Abstract
Radiotherapy is used in locally advanced pancreatic cancers where it can improve survival in combination with gemcitabine. However, prognosis is still poor in this setting where more effective therapies remain needed. MLN4924 is an investigational small molecule currently in Phase I clinical trials. MLN4924 inhibits NAE (NEDD8 Activating Enzyme), a pivotal regulator of the E3 ubiquitin ligase SCF (SKP1, Cullins, and F-box protein), that has been implicated recently in DNA repair. In this study, we provide evidence that MLN4924 can be used as an effective radiosensitizer in pancreatic cancer. Specifically, MLN4924 (20–100 nM) effectively inhibited cullin neddylation and sensitized pancreatic cancer cells to ionizing radiation in vitro with a sensitivity enhancement ratio (SER) of ~1.5. Mechanistically, MLN4924 treatment stimulated an accumulation of several SCF substrates, including CDT1, WEE1 and NOXA, in parallel with an enhancement of radiation-induced DNA damage, aneuploidy, G2/M phase cell cycle arrest and apoptosis. RNAi-mediated knockdown of CDT1 and WEE1 partially abrogated MLN4924-induced aneuploidy, G2/M arrest, and radiosensization, indicating a causal effect. Further, MLN4924 was an effective radiosensitizer in mouse xenograft models of human pancreatic cancer. Our findings offer proof of concept for use of MLN4924 as a novel class of radiosensitizer for the treatment of pancreatic cancer.
Keywords: NAE inhibitor, MLN4924, CRL/SCF E3 ubiquitin ligase, radiosensitization, DNA damage, pancreatic cancer cells
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer-related deaths in the USA and one of the deadliest human malignancies with an overall 5-year survival rate of ~5% (1). Although gemcitabine is the standard systemic therapy for pancreatic cancer (2), the finding that as many as 1/3 of pancreatic cancer patients die of locally destructive disease underscores the importance of radiotherapy for local disease control (3). Indeed, the combination of radiation with gemcitabine has been shown to significantly prolong survival compared to gemcitabine alone (4), and intensifying radiation also appears to improve survival (5). Despite these improvements, the prognosis for patients with pancreatic cancer is still very poor, and new combinational therapies are urgently needed. Strategies utilizing small molecule inhibitors to sensitize pancreatic tumor cells to radiation (and gemcitabine) are well underway. We have recently shown that small molecule inhibitors of CHK1 (checkpoint kinase 1), sensitized pancreatic tumor cells and xenografts to radiation by G2 checkpoint abrogation and homologous recombination repair inhibition (6). Of the small molecule inhibitors tested in Phase III clinical trials in pancreatic cancer, erlotinib, an inhibitor of EGFR with only very modest activity in pancreatic cancer, has been shown to improve survival (7, 8). Thus, there is a demand for novel and more effective therapies. Our laboratory has recently shown that SCF E3 ubiquitin ligases are promising radiosensitizing targets in cancer cells (9, 10). Whether inhibition of SCF E3 ubiquitin ligases might also be a useful strategy for sensitizing pancreatic cancers to radiation has not previously been tested.
The SCF (SKP1-Cullin-F-box proteins) E3 ubiquitin ligases, consisting of an adaptor protein SKP1, a scafford protein cullin, a substrate receptor F-box protein, and a RING protein RBX1 or RBX2, are the largest multiunit ubiquitin ligases (11, 12), responsible for the ubiquitination of ~20% of all ubiquitinated proteins for targeted degradation by proteasome (13). The SCF E3s, therefore, regulate numerous biological processes, including cell cycle progression, signal transduction and DNA replication, among others (11, 12). It is well established that the core of SCF E3 ligase is a cullin-RING finger protein complex (14). In human and mouse, there are 7 cullins (cullins 1–3, 4A, 4B, 5 and 7) and two RING family members, RBX1 (RING box protein-1) and RBX2, also known as SAG (Sensitive to Apoptosis Gene) (11, 12, 15). The ligase activity of the SCF requires a) RBX1/RBX2, which binds to ubiquitin-loaded E2, and b) cullin neddylation (16–19), the addition of a ubiquitin-like protein NEDD8 to cullin, catalyzed by NEDD8 activating enzyme E1 (NAE), NEDD8 conjugating enzyme E2 (Ubc12) and NEDD8-E3 ligase (20).
We have recently shown that inactivation of SCF E3 ligase by siRNA knockdown of either RBX1 or RBX2/SAG induced apoptosis and senescence, and sensitized human cancer cells to radiation (9, 10, 21), and that Sag knockout also sensitized mouse embryonic stem cells to radiation via inducing apoptosis (22) Since MLN4924 effectively inactivates SCF E3 by blocking cullin neddylation (13), we tested our working hypothesis that MLN4924 could sensitize pancreatic cancer cells to radiation. We reported here that MLN4924 indeed acts as a radiosensitizing agent by enhancing radiation-induced DNA damage, aneuploidy, G2/M arrest and apoptosis via mechanism including accumulation of CDT1 and WEE1, two well-known substrates of SCF E3 ubiquitin ligase.
Materials and Methods
Cell Culture
Two human MiaPaCa-2 and BxPC-3 pancreatic cancer lines, and human H1299 lung cancer cell line were purchased from ATCC. MiaPaCa-2 and H1299 cells were grown in DMEM with 10% fetal bovine serum. BxPC-3 and human lung fibroblast MRC5 cells (a gift from Dr. A. Rehemtulla) were cultured in RPMI1640 with 10% FBS. All lines were tested and were free-of-mycloplasma contamination.
ATPlite growth assay and IC50 determination
Cells were seeded in 96well plates in triplicate and treated with MLN4924 (13), in various doses for 7 days. Cell viability was measured with ATPlite kit (Perkin Elmer) (23).
MLN4924 preparation for in vitro and in vivo assays
MLN4924 was synthesized by Millennium Pharmaceuticals, Inc. (13) and was dissolved in DMSO to make a 10 mM stock solution and kept in −20°C before use. For in vivo experiment, MLN4924 was freshly made every week as follows: To make a 8.32 mg/ml solution, the compound was dissolved in 10% 2-hydroxypropyl-β-cyclodextrin (HPBCD) in sterile water. The pH value was adjusted to 5.0 with 1 M KOH. The solution of MLN4924 was stored at room temperature before use.
Radiation exposure and clonogenic assay
Cells were seeded in 60-mm dishes in duplicate and exposed to different doses of radiation (Philips RT250, Kimtron Medical) after pre-treatment with MLN4924 for 6 hrs (at 100 nM) or 24 hrs (at 20–30 nM). MLN4924 was either washed away (at 100 nM) or stayed in the medium (at 20–30 nM) afterwards, followed by incubation at 37 °C for 7 to 9 days. Survival curves were fitted using the linear-quadratic equation, and the mean inactivation dose was calculated (24).
siRNA knockdown of CDT1 and WEE1
Two independent sets of siRNA oligonucleotides were used to target CTD1 or WEE1, respectively. Their sequences are as follows: siCTD1-1 (5’-CGUGGAUGAAGUACCCGACUU-3’) and siCTD1-2 (5’-GCAAUGUUGGCCAGAUCAAUU-3’), and siWEE1-1 (5’-GAGGCUGGAUGGAUGCAUUUU-3’) and siWEE1-2 (5’-CUCCGGGGUAGUUCUCUCUUU-3’), and scrambled control siRNA (siCONT: 5’-ATTGTATGCGATCGCAGACTT-3’). All oligoes were purchased from Dharmacon (USA). Cells were transfected with siRNA using X-tremeGENE (Roche) and split 48 hrs later. One portion was used for clonogenic assay and the other portions for immunoblotting (IB) or FACS profile (23).
