Abstract
Background
Urochloa humidicola is a forage grass that grows in tropical regions and is recognized for its tolerance to seasonal flooding. It is a polyploid and apomictic species with high phenotypic plasticity. As molecular tools are important in facilitating the development of new cultivars and in the classification of related species, the objectives of this study were to develop new polymorphic microsatellite markers from an enriched library constructed from U. humidicola and to evaluate their transferability to other Urochloa species.
Findings
Microsatellite sequences were identified from a previously constructed enriched library, and specific primers were designed for 40 loci. Isolated di-nucleotide repeat motifs were the most abundant followed by tetra-nucleotide repeats. Of the tested loci, 38 displayed polymorphism when screened across 34 polyploid Urochloa sp. genotypes, including 20 accessions and six hybrids of U. humidicola and two accessions each from U. brizantha, U. dictyoneura, U. decumbens and U. ruziziensis. The number of bands per Simple Sequence Repeat (SSR) locus ranged from one to 29 with a mean of 11.5 bands per locus. The mean Polymorphism Information Content (PIC) of all loci was 0.7136, and the mean Discrimination Power (DP) was 0.7873. Six loci amplified in all species tested. STRUCTURE analysis revealed six different allelic pools, and the genetic similarity values analyzed using Jaccard's coefficient ranged from 0.000 to 0.913.
Conclusions
This work reports new polymorphic microsatellite markers that will be useful for breeding programs for Urochloa humidicola and other Urochloa species as well as for genetic map development, germplasm characterization, evolutionary and taxonomic studies and marker-assisted trait selection.
Background
Urochloa humidicola (Rendle) Morrone & Zuloaga (syn. Brachiaria humidicola (Rendle) Schweick..) [1] is an out-crossed and wind-pollinated perennial tropical grass that is widely used for pasture in several tropical regions, including Central and South America, Southeast Asia and Oceania. Also known as koroniviagrass, it is particularly recognized for its tolerance to poorly draining soils, seasonal flooding and infertile acid soils [2], characteristics that led to the successful use of this species in the Amazon region.
U. humidicola is a polyploid species that has ploidy levels ranging from tetraploid to heptaploid. The basic chromosome number has been recently reported as x = 6 [3-6]. This species reproduces through a Panicum-type of apospory [7], which is an asexual mode of reproduction through seeds where somatic cells of the nucellus form unreduced embryo sacs [8].
The difficulty in classifying Urochloa grasses is related to subtle differences between species, which are distinguished by slight differences in the small morphological features of the flowers [1,9] and phenotypic plasticity. These subtle differences make the identification of species and intra- and interspecific hybrids problematic and uncertain. As little is known about the genetic makeup of U. humidicola, molecular markers represent important tools for elucidating the classification and genetics of the species as well as for applications in breeding programs. More particularly, microsatellite markers are widely used in genetic studies, and due to their high mutation rates [10], they can be especially helpful when comparing closely related individuals [11].
The identification of microsatellite markers depends on knowledge of the flanking region sequences to design appropriate primer pairs. These sequences are usually obtained from enriched libraries [12] or from public sequences. The flanking regions have lower mutation rates than the microsatellites themselves [11] and are often identical among phylogenetically related species, allowing the use of the same markers in these species [13-15].
In a previous study, 384 clones were sequenced and analyzed from a microsatellite-enriched library constructed for Urochloa humidicola, and 27 polymorphic microsatellites loci were described [16]. The population structure of the germplasm collection of U. humidicola was then analyzed using these loci along with morphological markers [17]. To continue the genetic studies of this species, the present study aimed to develop new microsatellite markers for U. humidicola, test their transferability to other Urochloa species and validate the newly developed SSRs by evaluating the genetic diversity and population structure among 34 Urochloa genotypes (26 of U. humidicola and two each of the following species: U. brizantha, U. decumbens, U. ruziziensis and U. dictyoneura). The results were compared with previously reported data [17].
Results
Forty primer pairs were designed and amplified successfully in U. humidicola, and 38 SSRs were polymorphic (Table 1). Polymorphism Information Content (PIC) values for each locus were obtained for the 26 U. humidicola genotypes as previously described [18]. Discrimination Power (DP) was also determined for each locus [19]. The mean PIC of all loci was 0.7136, and the mean DP was 0.7873. Between one and 29 bands were observed per locus with a mean of 11.5 bands per locus.
Table 1.
