Abstract
One of the signaling mechanisms mediated by nitric oxide (NO) is through S-nitrosylation, the reversible redox-based modification of cysteine residues, on target proteins that regulate a myriad of physiological and pathophysiological processes. In particular, an increasing number of studies have identified important roles for S-nitrosylation in regulating cell death. These roles include double-edged effects dependent on the levels, spatiotemporal distribution, and origins of NO in the brain: in general S-nitrosylation resulting from the basal low level of NO in cells exerts anti-cell death effects, whereas S-nitrosylation elicited by induced NO upon stressed conditions is implicated in either pro-cell death effects or serves as a negative feedback mechanism and inhibits cell death. Furthermore, in addition to these cascades, mainly associated with apoptosis, massive levels of NO can lead to necrotic cell death. This review focuses on the proteins that are regulated by S-nitrosylation during cell death, in particular neuronal cell death and apoptosis. These mechanisms are involved in the pathogenesis of several diseases including degenerative diseases of the central nervous system (CNS).
Keywords: Nitric oxide, S-nitrosylation, Apoptosis, Caspases, XIAP, GAPDH
1. Introduction
Apoptosis is a mode of cell death that regulates tissue homeostasis during normal development and maintains adult organism under various conditions, including adaptive responses to cellular stress. Dysregulation of apoptosis can lead to various pathological conditions [1, 2, 3, 4, 5]: insufficient apoptosis can lead to the development of cancer and autoimmune diseases, whereas excessive or inappropriate apoptosis leads to tissue damages, including stroke, myocardial infarction, ischemia, and neurodegenerative diseases.
Nitric oxide (NO) is an important signaling molecule that acts in many tissues to regulate a diverse range of physiological processes, which include vasodilation, neuronal function, inflammation and immune function [6, 7, 8]. The major enzymes that generate endogenous NO in mammalian cells are three types of nitric oxide synthases (NOS) [(the l-arginine-dependent neuronal nitric oxide synthase (nNOS, NOS1), inducible NOS (iNOS, NOS2), and endothelial NOS (eNOS, NOS3)] [9]. Excessive generation of NO and NO-derived reactive nitrogen species, mainly peroxynitrite (ONOO−), NO2 and N2O3, has been implicated neuronal cell death and apoptosis [10, 11, 12, 13, 14, 15, 16]. Reactive nitrogen species can elicit reversible S-nitrosylation of thiol groups [17, 18] and irreversible protein tyrosine nitration [19, 20] in target proteins. These roles include double-edged effects dependent on the levels, spatiotemporal distribution, and origins of NO in the brain.
Tyrosine nitration is a reaction of amino acid tyrosine of target proteins with peroxynitrite (ONOO−), which leads to a covalent addition of a nitro group (-NO2) to one of the two equivalent ortho-carbons of the aromatic ring of tyrosine residues. Tyrosine nitration affects protein function and structure, which includes a change in the rate of proteolytic degradation and a loss of protein activity [19, 20, 21, 22, 23, 24, 25]. Elevated levels of protein tyrosine nitration underlie various age-related neurodegenerative diseases and can be used as a marker of these conditions [22, 23, 24, 25].
One of the key cellular roles of NO is reversible protein modification, such as S-nitrosylation, a covalent addition of an NO group to a cysteine thiol/sulfhydryl (RSH or, more properly, thiolate anion, RS-), resulting in formation of an S-nitrosothiol derivative (RSNO) [17, 18, 26]. In analogy to phosphorylation by kinases, S-nitrosylation by NOSs influences protein activity, protein-protein interactions, and protein location, thus serving as the prototypical redox-based signal [27]. S-Nitrosylation is readily reversible with high spatial and temporal specificity. In addition, there are two major physiologically relevant denitrosylases to remove NO group from S-nitrosylated Cys thiol side chains [glutathione/S-nitrosoglutathione reductase (GSH/GSNOR) and the thioredoxin/thioredoxin reductase (Trx/TrxR)] [28, 29]. The level of S-nitrosylation of any cellular protein depends on the balance between nitrosylation and denitrosylation [28]. S-Nitrosylation upon excessive generation of NO and NO-derived reactive nitrogen species (“nitrosative stress”) affects cellular homeostasis and pathological conditions [4, 6, 30, 31, 32, 33, 34]. Thus, regulatory mechanisms of nitrosylation/denitrosylation are crucial in S-nitrosylation-mediated cellular physiology and pathophysiology. The present review focuses on different targets and functional consequences of S-nitrosylation during cell death and apoptosis.
2. Anti-cell death mechanism associated with NO and protein S-nitrosylation: caspase inactivation and anti-apoptotic effects
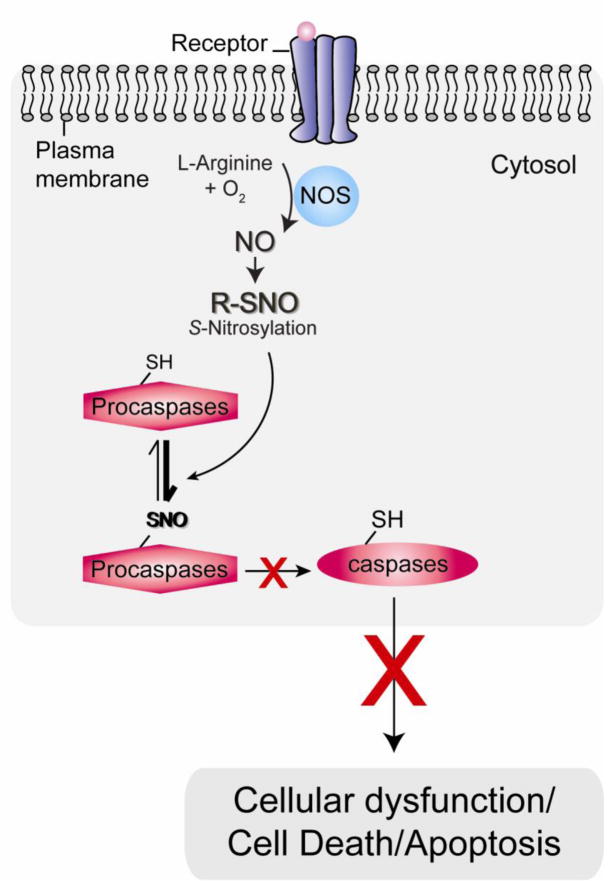
Apoptosis, characterized by cell shrinkage, membrane blebbing, and DNA fragmentation, is a highly regulated process leading to cell death [35]. The principal regulators of this process in the initiation and execution are the caspase family proteins, cysteine proteases. Caspases are expressed as inactive zymogens in resting cells, and upon exposure to a proapoptotic signal, the zymogen forms of caspases become proteolytically cleaved and activated. Initiator caspases (such as caspase-8, -9, and -10) can cleave other caspases, while executioner caspases (including caspase-3, -6, and -7) cleave substrates, which affect the death cascade [36]. Most caspases contain a single cysteine at the catalytic site, which is susceptible to redox modification and can be effectively modified by S-nitrosylation in the presence of NO with the subsequent inhibition of enzyme activity (Fig. 1) [10, 37, 38, 39].
