Summary of recent advances
The obligatory intracellular bacterial pathogens Anaplasma and Ehrlichia infect leukocytes by hijacking host-cell components and processes. The type IV secretion system is up-regulated during infection. Among type IV secretion candidate substrates, an ankyrin repeat protein of Anaplasma phagocytophilum, AnkA, is delivered into the host cytoplasm via a complex that includes VirD4. AnkA is highly tyrosine-phosphorylated and binds to the Abl interactor 1, SHP-1, and nuclear DNA fragments. Ehrlichia chaffeensis AnkA was recently reported to be translocated into host cell nucleus. The recent discovery of several ankyrin repeat proteins secreted via the type IV secretion system of different intracellular bacteria suggests that a common strategy evolved to subvert host-cell functions.
Introduction
Anaplasma phagocytophilum and Ehrlichia chaffeensis are Gram-negative cocci that cause human granulocytic anaplasmosis (HGA) and human monocytic ehrlichiosis (HME), respectively. They are obligatory intracellular bacteria most closely related to Rickettsia spp., for example, Rickettsia prowazekii that causes epidemic typhus, a disease that has been responsible for the deaths of millions of people during wartime or natural disasters. Wild animals, such as white-tailed deer and white-footed mice, are reservoirs for A. phagocytophilum and E. chaffeensis, and human infection occurs through the bite of infected ticks [1,2]. HGA and HME are acute flu-like illnesses characterized by fever, headache, myalgia, anorexia, and chills, and frequently are accompanied by leukopenia with the appearance of immature cells (or rebound leukocytosis), thrombocytopenia, anemia, and elevated levels of serum hepatic aminotransferases. The severity of the disease varies from asymptomatic seroconversion to severe morbidity or death [3-5]. Although these bacteria were initially culture-isolated from patients less than two decades ago, HGA and HME are among the most prevalent tick-borne zoonoses in North America. While A. phagocytophilum and E. chaffeensis do not have genes for the biosynthesis of lipopolysaccharides and peptidoglycans that activate host leukocytes, whole genome sequence data available in 2006 has been facilitating studies to find virulence determinants of these bacteria [6]. Based on NCBI Conserved Domain searches, bacterial type II, III, V, and VI secretion components have not been detected in A. phagocytophilum and E. chaffeensis; however, the type IV secretion (T4S) system has been identified in these bacteria, and the secreted products are expected to function as effectors in the host to facilitate infection and disease progression. Analysis showed that components of Agrobacterium tumefaciens VirB/D system are conserved in A. phagocytophilum and E. chaffeensis [6,7•]. In particular, A. phagocytophilum VirD4 exhibits high identity with A. tumefaciens VirD4, a component of the T4S apparatus. VirD4 is regarded as a coupling protein because it recognizes C-terminal sequences within T4S substrate proteins prior to delivery into the VirB transmembrane channel [7•]. Targeted gene insertion, knockout, or complementation in obligatory intracellular bacteria has not been feasible yet. However, using the CRAfT (Cre recombinase reporter assay for translocation) system developed in A. tumefaciens [8], we demonstrated that the A. phagocytophilum ankyrin repeat domain–containing protein, AnkA can be translocated from A. tumefaciens into plant cells in a VirD4-dependent manner [9••]. Bioinformatic analysis of whole genome sequences of these bacteria showed that A. phagocytophilum and E. chaffeensis encode several proteins with eukaryotic-like domains/motifs—a common theme of bacterial effector proteins that functionally mimic host cell proteins (Table 1) [10,11•]. Additionally, based on the characteristics of A. tumefaciens T4S substrate motifs [12] and/or using bacterial two-hybrid system, we have identified several putative T4S candidates [6,13] (W. Bao, MS thesis, The Ohio State University, 2008). Here, we discuss host subversion and exploitation events by these bacteria and the recent progress on regulation of T4S and the secreted effector proteins, in particular highlighting Ank proteins.
Table 1.
