Abstract
Background
Natural killer (NK) cells contribute to the defense against infected and transformed cells through the engagement of multiple germline-encoded activation receptors. Stimulation of the Fc receptor CD16 alone is sufficient for NK cell activation, whereas other receptors, such as 2B4 (CD244) and DNAM-1 (CD226), act synergistically. After receptor engagement, protein kinases play a major role in signaling networks controlling NK cell effector functions. However, it has not been characterized systematically which of all kinases encoded by the human genome (kinome) are involved in NK cell activation.
Results
A kinase-selective phosphoproteome approach enabled the determination of 188 kinases expressed in human NK cells. Crosslinking of CD16 as well as 2B4 and DNAM-1 revealed a total of 313 distinct kinase phosphorylation sites on 109 different kinases. Phosphorylation sites on 21 kinases were similarly regulated after engagement of either CD16 or co-engagement of 2B4 and DNAM-1. Among those, increased phosphorylation of FYN, KCC2G (CAMK2), FES, and AAK1, as well as the reduced phosphorylation of MARK2, were reproducibly observed both after engagement of CD16 and co-engagement of 2B4 and DNAM-1. Notably, only one phosphorylation on PAK4 was differentally regulated.
Conclusions
The present study has identified a significant portion of the NK cell kinome and defined novel phosphorylation sites in primary lymphocytes. Regulated phosphorylations observed in the early phase of NK cell activation imply these kinases are involved in NK cell signaling. Taken together, this study suggests a largely shared signaling pathway downstream of distinct activation receptors and constitutes a valuable resource for further elucidating the regulation of NK cell effector responses.
Introduction
Natural killer (NK) cells are lymphocytes belonging to the innate immune system. They can eliminate infected or transformed cells through direct killing of target cells [1]. Moreover, NK cells also influence cells of the adaptive immune system through release of chemokines and cytokines, as well as by contact-dependent killing of activated immune cells [2], [3]. The activation and effector functions of NK cells are controlled by signals from a multiplicity of germline-encoded activating and inhibitory receptors [4], [5]. Downstream of receptor engagement, the underlying signaling networks in NK cells are controlled by protein kinases [6], [7], [8], which constitute the largest family of enzymes in the human genome [9]. More than 500 different protein kinases coordinate steps in virtually all intra-cellular signaling pathways. Generally, protein kinases are expressed at relatively low levels and require targeted approaches for their direct characterization [10]. Dynamic post-translational modifications, including phosphorylations, regulate the enzymatic activity, localization, and substrate binding competence of kinases.
In terms of signaling by NK cell receptors, structurally distinct inhibitory receptors all contain immunoreceptor tyrosine-based inhibition motifs (ITIMs). The signaling by such motifs has been extensively studied and is mediated by activation of tyrosine phosphatases as well as the tyrosine kinase c-Abl [11], [12]. In contrast, activating NK cells receptors have highly divergent cytoplasmic signaling domains, and signaling pathways orchestrated by many activating NK cell receptors are not well defined. The low affinity Fc receptor CD16 (FcγRIIIA) is the prototypical NK cell activating receptor. Engagement of CD16 induces SRC tyrosine kinase-dependent phosphorylation of immunoreceptor tyrosine-based activation motifs (ITAM) on the adaptor chains CD3ζ and FcεRγ, which in turn recruit and activate SYK and ZAP-70 tyrosine kinases for downstream signaling [13]. Engagement of CD16 on NK cells is sufficient to induce NK cell degranulation for antibody-dependent cellular cytotoxicity (ADCC) [14]. Since several features of proximal CD16 signaling are shared with that of the T cell receptor (TCR), downstream events are thought to resemble those characterized in T cells. However, signaling induced by different ITAM-coupled receptors has not been rigorously compared. For natural cytotoxicity, engagement of individual receptors normally does not suffice, whereas co-engagement of specific pairwise combinations of receptors can induce synergistic intracellular Ca2+ mobilization as well as degranulation and cytokine production [15], [16]. Thus, receptors such as 2B4 (CD244) and DNAM-1 (CD226) have been termed “co-activating receptors” [15]. 2B4 binds CD48 [17], a ligand ubiquitously expressed on hematopoeitic cells. 2B4 contains cytoplasmic immunoreceptor tyrosine-based switch motifs (ITSMs). These motifs are phosphorylated and can recruit either phosphatases for negative regulation of NK cell functions, or, via the adaptor SAP, the SRC kinase FYN for the activation of NK cells [18], [19], [20]. In human NK cells, 2B4 is activating unless cells are deficient in SAP, which is the case in the primary immunodeficiency X-linked lymphoproliferative type 1 caused by mutations in SH2D1A whereupon engagement of 2B4 results in inhibitory signals [18], [21]. DNAM-1 binds the widely expressed nectins CD112 and CD155 [22]. The cytoplasmic domain of DNAM-1 is phosphorylated by protein kinase C (PKC) [23]. Further downstream, signaling by DNAM-1 in NK cells is not well understood but involves also serine and threonine phosphorylations rarely characterized in NK biology [24], [25]. In terms of co-activating receptor synergy, a recent report suggests that, rather than complementary signaling between different receptors, the basis for co-activation is dependent on a summation of signals from each receptor for VAV1 activation to overcome an inhibitory threshold set by c-Cbl [26].
In conclusion, further studies focusing at signaling downstream of co-activating receptors and the prototypical NK cell activation receptor CD16 are needed to identify common nodes as well as pathway specificities of receptors with distinct cytoplasmic domains, helping to define the core-signal network regulating NK cell activation.
Mass spectrometry (MS) can provide insights into protein phosphorylation events that coordinate signaling networks. In particular, Fourier transform mass spectrometry (FTMS) routinely achieves a very high data accuracy, facilitating unambiguous protein sequencing and identification of amino acid site-specific post-translational modifications [27]. Phosphoproteome studies have already been performed on T cell lines, shedding light on TCR signaling networks [28], [29], [30]. However, due to the requirement for large cell amounts, only a few MS-based studies have used primary lymphocytes [31]. Gaining access to signaling components, such as protein kinases, is more methodologically demanding and requires, in addition to significant amounts of starting cell material, efficient pre-enrichment of target proteins. Here, we use a kinase-selective phosphoproteome strategy to probe receptor-induced kinase phosphorylation in primary human NK cells. The results characterize the NK cell kinome including the first unbiased knowledge about its phosphorylation. Quantitative MS data reveal novel kinase phosphorylation sites that are modulated following CD16 activation or co-engagement of 2B4 and DNAM-1. Notably, the regulatory patterns were remarkably similar following activation through such distinct receptors, suggesting a largely shared signal network. To our knowledge, this study is the first proteome approach that specifically determines receptor-dependent kinase phosphorylation in primary lymphocytes.
Results
Expansion and characterization of primary NK cells
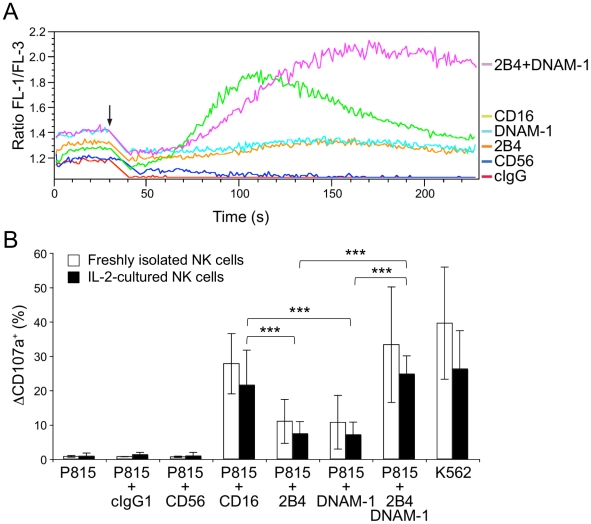
To obtain sufficient material for quantitative proteome studies, NK cells were isolated from the peripheral blood of healthy human donors and polyclonally expanded with irradiated, allogeneic feeder cells and recombinant IL-2 (Figure S1). To ensure that no gross perturbations in NK cell function occurred upon NK cell expansion, the readouts of early signaling events in IL-2–cultured NK cells were compared to those of freshly isolated NK cells (Figure 1). Initially, NK cells were labeled with Ca2+-sensitive fluorescent dyes and incubated either with IgG isotype control, anti-CD56 (both served as negative controls), anti-CD16, anti-2B4, or anti-DNAM-1, as well as co-incubated with anti-2B4 and anti-DNAM-1 monoclonal antibodies (mAbs) on ice. Thereafter, cells were pre-warmed for 5 minutes at 37°C and analyzed on a flow cytometer. The strength and dynamics of intracellular Ca2+-mobilization were assessed in NK cells following crosslinking with a secondary goat anti-mouse Fc antibody (Figure 1A). Whereas isotype control mAbs or crosslinking of CD56 did not elicit any Ca2+-mobilization in IL-2–cultured NK cells, crosslinking of CD16 induced a robust mobilization of intracellular Ca2+ similar, albeit diminished in magnitude, to that observed in freshly isolated NK cells [15]. Crosslinking of either 2B4 or DNAM-1 did not induce Ca2+-mobilization in IL-2–cultured NK cells, as previously observed for freshly isolated NK cells [15]. However, co-crosslinking of 2B4 and DNAM-1 synergized to produce intracellular Ca2+-mobilization in IL-2–cultured NK cells, again as previously described in freshly isolated NK cells [15]. Next, we compared receptor-induced degranulation in IL-2–cultured and freshly isolated NK cells (Figure 1B). NK cell-susceptible human K562 cells and murine P815 cells (Fc receptor-positive for redirected ADCC) were used as target cells. NK cells were incubated with target cells and mAbs, as indicated, for one hour followed by staining and analysis by flow cytometry. Degranulation was quantified in terms of induced CD107a surface expression. IL-2–cultured or freshly isolated NK cells did not degranulate in response to P815 cells or P815 cells co-incubated with isotype control or anti-CD56 mAb. However, engagement of CD16 induced degranulation of both IL-2–cultured and freshly isolated NK cells. Of note, degranulation was somewhat stronger in freshly isolated NK cells, consistent with stronger Ca2+-mobilization in freshly isolated NK cells. Co-engagement of 2B4 and DNAM-1 resulted in synergistic degranulation by both IL-2–cultured and freshly isolated NK cells, and degranulation induced by incubation with K562 cells was also similar between IL-2–cultured and freshly isolated NK cells. Moreover, comparison of receptor expression by IL-2–cultured and freshly isolated NK cells have previously revealed similar expression levels [15]. Taken together, the IL-2–cultured NK cells retained sufficient functional and phenotypic attributes of freshly isolated NK cells and were thus well suited for studies of signal transduction events by quantitative proteomics.
Figure 1. Functional characterization of IL-2–cultured primary NK cells.
(A) Intracellular Ca2+-mobilization induced by activating NK cell receptors. The following monoclonal antibodies (mAbs) were used for NK cell stimulation: negative controls, IgG isotype (cIgG, red) and anti-CD56 (dark blue); receptor stimulation, anti-2B4 (orange), anti-DNAM-1 (light blue), anti-CD16 (green) and anti-2B4 and anti-DNAM-1 (purple). Polyclonal IL-2–cultured NK cells were pre-incubated with the indicated mAbs on ice and loaded with Fluo-4 and Fura Red dyes. Ca2+-mobilization was assessed by flow cytometry. After 30 sec, secondary F(ab′)2 goat anti-mouse IgG crosslinking antibody was added (black arrow). FL-1/FL-3 ratios are plotted as a function of time. (B) Comparison of degranulation by freshly isolated (open bars) and IL-2–cultured (black bars) NK cells. NK cells were incubated with the following target cells: murine P815 cells (negative control), P815 cells coated with mAbs to the indicated receptors (Fc receptor-positive for redirected ADCC; IgG isotype and anti-CD56 mAb coated P815 cells were used as negative controls) or with NK cell-susceptible human K562 cells (positive control). Subsequently, NK cells were stained with fluorochrome-conjugated anti-CD56 and anti-CD107a mAbs and analyzed by flow cytometry. The percentage of CD107a-positive NK cells is shown as mean ± standard deviation (SD) of at least eight independent experiments. Differences between two groups were examined using the Student's t-test (***, p<0.001). Correlation between degranulation by freshly isolated (open bars) and IL-2–cultured (black bars) NK cells were determined according to Pearson (r = 0.95).