Description of SSR markers developed for Urochloa humidicola (Rendle) Morrone & Zuloaga
| Characteristics in five species* | Characteristics in U. humidicola | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SSR Locus | Genbank accession number | Repeat Motif | Ta (°C) | Predicted product size (pb) | Primer Sequences (5'- 3') | N° of bands | Size range (pb) | PIC | N° of bands | PIC | DP |
| BhUNICAMP028 | JF812604 | (GT)4GG(GT)3 | 57 | 228 | TCTTTTGGCTCTGAATGTGCT TTGATGCCGAATGGAACC |
6 | 195-212 | 0.7364 | 5 | 0.6845 | 0.9324 |
| BhUNICAMP029 | JF812605 | (TC)3(TG)8 | 55 | 176 | AAGGGATATTTGTGTTTCT TTTTTCAGGATTGCTAAG |
7 | 141-180 | 0.7363 | 4 | 0.6492 | 0.8708 |
| BhUNICAMP030 | JF812606 | (AT)3TGC(AT)4 | 60 | 133 | GGAATATTGTTGCTGAGAGTGG GCGACGACAGAATAAAAATGAT |
3 | 135-181 | 0.2211 | 2 | 0.1638 | 0.2123 |
| BhUNICAMP031 | JF812607 | (GT)7 | 60 | 126 | AGGATTTAAAGGACCCACCAT TCCGCTCGGACTGTGATT |
3 | 126-131 | 0.3749 | 3 | 0.3749 | 0.6554 |
| BhUNICAMP032 | JF812608 | (GT)7 | 60 | 212 | GCATATGCAGAGTTTTTGTT TGACCATTTTTCTTATCTTTCT |
10 | 196-222 | 0.8305 | 10 | 0.8305 | 0.9292 |
| BhUNICAMP033 | JF812609 | (TG)5GG(TG)3 | 60 | 255 | TGAGGTCTTCCGTTCTTGTAGGT ACGAGGCTGCCCGAGTAATC |
5 | 212-285 | 0.6965 | 4 | 0.6356 | 0.8246 |
| BhUNICAMP034 | JF812610 | (GA)20 | 51 | 204 | TGTAGTGTTGCTGTAGAGTTA CATTGTTTTGAAGATTTG |
3 | 176-182 | 0.5894 | 3 | 0.5916 | 0.2800 |
| BhUNICAMP035 | JF812611 | (AC)4(CA)5CG(CA)3...(CA)5 | 51 | 248 | GATGCATCTCCCTCCCTTACTC AGACATTCATCCCGTTTCACAA |
15 | 160-263 | 0.8767 | 13 | 0.8666 | 0.9784 |
| BhUNICAMP036 | JF812612 | (TTG)9(TG)6TT(TG)5CG(TG)5 | 60 | 265 | CGATAGTTAGGCGAGAGTTTTG TATTGTCGTATGGCAGAGTTCA |
9 | 204-282 | 0.8127 | 8 | 0.8052 | 0.8800 |
| BhUNICAMP037 | JF812613 | (TG)8 | 55 | 277 | CCGTGGAATCCGACAGGTAG CCGGGAGGAGAGTTAGAAGATG |
21 | 118-302 | 0.9270 | 19 | 0.9238 | 0.9846 |
| BhUNICAMP038 | JF812614 | (AG)13 | 60 | 294 | TCTCTTAAGCGACCAGTA CAGGAGATAAGTAAAATGAAT |
14 | 286-321 | 0.8955 | 14 | 0.8975 | 0.9785 |
| BhUNICAMP039 | JF812615 | (TC)9...(TC)10 | 55 | 231 | CATACTTGCATTCTTTTGAT TGTATGAATTTATTGTTTGACT |
22 | 183-263 | 0.9252 | 21 | 0.9254 | 0.9938 |
| BhUNICAMP040 | JF812616 | (CTTG)3 | 60 | 257 | TGTAAGCATATCATTTCGTCTA ACTGCCCTGTTTTCCTG |
2 | 258-261 | 0.2772 | 2 | 0.2784 | 0.4092 |
| BhUNICAMP041 | JF812617 | (TC)3(AAAAT)3 | 62 | 178 | GCTAGGATGGTGGGCTGTGCT CGACGTTTCCGGAATGATGTTT |
6 | 173-270 | 0.4797 | 3 | 0.3966 | 0.5662 |
| BhUNICAMP042 | JF812618 | (TG)7 | 60 | 223 | CCGCTGAGCTGTATAGGAAGTT AAGAGGCGGGACATTTAGGA |
6 | 210-226 | 0.6465 | 6 | 0.6465 | 0.8400 |
| BhUNICAMP043 | JF812619 | (GT)3(TG)5GG(TG)4 | 60 | 216 | TGTGTTTGTGTTCTCTATGTGT TATGTGATCCAAAAGTGAAGTG |
5 | 212-224 | 0.697 | 5 | 0.7367 | 0.9046 |
| BhUNICAMP044 | JF812620 | (CA)11 | 60 | 132 | TAACACAAGTGCAGGGCGTAAT TGAACACAGCGACACAAGACAC |
17 | 96-130 | 0.8913 | 17 | 0.8918 | 0.9815 |
| BhUNICAMP045 | JF812621 | (AC)11 | 60 | 245 | ACACCACACCAAATTCTTACCC TCTCGTTTCATGGCACTGTCTA |
14 | 225-300 | 0.8956 | 14 | 0.8994 | 0.9600 |
| BhUNICAMP046 | JF812622 | (TG)10 | 60 | 262 | ACGCCGCAGCAGTTGGTG TCAGGACGTGCCGATGGTAAT |
22 | 230-284 | 0.9268 | 20 | 0.9192 | 0.9877 |
| BhUNICAMP047 | JF812623 | (TC)20 | 57 | 284 | TACATGCAGCAACTAAGATA GCACTAACAAGAAAAGATT |
26 | 211-330 | 0.9231 | 23 | 0.9307 | 0.9969 |
| BhUNICAMP048 | JF812624 | (AG)20 | 57 | 286 | GCACTAAACAAGAAAAGATT TACATGCAGCAACTAAGATA |
29 | 212-350 | 0.8864 | 23 | 0.9200 | 0.9754 |
| BhUNICAMP049 | JF812625 | (AG)3A(AG)4 | 60 | 285 | GGGCCCGGCACAACAGTAG AGGCCCACACGCAGAGAACA |
8 | 184-287 | 0.5597 | 2 | 0.3698 | 0.4277 |
