Abstract
With a large percentage of clinical trials still using paper forms as the primary data collection tool, there is much potential for increasing efficiency through web-based data collection systems, especially for large-scale multi-center trials. This paper presents OnWARD, an ontology-driven, secure, rapidly-deployed, web-based framework supporting data capture for large-scale multi-center clinical research. Our approach is developed using the agile methodology to provide a flexible, user-centered dynamic form generator, which can be quickly deployed and customized for any clinical study without the need of deep technical expertise. Because of the flexible framework, the data management system can be extended to accommodate a large variety of data types, including genetic, genomic and proteomic data. In this paper, we demonstrate the initial deployment of OnWARD for a Phase II multi-center clinical trial after a development period of merely three months. The study utilizes 23 clinical report forms containing more than 1500 data points. Preliminary evaluation results show that OnWARD exceeded expectations of the clinical investigators in efficiency, flexibility and ease in setting up.
Keywords: ontology, clinical research form, dynamic form, web-based data entry
1. Introduction
Web-based data management systems offer great potential for facilitating the conduct of large scale or multi-center clinical studies [1, 2]. These systems are well suited for supporting real-time access to data by investigators working across multiple sites with varying infrastructure, minimizing the logistical challenges in conducting multi-center collaboration, providing improved monitoring capability, and facilitating new mechanisms for producing high quality validated data. They also prepare for linking the collected data with additional data resources to allow harmonization with the results of other studies or allow extension to include richer clinical and biological contexts available through other data sources. In addition, optimal systems would lead to new levels of operational simplicity and efficiency for conducting clinical and translational research.
In this paper we present the design, implementation and preliminary usage results of OnWARD, an Ontology-driven Web-based Architecture for Research Data Management. This framework supports the entire clinical research data life-cycle: data specification, data collection, and data reviewing, reporting and analysis in a secure web-based environment well suited for multi-site trials and multi-institution studies. The OnWARD architecture consists of three main modules, corresponding to the main phases of clinical research data life-cycle: Data Specification (DS), Data Capture (DC), and Data Exploration (DE). During the study development phase, key investigators or database administrators use OnWARD-DS to specify projects and develop collection forms by providing database structure and other metadata and mapping the database fields to terms in the ontology. During active data collection, study personnel such as data entry staff interact with OnWARD-DC through a dynamically generated web interface. Collected data are stored in a separate relational database, which can be viewed or edited in a distributed mode by personnel at any location. In the review and reporting phase, OnWARD-DE generates customizable reports using real-time data in the research database. All three modules sit inside a secured environment with role-based access control (RBAC [3]).
The developmental use case of OnWARD has been the Heart Biomarker Evaluation in Apnea Treatment trial (HeartBEAT, 1RC2HL101417), a multi-institution Phase II clinical trial funded through the American Recovery and Revitalization Act (ARRA). The features of OnWARD described in this paper make it ideally suited to serve as a data management tool for clinical and translational research projects of a similar magnitude.
2. Background on the HeartBEAT Study
HeartBEAT is a randomized, controlled multi-center study designed to assess the effects nocturnal supplemental oxygen and positive airway pressure therapy for reduction of cardiovascular risk in high-risk patients with obstructive sleep apnea. HeartBEAT involves four Clinical Centers (CCs), each of which will screen approximately 350 subjects with portable monitoring in order to enroll 88-89 subjects for a total enrollment of 354 subjects (118 per treatment arm). As an ARRA-funded initiative the total duration of the study is limited to 2 years, with only a three-month start-up phase.
This project provides a primary use case for the OnWARD framework because it features some 23 clinical research forms (CRFs) collected at one or more time points as part of the protocol. Further, the nature of the study requires remote entry from four distinct locations across the U.S. into a central database, and the accelerated time line of this study necessitated a rapidly deployed solution. In addition, the targeted data collection includes a wide variety of data types, such as complex physiological data captured using overnight polysomnographic studies and 24 hour blood pressure recordings, measurements of vascular function, and biomarker data obtained from blood and urine assays. Data are collected from a number of sources, including the study participants’ homes (polysomnography and 24 hour blood pressure), each clinical site’s Clinical Research Unit (vascular measurements, ECGs, anthropometry), and a Core Biochemistry Laboratory.
