Abstract
Peroxisomes produce reactive oxygen species (ROS), which may participate in biotransformations of innate biomolecules and xenobiotics. Isolating functional peroxisomes with low levels of contaminants would be a useful tool to investigate biotransformations occurring in these organelles that are usually confounded with biotransformations occurring in other co-isolated organelles. Here we immunoisolate peroxisomes, demonstrate that the impurity level after isolation is low and that peroxisomes retain their biological activity. In this method, an antibody targeting a 70 kDa peroxisomal membrane protein was immobilized to silanized magnetic iron oxide beads (1–4 μm in diameter) coated with Protein A. Peroxisomes from L6 rat myoblast homogenates were magnetically captured, washed and then analyzed for subcellular composition using enzymatic assays. Based on the ratio of peroxisomal to lysosomal activity, the retained fraction is 70-fold enriched relative to the unretained fraction. Similarly, the ratio of peroxisomal activity to mitochondrial content suggests that the retained fraction is >30-fold enriched relative to the unretained fraction. H2O2 production from the β-oxidation of palmitoyl-CoA demonstrated that the isolated peroxisomal fraction was biologically active. Capillary electrophoresis with laser-induced fluorescence detection (CE-LIF) analysis confirmed that the immunopurified fractions were capable of transforming the anti-cancer drug doxorubicin and the fatty acid analog, BODIPY 500/510 C1C12. Besides its use to investigate peroxisome biotransformations in health and disease, the combination of magnetic immunoisolation with CE-LIF could be widely applicable to investigate subcellular specific biotransformations of xenobiotics occurring at immunoisolated subcellular compartments.
Keywords: Peroxisome, fatty acid, doxorubicin, magnetic, MEKC, fluorescence, LIF
1. Introduction
Peroxisomes are organelles involved in drug metabolism and physiological detoxification.[1] About 50 enzymes identified in peroxisomes are responsible for these processes.[1] The metabolic functions of peroxisomes include balancing cellular reactive oxygen species (ROS),[2] α- and β-oxidation of fatty acids,[3] and biosynthesis of bile acids, docosahexaenoic acids and ether glycerolipids.[4] The deficiencies in peroxisomal function lead to inherited diseases such as Zellweger syndrome.[5] Peroxisomes are also involved in the metabolism of diverse drugs and xenobiotics such as analgesics, anti-inflammatories, nutritional ingestants, insecticides and plasticisers,[6] suggesting that they play an important role in the metabolism of drugs and xenobiotics.
To investigate the peroxisomal-specific biotransformation of drugs and xenobiotics, highly enriched peroxisomes free from interfering organelles such as mitochondria and lysosomes are necessary. The purifications of peroxisomes are usually done by differential or density gradient centrifugation or a combination of both.[7] However, the purity of the peroxisomes obtained by differential centrifugation is usually low because the sedimentation characteristics of peroxisomes are close to that of mitochondria and lysosomes. A considerable amount (~23%) of peroxisomes was detected in a light mitochondria fraction obtained by differential centrifugation at 25,300g for 20 min.[8] In density gradient centrifugation, the biological function of peroxisomes is impaired due to the long centrifugation time (e.g., up to 16 hours) and density gradient media such as sucrose, which exert osmotic pressure at the high-density end of the gradient. It is necessary to develop purification methods that can isolate peroxisomes with high selectivity and functional integrity. Luers et al. reported the immunoisolation of peroxisomes with an antibody conjugated to magnetic beads.[9] Kikuchi used this method to isolate peroxisomes for peroxisomal protein profiling.[10] There are no previous reports describing the combination of immunoisolated peroxisomes with capillary electrophoretic techniques to investigate subcellular specific biotransformations of drugs and xenobiotics such as doxorubicin (DOX).
DOX widely used in treating solid tumors and leukemia[11] is susceptible to oxidation by H2O2-activated peroxidases[12] or by direct exposure to H2O2.[13] Capillary electrophoresis (CE) with laser-induced fluorescence detection (LIF) has been used to analyze both DOX and its metabolites in plasma,[14, 15] cells,[16] subcellular fractions,[17, 18] and individual organelles.[19, 20] The requirement of small volume (i.e., nanoliters) of samples by CE and the high sensitivity of LIF detection make it possible to analyze fluorescent analytes in limited sample volumes such as those obtained from immunoisolated organelles.
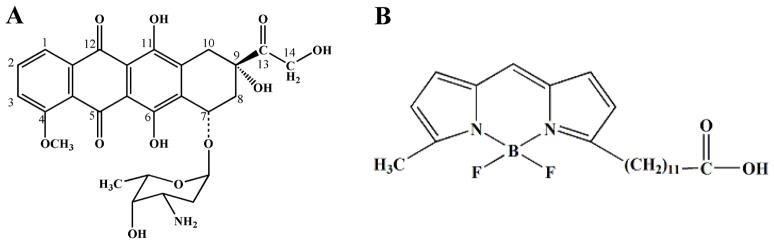
In this report, we immunoisolate peroxisomes from L6 rat myoblasts with low levels of contaminating organelles. The isolated peroxisomes were bioactive as confirmed by monitoring H2O2 production from β-oxidation of palmitoyl-CoA. The biotransformations of an anthracycline, DOX (Figure 1A) and a fluorescent fatty acid analog, BODIPY 500/510 C1C12 (B12FA, Figure 1B) in these organelles were monitored by micellar electrokinetic chromatography (MEKC), a mode of CE, with LIF detection (i.e., MEKC-LIF). These results confirmed that there is a modest biotransformation of DOX and fluorescent fatty acid analogs in the immunoisolated peroxisomes.
Figure 1.