| BhUNICAMP050 | JF812626 | (TGCG)3 | 60 | 236 | GTGTGGTTGCAGGACGGATTG TGAGTGCATGACAGGTGACGAA |
7 | 225-263 | 0.7144 | 6 | 0.7037 | 0.9692 |
| BhUNICAMP051 | JF812627 | (AC)17GC(AC)6AT(AG)7 | 60 | 294 | TAGCAATGCATGGATAAGACT TGGAGCTCACCCTAAGAAG |
1 | 297 | 0 | 1 | 0 | 0 |
| BhUNICAMP052 | JF812628 | (TG)5 | 60 | 268 | ATAACACGGCCAGAACTA ATGAACAATCGGGGTAT |
12 | 230-280 | 0.8663 | 12 | 0.8622 | 0.9446 |
| BhUNICAMP053 | JF812629 | (CA)12CG(CA)9 | 60 | 291 | GAGTAAGCTTCTAGGACAGGAT GCTCAAACAACTCGATAATAAC |
18 | 224-320 | 0.8887 | 10 | 0.8603 | 0.7692 |
| BhUNICAMP054 | JF812630 | (TG)9 | 60 | 230 | CCATATGTGAAGGCTGCGTGAA GTGGCGGGCTAGTGGCTTATC |
15 | 190-290 | 0.8262 | 14 | 0.8281 | 0.9385 |
| BhUNICAMP055 | JF812631 | (TC)7 | 60 | 261 | GGAAAAAGAAAAGCGGACTGAA ACGCAAAATAAATGGCAATGGA |
12 | 240-310 | 0.8829 | 10 | 0.8578 | 0.9538 |
| BhUNICAMP056 | JF812632 | (TGTT)4 | 60 | 239 | GCCACAACACGCAAATC ATGTATGAGGACCCAAGTTATG |
4 | 234-249 | 0.269 | 4 | 0.2841 | 0.2861 |
| BhUNICAMP057 | JF812633 | (AG)22 | 60 | 219 | AGCGACCTCCAGCAACCTT TTTCCCACTCTTTCCCTCTCAC |
21 | 169-239 | 0.9091 | 21 | 0.9091 | 0.9846 |
| BhUNICAMP058 | JF812634 | (TC)18 | 55 | 279 | CTAAACAGGTAAACAGACAAT CAAAACGTGAATACATAACA |
9 | 250-340 | 0.7913 | 9 | 0.7913 | 0.9015 |
| BhUNICAMP059 | JF812635 | (ATGT)3 | 55 | 290 | CAATCCATTTTAACAAGAAGTC GCAACAGTCCGTAGTAAGTATC |
5 | 288-305 | 0.5747 | 5 | 0.5783 | 0.5538 |
| BhUNICAMP060 | JF812636 | (TTTTG)3 | 55 | 279 | AATCCAAAGTCATCCCCACAAT TTTTTCGGCAACAGACAGGTAA |
8 | 270-290 | 0.7747 | 7 | 0.7721 | 0.9415 |
| BhUNICAMP061 | JF812637 | (GT)14 | 60 | 165 | TGATTCAAAACGCCACGATAGG GGACCGGAACACTGCTTACGA |
22 | 147-192 | 0.9139 | 19 | 0.9067 | 0.9846 |
| BhUNICAMP062 | JF812638 | (CA)8 | 60 | 155 | CAAACCTCGTGCTCGTG AGATGGGTTCGGCTGTC |
18 | 139-193 | 0.8919 | 15 | 0.8779 | 0.9631 |
| BhUNICAMP063 | JF812639 | (GA)8G(GA)14 | 60 | 199 | CAAGAAAGCGCGATGAAAAA GAACACAATGGAGAAGCAG GTC |
14 | 173-230 | 0.8967 | 14 | 0.8967 | 0.9508 |
| BhUNICAMP064 | JF812640 | (TC)19 | 60 | 175 | CCCCTACTTTTATACGACACAT GAACGAGAGTAGTAGCATTGGT |
13 | 145-180 | 0.8941 | 13 | 0.8941 | 0.9538 |
| BhUNICAMP065 | JF812641 | (AATA)3 | 55 | 198 | ATGTCACGTTATCAGCAGAAG GGGCCACATCACCTTTT |
1 | 200 | 0 | 1 | 0 | 0 |
| BhUNICAMP066 | JF812641 | (TCTT)3 | 55 | 218 | ATGACAAACTGACCGTATC TAGCAATTTTCTTTATCAACT |
10 | 217-231 | 0.7489 | 9 | 0.7598 | 0.8923 |
| BhUNICAMP067 | JF812642 | (CT)17...(CT)5 | 60 | 301 | ACCCCCTGTAATTGTTGTCC GATTCAGATGGTTAGCGTGTTA |
15 | 245-335 | 0.8961 | 14 | 0.8941 | 0.9354 |
* Species evaluated: Urochloa humidicola (Rendle) Morrone & Zuloaga, Urochloa brizantha (Hochst. ex A. Rich.) R.D. Webster, Urochloa decumbens (Stapf) R.D. Webster, Urochloa dictyoneura (Figure & De Not.) Veldkamp, Urochloa ruziziensis (R. Germ. & C.M. Evrard) Crins
Transferability of the developed SSR primer pairs was tested in two genotypes each of U. brizantha, U. decumbens, U. ruziziensis and U. dictyoneura for all the loci under the same PCR conditions used for U. humidicola. The number of successfully amplified genotypes per number of genotypes tested per species is shown in Table 2. The following loci did not amplify in any of the tested genotypes of Urochloa spp: BhUNICAMP031, BhUNICAMP032, BhUNICAMP042, BhUNICAMP051, BhUNICAMP052, BhUNICAMP057, BhUNICAMP058, BhUNICAMP063 and BhUNICAMP064. Twenty-one loci were amplified in at least one U. brizantha genotype, 24 were amplified in U. decumbens, six were amplified in U. ruziziensis, and 25 were amplified in U. dictyoneura.