Kinase-selective phosphoproteome analyses of activated NK cells
Kinase phosphorylation in receptor-activated and IgG isotype control-treated NK cells were comparatively analyzed by the quantitative proteome workflow depicted in Figure 2. In total, three independent phosphokinome analyses were performed (two included CD16 engagement, all three included 2B4 and DNAM-1 co-engagement; Table S1). IL-2–cultured NK cells were incubated with IgG isotype control or indicated receptor-specific mAbs on ice, pre-warmed to 37°C and crosslinked for 2 minutes (Figure 2A). Notably, at this time-point the strength of intracellular Ca2+-mobilization induced by engagement of CD16 or co-engagement of 2B4 and DNAM-1 was similar in magnitude (Figure 1A). After stimulation, cell lysates were generated and protein kinases were enriched as described in the following.
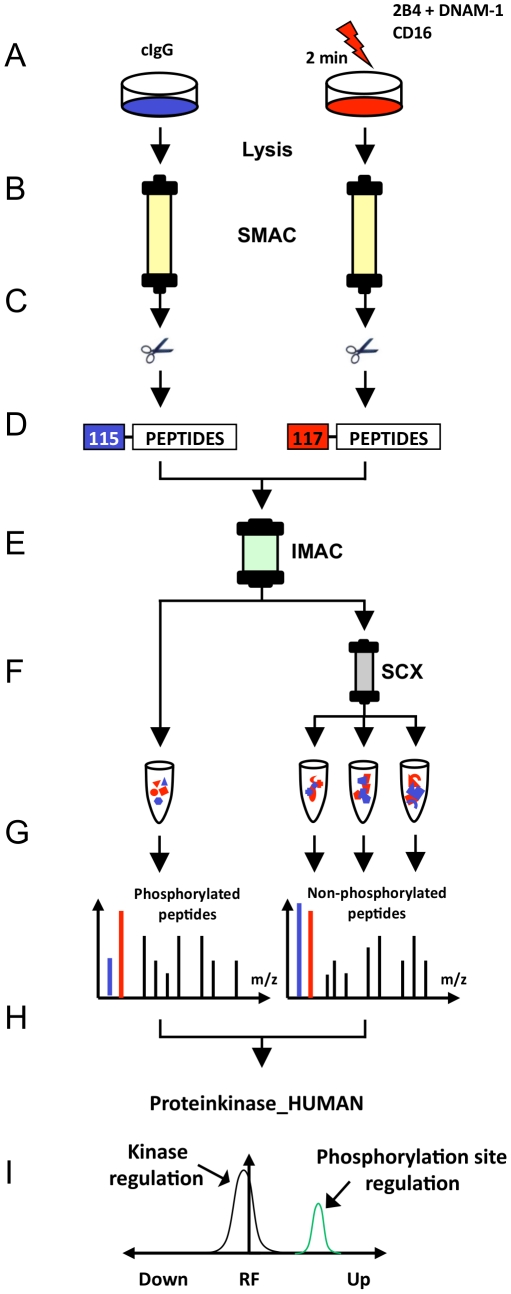
Figure 2. Proteome workflow for the detection of receptor-induced kinase phosphorylation in NK cells.
(A) Human, polyclonal, IL-2–cultured NK cells were stimulated each with the indicated receptor-specific monoclonal antibodies (mAbs) for 2 minutes (right side) and in all cases were comparatively analyzed with non-stimulated cells treated with control IgG isotype mAbs (cIgG) (left side). (B) Protein kinases were purified from total lysates using VI16743/Purvalanol-B-based small molecule affinity chromatography (SMAC). (C) Kinase elution and digestion into tryptic peptides comprising all non-modified and phosphorylated peptides. (D) Differential peptide labeling with iTRAQ reagents for MS-based quantification of phosphorylation site regulation and combination of samples. (E) Enrichment of phosphorylated peptides (phosphopeptides) by immobilized metal affinity chromatography (IMAC). (F) Further fractionation of non-phosphorylated peptide samples by strong cation exchange chromatography (SCX). (G) Peptide sequence analysis (LC-MS/MS) of all peptide fractions. (H) Peptide, kinase and phosphorylation site identification by Mascot database search and manual MS data inspection. (I) Statistical evaluation of significantly regulated phosphorylation sites on protein kinases by the MS-specific noise model iTRAQassist [38].
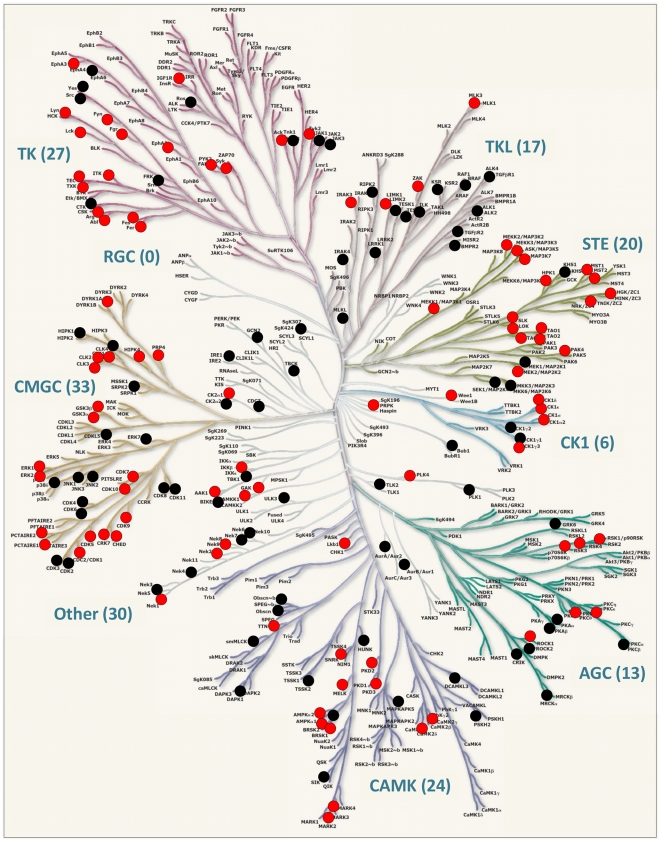
Sample complexity and high abundant proteins usually hamper the systematic characterization of protein kinases by MS. Thus, pre-enrichment of protein kinases from total cell lysates is required. Immobilized ATP-competitive small kinase inhibitors with a broad inhibitory spectrum can facilitate this step prior to proteomic characterization [10], [32], [33], [34]. Here, we used a serial combination of the kinase inhibitors VI16743 and Purvalanol-B that target a significant portion of the human kinome [10], [32], [35], [36]. Lysates generated from IL-2–cultured NK cells stimulated by indicated receptors were subjected to small molecule affinity chromatography (SMAC) for enrichment of protein kinases (Figure 2B). Proteome analyses of the SMAC eluate fractions revealed that approximately 30% of all identified proteins could be classified as kinases according to the Swiss-Prot protein knowledgebase (Figure S2A). Thus, kinases were the most abundant proteins in the eluted fractions and were generally characterized by a significant higher number of MS spectra and corresponding peptide sequences than non-kinases (Figure S2B). These results underline the efficient enrichment of protein kinases from total cell lysates using immobilized kinase inhibitors VI16743 and Purvalanol-B. In total, the expression of 175 protein kinases and 13 non-protein kinases (NPKs, kinases that have low molecular weight phosphate acceptors as substrates) was unambiguously determined in human NK cells (Table S2). According to a large-scale analysis of the human transcriptome suggesting the expression of around 300 distinct kinases in human cells [37], this chemical proteome approach characterized approximately 60% of the kinases expressed in human NK cells. Moreover, an alignment with the human kinome dendrogram [9] showed that kinases from almost all branches of the human kinome were detected in IL-2–cultured NK cells (Figure 3).
Figure 3. Protein kinases (kinome) expressed in human NK cells.
The figure shows the human kinome dendrogram [9]. Kinase-selective proteomics revealed the expression of 170 protein kinases (black and red circles), 5 atypical kinases (not depicted) and 13 non-protein kinases (NPKs, not depicted). Phosphorylation site-specific information was obtained for 95 kinases (red circles), whereas no such information was obtained for the remaining kinases (black circles). Abbreviations: AGC, PKA/PKG/PKC-family kinases; CAMK, calcium/calmodulin-dependent kinases; CK1, casein kinases; CMGC, CDK/MAPK/GSK3/CLK-family kinases; RCG, receptor guanylate cyclases; STE, sterile homologue kinases; TK, tyrosine kinases; TKL, tyrosine kinase-like kinases; atypical protein kinases; Other, belonging to non of the mentioned groups. Human kinome provided courtesy of Cell Signaling Technology, Inc. www.cellsignal.com.
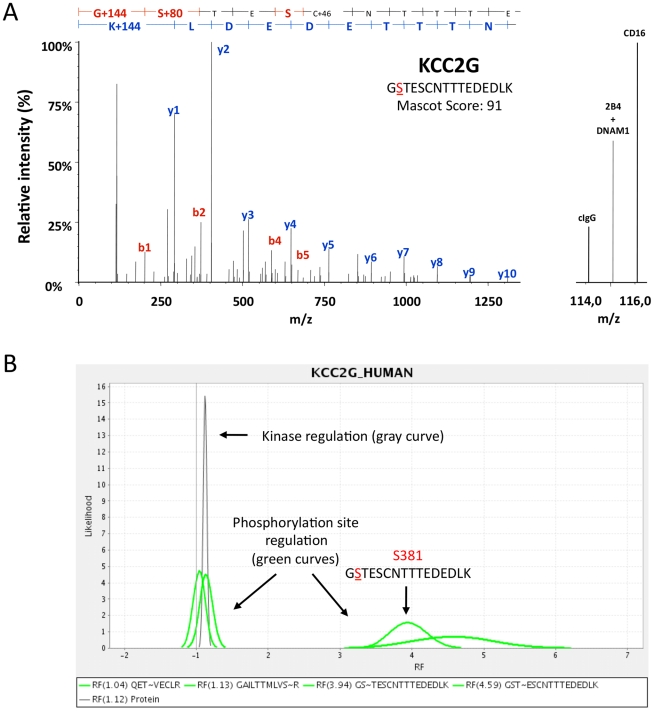
Next, all proteins of kinase-enriched SMAC-fractions were digested into peptides and differentially labeled with isobaric iTRAQ reagents (Figure 2C, D) required for peptide quantification by MS (for details, see Materials and methods). Differentially iTRAQ-labeled samples obtained from the respective stimulations were combined and phosphopeptides were purified from kinome-derived peptide mixtures by immobilized metal affinity chromatography (IMAC) to ascertain their optimal characterization by MS (Figure 2E). Phosphopeptide-depleted fractions comprising IMAC flow-through and wash fractions were further sub-fractionated by strong cation exchange (SCX) chromatography (Figure 2F) to reduce ion suppression effects in MS and to improve kinase sequence coverage. LC-MS/MS analyses (Figure 2G) were performed with all fractions and provided fragmentation data on both non-modified peptide sequences as well as those carrying phosphorylations on serine, threonine, or tyrosine residues (see Table S1 – Performed phosphokinome experiments). Since iTRAQ reporter ions are cleaved from each peptide under the condition of peptide sequencing simultaneously, each individual tandem MS fragment ion spectrum also provides quantitative information (Figure 2H, I). Figure 4 shows a representative MS/MS fragmentation spectra that depicts the phosphorylation of KCC2G on S381 induced by engagement of CD16 or co-engagement of 2B4 and DNAM-1: The detected molecular masses of b- (red) and y- (blue) fragment ions identify the phosphopeptide GS∼TESCNTTTEDEDLK, which can be exclusively assigned to the kinase KCC2G (Figure 4A). iTRAQ-reporter ions appeared in the low molecular mass region of this MS/MS spectrum indicating the relative abundance of this phosphorylated peptide following the activation of NK cells by engagement of CD16 (iTRAQ reporter 116) or co-engagement of 2B4 and DNAM-1 (iTRAQ reporter 115) in comparison to the isotype control stimulation (iTRAQ reporter 114). The intensities of the iTRAQ reporter masses 115 and 116 Dalton (Da) confirmed that phosphorylation on S381 was induced by CD16 engagement as well as 2B4 and DNAM-1 co-engagement. An example of the statistical evaluation of receptor-induced kinase phosphorylations by the MS-specific noise model iTRAQassist [38] is shown for CD16-induced phosphorylation of KCC2G (Figure 4B). Curves indicating most and less likely regulations, which must be considered based on the experimental results. Distinct and non-overlapping curves basically indicated significant regulations in stimulated NK cells. These so-called likelihood curves were calculated for each individual phosphopeptide (green curve) and comparatively inspected with a protein regulation curve summarizing quantitative data of all non-phosphorylated KCC2G peptides (gray curve). Phosphopeptides (phosphorylation sites) were only accepted as differentially regulated upon receptor engagement if their likelihood curves were clearly separated from the protein regulation curve (Figure S3).
