Abstract
AmyR is commonly considered a regulator of starch degradation whose activity is induced by the presence of maltose, the disaccharide building block of starch. In this study, we demonstrate that the role of AmyR extends beyond starch degradation. Enzyme activity assays, genes expression analysis and growth profiling on d-glucose- and d-galactose-containing oligo- and polysaccharides showed that AmyR regulates the expression of some of the Aspergillus niger genes encoding α- and β-glucosidases, α- and β- galactosidases, as well as genes encoding α-amlyases and glucoamylases. In addition, we provide evidence that d-glucose or a metabolic product thereof may be the inducer of the AmyR system in A. niger and not maltose, as is commonly assumed.
Electronic supplementary material
The online version of this article (doi:10.1007/s00253-011-3550-6) contains supplementary material, which is available to authorized users.
Keywords: Aspergillus niger, AmyR, Polysaccharide degradation, Gene regulation
Introduction
AmyR was the first transcriptional activator of the GAL4 type identified in filamentous fungi that is involved in the degradation of plant polysaccharides (Petersen et al. 1999). Its role in starch degradation has been studied in detail in different Aspergillus species, such as Aspergillus nidulans and Aspergillus oryzae, and AmyR was shown to activate expression of glucoamylase, α-amylase and α-glucosidase in the presence of starch or maltose (Gomi et al. 2000; Petersen et al. 1999; Tani et al. 2001). The amylolytic system has also been studied in Aspergillus niger, and a recent study compared the expression of genes related to starch degradation using micro-array analysis (Yuan et al. 2008) but did not study in detail other genes that may be under AmyR control.
Induction of the amylolytic system in Aspergillus is reported to be caused by the presence of starch or its disaccharide building block, maltose (Barton et al. 1972; Carlsen and Nielsen 2001; Pedersen et al. 2000; Santerre Henriksen et al. 1999), and the latter compound is often suggested to be the low-molecular-weight inducer of the system. However, the actual nature of the inducer has not been studied in detail. It has also been claimed that iso-maltose is the inducing compounds (Kato et al. 2002), but no conclusive evidence has been presented to verify this. d-glucose has been shown to repress the expression of the amylolytic genes (Tsukagoshi et al. 2001) but was also reported as an inducer for the amylolytic genes of A. oryzae (Carlsen and Nielsen 2001). However, no studies have been reported in which low levels of d-glucose were tested for induction of the amylolytic system in Aspergillus.
A role in both induction and repression has been observed previously for d-xylose and the xylanolytic system of A. niger (de Vries et al. 1999b). In this fungus, d-xylose induces the xylanolytic regulator XlnR that activates expression of xylanolytic and cellulolytic genes. The concentration of d-xylose in the medium affects the expression level of the genes. High d-xylose concentrations result in activation of the carbon catabolite repressor protein CreA that represses the expression of the XlnR-regulated genes.
Although XlnR was originally described as a xylanolytic activator, later studies indicated that it also controlled genes that were not involved in xylan degradation and utilization. These genes encoded enzymes involved in cellulose, xyloglucan and galactomannan degradation (de Vries et al. 1999a; Gielkens et al. 1999; Hasper et al. 2002).
Here, we present a study into the A. niger amyR gene in which we constructed deletion and multicopy strains. Using these strains, we have found that d-glucose induces the amylolytic system depending on the concentration in the medium. In addition, it became clear that AmyR has a broader physiological role than starch degradation in that it controls the production of d-glucose and d-galactose releasing enzymes.
Materials and methods
Strains and media
All A. niger strains used are derivatives of N402 (FGSC A733; Table 1). Inocula for shake flask cultivations were obtained from cultures grown on complete medium agar plates (de Vries et al. 2004). Minimal medium (MM; de Vries et al. 2004) was inoculated with harvested conidia to give a concentration of 1.0 × 106 conidia per milliliter. Supplements to complement the auxotrophic requirements were added when required and were 200 mg/l leucine, 122 mg/l uridine and 1 mg/l nicotinic acid (niacin). The liquid pre-cultures contained 2% d-fructose as the sole carbon source. Sugars were not autoclaved but filter-sterilised and added to the autoclaved medium. Maltose, d-glucose (G8270, 99.5%), d-fructose, lactose and d-xylose were from Sigma. Soluble starch was from Difco. Cultures were grown at 30°C, with 250 rpm rotary shaking. Precultures were cultivated for 16 h, mycelia were washed with MM (without carbon source), and 1.5 g wet weight filamentous mycelium was transferred to shake flasks containing 70 ml MM with the carbon sources, as stated in the results. Transferred mycelium was then incubated at 30°C with 250 rpm rotary shaking for 2–24 h (see “Results” for details of specific experiments) before mycelia were harvested and/or culture filtrate samples taken. A. niger transformations were performed as described previously (Kusters-van Someren et al. 1991).
Table 1.
A. niger strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| N402 | cspA1 | Bos et al. (1988) |
| NW249 | ΔargB, pyrA6, leuA1, nicA1 | vanKuyk et al. (2004) |
| UU-A049.1 | ΔargB, pyrA6, leuA1, nicA1, argB + | This study |
| UU-A101.1 | ΔargB, pyrA6, leuA1, nicA1, ΔamyR:: argB + | This study |
| UU-A001.13 | ΔargB, pyrA6, leuA1, nicA1, argB +, mcamyR | This study |
| UU-A001.24 | argBΔ, pyrA6, leuA1, nicA1, argB +, mcamyR | This study |
Molecular methods
DNA manipulations and molecular biology techniques were performed using standard methods (Sambrook et al. 1989) and Escherichia coli DH5α. Gel extractions were performed using the QIAgen QIAquick gel extraction kit.