Table 2.
Transferability of SSR markers developed for Urochloa humidicola in other Urochloa species
| Transferabilitya,b | ||||
|---|---|---|---|---|
| SSR Locus | U. brizantha | U. decumbens | U. ruziziensis | U. dictyoneura |
| BhUNICAMP028 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP029 | 1/2 | 1/2 | 0/2 | 2/2 |
| BhUNICAMP030 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP031 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP032 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP033 | 0/2 | 2/2 | 0/2 | 0/2 |
| BhUNICAMP034 | 2/2 | 2/2 | 0/2 | 2/2 |
| BhUNICAMP035 | 2/2 | 2/2 | 1/2 | 2/2 |
| BhUNICAMP036 | 0/2 | 1/2 | 0/2 | 0/2 |
| BhUNICAMP037 | 2/2 | 2/2 | 0/2 | 2/2 |
| BhUNICAMP038 | 0/2 | 1/2 | 0/2 | 0/2 |
| BhUNICAMP039 | 2/2 | 2/2 | 0/2 | 1/2 |
| BhUNICAMP040 | 2/2 | 2/2 | 0/2 | 0/2 |
| BhUNICAMP041 | 2/2 | 1/2 | 0/2 | 2/2 |
| BhUNICAMP042 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP043 | 0/2 | 2/2 | 0/2 | 0/2 |
| BhUNICAMP044 | 2/2 | 2/2 | 0/2 | 2/2 |
| BhUNICAMP045 | 2/2 | 1/2 | 0/2 | 0/2 |
| BhUNICAMP046 | 2/2 | 2/2 | 0/2 | 2/2 |
| BhUNICAMP047 | 2/2 | 1/2 | 0/2 | 2/2 |
| BhUNICAMP048 | 2/2 | 1/2 | 0/2 | 2/2 |
| BhUNICAMP049 | 2/2 | 1/2 | 0/2 | 1/2 |
| BhUNICAMP050 | 0/2 | 0/2 | 0/2 | 1/2 |
| BhUNICAMP051 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP052 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP053 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP054 | 0/2 | 0/2 | 0/2 | 2/2 |
| BhUNICAMP055 | 2/2 | 2/2 | 2/2 | 2/2 |
| BhUNICAMP056 | 0/2 | 0/2 | 0/2 | 2/2 |
| BhUNICAMP057 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP058 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP059 | 0/2 | 0/2 | 0/2 | 1/2 |
| BhUNICAMP060 | 2/2 | 2/2 | 0/2 | 2/2 |
| BhUNICAMP061 | 2/2 | 0/2 | 0/2 | 2/2 |
| BhUNICAMP062 | 2/2 | 2/2 | 2/2 | 1/2 |
| BhUNICAMP063 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP064 | 0/2 | 0/2 | 0/2 | 0/2 |
| BhUNICAMP065 | 0/2 | 0/2 | 0/2 | 1/2 |
| BhUNICAMP066 | 2/2 | 1/2 | 0/2 | 2/2 |
| BhUNICAMP067 | 0/2 | 0/2 | 0/2 | 1/2 |
| Total | 21 | 24 | 6 | 25 |
a Number of successfully amplified genotypes/Number of tested genotypes
b Nomenclatural classification: Urochloa humidicola (Rendle) Morrone & Zuloaga, Urochloa brizantha (Hochst. ex A. Rich.) R.D. Webster, Urochloa decumbens (Stapf) R.D. Webster, Urochloa dictyoneura (Figure & De Not.) Veldkamp, Urochloa ruziziensis (R. Germ. & C.M. Evrard) Crins
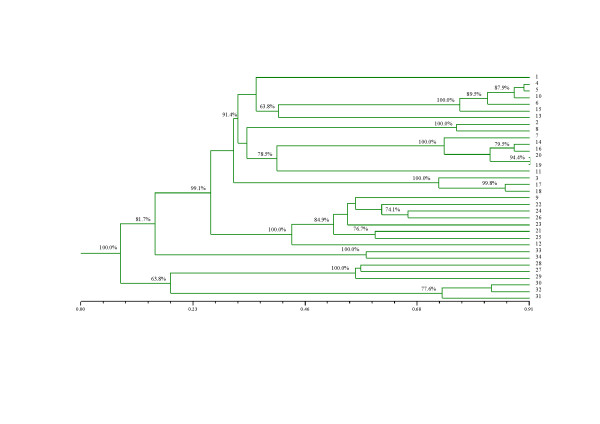
The genetic similarity values analyzed using Jaccard's coefficient ranged from 0.000 (H125 and H126) to 0.913. See Additional File 1: Genetic similarity based on Jaccard's coefficient. A dendrogram was constructed using the Unweighted Pair-Group Method with the Arithmetic Mean (UPGMA) that successfully discriminated all tested accessions (Figure 1).
Figure 1.
UPGMA tree representing the relationship among 34 genotypes of the Urochloa species using Jaccard's similarity coefficient. Bootstrap values (p < 0.0001) above 60% are indicated, and genotypes are named according to the annotated numbers listed in Table 3.