Figure 4. Quantification of kinase phosphorylation induced by activating NK cell receptors.
ITRAQ-based quantification and statistical analysis of kinase phosphorylation by iTRAQassist is exemplified for CD16- or 2B4 and DNAM-1-mediated phosphorylation of calcium/calmodulin-dependent protein kinase II gamma (KCC2G). (A) MS/MS fragmentation spectrum of a tryptic phosphopeptide derived from KCC2G. Peptide sequencing, protein identification and phosphorylation site annotation are based on fragment ions of the b- (red) and y- (blue) series. The sequence of the phosphopeptide (GSTESCNTTTEDEDLK) is shown in the upper part of the diagram. On the right, a magnification of the low molecular mass range is shown. The intensity of the iTRAQ reporter 114 correlates with the abundance of the respective phosphopeptide in IgG isotype control-treated (cIgG) NK cells, whereas the intensities of the 115 and 116 peaks represent 2B4 and DNAM-1 co-stimulated or CD16-stimulated cells. The MS/MS spectrum is derived from experiment II (see Table S1). (B) Statistical evaluation of CD16-induced KCC2G phosphorylation by iTRAQ assist. Statistical analyses of phosphopeptide regulation were performed by iTRAQassist as described previously [38]. Most and less likely peptide regulations (x-axis: regulation factors, RF) were calculated and depicted as likelihood curves (y-axis) for individual phosphopeptides (green curves). The regulation of non-phosphorylated peptides assigned to KCC2G were calculated cumulatively and the resulting protein curve characterizes the general kinase abundance (expression) under the condition of stimulation (gray curve, “kinase regulation”). KCC2G was equally expressed in IgG isotype control-treated and CD16-stimulated NK cells. Only phosphorylation sites having regulation curves clearly separated from the protein curve were considered as regulated. CD16 stimulation (2 min) led to KCC2G phosphorylation on S381 and T382, but not on T287 (QETVECLR) or S311 (GAILTTMLVSR).
In this way, all 313 phosphorylation sites identified at 95 distinct kinases (Table S3) were inspected quantitatively. Of those, 102 were detected in all experiments (Figure S4). In total, 22 tyrosine, 62 threonine, and 229 serine phosphorylation events were observed. Thereof, 47 phosphorylations could not be assigned unambiguously to an individual threonine/serine residue as indicated by alternative phosphopeptide sequence annotations. As expected, the majority of peptides, in particular the non-modified peptide sequences, were not regulated significantly following receptor activation. Cluster analyses of all regulation data obtained from non-phosphorylated peptides showed a normal distribution, whereas regulation factors of phosphorylated peptides were distributed non-normally and exhibited a clear shift toward positive regulation values (Figure S5).
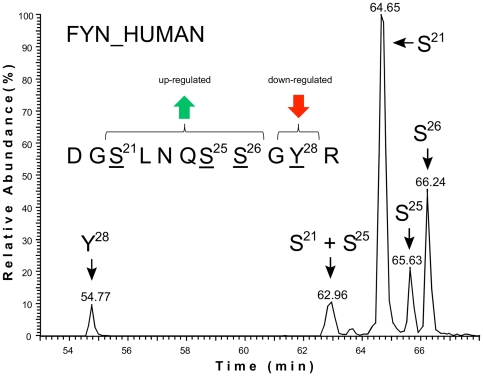
Particular attention was paid to the validation of significantly regulated phosphorylation sites determined by iTRAQassist [38], which also required a distinct separation of individual phosphopeptide populations. Nano ultra performance liquid chromatography (nano-UPLC) could separate peptides based on their sequence and number of modifications, but as is noteworthy, also according to the specific site of phosphorylation. Thus, subpopulations of isobaric phosphopeptides, such as the peptide DGSLNQSSGYR originating from FYN, could be quantified individually. Differentially phosphorylated peptide fractions representing the DGSLNQSSGYR region of FYN were clearly resolved by our chromatography and thus could be characterized separately by MS (Figure 5). This allowed the annotation and quantification of individual phosphorylation sites, even if modifications were located in close proximity. In the case of FYN, the discrimination of two N-terminal regions (S21-S25-S26 and Y28), inversely regulated by CD16 stimulation, was achieved (Figure 5 and Figure S6). Similarly, FYN was phosphorylated on the N-terminal amino acids S21, S25 and S26 following co-engagement of 2B4 and DNAM-1 (Figure S6). However, FYN exclusively phosphorylated at S21 constituted the most prominent part of the responding populations (Figure 5), whereas relatively small amounts of FYN were phosphorylated at S25, S26 and in particular Y28. Taken together, iTRAQassist-based statistical evaluation of quantitative MS data complemented by manual inspection of phosphopeptide elution profiles and fragmentation spectra assured the significance of NK cell receptor-induced kinase modifications in this study.
Figure 5. Identification of differentially phosphorylated FYN subsets in NK cells.
Nano ultra performance liquid chromatography (nano-UPLC) dissects distinct phosphorylation events at the same peptide DGSLNQSSGYR derived from the protein kinase FYN. MS analyses determined phosphorylations at the amino acid residues S21, S25, S26 and Y28. Successive elution of distinct phosphopeptides based on the same amino acid sequence facilitated the precise quantification of each phosphorylation event at FYN by MS (Figure S6). All phosphorylated serines showed a significantly induced phosphorylation following receptor engagements. However, the relative signal intensities of the distinct phosphopeptide populations indicate a predominant abundance of FYN phosphorylated on S21.
Kinase phosphorylations induced by CD16 and co-engagement of 2B4 and DNAM-1
Kinase-mediated signaling downstream of NK cell activation receptors had not been systematically investigated. Therefore, we used NK cells from two human donors and comparatively studied kinase phosphorylation induced by CD16 and co-engagement of 2B4 and DNAM-1. These experiments revealed 21 protein kinases with altered phosphorylation sites after 2 minutes of CD16 engagement (Table 1 and Figure S3). Several phosphorylation sites could not observed by MS under all conditions, which might either reflect the limited sensitivity of this study or indicate donor variations. However, the increased phosphorylation of FYN, KCC2G, FES, and AAK1, as well as the reduced phosphorylation of MARK2, were reproducibly observed in all experiments. 2B4 and DNAM-1 co-engagement generally led to a slightly weaker phosphorylation of these phosphorylation sites, and according to the statistical noise model iTRAQassist, some regulations did not achieve a significant regulation score. However, with few exceptions, phosphorylation induced by 2B4 and DNAM-1 co-engagement was in accordance with that induced by CD16 engagement (Figure 6).
Table 1. Kinase phosphorylation induced by CD16 (2 min).
| Kinase1 | Phosphorylation-site function2 and regulation3 | Function4 | References5 | |
| NK cell function$ | ||||
| FAK2T842, FAK2Y579 | N/K, N/K | down, down | Natural cytotoxicity, integrin signaling, IL15-mediated NK cell priming | [69] [70] |
| FGRY412 | (A) | up | IFNγ production | [71] |
| FYNS21-S25-S26, FYNY28 | A/N/K/*, (A) | up, down | 2B4 signaling, LFA-1-dependent DNAM-1 phosphorylation | [19] [45] [46] [25] |
| ITKY512 | A | donor-dependent | Fc receptor and NKG2D signaling | [72] |
| KCC2GT287, KCC2GS381–S384 | (A), N/K | up, up | LFA-1-dependent NK cell activation by DCs | [50] |
| KCC2DS330 | N/K | up | LFA-1-dependent NK cell activation by DCs | [50] |
| KPCTS685 | N/K | up | NK cell activation, CD16 and 2B4 signaling | [73] [74] [75] [76] [77] [78] [79] |
| LCKY192, LCKY394 or T395 | I, A/* | down, up | NK cell activation, CD16 signaling | [80] [81] [82] [83] |
| LYNS38, LYNS164 | *, * | up, down | CD16 signaling | [81] |
| Cytoskeleton rearrangement and degranulation§ | ||||
| FERY714 | A | down | Cytoskeleton dynamic, FcεRI signaling | [62] [63] |
| FESS716, FESY713 | N/K, A | up, down | Cytoskeleton dynamic, FcεRI signaling, vesicle trafficking and degranulation | [62] [63] [84] [64] |
| MARK2S456 | N/K | down | Mikrotubuli reorganization, granula exocytosis | [66] [85] |
| PAK4S181 | N/K | down | Actin cytoskeleton dynamic, adhesion | [43] [44] |
| Clathrin-mediated endocytosis§ | ||||
| AAK1T620, AAK1T672-S678 | N/K, N/K | up, down | Clathrin-mediated endocytosis | [86] [87] [88] |
| GAKS829 | N/K | up | Clathrin-mediated endocytosis | [86] [87] [88] |
| Other§ | ||||
| AAPK1S496 | (A) | up | ATP-dependent biosynthesis | [89] [90] |
| GSK3AS21 | I | up | PI3K/Akt-, NFAT signaling | [91] [92] |
| KPCD2S214 | N/K | up | DAG/phorbol ester, NFAT signaling | [93] [94] |
| NEK9S331-T333 | N/K | up | Cell cycle | [95] [96] |
| PCTK2S137, PCTK2S165 | N/K, N/K | up, up | Potential role in terminally differentiated neurons | [97] [98] |
| PRPKT8 | N/K | up | p53/TP53 regulation | [99] |
Footnote: Kinases listed here are ($) involved in or (§) so far not associated with NK cell function and/or CD16 signaling;
)UniProt name of kinase and phosphorylation site annotation;
)Function of phosphorylation site (A, activation; I, inhibition; brackets indicate possible function based on sequence homology; N/K, not known; *, novel phosphorylation site according to PhosphoSitePlus database);
)Direction of phosphorylation site regulation induced by CD16 (up: up-regulation, down: down-regulation, donor-dependent regulation;
)Kinase function and.
)PubMed references.