Immunoblotting
Cells were exposed to various treatments and harvested for IB as described (23) using antibodies against CDT1, WEE-1, CUL1, total CDC2 (Santa Cruz Biotechnology), ORC-1, p21, p27 (BD Biosciences), NOXA, BIM1, phospho-CHK1 (S345), phospho-CHK2 (T68), γ-H2AX (Ser139), phosphor-CDC2 (Y15), phospho-IκB, Cyclin B1 (Cell Signaling), and β-actin (Sigma).
FACS (Fluorescence-activated Cell Sorting)
Cells were treated with MLN4924, or exposed to radiation alone or in combination. Cells were harvested 24 or 48 hrs post radiation and analyzed by flow cytometry (23).
Immunofluorescence staining
Cells after exposure to various treatments were fixed with 4% paraformaldehyde, permeabilied with 0.5% triton, blocked with 0.5% BSA, and incubated with primary antibody against γ-H2AX (Millipore) at 1:1000, followed by incubation with Alexa Fluor 594 goat anti-mouse IgG at 1:2000. Cellular nuclei were stained with DAPI, as described (9). The stained cells were observed under fluorescent microscope (Olympus 1×71), using Olympus LCP lan F1 lens and Olympus DP70 cameras. The acquisition software used is Olympus DP Controller 2002 (Olympus Optical Co. LTD).
In vivo anti-tumor study
All animal studies were conducted in accordance with the guidelines established by the University Committee on Use and Care of Animals (UCUCA). Five million MiaPaCa-2 cells were inoculated s.c in both flanks of nude mice. The mice were randomized and the treatment started when the tumor size reached 100 mm3 at 14 days after inoculation. MLN4924 (30 mg/kg, s.c.) and radiation (1 Gy) were given once a day, 5 days a week for three weeks. Radiation was delivered directly to the tumor with the rest of the animal shielded. For combination treatment, MLN4924 was given 2 hrs prior to radiation exposure with the same schedule as for individual treatments. The growth of tumors (8 for control group, 12–16 for the other groups) was measured twice a week, and average tumor volumes were calculated, as estimated from the formula (L × W2)/2. Radiation was conducted in the University of Michigan Comprehensive Cancer Center Experimental Radiation Core.
Statistical analysis
ANOVA models were used with SPSS software for statistical comparisons involving multiple groups, followed by an SNK post hoc test to determine significance of each two groups (p < 0.05). Tumor volume doubling (tripling) was determined for each xenograft by identifying the earliest day on which it was at least twice (three times) as large as on the first day of treatment. A cubic smoothing spline was used to obtain the exact time of doubling (tripling). The Kaplan-Meier method was used to analyze the doubling (tripling) times derived from the smoothed growth curves, and the cox regression model was used for comparisons between any two treatment groups after adjusting the initial tumor sizes. Bayesian hierarchical changepoint model (BHC) was conducted to estimate the tumor growth profile characterized by a pre-nadir regression rate, a regression period, a nadir volume, and a post-nadir regrowth rate. The 90% highest probability density (HPD) confidence intervals were calculated to compare these features between different treatments. If the HPD interval of the difference in any feature between two treatments covers zero, the two treatments are not significantly different on that feature at the significant level of 0.1 (25).
Results
Sensitivity of pancreatic cancer lines to MLN4924 as a single agent
Our recent studies showed that inactivation of SCF E3 ubiquitin ligase by siRNA knockdown of its RING component, RBX1 or RBX2/SAG, sensitized human cancer cells to radiation (9, 10), whereas Sag knockout also sensitized mouse embryonic stem cells to radiation by inducing apoptosis (22), suggesting that SCF E3 is an attractive radiosensitizing target. Here we tested our working hypothesis that MLN4924, a small molecule inhibitor of NEDD8 activating enzyme (NAE) which suppresses cancer cell growth by inhibiting activity of SCF E3 ubiquitin ligase via cullin deneddylation (13), could be a novel radiosensitizing agent in pancreatic cancer cells.
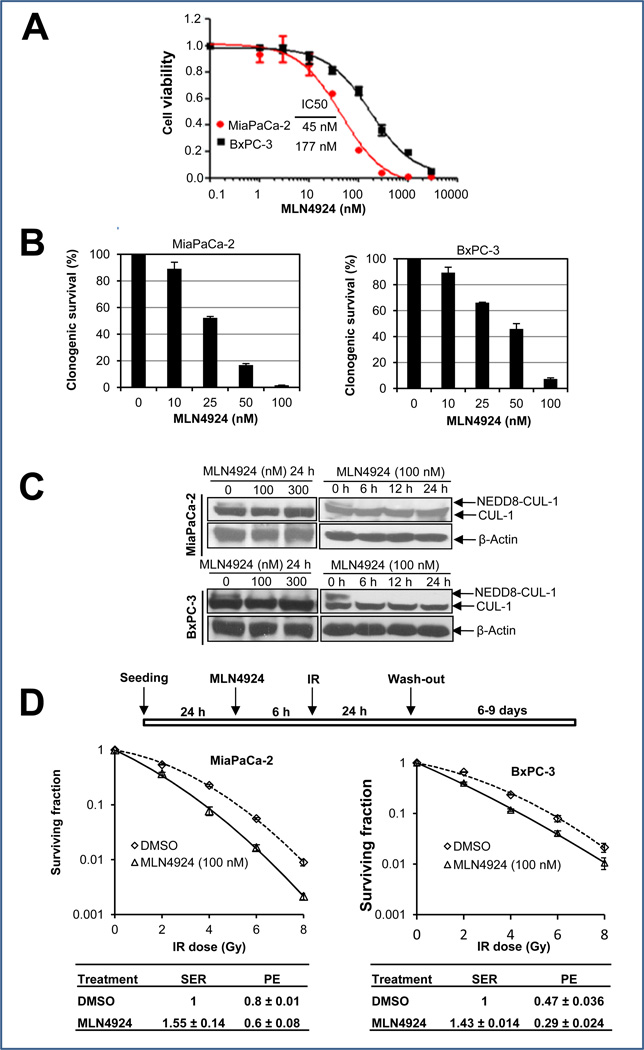
We first determined the sensitivity of two pancreatic cancer lines, MiaPaCa-2 and BxPC-3 to MLN4924 as a single agent. In an ATPlite-based 7-day cell proliferation assay, MLN4924 effectively suppressed growth of MiaPaCa-2 and BxPC-3 cells with an IC50 of 45 nM or 177 nM, respectively (Fig. 1A). In a standard clonogenic survival assay with MLN4924 in the culture medium for 7–9 days, MLN4924 caused a dose-dependent inhibition of colony formatin with an IC50 of ~25–50 nM (Fig. 1B). Thus, MLN4924 is a potent inhibitor of cell proliferation and survival in pancreatic cancer cell lines, with a higher efficacy against MiaPaCa-2 cells.
Figure 1. MLN4924 sensitizes pancreatic cancer cells to radiation.