The population structure inferred by a model-based Bayesian approach using the STRUCTURE software revealed K = 6 clusters. Each cluster was characterized by a set of allele frequencies at each locus and was represented by different colors (red, green, blue, yellow, magenta and light blue) as indicated in Figure 2a. If genotypes indicate admixture, they can be assigned to two or more clusters [20]. We used the term "Cluster" to refer to one or more individuals characterized by a distinguishable allelic set. The best K number of clusters was determined using the ΔK method [21], and its graphical representation is shown in Figure 2b.
Figure 2.
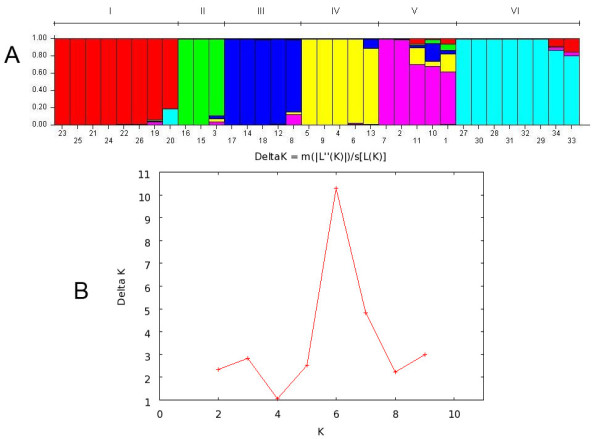
a Analysis performed with STRUCTURE 2.3 software using an admixture model with correlated allele frequencies. The clustering profile obtained for K = 6 (b) is displayed and is indicated by different colors. Each of the 34 genotypes is represented by a single column broken into colored segments with lengths proportional to each of the K inferred gene pools. The left-hand scale indicates the membership coefficients (Q) for allocating genotypes into clusters. Genotypes are named according to the annotated numbers listed in Table 3; four major Clusters of individuals were identified, and Clusters are indicated by numbers I-VI. b ΔK values for each K value, calculated according to Evanno et al. [21]. The highest ΔK value corresponds to the optimal K.
In the STRUCTURE analysis, Cluster I (CI-red) was composed of eight genotypes, Cluster II (CII-green) was composed of three genotypes, Cluster III (CIII-blue) was composed of five genotypes, Cluster IV (CIV-yellow) was composed of five genotypes, Cluster V (CV-magenta) was composed of five genotypes and Cluster VI (CVI-light blue) was composed of the last eight genotypes of the studied species. The estimated membership coefficients (Q) of each individual for each allelic pool are shown in Additional File 2: Inferred ancestry of individuals.
Discussion
A robust set of informative molecular markers for the species of interest is a prerequisite for marker-assisted breeding. Urochloa humidicola (or koroniviagrass) is an important tropical forage grass with limited genomic resources. As microsatellite markers are highly polymorphic, reproducible and distributed throughout the genome, they are the ideal marker system for genetic analysis and breeding applications [22]. However, only 27 SSR markers have been reported for koroniviagrass [16]. The present study reports a novel set of SSRs that adds to the existing repertoire of molecular markers in this species and validates the SSRs in some related species.
The majority of the SSRs isolated in the present study were comprised of di-nucleotide repeats (80%) followed by tetra-nucleotide (15%) and penta-nucleotide (5%) repeats. This distribution can be attributed to enrichment of the library for the two di-nucleotide repeats, (AC)n and (AG)n.
Of all the microsatellites analyzed, 38 out of 40 (95%) showed polymorphism. The most informative loci in this panel of SSRs were the ones with the highest PIC and DP values (BhUNICAMP037, BhUNICAMP039, BhUNICAMP046 and BhUNICAMP047). The BhUNICAMP051 and BhUNICAMP065 loci showed no polymorphism among the studied genotypes, but they may be useful in other studies. The BhUNICAMP030 locus resulted in low PIC and DP values (0.2211 and 0.2123, respectively) as expected because of its low observed polymorphism and its amplification in all other species, which may be a result of a conserved region among the Urochloa species studied here.
Of the 40 investigated loci, 18 cross-amplified in at least three other Urochloa species, and six cross-amplified in all the evaluated species. The highest success of transferability was obtained in U. dictyoneura, where 25 SSR primer pairs were cross-amplified in at least one genotype. These results were expected because the U. dictyoneura species is more closely related to U. humidicola than to the other three species [9,23]. U. ruziziensis was a scoreless species, as only six SSR primer pairs could be cross-amplified. These results are consistent with a previous study with different microsatellite loci [16].
Genetic distance and population structure analysis based on SSR allelic data showed differentiation among U. humidicola accessions, hybrids and other Urochloa species. Although the number of genotypes is limited, the analyses corroborate a previous study with 60 U. humidicola genotypes [17]. The STRUCTURE analysis showed that the genotypes distributed into six major groups, and such groupings were similarly observed by [17]. When examining the dendrogram based on Jaccard's similarity coefficient, the distribution of genotypes was similar to the STRUCTURE analysis, although the two analyses used different statistical approaches.
Indeed, as observed in the amplification profiles, the dendrogram and the allelic pools indicated a closer relationship between U. humidicola and U. dictyoneura than with the other species.
In the STRUCTURE analysis (Figure 2a), Clusters I-V contained only U. humidicola genotypes, and accessions from Clusters II, III and IV were grouped in the same way as has been previously reported [17]. The allelic pools were identified by different colors, and although a genotype might belong to a particular allelic pool, it can also represent a percentage of other allelic pools, as observed in genotypes H016, H31, H006, H013, H012, H044, H035, H030, H004, DT159 and DT157.