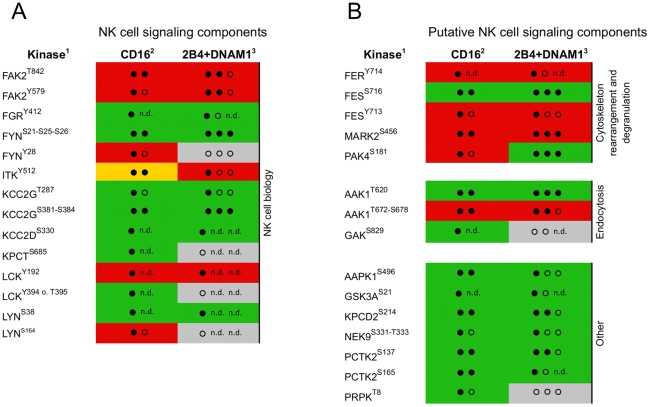
Figure 6. Kinase phosphorylation induced by engagement of CD16 or co-engagement of 2B4 and DNAM-1.
CD16- (two proteome experiments) or 2B4 and DNAM-1 co-activated (three proteome experiments) NK cells from two healthy human donors were analyzed in this study. (A) Kinases, which have previously been associated with NK cell signaling and/or function. (B) Putative novel CD16- and/or 2B4 and DNAM-1-dependent signaling components. 1)UniProt name of kinases and phosphorylation site annotation; regulation of phosphorylation site following 2)CD16 engagement or 3)2B4 and DNAM-1 co-activation is indicated as follows: green, up-regulated; red, down-regulated; gray, no statistical significant phosphorylation site regulation was detected; Number of (filled) circles indicates how often the significant regulation of a respective phosphorylation site was detected according to iTRAQassist (no overlap of protein and respective phosphopeptide regulation curves). Phosphorylation sites regulated with the same tendency, but not matching the criteria of statistical significance are marked as open circles. Proteome experiments that failed to detect a respective phosphorylation site are indicated by n.d. (not detected).
Only three of 21 kinases identified as significantly regulated have been previously described as components of the CD16-signaling pathway, i.e. LCK, LYN, and KPCT (PRKCQ/PKCθ). Our data indicate a CD16-induced dephosphorylation of LCK on the inhibitory tyrosine Y192, increased phosphorylation of LCK on the activating region Y394/T395, phosphorylation of LYN on S38, dephosphorylation of LYN on serine S164 and phosphorylation of KPCT on S685. The dephosphorylation of LCK on Y192 and phosphorylation of LYN on S38 were also observed in NK cells co-activated by 2B4 and DNAM-1. Furthermore, kinases implicated in NK cell function, but not specifically assigned to CD16 signaling, i.e. FAK2, FGR, FYN, ITK, and KCC2G/D, were phosphorylated upon CD16 stimulation. FAK2 was dephosphorylated on the amino acids T842 and Y579, which is part of the kinase domain, upon CD16 engagement or 2B4 and DNAM-1 co-engagement. Engagement of CD16 and co-stimulation of 2B4 and DNAM-1 induced phosphorylation of FGR on its supposed auto-phosphorylation site Y412. Triggering of CD16 led to the phosphorylation of FYN on the N-terminal serines S21, S25, and S26, whereas the adjacent Y28 showed a trend toward dephosphorylation (Figure S6). Phosphorylation of these serines was also induced by co-engagement of 2B4 and DNAM-1. Engagement of CD16 as well as co-engagement of 2B4 and DNAM-1 also induced phosphorylation of KCC2G on S381–S384 and KCC2D on S330, in addition to the tentative activation site KCC2G on position 287 (based on sequence homology). For ITK, phosphorylation of Y512 upon CD16 engagement was increased in one donor but decreased in the other one. In contrast, ITK Y512 phosphorylation was reduced following co-engagement of 2B4 and DNAM-1.
The majority of kinases identified by our study have not been characterized in NK cells before. Phosphorylation on the activating Y714 of FER was reduced, as was FES phosphorylation decreased on the activating Y713 by engagement of CD16 or co-engagement of 2B4 and DNAM-1. In contrast, phosphorylation of the proximate S716 residue of FES was significantly induced by engagement of CD16 or co-engagement of 2B4 and DNAM-1. Further, MARK2 phosphorylation was significantly reduced on S456 in NK cells following engagement of CD16 and co-engagement of 2B4 and DNAM-1. Remarkably, PAK4 phosphorylation on S181 was diminished after CD16 engagement, but significantly up-regulated upon co-engagement of 2B4 and DNAM-1. Moreover, NK cell receptor activation led to the phosphorylation of AAK1 and GAK, both of which are involved in clathrin-mediated endocytosis. AAK1 phosphorylation on T620 was significantly enhanced following engagement of CD16 and co-engagement of 2B4 and DNAM-1, whereas the region S672–S678 was reversely regulated. Our data also suggested the phosphorylation of GAK on S829 following CD16 activation. NK cell activation via CD16 and or 2B4 and DNAM-1 co-engagement also led to the phosphorylation of AAPK1 on its predicted auto-phosphorylation site S496, enhanced phosphorylation of GSK3A on the inhibitory site S21, phosphorylation of KPCD2 on S214, phosphorylation of NEK9 on S331-T333 and PCTK2 phosphorylation on S137 and S165. Furthermore, PRPK was phosphorylated on T8 in response to CD16 engagement. In summary, proteome analysis of receptor-induced kinase phosphorylation implicated several new kinases in the regulation of NK cell activation.
Discussion
The potential of kinase-selective phosphoproteomics (phosphokinomics) to provide comprehensive access to the human kinome has previously been demonstrated [10], [33], [39], [40]. Our results provide a snap-shot of kinase phosphorylation in NK cells shortly after crosslinking of different activating receptors and identify several kinases not previously implicated in NK cells signaling that are regulated downstream of receptor engagement. To our knowledge, this study is the first to employ phosphokinomics to systematically analyze proximal kinase signaling in primary immune cells.
Phosphokinomics is useful for the systematic characterization of kinase-based signaling on a minute time scale [41], [42]. MS-based characterization of phosphorylation events at the level of low abundant protein kinases currently still requires a large amount of cell material and therefore has been restricted to immortalized cell lines. An expansion protocol facilitated the generation of sufficient material (up to 1,5×109 NK cells) for phosphokinome analyses. Importantly, as compared to freshly isolated NK cells, IL-2–cultured NK cells were not significantly altered with respect to the expression of activation receptors, kinetics of intracellular calcium mobilization, or magnitude of degranulation following receptor stimulation. The non-specific kinase inhibitor VI16743 in combination with the commercially available ATP-competitor Purvalanol-B were used to capture the kinome and phosphorylation status of stimulated NK cells. The number of kinases identified by our workflow equaled the number of kinases detected by chemical proteome approaches using a pool of 5 different kinase inhibitors [33], [39], [40], [42] and exceeded the amount of kinases captured by VI16743 alone [33]. MS data verified the expression of 175 protein kinases (and 13 non-protein kinases) from virtually all branches of the human kinome. Although physiological kinase phosphorylation states and activities in primary cells are probably lower than compared to immortalized cell lines, we obtained quantitative information on more than 300 distinct phosphorylation sites. MS-based analysis of phosphopeptide regulation was assisted by a nano-UPLC system enabling the distinct separation of even isobaric phosphopeptides (identical amino acid sequence but different position of modification). This further supported the unambiguous characterization of phosphorylation sites by high-accuracy MS and the exclusion of compromised iTRAQ reporter signals for their quantitative analyses. Finally, statistical evaluation identified kinases significantly regulated by engagement of CD16 or co-engagement of 2B4 and DNAM-1. Our study revealed 21 kinases on which phosphorylation was modulated upon NK cell receptor stimulation. However, not all of the kinase regulations could be confirmed in each experiment. Besides potential variations in human donors, a limited sensitivity and randomized selection of low-abundant peptide ions during LC-MS experiments always cause some variations in the number of identified phosphopeptides, which then obligatory coincide with missing quantitative information.
Many of the regulated kinases presented in this study have not been previously described in NK cells (Table 1 and Figure 6). For most regulated sites, phosphorylation induced by CD16 engagement or 2B4 and DNAM-1 co-engagement demonstrated a similar pattern, in agreement with the notion of a convergence of signals for NK cell activation [26]. However, PAK4 phosphorylation on S181 was induced by co-stimulation of 2B4 and DNAM-1, but reduced following CD16 engagement (Figure 6). Preliminary experiments with single receptor stimulations suggest that PAK4 phosphorylation on S181 is induced by 2B4, but not DNAM-1. Thus, PAK4 may be downstream of 2B4 engagement. In T cells, 2B4 can recruit PIX (PAK interacting exchange factor), a PAK-Rac/Cdc42-specific guanine-exchange factor (GEF), via its adapter protein SAP [43], [44]. Thus, PAK4 might link proximal 2B4 signaling to actin cytoskeleton rearrangements and be dependent on the SAP/PIX adapter complex. In contrast to PAK4, the phosphokinome approach repeatedly revealed consistent regulation of phosphorylation sites on five kinases (FYN, KCC2, AAK1, FES and MARK2) following stimulation with different receptors.
FYN was consistently phosphorylated upon engagement of all receptors investigated on its N-terminal serines S21, S25 and S26, but phosphorylation of the adjacent tyrosine Y28 was unchanged or even reduced. FYN is known to play an important role in proximal signaling by 2B4 [19], [45], [46]. Additionally, FYN is involved in DNAM-1-mediated signaling, as has previously been demonstrated in T cells and mast cells [47], [48]. A role for FYN in NK cell signaling downstream of CD16 has not been described. FYN phosphorylation on S21 and S25 has been identified by other systematic phosphoproteome approaches, whereas FYN phosphorylation on S26 is novel. Interestingly, recent work suggests that FYN is phosphorylated by PKA on S21 and that this phosphorylation can regulate FYN activity and reduce focal adhesion dynamics [49]. Our data support the idea that S21 plays an important functional role: S21 showed a significantly regulated phosphorylation and constituted the most abundant FYN population in comparison to alternative phosphorylations at S25, S26 and Y28. Together, these data point to a central role for FYN in NK cell activation mediated by different activating NK cell receptors.
The calcium/calmodulin-dependent kinase 2 (KCC2, CAMK2) subunits gamma and delta were phosphorylated on S381–S384 and S330 following CD16 engagement or 2B4 and DNAM-1 co-engagement. Furthermore, our data suggests simultaneous phosphorylation of the S287 activation site on KCC2G. Poggi and colleagues have demonstrated that pharmacological inhibition of KCC2 blocks IL-2–cultured NK cell killing of autologous dendritic cells, but not of K562 cells [50]. In T cells, TCR engagement induces translocation of KCC2 to the immune synapse where KCC2 facilitates activation of NFκB through phosphorylation of CARMA1 and Bcl10 [51], [52]. Data also suggest that KCC2 can negatively regulate NFAT activation, diminishing cytokine transcription in T cells [53]. Taken together, although our data implicate activation of KCC2 downstream of activating NK cell receptor engagement, it is not clear how KCC2 modulates NK cell effector functions.
Results revealed that AAK1, the ubiquitously expressed AP2 associated kinase 1 implicated in clathrin-mediated endocytosis [54], [55], possesses a high basal serine/threonine phosphorylation in the region T606-S690. Following engagement of CD16 or co-engagement of 2B4 and DNAM-1, phosphorylation of T672-S678 was reduced. Conversely, stimulation induced AAK1 phosphorylation on T620. Thus, this regulation may potentially serve as molecular switch controlling AAK1 function. AAK1 function has not been studied in NK cells. However, following activation, several NK cell receptors have been shown to undergo SRC-dependent internalization [56]. Moreover, juxtaposed to an exocytic pathway, endocytosis is a prominent feature of the cytotoxic NK cell synapse [57]. The significance and molecular architecture of endocytosis in regard to NK cell cytotoxicity remains to be elucidated.
Stimulation of CD16 or 2B4 and DNAM-1 induced FES phosphorylation of S716 and dephosphorylation of the postulated auto-phosphorylation site Y173 [58], [59], suggesting a regulation of FES activity following NK cell receptor engagement. FES is a SRC family kinase involved in cytoskeleton rearrangements [60], [61]. In mast cells, FES participates in FcεRI-receptor-induced degranulation [62], [63], [64]. Hence, FES might also regulate NK cell degranulation.