3. Design and Implementation
The design of OnWARD involves three seamlessly integrated modules: DS, DC and DE. Agile development and agile project management methodologies were used to achieve a flexible and user-friendly web-based data management tool in the Ruby on Rails framework [4].
3.1. Clinical Research Ontology
The three modules of OnWARD (DS, DC and DE) are united by an underlying clinical research ontology. The research ontology includes standardized terminologies for items and response values. Conceptual level information about each individual item contained within each CRF and its associated range of acceptable values are captured in the ontology. Each project-specific data concept or variable is extended from the central ontology depending on the scope of the project. For example, the HeartBEAT study focused on patients between 40 and 70 years old. To account for this, we extended from the concept SDO:CurrentChronologicalAge in the Sleep Domain Ontology (SDO [6]) a variable called HBAge with the value range of (0,120) to describe the current age of a patient in the HeartBEAT study.
Unlike data dictionaries, an ontological system provides a formal model of a domain, consisting of standardized terms and named relationships between the terms. This allows the terms in an ontology to be interpreted more consistently and accurately by software applications as well as human users. Hence, the use of ontology in OnWARD allows it to provide a more flexible model to support a wide range of clinical trials and translational investigations by increasing the reusability of questionnaire elements. It also provides context-dependent concepts and relations with explicitly-specified semantics to facilitate data integration across studies.
For the purpose of the HeartBEAT study, we use the concepts from the Computer-based Patient Record (CPR) ontology [5] and an in-house-developed SDO [6]. HeartBEAT was the first study to use OnWARD. For this purpose, an additional set of ontological concepts was incorporated from an existing metadata repository containing a set of more than 1100 value domains with some 5300 permissible values. These permissible values determine which values are allowable in a specified domain (i.e. a field in a form). However, as more studies utilize OnWARD for data capture, the ontology is expected to expand with more concepts, improving their reusability for subsequent studies.
To support broader bioinformatics research, OnWARD provides the flexibility in incorporating ontologies from different domains with various data types and values. Relationships between concepts can be further exploited to facilitate the creation of new CRF in two ways:
calculated fields can be automatically derived from other concepts according to the relationship specified by the ontology, and
sub-concepts can be automatically added when a parent concept is selected in a CRF, creating a correspondence between CRFs and certain higher-level concepts.
These further enhancements, especially the second one, will make CRF design more robust and efficient.
3.2. Flexible Backend Database Selection
OnWARD-DS gives a researcher complete control of how and where they want their collected data to be stored. It maps each clinical study to a backend relational database (of the investigator’s choice) and connects to that database for data entry as well as retrieval, validation and other functionalities provided through the OnWARD-DC and OnWARD-DE modules. This architecture allows existing databases to be readily incorporated into the OnWARD framework without the need to translate and migrate data to a different platform.
To connect to a backend database, OnWARD uses ActiveRecord [7]. ActiveRecord turns an underlying relational database table into an object type. Each row of the table becomes an object instance, and each column becomes an access method for the object. Thus, ActiveRecord transforms the relational paradigm to the object-oriented paradigm, hiding many of the complexities of SQL query required to access the relational data. Adaptors are software plugins that activate the ActiveRecord interface to a relational database. For OnWARD, adapters for MySQL and Microsoft SQL Server are included as default, but additional adapters can be included easily.
3.3. Dynamic form generation
HeartBEAT required the development of a wide variety of data collection forms. These included forms designed to capture demographic, medical history and sleep study information needed to determine subject eligibility; forms needed to capture baseline physiological, functional status, symptom and biochemical data; and forms needed to track study process measures including intervention use. Data collection points included recorded physical measurements, technician-reported observations, measurements made using blood and urine specimens, and participant-reported responses. Data collected included continuous numeric responses, dates, categorical numeric responses, and free text responses.
A project that relies on the development of predesigned static HTML forms for web-based entry suffers from a relatively long start-up time line and cost, as well as a heavy maintenance burden as changes in forms or data fields are encountered during the course of the study. In contrast, a dynamic form generation engine as in OnWARD-DC provides flexible, customizable, dynamic generation of web-based entry screens based on the metadata specified in OnWARD-DS and informed by the ontology, with no programming required of the researcher. The form generator displays each item on each form with a descriptive label and a data control appropriate for the data type and distribution of the variable as specified in the ontology terms (text box, drop-down list, etc.). A simple general framework that includes a layout type and sections (pages and headings) is applied to every form, with the specific configurable parameters provided by the researcher and stored in OnWARD-DS.