RNA analysis was performed as described previously (de Vries et al. 2002). Internal fragments of lacA (A00968, An01g12150; Kumar et al. 1992), aglC (AJ251873, An09g00260/270; Ademark et al. 2001), agdA (An04g06920; Pel et al. 2007), glaA (X00548, An03g06550; Boel et al. 1984), amyA (An04g06930) and amyR (An04g06910; Pel et al. 2007) were used as probes for expression analysis. A 0.7-kb EcoRI fragment from the gene encoding the 18S rRNA subunit (Melchers et al. 1994) was used as an RNA loading control.
Sequence analysis
Nucleotide sequences were analysed with computer programs based on those of Devereux et al. (Devereux et al. 1984). Sequence alignments were performed by using the Blast programs (Altschul et al. 1990) at the server of the National Center for Biotechnology Information, Bethesda, USA (http://www.ncbi.nlm.nih.gov/blast/). Synteny analysis was performed at the AspGD website (http://www.aspgd.org/) using the Sybil programme.
Enzyme assays
Extracellular hydrolytic activities were assayed using 0.01% substrate, 20–40 μl sample and 25 mM sodium acetate pH 5.0 in a total volume of 100 μl. The mixtures were incubated for 1 h at 30°C after which the reaction was stopped by adding 100 μl 0.25 M Na2CO3. Absorbance was measured at 405 nm in a microtiter plate reader. The activity was calculated using a standard curve of p-nitrophenol. The substrates used for enzyme assays were all obtained from Sigma and were p-nitrophenol-α-arabinofuranoside, p-nitrophenol-α-xylopyranoside, p-nitrophenol-β-xylopyranoside, p-nitrophenol-α-galactopyranoside, p-nitrophenol-β-galactopyranoside, p-nitrophenol-α-glucopyranoside, p-nitrophenol-β-glucopyranoside, p-nitrophenol-α-fucopyranoside, p-nitrophenol-β-fucopyranoside, p-nitrophenol-β-glucuronoside and p-nitrophenol-β-mannopyranoside to measure α-arabinofuranosidase, α- and β-xylosidase, α- and β-galactosidase, α- and β-glucosidase, β-glucuronidase and β-mannosidase, respectively.
Results
Identification and cloning of amyR
A lambda phage clone was isolated from an A. niger gDNA library using a fragment of the A. oryzae amyR gene as a probe. A 4.3-kb NsiI fragment was subcloned into pGEM-11 and designated pJG01. Sequence analysis showed that this fragment contained the coding region of A. niger amyR as well as the 5′-flanking region and part of the A. niger agdA gene. Further analysis using the Sybil tool at www.aspgd.org (Crabtree et al. 2007) demonstrated that the amylolytic cluster (amyA–aglA–amyR) is highly conserved in all eight sequenced Aspergilli, with only a small insertion downstream of amyR in Aspergillus terreus (Suppl. Fig. 1). In all these species except A. nidulans, the proteolytic regulator PrtT (Punt et al. 2008) lies directly upstream from the cluster.
Construction and analysis of disruption and multicopy amyR strains
A deletion construct (pAmyRd) was made by introducing a PstI site 800 bp downstream of the endogenous PstI site of pJG01. Plasmid pJG01 was then digested with BamHI/NsiI and the liberated fragment cloned into pGEM9. The resulting construct (pJG10) was digested with PstI/BamHI, and a BglII/PstI fragment containing the argB gene of A. niger was inserted. Insertion of this construct at the amyR locus would result in the removal of 1,600 bp, consisting of approximately 600 bp 5′-region and 1,000 bp coding region (including the zinc finger). This fragment was used to transform A. niger NW249. In addition, pJG01 was used in a co-transformation with pIM2101 (containing the A. niger argB gene (Lenouvel et al. 2002)) to obtain amyR multicopy strains in A. niger NW249. Transformants were screened for their ability to grow on starch. Selected transformants from the disruption transformation with reduced growth and from the multicopy transformation with improved growth on starch were used for Southern analysis and demonstrated that multiple disruption and multicopy transformants were obtained (data not shown). Based on this analysis, UU-A001.24 was selected as amyR multicopy (overexpression) transformant and UU-A101.1 was selected as an amyR disruptant.
The influence of AmyR extends beyond amylolytic genes
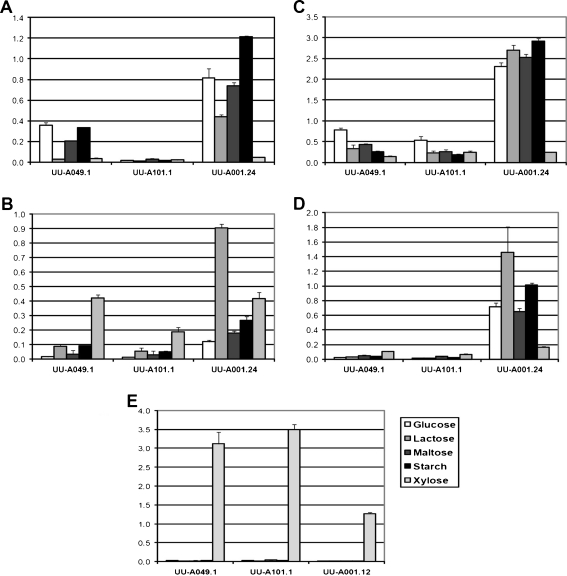
Strains UU-A001.24 (amyR overexpression) and UU-A101.1 (amyR disruptant) together with UU-A049.1 (reference strain) were grown overnight in complete medium (CM) with 2% fructose and transferred to MM with the carbon sources indicated in Fig. 1 for 6 h. Culture filtrate samples were used to determine the presence of extracellular enzymes. No activity was detected on any of the carbon sources for α- or β-fucosidase, α-xylosidase, α-arabinofuranosidase, β-mannosidase or β-glucuronidase (data not shown). However, α- and β-galactosidase and α- and β-glucosidase activity were detected on d-glucose, lactose, maltose and starch (Fig. 1). Compared with the reference strain (UU-A049.1), these activities were lower in the amyR disruptant and 2- to 15-fold increased in the amyR multicopy strain.
