Abstract
Candida grows on devices producing treatment resistant biofilms. A key tool for the study of biofilms includes an accurate assessment of viable cell growth. This study systematically tested seven techniques, among which the XTT assay provided the most reproducible, accurate, and efficient method for the quantitative estimation of C. albicans biofilms.
Keywords: Candida, biofilm, assay
Introduction
Infections caused by Candida albicans frequently occur in hospitalized patients, where the pathogenic fungus forms biofilms on medical devices [1–5]. Because these infections are difficult to treat, extensive research has focused on understanding genetic and biochemical aspects of biofilm development and the characteristic drug resistance phenotypes [6–10]. In vitro plate assays have become a key tool for propagating Candida biofilms and investigating their composition and properties. A necessary element of these experiments is the ability to accurately and reproducibly quantify the viable cells in the biofilm. Not only does this serve as an important study endpoint, but it provides the data needed for the normalization of biochemical assays. Through such normalizing based on levels of cells in the biofilms, accurate comparisons can be made between mutants and their parental strains, as well as among multiple clinical strains. Normalization can also minimize differences in growth rate as a factor when interpreting the assay’s results.
Numerous techniques have been employed for the measurement of the quantity of cells in biofilms. These procedures vary widely as to their time and cost requirements [10–18]. In addition, we have observed variations in assay performances that led us to systematically investigate the efficacy of these techniques. The goals of the current study were to: (1) identify quantitative biofilm assay(s) that provide a reproducible, accurate measurement of biofilm(s), and (2) compare time, efficiency, and cost of the quantitative assays. Studies included the use of XTT reduction assay, crystal violet staining, DNA quantification, qPCR, protein quantification, dry cell weight measurement, and viable colony counting.
Materials and methods
Three C. albicans strains were chosen for their varied abilities to form biofilms. The reference strain DAY185 produces extensive biofilms with abundant extracellular matrices and variable cell morphologies [19]. The two mutant strains lack adhesions, resulting in a partial biofilm defect for als3Δ/Δ and a more extensive biofilm defect in strain als1Δ/Δ als3Δ/Δ [8,20–22]. To evaluate biofilm formation, overnight cultures grown in yeast peptone dextrose (YPD) broth [8] at 30°C with orbital shaking at 200 rpm were enumerated by hemocytometer. Cells were resuspended in RPMI-MOPS [7] at 106 cells/ml and each well of a six-well polystyrene plate was inoculated with 1 ml of this suspension. After a 1 h adhesion period, the inoculum was removed and fresh media was applied. Biofilms were grown for 48 h at 37°C on an orbital shaker at 50 rpm. For comparative scanning electron microscopy (SEM), biofilms were formed on coverslips and processed and examined as previously described [23].
Protocols for the XTT and crystal violet assays have been described [7,24]. For the crystal violet assay, 4% aqueous crystal violet was added for 45 min and 100 µl aliquots were taken from each well for absorbance measurement (595 nm). Candida DNA quantification was accomplished using a commercially available kit (Genomic DNA Wizard Kit – Promega) following cellular disruption by bead-beating [25]. DNA samples were also used for qPCR with ACT1 primers and probes [23] and Quantitect Probe qPCR kit (Qiagen) performed with the CFX96 Real-Time PCR Detection System (Bio-Rad). The Protein Assay Kit (Pierce) was used to determine total cellular protein after biofilms were scraped from wells into 1 ml of ddH2O, sonicated for 20 min, disrupted by bead-beating, and boiled. Dry cell weight of each biofilm was determined following disruption, collection, and then dehydrated by vacuum centrifugation. Viable burdens were determined by plating serial dilutions following biofilm disruption, vortexing, and sonicated using a waterbath for 20 min to disperse cells [14].
Reproducibility was estimated by the standard deviation and coefficient of variation among replicates. At least six biofilms were grown for each strain and assay.