The form generation engine provides two basic layouts: List View and Tabular View. The List View consists of a single-record layout with all items displayed in a list, and the Tabular View displays a multi-row, multi-column layout with a single header. While the List View is used for most CRFs for OnWARD, the Tabular View is more desirable is situations such as Medication History (Figure 1), where the total number of medications is not predetermined.
Figure 1.
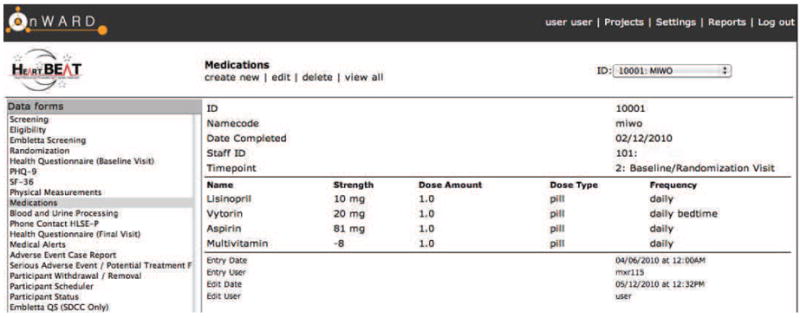
Medication form with all medications grouped by participant
3.3.1. Input validation
Input validation is a procedure to reduce user errors performed at the time of data entry and to prevent spurious values from entering the database. Because OnWARD-DC is based on a central ontology, all of the information pertaining to input validation is provided by the item and value domain mappings. This validation is handled at three levels:
Data Type. A first check of input data is to ensure that it is appropriate for the expected data type. Currently four general data types are supported: text, numeric, categorical, and date.
Hard Range Validation. For numeric value domains, a permissible range is specified outside of which no values are permitted; the user is presented with an error message and is prevented from submitting the record.
Soft Range Validation. For each numeric value domain, an expected range is also specified outside of which values are flagged as potentially out of range; the user is presented with a warning but is still allowed to submit the form upon confirming the value is correct. An example from HeartBEAT where both hard validation and soft validation have been useful is patient age. Patient age is expected to be from 45 to 75 years (soft range) but a patient’s age may never exceed the 18 to 100 range (hard range).
3.3.2. Skip patterns
To facilitate data entry, OnWARD-DC provides configurable logical skip patterns, allowing data entry personnel to skip certain questions depending on the answer to previous questions. This was especially helpful for the HeartBEAT Eligibility CRF in which inclusion and exclusion criteria were evaluated. Once one exclusion criterion is identified, there is not a need for further ascertainment of eligibility criteria. Thus, the remaining questions can be assigned default values and skipped, minimizing data entry effort.
OnWARD skip patterns are specified as metadata in the format of
For example, in the Eligibility Form, if the BMI field received a value outside the inclusion criterion (> 30), then the rest of the questions in the Eligibility Form will be skipped and receive an assigned default value (e.g. NA for not applicable). Each skipped question will have an entry in the above format, such as
This format allows different types of patterns, including skipping or specifying values of multiple responses.
3.4. Dynamic report generator
The demands of clinical trials require the regular monitoring and reporting of study recruitment benchmarks, adverse events, and other interim analyses that address study safety and progress. OnWARD-DE helps meeting the needs for the life cycle of a clinical study by providing capabilities to generate dynamic reports from real-time collected data.
The OnWARD-DE module allows researchers to search question labels across all CRFs within a study for inclusion in a report. Once a question is selected, a corresponding condition can be specified. Based on the set of (question, condition) pairs, SQL statements are constructed and executed to retrieve study participants’ data from the research database and the report contents are displayed on the web interface, and can be exported in CSV or PDF. This gives researchers the ability to generate a variety of types of reports depending on their requirements including, but not limited to, adverse event reports, patient demographic reports, and study progress reports. The dynamic report generating function is made available by the underlying relational database model, with each form stored in a separate table, and each record referenced by a unique patient identifier specific to a project.
In HeartBEAT, tracking of participant eligibility and randomization is essential. OnWARD-DE allows for custom reports to be generated by investigators to obtain counts of eligible participants, grouped by randomization arm or site, or stratified by physiological values. These reports require joining data from several distinct database tables, which is facilitated by OnWARD-DE, making the report easily created, reused, updated, and exported by research staff without knowledge of database structure or query language.