(A&B) Growth suppression of pancreatic cancer cells by MLN4924: Cells were seeded in 96-well plates in triplicates (A) or 60-mm dishes in duplicates (B), and treated with various concentrations of MLN4924 for 7 or 9 days, respectively. Cells were then lysed for ATPlite assay (A, mean ± SEM, n = 3) or the colonies with more than 50 cells were counted, following staining (B, mean ± SD, n = 2). (C) Inhibition of cullin-1 neddylation by MLN4924. Subconfluent cells were treated with MLN4924 at indicated concentrations or for indicated time periods, followed by immunoblotting (IB) with β-actin as a loading control. (D) Radiosensitization by MLN4924: Cells were seeded in 60-mm dishes in duplicate and treated with MLN4924 and radiation as indicated. The colonies with more than 50 cells were counted after 6–9 days. Surviving fraction was calculated as the proportion of seeded cells following irradiation to form colonies relative to that of untreated cells (mean ± SEM, n = 3). SER was calculated as the ratio of the mean inactivation dose under untreated control conditions divided by the mean inactivation dose after MLN4924 treatment. PE (plating efficiency was expressed as the percentage of the colonies formed out of cells seeded).
MLN4924 sensitized cancer cells, but not normal cells to radiation
We then determined the potential radiosensitizing effect of MLN4924 in pancreatic cancer cells. We first confirmed that MLN4924 treatment indeed caused deneddylation of cullin 1; MLN4924 at 100 nM for a 6-hr exposure was sufficient to block cullin neddylation (Fig. 1C). We next used two different dosing regimens to test MLN4924 radiosensitization. In the first regimen (higher MLN4924 dose with shorter exposure time), cells were pretreated with MLN4924 (100 nM) for 6 hrs, which effectively caused the accumulation of CDT1 and WEE1 (two critical substrates of SCF E3 ligase, see below) (Fig. S1), followed by radiation at different doses up to 8 Gy. MLN4924 was washed out 24 hrs post radiation and cells were cultured in MLN4924-free medium for additional 6–9 days, allowing colony formation. Under this condition, MLN4924 effectively sensitized both pancreatic cancer cell lines to radiation with a sensitivity enhancement ratio (SER) of 1.43–1.55, respectively (Fig. 1D). In the second regimen (lower MLN4924 dose with longer exposure time), we pretreated cells with MLN4924 at a lower concentration (20 nM for MiaPaCa-2, and 25 nM for BxPC-3) for 24 hrs, followed by radiation at different doses up to 8 Gy. Cells were then cultured for additional 7–9 days in the presence of MLN4924 at these low doses (without medium change). Under this condition, we confirmed that cullin-1 was deneddylated throughout the experimental periods (Fig. S2A), and observed MLN4924 radiosensitization with SER of ~1.4 (Fig. S2B). Thus, we conclude that MLN4924 is a potent radiosensizer in pancreatic cancer cells.
In order to determine whether radiosensitization by MLN4924 is tumor cell selective, we assessed the ability of MLN4924 to radiosensitize H1299 lung cancer cells, paired with human lung fibroblast MRC5 cells, since there are no “normal” pancreatic cells capable of forming colonies in stardard clonogenic assays. We found that in H1299 cancer cells, but not in MRC5 normal cells, that cullin-1 is highly neddylated, which is completely inhibited by MLN4924 at 100 nM (Fig. S3A). Consistent with targeting cullin neddylation, MLN4924 was 5-fold more potent in growth suppression of H1299 cancer cells (IC50 = 83 nM) than MRC5 normal cells (IC50 = 429 nM) (Fig. S3B), and was 4-fold more potent in inhibition of clonogenic survival of H1299 cells (IC50 = 50 nM) than MRC5 normal cells (IC50 = 200 nM) (Fig. S3C). Likewise, MLN4924 at as low as 15 nM sensitized radioresistant H1299 cells (23) to radiation with a SER of 1.39, but had no effect on MRC5 cells (SER = 1.13) (Fig. S3D). Thus, MLN4924 appears to selectively target cancer cells with high levels of cullin neddylation and SCF E3 ligase activity.
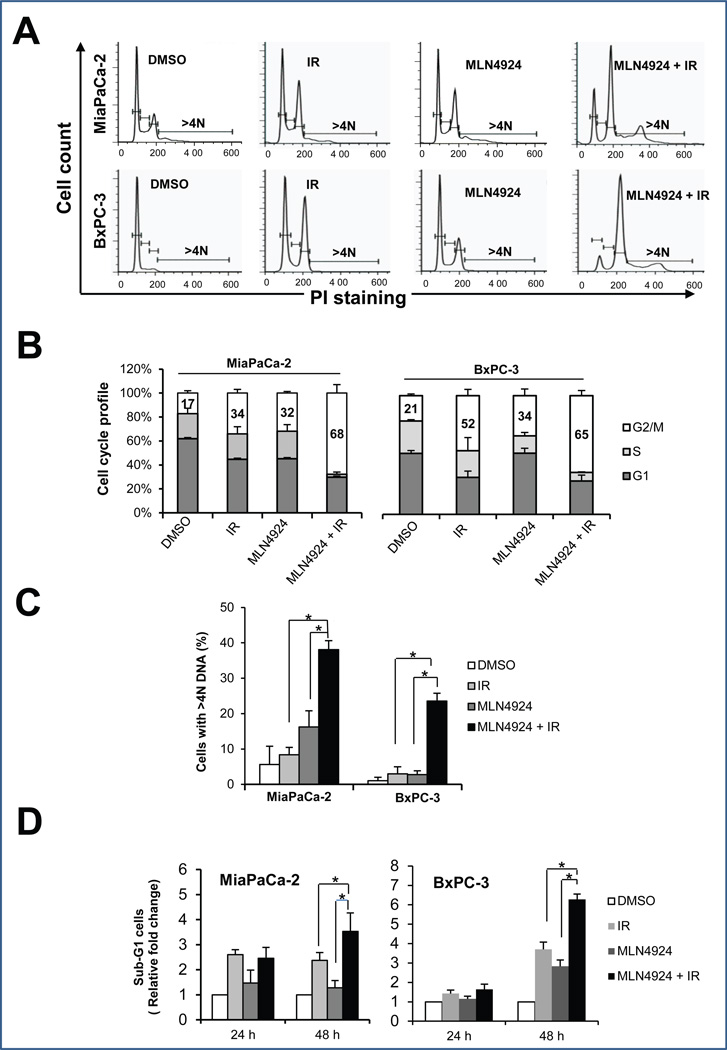
Radiosensitization by MLN4924 is attributable to enhanced G2/M arrest, aneuploidy, and induction of apoptosis
To determine the nature of MLN4924 radiosensitization, we performed cell cycle profile of two pancreatic cancer cell lines treated with MLN4924, radiation, or MLN4924 in combination with radiation (Fig. 2A), and found that MLN4924 remarkably enhanced the radiation-induced G2/M arrest in MiaPaCa-2 (IR at 34% vs. IR plus MLN4924 at 68%) and BxPC-3 cells (IR at 52% vs. IR plus MLN4924 at 65%) (Fig. 2B). MLN4924 also increased the aneuploid cell population (DNA content greater than 4N) from 10% in response to radiation alone to 40% in response to radiation plus MLN4924 in MiaPaCa-2 cells, and from 4% to 24% in BxPC-3 cells, respectively (Fig. 2C). In addition, MLN4924 increased the radiation-induced subG1 apoptotic population at later time points (48-hr; Fig. 2D). These results suggested that radiation-induced disruption of cell cycle progression and genomic instability, followed by apoptotic cell death, can be further enhanced by MLN4924.
