Abstract
Recent advances in massively parallel sequencing technology have created new opportunities to probe the hidden world of microbes. Taxonomy-independent clustering of the 16S rRNA gene is usually the first step in analyzing microbial communities. Dozens of algorithms have been developed in the last decade, but a comprehensive benchmark study is lacking. Here, we survey algorithms currently used by microbiologists, and compare seven representative methods in a large-scale benchmark study that addresses several issues of concern. A new experimental protocol was developed that allows different algorithms to be compared using the same platform, and several criteria were introduced to facilitate a quantitative evaluation of the clustering performance of each algorithm. We found that existing methods vary widely in their outputs, and that inappropriate use of distance levels for taxonomic assignments likely resulted in substantial overestimates of biodiversity in many studies. The benchmark study identified our recently developed ESPRIT-Tree, a fast implementation of the average linkage-based hierarchical clustering algorithm, as one of the best algorithms available in terms of computational efficiency and clustering accuracy.
Keywords: pyrosequencing, 16S rRNA, taxonomy-independent analysis, massive data, clustering, microbial diversity estimation, human microbiome
INTRODUCTION
Complex microbial communities contribute to many biological processes, including biogeochemical activities critical to life in all environments on earth and maintenance of human health. High-throughput sequencing technologies now allow researchers to circumvent earlier constraints of studying microbial communities via cultivation-based techniques. These recent technological developments can generate millions of sequences in a single sequencing run, and open new opportunities to probe the hidden world of microbes with unprecedented resolution [1–3].
Although shotgun metagenomic studies are also developing rapidly, 16S rRNA analysis remains a widely accepted and powerful tool for studying microbial community dynamics at high resolution. Existing algorithms for classifying microbes using 16S rRNA sequences can be generally categorized as taxonomy dependent or taxonomy independent. In taxonomy-dependent analysis, query sequences are compared against a database, then assigned to the organisms of the best-matched reference sequences (e.g. using BLAST [4]). Because the vast majority of microbes have not yet been formally described, these methods are inherently limited by incompleteness of reference databases [5]. Taxonomy-dependent analysis is usually performed for the purpose of sequence annotation. In contrast, taxonomy-independent analysis (TIA) compares query sequences against each other to form a distance matrix, followed by clustering to group sequences into operational taxonomic units (OTUs) with a specified amount of variability allowed within each OTU. Various ecological metrics can then be estimated from the frequency of each OTU in order to characterize a microbial community or to compare communities. The analysis does not rely on any reference database, and hence can enumerate novel uncultured and potentially pathogenic microbes, not just organisms that have already been cultured and sequenced.
Many different algorithms have been developed in the past decade for TIA [6–12]. Significant efforts were devoted to developing new algorithms that enable microbiologists to keep pace with the unprecedented growth of genomic data sets available today. However, many existing algorithms, although widely used by the biology community, have been benchmarked only informally or on limited data sets. A recent report suggests that different methodologies, and even small changes in algorithm parameters for the same methodology, can result in substantially different binning outcomes [13]. We have also observed major discrepancies in the number of bins, and of which sequences fall into the same bin, depending on the particular algorithm. The inconsistency in binning algorithms utilized in various studies makes it difficult to interpret and compare research findings from different research groups. A benchmark study that forms the basis for identifying the most appropriate algorithm for a particular application is therefore urgently needed.
A major obstacle to benchmarking is that for complex microbial communities there is no ground-truth information about what species are actually in the community. In addition, the criteria that have been previously used for quantitative assessment of algorithm performance have several important drawbacks. Traditionally, TIA was performed to estimate the biodiversity of a microbial community. Consequently, previous work mainly focused on the ability of different algorithms to recover the same number of OTUs (defined at certain distance levels) that were present in a mock community generated from a limited number of known 16S rRNA sequences [6, 12]. There are two problems with this criterion. First, the total number of clusters is a global statistics that provides no information on how each sequence is grouped (for example, one might recover 22 OTU clusters from 22 species, but sequences from different species might be grouped in the same OTU). In addition to microbial diversity estimation, TIA is used to analyze millions of sequences to identify lists of OTUs that separate clinically or biologically relevant states (e.g. OTUs that separate lean and obese individuals or different sites on the body) [14–16]. Incorrect grouping of sequences into OTUs can therefore have a major impact on downstream data analysis. Second, due to the use of different formulations to compute the distances between clusters, the numbers of OTUs generated by different algorithms at the same nominal distance levels are not directly comparable. Consequently, using OTU numbers as the sole criterion to evaluate TIA algorithms is limited and potentially misleading.
In this article, we present a review of existing algorithms in current, widespread use by the microbiology community for TIA, and conduct a comprehensive benchmark study that addresses several issues of concern. We develop an experimental protocol to compare different algorithms using the same platform, and introduce several criteria including normalized mutual information (NMI) [17] and F-score [18] to facilitate a quantitative evaluation of the clustering performance of each algorithm. Because this article is primarily intended for microbiologists, we present several toy examples and simulation studies performed on both artificially generated and real world data sets to illustrate the behavior of different algorithms. We find that existing methods, even when applied to the same data sets, can lead to substantially different results, and that many astoundingly high biodiversity estimates reported in the literature appear to be overestimates resulting from the inappropriate use of distance levels for defining OTUs. Our benchmark study identifies ESPRIT-Tree, a fast implementation of the average linkage-based hierarchical clustering algorithm, as one of the best TIA algorithms available in terms of computational efficiency and clustering accuracy. We hope that the results presented here will enable microbiologists to better understand the issues involved in analyzing large 16S rRNA data sets and provide them with a guideline for choosing a proper TIA algorithm for their particular research question.
LITERATURE SURVEY
Existing TIA algorithms generally consist of two major modules: (i) computing pairwise distances between sequence pairs and (ii) grouping sequences into OTUs at various distance levels. In some algorithms (e.g. ESPRIT [6], mothur [9], MUSCLE+DOTUR [19]) the two steps are performed sequentially, whereas in others (e.g. ESPRIT-Tree [8], CD-HIT [7] and UCLUST [10]) they are performed simultaneously. In this article, we mainly focus on sequence data sets where the number of sequences is on the order of 106 (about the number currently obtained in a single 454 Titanium run). The algorithms implemented sequentially generally cannot handle such massive data because, as an intermediate step, they compute a distance matrix, the size of which is proportional to the square of the number of sequences. However, computational efficiency is not the only consideration. Many existing algorithms are tradeoffs between computational efficiency and accuracy. We present below a brief description of how each algorithm works. Table 1 summarizes some existing algorithms commonly used by the microbial community for TIA.