3.5. Security
The security and integrity of study data are of major concern for any web-based data capture system and for any clinical trial. Clinical research not only requires that data be secure from outside access, but also requires levels of security within the study. For example, some protocols may require blinding of particular personnel to specific data points, or require that one recruitment site’s data only be accessible to personnel from that site. In addition, some users may need the ability to enter and edit data while others may only read data or only edit data. All of these considerations need to be addressed by any web-based clinical data capture system.
OnWARD adapts a project- and site-based parametric role-based access control (RBAC [3]) mechanism to the granularity of tables. Basic table privileges including view, edit and delete are associated with roles, and roles are assigned to users. This RBAC model helps to achieve the requirements listed above by allowing access to the system to only approved, registered users; limiting entry into a project to users assigned a role in that project; limiting privileges on specific tables to those specified by role assignment; and limiting access to records based on site assignment.
4. Results
4.1. System development
Initial development of the OnWARD framework was accomplished using agile methodologies involving close collaboration with the HeartBEAT investigator team and the OnWARD development team. This iterative process quickly produced a system that met the specific needs of the HeartBEAT study but which was also designed to be flexible and configurable for other projects.
The OnWARD framework provided a substantial reduction in development time. A study of this size, featuring 23 forms and over 1500 data points (see Figure 3 for a data entry form), typically requires 3-6 months of development to deploy and validate a custom web-based entry system, with continued web programming for changes and modifications during the course of the project. With OnWARD, deployment of a functional data capture system was accomplished in under 6 weeks with a team consisting of one developer, one data manager and one tester. After initial deployment, modifications of forms and data management approaches identified through piloting and by review by a Data Safety and Monitoring Committee were easily implemented by study staff through simply modifying values in the ontology, allowing the study to be initiated according to the ambitious ARRA-mandated timeline. Future modifications also will be possible and may be implemented by study personnel without any web programming expertise.
Figure 3.
A data entry screenshot
4.2. Validation and evaluation
OnWARD’s development was motivated by the needs of the HeartBEAT study, and designed to support research staff including investigators, data managers and nurses in their roles in study recruitment, data entry, and data monitoring and reporting. It was necessary to have a quickly deployed, easy to use and efficient system to facilitate the implementation and conduct of a study with an ambitious timeline.
After initial deployment it was easy for study staff to make modifications to forms by adjusting metadata and ontology mappings without involving the development team. Having been used since January 2010, research staff from the participating clinical sites accessed the system to input data more than 5000 screened subjects. Research staff also accessed the system to enter study inclusion and exclusion criteria for web-based randomization of eligible subjects. Randomization for the HeartBEAT study assigns participants to one of three treatment arms based on a stratified blocked design. Randomization is stratified based on field collection site (n = 4) and the presence or absence of diagnosis of Coronary Artery Disease (CAD), for a total of 8 strata. The randomization assignments were calculated using a custom program executed in SAS, and stored in a table in the research database. Over 120 randomization assignments were generated in each arm, far more than would be necessary during the execution of the study protocol.
A second database table was created to track randomization assignments. New records in this table include the information necessary to determine randomization stratum. Upon insertion of a record in this table, a database procedure is activated to determine and return the randomized assignment for that participant. The database procedure first determines the stratum using the site id and CAD variables. It then identifies the next available randomization assignment for the selected stratum and returns the value of the assignment. All randomization assignment values are saved in the table as they are determined by the procedure.
Although randomization is not directly supported, OnWARD allowed this process to be made web-based by providing a form to enter the randomization criteria, and then providing a web interface that would return the assigned randomization arm. By having the randomization assignment driven by the research database, randomization could be performed off-line when necessary, and the results of manual randomization (a backup procedure defined in the protocol to be used if the web-based system was not available) could be entered into the database manually.
Users were able to accomplish all data entry tasks with only minimal training, and the system meets or exceeds expectations of research staff and investigators for ease of configuration and use.
Although the development process was driven by an ARRA-funded Phase-2 multi-center clinical trial, the framework is built to be generic and flexible enough to be used in a wide range of clinical study, from small to medium scale.