Results
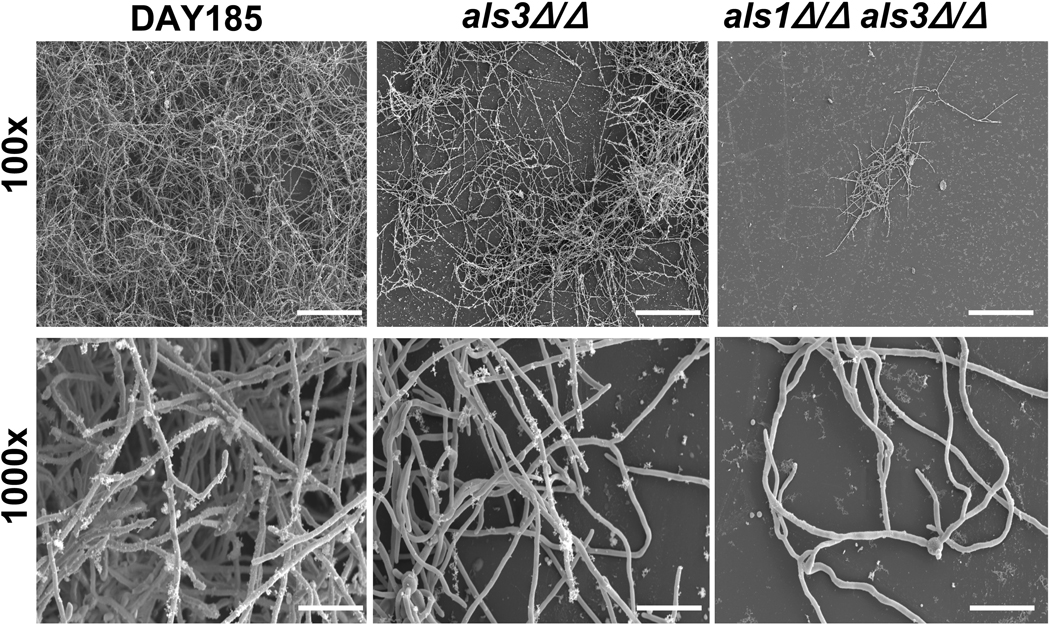
The differences in biofilm formation among the strains was observed visually and confirmed with SEM. The reference strain DAY185 produced extensive biofilms with abundant extracellular matrices and variable cell morphologies (Fig. 1). An intermediate biofilm defect was found for the als3Δ/Δ mutant, with few clumps of adherent hyphae. As previously described, a profound biofilm defect was noted for als1Δ/Δals3Δ/Δ mutant, with fewer adherent cells present. This was less apparent visually in the 6-well format, as als1Δ/Δals3Δ/Δ more closely resembled als3Δ/Δ, possibly due to a difference between the coverslip and 6-well growth models. [Fig. 1 near here]
Fig. 1.
SEM images of C. albicans mutant biofilms. DAY185, als3Δ/Δ, and als1Δ/Δ als3Δ/Δ biofilms were grown on coverslips. Biofilms were processed and imaged using SEM at 100× (top row, scale shows 200 µm) and 1000× (bottom row, scale shows 20 µm).