A. phagocytophilum and E. chaffeensis genes encoding potential eukaryotic-like motifs/domains
| Motif/Domains 1 | Predicted Roles | Gene Locus 2 |
|---|---|---|
| Ankyrin repeats | Protein-protein interactions | APH0259, APH0709, APH0740, APH0928; ECH0389, ECH0653, ECH0684, ECH0877. |
| Tetratricopeptide repeat domain (TPR) | Protein-protein interactions | APH1212, APH1316, ECH0048 |
| SNARE-associated Golgi Protein | Vesicular fusion | APH1021, ECH0947 |
| Ras-like GTPase | Vesicular trafficking | ECH0155 |
| Serine/Threonine protein kinases | Cellular Signaling | APH0984, ECH0840 |
| Protein Phosphatase | Cellular Signaling | ECH0964 |
| PKCI_related | Protein Kinase C Interacting protein related, HIT family of hydrolases | APH0860, ECH0826 |
| WD40 domain | Signal transduction and cytoskeleton assembly | APH1037 |
| Stomatin/prohibitin homologs, and Band 7 domain of flotillin/reggie like proteins | Lipid raft associated protein and regulated proteolysis | APH0260, APH1146, APH1147; ECH1050, ECH1051 |
| Breast Cancer Suppressor Protein (BRCA1) C-terminal domain (BRCT) | DNA damage repair | APH0138, ECH0301 |
The eukaryotic-like motifs/domains were determined by bioinformatic analysis of the genomes of A. phagocytophilum HZ strain (GenBank accession number: NC_007797) and E. chaffeensis Arkansas strain (NC_007799). Detailed domain information is available at GenBank Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml).
Gene locus abbreviations: APH, A. phagocytophilum; ECH, E. chaffeensis.
Subversion and exploitation of infected host cells
A. phagocytophilum and E. chaffeensis replicate in the membrane-bound compartments (inclusions, also called morulae [(morulae – Latin “morus”, mulberry)] as they look like mulberries under the light microscope) of human granulocytes and monocytes-macrophages, respectively, which are the primary immune defense cells that normally are responsible for powerful innate antimicrobial responses. These intracellular bacteria parasitize host cells by subverting their various innate immune responses, including inhibiting NADPH oxidase activation, lysosomal fusion with bacterial inclusions, and IFN-γ signaling, and by down-regulating Toll-like receptor 2/4, and CD14 expression [14]. Furthermore, this group of bacteria inhibits host cell apoptosis to maximize intracellular bacterial reproduction [15-17].
A. phagocytophilum and E. chaffeensis enter host cells via caveolae (lipid raft)-mediated endocytosis, which directs pathogens to an intracellular compartment secluded from late endosome or lysosome markers and NADPH oxidase components [18]. Both early and replicative A. phagocytophilum and E. chaffeensis inclusions co-localize with tyrosine-phosphorylated proteins and phospholipase C-γ2; activation of tyrosine kinases and phospholipase C-γ2 are required for infection of host cells [18]. E. chaffeensis inclusions retain the early endosome characteristics including Rab5 and early endosome antigen 1, and fuse with endosomes enriched with transferrin receptor [18]. In contrast, A. phagocytophilum inclusions are negative for these endosomal markers. Several hallmarks of early autophagosomes have been detected in A. phagocytophilum replicative inclusions, including a double lipid bilayer membrane, and ectopically expressed GFP-tagged LC3 and Beclin 1, the human homologs of Saccharomyces cerevisiae autophagy-related proteins Atg8 and Atg6, respectively [19]. Stimulation of autophagy by rapamycin favors A. phagocytophilum infection. Inhibition of the autophagosomal pathway by 3-methyladenine does not inhibit A. phagocytophilum internalization, but reversibly arrests its growth [19].
A. phagocytophilum and E. chaffeensis require cholesterol for survival and growth, but they lack genes for cholesterol biosynthesis or modification and thus must acquire cholesterol from host cells [20]. A. phagocytophilum infection significantly up-regulates host cellular cholesterol levels by enhancing low-density lipoprotein uptake through stabilization of host LDL receptor mRNA [21]. A. phagocytophilum inclusions become enriched with cholesterol by hijacking non-esterified free cholesterol from the host low-density lipoprotein uptake pathway [21].