Fig. 1.
Extracellular activities in UU-A049.1 (reference strain), UU-A101.1 (ΔamyR) and UU-A001.24 (amyR multicopy strain) on five carbon sources (25 mM for mono- and disaccharides, 1% for starch). Activity (nanomoles per minute per milliliter culture filtrate) was assayed in culture filtrates obtained 6 h after transfer. Enzyme activities measured were A α-glucosidase (AGD), B β-glucosidase (BGL), C α-galactosidase (AGL), D β-galactosidase (LAC), and (E) β-xylosidase (BXL)
Based on recent A. niger genome annotations (Coutinho et al. 2009; Pel et al. 2007), genes encoding α-amylase (AMY), glucoamylase (GLA), α-glucosidase (AGD), β-glucosidase (BGL), α-galactosidase (AGL) and β-galactosidase (LAC) were analysed for their expression in an A. niger wild-type and amyR disruptant on maltose using previously published micro-array data (Yuan et al. 2008). This demonstrated that AmyR controls only a small subset of these genes (Suppl. Table 1), but this subset included two BGL-, two AMY-, two AGD-, two GLA-, two AGL and one LAC-encoding genes, supporting the activity assays performed in our study. All these genes contained at least one putative AmyR binding site in their promoter.
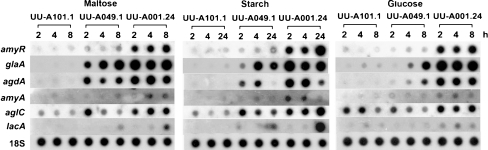
To confirm these data, a transfer experiment was performed in which UU-A049.1, UU-A001.24 and UU-A101.1 were pre-grown O/N in CM with 2% fructose. Mycelial aliquots were transferred to 25 mM d-glucose, 25 mM maltose and 1% starch and incubated for the time points indicated in Fig. 2. Northern analysis was performed for the three genes of the AmyR cluster (amyR, agdA, amyA), the main glucoamylase (glaA) and genes encoding an α-galactosidase (aglC; Ademark et al. 2001) and a β-galactosidase (lacA; de Vries et al. 1999a; Kumar et al. 1992). Expression of glaA, amyA and agdA on maltose and starch was reduced in the amyR disruption strain (UU-A101.1) and increased in the amyR multicopy strain (UU-A001.24; Fig. 2). A similar effect was observed for lacA and aglC, although the expression of the genes peaked at the latest and earliest timepoint, respectively.
Fig. 2.
Expression analysis of amyR (amylolytic activator), glaA (glucoamylase), agdA (α-glucosidase), amyA (α-amylase), aglC (α-galactosidase) and lacA (β-galactosidase) in UU-A049.1 (ref), UU-A101.1 (ΔamyR) and UU-A001.24 (amyR multicopy strain) during growth on 25 mM maltose (a), 1% starch (b) and 25 mM d-glucose (c) at various time points (in hours)
A similar expression pattern was observed for glaA, agdA and amyA on d-glucose, and expression was highest at the latest time point (Fig. 2). Expression of aglC and lacA was not reduced in the amyR disruptant compared to the reference on d-glucose but was increased in the amyR multicopy strain.