The system has also been used to support a small-scale chart review-based research project designed by a medical student (Placental Insufficiency and Pediatric Sleep Disordered Breathing Study or PIPS). This project featured two forms and 150 questions, and data was entered for 32 patients. PIPS is an example of OnWARD’s ability to use an existing research database. The PIPS data can be entered directly into an existing research database without the need for post-entry data translation and transfer. The specification and configuration of the forms for this study required minimum effort for a data manager to construct questionnaires taking less than 1 hour, and no further interface development or programing was required, facilitating a quick and low-cost approach for a student-oriented research project.
5. Discussion
There have been similar frameworks implemented to support clinical trials. For instance, Epoch [8] is a knowledge-based framework supporting the management of clinical trials at Immune Tolerance Network (ITN) including two main applications: (1) tracking participants, and (2) tracking clinical specimens. It was aimed at providing ITN and research organizations with a set of ontology to define clinical trial concepts to be used in clinical research tools.
Differently, Mirhaji et al [9] focus on using Semantic Web technology to define different data models to integrate both non-structure and structured information from different sources. An application called Survey on Demand System (SODS) is developed for designing questionnaires and automatically mapping the answers to the ontological concepts. Although many tools have been developed to support clinical studies, at the beginning of HeartBEAT project, we were not able to find a system that addressed our needs with the ability and flexibility to
use an existing database to store captured data,
give the investigators the option to control how collected data is stored in the database,
generate reports dynamically using real-time data, and
support a comprehensive set of forms used in a large-scale clinical study.
The above are some of the contributions that we made in OnWARD to provide a novel, flexible and robust framework.
5.1. Comparison with REDCap
REDCap (Research Electronic Data Capture [10]) is a notable web-based, metadata-driven data capture tool successfully adopted worldwide. While both OnWARD and REDCap share a number of features such as web-based data entry, flexible form design and automatic data validation, here are some unique aspects of each system:
Ontology support. OnWARD’s use of an underlying ontology sets it apart from other data capture systems in its ability for collected data to be understood, interpreted, and linked between projects by both software applications and human users. The ontology allows the creation of calculated data variables by using concept relationships defined within the ontology without having to explicitly specify calculations in each project’s metadata nor to store the calculated values in the database. The ontology also allows for the relationships between similar (but not identical) data fields to be understood and interpreted, facilitating data sharing and collaboration among projects. Although REDCap is not strictly ontology-driven, it achieves a similar benefit by sharing a library of standard forms for reuse by the community.
Data storage. Researchers can choose to use the OnWARD resident database in MySQL, or alternatively use an existing database hosted elsewhere. In the case of HeartBEAT, the investigators chose to use an existing research database in Microsoft SQL Server as the data repository. The database system featured stored procedures, which implemented the randomization algorithm, and was the backend for other real-time reporting and analytic functionalities outside of OnWARD. This flexibility also provides researchers with the capability to plug in their existing databases and continue to collect data instead of having to migrate data. Storage of all projects in a centralized database as in REDCap may alleviate the burden for researchers to host and maintain databases themselves, but may limit investigators who prefer to have the ability to access the databases from the backend and incorporate external functionalities, as is the case for HeartBEAT.
Tabular form. Tabular form provides an efficient way to collect data where the number of entries for each study participant is not statically predetermined. An example is medication history. Medication History for HeartBEAT allows a maximum of 30 medications in the paper form. Five attributes are captured per each medication: medication name, dosage, type, strength, frequency. If this were implemented in REDCap, 150 rows (30 × 5) would be needed, resulting in a long form and clumsy data-entry process. For OnWARD (see Figure 1), because of the relational data model, medication history data can be entered one medication at a time, with all the five attributes captured in the same row. The number of rows grows as needed per each study participant during data entry.
There are also subtle differences in the implementation of the role-based access control mechanism in the two systems, in that OnWARD completely separates the definition of roles from the users, making it easier to manage access control. On the other hand, REDCap provides robust export functionality that can be seamlessly pipelined to statistical analysis packages (SPSS, SAS, R, Stata, and Excel), a useful feature that is currently beyond the scope of OnWARD.
5.2. Future work
The initial experience with the development of OnWARD provides evidence that a user-friendly tool for rapid development of ontology-driven web-based data management for clinical trials can be accomplished. There are features that we anticipate will be added to the current framework: (1) enhancing the reporting interface, (2) providing data sharing mechanisms across projects to take advantage of the structured relationships provided by the ontology, (3) expanding the current sleep domain ontology to support a wider range of clinical studies.