T4S apparatus
There are at least two ancestral lineages for the T4S system: the virB/virD system of A. tumefaciens and the dot/icm system of Legionella pneumophila, referred to as T4aS and T4bS systems, respectively. A. phagocytophilum and E. chaffeensis have genes encoding the T4aS system. In A. tumefaciens, the single virB operon, along with virD4, encodes 12 membrane-associated proteins that form a transmembrane channel complex [7•]. In the A. phagocytophilum and E. chaffeensis genomes, several virB/D genes are duplicated and distributed in five clusters: sodB-virB3–virB4-virB6-1-virB6-2-virB6-3-virB6-4, virB8-1-virB9-1-virB10-virB11–virD4, virB4-2-tandem virB2s (eight nonidentical virB2s in A. phagocytophilum and four nonidentical virB2s in E. chaffeensis), virB8-2, and virB9-2 [13]. Interestingly, all five virB/D loci are up-regulated during the exponential growth stage of E. chaffeensis synchronously cultured in THP-1 human monocytic leukemia cells, and down-regulated prior to the release of E. chaffeensis from host THP-1 cells [22•]. Proteomic analysis identified an E. chaffeensis hypothetical protein, named EcxR for E. chaffeensis expression regulator in this study, which binds promoter regions of all five operons and genes, and transactivates them in a lacZ reporter assay [22•]. This may be a mechanism by which transcription of the scattered virB/D loci is temporarily coordinated. During the infection of human neutrophils in vitro, both virB9-1 and virB6-1 of A. phagocytophilum, which are present in separate genomic loci, are up-regulated at the mRNA level as is VirB9-1 at the protein level [23]. In contrast, the majority of A. phagocytophilum spontaneously released from infected host cells expresses only low levels of VirB9 protein [23]. This modulation of virB/D expression during the establishment of bacterial infection may promote intracellular survival and replication as well as resistance upon exposure to the extracellular environment. In addition to mammalian hosts, the T4S system is also expected to function during tick infection given that virB9 is expressed by Ehrlichia canis in tick tissues [24], and four E. chaffeensis virB6 paralogs and E. chaffeensis virB9-1 are expressed in ISE6 tick cell culture [25•]. Some of the duplicated virB paralogs of these bacteria may be reserved to function specifically in ticks, as differential transcription of several A. phagocytophilum virB2 paralogs in mammalian and ISE6 tick cells have been reported [26]. Interestingly, double immunofluorescence labeling of host cell–free Neorickettsia risticii, which is closely related to Ehrlichia and Anaplasma spp., showed bipolar surface localization of the primary T4S pilus component VirB2, despite the round shape of bacteria, suggesting an underlying subcellular structure that dictates the spatial distribution of the T4S pili [27]. VirB9 is bacterial surface-exposed in both E. chaffeensis and A. phagocytophilum [23,28]. In addition, the assembled macromolecular structure is expected to differ from the prototypical T4aS apparatus of A. tumefaciens, considering that all (total four) VirB6 homologs of E. chaffeensis are 3- to 10-fold larger than A. tumefaciens VirB6 and coexpressed. Coimmunoprecipitation analysis and far-western blotting of E. chaffeensis revealed that E. chaffeensis VirB9-1 and the four VirB6 proteins form a unique complex [25•].