Influence of AmyR on growth of A. niger on oligo- and polysaccharides
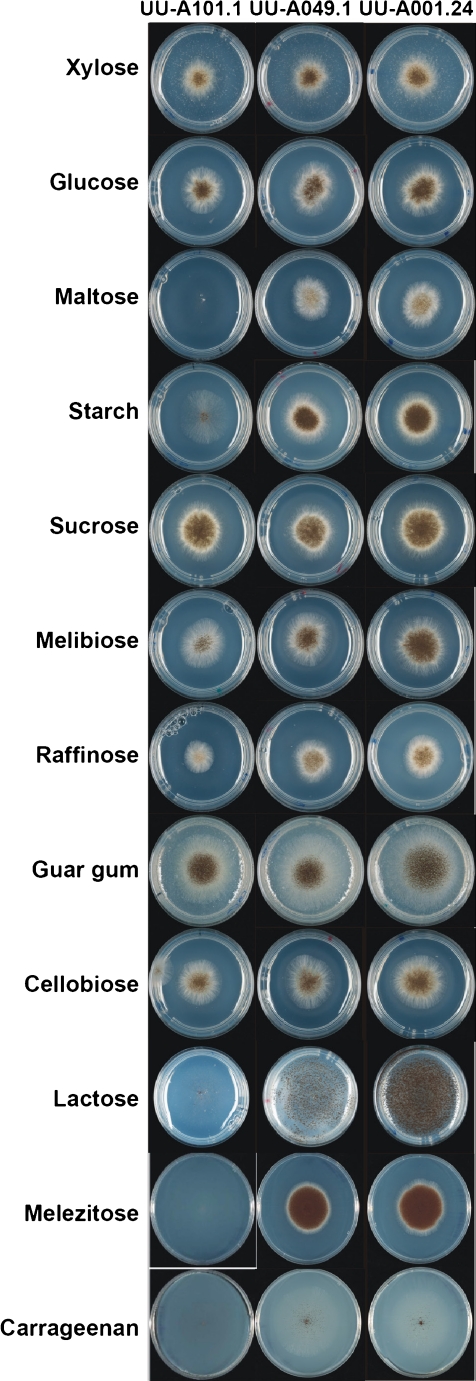
To study the physiological effect of this putative broader role for AmyR, a growth experiment was performed using UU-A049.1, UU-A001.24 and UU-A101.1 on a variety of carbon sources, including controls (xylose, d-glucose) and several oligo- and polysaccharides containing α- or β-linked d-glucose or d-galactose residues (Fig. 3). Reduced growth was observed for the amyR disruptant (UU-A101.1) on maltose, starch, lactose, melibiose, raffinose, melezitose and carrageenan compared with the reference strain (UU-A049.1). No clearly improved growth could be seen on most of these substrates for the amyR multicopy strain (UU-A001.24), with the exception of lactose, where some increase in vegetative growth was visible, but especially a significant increase in sporulation was observed. Growth and, in particular, sporulation of the reference strain and the amyR multicopy strain on carrageenan was poor on these substrates. No increase in sporulation was observed in the amyR multicopy strain compared with the reference strain.
Fig. 3.
Growth profiles of UU-A049.1 (reference strain), UU-A101.1 (ΔamyR) and UU-A001.24 (amyR multicopy strain) on a variety of carbon sources. Carbon source concentrations were 25 mM monosaccharides (d-glucose, d-xylose), 12.5 mM disaccharides (maltose, sucrose, lactose, cellobiose, melibiose), 7.57 mM trisaccharides (raffinose, melezitose), 1% polysaccharides (starch, carrageenan)
No difference in growth was observed between the strains using sucrose, d-xylose or d-glucose as a carbon source and only a slight reduction in growth of the amyR disruptant on cellobiose and Guar gum. Furthermore, no difference in growth was observed on chitin, N-acetylglucosamine and inulin (data not shown).
Increased concentrations of d-glucose results in altered morphology in amyR multicopy strains
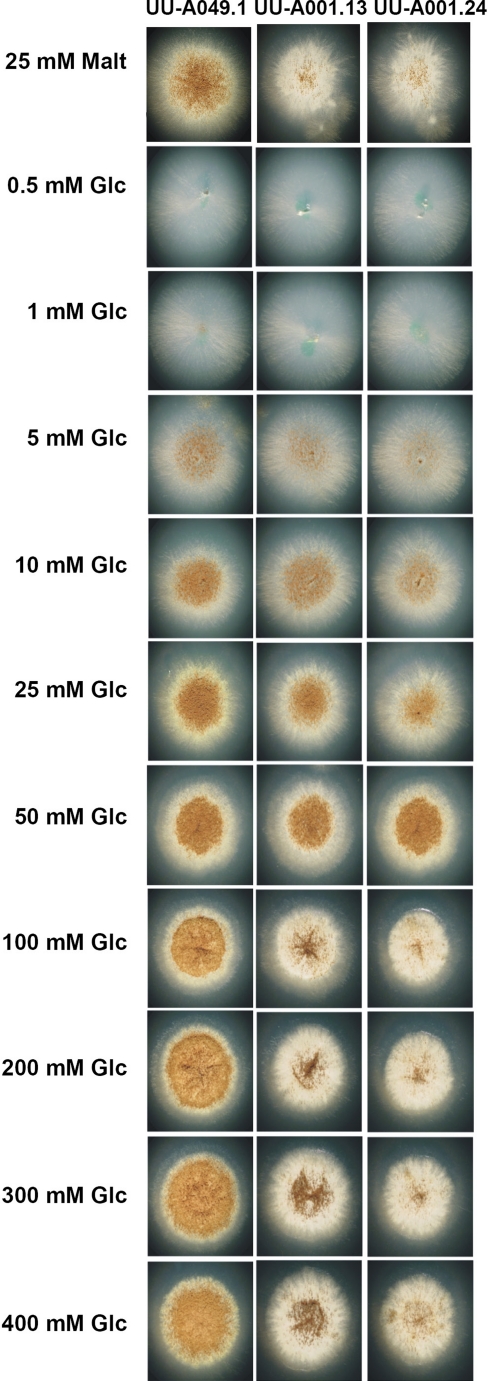
Early in the growth experiments, an unusual morphology (dense colonies with reduced sporulation) was observed for the amyR multicopy strain on maltose and starch. In this strain, it can be expected that maltose is very quickly converted extracellularly to d-glucose by the overproduction of glucoamylase, and therefore a second growth experiment was performed at different d-glucose concentrations to determine whether this morphology was indeed caused by the d-glucose level. The reference (UU-A049.1) and two multicopy strains (UU-A001.24 and UU-A001.13; a second multicopy strain was included to verify that this is a general effect) were grown on MM plates with d-glucose ranging from 0.5 to 400 mM. No significant difference was observed between the strains at d-glucose concentration of 25 mM or lower. At 50 mM, a small difference can be observed in that the mycelium of the amyR multicopy strains is denser. At 100 mM and higher, the same morphology is observed as seen for maltose in the multicopy strains: dense mycelium and reduced sporulation (Fig. 4).