Table 1:
Some existing algorithms commonly used by the microbiology community for TIA
| Method | Function supported | Alignment method | Clustering methoda | Using reference database | Generating distance matrix | Computational complexity | Space complexity |
|---|---|---|---|---|---|---|---|
| DOTUR | Clustering | N/A | HC | N | Y | O(N2) | O(N2) |
| Mothur | Sequence alignment + clustering | Profile based MSA method | HC | Y | Y | O(N2) | O(N2) |
| ESPRIT | Sequence alignment + clustering | PSA | HC | N | Y | O(N2) | O(N2) |
| ESPRIT-Tree | Sequence alignment + clustering | PSA | HC | N | N | O(N1.2) | O(N) |
| NASTb | Sequence alignment | Profile based MSA method | N/A | Y | Y | O(N) | O(N2) |
| RDP/Pyro | Sequence alignment + clustering | Infernal aligner | HC | Y | Y | O(N2) | O(N2) |
| CD-HIT | Sequence alignment + clustering | PSA | Greedy heuristic clustering | N | N | O(N1.2) | O(N) |
| UCLUST | Sequence alignment + clustering | PSA | Greedy heuristic clustering | N | N | O(N1.2) | O(N) |
| MUSCLE | Sequence alignment | MSA | N/A | N | Y | O(N4) | O(N2) |
aComplete linkage is the default method in DOTUR, mothur, ESPRIT and RDP/Pyro. ESPRIT-Tree supports only average linkage. bNAST only supports the sequence-alignment step. By aligning query sequences against a database, its computational complexity grows linearly with respect to the number of sequences. However, according to the NAST website, it aligns at a rate of approximately 10 sequences per minute. N/A = not applicable; N = no; Y = yes.
One of the most commonly used methods in the first step (pairwise distance calculations) is multiple sequence alignment (MSA), which incorporates information about sequence homology into the distance calculation [19, 20]. The optimal MSA is an NP-complete problem (i.e. it has been proven to be a ‘hard’ problem that cannot be solved in reasonable time for large numbers of sequences). Although significant improvement has been made in the last decade to reduce computational complexity of MSA (e.g. MUSCLE [21] and MAFFT [22]), it remains computationally intractable to perform MSA on millions of sequences. In addition, the use of MSA to align hypervariable regions of 16 S rRNA gene has not yet been well justified. MSA was originally designed to infer homologous segments of input sequences with the underlying assumption that input sequences share recognizable evolutionary similarities at the level of the primary sequences. This assumption may not hold for 16 S rRNA-based studies that target hypervariable regions of rRNA genes from highly diverse microbial communities. In a simulation study presented below using real 16 S rRNA sequences derived from a seawater sample, we observed that many sequences are from distantly related OTUs with large genetic distances. MSA aims to minimize the sum of pairwise alignment scores. In order to align a large number of highly diverse sequences, the alignment quality of closely related sequences is sacrificed, leading to a severe overestimation of genetic distances and microbial diversities. Moreover, the harder MSA tries to minimize the sum of pairwise scores by aligning unrelated sequences, the larger distances between closely related sequences can be. By definition, the optimal MSA algorithm yields the lowest score. This suggests that MSA is not suitable for analyzing hypervariable regions of rRNA genes to estimate microbial diversity, even if the optimal MSA could be performed.
One possible way to address these issues with MSA is to align input sequences against a prealigned reference database. The representative methods include RDP/Pyro [5], NAST [23] and a profile-based algorithm included in the well-known mothur pipeline [9, 24]. Because a reference database can be maintained off-line, the computational complexity of these algorithms grows only linearly with respect to the number of input sequences. However, unlike generic MSA algorithms, these algorithms share a problem with those used for taxonomy-dependent analysis: their performance depends heavily on the completeness of a reference database, and also on the quality of the alignment of the sequences in that database. Since most bacterial genomes have not been sequenced yet, a large proportion of input sequences from unknown microorganisms may not be able to find significant hits and can only be aligned to distantly related reference sequences, leading to inaccurate estimates of pairwise distances. We performed a simulation study that suggests that although fixed aligners work well for known sequences, they perform poorly for novel sequences that are not closely related to those already represented in a reference database.
CD-HIT, UCLUST and ESPRIT use pairwise sequence alignment (PSA) to obtain optimal pairwise alignments of 16 S rRNA sequences and to compute similarities between sequence pairs. Whereas many MSA algorithms (e.g. MUSCLE) can only be deployed using a single processor, PSA allows for parallel computing and provides much more flexibilities in algorithm design. Moreover, simulation studies have shown that by eliminating heuristics in sequence comparison, PSA provides a much more accurate estimate of microbial richness than MSA [6, 12]. A frequent criticism of PSA is that currently implemented methods do not take RNA secondary structure information into consideration. However, in the benchmark study presented below, we found that, compared to profile-based MSA algorithms, excluding secondary-structure information does not have a significant impact on clustering performance.
Existing algorithms can also be categorized based on the clustering method they use to group sequences into OTUs at various levels of sequence identity. The two most commonly used methods are hierarchical clustering (HC) and greedy heuristic clustering (GHC). HC is a classic unsupervised learning technique that has been used in numerous biomedical applications [25]. The major drawback of HC is its high computational and space complexity. The standard implementation has an O(N2) complexity, where N is the number of input sequences. DOTUR is probably the first published HC algorithm widely used by the microbiology community [11]. DOTUR, however, cannot handle the extremely large data sets available today. The main reason it does not scale is that it needs to load a distance matrix into memory before clustering. Given one million reads, a full distance matrix can be as large as 7500 GB. Even if we remove duplicate sequences and sequence pairs that have a pairwise distance larger than a specified value (say 0.1), the resulting distance matrix in a sparse format can still be several hundred gigabytes in size, which is too large to be directly loaded into the memory of most computers. To address this issue, we recently developed a new clustering algorithm, referred to as Hcluster, within the ESPRIT framework to handle large-scale complete-linkage and single-linkage HC operations. Unlike conventional methods, Hcluster groups sequences into OTUs on the fly (i.e. reading one distance at a time), whereas keeping track of linkage information. A large-scale experiment was conducted that showed that Hcluster works well with one million reads [14]. Hcluster has already been incorporated in the mothur pipeline. One limitation of Hcluster is that one still needs to generate a full or sparse distance matrix before clustering. Although ESPRIT uses complete linkage as the default option, it provides a separate function that allows users to perform average linkage, but its memory footprint is much higher than Hcluster. ESPRIT uses kmer statistics (i.e. frequencies of ‘words’ of a specified length in the sequence) to remove unnecessary sequence alignments. However, its space and computational complexities remain O(N2).