Structures of B12FA and DOX
2. Material and methods
2.1 Chemicals and reagents
MagnaBind Protein A Beads and BCA kit were purchased from Thermo Fisher Scientific (Rockford, IL). Anti-PMP70 antibody (produced in rabbit), coenzyme A trilithium salt (CoA), β-Nicotinamide adenine dinucleotide (NAD), flavin adenine dinucleotide disodium salt hydrate (FAD), palmitoyl coenzyme A (palmitoyl-CoA), horseradish peroxidase (Type I), protease inhibitor cocktail, gentamicin, imidazole, Triton X-100, titanium (IV) oxysulfate and p-nitrophenyl-N-acetyl-β-D-glucosamide were purchased from Sigma-Aldrich (St. Louis, MO). BODIPY® 500/510 C1C12, BODIPY® FL C3, BODIPY® FL C5 and BODIPY® FL C11 (abbreviated as B12FA, B3FA, B5FA and B11FA, respectively; see structures in Figure S1 in the Electronic Supplementary Material), 10-N-nonyl-Acridine Orange (NAO), fluorescein, Amplex Red reagent, Dulbecco’s modified Eagle’s medium (DMEM), trypsin (5.0 g/L, 10×)-EDTA (2.0 g/L) and bovine serum were purchased from Invitrogen (Carlsbad, CA). Adenosine-5′-triphosphate (ATP) was from Roche Scientific (Indianapolis, IN). Doxorubicin hydrochloride was a generous gift from Meiji Seika Kaisha Ltd. (Tokyo, Japan). Sodium dodecyl sulfate (SDS), sodium citrate, sodium phosphate monobasic and dibasic, sodium carbonate, sodium bicarbonate, sulfuric acid (H2SO4), hydrochloric acid (HCl) and hydrogen peroxide (H2O2, 30% solution) were purchased from Mallinckrodt Chemicals (Phillipsburg, NJ). Sodium borate decahydrate, sodium hydroxide (NaOH) and sucrose were from Fisher Scientific (Fair Lawn, NJ). γ-cyclodextrin was purchased from TCI America (Portland, OR). 10× phosphate-buffer saline (1.37 M NaCl, 14.7 mM KH2PO4, 78.1 mM Na2HPO4, and 26.8 mM KCl) was purchased from EMD Chemicals (Gibbstown, NJ). Rat IgG control (Catalogue # NBP1-71666) was purchased from Novus Biologicals (Littleton, CO). Mouse IgG1 control (Catalogue # ALX-804-870TD-C050) was purchased from Enzo Life Sciences (Farmingdale, NY).
The cell homogenization buffer was made of 250 mM sucrose, 3 mM imidazole, 1 mM EDTA and 1% protease inhibitor cocktail, pH 7.4. The binding buffer used in immunoisolation was 100 mM sodium phosphate buffer, pH 8.0. The MEKC separation buffer for DOX and metabolites contained 10 mM borate, 10 mM SDS, pH 9.3 (BS10). The MEKC separation buffer for B12FA and metabolites contained 30 mM borate, 30 mM SDS, 5 mM γ-cyclodextrin, pH 9.3 (BS30-γCD5). All the buffers were made using 18 MΩ water purified from a Millipore water purification system (Millipore, Billerica, MA) and the pH was adjusted with 0.1 M HCl or NaOH.
The stock solutions of DOX and BODIPY fatty acid analogs were prepared by dissolving them in methanol to a concentration of 1 mM.
2.2 Cell culture and sample preparation
L6 myoblasts (ATCC, Manassas, VA) were cultured in DMEM containing 10% (v/v) bovine serum and 10 μg/ml gentamicin in 75-cm2 vented culture flasks at 37 °C and 5% CO2. The cells were split every 3 to 4 days before they reached confluence. For splitting, the cells were rinsed with PBS, lifted with 5 ml 0.25 g/L trypsin and diluted in fresh growth medium to 20 ml.
In sample preparation, L6 cells were pelleted by centrifugation at 1000g for 10 min, washed twice with 1×PBS and reconstituted in the homogenization buffer to a cell density of 2×107 cells/ml. The cells were then disrupted by 30 strokes in an ice-chilled Dounce homogenizer (0.00025″ clearance, Kontes Glass, Vineland, NJ). Trypan blue stain was used to ensure that more than 95% of the cells were disrupted. The cell homogenate was centrifuged at 600g for 10 min to remove unbroken cells, nuclei and cell debris. The post nuclear fraction (PNF) was used for immunoisolation of peroxisomes.
2.3 Immunoisolation of peroxisomes
Protein A beads were first incubated in binding buffer containing 1% BSA for 1 h to reduce non-specific binding. An anti-PMP70 antibody or a control (rat IgG or mouse IgG1) antibody was then bound to the protein A beads (4×10−10 mg IgG per bead) at 4 °C for 1 h while the mixture was gently vortexed. After incubation, the antibody-coated beads were washed three times with binding buffer to remove any unbound antibody. PNF was added to the coated beads at a ratio of 25 beads per cell and incubated at 4 °C for 1 h. The supernatant (unretained fraction) containing cytosol and unbound organelles was removed and saved for further analysis; the beads with bound peroxisomes (retained fraction) was washed 3 times in binding buffer and reconstituted in homogenization buffer.
2.4 Protein quantification and enzyme assays
Protein amounts in the retained and unretained fractions were quantified with a BCA kit according to the manufacture’s protocol. For the retained fraction, the proteins were first eluted from the beads with 10% SDS and the eluent was used for protein quantification. The difference in protein contents of antibody-coated beads with and without PNF exposure was used to determine the amount of PNF protein that is not specifically adsorbed to the beads. Non-specifically bound PNF protein was always present regardless of antibody used. For instance, in Trial 1 the non-specifically bound PNF protein was 0.26±0.02 and 0.26±0.04 μg/μl of bead suspension for rabbit anti-PMP70 and mouse IgG1, respectively; in Trial 2 the non-specifically bound PNF protein was 0.11±0.03 and 0.15±0.03 μg/μl of bead suspension for rabbit anti-PMP70 and rat IgG, respectively. These results show that protein amounts are not sufficient to assess specific binding and that organelle specific markers (i.e. enzymatic assays) are needed to characterize the isolation of peroxisomes.