Fig. 4.
Growth differences of UU-A049.1 (reference strain), and UU-A001.13 and UU-A001.24 (amyR multicopy strains) on different concentrations of d-glucose (Glc). Maltose (Malt) is included at the top of the figure for comparison
Discussion
AmyR has been studied in detail in several Aspergilli, with emphasis on genes involved in starch and maltose hydrolysis, although a recent study (Yuan et al. 2008) already indicated that other genes may also be controlled by AmyR. Analysis of enzyme activity in our study provided clear indications for a broader role of this regulator, in that we observed strongly elevated levels of both α- and β-glucosidase as well as α- and β-galactosidase in the amyR multicopy strain compared with the reference. The reduction in enzyme activity in the amyR deletion strain was less obvious, possibly due to the fact that the activities in the reference were already very low. Analysis of publicly available micro-array data of an A. niger wild-type and amyR disruptant during growth on maltose (Yuan et al. 2008) demonstrated that some, but not all genes encoding these enzymes (Coutinho et al. 2009), were downregulated in the disruptant strain, suggesting regulation by AmyR. All the downregulated genes contained at least one putative AmyR binding site, which would be a pre-requisite for direct AmyR regulation. For one putative α-galactosidase encoding gene (An04g02700), no putative AmyR binding site was identified in the DSM genome strain (CBS 513.88) (Coutinho et al. 2009). However, a putative binding site was detected in the genome of A. niger ATCC 1015 that was sequenced by JGI (Andersen et al. 2011). As we used the laboratory strain N402, this result suggests that this strain resembles ATCC 1015 with respect to the promoter of this gene. A similar difference was observed previously for the acetyl xylan esterase encoding gene axeA (R.P. de Vries, unpublished data). In both N402 and ATCC 1015, the promoter of this gene contains an XlnR consensus sequence, while CBS 513.88 does not. Also, several of the genes whose expression is not affected in the amyR disruption strain contain putative AmyR binding sites in their promoter. This confirms that the presence of such a sequence is no direct indication for regulation by AmyR, as was concluded previously for AmyR, XlnR and other regulators of Aspergillus (Coutinho et al. 2009). However, the absence of such binding sites strongly suggests that the gene is not (directly) regulated.
These data were confirmed for one α-galactosidase encoding gene (aglC) and one β-galactosidase encoding gene (lacA) using Northern analysis. On maltose and starch these two genes followed a similar pattern as the established AmyR regulated genes glaA, amyA and agdA. However, some differences were observed. The expression of lacA was only observed at late time points (8 h on maltose and 24 h on starch), while aglC expression peaked at 2 h, and the other genes were expressed on all time points. The reduced expression of aglC at later time points is not due to a reduction in expression compared with the pre-culture, as micro-array data from another study (R.P. de Vries, unpublished results) demonstrates lower aglC expression for the pre-culture than after 2 h on maltose or glucose. The expression of aglC in the amyR deletion strain was higher than that observed for the other genes tested, suggesting an AmyR-independent induction of aglC. Expression of aglC on d-glucose was observed previously (Ademark et al. 2001) and was shown to be independent of the major carbon catabolite repression system CreA (Ruijter and Visser 1997) while d-galactose induction of this gene was not observed (Ademark et al. 2001).
Expression of agdA in the reference strain on glucose was only observed after prolonged incubation, which correlates well with detectable α-glucosidase activity during growth of this strain on glucose as these activities were measured after 6 h. The activity level is higher than expected based on the expression of agdA, but this is likely due to the presence of additional α-glucosidases in the medium that are also produced under this condition.
Regulation of α- and β-glucosidase and α- and β-galactosidase activity by AmyR was further supported by growth profiling. Reduced growth of the amyR deletion strain compared with the reference was observed on substrates containing α- (starch, maltose, melibiose, melezitose, raffinose, sucrose) and β-linked d-glucose (cellobiose) as well as α- (melibiose, raffinose, carrageenan) and β-linked d-galactose (lactose, carrageenan), while improved growth on several of these substrates was observed for the amyR multicopy strain. This provides in vivo evidence for a function for AmyR in utilization of these oligo- and polysaccharides by A. niger. This situation is similar to that for another Aspergillus transcriptional regulator related to polysaccharide degradation: XlnR. This protein was first described as a xylanolytic regulator (van Peij et al. 1998b) but was later shown to also regulate genes involved in cellulose degradation (van Peij et al. 1998a) and galactomannan degradation (de Vries et al. 1999a).