Greedy heuristic clustering (e.g. CD-HIT [7] and UCLUST [10]) processes input sequences one at a time, avoiding the expensive step of comparing all pairs of sequences. Given a predefined threshold, an input sequence is either assigned to an existing cluster if the distance between the sequence and a seed (the sequence representing that cluster) is smaller than the threshold, or becomes a new seed for a new cluster otherwise. Consequently, the computational complexity of greedy heuristic clustering is O(MN), where M is the number of seeds. Usually M<<N, and hence greedy heuristic clustering is computationally much more efficient than HC. CD-HIT and UCLUST are the only two algorithms that we are aware of that can process millions of reads on a desktop computer. Mathematically speaking, greedy heuristic clustering partitions the input space into a set of closed balls where the distances between sequences and their associated seeds are smaller than a predefined threshold. However, there is no guarantee that the true clustering structure can be recovered from such partitions. Also, because sequences are sequentially processed, adjacent clusters may be overlapped. That is, the distance between a sequence and its assigned seed is not necessarily smaller than the distance between that sequence and any other seed (i.e. if the search were continued, a better match might be found). CD-HIT was originally designed to reduce the size of a large database to speed up a database search [7]. However, it is not designed for uncovering clustering structures, and the performance for that purpose has not yet been benchmarked. In the study presented below, we show that although CD-HIT and UCLUST run several orders of magnitude faster than a HC algorithm, their ability to group sequences into the correct clusters is much worse.
We recently developed a new online learning-based algorithm, referred to as ESPRIT-Tree [8], which simultaneously addressed the space and computational issues with conventional HC algorithms. The basic idea is to partition an input sequence space into a set of subspaces using a partition tree constructed using a pseudo-metric, then to recursively refine a clustering structure in these subspaces. As with CD-HIT and UCLUST, ESPRIT-Tree does not need to generate a distance matrix. All of the operations are executed on the fly, and the distances are computed only when they are needed. ESPRIT-Tree achieves a similar accuracy to the standard average linkage HC algorithm but with a computational complexity comparable to CD-HIT and UCLUST.
EXPERIMENTS
Multiple sequence alignment versus pairwise sequence alignment
We performed two simulation studies to investigate how different sequence alignment methods affect the performance of a TIA algorithm.
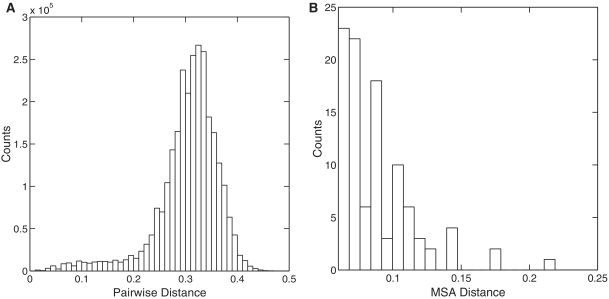
First, we tested how divergent sequences are in a representative biological sample. MSA assumes that input sequences share recognizable evolutionary homology at the level of primary sequences along their whole length. It has been reported that microbial communities are much more diverse than expected [19, 20], which suggests that the underlying assumption of MSA might not be valid because parts of the sequence are not homologous due to insertions and deletions, or because so much change has occurred that bases that are the same are as likely to be the same by coincidence rather than because they are derived from a common ancestor. This effect is especially severe with short sequences that focus on the most variable parts of the 16 S rRNA gene. To demonstrate this effect, we performed a simulation study where we calculated the pairwise distances of the V6 sequences of a seawater sample by using the Needleman–Wunsch algorithm, and plotted a histogram of the distances in Figure 1A. We report that only about 2% of sequence pairs have a genetic distance smaller than 0.1. In other words, for every two randomly selected sequences, there is a 98% probability that they are from two distantly related OTUs. Note that if MSA were used, the histogram would be further skewed towards the right.
Figure 1:
(A) Sequence pairs with distances less than 0.10 only account for a small fraction of all possible pairs (2.25% in this example). (B) Pairwise distances between the same pair of sequences computed based on multiple sequence alignments containing different sequences in the rest of the alignment are much larger than the constant value of 0.06 computed by using pairwise sequence alignment, and vary over a wide range, from 0.06 to 0.22 (i.e. sequences that are really 6% different can appear 22% different due to the MSA procedure). The experiment was performed on the 53R seawater sample downloaded from [19].
Second, we performed a simulation study to demonstrate how the presence of a large proportion of highly diverse sequences affects the alignment of sequences with small genetic distances. We randomly selected two sequences with a distance of 0.06 computed based on pairwise alignment, then performed a multiple sequence alignment of the two sequences together with 100 sequences randomly selected from the seawater data using MUSCLE with default parameter settings, and recorded the pairwise distance calculated between the first two sequences based on their alignment in the resulting MSA. The experiment was repeated 100 times using the same initial pair of sequences. The distances are plotted in Figure 1B. We can see that the pairwise distances computed based on MSA are much larger than 0.06. The maximum distance is 0.22 and the average is 0.09 (SD = 0.03). This can be explained by the fact that MSA is designed to minimize the sum of pairwise scores. In order to align the 100 highly diverse sequences, the alignment quality of the first two sequences had to be sacrificed, leading to an inflated estimate of genetic distances. Note that we include only 100 randomly selected sequences in this experiment. With additional distantly related sequences and/or if these sequences were even more diverse, the calculated distance between the first two sequences would be even larger. This leads to an interesting observation: whether two sequences are from the same OTU should be physically determined by their sequence composition. Now by using MSA, the distance between two sequences, and also their assignment to the same or different OTUs, also depends on sampling depth (i.e. the number of sequences sampled) and the environment from which they are extracted (i.e. how diverse other sequences are), which is highly undesirable. In ‘Benchmark study on human gut data’ section, we performed additional tests on a human gut microbiota data using other criteria (NMI score and number of OTUs), and found that due to its inability to align highly diverse sequences MSA performed significantly worse than PSA.