Figure 2. MLN4924 alters cell cycle progression and induces apoptosis.
Cells were treated with MLN4924 at 100 nM for 24 hrs (A-C) or up to 48 hrs (D), alone or in combination with radiation (6 Gy), followed by FACS analysis for cell cycle profiling (A), cell cycle distribution (B), cell population with >4N DNA content (C), and sub-G1 population for apoptosis (D). Shown is mean ± SEM, (n = 3): *, p<0.05. One representative result is shown for (A).
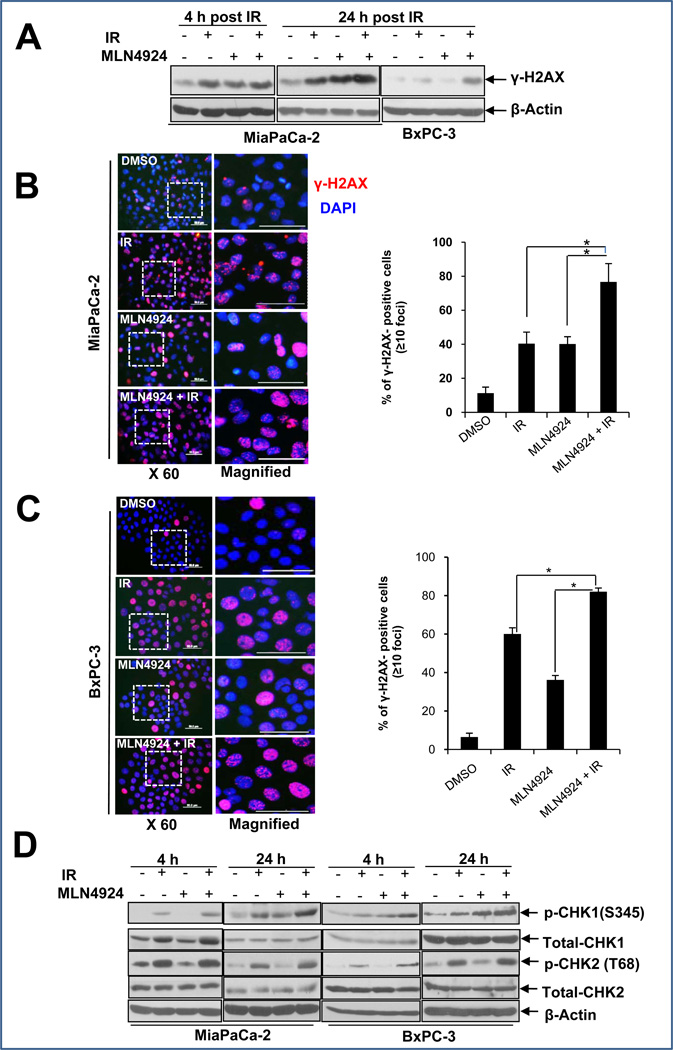
MLN4924 enhanced radiation-induced DNA damage
Since the major cellular effect of ionizing radiation is to cause DNA damage and trigger the DNA damage response (26), we, therefore, examined if MLN4924 treatment would enhance radiation-induced DNA damage and interfere with the the DNA damage repair process. We determined DNA double-strand breaks (DSBs) by measuring the overall levels of γ-H2AX protein and formation of nuclear foci upon radiation and MLN4924 treatment, alone or in combination. We found the levels of γ-H2AX increased regardless of single or combination treatment at an earlytime point (4-hr). However, a higher protein level of γ-H2AX was seen in the combination treatment group at 24 hrs for both pancreatic cancer cell lines (Fig. 3A). Consistently, we observed a significant increase in the population of γ-H2AX foci positive cells in combination treatement group (Fig. 3B&C). We also determined the DNA damage response upon MLN4924-radiation treatment by measuring phosphorylation of CHK1 and CHK2 and found that while MLN4924 did not enhance CHK2 phosphorylation, MLN4924 did enhance CHK1 phosphorylation in response to radiation, particularly at the later 24-hr time point (Fig. 3D). Similar enhancement of radiation-induced γ-H2AX levels and CHK1 phosphorylation by MLN4924 was also observed in H1299 lung cancer cells, which are sensitive to MLN4924 radiosensitization, b u t n o t i n MRC5 lung fibroblast, which are resistant to MLN4924 radiosensitization (Fig. S4). Taken together, these results suggested that MLN4924 enhances radiation-induced DNA damage and prolongs the process of DNA repair, which is likely attributable to its radiosensitizing effects.
Figure 3. MLN4924 induces DNA damage and prolongs DNA damage response.
Cells were treated with MLN4924 (100 nM) alone or in combination with radiation (6 Gy) for indicated periods of time, followed by IB analysis (A&D), or by immunofluorescence staining for γ-H2AX foci (B&C, left panels). Cells with more than 10 foci were counted and quantified data is plotted (B&C, right panel). Shown is mean ± SEM, (n = 5): *, p<0.05.
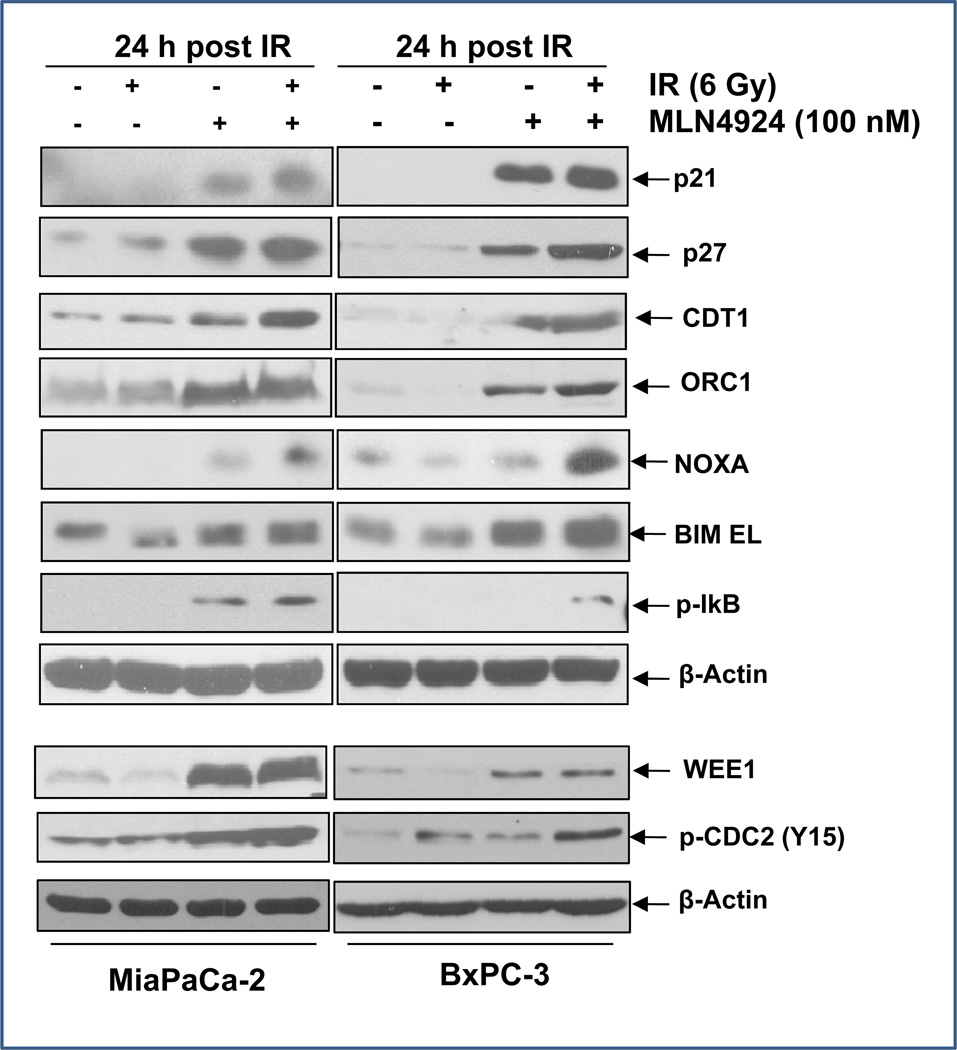
MLN4924 caused accumulation of SCF E3 ligase substrates, which is responsible for G2/M arrest and DNA damage response
To identify the mediators of MLN4924 which lead to the enhanced radiation effects, we measured the levels of proteins known to be a) the substrates of SCF E3 ligases and b) involved in the regulation of cell cycle progression, DNA damage response, and apoptosis (10, 12, 27). We found that as expected, the levels of cell cycle regulators, including p21, p27, WEE1, DNA licencing proteins CDT1 and ORC1, and apoptosis regulators NOXA, BIM EL, and pIκBα, increased substantially upon treatment with MLN4924, but not radiation (Fig. 4). The MLN4924-radiation combination further increased the levels of CDT1 and NOXA (Fig. 4), as well as WEE1 activity, as reflected by enhanced phosphorylation of its substrate, CDC2 (Y15) (Fig. 4, bottom panels), suggesting these three proteins may contribute to MLN4924 radiosensitization.