T4S secretion substrates
The A. tumefaciens VirB/D T4S system secretes approximately 10 substrates into the host cell [7•]. In contrast, the Legionella T4S system can secrete over 100 effector molecules, and it is not yet clear whether and why so many effectors are required for the survival of this bacterium in the host cells [29]. The T4aS effector molecules, CagA of Helicobacter pylori, pertussis toxin, Beps of Bartonella henselae, and the T-DNA complex have been shown to play a major role in disease pathogenesis [30-33]. The T4S system is essential for the biogenesis of cytoplasmic replicative compartments unique to each of several facultative intracellular bacteria, such as Brucella, Bartonella, and Legionella [29,34,35]. Although there are no conserved protein sequence motifs, certain C-terminal sequences are critical for secretion via T4S apparatus [7•]. In A. tumefaciens, basic amino acids, net positive charge, and a characteristic hydropathy profile at C-terminus are critical for recruitment and T4aS translocation of effector proteins [7•,12]. Some T4aS substrates, such as H. pylori CagA and B. henselae Beps, require positively charged C-terminal tail and certain N-terminal or internal domains for the translocation [31,36]. For RalF and several other T4bS substrates in L. pneumophila, a hydrophobic residue at -3 or -4 position is critical for the translocation [37]. In A. phagocytophilum, an ankyrin-repeats (Ank) protein, AnkA and a hypothetical protein, named Anaplasma translocated substrate 1 (Ats-1), are known to be secreted into the host cell cytoplasm [9••,13]. Both AnkA and Ats-1 have C-terminal positive charged residues and hydropathy profiles similar to those of T4aS substrates [9••,12,13].
A. phagocytophilum AnkA is one of strain-variable proteins [38,39]. As shown in Figure 1, different numbers of ankyrin repeats and tyrosine kinase domains are found in four A. phagocytophilum strains [MRK, HZ, NCH-1, and BDS (BDS strain is similar to the Webster strain for which the sequence is currently unavailable)]. A. phagocytophilum HZ strain AnkA is one of most abundantly expressed proteins that shows apparent molecular mass of 160 kDa, and is highly tyrosine phosphorylated after delivery into the host leukocyte cytoplasm [9••]. AnkA tyrosine phosphorylation occurs 2 min after infection, and phosphorylated AnkA accumulates during infection in the host cell cytoplasm, whereas very little AnkA is retained within the bacteria or the inclusion as demonstrated by immunofluorescence labeling [9••]. AnkA binds to Abl-interactor 1 that interacts with Abl-1 tyrosine kinase, thus mediating AnkA phosphorylation. AnkA and Abl-1 are critical for A. phagocytophilum infection, as infection is inhibited upon host cytoplasmic delivery of an antibody against AnkA, Abl-1 knockdown via a targeted short interfering RNA, or treatment with Gleevec, a specific pharmacological inhibitor of Abl-1, suggesting a potential supplement to the antibiotic doxycycline to enhance the efficacy of HGA therapy [9••]. This is in contrast to Legionella in which knockout of a single T4S substrate does not result in phenotype change due to functional redundancy of multiple T4S substrates [29, 35], suggesting that in A. phagocytophilum each T4S substrate has significant weight in regulating host cell functions. A study showed that the infection by A. phagocytophilum strain NCH-1 specifically induces tyrosine phosphorylation of a 190-kDa AnkA, which then interacts with the host tyrosine phosphatase SHP-1 through the SH2 domain [40•• ]. AnkA of the A. phagocytophilum Webster strain was reported to localize within nuclei of infected HL-60 cells and bind to the inter-nucleosomal region of chromosomes as well as transcriptional regulatory regions of the CYBB locus, which suppresses the host cell innate immune response [41,42••]. However, it remains to be determined whether and how AnkA can be imported topologically inside host cell nucleus. It is currently unclear whether AnkA has these multiple biological activities or these are strain-specific phenotypes. Different from AnkA, a large proportion of expressed Ats-1 colocalizes with A. phagocytophilum inclusions, and Ats-1 translocation to the host cell cytoplasm becomes evident 32 h post-infection (early exponential growth stage) [13]. Interestingly, Ats-1 has an N-terminal mitochondria localization signal that allows it to translocate into the mitochondria of infected-human neutrophils and HL-60 cells, and when ectopically expressed, it inhibits etoposide-induced apoptosis in mammalian cells [13] (H Niu & Y Rikihisa. Abstract D-146, 108th Amer. Soc. Microbiology General Meeting. Boston, MA. June 2008).