MARK2 was consistently dephosphorylated on S456 following engagement of CD16 or co-engagement of 2B4 and DNAM-1. MARK family kinases phosphorylate microtubule-associated proteins, regulate microtubule-based intracellular transport, and are implicated in cellular polarity [65]. In T cells, MARK2 has been shown to become phosphorylated on S400 and T595 following TCR engagement and be required for microtubule-organizing center polarization [66]. Thus, it will be interesting to investigate how MARK2 might contribute to NK cell cytotoxicity.
This study characterizes a significant portion of the NK cell kinome and provides a first unbiased and systematic view into kinase signaling in primary NK cells. Proteomics detected receptor-dependent kinase phosphorylations already after 2 minutes of receptor engagement and allowed the discrimination of differentially phosphorylated kinase populations as demonstrated by FYN. Several kinases were consistently regulated following the engagement of activating receptors. Co-engagement of DNAM-1 and 2B4 generates a phosphorylation pattern at kinases, which was rather similar to that of CD16-stimulated cells, supporting the idea of a core signal network for NK cell activation. Kinases and phosphorylations, which have not previously been implicated in NK cell signaling were regulated and may thus contribute to the process of NK cell activation. Regulation of kinases such as MARK2 and AAK1 are interesting in terms of understanding the molecular pathways for cellular polarization and endocytosis, processes that are induced upon NK cell activation. Thus, contemplating data presented here will direct future studies focused on elucidating the molecular regulation of NK cell effector functions.
Materials and Methods
Cells
This study was approved by The Regional Ethics Review Board in Stockholm. Peripheral blood was obtained with informed consent from healthy volunteers. Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation (Lymphoprep, Axis-Shield). NK cells were subsequently purified by negative magnetic selection (NK cell isolation kit, Miltenyi Biotec). Freshly isolated NK cells were resuspended in complete medium (RPMI 1640 supplemented with 2 mM L-glutamine and 10% fetal bovine serum [FBS], all Invitrogen) and analyzed within two days after isolation. For polyclonal expansion, purified NK cells (105 per well) and irradiated, allogeneic PBMCs (105 per well) were resuspended in complete medium supplemented with 500 U/ml recombinant IL-2 (Proleukin, Roche) and 10 µg/ml PHA-L (Sigma). Cells were co-cultured in 96-well plates for one week. NK cells were transferred to culture flasks and cultured further for 2 weeks in complete medium supplemented with 50 U/ml IL-2. The purity and viability of NK cells were assessed by flow cytometry prior to analysis. The percentage of CD3−CD56+ NK cells was greater than 95% in all experiments.
The human erythroleukemia cell line K562 and murine mastocytoma cell line P815 (both American Type Culture Collection) were maintained in complete medium.
Antibodies and fluorescent reagents
For flow cytometry, NK cell activation, calcium flux analyses, and CD107a degranulation assays, the following mouse monoclonal antibodies (mAbs) were used: anti-CD107a (clone H4A3, IgG1), anti-CD16 (clone 3G8, IgG1), anti-CD56 (clone NCAM16.2, IgG2b, or B159, IgG1), anti-DNAM-1 (clone DX11, IgG1), anti-NKG2D (clone 1D11, IgG1), and anti-CD3 (clone UCHT1, IgG1; all BD Biosciences), anti-2B4 (clone C1.7, IgG1, Beckman Coulter), and isotype control mAb (clone MOPC-21, Sigma-Aldrich). For determination of intracellular calcium mobilization, the calcium-sensitive dyes Fluo-4 and FuraRed (both Molecular Probes) were used.
Calcium flux analysis
NK cells were resuspended in 50 µl Hanks buffered salt solution (HBSS, Biosource) and incubated with the primary mAbs indicated (1 µg mAb per 107 NK cells) for 30 min on ice. 300 µl staining solution (HBSS supplemented with 1% FBS, 2 µM Fluo-4 AM, 5 µM Fura Red AM, and 5 mM Probenecid) were added, and cells were incubated for an additional 30 minutes at 4°C. Dye-loaded NK cells were washed, resuspended in HBSS, and kept on ice. Prior to analysis NK cells were resuspended in 300 µl HBSS supplemented with 1% FBS, pre-warmed at 37°C in a water bath, and analyzed by flow cytometry. After 30 seconds, samples were removed from the flow cytometer, and F(ab′)2-anti-mouse IgG crosslinking antibodies (2 µg Ab per 107 NK cells; Jackson ImmunoResearch) were added. Thereafter, samples were measured for 4 minutes by flow cytometry. Acquired flow cytometric data were analyzed using the FlowJo software (Treestar). NK cells were gated on forward/side scatter, and the ratios of FL-1 and FL-3 intensities were displayed as a function of time.
CD107a degranulation assay
For assessment of NK cell degranulation, 2×105 K562 cells or 2×105 P815 target cells pre-incubated with 5 µg/ml of the indicated mAbs for 30 minutes at room temperature were mixed with 1×105 NK cells for one hour at 37°C in complete medium. Thereafter, the cells were centrifuged and resuspended in FACS buffer (PBS supplemented with 2% FBS and 2 mM EDTA) containing fluorochrome-conjugated anti-CD56 and anti-CD107a mAbs, and analyzed by flow cytometry. Data were analyzed with FlowJo software.
IL-2–cultured NK cell stimulation for phosphokinome analysis
For kinome experiments, up to 1,5×109 IL-2-cultured NK cells were used (5×108 NK cells per stimulation). IL-2-cultured NK cells were starved overnight in complete medium without IL-2. Prior to stimulation, cells were resuspended in complete medium at 2×107 cells/ml. NK cells were incubated with primary mAbs directed against CD16, DNAM-1 and 2B4 for 30 minutes on ice (1 µg mAb per 107 NK cells). MAb-coated NK cells were pre-warmed at 37°C in a water bath, thereafter added F(ab′)2-anti-mouse-IgG crosslinking antibody (Jackson ImmunoResearch), and stimulated for the indicated time.
Kinase enrichment from NK cell lysates
Following stimulation, NK cells were lysed in ice-cold lysis buffer containing 50 mM HEPES pH 7.5, 1 M NaCl, 1 mM EGTA, protease inhibitor cocktail (Roche), and 1% Triton-X100. Lysates were centrifuged at 70000× g for 30 minutes at 4°C and filtered using a 0.45 µm syringe membrane (Millipore). The protein concentrations of lysates were determined using a Bradford protein assay (BioRad) to ensure equal loading of kinase affinity columns. For small molecule affinity chromatography (SMAC), the kinase inhibitors VI16743 and Purvalanol-B (Tocris) were used. Synthesis of V16743 and generation of both kinase affinity materials were conducted as previously described [10], [41]. Columns (5/50 Tricorn, GE Healthcare) were equilibrated with buffer A (50 mM HEPES-NaOH pH 7.5, 1 M NaCl, 1 mM EGTA, 1 mM EDTA, 0.1% Triton-X100), loaded with NK cell lysates (3 ml/h), washed with 60 column volumes of buffer A (6 ml/h), and equilibrated (2 h, 6 ml/h) with buffer B (50 mM HEPES-NaOH pH 7.5, 1 mM EGTA, 1 mM EDTA, 0.1% Triton-X100). Kinase-enriched fractions were eluted (6 ml/h) separately with 0.5% SDS at room temperature. Eluates were pooled and concentrated by vacuum centrifugation.
Peptide generation and iTRAQ labeling
Proteins were extracted from concentrated SMAC eluate fractions by chloroform/methanol precipitation [67]. Protein dissolution, denaturation, tryptic digestion, and quantitative peptide labeling by iTRAQ were performed according to the manufacturer's guidelines (Applied Biosystems). Following labeling, peptides derived from IgG isotype control-, CD16 or 2B4 and DNAM-1 co-stimulated NK cells were combined and pooled samples were vacuum-dried, and peptides were desalted using self-packed LiChroprep RP-18 SPE columns (Merck). To quantify kinase phosphorylation induced by CD16, individually, or the co-engagement of 2B4 and DNAM-1, the following iTRAQ-labels were used: experiments I and II (cIgG:114, 2B4/DNAM-1:115, CD16:116), and experiment III (cIgG:114, 2B4/DNAM-1: 117).
Phosphopeptide enrichment and peptide fractionation
Phosphorylated peptides (phosphopeptides) were enriched from complex peptide samples using the following immobilized metal affinity chromatography (IMAC) protocol: iTRAQ-labeled and desalted peptide mixtures were resolved in IMAC binding buffer (1∶1∶1 methanol∶acetonitrile∶H2O containing 2% acetic acid, pH 2.8) and incubated with two Ga3+-chelated gel disks (Pierce) for one hour at room temperature or overnight at 4°C. The Ga3+-matrix was washed 3 times with 100 µl IMAC-binding buffer to remove unmodified peptides. Retained phosphopeptides were eluted with 600 µl IMAC elution buffer (100 mM ammonium phosphate buffer, pH 4.5). The phosphopeptide-enriched fraction was vacuum-dried, desalted and analyzed by LC-MS/MS. IMAC flow-through and washing fractions were collected and constituted the phosphopeptide-depleted fraction. The phosphopeptide-depleted fraction was further sub-fractioned by SCX chromatography. Thereafter, desalted peptides were resolved and separated on a MonoS PC1.6/5 column (GE Healthcare) using an Ettan micro LC system (GE Healthcare). SCX-separated peptide fractions were vacuum-dried, desalted, and analyzed separately by LC-MS/MS.
LC-MS/MS analysis and database search
LC-MS/MS analyses of phosphopeptide-enriched and -depleted fractions were performed on an Acquity Ultra Performance LC system (Waters Corp.) connected to an LTQ Orbitrap XL Fourier transform mass spectrometer (Thermo Scientific). Raw MS data were converted to data formats compatible with Mascot search engine (Matrix Science). Data from all peptide fractions were merged using Mascot Daemon (version 2.1.6). The UniProtKB/Swiss-Prot primary sequence database was used for protein identification (release 2011_03, with 525,997 entries; taxonomy Homo sapiens with 20,226 entries). In this study, proteins were only considered if they were identified at least with one unique peptide having an individual Mascot peptide score above 20, which indicated identity or extensive homology (p<0.05) using the following Mascot search parameters: enzyme, trypsin (specificity: K/R); maximum missed cleavages, 1; fixed modifications, iTRAQ 4-plex (K), iTRAQ (N terminus), Methylthio (C); variable modifications, phosphorylation (S, T, Y), oxidation (M); peptide tolerance, 10 ppm; MS/MS tolerance, 0.1 Da. Proteins identified by one peptide and Mascot-aided assignment of tyrosine (Y) phosphorylation (n = 21) were checked by manual inspection of corresponding MS/MS spectra, whereas annotation of threonine (T, n = 63) and serine (S, n = 209) phosphorylation was statistically validated by the AScore algorithms [68]. False discovery rates (FDRs) were calculated using the software Scaffold (version Scaffold_3_00_06). On average peptide FDRs less than 1.4% were determined. All MS-data associated with this manuscript are published in the PROteomics IDEntifications Database (PRIDE) (www.ebi.ac.uk/pride/).