Fig. 1. Anti-cell death mechanism associated with NO and protein S-nitrosylation: caspase inactivation and anti-apoptotic effects.
Under basal conditions, nitric oxide synthase (NOS) activity is low and basal levels of nitric oxide (NO) contribute to anti-cell death mechanisms. For example, low dose of NO, S-nitrosylates (SNO) procaspases/caspases and inhibits their activities.
NO reportedly inhibits the enzymatic activity of caspase-3 and -8 via S-nitrosylation of active-site cysteine residues and suppresses apoptosis of hepatocytes in vitro and in vivo [40, 41, 42]. Furthermore, other studies showed that NO prevents cells from apoptotic cell death by basal S-nitrosylation of caspases [43, 44, 45]. S-Nitrosylation has also been shown to reduce the activity of caspases in neurons [46, 47, 48, 49, 50]. Furthermore, cortical neurons treated with several NO donors, including S-nitrosothiols, exhibited a significant reduction in staurosporin-induced caspase-3 and caspase-9 activation, probably owing to the NO-mediated S-nitrosylation of the cysteine residue at the catalytic site of these caspases [51]. These studies indicate that endogenous NO generated by NOS exerts an antiapoptotic function by S-nitrosylated inhibition of caspase activity (Fig. 1).
3. Pro-cell death mechanisms associated with NO and protein S-nitrosylation
3.1 Caspase activation by denitrosylation and XIAP S-nitrosylation
In several elegant studies Mannick et al [52, 53] documented that that S-nitrosylation-denitrosylation of caspase-3 were mechanisms by which this ligand was controlling the general process of apoptosis: in human B lymphocytes caspase-3 is constitutively S-nitrosylated under basal conditions and keeps the zymogen in an inactive state, which protects cells from apoptosis, whereas activation of the pro-apoptotic FAS receptor promotes denitrosylation of the catalytic cysteine as well as proteolytic cleavage of caspase-3 and induces apoptosis (Fig. 2) [52, 53]. Further study has clarified that cytosolic caspase-3 is largely present in its reduced and unnitrosylated state, whereas mitochondria-associated pool of caspase-3 that is constitutively S-nitrosylated in resting cells, which is the target of denitrosylation upon FAS stimulation [45]. Under pro-apoptotic conditions Trx/Trxr proteins denitrosylate SNO-caspase-3, which results in activation of caspase-3 [53, 54]. More studies are needed to show that similar mechanisms operate in neuronal cells.
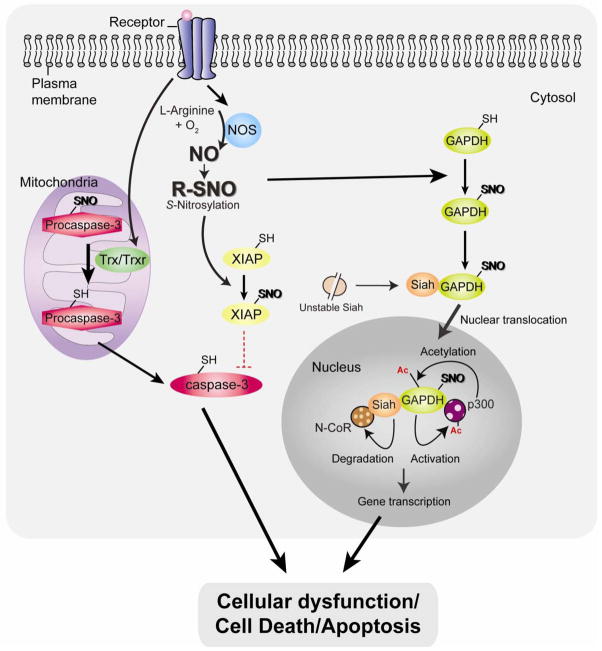
Fig. 2. Pro-cell death mechanisms associated with NO and protein S-nitrosylation: caspase activation and nuclear translocation of GAPDH.
Under stressed condition, activation of thioredoxin (Trx)/Trx reductase (TrxR) system (specifically in mitochondria) leads to procaspase3 denitrosylation, resulting in caspase-3 activation and apoptosis. In addition, NO inactivates the E3 ligase activity of X-linked inhibitor of apoptosis (XIAP) via S-nitrosylation (SNO), thus stabilizing caspases. Excessive nitrosative stress stimulates the formation of NO, which S-nitrosylates glyceraldehyde-3-phosphate dehydrogenase (GAPDH) enabling it to bind and stabilize Siah. Siah’s nuclear localization signal mediates nuclear translocation of the GAPDH-Siah complex. Stabilized Siah in the protein complex with S-nitrosylated GAPDH facilitate degradation of nuclear co-repressor N-CoR. Nuclear translocated GAPDH is further acetylated at by the histone acetyltransferase p300 via direct protein interaction, which in turn stimulates the catalytic activity of p300. Both of these mechanisms by the nuclear GAPDH-Siah complex regulate gene expression, which results in cellular dysfunction/cell death/apoptosis.
Caspase activation during NO stimulation also occurs as a result of down-regulation of X-linked inhibitor of apoptosis protein (XIAP) [55]. XIAP interacts with active caspases-3/-7/-9 in the cytosol and is thought to be the most potent endogenous caspase inhibitor. Under normal conditions, XIAP efficiently bind and inhibit the catalytic activity of apoptotic caspases [56]. Additionally, XIAP serves as an E3 ligase that ubiquitinates caspases and thus targets caspases for proteasomal degradation [57, 58, 59, 60, 61]. Under nitrosative stress conditions, NO suppresses the E3 ligase activity of XIAP by S-nitrosylation of the RING figure domain of the protein (cysteine 450), thus stabilizing caspases and sensitizing the neurons to apoptotic stimuli (Fig. 2) [62]. Constitutively S-nitrosylated caspases (e.g., caspase-3) as described above [53] can be a source to transfer of NO to XIAP via transnitrosylation and contribute to an additional mechanism to produce SNO-XIAP, inhibit its E3 ubiquitin ligase activity, leading to neuronal death [62]. In addition, XIAP is also S-nitrosylated in the BIR domain (baculoviral IAP repeat) by high concentrations of NO, leading to XIAP unable to bind and activate caspase-3 [50].