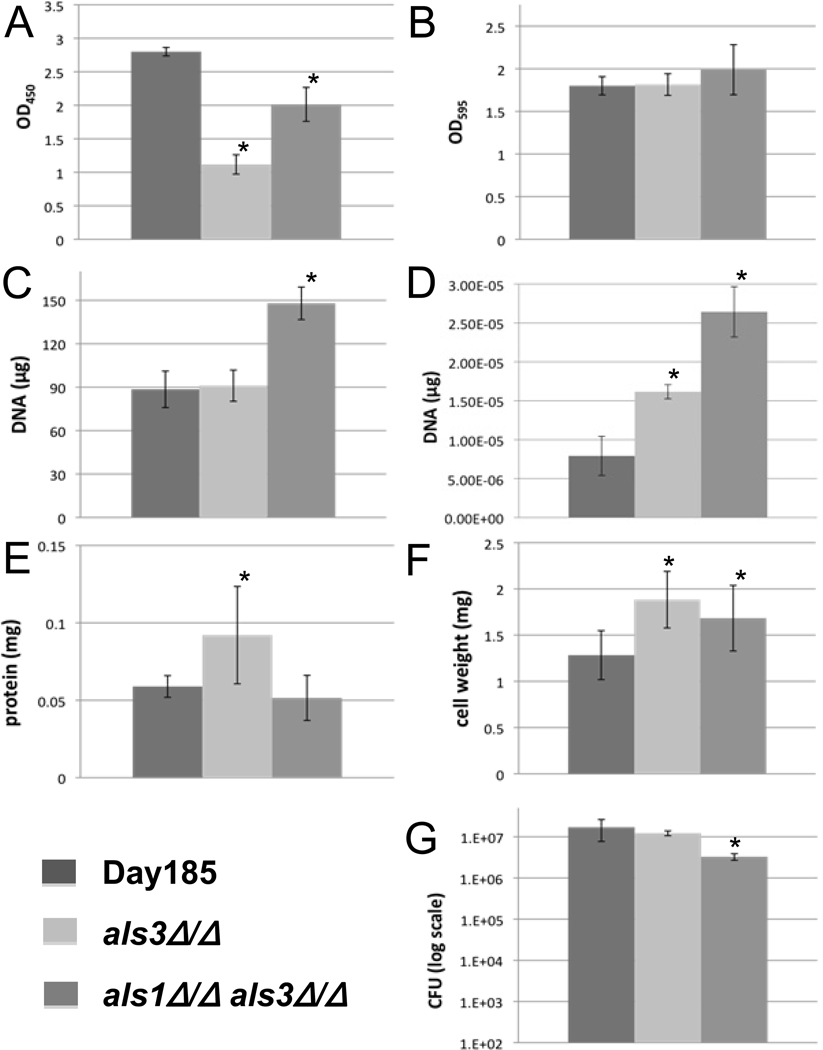
The XTT assay accurately detected the observed differences in the extent of biofilm formation between the parent and the two biofilm deficient strains (Fig. 2A). In addition, the measurements were reproducible. However, the assay did not differentiate between the more modest differences in the biofilms of the two mutants. Although reproducible, the crystal violet assay did not detect the anticipated differences in the biofilms among the strains (Fig. 2B). Total DNA quantification and ACT1 qPCR failed to detect the biofilm defects of the mutants (Fig. 2C and 2D). In fact, a higher concentration of DNA was detected for the poor biofilm forming mutants for reasons that are unclear. For the total cellular protein assay, we observed significant variability in assay replicates and the measurements did not accurately reflect the differences in the extent of biofilms among strains (Fig. 2E). [Fig. 2 near here]
Fig. 2.
Results of seven in vitro biofilm quantification protocols. Each graph represents data of the mean of individual biofilms from a 6-well plate unless otherwise specified. All error bars represent the standard deviations. An asterisk (*) indicates a significant difference between the mutant and parent strain, with a P < 0.05 using ANOVA. (A) and (B) OD490 and OD595 of biofilms using the XTT reduction assay and crystal violet staining, respectively. Six-well bars are the overall average of seven (A) or eight (B) replicates each from six individual wells of biofilm. (C) Biofilm quantification via DNA extraction and quantification. Each bar represents the mean of three extractions. (D) qPCR results measuring the Act1 gene. Bars represent the mean of three DNA extractions, amplified by qPCR each in triplicate. (E) Quantification of the protein in each Candida strain. Bars represent the mean of five wells, each done in triplicate. (F) Dry cell weight of biofilms. The higher values for mutants in E and F were possibly due to the values being near their protocols’ limits of detection. (G) Biofilm quantification by cell plating and viable colony counting. Each bar represents the overall average of four dilutions from each of five wells, each done in triplicate.