Evaluation of quantitative LC-MS/MS data
Statistical evaluation of quantitative peptide data was conducted by iTRAQassist, a MS device-specific noise algorithm, as previously described [38]. Briefly, statistical evaluation was performed on the basis of spectral data derived from Mascot result files (.dat files) and restricted to peptides uniquely and unambiguously identified within the particular MS/MS data set (peptide tolerance: 10 ppm; MS/MS tolerance: 0.1 Da; iTRAQ reporter mass delta: 0.02 Da). ITRAQassist combines iTRAQ by-product correction and normalization of iTRAQ intensities with a noise-specific algorithm allowing determination of a weighted cumulative regulation factor for each unique peptide and prediction of possible deviations from the calculated regulation values dependent on iTRAQ reporter quality. ITRAQ reporter intensities were re-calculated according to certified and experimentally confirmed ratios of isotopic impurities of the different iTRAQ reporter molecules. The subsequent normalization was accomplished by comparing the trimmed mean of reporter intensities in all samples and discarding the upper and lower 20% of reporter intensities. Peptide regulations were depicted as likelihood curves for every peptide. Peptides containing phosphorylated amino acids were accepted as differentially regulated if their corresponding likelihood curves (phosphopeptide curve) were clearly separated from the cluster of non-phosphorylated peptides belonging to the same protein (protein curve). Regulated and discussed phosphopeptide sequence qualities and phosphorylation site annotations were examined manually.
Supporting Information
Purity of IL-2–cultured primary NK cells. (A) PBMC obtained from human peripheral blood by Ficoll gradient centrifugation and (B) IL-2–cultured NK cell were stained with fluorochrome-conjugated anti-CD3 and anti-CD56 mAbs and analyzed by flow cytometry. IL-2–cultured NK cell were 95 to 99% CD3−CD56+ as determined by flow cytometry. Data represent IL-2–cultured NK cell populations used in the phosphoproteome studies.
(PDF)
VI16743/Purvalanol-B-based affinity chromatography permits comprehensive enrichment of human protein kinases. (A) Kinases were significantly enriched by VI16743/Purvalanol-B-based affinity chromatography. Percentages of kinases and non-kinases (others: proteins without kinase activity) after VI16743/Purvalanol-B-mediated affinity purification are shown. (B) Kinases are identified based on a significantly higher number of MS spectra reflecting a better peptide identification and coverage of kinases achieved by MS. Assigned MS spectra per kinase and non-kinases (others) are depicted as mean values ± standard deviation (SD) of all conducted kinase-selective phosphoproteome experiments. Differences between two groups were examined using the Student's t-test (***, p<0.001).
(PDF)
Statistical validation of phosphorylation site regulation at protein kinases by iTRAQassist. Statistical evaluation of quantitative peptide data was conducted by iTRAQassist, a MS device specific noise algorithm, as previously described [38]. Phosphorylation sites were accepted as differentially regulated if their corresponding likelihood curves (phosphopeptide curve, green) were clearly separated from the cluster of non-phosphorylated peptides belonging to the same protein (protein curve, gray).
(PDF)
Number of kinase phosphorylation sites identified in respective experiments.
(PDF)
Distribution of iTRAQ-based protein and phosphorylation site regulation factors. This figure provides a general overview of all protein (black curve) and phosphorylation site (red curve) regulation factors calculated for two representative proteome experiments (I and II, see Table S1). Protein regulations are given as cumulative regulation values calculated on the basis of all non-phosphorylated peptides belonging to the same kinase. The majority of non- and phosphorylated peptides were expectedly not regulated. Protein regulation factors showed a normal distribution, whereas the phosphorylation site curve exhibits a non-normal distribution and was clearly shifted towards positive regulation values.
(PDF)
ITRAQ-MS-based quantification of N-terminal FYN phosphorylation induced by the engagement of CD16 or 2B4 and DNAM-1. Distinct phosphopeptide populations were resolved by nano-UPLC (Figure 5) and separately analyzed by mass spectrometry. The figure shows MS/MS fragmentation spectra of distinct FYN phosphopeptides originating from the same peptide sequence (DGSLNQSSGYR). FYN identification and phosphorylation site annotation are based on fragment ions of the b- and y-series (color code: red, b-ion; purple, b-ions minus 98; blue, y-ions; turquoise, y-ions minus 98; green, intensities of iTRAQ reporters or parent ions or b-ions minus H2O). Relative iTRAQ intensities of the corresponding phosphopeptides are shown at the right. FYN was differentially phosphorylated at S21, S25, S28 and Y28 following engagement of CD16 or co-engagement of 2B4 and DNAM-1.
(PDF)
Performed phosphokinome experiments. Footnote: In experiment I and II cIgG-, CD16- and 2B4 and DNAM-1 co-activated NK cells were analyzed. In experiment III cIgG- and 2B4 and DNAM-1 co-activated NK cells were analyzed. §Mascot-based annotation of serine and threonine phosphorylation was checked by the AScore algorithm. 47 phosphorylation sites could not be assigned unambiguously.
(PDF)
Kinases expressed in human NK cells. Footnote: *Kinase grouping according to Manning et al., 2002.
(PDF)
Kinase phosphorylation in human NK cells. Footnote: 1Serine/threonine annotations by Mascot were validated using AScore algorithm [68]; apredicted phosphorylation site (UniProt); *not listed in UniProt database; when phosphorylation sites could not be assigned unambiguously, alternative sequences are given. Note: Only for peptides containing more than one single serine/threonine residue AScore is required and annotation of tyrosine phosphorylation is not evaluated by AScore algorithm, but was inspected manually.
(PDF)
Acknowledgments
We thank Kirsten Minkhart for technical assistance, Dr. Frank Klawonn, Christoph Gernert for statistical evaluations and Thorsten Johl for support in bioinformatics, and Dr. Victor Wray for helpful discussions (all HZI).
Footnotes
Competing Interests: The authors Dr. König, Dr. Nimtz, Dr. Scheiter and Dr. Jänsch declare their affiliation to Helmholtz-Zentrum fuer Infektionsforschung GmbH. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: This work was supported by the International Research Training Group 1273 funded by the German Research Foundation (Dr. König, Dr. Scheiter, Dr. Jänsch), the Histocytosis Association of America (Dr. Bryceson), Mary Beve's Foundation (Dr. Bryceson), Clas Groschinsky's Memorial Fund (Dr. Bryceson), Shizu Matsumaras Donation (Dr. Bryceson), the Karolinska Institute Research Foundation (Dr. Bryceson and Dr. Ljunggren), the Swedish Research Council (Dr. Bryceson and Dr. Ljunggren), and the Swedish Cancer Society (Dr. Ljunggren). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 2.Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strowig T, Brilot F, Munz C. Noncytotoxic functions of NK cells: direct pathogen restriction and assistance to adaptive immunity. J Immunol. 2008;180:7785–7791. doi: 10.4049/jimmunol.180.12.7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryceson YT, March ME, Ljunggren HG, Long EO. Activation, coactivation, and costimulation of resting human natural killer cells. Immunol Rev. 2006;214:73–91. doi: 10.1111/j.1600-065X.2006.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vivier E, Nunes JA, Vely F. Natural killer cell signaling pathways. Science. 2004;306:1517–1519. doi: 10.1126/science.1103478. [DOI] [PubMed] [Google Scholar]
- 7.Zompi S, Colucci F. Anatomy of a murder–signal transduction pathways leading to activation of natural killer cells. Immunol Lett. 2005;97:31–39. doi: 10.1016/j.imlet.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Watzl C, Long EO. Signal transduction during activation and inhibition of natural killer cells. Curr Protoc Immunol. 2010;Chapter 11:Unit 11 19B. doi: 10.1002/0471142735.im1109bs90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 10.Wissing J, Jansch L, Nimtz M, Dieterich G, Hornberger R, et al. Proteomics analysis of protein kinases by target class-selective prefractionation and tandem mass spectrometry. Mol Cell Proteomics. 2007;6:537–547. doi: 10.1074/mcp.T600062-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Long EO. Negative signaling by inhibitory receptors: the NK cell paradigm. Immunol Rev. 2008;224:70–84. doi: 10.1111/j.1600-065X.2008.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson ME, Long EO. Inhibitory Receptor Signaling via Tyrosine Phosphorylation of the Adaptor Crk. Immunity. 2008 doi: 10.1016/j.immuni.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ting AT, Dick CJ, Schoon RA, Karnitz LM, Abraham RT, et al. Interaction between lck and syk family tyrosine kinases in Fc gamma receptor-initiated activation of natural killer cells. J Biol Chem. 1995;270:16415–16421. doi: 10.1074/jbc.270.27.16415. [DOI] [PubMed] [Google Scholar]
- 14.Bryceson YT, March ME, Barber DF, Ljunggren HG, Long EO. Cytolytic granule polarization and degranulation controlled by different receptors in resting NK cells. J Exp Med. 2005;202:1001–1012. doi: 10.1084/jem.20051143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bryceson YT, March ME, Ljunggren HG, Long EO. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood. 2006;107:159–166. doi: 10.1182/blood-2005-04-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryceson YT, Ljunggren HG, Long EO. Minimal requirement for induction of natural cytotoxicity and intersection of activation signals by inhibitory receptors. Blood. 2009;114:2657–2666. doi: 10.1182/blood-2009-01-201632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown MH, Boles K, van der Merwe PA, Kumar V, Mathew PA, et al. 2B4, the natural killer and T cell immunoglobulin superfamily surface protein, is a ligand for CD48. J Exp Med. 1998;188:2083–2090. doi: 10.1084/jem.188.11.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tangye SG, Cherwinski H, Lanier LL, Phillips JH. 2B4-mediated activation of human natural killer cells. Mol Immunol. 2000;37:493–501. doi: 10.1016/s0161-5890(00)00076-6. [DOI] [PubMed] [Google Scholar]
- 19.Eissmann P, Beauchamp L, Wooters J, Tilton JC, Long EO, et al. Molecular basis for positive and negative signaling by the natural killer cell receptor 2B4 (CD244). Blood. 2005;105:4722–4729. doi: 10.1182/blood-2004-09-3796. [DOI] [PubMed] [Google Scholar]
- 20.Veillette A. NK cell regulation by SLAM family receptors and SAP-related adapters. Immunol Rev. 2006;214:22–34. doi: 10.1111/j.1600-065X.2006.00453.x. [DOI] [PubMed] [Google Scholar]
- 21.Parolini S, Bottino C, Falco M, Augugliaro R, Giliani S, et al. X-linked lymphoproliferative disease. 2B4 molecules displaying inhibitory rather than activating function are responsible for the inability of natural killer cells to kill Epstein-Barr virus-infected cells. J Exp Med. 2000;192:337–346. doi: 10.1084/jem.192.3.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bottino C, Castriconi R, Pende D, Rivera P, Nanni M, et al. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med. 2003;198:557–567. doi: 10.1084/jem.20030788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibuya A, Lanier LL, Phillips JH. Protein kinase C is involved in the regulation of both signaling and adhesion mediated by DNAX accessory molecule-1 receptor. J Immunol. 1998;161:1671–1676. [PubMed] [Google Scholar]