Catalase and β-hexosaminidase assays were used to assess the enzyme activities of retained and unretained fractions. The catalase assay, an peroxisomal enzyme activity assay, was carried out according to Storrie’s protocol.[22] Briefly, H2O2 (11.7 mM) in 2 mM imidazole buffer containing 5% BSA and 0.2% Triton X-100 was incubated with both fractions for 10 min at room temperature. After incubation, titanium (IV) oxysulfate in 0.5 M H2SO4 was added to form a yellow complex with the unreacted H2O2. The absorbance of the complex was measured at 405 nm with a Synergy 2 plate reader (Biotek, Winooski, VT). Catalase activity was reported as the rate of decomposition of H2O2 (μmol/min). The β-hexosaminidase assay was used to assess the lysosomal activity in both retained and unretained fractions. The assay was modified from that described by Barret.[23] Both fractions were mixed with p-nitrophenyl-N-acetyl-β-D-glucosamine (7.5 mM) in citrate buffer (0.3 M sodium citrate and 0.3 M NaCl, pH 4.3) and incubated at 37 °C for 45 min. After incubation, terminating solution (0.5 M Na2CO3 and 0.5 M NaHCO3) was added to the mixture to stop the reaction. The absorbance of the product, p-nitrophenol, was measured at 410 nm. β-hexosaminidase activity was reported as the rate of formation of p-nitrophenol (μmol/min).
The abundance of mitochondria in both fractions was determined by NAO labeling. NAO is a fluorescent marker labeling cardiolipin in the mitochondria inner membrane.[24] In this assay, cells was labeled with 5 μM NAO on ice for 10 min and washed twice with 1×PBS before homogenization. After immunoisolation, the unretained fraction was centrifuged at 16,000g and the resulting pellet was dissolved in 10% SDS. The retained fraction was eluted from the beads with 10% SDS. The fluorescence intensities of dissolved pellet (unretained fraction) and eluent (retained fraction) were measured with a Synergy 2 plate reader (λex = 480 nm and λem = 528 nm). The abundance of mitochondria was reported in μmole of NAO.
The ratios of catalase activity to β-hexosaminidase activity and catalase activity to NAO amount were used to determine the relative enrichment of peroxisomes in the various fractions. Enzymatic activities or NAO abundance were not converted to specific activities (i.e. activity/mg protein) to avoid complications that may result from non-selective protein absorption observed in the protein A-antibody conjugates. The null hypothesis “there is no significant difference of ratios (mentioned above) between the retained and unretained fractions” was tested by Student’s t-test using statistical software “R”. The null hypothesis was rejected when the p-value (p) <0.02 (98% confidence level).
2.5 Amplex Red H2O2 assay
After peroxisomes were immunoisolated, their β-oxidation activity was assessed by measuring H2O2 resulting from oxidation of palmitoyl-CoA using the Amplex Red reagent. The retained fractions were incubated with Amplex Red reagent (50 μM), horseradish peroxidase (2 units/ml) and palmitoyl-CoA (10 μM). A preparation not containing peroxisomes was used as a control. After 30 min incubation at room temperature, the fluorescence intensities of the mixtures were measured with a Synergy 2 plate reader (λex = 530 nm and λem = 590 nm).
2.6 In vitro metabolism of B12FA and DOX in isolated peroxisomes
After immunoisolation, the retained and unretained fractions were incubated with 5 μM B12FA, and cofactors for β-oxidation, i.e., 0.5 mM Coenzyme A, 1mM NAD, 0.5 mM FAD and 5 mM ATP at 37 °C.[25] Aliquots was removed from the incubation mixture at 15, 30, 60 and 120 min and frozen at −80 °C until MEKC analysis.
For DOX biotransformations, after immunoisolation the retained and unretained fractions were incubated with DOX (10 μM), palmitoyl-CoA (0.5 mM), FAD (0.5 mM) and NAD (1 mM) at 37 °C. Palmitoyl-CoA was added to induce the production of H2O2.[26] FAD and NAD were added as cofactors for peroxisomal β-oxidation of palmitoyl-CoA.[27] Samples from the reaction mixture were taken at 15, 30, 60 and 120 min and frozen at −80 °C until MEKC analysis.
2.7 MEKC analysis of B12FA or DOX in vitro metabolites
The MEKC-LIF separation of DOX and metabolites was reported previously,[16] while the separation system of B12FA used a new buffer system containing borate, SDS and γ-cyclodextrin.
After incubation with xenobiotics, both the retained and unretained fraction were directly diluted with the MEKC-LIF separation buffer. The unretained fraction did not require further preparation prior to MEKC-LIF analysis, while the retained fraction required gently mixing to dissolve the retained material and removal of the magnetic beads with a magnet. Samples were introduced into a fused silica capillary (50 μm I.D. and 150 μm O.D., Polymicro Technologies, Phoenix, AZ) by hydrodynamic injection at 10.8 kPa for 1 s. Then the capillary was then brought into a vial containing separation buffer and MEKC was performed under a +400 V/cm electric field in a custom-built instrument equipped with post-column LIF detection, previously described.[18] Briefly, a sheath flow cuvette encased the detector end of the capillary. The last 2-mm of this end had the polyimide coating burned off to reduce the background fluorescence caused by this material. As fluorescent analytes migrated out from the capillary, they were excited at 488 nm with an argon ion laser (JDS Uniphase, San Jose, CA). Fluorescence was collected at a 90° angle with respect to the laser beam by a 60×, N.A. 0.7 microscope objective (Universe Kogaku, Inc., Oyster Bay, NY). A 505 nm long-pass filter (505 AELP, Omega Optical, Brattleboro, VT) and a 1.4 mm pinhole were used to reduce light scattering. A 635 ± 27.5 nm (XF3015) or a 520 ± 17.5 nm (XF3007) band-pass filter (Omega Optical, Brattleboro, VT) was used to select fluorescence from DOX and its products or B12FA and its products, respectively. Fluorescence was then detected in a photomultiplier tube (PMT) (Hamamatsu, Bridgewater, NJ) biased at 1000 V. The PMT output was sampled at 10 Hz and processed with a Labview program (National Instruments, Austin, TX).