Although the viable counts were reproducible for an individual strain, the degree of the expected biofilm defects of the mutants was not appreciated using this method (Fig. 2F).
Among the assays tested, the most time consuming included the total DNA and qPCR methods (Table 1). The most costly technique was the qPCR. The least expensive viable option both from the standpoint of time and supplies was the XTT assay. [Table 1 near here]
Table 1.
Summarization of the various factors influencing each quantification protocol
| Accuracy | Reproducibility | Time | Cost | |
|---|---|---|---|---|
| XTT | +++ | ++++ | + | ++ |
| Crystal Violet | + | ++++ | ++ | + |
| CFU | ++ | ++++ | +++ | ++ |
| DNA | − | +++ | ++ | +++ |
| Amplification | − | ++ | +++ | ++++ |
| BCA Protein | + | + | + | + |
| Dry Cell Weight | − | ++ | ++ | + |
Accuracy represents whether the data matches with the pattern seen in SEM images, as well as whether the differences between the strains was significant (P < 0.05). Reproducibility is determined by the coefficient of variation (CV) for each set of data. ++++ = CV <0.1, +++ = CV of 0.1–0.15, ++ = CV of 0.15–0.2, + = CV of >0.2. Time accounts for both the total lengths of the protocols, and the amount of labor required for each, with ++++ representing the most time consuming protocols. Cost is based on the amount needed per 6-well plate, and presumes that researchers already have access to any large devices required. ++++ = >$100 per plate, +++ = $20–100, ++ = $10–20, + = <$10.
Discussion
There are several possible explanations for the variability observed with several of the assays. First, those that require removal of the adherent biofilms (total DNA, qPCR, total protein, and viable burden) may create inconsistent cell collection. We have confirmed this hypothesis by performing XTT assays on disrupted biofilms and finding that up to 30% may remain in the well despite multiple washes and the appearance that the films were removed (data not shown). Secondly, for several of the assays (total protein and cell weight assays) the measured values for biofilms in the 6-well format were small relative to the limit of detection.
For the C. albicans strains tested, we found the XTT assay to be the most reproducible, accurate, and efficient method for measurement of biofilm extent. Measurement of viable burden was also a fairly reproducible method of quantifying biofilm formation.
Acknowledgements
The authors would like to thank A. Mitchell and C. Nobile for the strains. This work was supported by the National Institutes of Health (RO1 AI073289-01).
Footnotes
Declaration of interest: The authors declare that they have no conflict of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Douglas LJ. Candida biofilms and their role in infection. TIM. 2003;11:30–36. doi: 10.1016/s0966-842x(02)00002-1. [DOI] [PubMed] [Google Scholar]
- 2.Kojic EM, Darouiche RO. Candida infections of medical devices. CMR. 2004;17:255–267. doi: 10.1128/CMR.17.2.255-267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumamoto CA, Vinces MD. Alternative Candida albicans lifestyles: growth on surfaces. Annu Rev Microbiol. 2005;59:113–133. doi: 10.1146/annurev.micro.59.030804.121034. [DOI] [PubMed] [Google Scholar]
- 4.Mermel LA, Allon M, Bouza E, et al. Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis. 2009;49:1–45. doi: 10.1086/599376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramage G, Martinez JP, Lopez-Ribot JL. Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res. 2006;6:979–986. doi: 10.1111/j.1567-1364.2006.00117.x. [DOI] [PubMed] [Google Scholar]
- 6.Khot PD, Suci PA, Tyler BJ. Candida albicans viability after exposure to amphotericin B: assessment using metabolic assays and colony forming units. J Microbiol Meth. 2008;72:268–274. doi: 10.1016/j.mimet.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Nett J, Lincoln L, Marchillo K, et al. Putative role of beta-1,3 glucans in Candida albicans biofilm resistance. Antimicrob Agents Chemother. 2007;51:510–520. doi: 10.1128/AAC.01056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nobile CJ, Andes DR, Nett JE, et al. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Path. 2006;2:0636–0649. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramage G, Mowat E, Jones B, Williams C, Lopez-Ribot J. Our current understanding of fungal biofilms. Crit Rev Microbiol. 2009;35:340–355. doi: 10.3109/10408410903241436. [DOI] [PubMed] [Google Scholar]