Figure 4. MLN4924 induces accumulation of SCF E3 ligase substrates.
Subconfluent cells were treated with MLN4924 (100 nM) or radiation (6 Gy) alone or in combination for 24 hr, followed by IB analysis using indicated antibodies.
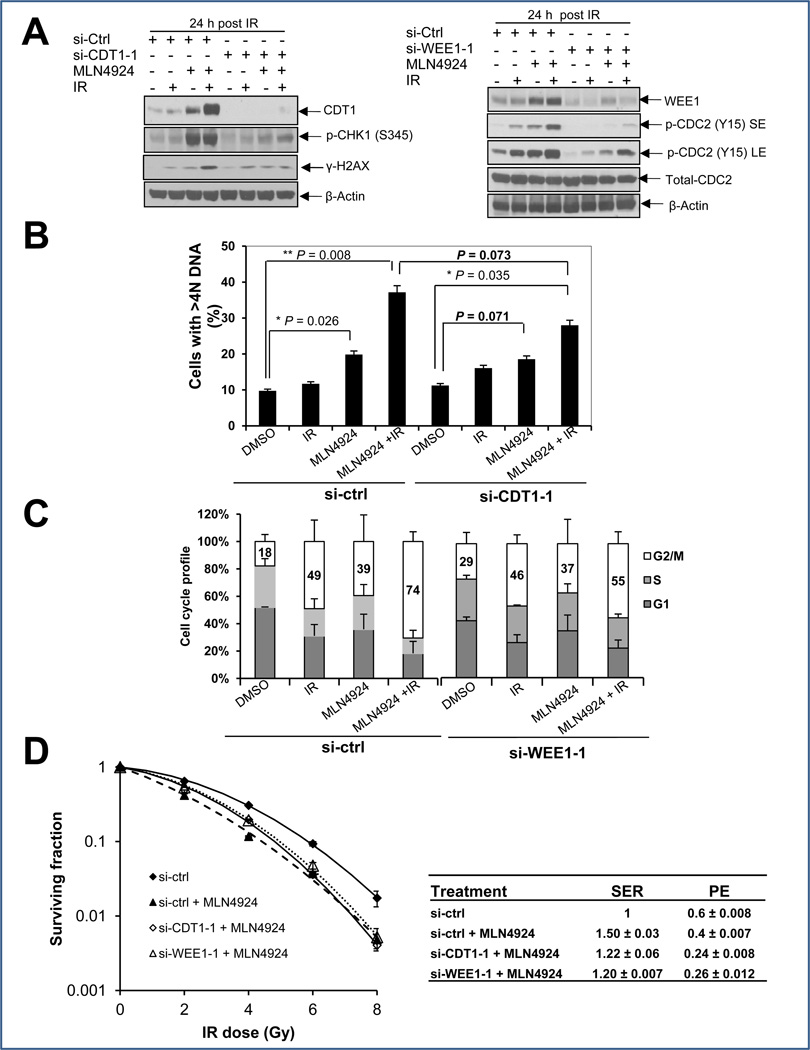
Partial rescue of aneuploidy, G2/M arrest, and radiosensitization by siRNA knockdown of CDT1 and WEE1
Since apoptosis appears to be a late and secondary effect of MLN4924-radiation (Fig. 2D), likely resulting from enhanced DNA damage and prolonged G2/M arrest, we focused our attention on CDT1, a DNA licencing protein whose overexpression is known to cause DSB and trigger DNA damage (9, 28), and WEE1, a protein tyrosine kinase that phosphorylates CDC2 on Tyr15 to arrest cells at the G2 phase of cell cycle (29, 30). We reasoned that if CDT1 and/or WEE1 were critical to MLN4924-induced enhancement of radiation effects, we should be able to abrogate these effects, at least in part by their simultaneous knockdown. Indeed, in MLN4924-radiation-treated cells, CDT1 knockdown, using two independent siRNA oligoes significantly reduced CHK1 phosphorylation (Figs. 5A&S5A) with a consequent reduction of the aneuploid population from ~40% to ~30% (Fig. 5B) or from 34% to 19% (Fig. S5B). Likewise, WEE1 knockdown using two independent siRNA oligoes abrogated CDC2 phosphorylation at Y15 (Fig. 5A, Fig. S5A) and reduced the G2/M population from 74% to 55% (Fig. 5C) or 72% to 56% (Fig. S5C). Importantly, MLN4924 radiosensitization was largely abrogated upon knockdown of CDT1 or WEE1, as assayed by the first regimen (Figs. 5D&S5D), as well as the second regimen (Fig. S2C). These results clearly suggest a causal effect of CDT1 and/or WEE1 accumulation (upon SCF E3 inactivation by MLN4924) on MLN4924 radiosensitization in MiaPaCa-2 cells. On the other hand, the incomplete rescue of MLN4924 radiosensitization upon CDT1 and/or Wee1 knockdown suggests that other downstream targets (such as p21 or p27, Figure 4) may also mediate the efficacy of MLN4924.
Figure 5. Radiation-enhancing activity of MLN4924 is inhibited at least in part by siRNA knockdown of CDT1 or WEE1.
MiaPaCa-2 cells were transfected with siRNA oligonucleotides targeting CDT1 or WEE1. Forty-eight hrs later, one portion of cells was subjected for IB analysis (A), the other portion was for FACS analysis (B&C), and still other portion was plated for clonogenic assay (D). Shown (B-D) is mean ± SD (n = 2).