Daily, prior any MEKC-LIF experiment, the capillary was conditioned with sequential flushes of 0.1 mM NaOH, water and separation buffer for 30 min each using 150 kPa pressure at the inlet. For conditioning capillaries between samples, the same conditioning sequence was followed, but for the flushes lasted only 2 min each. The separation buffer was replaced every 2 h with a new vial to avoid buffer contamination and electrolyte depletion which causes migration time drift.[28] Prior to sample analysis, the instrument was aligned by maximizing the response of 5 ×10−10 M fluorescein that continuously flowed through the detector while applying a +400 V/cm electric field.
2.8 MEKC data analysis
Electropherograms were processed using Igor Pro (Wavemetrics, Lake Oswego, OR) to determine peak intensities and areas.
In the study of DOX transformation, corrected mobilities were used to compare peaks in retained and unretained fractions. The correction was done according to the two-marker method described by Li et al.[29] that uses the expression
| (Equation 1) |
| (Equation 2) |
where tX,corrected is the corrected migration time of a peak X in a sample and t̂x is its observed migration time; t̂m1 and t̂m2 are the migration times of the markers, and tm1 and tm2 are the migration times of the markers in the reference run. The corrected mobility of the peak X was calculated by
| (Equation 3) |
where L is the length of the capillary, and V is the separation voltage.
The corrected mobilities of the peaks in the retained and unretained fractions were compared using Student’s t-test. The null hypothesis “there is no significant difference in the mobility of peak X in the retained and unretained fractions” was rejected when the p-value <0.02 (98% confidence level).
3. Results and Discussion
3.1 Purity of the immunoisolated peroxisomes
Catalase and β-hexosaminidase are established enzyme markers of peroxisomes and lysosomes, respectively.[22, 30] We measured their activities in the retained and unretained fractions to determine the relative abundance of their respective organelles. NAO, which is a mitochondrion-specific fluorescent probe, labels cadiolipins in the mitochondria inner membrane.[24] We measured NAO fluorescence intensity in the retained and unretained fractions to determine their relative mitochondrial contents. Conventional mitochondrial enzymatic assays could not be used because the magnetic beads used for immunoisolation interfered with these assays. Western blots, commonly used to assess subcellular composition, were not suitable because (1) they are less quantitative than enzymatic assays and (2) the antibodies used for immunopurification displayed bands that interfered with the reliable identification of the subcellular targets (data not shown). This problem is particularly critical when the same antibody (anti-PMP70 antibody) is used in both immunoisolation and Western blots. [31]
Table 1 shows the relative enrichment of peroxisome in various fractions. The ratios of peroxisomal catalase activity to the activity of lysosomal marker enzyme, β-hexosaminidase, increases ~ 60-fold in the retained fraction compared to the unretained one and PNF, indicating a significant enrichment of the peroxisome in the immunoisolation procedure. The mitochondrial marker, NAO is not detected in the enriched fraction indicating the mitochondrial contamination is minimal. The catalase activity in unretained fraction suggests that the bead-antibody conjugates are not sufficient to capture all peroxisomes in the cell homogenate. This is not critical in our studies that focus on the immunoisolation of peroxisomes and not on their recovery yield. Based on these results, the immunoisolated peroxisomes have enough purity to study peroxisomal biotransformations.
Table 1. Relative catalase activities.
Results are the average ± standard deviation (n=3). The Catalase/β-hexosaminidase ratio is dimensionless. The Catalase/NAO ratio is given as μmole H2O2/min•μmole NAO
| Catalase/β-hexosaminidase | Catalase/NAO | |
|---|---|---|
| PNF | 6.1 ± 0.6 | (8 ±2)×102 |
| Unretained | 5.4 ± 0.2 | (8±1)×102 |
| Retained | 383 ± 24 | >2.6×104* |
NAO in the retained fraction is below the LOD of the NAO assay which is 7.16×10−11 M of NAO. The calculation is based on the LOD of the NAO assay.
3.2 Peroxisomal bioactivity
The major process for breaking down fatty acids is β-oxidation which can occur both in peroxisomes and mitochondria.[32] One major difference is that in peroxisomes, β-oxidation of (activated) fatty acids is accompanied by the formation of H2O2,[26] which is a marker of peroxisomal bioactivity. In this study we monitored H2O2 formation upon addition of palmitoyl-CoA (activated fatty acid) with the fluorogenic reagent, Amplex Red.[33] Amplex Red is oxidized by H2O2 to its fluorescence product, resorufin in the presence of peroxidase. The LOD of H2O2 in this study was (4.0 ± 0.8) ×10−9 M (n = 3). Without addition of palmitoyl-CoA, retained peroxisomal fractions had low H2O2 production (0.35 ± 0.09 nM/min, n = 3). With the addition of palmitoyl-CoA, H2O2 production in the retained peroxisomal fraction increased seven-fold (2.4 ± 0.1 nM/min, n = 3). These results suggest that the retained peroxisomal fraction maintains the β-oxidation activity, which is indicative of biofunctional peroxisomes.