- 24.Alvarez-Arias DA, Campbell KS. Protein kinase C regulates expression and function of inhibitory killer cell Ig-like receptors in NK cells. J Immunol. 2007;179:5281–5290. doi: 10.4049/jimmunol.179.8.5281. [DOI] [PubMed] [Google Scholar]
- 25.Shibuya K, Lanier LL, Phillips JH, Ochs HD, Shimizu K, et al. Physical and functional association of LFA-1 with DNAM-1 adhesion molecule. Immunity. 1999;11:615–623. doi: 10.1016/s1074-7613(00)80136-3. [DOI] [PubMed] [Google Scholar]
- 26.Kim HS, Das A, Gross CC, Bryceson YT, Long EO. Synergistic signals for natural cytotoxicity are required to overcome inhibition by c-Cbl ubiquitin ligase. Immunity. 2010;32:175–186. doi: 10.1016/j.immuni.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng F, Forbes AJ, Miller LM, Kelleher NL. Detection and localization of protein modifications by high resolution tandem mass spectrometry. Mass Spectrom Rev. 2005;24:126–134. doi: 10.1002/mas.20009. [DOI] [PubMed] [Google Scholar]
- 28.Kim JE, White FM. Quantitative analysis of phosphotyrosine signaling networks triggered by CD3 and CD28 costimulation in Jurkat cells. J Immunol. 2006;176:2833–2843. doi: 10.4049/jimmunol.176.5.2833. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen V, Cao L, Lin JT, Hung N, Ritz A, et al. A new approach for quantitative phosphoproteomic dissection of signaling pathways applied to T cell receptor activation. Mol Cell Proteomics. 2009;8:2418–2431. doi: 10.1074/mcp.M800307-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, et al. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal. 2009;2:ra46. doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- 31.Oberprieler NG, Lemeer S, Kalland ME, Torgersen KM, Heck AJ, et al. High-resolution mapping of prostaglandin E2-dependent signaling networks identifies a constitutively active PKA signaling node in CD8+CD45RO+ T cells. Blood. 2010;116:2253–2265. doi: 10.1182/blood-2010-01-266650. [DOI] [PubMed] [Google Scholar]
- 32.Reinl T, Nimtz M, Hundertmark C, Johl T, Keri G, et al. Quantitative phosphokinome analysis of the Met pathway activated by the invasin internalin B from Listeria monocytogenes. Mol Cell Proteomics. 2009;8:2778–2795. doi: 10.1074/mcp.M800521-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, et al. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 35.Knockaert M, Gray N, Damiens E, Chang YT, Grellier P, et al. Intracellular targets of cyclin-dependent kinase inhibitors: identification by affinity chromatography using immobilised inhibitors. Chem Biol. 2000;7:411–422. doi: 10.1016/s1074-5521(00)00124-1. [DOI] [PubMed] [Google Scholar]
- 36.Becker F, Murthi K, Smith C, Come J, Costa-Roldan N, et al. A three-hybrid approach to scanning the proteome for targets of small molecule kinase inhibitors. Chem Biol. 2004;11:211–223. doi: 10.1016/j.chembiol.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 37.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hundertmark C, Fischer R, Reinl T, May S, Klawonn F, et al. MS-specific noise model reveals the potential of iTRAQ in quantitative proteomics. Bioinformatics. 2009;25:1004–1011. doi: 10.1093/bioinformatics/btn551. [DOI] [PubMed] [Google Scholar]
- 39.Dulla K, Daub H, Hornberger R, Nigg EA, Korner R. Quantitative site-specific phosphorylation dynamics of human protein kinases during mitotic progression. Mol Cell Proteomics. 2010;9:1167–1181. doi: 10.1074/mcp.M900335-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, et al. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics. 2009;8:1751–1764. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinl T, Nimtz M, Hundertmark C, Johl T, Keri G, et al. Quantitative phosphokinome analysis of the Met pathway activated by the invasin InlB from listeria monocytogenes. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M800521-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma K, Kumar C, Keri G, Breitkopf SB, Oppermann FS, et al. Quantitative analysis of kinase-proximal signaling in lipopolysaccharide-induced innate immune response. J Proteome Res. 2010;9:2539–2549. doi: 10.1021/pr901192p. [DOI] [PubMed] [Google Scholar]
- 43.Gringel A, Walz D, Rosenberger G, Minden A, Kutsche K, et al. PAK4 and alphaPIX determine podosome size and number in macrophages through localized actin regulation. J Cell Physiol. 2006;209:568–579. doi: 10.1002/jcp.20777. [DOI] [PubMed] [Google Scholar]
- 44.Gu C, Tangye SG, Sun X, Luo Y, Lin Z, et al. The X-linked lymphoproliferative disease gene product SAP associates with PAK-interacting exchange factor and participates in T cell activation. Proc Natl Acad Sci U S A. 2006;103:14447–14452. doi: 10.1073/pnas.0606624103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bloch-Queyrat C, Fondaneche MC, Chen R, Yin L, Relouzat F, et al. Regulation of natural cytotoxicity by the adaptor SAP and the Src-related kinase Fyn. J Exp Med. 2005;202:181–192. doi: 10.1084/jem.20050449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen R, Latour S, Shi X, Veillette A. Association between SAP and FynT: Inducible SH3 domain-mediated interaction controlled by engagement of the SLAM receptor. Mol Cell Biol. 2006;26:5559–5568. doi: 10.1128/MCB.00357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shibuya K, Shirakawa J, Kameyama T, Honda S, Tahara-Hanaoka S, et al. CD226 (DNAM-1) is involved in lymphocyte function-associated antigen 1 costimulatory signal for naive T cell differentiation and proliferation. J Exp Med. 2003;198:1829–1839. doi: 10.1084/jem.20030958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bachelet I, Munitz A, Mankutad D, Levi-Schaffer F. Mast cell costimulation by CD226/CD112 (DNAM-1/Nectin-2): a novel interface in the allergic process. J Biol Chem. 2006;281:27190–27196. doi: 10.1074/jbc.M602359200. [DOI] [PubMed] [Google Scholar]
- 49.Yeo MG, Oh HJ, Cho HS, Chun JS, Marcantonio EE, et al. Phosphorylation of Ser 21 in Fyn regulates its kinase activity, focal adhesion targeting, and is required for cell migration. J Cell Physiol. 2011;226:236–247. doi: 10.1002/jcp.22335. [DOI] [PubMed] [Google Scholar]
- 50.Poggi A, Carosio R, Spaggiari GM, Fortis C, Tambussi G, et al. NK cell activation by dendritic cells is dependent on LFA-1-mediated induction of calcium-calmodulin kinase II: inhibition by HIV-1 Tat C-terminal domain. J Immunol. 2002;168:95–101. doi: 10.4049/jimmunol.168.1.95. [DOI] [PubMed] [Google Scholar]
- 51.Ishiguro K, Green T, Rapley J, Wachtel H, Giallourakis C, et al. Ca2+/calmodulin-dependent protein kinase II is a modulator of CARMA1-mediated NF-kappaB activation. Mol Cell Biol. 2006;26:5497–5508. doi: 10.1128/MCB.02469-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishiguro K, Ando T, Goto H, Xavier R. Bcl10 is phosphorylated on Ser138 by Ca2+/calmodulin-dependent protein kinase II. Mol Immunol. 2007;44:2095–2100. doi: 10.1016/j.molimm.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 53.Hama N, Paliogianni F, Fessler BJ, Boumpas DT. Calcium/calmodulin-dependent protein kinase II downregulates both calcineurin and protein kinase C-mediated pathways for cytokine gene transcription in human T cells. J Exp Med. 1995;181:1217–1222. doi: 10.1084/jem.181.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ricotta D, Conner SD, Schmid SL, von Figura K, Honing S. Phosphorylation of the AP2 mu subunit by AAK1 mediates high affinity binding to membrane protein sorting signals. J Cell Biol. 2002;156:791–795. doi: 10.1083/jcb.200111068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conner SD, Schmid SL. Identification of an adaptor-associated kinase, AAK1, as a regulator of clathrin-mediated endocytosis. J Cell Biol. 2002;156:921–929. doi: 10.1083/jcb.200108123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masilamani M, Peruzzi G, Borrego F, Coligan JE. Endocytosis and intracellular trafficking of human natural killer cell receptors. Traffic. 2009;10:1735–1744. doi: 10.1111/j.1600-0854.2009.00973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu D, Bryceson YT, Meckel T, Vasiliver-Shamis G, Dustin ML, et al. Integrin-dependent organization and bidirectional vesicular traffic at cytotoxic immune synapses. Immunity. 2009;31:99–109. doi: 10.1016/j.immuni.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fang F, Ahmad S, Lei J, Klecker RW, Trepel JB, et al. Effect of the mutation of tyrosine 713 in p93c-fes on its catalytic activity and ability to promote myeloid differentiation in K562 cells. Biochemistry. 1993;32:6995–7001. doi: 10.1021/bi00078a026. [DOI] [PubMed] [Google Scholar]
- 59.Rogers JA, Read RD, Li J, Peters KL, Smithgall TE. Autophosphorylation of the Fes tyrosine kinase. Evidence for an intermolecular mechanism involving two kinase domain tyrosine residues. J Biol Chem. 1996;271:17519–17525. doi: 10.1074/jbc.271.29.17519. [DOI] [PubMed] [Google Scholar]
- 60.Laurent CE, Delfino FJ, Cheng HY, Smithgall TE. The human c-Fes tyrosine kinase binds tubulin and microtubules through separate domains and promotes microtubule assembly. Mol Cell Biol. 2004;24:9351–9358. doi: 10.1128/MCB.24.21.9351-9358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takahashi S, Inatome R, Hotta A, Qin Q, Hackenmiller R, et al. Role for Fes/Fps tyrosine kinase in microtubule nucleation through is Fes/CIP4 homology domain. J Biol Chem. 2003;278:49129–49133. doi: 10.1074/jbc.C300289200. [DOI] [PubMed] [Google Scholar]
- 62.McPherson VA, Everingham S, Karisch R, Smith JA, Udell CM, et al. Contributions of F-BAR and SH2 domains of Fes protein tyrosine kinase for coupling to the FcepsilonRI pathway in mast cells. Mol Cell Biol. 2009;29:389–401. doi: 10.1128/MCB.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Udell CM, Samayawardhena LA, Kawakami Y, Kawakami T, Craig AW. Fer and Fps/Fes participate in a Lyn-dependent pathway from FcepsilonRI to platelet-endothelial cell adhesion molecule 1 to limit mast cell activation. J Biol Chem. 2006;281:20949–20957. doi: 10.1074/jbc.M604252200. [DOI] [PubMed] [Google Scholar]
- 64.Zirngibl R, Schulze D, Mirski SE, Cole SP, Greer PA. Subcellular localization analysis of the closely related Fps/Fes and Fer protein-tyrosine kinases suggests a distinct role for Fps/Fes in vesicular trafficking. Exp Cell Res. 2001;266:87–94. doi: 10.1006/excr.2001.5217. [DOI] [PubMed] [Google Scholar]
- 65.Marx A, Nugoor C, Panneerselvam S, Mandelkow E. Structure and function of polarity-inducing kinase family MARK/Par-1 within the branch of AMPK/Snf1-related kinases. FASEB J. 2010;24:1637–1648. doi: 10.1096/fj.09-148064. [DOI] [PubMed] [Google Scholar]
- 66.Lin J, Hou KK, Piwnica-Worms H, Shaw AS. The polarity protein Par1b/EMK/MARK2 regulates T cell receptor-induced microtubule-organizing center polarization. J Immunol. 2009;183:1215–1221. doi: 10.4049/jimmunol.0803887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 68.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 69.Gismondi A, Jacobelli J, Mainiero F, Paolini R, Piccoli M, et al. Cutting edge: functional role for proline-rich tyrosine kinase 2 in NK cell-mediated natural cytotoxicity. J Immunol. 2000;164:2272–2276. doi: 10.4049/jimmunol.164.5.2272. [DOI] [PubMed] [Google Scholar]
- 70.Lee SH, Yun S, Piao ZH, Jeong M, Kim DO, et al. Suppressor of cytokine signaling 2 regulates IL-15-primed human NK cell function via control of phosphorylated Pyk2. J Immunol. 2010;185:917–928. doi: 10.4049/jimmunol.1000784. [DOI] [PubMed] [Google Scholar]