Several reports have provided evidence for different inducing compounds of the amylolytic system in Aspergillus. Most commonly, maltose is suggested to be the inducer of AmyR (Barton et al. 1972; Carlsen and Nielsen 2001; Pedersen et al. 2000; Santerre Henriksen et al. 1999), but also glucose (Carlsen and Nielsen 2001). Another study claimed that the inducer is iso-maltose (Kato et al. 2002), but a conclusive study has so far not been published. A. niger has been shown to produce high levels of glucoamylase in the presence of maltose or starch (Barton et al. 1972; Gouka et al. 1997a, b; Pedersen et al. 2000; Schrickx et al. 1995). In natural biotopes, high levels of maltose are uncommon, and it is therefore unlikely that the maltose liberated by A. niger during hydrolysis of starch is able to avoid the high levels of glucoamylase and get imported into the cell. It is more likely that (nearly) all maltose is hydrolysed extracellularly to d-glucose. Our study demonstrates that AmyR-regulated genes in A. niger are induced during growth on low levels of d-glucose, as their expression increases during the cultivation, which is similar to what was shown previously in A. oryzae (Carlsen and Nielsen 2001). As d-glucose has been shown to act as a repressor through the carbon catabolite repressor protein CreA (Ruijter and Visser 1997), the expression levels are likely a balance between induction through AmyR and repression through CreA. A previous study showed a similar effect for d-xylose and XlnR, in which higher d-xylose concentrations resulted in reduced expression of xylanolytic genes, mediated by CreA (de Vries et al. 1999b). In most studies reported previously (reviewed in Tsukagoshi et al. 2001), d-glucose is considered a repressing rather than an inducing carbon source. However, usually 1–3% (67–200 mM) of d-glucose is used in these studies, which are concentrations at which the repression through CreA probably overrules induction through AmyR. These data suggests that in A. niger d-glucose or a metabolic product thereof is the inducer of AmyR as was shown previously for A. oryzae (Carlsen and Nielsen 2001). Higher induction of the AmyR-regulated genes on maltose and starch than on d-glucose supports this hypothesis, as growth on these substrates would give a gradual release of d-glucose, resulting in very low steady-state levels. Based on studies in other Aspergilli, differences in the possible inducing compounds may exist between species of this genus. The presence of the recently discovered mal-cluster (Hasegawa et al. 2010; Vongsangnak et al. 2009) in A. oryzae that consists of a putative second maltose-responsive regulator, a maltose transporter and a maltase, suggests that, in this species, at least part of the maltose is transported into the fungal cell. This cluster is also present in Aspergillus flavus, Aspergillus clavatus, Neosartorya fischeri and Aspergillus fumigatus but not in A. niger, A. nidulans and A. terreus (Suppl. Fig. 2), suggesting a different approach to starch utilisation in these two groups of Aspergilli.
The increased hydrolysis of maltose to d-glucose in the amyR multicopy strains is supported by similar morphology of these strains on maltose and on high concentrations of d-glucose. This morphology is likely due to the presence of a high intracellular concentration of d-glucose or a metabolite thereof as it is not observed in the reference strain when it is exposed to high extracellular d-glucose concentrations. This implies that d-glucose transport is upregulated in the multicopy strains, resulting in this higher intracellular concentration of d-glucose or a metabolic product of d-glucose. Analysis of the expression of the three A. niger d-glucose transporters reported previously (Jorgensen et al. 2007; vanKuyk et al. 2004) in the published AmyR micro array data set (Yuan et al. 2008) demonstrated that two of these genes (both encoding high affinity d-glucose transporters) are downregulated in the amyR deletion strain, suggesting control by AmyR. However, considering the presence of multiple putative d-glucose transporters in the A. niger genome (Pel et al. 2007), more functional data is needed on these other putative d-glucose transporters before any firm conclusion can be drawn. A similar growth phenotype was observed for strains under secretion stress (Carvalho et al. 2011), but it is unlikely that this can explain our results. It has been well-documented that AmyR-regulated genes are subject to carbon catabolite repression (Tsukagoshi et al. 2001), indicating that an increase in glucose concentration would reduce expression of these genes, resulting in lower protein production and likely also reduced rather than increased secretion stress
In conclusion, our study has shown that the influence of AmyR in A. niger extends beyond starch hydrolysis and suggests a role for d-glucose or a metabolic product thereof as the inducer of the AmyR system in this fungus.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 220 kb)
Acknowledgements