Figure 1. Domain structures of Anaplasma phagocytophilum AnkA proteins from various strains.
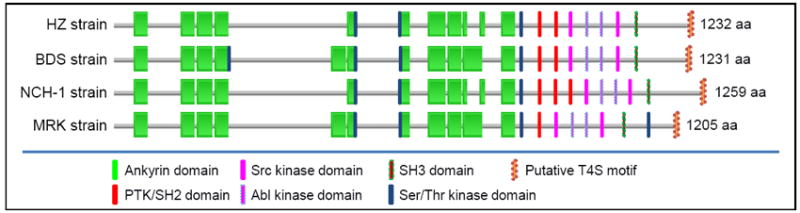
Domain structures and putative motifs were predicted by searching against the Pfam database (http://pfam.sanger.ac.uk) and ScanSite (http://scansite.mit.edu), respectively, using default parameters. The presence of a T4S motif was determined as shown in Table 2. Protein lengths [amino acids (AA) numbers] are shown on the right, and domains are not drawn proportional to the actual size. All strains are human isolates with the exception of MRK, which is a horse isolate. Isolates collection locations: HZ, New York; BDS, Wisconsin; NCH-1, Massachusetts; MRK, California.
Ank proteins
Eukaryotic Ankyrin-repeat (Ank) proteins mediate protein-protein interactions involved in a multitude of host processes including cytoskeletal motility, tumor suppression, and transcriptional regulation [43]. Ankyrin domain contains a 33-residue repeating motif, which consists of two anti-parallel α-helices connected to the next repeat via a loop region [43]. A series of Ank-repeats are arranged in a curved structure with protein-protein interactions occurring in the loop regions. Several ank genes encoding heterogeneous Ank proteins are present in the facultative or obligate intracellular bacteria, including L. pneumophila (15 copies) [44], Coxiella burnetii (15 copies) [11•], Orientia tsutsugamushi (40 copies) [45], Wolbachia pipientis (23 copies) [46], Wolbachia from Culex pipiens group (60 copies) [47], and Rickettsia prowazekii (1 copy) [48]. The L. pneumophila AnkX protein prevents microtubule-dependent vesicular transport, which interferes with fusion of the L. pneumophila–containing vacuole with late endosomes after infection of macrophages [49••]. In C. burnetii, ank genes show remarkable heterogeneity [11•]. Ectopically expressed Ank proteins localize to a variety of subcellular regions in mammalian cells including microtubules, mitochondria, and the parasitophorous vacuole membrane [11•]. Using L. pneumophila as a surrogate host, AnkG of C. burnetii was recently reported to be secreted by the L. pneumophila T4S system. Once secreted into host cells, AnkG can bind to mammalian protein p32 (gC1qR) and interfere with the ability of p32 to modulate RNA splicing of Bcl-x, thereby preventing host cell apoptosis (A Luhrmann et al., abstract 137, 23rd Meeting of the American Society for Rickettsiology, Hilton Head Island, SC, August 2009).
As shown in Table 2 and Figure 2, Anaplasma and Ehrlichia spp. each have three to five Ank proteins, some of which contain multiple potential phosphorylation sites for host signaling and regulation. AnkA of A. phagocytophilum strain HZ has 11 ankyrin repeats, and several tyrosine phosphorylation and/or SH2 domains are concentrated near the C terminus (Figure 1). In contrast, AnkA of E. chaffeensis strain Arkansas (ECH0684) has 19 ankyrin repeats, and SH2 domains are concentrated near the N terminus (Figure 2). Both AnkA proteins have basic amino acids, net positive charges, and hydropathy profiles at the C terminus similar to those of Agrobacterium T4S substrates (Table 2) [12]. E. chaffeensis AnkA is also secreted and tyrosine phosphorylated in host cells (M Lin and Y Rikihisa, unpublished data). Recently, E. chaffeensis AnkA was reported to be translocated into host cell nucleus and bind to Alu elements, suggesting global modulation of host genes [50••]. AnkA of Ehrlichia canis and Anaplasma marginale have only two to three basic residues within the 25-residue C-terminal region (Table 2). However, net positive charge and the hydropathy profiles of these proteins suggest that they are potential T4S substrates (Table 2). The largest Ank proteins of >300-kDa are conserved among Ehrlichia and Anaplasma species. These proteins have seven to ten ankyrin repeats, several serine/threonine phosphorylation sites, and possess T4S motifs similar to those of Agrobacterium T4S substrates (Table 2, Figure 2). It will be interesting to learn whether these proteins are secreted by the T4S system to induce some of the above-mentioned cellular phenotypes.