- 71.Sasanuma H, Tatsuno A, Hidano S, Ohshima K, Matsuzaki Y, et al. Dual function for the adaptor MIST in IFN-gamma production by NK and CD4+NKT cells regulated by the Src kinase Fgr. Blood. 2006;107:3647–3655. doi: 10.1182/blood-2005-10-4102. [DOI] [PubMed] [Google Scholar]
- 72.Khurana D, Arneson LN, Schoon RA, Dick CJ, Leibson PJ. Differential regulation of human NK cell-mediated cytotoxicity by the tyrosine kinase Itk. J Immunol. 2007;178:3575–3582. doi: 10.4049/jimmunol.178.6.3575. [DOI] [PubMed] [Google Scholar]
- 73.Ito M, Tanabe F, Sato A, Takami Y, Shigeta S. A potent inhibitor of protein kinase C inhibits natural killer activity. Int J Immunopharmacol. 1988;10:211–216. doi: 10.1016/0192-0561(88)90051-3. [DOI] [PubMed] [Google Scholar]
- 74.Leibson PJ, Midthun DE, Windebank KP, Abraham RT. Transmembrane signaling during natural killer cell-mediated cytotoxicity. Regulation by protein kinase C activation. J Immunol. 1990;145:1498–1504. [PubMed] [Google Scholar]
- 75.Tassi I, Cella M, Presti R, Colucci A, Gilfillan S, et al. NK cell-activating receptors require PKC-theta for sustained signaling, transcriptional activation, and IFN-gamma secretion. Blood. 2008;112:4109–4116. doi: 10.1182/blood-2008-02-139527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chuang SS, Kumaresan PR, Mathew PA. 2B4 (CD244)-mediated activation of cytotoxicity and IFN-gamma release in human NK cells involves distinct pathways. J Immunol. 2001;167:6210–6216. doi: 10.4049/jimmunol.167.11.6210. [DOI] [PubMed] [Google Scholar]
- 77.Saborit-Villarroya I, Martinez-Barriocanal A, Oliver-Vila I, Engel P, Sayos J, et al. The adaptor 3BP2 activates CD244-mediated cytotoxicity in PKC- and SAP-dependent mechanisms. Mol Immunol. 2008;45:3446–3453. doi: 10.1016/j.molimm.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 78.Aguilo JI, Garaude J, Pardo J, Villalba M, Anel A. Protein kinase C-theta is required for NK cell activation and in vivo control of tumor progression. J Immunol. 2009;182:1972–1981. doi: 10.4049/jimmunol.0801820. [DOI] [PubMed] [Google Scholar]
- 79.Abraha AB, Rana K, Whalen MM. Role of protein kinase C in TBT-induced inhibition of lytic function and MAPK activation in human natural killer cells. Arch Environ Contam Toxicol. 2010;59:661–669. doi: 10.1007/s00244-010-9520-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pignata C, Prasad KV, Robertson MJ, Levine H, Rudd CE, et al. Fc gamma RIIIA-mediated signaling involves src-family lck in human natural killer cells. J Immunol. 1993;151:6794–6800. [PubMed] [Google Scholar]
- 81.Manciulea M, Rabinowich H, Sulica A, Lin WC, Whiteside TL, et al. Divergent phosphotyrosine signaling via Fc gamma RIIIA on human NK cells. Cell Immunol. 1996;167:63–71. doi: 10.1006/cimm.1996.0008. [DOI] [PubMed] [Google Scholar]
- 82.Azzoni L, Kamoun M, Salcedo TW, Kanakaraj P, Perussia B. Stimulation of Fc gamma RIIIA results in phospholipase C-gamma 1 tyrosine phosphorylation and p56lck activation. J Exp Med. 1992;176:1745–1750. doi: 10.1084/jem.176.6.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ting AT, Karnitz LM, Schoon RA, Abraham RT, Leibson PJ. Fc gamma receptor activation induces the tyrosine phosphorylation of both phospholipase C (PLC)-gamma 1 and PLC-gamma 2 in natural killer cells. J Exp Med. 1992;176:1751–1755. doi: 10.1084/jem.176.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Truesdell PF, Zirngibl RA, Francis S, Sangrar W, Greer PA. fps/fes knockout mice display a lactation defect and the fps/fes tyrosine kinase is a component of E-cadherin-based adherens junctions in breast epithelial cells during lactation. Exp Cell Res. 2009;315:2929–2940. doi: 10.1016/j.yexcr.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 85.Beal AM, Anikeeva N, Varma R, Cameron TO, Vasiliver-Shamis G, et al. Kinetics of early T cell receptor signaling regulate the pathway of lytic granule delivery to the secretory domain. Immunity. 2009;31:632–642. doi: 10.1016/j.immuni.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Flett A, Semerdjieva S, Jackson AP, Smythe E. Regulation of the clathrin-coated vesicle cycle by reversible phosphorylation. Biochem Soc Symp. 2005:65–70. doi: 10.1042/bss0720065. [DOI] [PubMed] [Google Scholar]
- 87.Ungewickell EJ, Hinrichsen L. Endocytosis: clathrin-mediated membrane budding. Curr Opin Cell Biol. 2007;19:417–425. doi: 10.1016/j.ceb.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 88.Eisenberg E, Greene LE. Multiple roles of auxilin and hsc70 in clathrin-mediated endocytosis. Traffic. 2007;8:640–646. doi: 10.1111/j.1600-0854.2007.00568.x. [DOI] [PubMed] [Google Scholar]
- 89.Stapleton D, Mitchelhill KI, Gao G, Widmer J, Michell BJ, et al. Mammalian AMP-activated protein kinase subfamily. J Biol Chem. 1996;271:611–614. doi: 10.1074/jbc.271.2.611. [DOI] [PubMed] [Google Scholar]
- 90.Stapleton D, Woollatt E, Mitchelhill KI, Nicholl JK, Fernandez CS, et al. AMP-activated protein kinase isoenzyme family: subunit structure and chromosomal location. FEBS Lett. 1997;409:452–456. doi: 10.1016/s0014-5793(97)00569-3. [DOI] [PubMed] [Google Scholar]
- 91.Liang J, Slingerland JM. Multiple roles of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell Cycle. 2003;2:339–345. [PubMed] [Google Scholar]
- 92.Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109(Suppl):S67–79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- 93.Irie A, Harada K, Tsukamoto H, Kim JR, Araki N, et al. Protein kinase D2 contributes to either IL-2 promoter regulation or induction of cell death upon TCR stimulation depending on its activity in Jurkat cells. Int Immunol. 2006;18:1737–1747. doi: 10.1093/intimm/dxl108. [DOI] [PubMed] [Google Scholar]
- 94.Li Q, Sun X, Wu J, Lin Z, Luo Y. PKD2 interacts with Lck and regulates NFAT activity in T cells. BMB Rep. 2009;42:35–40. doi: 10.5483/bmbrep.2009.42.1.035. [DOI] [PubMed] [Google Scholar]
- 95.Quarmby LM, Mahjoub MR. Caught Nek-ing: cilia and centrioles. J Cell Sci. 2005;118:5161–5169. doi: 10.1242/jcs.02681. [DOI] [PubMed] [Google Scholar]
- 96.O'Connell MJ, Krien MJ, Hunter T. Never say never. The NIMA-related protein kinases in mitotic control. Trends Cell Biol. 2003;13:221–228. doi: 10.1016/s0962-8924(03)00056-4. [DOI] [PubMed] [Google Scholar]
- 97.Hirose T, Tamaru T, Okumura N, Nagai K, Okada M. PCTAIRE 2, a Cdc2-related serine/threonine kinase, is predominantly expressed in terminally differentiated neurons. Eur J Biochem. 1997;249:481–488. doi: 10.1111/j.1432-1033.1997.t01-1-00481.x. [DOI] [PubMed] [Google Scholar]
- 98.Hirose T, Kawabuchi M, Tamaru T, Okumura N, Nagai K, et al. Identification of tudor repeat associator with PCTAIRE 2 (Trap). A novel protein that interacts with the N-terminal domain of PCTAIRE 2 in rat brain. Eur J Biochem. 2000;267:2113–2121. doi: 10.1046/j.1432-1327.2000.01218.x. [DOI] [PubMed] [Google Scholar]
- 99.Abe Y, Matsumoto S, Wei S, Nezu K, Miyoshi A, et al. Cloning and characterization of a p53-related protein kinase expressed in interleukin-2-activated cytotoxic T-cells, epithelial tumor cell lines, and the testes. J Biol Chem. 2001;276:44003–44011. doi: 10.1074/jbc.M105669200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Purity of IL-2–cultured primary NK cells. (A) PBMC obtained from human peripheral blood by Ficoll gradient centrifugation and (B) IL-2–cultured NK cell were stained with fluorochrome-conjugated anti-CD3 and anti-CD56 mAbs and analyzed by flow cytometry. IL-2–cultured NK cell were 95 to 99% CD3−CD56+ as determined by flow cytometry. Data represent IL-2–cultured NK cell populations used in the phosphoproteome studies.
(PDF)
VI16743/Purvalanol-B-based affinity chromatography permits comprehensive enrichment of human protein kinases. (A) Kinases were significantly enriched by VI16743/Purvalanol-B-based affinity chromatography. Percentages of kinases and non-kinases (others: proteins without kinase activity) after VI16743/Purvalanol-B-mediated affinity purification are shown. (B) Kinases are identified based on a significantly higher number of MS spectra reflecting a better peptide identification and coverage of kinases achieved by MS. Assigned MS spectra per kinase and non-kinases (others) are depicted as mean values ± standard deviation (SD) of all conducted kinase-selective phosphoproteome experiments. Differences between two groups were examined using the Student's t-test (***, p<0.001).
(PDF)
Statistical validation of phosphorylation site regulation at protein kinases by iTRAQassist. Statistical evaluation of quantitative peptide data was conducted by iTRAQassist, a MS device specific noise algorithm, as previously described [38]. Phosphorylation sites were accepted as differentially regulated if their corresponding likelihood curves (phosphopeptide curve, green) were clearly separated from the cluster of non-phosphorylated peptides belonging to the same protein (protein curve, gray).
(PDF)
Number of kinase phosphorylation sites identified in respective experiments.
(PDF)
Distribution of iTRAQ-based protein and phosphorylation site regulation factors. This figure provides a general overview of all protein (black curve) and phosphorylation site (red curve) regulation factors calculated for two representative proteome experiments (I and II, see Table S1). Protein regulations are given as cumulative regulation values calculated on the basis of all non-phosphorylated peptides belonging to the same kinase. The majority of non- and phosphorylated peptides were expectedly not regulated. Protein regulation factors showed a normal distribution, whereas the phosphorylation site curve exhibits a non-normal distribution and was clearly shifted towards positive regulation values.
(PDF)
ITRAQ-MS-based quantification of N-terminal FYN phosphorylation induced by the engagement of CD16 or 2B4 and DNAM-1. Distinct phosphopeptide populations were resolved by nano-UPLC (Figure 5) and separately analyzed by mass spectrometry. The figure shows MS/MS fragmentation spectra of distinct FYN phosphopeptides originating from the same peptide sequence (DGSLNQSSGYR). FYN identification and phosphorylation site annotation are based on fragment ions of the b- and y-series (color code: red, b-ion; purple, b-ions minus 98; blue, y-ions; turquoise, y-ions minus 98; green, intensities of iTRAQ reporters or parent ions or b-ions minus H2O). Relative iTRAQ intensities of the corresponding phosphopeptides are shown at the right. FYN was differentially phosphorylated at S21, S25, S28 and Y28 following engagement of CD16 or co-engagement of 2B4 and DNAM-1.
(PDF)
Performed phosphokinome experiments. Footnote: In experiment I and II cIgG-, CD16- and 2B4 and DNAM-1 co-activated NK cells were analyzed. In experiment III cIgG- and 2B4 and DNAM-1 co-activated NK cells were analyzed. §Mascot-based annotation of serine and threonine phosphorylation was checked by the AScore algorithm. 47 phosphorylation sites could not be assigned unambiguously.
(PDF)
Kinases expressed in human NK cells. Footnote: *Kinase grouping according to Manning et al., 2002.
(PDF)
Kinase phosphorylation in human NK cells. Footnote: 1Serine/threonine annotations by Mascot were validated using AScore algorithm [68]; apredicted phosphorylation site (UniProt); *not listed in UniProt database; when phosphorylation sites could not be assigned unambiguously, alternative sequences are given. Note: Only for peptides containing more than one single serine/threonine residue AScore is required and annotation of tyrosine phosphorylation is not evaluated by AScore algorithm, but was inspected manually.
(PDF)