The authors thank Ec Agbo, Joep Geerlings and Dirk Blom for technical assistance.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Ademark P, de Vries RP, Hagglund P, Stalbrand H, Visser J. Cloning and characterization of Aspergillus niger genes encoding an α-galactosidase and a β-mannosidase involved in galactomannan degradation. Eur J Biochem. 2001;268:2982–2990. doi: 10.1046/j.1432-1327.2001.02188.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andersen MR, Salazar MP, Schaap PJ, van de Vondervoort PJ, Culley D, Thykaer J, Frisvad JC, Nielsen KF, Albang R, Albermann K, Berka RM, Braus GH, Braus-Stromeyer SA, Corrochano LM, Dai Z, van Dijck PW, Hofmann G, Lasure LL, Magnuson JK, Menke H, Meijer M, Meijer SL, Nielsen JB, Nielsen ML, van Ooyen AJ, Pel HJ, Poulsen L, Samson RA, Stam H, Tsang A, van den Brink JM, Atkins A, Aerts A, Shapiro H, Pangilinan J, Salamov A, Lou Y, Lindquist E, Lucas S, Grimwood J, Grigoriev IV, Kubicek CP, Martinez D, van Peij NN, Roubos JA, Nielsen J, Baker SE. Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Res. 2011;21:885–987. doi: 10.1101/gr.112169.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton LL, Georgi CE, Lineback DR. Effect of maltose on glucoamylase formation by Aspergillus niger. J Bacteriol. 1972;111:771–777. doi: 10.1128/jb.111.3.771-777.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boel E, Hjort I, Svensson B, Norris F, Norris KE, Fiil NP. Glucoamylases G1 and G2 from Aspergillus niger are synthesized from two different but closely related mRNAs. EMBO J. 1984;3:1097–1102. doi: 10.1002/j.1460-2075.1984.tb01935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos CJ, Debets AJM, Swart K, Huybers A, Kobus G, Slakhorst SM. Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr Genet. 1988;14:437–443. doi: 10.1007/BF00521266. [DOI] [PubMed] [Google Scholar]
- Carlsen M, Nielsen J. Influence of carbon source on α-amylase production by Aspergillus oryzae. Appl Microbiol Biotechnol. 2001;57:346–349. doi: 10.1007/s002530100772. [DOI] [PubMed] [Google Scholar]
- Carvalho ND, Arentshorst M, Kooistra R, Stam H, Sagt CM, van den Hondel CA, Ram AF. Effects of a defective ERAD pathway on growth and heterologous protein production in Aspergillus niger. Appl Microbiol Biotechnol. 2011;89:357–373. doi: 10.1007/s00253-010-2916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho PM, Andersen MR, Kolenova K, vanKuyk PA, Benoit I, Gruben BS, Trejo-Aguilar B, Visser H, van Solingen P, Pakula T, Seiboth B, Battaglia E, Aguilar-Osorio G, de Jong JF, Ohm RA, Aguilar M, Henrissat B, Nielsen J, Stalbrand H, de Vries RP. Post-genomic insights into the plant polysaccharide degradation potential of Aspergillus nidulans and comparison to Aspergillus niger and Aspergillus oryzae. Fungal Genet Biol. 2009;46(Suppl 1):S161–S169. doi: 10.1016/j.fgb.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Crabtree J, Angiuoli SV, Wortman JR, White OR (2007) Sybil: methods and software for multiple genome comparison and visualization. Methods Mol Biol 408:93–108 [DOI] [PubMed]
- de Vries RP, van den Broeck HC, Dekkers E, Manzanares P, de Graaff LH, Visser J. Differential expression of three α-galactosidase genes and a single β-galactosidase gene from Apergillus niger. Appl Environ Microbiol. 1999;65:2453–2460. doi: 10.1128/aem.65.6.2453-2460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RP, Visser J, de Graaff LH. CreA modulates the XlnR-induced expression on xylose of Aspergillus niger genes involved in xylan degradation. Res Microbiol. 1999;150:281–285. doi: 10.1016/S0923-2508(99)80053-9. [DOI] [PubMed] [Google Scholar]
- de Vries RP, van de Vondervoort PJI, Hendriks L, van de Belt M, Visser J. Regulation of the α-glucuronidase encoding gene (aguA) from Aspergillus niger. Mol Gen Genet. 2002;268:96–102. doi: 10.1007/s00438-002-0729-7. [DOI] [PubMed] [Google Scholar]
- de Vries RP, Burgers K, van de Vondervoort PJI, Frisvad JC, Samson RA, Visser J. A new black Aspergillus species, A. vadensis, is a promising host for homologous and heterologous protein production. Appl Environ Microbiol. 2004;70:3954–3959. doi: 10.1128/AEM.70.7.3954-3959.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucl Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1Part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielkens MM, Dekkers E, Visser J, de Graaff LH. Two cellobiohydrolase-encoding genes from Aspergillus niger require d-xylose and the xylanolytic transcriptional activator XlnR for their expression. Appl Environ Microbiol. 1999;65:4340–4345. doi: 10.1128/aem.65.10.4340-4345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi K, Akeno T, Minetoki T, Ozeki K, Kumagai C, Okazaki N, Iimura Y. Molecular cloning and characterization of a transcriptional activator gene, amyR, involved in the amylolytic gene expression in Aspergillus oryzae. Biosci Biotechnol Biochem. 2000;64:816–827. doi: 10.1271/bbb.64.816. [DOI] [PubMed] [Google Scholar]
- Gouka RJ, Punt PJ, van den Hondel CA. Efficient production of secreted proteins by Aspergillus: progress, limitations and prospects. Appl Microbiol Biotechnol. 1997;47:1–11. doi: 10.1007/s002530050880. [DOI] [PubMed] [Google Scholar]
- Gouka RJ, Punt PJ, van den Hondel CA. Glucoamylase gene fusions alleviate limitations for protein production in Aspergillus awamori at the transcriptional and (post) translational levels. Appl Environ Microbiol. 1997;63:488–497. doi: 10.1128/aem.63.2.488-497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa S, Takizawa M, Suyama H, Shintani T, Gomi K. Characterization and expression analysis of a maltose-utilizing (MAL) cluster in Aspergillus oryzae. Fungal Genet Biol. 2010;47:1–9. doi: 10.1016/j.fgb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Hasper AA, Dekkers E, van Mil M, van de Vondervoort PJ, de Graaff LH. EglC, a new endoglucanase from Aspergillus niger with major activity towards xyloglucan. Appl Environ Microbiol. 2002;68:1556–1560. doi: 10.1128/AEM.68.4.1556-1560.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen TR, vanKuyk PA, Poulsen BR, Ruijter GJ, Visser J, Iversen JJ. Glucose uptake and growth of glucose-limited chemostat cultures of Aspergillus niger and a disruptant lacking MstA, a high-affinity glucose transporter. Microbiology. 2007;153:1963–1973. doi: 10.1099/mic.0.2006/005090-0. [DOI] [PubMed] [Google Scholar]