Cluster I in the STRUCTURE analysis separated the H016 and H031 accessions (Figure 2a), which were found along with their six hybrids in the same cluster in a previous study [17]. The H031 and H016 accessions were the parents of the first and single mapping population of the species and were the originators of the hybrids used in this study. It is also important to note that these genotypes presented a high degree of divergence, corroborating previous results [17]. Mapping studies are currently underway with the SSR loci reported in this and a previous study [16].
When examining Cluster VI (Figure 2a), which was formed by the other Urochloa species, a different allelic pool was found (light blue), and the U. dictyoneura accessions (DT159 and DT157) showed some percentage of the red and magenta allelic pools, corroborating what was observed in the dendrogram. When analyzing the grouping pattern of the other Urochloa species, it is important to note that only two genotypes of each species were used in contrast to the 26 U. humidicola genotypes; this could be biasing the analysis.
As previously observed [17], the H031 accession, which is the sole sexual accession in the germplasm collection, presented a different allelic pool composition. However, when compared to other species, the present study revealed that this accession might have the same origin as the other species based on the high proportion of the blue allelic pool in the genotype.
Conclusions
The data reported herein indicate that the newly developed SSR markers from U. humidicola represent a powerful set of genetic resources for genetic diversity studies and are potentially useful for further studies, including molecular mapping, species and hybrid identification, gene flow and seed purity, in U. humidicola and other Urochloa species.
Methods
Thirty-four Urochloa genotypes were used in this study. Twenty are U. humidicola accessions maintained in the germplasm collection of Embrapa Beef Cattle, six are hybrids from the same species and the other eight are represented by two different accessions from each of the following species: U. brizantha, U. decumbens, U. ruziziensis and U. dictyoneura. The annotation numbers, accession numbers (as recorded in Embrapa Beef Cattle (EBC) and Center for Tropical Agriculture (CIAT), genotypes and species identifications are shown in Table 3. Freeze-dried leaf samples were used for DNA extraction following the cetyl trimethyl ammonium bromide (CTAB) method previously described [24].
Table 3.
List of all Urochloa genotypes used in this study
| AN | CIAT | BRA | EBC | Genotype | Species |
|---|---|---|---|---|---|
| 1 | 16181 | 4821 | H004 | germplasm accession |
U. humidicola |
| 2 | 16182 | 4839 | H005 | germplasm accession |
U. humidicola |
| 3 | 16867 | 4863 | H006 | germplasm accession |
U. humidicola |
| 4 | 16871 | 4901 | H008 | germplasm accession |
U. humidicola |
| 5 | 16880 | 4952 | H010 | germplasm accession |
U. humidicola |
| 6 | 16882 | 4979 | H012 | germplasm accession |
U. humidicola |
| 7 | 16886 | 5011 | H013 | germplasm accession |
U. humidicola |
| 8 | 26141 | 5088 | H015 | germplasm accession |
U. humidicola |
| 9 | 26149 | 5118 | H016 | germplasm accession |
U. humidicola |
| 10 | 16877 | 4928 | H023 | germplasm accession |
U. humidicola |
| 11 | 16894 | 5070 | H030 | germplasm accession |
U. humidicola |
| 12 | 26146 | 5100 | H031 | germplasm accession |
U. humidicola |
| 13 | 26413 | 6131 | H035 | germplasm accession |
U. humidicola |
| 14 | 26432 | 6203 | H041 | germplasm accession |
U. humidicola |
| 15 | 16884 | 4995 | H044 | germplasm accession |
U. humidicola |
| 16 | NA | NA | H048 | germplasm accession |
U. humidicola |
| 17 | NA | 1929 | H107 | germplasm accession |
U. humidicola |
| 18 | 6705 | 2208 | H112 | germplasm accession |
U. humidicola |
| 19 | 6133 | 1449 | H125 | germplasm accession |
U. humidicola |
| 20 | 6369 | 0370 | H126 | germplasm accession |
U. humidicola |
| 21 | - | - | 20 | hybrid | U. humidicola |
| 22 | - | - | 45 | hybrid | U. humidicola |
| 23 | - | - | 184 | hybrid | U. humidicola |
| 24 | - | - | 215 | hybrid | U. humidicola |
| 25 | - | - | 264 | hybrid | U. humidicola |
| 26 | - | - | 320 | hybrid | U. humidicola |
| 27 | 16162 | B057 | germplasm accession |
U. brizantha | |
| 28 | 16467 | B166 | germplasm accession |
U. brizantha | |
| 29 | 16499 | 004481 | D009 | germplasm accession |
U. decumbens |
| 30 | 26300 | 004707 | D028 | germplasm accession |
U. decumbens |
| 31 | 26163 | 005614 | R102 | germplasm accession |
U. ruziziensis |
| 32 | 26174 | 005614 | R104 | germplasm accession |
U. ruziziensis |
| 33 | 16186 | 007889 | DT157 | germplasm accession |
U. dictyoneura |
| 34 | 16188 | 007901 | DT159 | germplasm accession |
U. dictyoneura |
NA: not available, AN: annotation numbers, CIAT: Center for Tropical Agriculture, BRA and EBC (Embrapa Beef Cattle): codes from each of the Institutions
In a previous study, a microsatellite-enriched library was constructed for Urochloa humidicola, and 384 clones were sequenced. The sequences were then treated as described [16], and the microsatellites were identified using the Simple Sequence Repeat Identification Tool (SSRIT) [25]. Only di-nucleotides with five or more repeats, tri-nucleotides with four or more repeats, and tetra-, penta- and hexanucleotides with three or more repeats were considered. Primer pairs were designed using the Primer Select 5.01 (DNASTAR Inc.) and Primer3Plus software [26].