Figure 2.
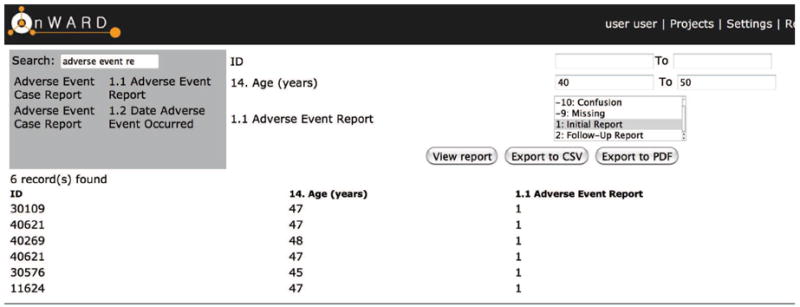
A report generation screenshot
Acknowledgments
This publication was made possible by the Case Western Reserve University/Cleveland Clinic CTSA (UL1 RR024989) from the National Center for Research Resources (NCRR), by the Physio-MIMI project (NCRR-94681DBS78), and by an ARRA grant (RC2HL101417) to the Brigham and Women’s Hospital. VT also acknowledges support from the Choose Ohio First Scholarship (UT15439) from the Ohio Board of Regents through the Ohio Consortium for Bioinformatics. We would like to thank our colleagues Satya Sahoo, Sheree Hemphill, John Sharp and Michael Rueschman for their discussions and feedback.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Avidan A, Weissman C, Sprung CL. An internet web site as a data collection platform for multicenter research. Anesth Analg. 2005;100(2):506–11. doi: 10.1213/01.ANE.0000142124.62227.0F. [DOI] [PubMed] [Google Scholar]
- 2.Cooper CJ, Cooper SP, del Junco DJ, Shipp EM, Whitworth R, Cooper SR. Web-based data collection: detailed methods of a questionnaire and data gathering tool. Epidemiologic Perspectives & Innovations. 2006;3:1. doi: 10.1186/1742-5573-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mueller R, Tran V, Zhang GQ. A Scalable Parametric-RBAC Architecture for the Propagation of MIMI. The 11th International Conference on Enterprise Information Systems, Lecture Notes in Business Information Processing; Springer-Verlag; 2009. pp. 114–124. [Google Scholar]
- 4. [August 15, 2011];Ruby on Rails. http://rubyonrails.org.
- 5.Ogbuji C, Arabandi S. CPR ontology. [August 15, 2011]; http://code.google.com/p/cpr-ontology/
- 6.Arabandi S, Ogbuji C, Redline S, Chervin R, Boero J, Benca R, Zhang GQ. Developing a Sleep Domain Ontology (Abstract). Proceedings of the 2010 AMIA Clinical Research Informatics Summit; San Francisco. March 12-13. [Google Scholar]
- 7.Martin F. Patterns of enterprise application architecture. Addison-Wesley; 2003. [Google Scholar]
- 8.Shankar RD, O’Connor MJ, Parrish DB, Martins SB, Das AK. Epoch: an Ontological Framework to Support Clinical Trials Management. International Workshop on Health Information and Knowledge Management (HIKM’06) 2006:25–32. doi: 10.1145/1183568.1183574. [DOI] [Google Scholar]
- 9.Mirhaji P, Zhu M, Vagnoni M, Bernstam EV, Zhang J, Smith J. Ontology driven integration platform for clinical and translational research. BMC Bioinformatics. 2008 2009 Feb 5;10(Suppl 2):S2. doi: 10.1186/1471-2105-10-S2-S2. PMCID: PMC2646248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris P, Taylor R, Thielke R, Payne J, Gonzalez N, Conde J. Research electronic data capture (REDCap) - A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009 2009 Apr;42(2):377–81. doi: 10.1016/j.jbi.2008.08.010. PMCID: PMC2700030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang GQ, Siegler T, Saxman P, Sandberg N, Mueller R, Johnson N, Hunscher D, Arabandi S. VISAGE: A Query Interface for Clinical Research. Proceedings of the 2010 AMIA Clinical Research Informatics Summit; San Francisco. March 12-13, 2010; pp. 76–80. PMCID: PMC3041531. [PMC free article] [PubMed] [Google Scholar]