HC versus greedy heuristic clustering
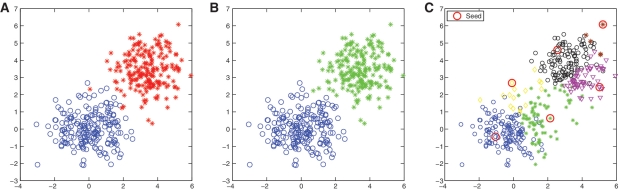
We performed a simulation study using a toy example to illustrate the algorithmic behaviors of hierarchical and greedy clustering methods. The data set was generated from two distinct Gaussian distributions (Figure 2A). The data points are distributed in a two-dimensional space, and the pairwise distances can be computed precisely, which enables us to visualize clustering results and to remove the compounding factor of using different alignment methods. By using Euclidean distance and average linkage, HC successfully recovered the true clustering structure as expected (Figure 2B). In contrast, greedy heuristic clustering performed poorly, although it did group samples into clusters (Figure 2C). For example, the green and yellow groups contain data points that originally came from the two different distributions. Also, we notice that the blue and green clusters have some overlaps. In other words, the blue points in the ‘green’ territory have smaller distances to the ‘green’ seed than to the ‘blue’ seed. In summary, the two methods have their own advantages and disadvantages: greedy heuristic clustering is computationally very efficient whereas HC is more accurate. However, if computational resource allows, HC is clearly the method of choice. These effects are especially important when one tries to determine the significance of relatively rare taxa that distinguish between physiological states: incorrect cluster assignment could cause an investigator to miss taxa that really do associate with a disease if they are assigned to the wrong cluster frequently enough to obscure the signal.
Figure 2:
A toy example that illustrates the algorithmic behaviors of the HC and greedy heuristic clustering methods. (A) The data set was generated from two distinct Gaussian distributions; (B) HC successfully recovered the true clustering structure; (C) greedy heuristic clustering performed poorly, and the result depended on selected seeds. At the same dissimilarity level, the two approaches behave differently.
Complete linkage versus average linkage
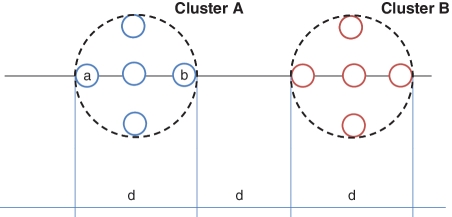
If HC is considered, one needs to decide which linkage function is used to define distances between clusters. Single linkage is rarely used in TIA due to its chaining effect. Complete linkage (CL) is the default setting in DOTUR and ESPRIT, whereas ESPRIT-Tree uses average linkage (AL). Huse et al. [12] conducted a simulation study to compare the number of OTUs generated by the two linkage functions at the 0.03 distance level against the species-level ground truth of an artificially generated data set, and found that AL yielded a much more accurate estimate of the number of OTUs than CL. However, as we mentioned in the introduction, the total number of OTUs is not a proper criterion to evaluate a TIA algorithm. CL differs from AL in how distances between clusters are computed when two clusters are merged. However, the numbers of OTUs generated by the two methods at the same distance level are not directly comparable. To illustrate this, we present a two-dimensional example in Figure 3, where each node represents a sequence. In order to merge the two clusters, the distance levels required in CL and AL are 3d and 2d, respectively. This problem is not straightforward for UCLUST and CD-HIT, as the result depends on the order of the sequences presented to the algorithms. If sequence ‘a’ is selected as a seed, the threshold is 3d, and if sequence ‘b’ is used as a seed, the threshold is 2d. Although UCLUST and CD-HIT do not support CL and AL, the distance levels required by the two methods to merge two clusters are somewhere between those used by CL and AL, which we verified in a benchmark study (Figures 5 and 7). The above example suggests that there is no single threshold that works for all methods. This is a crucial issue when comparing the microbial diversities detected in different studies, but has largely been ignored by the microbiology community.
Figure 3:
A toy example illustrates that distance levels required to merge the same pair of clusters are different for AL, CL and greedy heuristic clustering. Each node represents a sequence.
Figure 5:
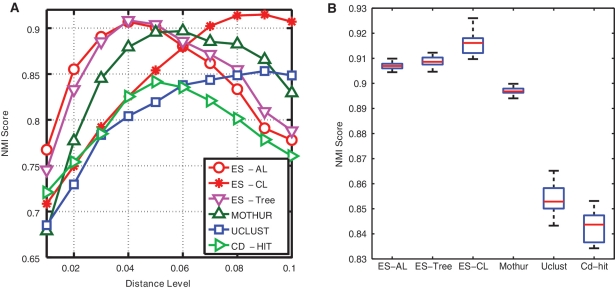
(A) NMI scores of six methods evaluated at ten distance levels. (B) Boxplots of the maximum NMI scores of six methods. Species assignments of input sequences were used as the ground truth. MUSCLE+AL performed much worse than all other methods, and its results are omitted so that the remainder can usefully be compared on the same scale.
Figure 7:
(A) NMI scores of six methods evaluated at 10 distance levels. (B) Boxplots of the maximum NMI scores of six methods. Genus assignments of input sequences were used as the ground truth.
Benchmark study on human gut data
We performed a benchmark study to evaluate the performance of seven existing TIA algorithms using a real-world data set. The data set was originally used to study the connection between obesity and altered composition of the human gut flora [26]. It contains about 1 100 000 sequences with an average length of 219 nucleotides, covering the V2 hypervariable region of the 16 S rRNA gene collected from the stool samples of 154 individuals. This is the most comprehensive 16 S rRNA based survey of the human gut microbiota published as of this writing.
The seven algorithms we compared include most TIA algorithms currently used by the microbiology community: CD-HIT, UCLUST, ESPRIT with CL and AL, mothur (profile-based MSA+AL using the SILVA database), MUSCLE+AL and ESPRIT-Tree. We did not include tests of pipelines that wrap these methods as the results are the same. In order to examine how PSA + CL and PSA+AL perform compared to other methods, we used a loose kmer threshold of 0.8 in ESPRIT to remove unnecessary sequence alignments. We also tested ESPRIT with the default kmer threshold of 0.5, and found that both thresholds yielded almost identical results (P > 0.3). For CD-HIT and UCLUST, the clustering outcomes typically depend on the order of sequences presented to the algorithms. One default setting is to order input sequences based on their lengths, although a more biologically plausible choice is to order them based on their abundances (with prefix matching so that a sequence and shorter sequences that occur entirely within that sequence are considered to be the same sequence). Both choices yielded similar results in our simulations. Here, we only present the results obtained by using the default settings. For all other algorithms, the default parameters were used.