3.2 GAPDH nuclear translocation by S-nitrosylation
We reported that a small pool of GAPDH is translocated from cytosol to the nucleus upon exposure to stressors and participates in cellular dysfunction, death, and apoptosis [63], with other groups also replicating this observation [64, 65, 66, 67, 68, 69, 70, 71, 72]. This indicates that GAPDH may act as a relay molecule between distinct cellular compartments, such as the cytosol and nucleus, upon stressed conditions. The signaling is mediated by S-nitrosylated GAPDH at the catalytic site cysteine-150, allowing GAPDH to interact with the Siah (an E3 ubiquitin ligase, which possesses a nucleus localization signal), which leads to nuclear translocation of the GAPDH-Siah protein complex [73]. Inside the nucleus, GAPDH stabilizes Siah, which together with S-nitrosylated GAPDH is likely to facilitate ubiquitination and degradation of the nuclear co-repressor N-CoR [73, 74]. Further studies have also shown that nuclear translocated GAPDH is acetylated at lysine-160 by the histone acetyltransferase p300/CBP via direct protein interaction, which in turn stimulates the catalytic activity of p300/CBP. This nuclear event leads to the acetylation of downstream targets, including the tumor suppressor p53 [75]. By both of these mechanisms, the nuclear GAPDH-Siah complex may regulate gene expression associated with cellular dysfunction and death (Fig. 2).
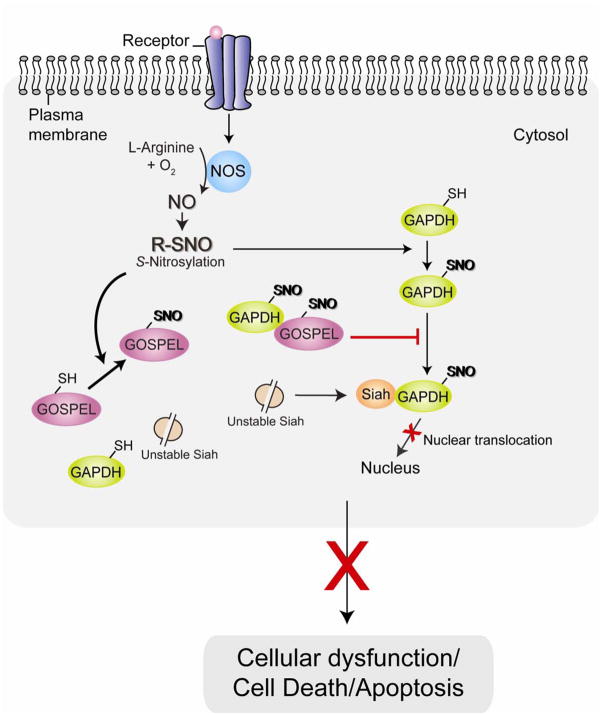
We have recently reported a novel protein, GOSPEL (GAPDH’s competitor Of Siah Protein Enhances Life) through the yeast two-hybrid screening of GAPDH’s interactors, which can antagonize cell death induced by S-nitrosylated GAPDH in neurons [76]. GOSPEL, a cytosolic protein, is highly expressed in organs with high expression levels of GAPDH, such as skeletal muscle, lung, heart, and brain. Under basal conditions GOSPEL physiologically binds GAPDH, retaining GAPDH in the cytosol [76]. GOSPEL is also S-nitrosylated at cysteine-47, and S-nitrosylated GOSPEL competes with Siah for binding to GAPDH and retaining GAPDH within the cytoplasm. Thus, the competitive binding between GOSPEL and Siah for GAPDH is likely to block cell death and maintain cellular homeostasis when cells are initially exposed to stressors and cellular NO levels are elevated but not massive yet (Fig. 3). However, once the level of nitrosative stress and cellular NO exceed a threshold, the GAPDH-Siah binding driven by S-nitrosylation of GAPDH predominates over GAPDH-GOSPEL interaction and leads to cellular dysfunction and death [76]. Therefore, it is likely that the protein GOSPEL can act as a regulator of this pro-cell death pathway by preventing the initial formation of the GAPDH-Siah1 complex.
Fig. 3. GOSPEL as the regulator of the cell death mechanism associated with GAPDH nuclear translocation.
GOSPEL (GAPDH’s competitor Of Siah Protein Enhances Life) was recently identified as a physiological inhibitor against GAPDH-Siah binding. At the initial phase of nitrosative stress when NO levels are low, S-nitrosylation (SNO) of GOSPEL augments binding of GOSPEL and GAPDH, competing with Siah for binding to GAPDH, leading to retention of GAPDH in the cytosol and preventing its nuclear translocation and inhibiting cell death.
The involvement of S-nitrosylation in GAPDH-mediated cell death is further supported by the study with thyroid carcinoma cells in which S-nitrosylation and nuclear translocation of GAPDH is observed after the exposure to TNF-related apoptosis-inducing ligand (TRAIL) [77].
4. Contribution of NO and protein S-nitrosylation in the brain diseases
Increasing evidence suggests roles for nNOS-derived NO and protein S-nitrosylation in brain disorders[1, 4, 5, 26, 31, 33, 34]. In addition, a massive induction of iNOS, excessive amount of NO, and S-nitrosylation of protein targets, such as parkin [78], are elicited in microglia and astrocytes in inflammation, especially in association with neuronal loss in brain disorders, including Alzheimer’s and Parkinson’s diseases involves inflammation [79].
Recent studies have shown dysregulated S-nitrosylation of many proteins involved in neuronal death and neurodegenerative disorders, including Parkin, protein disulfide isomerase (PDI), peroxiredoxin 2 (Prx2), XIAP, Dynamin-related protein 1 (Drp1), Heat Shock Protein 90 (HSP90), matrix metalloproteinase-9 (MMP-9), phosphatase and tensin homolog (PTEN), and GAPDH (Table 1) [50, 62, 78, 80, 81, 82, 83, 84, 85, 86, 87, 88]. Moreover, alterations in the activity of denitrosylases that remove NO groups may also contribute to diseases. A subset of patients with familial amyotrophic lateral sclerosis (ALS) that have mutations in superoxide dismutase 1 (SOD1) are reported to result in elevated denitrosylating activity of SOD1, leading to the depletion of cellular SNOs, including nuclear SNO-GAPDH, which might be linked to disease pathogenesis [89].
Table 1.
S-Nitrosylation of protein targets implicated in neurodegenerative diseases.
| Protein targets | Neurodegenerative disease relevance | Reference |
|---|---|---|
| Dynamin-related protein 1 (Drp1) | Alzheimer’s disease | [85] |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | Alzheimer’s disease | [86] |
| Heat shock protein 90 (HSP90) | Alzheimer’s disease | [100,101] |
| Matrix metalloproteinase 9 (MMP-9) | Stroke | [80,102,103] |
| Parkin | Parkinson’s disease, Alzheimer’s disease | [78,81,91] |
| Peroxiredoxin 2 (Prx2) | Alzheimer’s disease, Parkinson’s disease | [84,95,96] |
| Phosphatase and tensin homolog (PTEN) | Alzheimer’s disease, Parkinson’s disease, Stroke | [87,106,108] |
| Protein disulfide isomerase (PDI) | Alzheimer’s disease, Parkinson’s disease, Amyotrophic lateral sclerosis | [88,92] |
| X-linked inhibitor of apoptosis (XIAP) | Alzheimer’s disease, Parkinson’s disease | [50,62,93] |
Parkin, an ubiquitin E3 ligase whose mutation is known to cause an autosomal recessive juvenile Parkinson’s disease (PD) [90], has been identified as a target of S-nitrosylation (in Zn-binding cysteine residues in the RING domain of parkin) [81]. S-Nitrosylation of parkin increases its E3 ligase activity [81], promotes its auto-ubiquitination, inhibits its enzymatic activity, and compromises its neuroprotective function [78, 81]. Recently parkin was shown to colocalize with classic senile plaques, amyloid-laden vessels, and astrocytes associated with both lesions, in brains from patients with Alzheimer’s disease (AD) [91].