MLN4924 sensitized MiaPaCa-2 cells to radiation in an in vivo xenograft tumor model
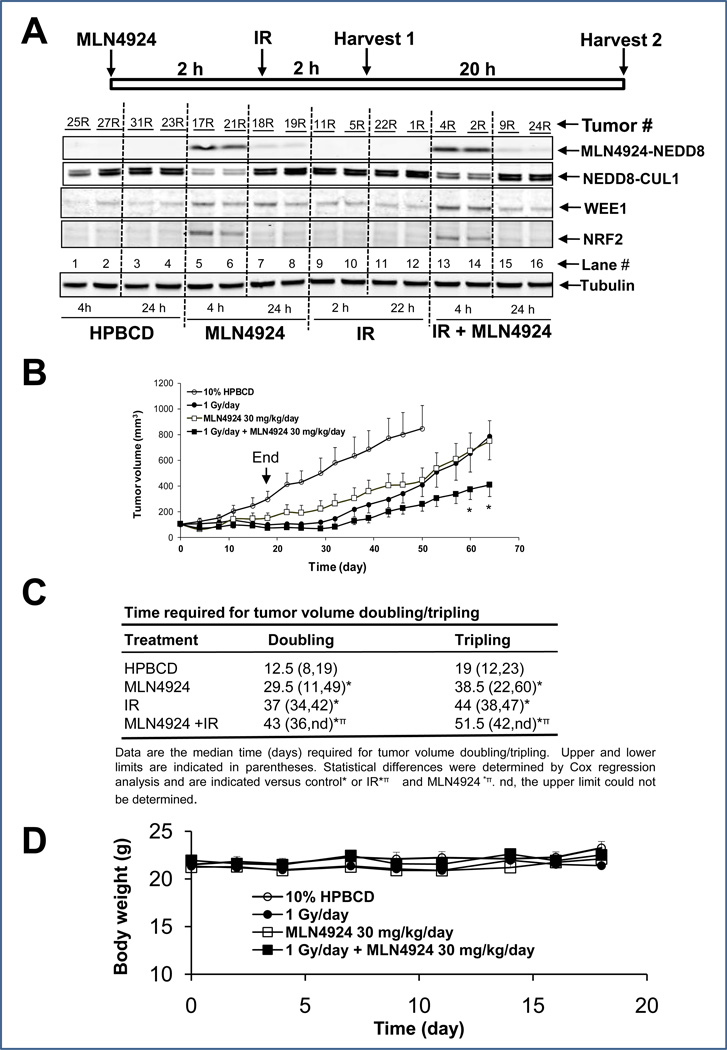
Finally, we assessed MLN4924-mediated radiosensitization in vivo using the MiaPaCa-2 xenograft model. To ensure that MLN4924 “hits” its target cullin-1 in in vivo tumors and to determine when radiation should be delivered post MLN4924 administration, we measured the levels of MLN4924-conjugated NEDD8, Neddylated cullin-1, SCF substrates, WEE1 and NRF2 at the indicated times after a single administration of vehicle, MLN4924, radiation, or MLN4924 plus radiation (two representive tumors per group). As shown in Figure 6A, MLN4924-NEDD8 adduct was detectable only in groups with MLN4924 treatment, alone (lanes 5&6) or in combination (lanes 13&14) at the early time point (4 hr), which nearly disappeard at 24 hr post treatment (lanes 7&8 and 15&16). Similarly, cullin-1 neddylation was inhibited with a corresponding increase in the SCF E3 substrates WEE1 and NRF2 in MLN4924-treated groups at the early 4 hr time point (lanes 5&6 and 13&14). At later time point (24 hr), both neddylated cullin-1 and NRF2 levels returned to the control levels with WEE1 level remaining slightly higher (lanes 7&8 and 15&16) (Fig. 6A). As expected, radiation alone had no effect on cullin-neddylation or the levels of selected SCF E3 substrates (lanes 9–12). Our results clearly demonstrate that MLN4924 indeed “hits” its target and suggest that radiation should be delivered 2 hr post-administration of MLN4924, when SCF E3 is inactivated due to cullin deneddylation.
Figure 6. MLN4924 radiosensitization in MiaPaCa-2 in vivo xenograft tumor model.
(A) MLN4924 inhibits cullin-1 neddylation and causes accumulation of SCF E3 substrates in tumor tissues: In this biomarker experiment, 5×106 MiaPaCa-2 cells were inoculated s.c in both flank sides (R, right; L, left) of nude mice. The mice were randomized when the tumor size reached 100 mm3 at 14 days after inoculation and were treated with single dose of HPBCD (vehicle control) or with MLN4924, radiation, or MLN4924-radiation combination, as indicated. Tumor tissues (n=2 per group) were harvested at indicated time points for IB. (B &C) In vivo radiosensitizing activity of MLN4924 in the MiaPaCa-2 xenograft model: The tumor inoculation and animal randomization were as described above. The number of tumors for each group are as follows: Control (HPBCD), n=12, radiation (1 Gy), n=12, MLN4924 (30 mg/kg, s.c, once a day for 5 day/week for 3 weeks), n=16, or combination (MLN4924 was given 2–3 hrs prior to radiation), n=16. The tumor growth was monitored up to 65 days and growth curve plotted. The arrow indicates the treatment end time. Student’s t test was used to compare each treatment group with the control group. Shown are mean ± SEM, * indicates p<0.05 (B), and average time periods required for tumors to double or triple in size (C). (D). MLN4924 is well tolerated by mice: Body weight was measured during the treatment and plotted (mean ± SEM).
We next determined the in vivo radiosensitizing activity of MLN4924. As shown in Figure 6B&C, administration of MLN4924 alone at a dose of 30 mg/kg s.c./day, 5 days/week for 3 weeks had a moderate inhibitory effect on tumor growth in nude mice. Radiation treatment at a clinically relevant dose of 1 Gy/day, 5 days/week for 3 weeks also had moderate anti-tumor activity. In response to treatment with the combination of MLN4924 and radiation, tumor growth was significantly inhibited compared to either treatment alone (at day 60; Fig 6B). Similarly, the time required for tumor volume doubling or tripling was significantly increased in response to MLN4924-radiation treatment compared to vehicle or radiation treatment (Fig. 6C). Consistent with this, we also observed a signicantly reduced tumor volume nadir corresponding with a marginally significant increase in the tumor regression rate in response to MLN4924-radiation (compared to radiation) (Supplemental Table 1). Importantly, the combination treatment was well-tolerated by the animals with a minimal loss of body weight (Fig 6D). Taken together, our results demonstrate that MLN4924 is a radiosensitizer in pancreatic cancer, as assessed in both in vitro cell culture and in vivo tumor xenograft models, likely by causing accumulation of CDT1 and WEE1, among with other substrates of SCF E3 ubiquitin ligase.