- Kato N, Murakoshi Y, Kato M, Kobayashi T, Tsukagoshi N. Isomaltose formed by α-glucosidases triggers amylase induction in Aspergillus nidulans. Curr Genet. 2002;42:43–50. doi: 10.1007/s00294-002-0325-8. [DOI] [PubMed] [Google Scholar]
- Kumar V, Ramakrishnan S, Teeri TT, Knowles JKC, Hartley BS. Saccharomyces cerevisiae cells secreting an Aspergillus niger β-galactosidase grown on whey permeate. Bio/Technology. 1992;10:82–85. doi: 10.1038/nbt0192-82. [DOI] [PubMed] [Google Scholar]
- Kusters-van Someren MA, Harmsen JAM, Kester HCM, Visser J. The structure of the Aspergillus niger pelA gene and its expression in Aspergillus niger and Aspergillus nidulans. Curr Genet. 1991;20:293–299. doi: 10.1007/BF00318518. [DOI] [PubMed] [Google Scholar]
- Lenouvel F, van de Vondervoort PJ, Visser J. Disruption of the Aspergillus niger argB gene: a tool for transformation. Curr Genet. 2002;41:425–432. doi: 10.1007/s00294-002-0320-0. [DOI] [PubMed] [Google Scholar]
- Melchers WJG, Verweij PE, van den Hurk P, van Belkum A, de Pauw BE, Hoogkamp-Korstanje AA, Meis JFGM. General primer-mediated PCR for detection of Aspergillus species. J Clin Microbiol. 1994;32:1710–1717. doi: 10.1128/jcm.32.7.1710-1717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen H, Beyer M, Nielsen J. Glucoamylase production in batch, chemostat and fed-batch cultivations by an industrial strain of Aspergillus niger. Appl Microbiol Biotechnol. 2000;53:272–277. doi: 10.1007/s002530050020. [DOI] [PubMed] [Google Scholar]
- Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JA, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EG, Debets AJ, Dekker P, van Dijck PW, van Dijk A, Dijkhuizen L, Driessen AJ, d’Enfert C, Geysens S, Goosen C, Groot GS, de Groot PW, Guillemette T, Henrissat B, Herweijer M, van den Hombergh JP, van den Hondel CA, van der Heijden RT, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel MJ, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij NN, Ram AF, Rinas U, Roubos JA, Sagt CM, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, van de Vondervoort PJ, Wedler H, Wosten HA, Zeng AP, van Ooyen AJ, Visser J, Stam H. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- Petersen KL, Lehmbeck J, Christensen T. A new transcriptional activator for amylase genes in Aspergillus. Mol Gen Genet. 1999;262:668–676. doi: 10.1007/s004380051129. [DOI] [PubMed] [Google Scholar]
- Punt PJ, Schuren FH, Lehmbeck J, Christensen T, Hjort C, van den Hondel CA. Characterization of the Aspergillus niger prtT, a unique regulator of extracellular protease encoding genes. Fungal Genet Biol. 2008;45:1591–1599. doi: 10.1016/j.fgb.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Ruijter GJG, Visser J. Carbon repression in aspergilli. FEMS Microbiol Lett. 1997;151:103–114. doi: 10.1111/j.1574-6968.1997.tb12557.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning—a laboratory manual. 2. Cold Spring Harbour: Cold Spring Harbour Laboratory; 1989. [Google Scholar]
- Santerre Henriksen AL, Even S, Muller C, Punt PJ, van den Hondel CA, Nielsen J. Study of the glucoamylase promoter in Aspergillus niger using green fluorescent protein. Microbiology. 1999;145:729–734. doi: 10.1099/13500872-145-3-729. [DOI] [PubMed] [Google Scholar]
- Schrickx JM, Stouthamer AH, van Verseveld HW. Growth behaviour and glucoamylase production by Aspergillus niger N402 and a glucoamylase overproducing transformant in recycling culture without a nitrogen source. Appl Microbiol Biotechnol. 1995;43:109–116. doi: 10.1007/BF00170631. [DOI] [PubMed] [Google Scholar]
- Tani S, Katsuyama Y, Hayashi T, Suzuki H, Kato M, Gomi K, Kobayashi T, Tsukagoshi N. Characterisation of the amyR gene encoding a transcriptional activator for the amylase genes in Aspergillus nidulans. Curr Genet. 2001;39:10–15. doi: 10.1007/s002940000175. [DOI] [PubMed] [Google Scholar]
- Tsukagoshi N, Kobayashi T, Kato M. Regulation of the amylolytic and (hemi-) cellulolytic genes in aspergilli. J Gen Appl Microbiol. 2001;47:1–19. doi: 10.2323/jgam.47.1. [DOI] [PubMed] [Google Scholar]
- van Peij N, Gielkens MMC, de Vries RP, Visser J, de Graaff LH. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl Environ Microbiol. 1998;64(10):3615–3619. doi: 10.1128/aem.64.10.3615-3619.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Peij NN, Visser J, de Graaff LH. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol Microbiol. 1998;27:131–142. doi: 10.1046/j.1365-2958.1998.00666.x. [DOI] [PubMed] [Google Scholar]
- vanKuyk PA, Diderich JA, MacCabe AP, Hererro O, Ruijter GJG, Visser J. Aspergillus niger mstA encodes a high-affinity sugar/H+ symporter which is regulated in response to extracellular pH. Biochem J. 2004;379:375–383. doi: 10.1042/BJ20030624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongsangnak W, Salazar M, Hansen K, Nielsen J. Genome-wide analysis of maltose utilization and regulation in aspergilli. Microbiology. 2009;155:3893–3902. doi: 10.1099/mic.0.031104-0. [DOI] [PubMed] [Google Scholar]
- Yuan XL, van der Kaaij RM, van den Hondel CA, Punt PJ, van der Maarel MJ, Dijkhuizen L, Ram AF. Aspergillus niger genome-wide analysis reveals a large number of novel α-glucan acting enzymes with unexpected expression profiles. Mol Genet Genomics. 2008;279:545–561. doi: 10.1007/s00438-008-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 220 kb)