3.3 Peroxisomal biotransformations of the fatty acid analog B12FA
The most common biotransformation of fatty acids in peroxisomes is β-oxidation, which shortens the alkyl chain by two carbons each cycle thereby forming a shorter CoA derivative (c.f. Figure 2). In order to explore fatty acid biotransformations in the retained peroxisomal fraction we selected B12FA as fatty acid model molecule. The fluorophore BODIPY in this model molecule is lipophilic and partitions into the cell membrane easily.[34] Furthermore, the fluorophore is on the C1 position of B12FA, away from the carboxylic acid group which decreases possible perturbations on fatty acid processing.[35]
Figure 2.
First cycle of the β-oxidation of B12FA. B12FA is first activated to the acyl-CoA ester (compound A). The acyl-CoA ester then undergoes β-oxidation that shortens the fatty acid chain by two carbons (compound B).
Because CE-LIF has been previously used to analyze fluorescent fatty acid derivatives,[36] here we designed an MEKC-LIF separation to monitor peroxisomal biotransformations of B12FA. The best separation buffer was BS30-γCD (30 mM borate, 30 mM SDS and 5 mM γ-cyclodextrin, pH 9.3) that provided excellent resolution in the separation of four BODIPY standards B3FA, B5FA, B11FA and B12FA (Figure S1 in the Electronic Supplementary Material).
Incubation of the retained or the unretained fractions with B12FA, Coenzyme A, NAD, FAD and ATP for 2 hours did not show formation of any free fatty acid analogs with shorter alkyl chains (Figure 3, Traces b and c). Compared to the control (Figure 3, Trace a), two changes were observed: one new peak (Peak 3) appeared and the B12FA peak (Peak 1) intensity increased with incubation time (Table 2).
Figure 3.
In vitro biotransformation of B12FA in retained peroxisomal and unretained fractions. Trace a: Control containing B12FA (5 μM), CoA (0.5 mM), FAD (0.5 mM), NAD (1 mM) and ATP (5 mM). Trace b: Unretained fraction incubated with the reaction mixture used in Trace a. Trace c: Retained peroxisomal fraction incubated with the reaction mixture used in Trace a. Incubations were done for 2 h at 37 °C. Traces b and c were y-axis offset for clarity. The MEKC separations were performed in a 43.3 cm fused silica capillary under a 400 V/cm electric field in a buffer containing 30 mM borate, 30 mM SDS and 5 mM γ-cyclodextrin (pH 9.3). Samples were injected at 10.8 kPa for 1 s. For LIF detection, fluorescence was selected by a 520 ± 17.5 nm bandpass filter. Insert is the enlargement of the electropherogram in 400 – 550 s range.
Table 2.
Peak areas in MEKC-LIF electropherograms of retained peroxisomal and unretained fractions. A.U.: Arbitrary Units. For peak assignments and the reaction mixture composition see Figure 3 caption. Peak 1 corresponds to B12FA in the control, but may overlap with other compounds formed by the retained or the unretained fraction. Results are represented in average ± standard deviation (n=3 replicate MEKC-LIF runs of the same mixture).
| Peak 1 Area (A.U.) | Peak 2 Area (A.U.) | Peak 3 Area (A.U.) | ||||
|---|---|---|---|---|---|---|
| Incubation time | 30 min | 2 h | 30 min | 2 h | 30 min | 2 h |
| control | 9.7±0.3 | 9.7±0.3 | 0.33±0.03 | 0.33±0.03 | - | |
| retained | 10.0±0.1 | 14.2±0.2 | 0.34±0.02 | 0.40±0.10 | 0.26±0.04 | 0.27±0.01 |
| unretained | 10.9±0.1 | 17.6±0.7 | 0.36±0.08 | 0.54±0.02 | 0.60±0.10 | 0.78±0.03 |
The fact that Peak 3 migrates more slowly than the B12FA peak in the MEKC separation may imply that this peak corresponds to a more hydrophobic product, larger size, decreased net charge, or a combination of these factors. This product could either be Coenzyme A fatty acid esters with different alkyl chains lengths resulting from β-oxidation of B12FA or triacylglycerol synthesis from B12FA. The second possibility is supported by previous observations that indicate large accumulation of triacylglycerol in L6 cells cultured in fatty acid-supplemented growth medium.[37] We also observed peaks that migrate after the B12FA peak when L6 myoblasts were cultured in medium containing B12FA (Figure S2 in the Electronic Supplementary Material), supporting this possibility.
The increase in the area of Peak 1 (Table 2) suggests that B12FA is biotransformed into other compounds that might have enhanced fluorescence resulting from variations in the molecular environment where BODIPY is found.[38] Such co-migrating species may include Coenzyme A fatty acid esters with different alkyl chains lengths resulting from β-oxidation of B12FA or triacylglycerol synthesis from B12FA. Identifying conditions that separate unknown co-migrating species is better accomplished if the species are indentified first and then pure standards are used to test new separation conditions. Although it is beyond the scope of this report, further identification of the products comigrating with the B12FA peak or formed as Peak 3 would require the use of complementary techniques such as HPLC-(LIF)-MS[39] that has been used to detect free fatty acids[40] and fatty acyl-CoAs[41, 42] in cell and tissue extracts.
The relevance of the similarity of the changes observed in both the peroxisomal retained and unretained fraction cannot be left unnoticed. The immunoisolation procedure described here is effective at removing mitochondrial contaminants from the retained fraction, while peroxisomal activity is detected along with mitochondrial and lysosomal activities in the unretained fractions (c.f. Table 1). The changes in the electropherograms of the biotransformations observed in the unretained fraction clearly exemplify current limitations at investigating subcellular biotransformations when more than one type of organelles may have similar biotransformation pathways. For instance, β-oxidation of fatty acids and fatty acid analogs (e.g., 12-(1-pyrene)dodecanoic acid) occurs both at mitochondria and peroxisomes.[43] In contrast, the MEKC-LIF analysis of the peroxisomal retained fraction reported here provides peroxisomal specific biotransformations of B12FA.