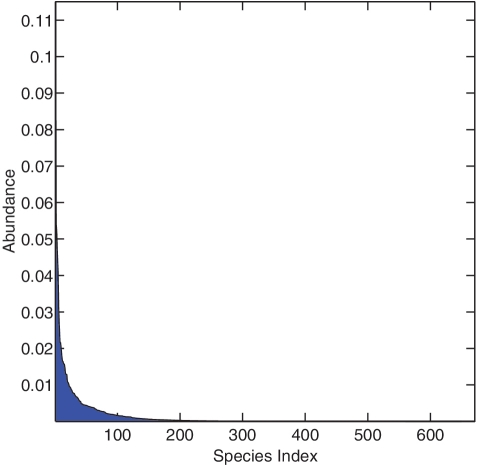
One of the major obstacles of a benchmark study is the lack of ground-truth information for performance evaluation. To overcome this difficulty, we constructed a reference database from the RDP-II database [5] using TaxCollector [16], where each reference sequence was fully annotated. We then ran a MegaBlast search of the gut data against the reference database, and used a stringent criterion to retain the annotated sequences: >97% identity over an aligned region >97% of the total length of the sequences. This resulted in a total of about 750 000 reads classified into 671 species and 283 genera. We then applied the seven methods to the annotated sequences, and used the commonly used NMI criterion [17] to evaluate how the outcome of a TIA algorithm agreed with the ground truth. NMI penalizes two types of errors: wrongly assigning sequences with the same species label into different OTUs, and assigning sequences with different species labels into the same OTUs. NMI = 1 means that a clustering result completely agrees with the ground truth, and NMI = 0 means that sequences are randomly assigned. In order to minimize statistical variations, the experiment was repeated 20 times. In each iteration, 30 000 reads were randomly extracted from the annotated data set, the seven methods were used to group the sequences into OTUs at various distance levels ranging from 0.01 to 0.15, a NMI score was computed at each distance level by using the species or genus labels of the input sequences as the ground truth, and the maximum NMI scores and the number of species and genera of the 30 000 reads were recorded. Although retaining only the sequences that can be confidently annotated somewhat simplified the problem, the experimental protocol is unbiased toward any particular clustering method. Figure 4 depicts the abundance of the species represented by one of the test data sets. The simulated data contains high, medium and low abundance components (i.e. a long tail), which is similar to those observed in a real microbial community, and is much more complicated than our previously used mock community generated from 43 known 16 S rRNA sequences [6, 12].
Figure 4:
The species abundance distribution represented by one of the test data sets. The simulated data contains high, medium and low abundance components, which is similar to those observed in a real microbial community and much more complicated than our previously used mock community generated from 43 known 16 S rRNA sequences.
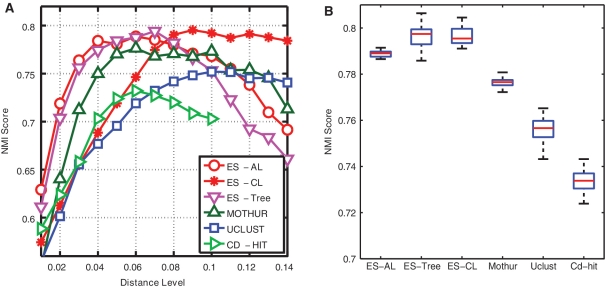
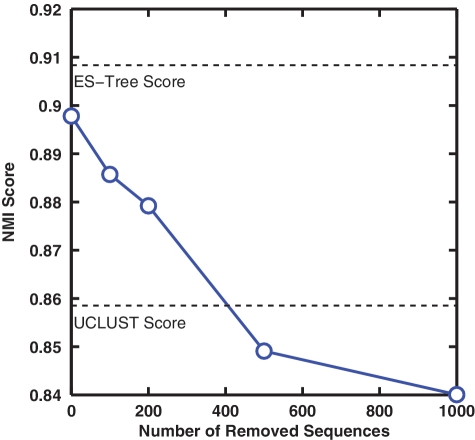
Figure 5A depicts the NMI scores of six methods as a function of ten distance levels, averaged over the 20 runs. The maximum NMI score of MUSCLE+AL is 0.66, which is much worse than all other methods, and its results were omitted to better visualize the results of other methods. We observe that all curves have a bell shape, which can be explained by the fact that when a distance level is small, sequences belonging to the same species are partitioned into different clusters, and when a distance level is large, sequences belonging to different species are grouped into the same clusters. By definition, both result in lower NMI scores. We also observe that the NMI curves obtained by different methods peak at different distance levels, for instance, 0.04 for ESPRIT-Tree and ESPRIT-AL, 0.08 for ESPRIT-CL, 0.05 for CD-HIT and 0.06 for UCLUST. These differences are due to the different methods used in each algorithm to define the distance between two clusters. The peak positions of CD-HIT and UCLUST are somewhere between those of ESPRIT-AL and ESPRIT-CL, which is consistent with the discussion presented in ‘Complete linkage versus average linkage’ section (Figure 3). The above observation suggests that the NMI scores obtained at the same distance level are not directly comparable. We thus proceeded to compare the maximum NMI score of each method (Figure 5B), which by definition corresponds to the best clustering result that a method can achieve. We observe that (i) ESPRIT-AL performed similarly to ESPRIT-CL, although they peaked at the different distance levels; (ii) ESPRIT-Tree performed similarly to ESPRIT-AL, and significantly better than CD-HIT and UCLUST (P-value <10–5 based on a Student's t-test), which is consistent with the result presented in ‘HC versus greedy heuristic clustering’ section; (iii) mothur+AL performed much better than CD-HIT and UCLUST, but slightly worse than ESPRIT+AL. We should emphasize that because the test data sets contain only sequences that can be confidently annotated by the RDP database, the experimental procedure favors mothur (which calculates pairwise distances by aligning input sequences against a prealigned reference database). The underlying assumption of mothur is that pairwise distances between novel sequences can be accurately estimated from the reference sequences. The following simulation study shows that this might not be the case. We used one of the V2 annotated data sets. We first mapped each query sequence to its closest reference sequence in the SILVA database and removed the top 100, 200, 500 and 1000 best-matched sequences from the database. We then applied mothur to the test data set by using these incomplete databases, and computed the corresponding NMI scores. We report that the clustering quality of mothur drops substantially as the number of removed sequences increases (Figure 6). It should be noted that by removing 1000 out of 14 900 reference sequences, only 6.7% of the database is missing. The above results suggest that although the profile-based MSA algorithm may be able to capture homologous and secondary-structure information, it does not work well for novel sequences that are not well represented in a reference database. Moreover, even for the annotated data, ESPRIT+AL performed slightly better than mothur+AL, suggesting that, compared to profile-based MSA approaches, the information loss due to excluding secondary-structure information in PSA does not have a detrimental impact on clustering results.