Table 2.
Ankyrin-repeats containing proteins in Ehrlichia and Anaplasma spp.
| Organisms (Locus ID/GenBank #) | Protein Properties | # of Ankyrin Repeats 1 | T4S Motif (pI, Hydropathy Profile)2 (C-Term. 25 aa) |
|---|---|---|---|
| A. phagocytophilum HZ: | |||
| APH_0259 (YP_504874) | 26.4 kDa (239 aa), pI 4.2 | 2 |
|
| APH_0709 (YP_505290) | 367.3 kDa (3373 aa), pI 5.4 | 10 |
|
| APH_0740 (YP_505319), AnkA | 131.2 kDa (1232 aa), pI 6.5 | 11 |
|
| APH_0928 (YP_505501) | 97.0 kDa (886 aa), pI 7.7 | 9 |
|
| A. marginale St Maries: | |||
| AM638 (AAV86627) | 347.8 kDa (3194 aa), pI 5.5 | 9 |
|
| AM705 (AAV86672), AnkA | 145.7 kDa (1387 aa), pI 6.3 | 13 |
|
| AM926 (AAV86836) | 31.0 kDa (282 aa), pI 5.0 | 2 |
|
| E. chaffeensis Arkansas: | |||
| ECH_0389 (YP_507209) | 16.0 kDa (140 aa), pI 4.7 | 2 |
|
| ECH_0653 (YP_507462) | 467.8 kDa (4313 aa), pI 5.6 | 9 |
|
| ECH_0684 (YP_507490), AnkA | 156.7 kDa (1463 aa), pI 4.6 | 19 |
|
| ECH_0877 (YP_507672) | 96.8 kDa (874 aa), pI 6.8 | 9 |
|
| E. canis Jake: | |||
| Ecaj_0052 (AAZ68103) | 70.5 kDa (613 aa), pI 6.6 | 3 |
|
| Ecaj_0221 (AAZ68270) | 104.4 kDa (933 aa), pI 6.9 | 7 |
|
| Ecaj_0365 (AAZ68408), AnkA | 152.9 kDa (1421 aa), pI 5.3 | 21 |
|
| Ecaj_0387 (AAZ68430) | 471.4 kDa (4245 aa), pI 5.2 | 7 |
|
| Ecaj_0627 (AAZ68661) | 173.4 kDa (151 aa), pI 8.6 | 2 |
|
| E. ruminantium Gardel: | |||
| Erga_CDS_03830 (CAI27668) | 98.8 kDa (877 aa), pI 6.5 | 9 |
|
| Erga_CDS_03830 (CAI27835), AnkA | 175.9 kDa (1640 aa), pI 4.9 | 18 |
|
| Erga_CDS_04060 (CAI27858) | 329.1 kDa (2992 aa), pI 6.1 | 7 |
|
| Erga_CDS_06440 (CAI28096) | 141.5 kDa (125 aa), pI 5.1 | 2 |
|
The numbers of ankyrin repeats were determined by searching against PFAM domain database with E-value < 1 (http://pfam.sanger.ac.uk/).
The presence of a T4S motif was determined by the characteristics of Agrobacterium tumefaciens T4S substrates, including three or more basic amino acids (K, H, or R shown in red), net positive charges, and hydropathy profiles at its C-terminal 25 amino acids [12].
Figure 2. Domain structure of ankyrin repeat proteins in Anaplasma phagocytophilum HZ and Ehrlichia chaffeensis Arkansas.