3.4 Peroxisomal metabolism of DOX
H2O2 is the major byproduct of the oxidation of activated fatty acids in peroxisomes.[26] This reaction was used to confirm that immunoisolated peroxisomes mixed with palmitoyl-CoA produce H2O2 (c.f. Section 3.2). The oxidation of the anti-cancer drug DOX by H2O2-activated peroxidases[12] or H2O2 alone[13] (see Figure S3 in the Electronic Supplementary Material) suggests that DOX and other anthracyclines may be biotransformed in peroxisomes.
In this study, the MEKC-LIF analysis of retained or unretained fractions incubated with DOX, revealed new DOX-associated peaks (Figure 4). Even when the MEKC-LIF electropherogram of the control shows a complex background, which is attributed to FAD, NAD and other impurities in the mixture (Figure 4, Trace a), the corresponding electropherograms for the DOX treated fractions show distinctive peaks (indicated by arrows in Traces b and c of Figure 4). After correcting their migration times with the equations 1 and 2 and calculating their electrophoretic mobilities with equation 3, peaks r1 and u1 in the retained peroxisomal and unretained fractions, respectively, have statistically different electrophoretic mobilities (p = 3.3×10−4). None of these peaks overlap with the peak b1 in the control. (p = 0.018 and 1.8×10−4 for r1 and u1, respectively). Similarly, peaks r2 and u2 in the retained peroxisomal and unretained fractions, respectively, have statistically different electrophoretic mobilities (p = 1.1×10−3).
Figure 4.
In vitro metabolism of DOX in retained peroxisomal and unretained fractions. The Trace a: Control. Trace b: unretained fraction. Trace c: Peroxisomal retained fraction. New peaks relative to the control are indicated by arrows. The reaction mixture contained DOX (10 μM) palmitoyl CoA (0.5 mM), FAD (0.5 mM) and NAD (1 mM). This mixture was incubated with only buffer (control) or either one of the fractions for 2 h at 37 °C. The MEKC separation was performed in a 43.5 cm fused silica capillary under a 400 V/cm electric field in a buffer containing 10 mM borate and 10 mM SDS (pH 9.3). Other conditions were same as that described in Figure 3, except for the 635±27.5 band pass filter that was used here. The DOX peak (380 s) was off-scale and therefore not labeled. The two peaks labeled with stars were used to correct migration times using Equation 1. Trace b and c were y-axis offset for clarity.
The relatively small size of the DOX-associated in the electropherograms is expected. The H2O2 levels in peroxisomes (c.f. Table 2) are much lower than the 1 mM H2O2 treatment of DOX that lead to observation of more prominent peaks associated with DOX oxidation products (See Figure S3 in the Electronic Supplementary Material). Similarly, the small size of the peaks associated with the DOX incubation of the unretained fraction is expected as a result of the lack of external addition of cofactors (e.g. NADPH for cytochrome P450 reactions) that are needed to drive metabolic transformations of xenobiotics that occur at other subcellular environments. Most importantly, the ability to separate functional peroxisomes from other subcellular components and observe biotransformations of xenobiotics such as DOX, opens up opportunities to explore metabolism in peroxisomes that is largely unknown.
4. Conclusions
Immunoisolation of functional peroxisomes made possible to investigate in vitro peroxisomal associated biotransformations of two xenobiotics: a BODIPY fatty acid analog and doxorubicin. MEKC-LIF analysis of the biotransformation products revealed subtle transformation of such xenobiotics, which is not biased by the presence of other subcellular compartments. Use of a catalase inhibitor (e.g., NaN3) may increase the H2O2 steady state levels in peroxisomes, which in turn would increase the yield of H2O2 dependent biotransformations. Future studies may identify such products using techniques such as HPLC-LIF-MS.[39] The reported developments may also enhance studies on peroxisome deficient diseases at the metabolic level,[5] provide the tools to investigate metabolic exchanges between peroxisomes and other subcellular regions,[44] and pave the way to explore other subcellular specific biotransformations.
Supplementary Material
Acknowledgments
YW acknowledges support through a 2008–2009 Merck Research Laboratories Fellowship in Analytical/Physical Chemsitry; THT acknowledges support from NIH grant T32GM008700; EAA acknowledges support from NIH grant AG020866.