- 10.Ramage G, Vande Walle K, Wickes BL, Lopez-Ribot JL. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob Agents Chemother. 2001;45:2475–2479. doi: 10.1128/AAC.45.9.2475-2479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandra J, Mukherjee PK, Ghannoum MA. In vitro growth and analysis of Candida biofilms. Nat Protoc. 2008;03:1909–1924. doi: 10.1038/nprot.2008.192. [DOI] [PubMed] [Google Scholar]
- 12.Gobor T, Corol G, Ferreira LE, et al. Proposal of protocols using D-glutamine to optimize the 2,3-bis(2-methoxy-4-nitro-5-sulfophenly)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide (XTT) assay for indirect estimation of microbial loads in biofilms of medical importance. J Microbiol Meth. 2010;84:299–306. doi: 10.1016/j.mimet.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 13.Hawser SP, Douglas LJ. Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect Immun. 1994;62:915–921. doi: 10.1128/iai.62.3.915-921.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honraet K, Goetghebeur E, Nelis HJ. Comparison of three assays for the quantification of Candida biomass in suspension and CDC reactor grown biofilms. J Microbiol Meth. 2005;63:287–295. doi: 10.1016/j.mimet.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 15.Kuhn DM, Balkis M, Chandra J, Mukherjee PK, Ghannoum MA. Uses and limitations of the XTT assay in studies of Candida growth and metabolism. J Clin Microbiol. 2003;41:506–508. doi: 10.1128/JCM.41.1.506-508.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nailis H, Coenye T, Van Nieuwerburgh F, Deforce D, Nelis HJ. Development and evaluation of different normalization strategies for gene expression studies in Candida albicans biofilms by real-time PCR. BMC Mol Biol. 2006;7:1471–2199. doi: 10.1186/1471-2199-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peeters E, Nelis HJ, Coenye T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J Microbiol Meth. 2008;72:157–165. doi: 10.1016/j.mimet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 18.Silva S, Henriques M, Martins A, et al. Biofilms of non-Candida albicans Candida species: quantification, structure and matrix composition. Med Mycol. 2009;47:681–689. doi: 10.3109/13693780802549594. [DOI] [PubMed] [Google Scholar]
- 19.Davis D, Wilson RB, Mitchell AP. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol Cell Biol. 2000;20:971–978. doi: 10.1128/mcb.20.3.971-978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Filler SG. Candida albicans Als3: a multifunctional adhesin and invasin. Eukaryot Cell. 2011;10:168–173. doi: 10.1128/EC.00279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nobile CJ, Schneider HA, Nett JE, et al. Complementary adhesin function in C. albicans biofilm formation. Curr Biol. 2008;18:1017–1024. doi: 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X, Daniels KJ, Oh SH, et al. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology. 2006;152:2287–2299. doi: 10.1099/mic.0.28959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andes D, Nett J, Oschel P, et al. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun. 2004;72:6023–6031. doi: 10.1128/IAI.72.10.6023-6031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin Y, Yip HK, Samaranayake YH, Yau JY, Samaranayake LP. Biofilm-forming ability of Candida albicans is unlikely to contribute to high levels of oral yeast carriage in cases of human immunodeficiency virus infection. J Clin Microbiol. 2003;41:2961–2967. doi: 10.1128/JCM.41.7.2961-2967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uppuluri P, Perumal P, Chaffin WL. Analysis of RNA species of various sizes from stationary-phase planktonic yeast cells of Candida albicans. FEMS Yeast Res. 2007;7:110–117. doi: 10.1111/j.1567-1364.2006.00143.x. [DOI] [PubMed] [Google Scholar]