PDI is an endoplasmic reticulum (ER)-associated chaperone protein that prevents neurotoxicity caused by ER stress and protein misfolding [92]. PDI is S-nitrosylated at active site cysteine residues, resulting in the inhibition of its disulfide isomerase activity and accumulation of misfolded protein in the ER [92]. Increased S-nitrosylation of PDI has been found in the brain from patients with PD and AD [92] and the spinal cord from patients with sporadic amyotrophic lateral sclerosis [88]. Cumulative S-nitrosylation of parkin and PDI might underlie the accumulation of misfolded and ubiquitylated proteins that ultimately leads to cell death.
XIAP, a well-known anti-apoptotic protein, is reportedly overexpressed in patients with PD and AD [50, 62, 93]. The increased level of XIAP S-nitrosylation is reported in PD patients [50, 62]. As described in subsection 3.1, S-nitrosylation of XIAP is involved in regulating neuronal death [50, 62].
Prx2, a 2-Cys peroxiredoxin, a member of a family of abundant antioxidants is known to protect against oxidative stress in neurons [94]. The levels of Prx2 are increased in the frontal cortex of patients with AD and PD [95, 96]. Prx2 isS -nitrosylated at cysteine residues, which impairs its protective function against oxidative stress. The increased level of Prx2 S-nitrosylation is reported in PD patients [84].
The mitochondrion undergoes consistent fusion and fission to maintain its proper function, and dysregulation of mitochondrial fusion or fission has also been implicated in neurodegeneration [97, 98, 99]. Drp1, one of the important proteins that regulate mitochondrial fission, was shown to be S-nitrosylated at cysteine 644, which promotes its multimerization and thus mitochondrial fission, which causes neuronal damage [85]. S-Nitrosylated Drp1 was enhanced in brains from patients with AD [85]. Furthermore, exposure of nNOS-expressing cells to β-amyloid peptide results in the S-nitrosylation of Drp1 [85].
HSP90, a chaperone protein and co-activator of eNOS, is another S-nitrosylated protein. S-Nitrosylation of HSP90 abolishes its ATPase activity, which is required for its function as molecular chaperone [83]. The brain from patients with AD exhibits increased levels of HSP90 where it is thought to maintain tau and Aβ in a soluble conformation, thereby averting their aggregation [100, 101].
MMP-9, a protein involved in remodeling of extracellular matrix, is induced [102, 103] and S-nitrosylated during cerebral ischemia (stroke) [80]. S-Nitrosylation results in pathological activation of MMP-9, contributing to neuronal injury and death during stroke [80]. However, a study by McCarthy and co-workers [104] demonstrated that NO is incapable of directly activating pro-MMP-9 and that S-nitrosylation of MMP-9 propeptide by NO donors is unrelated to their ability to regulate MMP-9 activity.
PTEN governs the phosphoinositide 3-kinase (PI3K)/Akt signaling pathway which may be a pro-survival pathway in neurons [105]. It has been reported that PTEN protein levels are reduced in AD brains accompanied by elevated Akt phosphorylation [106, 107, 108]. It was recently shown that S-nitrosylation of PTEN was markedly elevated in the brains in the early stages of AD [87]. Excess NO leads to S-nitrosylation and ubiquitination of PTEN, resulting in loss of its enzymatic activity and degradation [87].
A pathological role for nuclear GAPDH has been suggested in several neurodegenerative disorders. Nuclear accumulation of GAPDH has been found in fibroblasts and in postmortem brains from patients with polyglutamine diseases (such as Huntington’s disease or dentatorubral-pallidoluysian atrophy) [68, 109], PD [66], and AD [70, 110]. Several studies have demonstrated that GAPDH is subject to many different types of oxidative modification in AD brain, which drastically affect its structure and function, including S-glutathionylation and nitration [111, 112] and is comprehensively reviewed by Butterfield et al [86]. Moreover, promising pharmacological evidence provided by Deprenyl and TCH346 that may inhibit GAPDH-Siah binding and nuclear translocation of the GAPDH-Siah protein complex [113], further supports a role for nuclear GAPDH in cell dysfunction and death, especially in association with neurodegenerative disorders.
6. Concluding remarks
NO signaling, through S-nitrosylation and denitrosylation, can regulate cellular homeostasis in order to maintain the balance between induction and prevention of cell death/apoptosis, in a context-dependent manner. In general S-nitrosylation resulting from basal low level NO in cells exerts anti-cell death effects, whereas S-nitrosylation elicited by induced NO upon stressors leads to pro-cell death effects. It is increasingly evident that dysregulated S-nitrosylation/denitrosylation could contribute to pathophysiologies characteristic of a wide range of disorders, in particular degenerative diseases of the CNS. Therefore, understanding of their regulatory mechanisms and identifying potential targets may aid therapeutic intervention in a wide range of brain diseases.
Research Highlights.
The present review focuses on different targets and functional consequences associated with nitric oxide and protein S-nitrosylation during neuronal cell death.