References
- 1.Schrader M, Fahimi HD. The peroxisome: still a mysterious organelle. Histochem Cell Biol. 2008;129 (4):421–440. doi: 10.1007/s00418-008-0396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schrader M, Fahimi HD. Mammalian peroxisomes and reactive oxygen species. Histochem Cell Biol. 2004;122 (4):383–393. doi: 10.1007/s00418-004-0673-1. [DOI] [PubMed] [Google Scholar]
- 3.Wanders RJ, Vreken P, Ferdinandusse S, Jansen GA, Waterham HR, van Roermund CW, Van Grunsven EG. Peroxisomal fatty acid alpha- and beta-oxidation in humans: enzymology, peroxisomal metabolite transporters and peroxisomal diseases. Biochem Soc Trans. 2001;29 (Pt 2):250–267. doi: 10.1042/0300-5127:0290250. [DOI] [PubMed] [Google Scholar]
- 4.Wanders RJ, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 5.Wanders RJ, Waterham HR. Peroxisomal disorders: the single peroxisomal enzyme deficiencies. Biochim Biophys Acta. 2006;1763 (12):1707–1720. doi: 10.1016/j.bbamcr.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Masters CJ. On the role of the peroxisome in the metabolism of drugs and xenobiotics. Biochem Pharmacol. 1998;56 (6):667–673. doi: 10.1016/s0006-2952(98)00116-6. [DOI] [PubMed] [Google Scholar]
- 7.Graham JM. Isolation of peroxisomes from tissues and cells by differential and density gradient centrifugation. Curr Protoc Cell Biol. 2001;Chapter 3(Unit 3):5. doi: 10.1002/0471143030.cb0305s06. [DOI] [PubMed] [Google Scholar]
- 8.Volkl A, Fahimi HD. Isolation and characterization of peroxisomes from the liver of normal untreated rats. Eur J Biochem. 1985;149 (2):257–265. doi: 10.1111/j.1432-1033.1985.tb08920.x. [DOI] [PubMed] [Google Scholar]
- 9.Luers GH, Hartig R, Mohr H, Hausmann M, Fahimi HD, Cremer C, Volkl A. Immuno-isolation of highly purified peroxisomes using magnetic beads and continuous immunomagnetic sorting. Electrophoresis. 1998;19 (7):1205–1210. doi: 10.1002/elps.1150190722. [DOI] [PubMed] [Google Scholar]
- 10.Kikuchi M, Hatano N, Yokota S, Shimozawa N, Imanaka T, Taniguchi H. Proteomic analysis of rat liver peroxisome: presence of peroxisome-specific isozyme of Lon protease. J Biol Chem. 2004;279 (1):421–428. doi: 10.1074/jbc.M305623200. [DOI] [PubMed] [Google Scholar]
- 11.Blum RH, Carter SK. Adriamycin. A new anticancer drug with significant clinical activity. Ann Intern Med. 1974;80 (2):249–259. doi: 10.7326/0003-4819-80-2-249. [DOI] [PubMed] [Google Scholar]
- 12.Menna P, Salvatorelli E, Minotti G. Anthracycline degradation in cardiomyocytes: a journey to oxidative survival. Chem Res Toxicol. 2010;23 (1):6–10. doi: 10.1021/tx9003424. [DOI] [PubMed] [Google Scholar]
- 13.Taatjes DJ, Gaudiano G, Resing K, Koch TH. Redox pathway leading to the alkylation of DNA by the anthracycline, antitumor drugs adriamycin and daunomycin. J Med Chem. 1997;40 (8):1276–1286. doi: 10.1021/jm960835d. [DOI] [PubMed] [Google Scholar]
- 14.Reinhoud NJ, Tjaden UR, Irth H, van der Greef J. Bioanalysis of some anthracyclines in human plasma by capillary electrophoresis with laser-induced fluorescence detection. J Chromatogr. 1992;574 (2):327–334. doi: 10.1016/0378-4347(92)80047-t. [DOI] [PubMed] [Google Scholar]
- 15.Simeon N, Chatelut E, Canal P, Nertz M, Couderc F. Anthracycline analysis by capillary electrophoresis. Application to the analysis of daunorubicine in Kaposi sarcoma tumor. J Chromatogr A. 1999;853 (1–2):449–454. doi: 10.1016/s0021-9673(99)00383-0. [DOI] [PubMed] [Google Scholar]
- 16.Anderson AB, Gergen J, Arriaga EA. Detection of doxorubicin and metabolites in cell extracts and in single cells by capillary electrophoresis with laser-induced fluorescence detection. J Chromatogr B. 2002;769 (1):97–106. doi: 10.1016/s1570-0232(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 17.Anderson AB, Arriaga EA. Subcellular metabolite profiles of the parent CCRF-CEM and the derived CEM/C2 cell lines after treatment with doxorubicin. J Chromatogr B. 2004;808 (2):295–302. doi: 10.1016/j.jchromb.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 18.Eder AR, Arriaga EA. Capillary electrophoresis monitors enhancement in subcellular reactive oxygen species production upon treatment with doxorubicin. Chem Res Toxicol. 2006;19 (9):1151–1159. doi: 10.1021/tx060083i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson AB, Xiong G, Arriaga EA. Doxorubicin accumulation in individually electrophoresed organelles. J Am Chem Soc. 2004;126 (30):9168–9169. doi: 10.1021/ja0492539. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Walsh RJ, Arriaga EA. Selective determination of the doxorubicin content of individual acidic organelles in impure subcellular fractions. Anal Chem. 2005;77 (8):2281–2287. doi: 10.1021/ac0480996. [DOI] [PubMed] [Google Scholar]
- 21.Lu L, Wang Y. Immunoprecipitation alert: DNA binding proteins directly bind to protein A/G without any antibody as the bridge. Cell Cycle. 2008;7 (3):417–418. doi: 10.4161/cc.7.3.5359. [DOI] [PubMed] [Google Scholar]
- 22.Storrie B, Madden EA. Isolation of subcellular organelles. Methods Enzymol. 1990;182:203–225. doi: 10.1016/0076-6879(90)82018-w. [DOI] [PubMed] [Google Scholar]
- 23.Barrett AJ, Heath MF. Lysosomal enzymes. In: Dingle JT, editor. Lysosomes: A Laboratory Handbook. Elsevier/North-Holland Biomedical Press; Amsterdam: 1977. [Google Scholar]
- 24.Petit JM, Maftah A, Ratinaud MH, Julien R. 10N-nonyl acridine orange interacts with cardiolipin and allows the quantification of this phospholipid in isolated mitochondria. Eur J Biochem. 1992;209 (1):267–273. doi: 10.1111/j.1432-1033.1992.tb17285.x. [DOI] [PubMed] [Google Scholar]