Figure 6:
NMI scores of mothur evaluated on a V2 annotated data set dropped significantly when the top 1000 best-matched reference sequences of query sequences were removed from the SILVA database.
We repeated the above analysis by using the genus assignments as ground truth, and observed similar results (Figure 7). Again, MUSCLE+AL performed much worse than other methods (the maximum NMI score = 0.53), and its results was omitted for display purposes.
One of the main purposes of TIA is to estimate the biodiversity of a microbial community. In the microbiology literature, sequences with <3% and 5% dissimilarity are typically assigned to the same species and genus, respectively [26–28]. When comparing microbial diversities from different studies, microbiologists rarely pay sufficient attention to the computational algorithms employed. We found that the use of different methods can have a drastic impact on diversity estimates. As shown in Table 2, the numbers of OTUs observed at the 0.03 distance level are much larger than the ground truth, and vary over a wide range depending on the method employed. Sequencing errors were previously considered to be the main source for severe overestimation of microbial diversity [12,29]. However, we observe from Table 2 that except for MUSCLE+AL, the typical numbers of OTUs obtained by each method at the peak positions are always much closer to the ground truth than those obtained at the 0.03 and 0.05 distance levels. This suggests that the overestimation is not mostly due to sequencing error, but to incorrect interpretation and use of sequence identity thresholds and methods for measuring distance. The above observation supports a previous result reported in [13] that the commonly used 3% and 5% are not optimal for defining species and genus-level OTUs. Among the seven methods, ESPRIT-Tree and ESPRIT-AL yielded the most accurate estimates. Interestingly, although ESPRIT-CL has the best NMI score, the number of OTUs and NMI scores obtained by ESPRIT-CL have larger variations than those obtained by ESPRIT-AL (P < 10–6 and <2 x 10–5, respectively, by F-test). This surprising result may arise because intercluster distances calculated using an average are typically more robust to outliers and to random variation than those calculated using a maximum.
Table 2:
The numbers of OTUs observed at the 0.03 and 0.05 distance levels and at the peak positions for the seven methods
| ES-CL | ES-AL | ES-Tree | UCLUST | CD-HIT | mothur + AL | MUSCLE + AL | |
|---|---|---|---|---|---|---|---|
| 0.03 level | 2139 (31) | 1045 (19) | 1137 (30) | 1446 (28) | 920 (23) | 2893 (37) | 9508 (533) |
| 0.05 level | 702 (10) | 241 (7) | 268 (6) | 392 (13) | 314 (9) | 729 (17) | 8523 (377) |
| Peak position species | 270 (52) | 402 (9) | 400 (9) | 183 (16) | 243 (7) | 444 (12) | 13712 (301) |
| Peak position genus | 204 (14) | 190 (5) | 176 (7) | 163 (5) | 243 (7) | 444 (12) | 13712 (301) |
The numbers of species and genera averaged over the 20 test data sets are 371 ± 7 and 170 ± 5, respectively. The number in the parenthesis is one SD. ESPRIT-Tree and ESPRIT-AL yielded the most accurate estimates of microbial diversity among the seven methods.
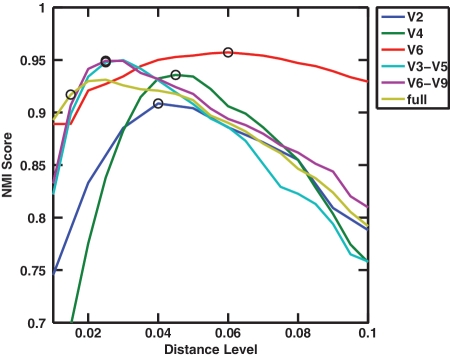
In addition to employed computational algorithms, the distance levels for defining OTUs are also dependent on targeted hypervariable regions, as different parts of the ribosome evolve at different rates. To demonstrate this, we applied ESPRIT-Tree to the reads extracted from V2 [30], V4 [31], V6 [30], V3–5 [32], V6–9 [32] and near full-length 16 S rRNA genes [33]. We found that the NMI scores peak at different distance levels ranging from 0.02 to 0.06 (Figure 8). Although the peak positions do not always coincide with the locations where the estimated numbers of OTUs equal to the numbers of species in the test data sets, they are quite close. This interesting observation merits further investigation, as it suggests that NMI scores could be used to find thresholds that lead to groups that comply optimally with known taxonomy (rather than groups that are based on guesses about rates of evolution in different regions, which may be taxon specific).
Figure 8:
The NMI scores of ESPRIT-Tree applied to simulated reads extracted from various hypervariable regions including V2, V4, V6, V3–5, V6–9 and near full-length 16 S rRNA gene. The species assignments were used as ground truth. The scores peak at different distance levels. The circles indicate the distance levels where the estimated numbers of OTUs equal to the numbers of species in the test data sets. Colour version of this figure available at http://bib.oxfordjournals.org.
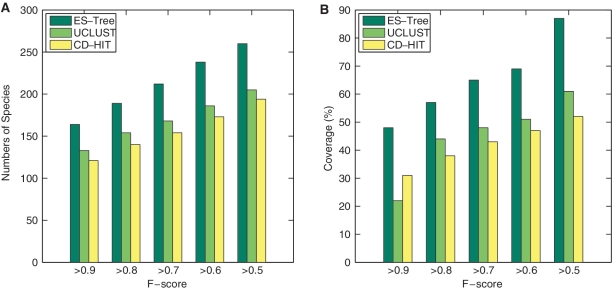
Although the number of OTUs is one criterion to be considered when comparing different methods, it is more informative to examine how the sequences originating from the same species are grouped. Here, we used the F-score [18] to compare clustering quality. The F-score considers both the precision p and the recall r, where p is the number of correct assigned sequences divided by the number of all sequences grouped into one OTU and r is the number of correct assigned sequences divided by the number of sequences that should be grouped into one OTU. Mathematically, the F-score is computed as F = 2pr/(p+r). The F-score can be interpreted as a weighted average of the precision and recall, where an F-score reaches its best value at 1 and worst score at 0. Although a NMI score grades the overall clustering quality, an F-score shows the quality of individual clusters. Since the sequences labeled as the same species can be scattered into multiple clusters, an F-score was calculated for each cluster and the maximum score was used as the F-score for a species. Figure 9 reports the number of species identified by ESPRIT-Tree, CD-HIT and ULCUST with F-scores exceeding 0.9, 0.8, 0.7, 0.6 and 0.5, respectively. The test data set we used here contains 366 species. ESPRIT-Tree recovered 164 species with a high accuracy (F-score >0.9) covering 48% of the total sequences, whereas UCLUST identified 133 species covering only 22% of the total input sequences at this level. A total of 260 species (87% of the total sequences) were identified with an F-score >0.5 for ESPRIT-Tree, which outperformed UCLUST by 55 in the number of species and 26% in total coverage. These results are in agreement with that presented in the toy example in Figure 2, where GHC partitioned the samples from the same cluster into several sub-clusters dependent on selected seeds.