Double-edged effects of S-nitrosylation depends on the levels, spatiotemporal distribution, and origins of NO in the brain
These mechanisms are involved in the pathogenesis of several diseases including degenerative diseases of the central nervous system
Acknowledgments
This work was supported by USPHS grants of MH-084018 Silvo O. Conte center (A.S.), MH-069853 (A.S.), MH-085226 (A.S.), MH-088753 (A.S.), as well as grants from Stanley (A.S.), S-R (A.S.), RUSK (A.S.), and NARSAD (N.S., A.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mattson MP. Neuronal life-and-death signaling, apoptosis, and neurodegenerative disorders. Antioxid Redox Signal. 2006;8:1997–2006. doi: 10.1089/ars.2006.8.1997. [DOI] [PubMed] [Google Scholar]
- 2.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cotter TG. Apoptosis and cancer: the genesis of a research field. Nat Rev Cancer. 2009;9:501–507. doi: 10.1038/nrc2663. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura T, Lipton SA. Cell death: protein misfolding and neurodegenerative diseases. Apoptosis. 2009;14:455–468. doi: 10.1007/s10495-008-0301-y. [DOI] [PubMed] [Google Scholar]
- 5.Zhivotovsky B, Orrenius S. Cell death mechanisms: cross-talk and role in disease. Exp Cell Res. 2010;316:1374–1383. doi: 10.1016/j.yexcr.2010.02.037. [DOI] [PubMed] [Google Scholar]
- 6.Calabrese V, Cornelius C, Rizzarelli E, Owen JB, Dinkova-Kostova AT, Butterfield DA. Nitric oxide in cell survival: a janus molecule. Antioxid Redox Signal. 2009;11:2717–2739. doi: 10.1089/ars.2009.2721. [DOI] [PubMed] [Google Scholar]
- 7.Knott AB, Bossy-Wetzel E. Nitric oxide in health and disease of the nervous system. Antioxid Redox Signal. 2009;11:541–554. doi: 10.1089/ars.2008.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez MC, Andriantsitohaina R. Reactive nitrogen species: molecular mechanisms and potential significance in health and disease. Antioxid Redox Signal. 2009;11:669–702. doi: 10.1089/ars.2007.1993. [DOI] [PubMed] [Google Scholar]
- 9.Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: structure, function and inhibition. Biochem J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung HT, Pae HO, Choi BM, Billiar TR, Kim YM. Nitric oxide as a bioregulator of apoptosis. Biochem Biophys Res Commun. 2001;282:1075–1079. doi: 10.1006/bbrc.2001.4670. [DOI] [PubMed] [Google Scholar]
- 11.Choi BM, Pae HO, Jang SI, Kim YM, Chung HT. Nitric oxide as a pro-apoptotic as well as anti-apoptotic modulator. J Biochem Mol Biol. 2002;35:116–126. doi: 10.5483/bmbrep.2002.35.1.116. [DOI] [PubMed] [Google Scholar]
- 12.Kim KM, Kim PK, Kwon YG, Bai SK, Nam WD, Kim YM. Regulation of apoptosis by nitrosative stress. J Biochem Mol Biol. 2002;35:127–133. doi: 10.5483/bmbrep.2002.35.1.127. [DOI] [PubMed] [Google Scholar]
- 13.Razavi HM, Hamilton JA, Feng Q. Modulation of apoptosis by nitric oxide: implications in myocardial ischemia and heart failure. Pharmacol Ther. 2005;106:147–162. doi: 10.1016/j.pharmthera.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Hara MR, Snyder SH. Nitric oxide-GAPDH-Siah: a novel cell death cascade. Cell Mol Neurobiol. 2006;26:527–538. doi: 10.1007/s10571-006-9011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leon L, Jeannin JF, Bettaieb A. Post-translational modifications induced by nitric oxide (NO): implication in cancer cells apoptosis. Nitric Oxide. 2008;19:77–83. doi: 10.1016/j.niox.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 17.Lipton SA, Choi YB, Pan ZH, Lei SZ, Chen HS, Sucher NJ, Loscalzo J, Singel DJ, Stamler JS. A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature. 1993;364:626–632. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- 18.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 19.Ischiropoulos H. Biological tyrosine nitration: a pathophysiological function of nitric oxide and reactive oxygen species. Arch Biochem Biophys. 1998;356:1–11. doi: 10.1006/abbi.1998.0755. [DOI] [PubMed] [Google Scholar]
- 20.Radi R, Peluffo G, Alvarez MN, Naviliat M, Cayota A. Unraveling peroxynitrite formation in biological systems. Free Radic Biol Med. 2001;30:463–488. doi: 10.1016/s0891-5849(00)00373-7. [DOI] [PubMed] [Google Scholar]
- 21.Mattson MP. Methylation and acetylation in nervous system development and neurodegenerative disorders. Ageing Res Rev. 2003;2:329–342. doi: 10.1016/s1568-1637(03)00013-8. [DOI] [PubMed] [Google Scholar]
- 22.Rubbo H, Radi R. Protein and lipid nitration: role in redox signaling and injury. Biochim Biophys Acta. 2008;1780:1318–1324. doi: 10.1016/j.bbagen.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Souza JM, Peluffo G, Radi R. Protein tyrosine nitration--functional alteration or just a biomarker? Free Radic Biol Med. 2008;45:357–366. doi: 10.1016/j.freeradbiomed.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 24.Ischiropoulos H. Protein tyrosine nitration--an update. Arch Biochem Biophys. 2009;484:117–121. doi: 10.1016/j.abb.2008.10.034. [DOI] [PubMed] [Google Scholar]
- 25.Lee JR, Kim JK, Lee SJ, Kim KP. Role of protein tyrosine nitration in neurodegenerative diseases and atherosclerosis. Arch Pharm Res. 2009;32:1109–1118. doi: 10.1007/s12272-009-1802-0. [DOI] [PubMed] [Google Scholar]
- 26.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stamler JS, Lamas S, Fang FC. Nitrosylation. the prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 28.Benhar M, Forrester MT, Stamler JS. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat Rev Mol Cell Biol. 2009;10:721–732. doi: 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- 29.Forrester MT, Seth D, Hausladen A, Eyler CE, Foster MW, Matsumoto A, Benhar M, Marshall HE, Stamler JS. Thioredoxin-interacting protein (Txnip) is a feedback regulator of S-nitrosylation. J Biol Chem. 2009;284:36160–36166. doi: 10.1074/jbc.M109.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guix FX, Uribesalgo I, Coma M, Munoz FJ. The physiology and pathophysiology of nitric oxide in the brain. Prog Neurobiol. 2005;76:126–152. doi: 10.1016/j.pneurobio.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura T, Lipton SA. Emerging roles of S-nitrosylation in protein misfolding and neurodegenerative diseases. Antioxid Redox Signal. 2008;10:87–101. doi: 10.1089/ars.2007.1858. [DOI] [PubMed] [Google Scholar]