Discussion
Our previous work has demonstrated that SCF E3 ubiquitin ligase is an attractive anti-cancer target [for review, see (15, 31–33)]. However, due to the complexity of establishing a high-throughput (HTS) screen for multi-components SCF E3 ubiquitin ligase, there is no single small molecule discovered that directly inhibits its ligase activity, although a number of HTS methods have been established and optimized to screen for small molecule inhibitors of single peptide E3 ligases (31, 34). MLN4924 is a newly discovered small molecule inhibitor of NAE (13). MLN4924 binds to NAE at its active site to create a covalent NEDD8-MLN4924 adduct, which resembles NEDD8 adenylate, but cannot be further utilized in subsequent intraenzyme reactions, thus inhibiting NAE activity and blocking cullin neddylation (13, 35). Since cullin neddylation is required for SCF E3 activity (16, 18, 19), MLN4924 becomes the first of a class of “indirect” inhibitors of SCF E3. By inhibiting SCF E3 ligase activity, MLN4924 causes the accumulation of a number of SCF E3 substrates to suppress the growth of acute myeloid leukemia (36), diffuse large B-cell lymphoma (37), and many cancer cell lines derived from solid tumors both in vitro and in vivo by inducing apoptosis (13, 36, 37) and senescence (38–40). Importantly, in vivo xenograft assays showed that MLN4924 was well tolerated in mice at various doses and treatment regimens (13), showing a selective killing of cancer cells. With all these preclinical efficacies, MLN4924 has been advanced to several Phase I clinical trials for solid tumors and hematological malignancies (41).
In the present study, we tested our hypothesis that MLN4924 is a potent radiosensitizing agent for pancreatic cancer cells, based upon our target validation work using both siRNA knockdown and gene knockout approaches for RBX1 and SAG, the RING components of SCF E3, required for its ligase activity (9, 10, 21, 22). For the first time, we demonstrated here that in in vitro cell culture models, MLN4924 was a potent growth inhibitor as a single agent, as well as a radiosensitizer in two pancreatic cancer cell lines and in one lung cancer cell line, but not normal lung fibroblast cells. The resistance of normal fibroblasts to MLN4924 radiosensitization is likely due to the lack of cullin neddylation. We also demonstrated in vivo using a MiaPaCa-2 xenograft model, that MLN4924 inhibited tumor growth and conferred radiosensitization with minimal toxicity to the mice. Mechanistically, we showed that MLN4924 enhanced radiation-induced G2/M arrest, aneuploidy, and eventually apoptosis by inhibiting cullin neddylation to trigger the accumulation of a number of SCF E3 substrates, related to DNA damage response and cell cycle regulation. Among these accumulated substrates, CDT1 and WEE1 were further increased when combined with radiation, and were critical for enhanced DNA damage and aneuploidy or G2/M arrest, respectively, and eventually for MLN4924 radiosensitization, as evidenced by siRNA knockdown-based rescuing experiments.
Precise duplication of the genome at the S phase of each cell cycle requires initiation of DNA replication from thousands of origins. Initiation of too few origins would cause collapse of replication forks, leading to DNA damage and incomplete replication of the genome, whereas more than one intiation of DNA replication per cell cycle would cause DNA “hyper-replication” or “rereplication” and subsequent DNA damage (42), leading to genomic instability and cancer (43). CDT1 is a key licensing factor which, along with the protein CDC6, functions to form the pre-replicative complex (pre-RC) to initiate DNA replication (44). It was recently reported that overexpression of Double-arked (Dup), the Drosophila ortholog of CDT1 caused DNA rereplication and DNA damage, followed by caspase activation and apoptotic cell death (45). In human U2-OS cancer cells, CDT1 overexpression caused double strand breaks to trigger DDR, followed by induction of senescence and apoptosis (28). Induction of CDT1 accumulation by MLN4924 was found to be attributable to DNA rereplication, aneuploidy, and apoptotic death in HCT116 cells (13, 40, 46). Our data presented here indicated that CDT1 accumulation is also found in two pancreatic cancer cell lines and is critical for enhanced aneuploidy and MLN4924 radiosensitization.
The WEE1 kinase negatively regulates G2/M transition by directly phosphorylating Tyr15 of CDC2, which inhibits the CDC2 activity (30). We have previously shown that WEE1 can be stabilized and activated by binding to 14-3-3β (47), and is a valid radiosensitizing target whose inhibition abrogates the radiation-induced G2 checkpoint (48, 49). In this study, we found that the level of WEE1, a substrate of SCF E3 ligase (50), increased, expectedly upon MLN4924 treatment. The WEE1 activity was further enhanced when MLN4924 was combined with radiation. Our rescue experiment demonstrated that accumulated WEE1 is responsible, at least in part, for a remarkable increase in the G2/M population in response to MLN4924 and radiation, which appears causally related to MLN4924 radiosensitization.
In summary, our study revealed radiosensitizing activity of MLN4924, a small molecule inhibitor of SCF E3, in pancreatic cancer as well as in lung cancer cells, and elucidated its mechanisms of action which include accumulation of CDT1 and WEE1, among other cell cycle and apoptosis regulators, which trigger DNA damage, aneuploidy and G2/M arrest, followed by apoptotic death. Our study, therefore, provides the first preclinical proof-of-concept evidence for future development of MLN4924 as a novel radiosensitizing agent against pancreatic cancer and possibly other types of human cancers.
Supplementary Material
Acknowledgements
We thank Millennium Pharmaceuticals, Inc. for providing us MLN4924. This work is supported by the NCI grants, CA111554, CA118762 and CA156744 to YS, and CA78554 and CA78554-10S231071204 to TSL. This work was also partially supported by Millennium Pharmaceuticals, Inc.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Sharma C, Eltawil KM, Renfrew PD, Walsh MJ, Molinari M. Advances in diagnosis, treatment and palliation of pancreatic carcinoma: 1990–2010. World J Gastroenterol. 2011;17:867–897. doi: 10.3748/wjg.v17.i7.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iacobuzio-Donahue CA, Fu B, Yachida S, Luo M, Abe H, Henderson CM, et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol. 2009;27:1806–1813. doi: 10.1200/JCO.2008.17.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loehrer PJ, Powell ME, Cardenes HR, Wagner L, Brell JM, Ramanathan RK, et al. A randomized phase III study of gemcitabine in combination with radiation therapy versus gemcitabine alone in patients with localized, unresectable pancreatic cancer: E4201. J Clin Oncol. 2008 ASCO Meeting Abstracts May 20 4506. [Google Scholar]
- 5.Ben-Josef E, Griffith K, Francis IR, Khan G, Lawrence T, Abrams R, et al. Phase I radiation dose-escalation trial of intensity-modulated radiotherapy (IMRT) with concurrent fixed dose-rate gemcitabine (FDR-G) for unresectable pancreatic cancer. J Clin Oncol. 2009;27:15s. (suppl; abstr 4602) [Google Scholar]