Figure 9:
The numbers of species identified by ESPRIT-Tree, CD-HIT and UCLUST that have F-scores exceeding 0.9, 0.8, 0.7, 0.6 and 0.5, respectively. The corresponding coverage is also reported. ESPRIT-Tree recovered 260 species with F-score >0.5 covering 87% of the total sequences, which is significantly better than CD-HIT and UCLUST. Colour version of this figure available at http://bib.oxfordjournals.org.
Computational complexity
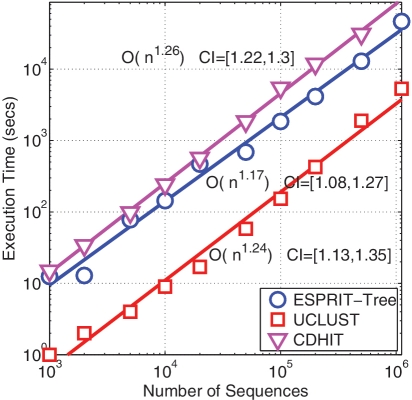
The massive amount of data generated by high-throughput sequencing technologies poses serious challenges to existing algorithms. In addition to accuracy, computational complexity is an important issue. Figure 10 reports the CPU times of ESPRIT-Tree, UCLUST and CD-HIT, applied to gut data sets with numbers of sequences ranging from 1000 to 1 100 000. The analyses were performed on an Intel E5462 2.8 GHz processor. Due to the need to generate an intermediate distance matrix, it is computationally expensive for all other methods to process one million sequences on a desktop computer. In terms of computational efficiency, UCLUST performs the best, closely followed by ESPRIT-Tree. However, all three methods have a quasilinear computational complexity of O(N1.2). It took ESPRIT-Tree about 11 h to process 1 100 000 reads to generate OTUs at 10 distance levels (0.01–0.1). We have previously applied ESPRIT-CL to the same gut data set using 100 processors, which required almost 100 h.
Figure 10:
CPU times of ESPRIT-Tree, UCLUST and CD-HIT performed on gut data sets with a varying number of sequences (1000–1 100 000). The empirical complexity and confidence interval (CI) are also reported. ESPRIT-Tree, UCLUST and CD-HIT have a quasilinear computational complexity of O(N1.2).
CONCLUSION
Although there is an urgent need to develop advanced algorithms to process massive sequence data, it is equally important to validate the performance of new approaches. Because there have been few comprehensive benchmark studies using the same data sets, the shortcomings of many existing methods have not yet been fully recognized by the microbiology community. In this article, we presented a survey of existing algorithms currently used by the community, and conducted a comprehensive benchmark study that compared seven representative methods using both real and simulated date. We showed that (i) existing methods can yield vastly different results and (ii) many microbial diversity overestimates reported in the literature are due to the inappropriate use of distance levels for defining taxonomies. When we evaluated sequence alignment methods, both MSA- and PSA-based methods appeared to have advantages and disadvantages. Due to computational constraints and their inability to align highly diverse sequences, generic MSA algorithms are not suitable for analyzing massive 16 S rRNA tag sequence data sets. Although the fixed-alignment methods overcome the computational burdens of sequence alignment by aligning input sequences against a reference database, their performance is limited by the incompleteness and alignment quality of existing databases (although these methods will improve as these reference databases improve, and are still useful for other analyses, such as phylogenetic tree building, not considered here). Although currently available PSA-based clustering methods ignore secondary structures, they provide more reliable estimates of pairwise distances by removing problems associated with heuristics involved in sequence comparison. When we compared clustering methods, we found that classic HC is superior to greedy heuristic clustering in terms of clustering accuracy. The standard HC algorithm, however, does not scale well for handling millions of sequences available to date. We have recently developed a new online learning-based HC algorithm, referred to as ESPRIT-Tree, which simultaneously addresses the space and computational issues. The algorithm exhibited a close-to-linear time and space complexity comparable to greedy heuristic algorithms, and achieved a similar accuracy to the standard HC algorithm. We further found that although both complete and average linkages perform similarly in terms of clustering accuracy, average linkage is numerically more stable (i.e. the number of OTUs and NMI scores estimated by AL have smaller variations). Our studies suggest that the distance levels for defining OTUs at various phylogenetic levels are moving targets, and depend on both the hypervariable region that is sequenced and the deployed algorithm. An indiscriminate application of the commonly used OTU definitions can lead to inaccurate estimates that could obscure real biological patterns, especially for rare taxa that can be critical in disease and in biogeochemical processes. We thus suggest that microbiologists perform an internal validation study using known sequences extracted from their own data to estimate a distance level that might offer a more accurate estimate.
Some caveats of the present analysis are worth mentioning. First, the field of metagenomics is evolving rapidly, and there are many other applications for sequence data and clustering approaches that remain to be compared, so the present study should not be seen as exhaustive. Although we tried to compare different algorithms fairly on the same platform, we did not vary the parameters, but simply relied on the default parameters chosen by the authors of each software package. Second, the BLAST-based bootstrapping approach for assessing accuracy seems to be a good compromise between real-world and simulated data. A generic TIA algorithm, by definition, does not rely on any database (except for profile-based methods). Hence, the results reported here should be able to generalize well on real-world data. However, we need to emphasize that there is currently no way to directly assess the performance of existing methods on unknown reads. We hope that the observations presented in this article will help researchers to better understand the complexities involved in various sequence analysis methods, and to choose one appropriate to their study.
Key Points.
TIA is generally considered as the first step in performing microbial community analysis. Many existing algorithms, though widely used by the biology community, have not yet been fully benchmarked and vary widely in their outputs.
This article presents a comprehensive benchmark study that addresses several issues of concern. Multiple sequence alignment is not suitable for analyzing massive 16 S rRNA tag sequences, and pairwise sequence alignment yields much more reliable estimates of microbial diversities; the performance of fixed alignment-based methods is limited by the incompleteness and the alignment quality of existing databases; classic HC is superior to greedy heuristic clustering in terms of clustering accuracy; the average linkage method is numerically more stable than the complete linkage method.