- 32.Chung KK, David KK. Emerging roles of nitric oxide in neurodegeneration. Nitric Oxide. 2010 doi: 10.1016/j.niox.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Chung KK. Modulation of pro-survival proteins by S-nitrosylation: implications for neurodegeneration. Apoptosis. 2010 doi: 10.1007/s10495-010-0464-1. [DOI] [PubMed] [Google Scholar]
- 34.Shahani N, Sawa A. Nitric oxide signaling and nitrosative stress in neurons: role for S-nitrosylation. Antioxid Redox Signal. 2010 doi: 10.1089/ars.2010.3580. [DOI] [PubMed] [Google Scholar]
- 35.Wyllie AH, Kerr JF, Currie AR. Cell death: the significance of apoptosis. Int Rev Cytol. 1980;68:251–306. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- 36.Thornberry NA, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 37.Li J, Billiar TR, Talanian RV, Kim YM. Nitric oxide reversibly inhibits seven members of the caspase family via S-nitrosylation. Biochem Biophys Res Commun. 1997;240:419–424. doi: 10.1006/bbrc.1997.7672. [DOI] [PubMed] [Google Scholar]
- 38.Melino G, Bernassola F, Knight RA, Corasaniti MT, Nistico G, Finazzi-Agro A. S-nitrosylation regulates apoptosis. Nature. 1997;388:432–433. doi: 10.1038/41237. [DOI] [PubMed] [Google Scholar]
- 39.Liu L, Stamler JS. NO: an inhibitor of cell death. Cell Death Differ. 1999;6:937–942. doi: 10.1038/sj.cdd.4400578. [DOI] [PubMed] [Google Scholar]
- 40.Kim YM, Talanian RV, Billiar TR. Nitric oxide inhibits apoptosis by preventing increases in caspase-3-like activity via two distinct mechanisms. J Biol Chem. 1997;272:31138–31148. doi: 10.1074/jbc.272.49.31138. [DOI] [PubMed] [Google Scholar]
- 41.Saavedra JE, Billiar TR, Williams DL, Kim YM, Watkins SC, Keefer LK. Targeting nitric oxide (NO) delivery in vivo. Design of a liver-selective NO donor prodrug that blocks tumor necrosis factor-alpha-induced apoptosis and toxicity in the liver. J Med Chem. 1997;40:1947–1954. doi: 10.1021/jm9701031. [DOI] [PubMed] [Google Scholar]
- 42.Kim YM, Kim TH, Chung HT, Talanian RV, Yin XM, Billiar TR. Nitric oxide prevents tumor necrosis factor alpha-induced rat hepatocyte apoptosis by the interruption of mitochondrial apoptotic signaling through S-nitrosylation of caspase-8. Hepatology. 2000;32:770–778. doi: 10.1053/jhep.2000.18291. [DOI] [PubMed] [Google Scholar]
- 43.Dimmeler S, Haendeler J, Nehls M, Zeiher AM. Suppression of apoptosis by nitric oxide via inhibition of interleukin-1beta-converting enzyme (ICE)-like and cysteine protease protein (CPP)-32-like proteases. J Exp Med. 1997;185:601–607. doi: 10.1084/jem.185.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohr S, Zech B, Lapetina EG, Brune B. Inhibition of caspase-3 by S-nitrosation and oxidation caused by nitric oxide. Biochem Biophys Res Commun. 1997;238:387–391. doi: 10.1006/bbrc.1997.7304. [DOI] [PubMed] [Google Scholar]
- 45.Mannick JB, Schonhoff C, Papeta N, Ghafourifar P, Szibor M, Fang K, Gaston B. S-Nitrosylation of mitochondrial caspases. J Cell Biol. 2001;154:1111–1116. doi: 10.1083/jcb.200104008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tenneti L, D’Emilia DM, Lipton SA. Suppression of neuronal apoptosis by S-nitrosylation of caspases. Neurosci Lett. 1997;236:139–142. doi: 10.1016/s0304-3940(97)00780-5. [DOI] [PubMed] [Google Scholar]
- 47.Lipton SA. Neuronal protection and destruction by NO. Cell Death Differ. 1999;6:943–951. doi: 10.1038/sj.cdd.4400580. [DOI] [PubMed] [Google Scholar]
- 48.Kang YC, Kim PK, Choi BM, Chung HT, Ha KS, Kwon YG, Kim YM. Regulation of programmed cell death in neuronal cells by nitric oxide. In Vivo. 2004;18:367–376. [PubMed] [Google Scholar]
- 49.Tannenbaum SR, White FM. Regulation and specificity of S-nitrosylation and denitrosylation. ACS Chem Biol. 2006;1:615–618. doi: 10.1021/cb600439h. [DOI] [PubMed] [Google Scholar]
- 50.Tsang AH, Lee YI, Ko HS, Savitt JM, Pletnikova O, Troncoso JC, Dawson VL, Dawson TM, Chung KK. S-nitrosylation of XIAP compromises neuronal survival in Parkinson’s disease. Proc Natl Acad Sci U S A. 2009;106:4900–4905. doi: 10.1073/pnas.0810595106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou P, Qian L, Iadecola C. Nitric oxide inhibits caspase activation and apoptotic morphology but does not rescue neuronal death. J Cereb Blood Flow Metab. 2005;25:348–357. doi: 10.1038/sj.jcbfm.9600036. [DOI] [PubMed] [Google Scholar]
- 52.Mannick JB, Miao XQ, Stamler JS. Nitric oxide inhibits Fas-induced apoptosis. J Biol Chem. 1997;272:24125–24128. doi: 10.1074/jbc.272.39.24125. [DOI] [PubMed] [Google Scholar]
- 53.Mannick JB, Hausladen A, Liu L, Hess DT, Zeng M, Miao QX, Kane LS, Gow AJ, Stamler JS. Fas-induced caspase denitrosylation. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- 54.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manderscheid M, Messmer UK, Franzen R, Pfeilschifter J. Regulation of inhibitor of apoptosis expression by nitric oxide and cytokines: relation to apoptosis induction in rat mesangial cells and raw 264.7 macrophages. J Am Soc Nephrol. 2001;12:1151–1163. doi: 10.1681/ASN.V1261151. [DOI] [PubMed] [Google Scholar]
- 56.Yang YL, Li XM. The IAP family: endogenous caspase inhibitors with multiple biological activities. Cell Res. 2000;10:169–177. doi: 10.1038/sj.cr.7290046. [DOI] [PubMed] [Google Scholar]
- 57.Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki Y, Nakabayashi Y, Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc Natl Acad Sci U S A. 2001;98:8662–8667. doi: 10.1073/pnas.161506698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Silke J, Kratina T, Chu D, Ekert PG, Day CL, Pakusch M, Huang DC, Vaux DL. Determination of cell survival by RING-mediated regulation of inhibitor of apoptosis (IAP) protein abundance. Proc Natl Acad Sci U S A. 2005;102:16182–16187. doi: 10.1073/pnas.0502828102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vaux DL, Silke J. IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol. 2005;6:287–297. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- 61.Schile AJ, Garcia-Fernandez M, Steller H. Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev. 2008;22:2256–2266. doi: 10.1101/gad.1663108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakamura T, Wang L, Wong CC, Scott FL, Eckelman BP, Han X, Tzitzilonis C, Meng F, Gu Z, Holland EA, Clemente AT, Okamoto S, Salvesen GS, Riek R, Yates JR, 3rd, Lipton SA. Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol Cell. 2010;39:184–195. doi: 10.1016/j.molcel.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sawa A, Khan AA, Hester LD, Snyder SH. Glyceraldehyde-3-phosphate dehydrogenase: nuclear translocation participates in neuronal and nonneuronal cell death. Proc Natl Acad Sci U S A. 1997;94:11669–11674. doi: 10.1073/pnas.94.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ishitani R, Tanaka M, Sunaga K, Katsube N, Chuang DM. Nuclear localization of overexpressed glyceraldehyde-3-phosphate dehydrogenase in cultured cerebellar neurons undergoing apoptosis. Mol Pharmacol. 1998;53:701–707. doi: 10.1124/mol.53.4.701. [DOI] [PubMed] [Google Scholar]