Polymerase chain reactions (PCRs) were carried out as previously described [16]. Amplification products were resolved by electrophoresis in 3% agarose gels prior to vertical electrophoresis in 6% denaturing polyacrylamide gels; gels were then silver stained as previously described [27]. Product sizes were determined by comparison to a 10-bp DNA ladder (Invitrogen, Carlsbad, CA).
The microsatellites were treated as dominant markers due to the polyploid nature of the genotypes. Accordingly, data were scored based on the presence (1) or absence (0) of a band for each of the Urochloa genotypes. Both PIC and DP values were calculated to estimate the polymorphism of each locus. PIC values were calculated based on [18] and DP values based on [19]. PIC was used as a tool to measure the information that a given marker locus could provide for the pool of genotypes, whereas DP was used as a quantification tool to measure the efficiency of a given marker for the discrimination of genotypes, i.e., the probability that two randomly chosen individuals have different patterns.
The genetic similarity among all the genotypes was estimated according to Jaccard's similarity coefficient [28] based on a binary matrix constructed with the polymorphic bands. The corresponding genetic similarity matrix was used to generate a dendrogram based on the Unweighted Pair Group Method with the Arithmetic Mean (UPGMA) algorithm as previously reported [29]. All analyses were carried out using NTSYSpc 2.11X [30]. A bootstrap analysis with 10,000 random samplings was applied to estimate the reliability of the dendrogram branches using BOOD version 3.0 [31].
A Bayesian clustering method was employed to assess population structure using the STRUCTURE software version 2.3.3 [20]. We performed 10 runs for each K (from K = 1 to K = 10) and ran the analysis assuming a model of admixture and correlated allele frequencies. We did not use any prior information about the population origin of the genotypes. A burn-in period of 500,000 generations and MCMC simulations of 700,000 iterations were used in all the above runs. The values of LnP(D) (the log probability of data) were estimated by assigning a prior from 1 to 10, and the optimal K was chosen based on the delta K (ΔK) value [21].
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
BBZV carried out computational searches for microsatellite identification, designed flanking primers, participated in microsatellite marker validation, performed the statistical analysis and drafted the manuscript. GCA participated in microsatellite marker validation and statistical analysis. LJ participated in the design and implementation of the study and the microsatellite identification and design of flanking primers. CBV and APS conceived of the study and participated in its design and coordination. APS helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Jaccard's similarity coefficients among 34 genotypes of Urochloa app evaluated through 40 microsatellite markers. Individuals are identified according to their EBC code (Table 3)
The membership coefficient (Q) from STRUCTURE analysis based on 40 microsatellite loci data.
Contributor Information
Bianca BZ Vigna, Email: bianca@cppse.embrapa.br.
Guilherme C Alleoni, Email: gc.alleoni@gmail.com.
Leticia Jungmann, Email: jungmann@cnpgc.embrapa.br.
Cacilda B do Valle, Email: cacilda@cnpgc.embrapa.br.
Anete P de Souza, Email: anete@unicamp.br.
Acknowledgements
The authors would like to thank Dr. Felipe Rodrigues da Silva for assisting with the bioinformatics. This work was supported by grants from the Brazilian Agricultural Research Corporation (Embrapa), the Brazilian National Council for Scientific and Technological Development (CNPq), the State of São Paulo Research Foundation (FAPESP, 2005/51010-0), and the Foundation for Science and Technological Development of the State of Mato Grosso do Sul (FUNDECT). BBZV and GCA were recipients of graduate (2007/57022-5; 2010/50032-8) and undergraduate (2009/53787-2) fellowships, respectively, from FAPESP; APS is recipient of a research fellowship from CNPq.
References
- Morrone O, Zuloaga FO. Revisión de las especies sudamericanas nativas y introducidas de los géneros Brachiaria y Urochloa (Poaceae: Panicoideae: Paniceae) Darwiniana. 1992;31:43–109. [Google Scholar]
- Keller-Grein G, Maass BL, Hanson J. In: Brachiaria: biology, agronomy and improvement. 1. Miles JW, Maass BL, Valle CB, editor. Cali: Embrapa/CIAT; 1996. Natural variation in Brachiaria and existing germplasm collections; pp. 16–42. [Google Scholar]
- Adamowski EV, Boldrini KR, Pagliarini MS, Valle CB. Abnormal cytokinesis in microsporogenesis of Brachiaria humidicola (Poaceae: Paniceae) Genet Mol Res. 2007;6:616–621. [PubMed] [Google Scholar]
- Boldrini KR, Pagliarini MS, do Valle CB. Meiotic behavior of a nonaploid accession endorses x = 6 for Brachiaria humidicola (Poaceae) Genet Mol Res. 2009;8:1444–1450. doi: 10.4238/vol8-4gmr679. [DOI] [PubMed] [Google Scholar]
- Boldrini KR, Micheletti PL, Gallo PH, Mendes-Bonato AB, Pagliarini MS, do Valle CB. Origin of a polyploid accession of Brachiaria humidicola (Poaceae: Panicoideae: Paniceae) Genet Mol Res. 2009;8:888–895. doi: 10.4238/vol8-3gmr617. [DOI] [PubMed] [Google Scholar]