The distance levels for defining taxonomies at various phylogenetic levels are a moving target and depend on both the hypervariable region that is sequenced and the deployed algorithm. Many astoundingly high biodiversity estimates reported in the literature appear to be overestimates resulting from the inappropriate use of distance levels for defining taxonomies. One possible way to alleviate this issue is to perform an internal validation study using known sequences to estimate a distance level that may offer a more accurate estimate.
The benchmark study identified ESPRIT-Tree, a fast implementation of the average linkage-based HC algorithm, as one of the best algorithms available in terms of computational efficiency and clustering accuracy.
Biographies
Yijun Sun is an Assistant Scientist at the Interdisciplinary Center for Biotechnology Research and an affiliated faculty member at the Department of Electrical and Computer Engineering at University of Florida. His research interests are mainly on machine learning, bioinformatics and their applications to cancer study and metagenomics.
Yunpeng Cai is a postdoctoral fellow at the Interdisciplinary Center for Biotechnology Research at University of Florida. His research interests are mainly on machine learning, bioinformatics and their applications to cancer study and metagenomics.
Susan M. Huse is an Assistant Research Scientist at Josephine Bay Paul Center for Comparative Molecular Biology and Evolution, Marine Biological Laboratory, Woods Hole, Massachusetts. She studies microbial diversities, population structures, evolution and bioinformatics.
Rob Knight is an HHMI Early Career Scientist and Associate Professor in the Department of Chemistry and Biochemistry, University of Colorado at Boulder. His research interests include the human microbiome, molecular evolution and the RNA World.
William G. Farmerie is an Assistant Scientist and Associate Director at the Interdisciplinary Center for Biotechnology Research at the University of Florida.
Xiaoyu Wang is a postdoctoral fellow and Head of bioinformatics in the Microbial Epidemiology Laboratory at the Emerging Pathogens Institute. He has been trained as a computer scientist and expanded his skills to bioinformatics.
Volker Mai is an Assistant Professor in the Department of Microbiology and Cell Science and Director of the Microbial Epidemiology Laboratory at the Emerging Pathogens Institute. He has been trained as both a microbiologist as well as an epidemiologist.
FUNDING
This work was supported by a subcontract to Mitchell L. Sogin from the NIH Human Microbioime 1UH2DK083993-01 awarded to the University of Michigan and foundation awards from the W.M. Keck Foundation to M.L.S. and the Alfred P. Sloan Foundation to M.L.S. and David Mark Welch; Howard Hughes Medical Institute [to R.K.]; American Cancer Society [grant MRSGT CCE-107301 to V.M.].
References
- 1.Eisen JA. Environmental shotgun sequencing: its potential and challenges for studying the hidden world of microbes. PLoS Biol. 2007;5:e82. doi: 10.1371/journal.pbio.0050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothberg J, Leamon J. The development and impact of 454 sequencing. Nat Biotechnol. 2008;26:1117–24. doi: 10.1038/nbt1485. [DOI] [PubMed] [Google Scholar]
- 3.Peterson J, Garges S, Giovanni M, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–23. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Cole JR, Wang Q, Cardenas E, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–5. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun Y, Cai Y, Liu L, et al. ESPRIT: Estimating species richness using large collections of 16 S rRNA pyrosequences. Nucleic Acids Res. 2009;37(10):e76. doi: 10.1093/nar/gkp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W, Jaroszewski L, Godzik A. Clustering of highly homologous sequences to reduce the size of large protein database. Bioinformatics. 2001;17:282–3. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- 8.Cai Y, Sun Y. ESPRIT-Tree: taxonomy independent analysis of millions of 16 S rRNA pyrosequences in quasilinear computational time. Technical Report. doi: 10.1093/nar/gkr349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 11.Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–6. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huse SM, Welch DM, Morrison HG, et al. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2009;12(7):1889–98. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White JR, Navlakha S, Nagarajan N, et al. Alignment and clustering of pylogenetic markers - implications for microbial diversity studies. BMC Bioinfomatics. 2010;11(1):152. doi: 10.1186/1471-2105-11-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Y, Cai Y, Mai V, et al. Advanced computational algorithms for microbial community analysis using massive 16 S rRNA sequence data. Nucleic Acids Res. 2010;38(22):e205. doi: 10.1093/nar/gkq872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME: allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giongo A, Crabb DB, Davis-Richardson AG, et al. PANGEA: pipeline for analysis of next generation amplicons. ISME J. 2010;4(7):852–61. doi: 10.1038/ismej.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Studholme C, Hill DLG, Hawkes DJ. An overlap invariant entropy measure of 3D medical image alignment. Pattern Recognition. 1999;32(1):71–86. [Google Scholar]
- 18.Van Rijsbergen CJ. Information Retrieval. 2nd edn. London: Butterworth-Heinemann; 1979. [Google Scholar]
- 19.Sogin ML, Morrison HG, Huber JA, et al. Microbial diversity in the deep sea and the underexplored ``rare biosphere". Proc Natl Acad Sci USA. 2006;103:12115–20. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keijser BJ, Zaura E, Huse SM, et al. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res. 2008;87:1016–20. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- 21.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh K, Kuma K, Toh H, et al. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–8. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeSantis TZ, Hugenholtz P, Keller K, et al. NAST: a multiple sequence alignment server for comparative analysis of 16 S rRNA genes. Nucleic Acids Res. 2006;34:W394–9. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schloss PD. The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16 S rRNA gene-based studies. PLoS Comput Biol. 2010;6(7):e1000844. doi: 10.1371/journal.pcbi.1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward JH. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58(301):236–44. [Google Scholar]
- 26.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, DiBaise JK, Zuccolo A, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA. 2009;106:2365–70. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward BB. How many species of prokaryotes are there? Proc Natl Acad Sci USA. 2002;99:10234–6. doi: 10.1073/pnas.162359199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quince C, Lanzén A, Curtis TP, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6(9):639–41. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 30.Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature. 2007;449:804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Claesson MJ, Cusack S, O’Sullivan O, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA. doi: 10.1073/pnas.1000097107. doi:10:1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Ellen. Effect of Crohn’s Disease Risk Alleles on Enteric Microbiota. NIH project No. 1UH2DK083994-01. NCBI accession NO. SRX021353 and SRX021354. [Google Scholar]
- 33.Frank DN, St, Amand AL, Feldman RA, et al. (2007) Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 104(34):13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]