- 6.Morgan MA, Parsels LA, Zhao L, Parsels JD, Davis MA, Hassan MC, et al. Mechanism of radiosensitization by the Chk1/2 inhibitor AZD7762 involves abrogation of the G2 checkpoint and inhibition of homologous recombinational DNA repair. Cancer Res. 2010;70:4972–4981. doi: 10.1158/0008-5472.CAN-09-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gallinger S, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 8.Morgan MA, Parsels LA, Kollar LE, Normolle DP, Maybaum J, Lawrence TS. The combination of epidermal growth factor receptor inhibitors with gemcitabine and radiation in pancreatic cancer. Clin Cancer Res. 2008;14:5142–5149. doi: 10.1158/1078-0432.CCR-07-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia L, Bickel JS, Wu J, Morgan MA, Li H, Yang J, et al. RBX1 (RING-box protein 1) E3 ubiquitin ligase is required for genomic integrity by modulating DNA replication licensing proteins. J Biol Chem. 2011;286:3379–3386. doi: 10.1074/jbc.M110.188425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia L, Yang J, Hao X, Zheng M, He H, Xiong X, et al. Validation of SAG/RBX2/ROC2 E3 Ubiquitin Ligase as an Anticancer and Radiosensitizing Target. Clin Cancer Res. 2010;16:814–824. doi: 10.1158/1078-0432.CCR-09-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 12.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 13.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 14.Wu K, Fuchs SY, Chen A, Tan P, Gomez C, Ronai Z, et al. The SCF(HOS/beta-TRCP)-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation. Mol Cell Biol. 2000;20:1382–1393. doi: 10.1128/mcb.20.4.1382-1393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia. 2006;8:645–654. doi: 10.1593/neo.06376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, et al. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 19.Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xirodimas DP. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem Soc Trans. 2008;36:802–806. doi: 10.1042/BST0360802. [DOI] [PubMed] [Google Scholar]
- 21.Jia L, Soengas MS, Sun Y. ROC1/RBX1 E3 ubiquitin ligase silencing suppresses tumor cell growth via sequential induction of G2-M arrest, apoptosis, and senescence. Cancer Res. 2009;69:4974–4982. doi: 10.1158/0008-5472.CAN-08-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan M, Zhu Y, Kovacev J, Zhao Y, Pan ZQ, Spitz DR, et al. Disruption of Sag/Rbx2/Roc2 induces radiosensitization by increasing ROS levels and blocking NF-kB activation in mouse embryonic stem cells. Free Radic Biol Med. 2010;49:976–983. doi: 10.1016/j.freeradbiomed.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng M, Morgan-Lappe SE, Yang J, Bockbrader KM, Pamarthy D, Thomas D, et al. Growth inhibition and radiosensitization of glioblastoma and lung cancer cells by small interfering RNA silencing of tumor necrosis factor receptor-associated factor 2. Cancer Res. 2008;68:7570–7578. doi: 10.1158/0008-5472.CAN-08-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fertil B, Dertinger H, Courdi A, Malaise EP. Mean inactivation dose: a useful concept for intercomparison of human cell survival curves. Radiat Res. 1984;99:73–84. [PubMed] [Google Scholar]
- 25.Zhao L, Morgan MA, Parsels LA, Maybaum J, Lawrence TS, Normolle D. Bayesian hierarchical changepoint methods in modeling the tumor growth profiles in xenograft experiments. Clin Cancer Res. 2011;17:1057–1064. doi: 10.1158/1078-0432.CCR-10-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Begg AC, Stewart FA, Vens C. Strategies to improve radiotherapy with targeted drugs. Nat Rev Cancer. 2011;11:239–253. doi: 10.1038/nrc3007. [DOI] [PubMed] [Google Scholar]
- 27.Skaar JR, D'Angiolella V, Pagan JK, Pagano M. SnapShot: F Box Proteins II. Cell. 2009;137:1358. doi: 10.1016/j.cell.2009.05.040. e1. [DOI] [PubMed] [Google Scholar]
- 28.Liontos M, Koutsami M, Sideridou M, Evangelou K, Kletsas D, Levy B, et al. Deregulated overexpression of hCdt1 and hCdc6 promotes malignant behavior. Cancer Res. 2007;67:10899–10909. doi: 10.1158/0008-5472.CAN-07-2837. [DOI] [PubMed] [Google Scholar]
- 29.Heald R, McLoughlin M, McKeon F. Human wee1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated Cdc2 kinase. Cell. 1993;74:463–474. doi: 10.1016/0092-8674(93)80048-j. [DOI] [PubMed] [Google Scholar]
- 30.Parker LL, Piwnica Worms H. Inactivation of the p34cdc2-cyclin B complex by the human WEE1 tyrosine kinase. Science. 1992;257:1955–1957. doi: 10.1126/science.1384126. [DOI] [PubMed] [Google Scholar]
- 31.Jia L, Sun Y. SCF E3 ubiquitin ligases as anticancer targets. Current Cancer Drug Targets. 2011;11:347–356. doi: 10.2174/156800911794519734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Y. Targeting E3 ubiquitin ligases for cancer therapy. Cancer Biol Therapy. 2003;2:623–629. [PubMed] [Google Scholar]
- 33.Wei D, Sun Y. Small RING finger proteins RBX1 and RBX2 of SCF E3 ubiquitin ligases: the role in cancer and as cancer targets. Genes & Cancer. 2010;1:700–707. doi: 10.1177/1947601910382776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Y. Overview of approaches for screening for ubiquitin ligase inhibitors. Methods Enzymol. 2005;399:654–663. doi: 10.1016/S0076-6879(05)99043-5. [DOI] [PubMed] [Google Scholar]
- 35.Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 36.Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115:3796–3800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- 37.Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood. 2010;116:1515–1523. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- 38.Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464:374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia L, Li H, Sun Y. Induction of p21-Dependent Senescence by an NAE Inhibitor, MLN4924, as a Mechanism of Growth Suppression. Neoplasia. 2011;13:561–569. doi: 10.1593/neo.11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin JJ, Milhollen MA, Smith PG, Narayanan U, Dutta A. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res. 2010;70:10310–10320. doi: 10.1158/0008-5472.CAN-10-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soucy TA, Smith PG, Rolfe M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clin Cancer Res. 2009;15:3912–3916. doi: 10.1158/1078-0432.CCR-09-0343. [DOI] [PubMed] [Google Scholar]
- 42.Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- 43.Dutta A. Chaotic license for genetic instability and cancer. Nat Genet. 2007;39:10–11. doi: 10.1038/ng0107-10. [DOI] [PubMed] [Google Scholar]
- 44.Rialland M, Sola F, Santocanale C. Essential role of human CDT1 in DNA replication and chromatin licensing. J Cell Sci. 2002;115:1435–1440. doi: 10.1242/jcs.115.7.1435. [DOI] [PubMed] [Google Scholar]
- 45.Mehrotra S, Maqbool SB, Kolpakas A, Murnen K, Calvi BR. Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes Dev. 2008;22:3158–3171. doi: 10.1101/gad.1710208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milhollen MA, Narayanan U, Soucy TA, Veiby PO, Smith PG, Amidon B. Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res. 2011;71:3042–3051. doi: 10.1158/0008-5472.CAN-10-2122. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Jacobs C, Hook KE, Duan H, Booher RN, Sun Y. Binding of 14-3-3beta to the carboxyl terminus of Wee1 increases Wee1 stability, kinase activity, and G2-M cell population. Cell Growth Differ. 2000;11:211–219. [PubMed] [Google Scholar]
- 48.Li J, Wang Y, Sun Y, Lawrence TS. Wild-type TP53 inhibits G(2)-phase checkpoint abrogation and radiosensitization induced by PD0166285, a WEE1 kinase inhibitor. Radiat Res. 2002;157:322–330. doi: 10.1667/0033-7587(2002)157[0322:wttigp]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Li J, Booher RN, Kraker A, Lawrence T, Leopold WR, et al. Radiosensitization of p53 mutant cells by PD0166285, a novel G(2) checkpoint abrogator. Cancer Res. 2001;61:8211–8217. [PubMed] [Google Scholar]
- 50.Watanabe N, Arai H, Nishihara Y, Taniguchi M, Hunter T, Osada H. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc Natl Acad Sci U S A. 2004;101:4419–4424. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.