; nineteenth informational supplement. CLSI document M100-S19. 3. Vol. 29. Wayne, Pa: Clinical and Laboratory Standards Institute; 2009. [Google Scholar]
- 83.Chain PS, Grafham DV, Fulton RS, Fitzgerald MG, Hostetler J, et al. Genomics. Genome project standards in a new era of sequencing. Science. 2009;326:236–237. doi: 10.1126/science.1180614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berriman M, Rutherford K. Viewing and annotating sequence data with Artemis. Brief Bioinform. 2003;4:124–132. doi: 10.1093/bib/4.2.124. [DOI] [PubMed] [Google Scholar]
- 85.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 88.Crabbe A, Sarker SF, Van Houdt R, Ott CM, Leys N, et al. Alveolar epithelium protects macrophages from quorum sensing-induced cytotoxicity in a three-dimensional co-culture model. Cell Microbiol. 2010;13:469–481. doi: 10.1111/j.1462-5822.2010.01548.x. [DOI] [PubMed] [Google Scholar]
- 89.O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 90.Dozois CM, Dho-Moulin M, Bree A, Fairbrother JM, Desautels C, et al. Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the Tsh genetic region. Infect Immun. 2000;68:4145–4154. doi: 10.1128/iai.68.7.4145-4154.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plasmids genomes comparison. Mauve pairwise nucleotide comparison of the complete pChi7122-2 DNA sequence to that of pEG356 (FN594520.1), pHK01 (HM355591.1), pAA (FN554767.1), and pKF3-70 (FJ494913.1) (A) and pChi7122-3 DNA sequence to that of R721 (AP002527.1). The colored boxes represent homologous segments completely free of genomic rearrangements. These boxes are connected by lines between genomes. Blocks below the center line indicate regions with inverse orientation. Regions outside blocks lack homology between genomes. White regions indicate the sequence specific to a genome.
(TIF)
Comparison of growth rates of bacteria in iron-restricted media. E. coli K-12 (χ6092) and its derivatives: pChi7122-1 (χ7346), pChi7122-2 (χ7347), and pChi7122-3 (χ7348) were grown in LB medium containing 2,2′-dipyridyl (- iron) or supplemented with either FeSO4 (control), Heme, or Hemoglobin at 37°C for 24 h.
(TIF)
Multiple amino acid sequence alignment. pChiD, pChiO, pChiT, and pChiA of pChi7122-2 were aligned with their homologous proteins from other bacteria. Arrows indicate β sheets; spirals α helixes and TT loops.
(TIF)
Comparison of growth rates of bacteria in the presence of different carbon sources. The wild-type strain χ7122 and its derivatives: No-plasmids (χ7368), pChi7122-1 (χ7394), pChi7122-2 (χ7392), pChi7122-3 (χ7367) were tested for growth in either strict MM9 (A) or MM9 containing thiamin and casamino acid (B) without sugar, or with different sugars (glucose, lactose, arabinose, mannose, galactose, glucoronic acid, or glucoronic acid).
(TIF)
Summary of information about the coding sequences of pChi7122-2. In this table, we present details of all coding sequences found in pChi7122-2.
(DOC)
Summary of information about the coding sequences of pChi7122-3. In this table, we present details of all coding sequences found in pChi7122-3.
(DOC)
Putative functions of pChi7122-2-encoded sugar pathways genes. In this table, we present the putative functions of pChiA, pChiD, pChiT, pChiO, and pChiR genes of the sugar pathway encoded by pChi7122-2.
(DOC)