- 65.Saunders PA, Chen RW, Chuang DM. Nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase isoforms during neuronal apoptosis. J Neurochem. 1999;72:925–932. doi: 10.1046/j.1471-4159.1999.0720925.x. [DOI] [PubMed] [Google Scholar]
- 66.Tatton NA. Increased caspase 3 and Bax immunoreactivity accompany nuclear GAPDH translocation and neuronal apoptosis in Parkinson’s disease. Exp Neurol. 2000;166:29–43. doi: 10.1006/exnr.2000.7489. [DOI] [PubMed] [Google Scholar]
- 67.Maruyama W, Oya-Ito T, Shamoto-Nagai M, Osawa T, Naoi M. Glyceraldehyde-3-phospate dehydrogenase is translocated into nuclei through Golgi apparatus during apoptosis induced by 6-hydroxydopamine in human dopaminergic SH-SY5Y cells. Neurosci Lett. 2002;321:29–32. doi: 10.1016/s0304-3940(01)02490-9. [DOI] [PubMed] [Google Scholar]
- 68.Mazzola JL, Sirover MA. Alteration of nuclear glyceraldehyde-3-phosphate dehydrogenase structure in Huntington’s disease fibroblasts. Brain Res Mol Brain Res. 2002;100:95–101. doi: 10.1016/s0169-328x(02)00160-2. [DOI] [PubMed] [Google Scholar]
- 69.Tanaka R, Mochizuki H, Suzuki A, Katsube N, Ishitani R, Mizuno Y, Urabe T. Induction of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression in rat brain after focal ischemia/reperfusion. J Cereb Blood Flow Metab. 2002;22:280–288. doi: 10.1097/00004647-200203000-00005. [DOI] [PubMed] [Google Scholar]
- 70.Mazzola JL, Sirover MA. Subcellular alteration of glyceraldehyde-3-phosphate dehydrogenase in Alzheimer’s disease fibroblasts. J Neurosci Res. 2003;71:279–285. doi: 10.1002/jnr.10484. [DOI] [PubMed] [Google Scholar]
- 71.Kusner LL, Sarthy VP, Mohr S. Nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase: a role in high glucose-induced apoptosis in retinal Muller cells. Invest Ophthalmol Vis Sci. 2004;45:1553–1561. [PubMed] [Google Scholar]
- 72.Kim CI, Lee SH, Seong GJ, Kim YH, Lee MY. Nuclear translocation and overexpression of GAPDH by the hyper-pressure in retinal ganglion cell. Biochem Biophys Res Commun. 2006;341:1237–1243. doi: 10.1016/j.bbrc.2006.01.087. [DOI] [PubMed] [Google Scholar]
- 73.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, Hayward SD, Snyder SH, Sawa A. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 74.Zhang J, Guenther MG, Carthew RW, Lazar MA. Proteasomal regulation of nuclear receptor corepressor-mediated repression. Genes Dev. 1998;12:1775–1780. doi: 10.1101/gad.12.12.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sen N, Hara MR, Kornberg MD, Cascio MB, Bae BI, Shahani N, Thomas B, Dawson TM, Dawson VL, Snyder SH, Sawa A. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat Cell Biol. 2008;10:866–873. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sen N, Hara MR, Ahmad AS, Cascio MB, Kamiya A, Ehmsen JT, Agrawal N, Hester L, Dore S, Snyder SH, Sawa A. GOSPEL: a neuroprotective protein that binds to GAPDH upon S-nitrosylation. Neuron. 2009;63:81–91. doi: 10.1016/j.neuron.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Du ZX, Wang HQ, Zhang HY, Gao DX. Involvement of glyceraldehyde-3-phosphate dehydrogenase in tumor necrosis factor-related apoptosis-inducing ligand-mediated death of thyroid cancer cells. Endocrinology. 2007;148:4352–4361. doi: 10.1210/en.2006-1511. [DOI] [PubMed] [Google Scholar]
- 78.Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 79.Brown GC. Nitric oxide and neuronal death. Nitric Oxide. 2010;23:153–165. doi: 10.1016/j.niox.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 80.Gu Z, Kaul M, Yan B, Kridel SJ, Cui J, Strongin A, Smith JW, Liddington RC, Lipton SA. S-nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science. 2002;297:1186–1190. doi: 10.1126/science.1073634. [DOI] [PubMed] [Google Scholar]
- 81.Yao D, Gu Z, Nakamura T, Shi ZQ, Ma Y, Gaston B, Palmer LA, Rockenstein EM, Zhang Z, Masliah E, Uehara T, Lipton SA. Nitrosative stress linked to sporadic Parkinson’s disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc Natl Acad Sci U S A. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chuang DM, Hough C, Senatorov VV. Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 2005;45:269–290. doi: 10.1146/annurev.pharmtox.45.120403.095902. [DOI] [PubMed] [Google Scholar]
- 83.Martinez-Ruiz A, Villanueva L, Gonzalez de Orduna C, Lopez-Ferrer D, Higueras MA, Tarin C, Rodriguez-Crespo I, Vazquez J, Lamas S. S-nitrosylation of Hsp90 promotes the inhibition of its ATPase and endothelial nitric oxide synthase regulatory activities. Proc Natl Acad Sci U S A. 2005;102:8525–8530. doi: 10.1073/pnas.0407294102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fang J, Nakamura T, Cho DH, Gu Z, Lipton SA. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson’s disease. Proc Natl Acad Sci U S A. 2007;104:18742–18747. doi: 10.1073/pnas.0705904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Butterfield DA, Hardas SS, Lange ML. Oxidatively modified glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Alzheimer’s disease: many pathways to neurodegeneration. J Alzheimers Dis. 2010;20:369–393. doi: 10.3233/JAD-2010-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kwak YD, Ma T, Diao S, Zhang X, Chen Y, Hsu J, Lipton SA, Masliah E, Xu H, Liao FF. NO signaling and S-nitrosylation regulate PTEN inhibition in neurodegeneration. Mol Neurodegener. 2010;5:49. doi: 10.1186/1750-1326-5-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Walker AK, Farg MA, Bye CR, McLean CA, Horne MK, Atkin JD. Protein disulphide isomerase protects against protein aggregation and is S-nitrosylated in amyotrophic lateral sclerosis. Brain. 2010;133:105–116. doi: 10.1093/brain/awp267. [DOI] [PubMed] [Google Scholar]