- 25.Mannaerts GP, Debeer LJ. Mitochondrial and peroxisomal beta-oxidation of fatty acids in rat liver. Ann N Y Acad Sci. 1982;386:30–39. doi: 10.1111/j.1749-6632.1982.tb21405.x. [DOI] [PubMed] [Google Scholar]
- 26.Mueller S, Weber A, Fritz R, Mutze S, Rost D, Walczak H, Volkl A, Stremmel W. Sensitive and real-time determination of H2O2 release from intact peroxisomes. Biochem J. 2002;363 (Pt 3):483–491. doi: 10.1042/0264-6021:3630483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wanders RJ, van Roermund CW, de Vries CT, van den Bosch H, Schrakamp G, Tager JM, Schram AW, Schutgens RB. Peroxisomal beta-oxidation of palmitoyl-CoA in human liver homogenates and its deficiency in the cerebro-hepato-renal (Zellweger) syndrome. Clin Chim Acta. 1986;159 (1):1–10. doi: 10.1016/0009-8981(86)90160-9. [DOI] [PubMed] [Google Scholar]
- 28.Hows MEP, Perrett D. Effects of buffer depletion in capillary electrophoresis: Development of a continuous flow cathode. Chromatographia. 1998;48 (5–6):355–359. [Google Scholar]
- 29.Li XF, Ren H, Le X, Qi M, Ireland ID, Dovichi NJ. Migration time correction for the analysis of derivatized amino acids and oligosaccharides by micellar capillary electrochromatography. J Chromatogr A. 2000;869 (1–2):375–384. doi: 10.1016/s0021-9673(99)00893-6. [DOI] [PubMed] [Google Scholar]
- 30.Wendeler M, Sandhoff K. Hexosaminidase assays. Glycoconj J. 2009;26 (8):945–952. doi: 10.1007/s10719-008-9137-5. [DOI] [PubMed] [Google Scholar]
- 31.Lal A, Haynes SR, Gorospe M. Clean Western blot signals from immunoprecipitated samples. Mol Cell Probes. 2005;19 (6):385–388. doi: 10.1016/j.mcp.2005.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy JK, Hashimoto T. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu Rev Nutr. 2001;21:193–230. doi: 10.1146/annurev.nutr.21.1.193. [DOI] [PubMed] [Google Scholar]
- 33.Zhou M, Diwu Z, Panchuk-Voloshina N, Haugland RP. A stable nonfluorescent derivative of resorufin for the fluorometric determination of trace hydrogen peroxide: applications in detecting the activity of phagocyte NADPH oxidase and other oxidases. Anal Biochem. 1997;253 (2):162–168. doi: 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]
- 34.Kasurinen J. A novel fluorescent fatty acid, 5-methyl-BDY-3-dodecanoic acid, is a potential probe in lipid transport studies by incorporating selectively to lipid classes of BHK cells. Biochem Biophys Res Commun. 1992;187 (3):1594–1601. doi: 10.1016/0006-291x(92)90485-4. [DOI] [PubMed] [Google Scholar]
- 35.Naylor BL, Picardo M, Homan R, Pownall HJ. Effects of fluorophore structure and hydrophobicity on the uptake and metabolism of fluorescent lipid analogs. Chem Phys Lipids. 1991;58 (1–2):111–119. doi: 10.1016/0009-3084(91)90117-t. [DOI] [PubMed] [Google Scholar]
- 36.Brando T, Pardin C, Prandi J, Puzo G. Analysis of aminofluorescein-fatty acid derivatives by capillary electrophoresis with laser-induced fluorescence detection at the attomole level: application to mycobacterial fatty acids. J Chromatogr A. 2002;973 (1–2):203–210. doi: 10.1016/s0021-9673(02)01216-5. [DOI] [PubMed] [Google Scholar]
- 37.Sauro VS, Strickland KP. Changes in oleic acid oxidation and incorporation into lipids of differentiating L6 myoblasts cultured in normal or fatty acid-supplemented growth medium. Biochem J. 1987;244 (3):743–748. doi: 10.1042/bj2440743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thumser AE, Storch J. Characterization of a BODIPY-labeled fluorescent fatty acid analogue. Binding to fatty acid-binding proteins, intracellular localization, and metabolism. Mol Cell Biochem. 2007;299 (1–2):67–73. doi: 10.1007/s11010-005-9041-2. [DOI] [PubMed] [Google Scholar]
- 39.Katzenmeyer JB, Eddy CV, Arriaga EA. Tandem laser-induced fluorescence and mass spectrometry detection for high-performance liquid chromatography analysis of the in vitro metabolism of doxorubicin. Anal Chem. 2010;82 (19):8113–8120. doi: 10.1021/ac1011415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chu XP, Zhao T, Zhang YY, Zhao AH, Zhou MM, Zheng XJ, Dan M, Jia W. Determination of 13 Free Fatty Acids in Pheretima Using Ultra-Performance LC-ESI-MS. Chromatographia. 2009;69 (7–8):645–652. [Google Scholar]
- 41.Haynes CA, Allegood JC, Sims K, Wang EW, Sullards MC, Merrill AH., Jr Quantitation of fatty acyl-coenzyme As in mammalian cells by liquid chromatography-electrospray ionization tandem mass spectrometry. J Lipid Res. 2008;49 (5):1113–1125. doi: 10.1194/jlr.D800001-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magnes C, Sinner FM, Regittnig W, Pieber TR. LC/MS/MS method for quantitative determination of long-chain fatty acyl-CoAs. Anal Chem. 2005;77 (9):2889–2894. doi: 10.1021/ac048314i. [DOI] [PubMed] [Google Scholar]
- 43.Yamada J, Ogawa S, Horie S, Watanabe T, Suga T. Participation of peroxisomes in the metabolism of xenobiotic acyl compounds: comparison between peroxisomal and mitochondrial beta-oxidation of omega-phenyl fatty acids in rat liver. Biochim Biophys Acta. 1987;921 (2):292–301. [PubMed] [Google Scholar]
- 44.Jakobs BS, Wanders RJ. Fatty acid beta-oxidation in peroxisomes and mitochondria: the first, unequivocal evidence for the involvement of carnitine in shuttling propionyl-CoA from peroxisomes to mitochondria. Biochem Biophys Res Commun. 1995;213 (3):1035–1041. doi: 10.1006/bbrc.1995.2232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.