- Boldrini KR, Pagliarini MS, do Valle CB. Evidence of natural hybridization in Brachiaria humidicola (Rendle) Schweick. (Poaceae: Panicoideae: Paniceae) J Genet. 2010;89:91–94. doi: 10.1007/s12041-010-0016-z. [DOI] [PubMed] [Google Scholar]
- Valle CB, Savidan YH. In: Brachiaria: biology, agronomy and improvement. 1. Miles JW, Maass BL, Valle CB, editor. Cali: Embrapa/CIAT; 1996. Genetics, cytogenetics and reproductive biology of Brachiaria; pp. 147–163. [Google Scholar]
- Nogler GA. Genetics of apospory in apomictic in Ranunculus auricomus. 5. Conclusion. Bot Helv. 1984;94:411–422. [Google Scholar]
- Renvoize SA, Clayton WD, Kabuye CHS. In: Brachiaria: biology, agronomy and improvement. 1. Miles JW, Maass BL, Valle CB, editor. Cali: Embrapa/CIAT; 1996. Morphology, taxonomy and natural distribution of Brachiaria (Trin.) Griseb; pp. 1–15. [Google Scholar]
- Li YC, Korol AB, Fahima T, Beiles A, Nevo E. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol. 2002;11:2453–2465. doi: 10.1046/j.1365-294X.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- Holmen J, Vollestad LA, Jakobsen KS, Primer CR. Cross-species amplification of 36 cyprinid microsatellite loci in Phoxinus phoxinus (L.) and Scardinius erythrophthalamus (L.) BMC Res Notes. 2009;2:248. doi: 10.1186/1756-0500-2-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billotte N, Lagoda PJR, Risterucci AM, Baurens FC. Microsatellite-enriched libraries: applied methodology for the development of SSR markers in tropical crops. Fruits. 1999;54:277–288. [Google Scholar]
- Sousa ACB, Jungmann L, Campos T, Sforça DA, Boaventura LR, Silva GMB, Zucchi MI, Jank L, de Souza AP. Development of microsatellite markers in Guineagrass (Panicum maximum Jacq.) and their transferability to other tropical forage grass species. Plant Breed. 2011;130:104–108. doi: 10.1111/j.1439-0523.2010.01779.x. [DOI] [Google Scholar]
- Laborda PR, Klaczko LB, de Souza AP. Drosophila mediopunctata II: cross-species amplification in the tripunctata group and other Drosophila species. Conserv Genet Resour. 2009;01:281–296. doi: 10.1007/s12686-009-9069-9. [DOI] [Google Scholar]
- Cidade FW. PhD thesis. University of Campinas, Genetics Department; 2011. Estudos genético-moleculares no gênero Paspalum L. (Poaceae: Panicoideae: Paniceae) [Google Scholar]
- Jungmann L, Vigna BBZ, Paiva J, Sousa ACB, do Valle CB, Laborda PR, Zucchi MI, de Souza AP. Development of microsatellite markers for Brachiaria humidicola (Rendle) Schweick. Conserv Genet Resour. 2009;01:475–479. doi: 10.1007/s12686-009-9111-y. [DOI] [Google Scholar]
- Jungmann L, Vigna BBZ, Boldrini KR, Sousa ACB, do Valle CB, Resende SMS, Pagliarini MS, Zucchi MI, de Souza AP. Genetic diversity and population structure analysis of the tropical pasture grass Brachiaria humidicola based on microsatellites, cytogenetics, morphological traits, and geographical origin. Genome. 2010;53:698–709. doi: 10.1139/G10-055. [DOI] [PubMed] [Google Scholar]
- Mateescu RG, Zhang Z, Tsai K, Phavaphutanon J, Burton Wursten NI, Lust G, Quaas R, Murphy K, Acland GM, Todhunter RJ. Analysis of Allele Fidelity, Polymorphic Information Content, and Density of Microsatellites in a Genome-Wide Screening for Hip Dysplasia in a Crossbreed Pedigree. J Hered. 2005;96:847–853. doi: 10.1093/jhered/esi109. [DOI] [PubMed] [Google Scholar]
- Tessier C, David J, This P, Boursiquot JM, Charrier A. Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theor Appl Genet. 1999;98:171–177. doi: 10.1007/s001220051054. [DOI] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Gupta PK, Varshney RK. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica. 2000;113:163–85. doi: 10.1023/A:1003910819967. [DOI] [Google Scholar]
- Gonzalez AMT, Morton CM. Molecular and morphological phylogenetic analysis of Brachiaria and Urochloa (Poaceae) Mol Phylogenet Evol. 2005;37:36–44. doi: 10.1016/j.ympev.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001;11:1441–1452. doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creste S, Tulmann Neto A, Figueira A. Detection of single sequence repeat polymorphisms in denature polyacrylamide sequencing gels by silver staining. Plant Mol Bio Rep. 2001;19:299–306. doi: 10.1007/BF02772828. [DOI] [Google Scholar]
- Jaccard P. Etude comparative de la distribution florale dans une portion des Alpes et du Jura. Bull Soc Vaud Sc Nat. 1901;37:547–579. [Google Scholar]
- Sneath PHA, Sokal RR. Numerical taxonomy. San Francisco: Freeman; 1973. [Google Scholar]
- Rohlf FJ. NTSYSpc: numerical taxonomy and multivariate analyses system. New York: Applied Biostatistics; 2000. Version 2.11X. [Google Scholar]
- Coelho ASG. Software BOOD version 3.0. Laboratório Genética Vegetal, DBG/ICB: UFG; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Jaccard's similarity coefficients among 34 genotypes of Urochloa app evaluated through 40 microsatellite markers. Individuals are identified according to their EBC code (Table 3)
The membership coefficient (Q) from STRUCTURE analysis based on 40 microsatellite loci data.