- 89.Schonhoff CM, Matsuoka M, Tummala H, Johnson MA, Estevez AG, Wu R, Kamaid A, Ricart KC, Hashimoto Y, Gaston B, Macdonald TL, Xu Z, Mannick JB. S-nitrosothiol depletion in amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2006;103:2404–2409. doi: 10.1073/pnas.0507243103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 91.Witte ME, Bol JG, Gerritsen WH, van der Valk P, Drukarch B, van Horssen J, Wilhelmus MM. Parkinson’s disease-associated parkin colocalizes with Alzheimer’s disease and multiple sclerosis brain lesions. Neurobiol Dis. 2009;36:445–452. doi: 10.1016/j.nbd.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 92.Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 93.Christie LA, Su JH, Tu CH, Dick MC, Zhou J, Cotman CW. Differential regulation of inhibitors of apoptosis proteins in Alzheimer’s disease brains. Neurobiol Dis. 2007;26:165–173. doi: 10.1016/j.nbd.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Qu D, Rashidian J, Mount MP, Aleyasin H, Parsanejad M, Lira A, Haque E, Zhang Y, Callaghan S, Daigle M, Rousseaux MW, Slack RS, Albert PR, Vincent I, Woulfe JM, Park DS. Role of Cdk5-mediated phosphorylation of Prx2 in MPTP toxicity and Parkinson’s disease. Neuron. 2007;55:37–52. doi: 10.1016/j.neuron.2007.05.033. [DOI] [PubMed] [Google Scholar]
- 95.Kim SH, Fountoulakis M, Cairns N, Lubec G. Protein levels of human peroxiredoxin subtypes in brains of patients with Alzheimer’s disease and Down syndrome. J Neural Transm Suppl. 2001:223–235. doi: 10.1007/978-3-7091-6262-0_18. [DOI] [PubMed] [Google Scholar]
- 96.Krapfenbauer K, Engidawork E, Cairns N, Fountoulakis M, Lubec G. Aberrant expression of peroxiredoxin subtypes in neurodegenerative disorders. Brain Res. 2003;967:152–160. doi: 10.1016/s0006-8993(02)04243-9. [DOI] [PubMed] [Google Scholar]
- 97.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 98.Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 99.Cho DH, Nakamura T, Lipton SA. Mitochondrial dynamics in cell death and neurodegeneration. Cell Mol Life Sci. 2010;67:3435–3447. doi: 10.1007/s00018-010-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kakimura J, Kitamura Y, Takata K, Umeki M, Suzuki S, Shibagaki K, Taniguchi T, Nomura Y, Gebicke-Haerter PJ, Smith MA, Perry G, Shimohama S. Microglial activation and amyloid-beta clearance induced by exogenous heat-shock proteins. FASEB J. 2002;16:601–603. doi: 10.1096/fj.01-0530fje. [DOI] [PubMed] [Google Scholar]
- 101.Dou F, Netzer WJ, Tanemura K, Li F, Hartl FU, Takashima A, Gouras GK, Greengard P, Xu H. Chaperones increase association of tau protein with microtubules. Proc Natl Acad Sci U S A. 2003;100:721–726. doi: 10.1073/pnas.242720499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Montaner J, Alvarez-Sabin J, Molina C, Angles A, Abilleira S, Arenillas J, Gonzalez MA, Monasterio J. Matrix metalloproteinase expression after human cardioembolic stroke: temporal profile and relation to neurological impairment. Stroke. 2001;32:1759–1766. doi: 10.1161/01.str.32.8.1759. [DOI] [PubMed] [Google Scholar]
- 103.Rosell A, Ortega-Aznar A, Alvarez-Sabin J, Fernandez-Cadenas I, Ribo M, Molina CA, Lo EH, Montaner J. Increased brain expression of matrix metalloproteinase-9 after ischemic and hemorrhagic human stroke. Stroke. 2006;37:1399–1406. doi: 10.1161/01.STR.0000223001.06264.af. [DOI] [PubMed] [Google Scholar]
- 104.McCarthy SM, Bove PF, Matthews DE, Akaike T, van der Vliet A. Nitric oxide regulation of MMP-9 activation and its relationship to modifications of the cysteine switch. Biochemistry. 2008;47:5832–5840. doi: 10.1021/bi702496v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chang N, El-Hayek YH, Gomez E, Wan Q. Phosphatase PTEN in neuronal injury and brain disorders. Trends Neurosci. 2007;30:581–586. doi: 10.1016/j.tins.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 106.Griffin RJ, Moloney A, Kelliher M, Johnston JA, Ravid R, Dockery P, O’Connor R, O’Neill C. Activation of Akt/PKB, increased phosphorylation of Akt substrates and loss and altered distribution of Akt and PTEN are features of Alzheimer’s disease pathology. J Neurochem. 2005;93:105–117. doi: 10.1111/j.1471-4159.2004.02949.x. [DOI] [PubMed] [Google Scholar]
- 107.Rickle A, Behbahani H, Ankarcrona M, Winblad B, Cowburn RF. PTEN, Akt, and GSK3beta signalling in rat primary cortical neuronal cultures following tumor necrosis factor-alpha and trans-4-hydroxy-2-nonenal treatments. J Neurosci Res. 2006;84:596–605. doi: 10.1002/jnr.20970. [DOI] [PubMed] [Google Scholar]
- 108.Zhang X, Li F, Bulloj A, Zhang YW, Tong G, Zhang Z, Liao FF, Xu H. Tumor-suppressor PTEN affects tau phosphorylation, aggregation, and binding to microtubules. FASEB J. 2006;20:1272–1274. doi: 10.1096/fj.06-5721fje. [DOI] [PubMed] [Google Scholar]
- 109.Shiozawa M, Fukutani Y, Arai N, Cairns NJ, Mizutani T, Isaki K, Lantos PL, Wada Y. Glyceraldehyde 3-phosphate dehydrogenase and endothelin-1 immunoreactivity is associated with cerebral white matter damage in dentatorubral-pallidoluysian atrophy. Neuropathology. 2003;23:36–43. doi: 10.1046/j.1440-1789.2003.00480.x. [DOI] [PubMed] [Google Scholar]
- 110.Tsuchiya K, Tajima H, Yamada M, Takahashi H, Kuwae T, Sunaga K, Katsube N, Ishitani R. Disclosure of a pro-apoptotic glyceraldehyde-3-phosphate dehydrogenase promoter: anti-dementia drugs depress its activation in apoptosis. Life Sci. 2004;74:3245–3258. doi: 10.1016/j.lfs.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 111.Sultana R, Poon HF, Cai J, Pierce WM, Merchant M, Klein JB, Markesbery WR, Butterfield DA. Identification of nitrated proteins in Alzheimer’s disease brain using a redox proteomics approach. Neurobiol Dis. 2006;22:76–87. doi: 10.1016/j.nbd.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 112.Newman SF, Sultana R, Perluigi M, Coccia R, Cai J, Pierce WM, Klein JB, Turner DM, Butterfield DA. An increase in S-glutathionylated proteins in the Alzheimer’s disease inferior parietal lobule, a proteomics approach. J Neurosci Res. 2007;85:1506–1514. doi: 10.1002/jnr.21275. [DOI] [PubMed] [Google Scholar]
- 113.Hara MR, Thomas B, Cascio MB, Bae BI, Hester LD, Dawson VL, Dawson TM, Sawa A, Snyder SH. Neuroprotection by pharmacologic blockade of the GAPDH death cascade. Proc Natl Acad Sci U S A. 2006;103:3887–3889. doi: 10.1073/pnas.0511321103. [DOI] [PMC free article] [PubMed] [Google Scholar]