Domain structures and putative motifs were predicted by searching against the Pfam database and ScanSite, respectively, using default parameters. The presence of a T4S motif was determined as shown in Table 2. ECH: E. chaffeensis Arkansas gene locus; APH: A. phagocytophilum HZ gene locus. Protein lengths (AA numbers) are shown on the right, and domains are not drawn proportional to actual size.
Conclusions
Although much remains to be learned about the mechanistic details of how effectors are secreted by the Anaplasma and Ehrlichia T4S apparatus and their subcellular sites of action, further studies on T4S effector candidates—including Anks and cognate host cell partners—are expected to provide a molecular basis for understanding pathogen subversion of host-cell functions and disease pathogenesis.
Acknowledgments
A portion of the studies in the author’s laboratory reported in this review was supported by grants R01 AI054476 and R01 AI30010 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
-
•
of special interest
-
••
of outstanding interest
- 1.Ewing SA, Dawson JE, Kocan AA, Barker RW, Warner CK, Panciera RJ, Fox JC, Kocan KM, Blouin EF. Experimental transmission of Ehrlichia chaffeensis (Rickettsiales: Ehrlichieae) among white-tailed deer by Amblyomma americanum (Acari: Ixodidae) J Med Entomol. 1995;32:368–374. doi: 10.1093/jmedent/32.3.368. [DOI] [PubMed] [Google Scholar]
- 2.Telford SR, 3rd, Dawson JE, Katavolos P, Warner CK, Kolbert CP, Persing DH. Perpetuation of the agent of human granulocytic ehrlichiosis in a deer tick-rodent cycle. Proc Natl Acad Sci USA. 1996;93:6209–6214. doi: 10.1073/pnas.93.12.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paddock CD, Childs JE. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin Microbiol Rev. 2003;16:37–64. doi: 10.1128/CMR.16.1.37-64.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakken JS, Dumler S. Human granulocytic anaplasmosis. Infect Dis Clin North Am. 2008;22:433–448. viii. doi: 10.1016/j.idc.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Thomas RJ, Dumler JS, Carlyon JA. Current management of human granulocytic anaplasmosis, human monocytic ehrlichiosis and Ehrlichia ewingii ehrlichiosis. Expert Rev Anti Infect Ther. 2009;7:709–722. doi: 10.1586/eri.09.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunning Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen J, Seshadri R, Ren Q, Wu M, Utterback TR, et al. Comparative Genomics of Emerging Human Ehrlichiosis Agents. PLoS Genetics. 2006;2:e21. doi: 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7•.Alvarez-Marinez CE, Christie PJ. Biological diversities of prokaryotic Type IV secretion systems. Microbiol Mol Biol Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. This is a comprehensive and updated review of comparative Type IV secretion systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CM, Regensburg-Tuink TJ, Hooykaas PJ. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- 9••.Lin M, den Dulk-Ras A, Hooykaas PJ, Rikihisa Y. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol. 2007;9:2644–2657. doi: 10.1111/j.1462-5822.2007.00985.x. This study demonstrated a tyrosine-phosphorylated protein throughout infection, is A. phagocytophilum AnkA. AnkA is shown to be secreted in a VirB/D4-dependent manner. AnkA binds to Abl-interactor 1 that interacts with Abl-1 tyrosine kinase, thus mediating AnkA phosphorylation. Infection was inhibited upon host cytoplasmic delivery of anti-AnkA antibody, Abl-1 knockdown with targeted siRNA, or treatment with a specific pharmacological inhibitor of Abl-1. [DOI] [PubMed] [Google Scholar]
- 10.Stebbins CE, Galan JE. Structural mimicry in bacterial virulence. Nature. 2001;412:701–705. doi: 10.1038/35089000. [DOI] [PubMed] [Google Scholar]
- 11•.Voth DE, Howe D, Beare PA, Vogel JP, Unsworth N, Samuel JE, Heinzen RA. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous with C-terminal truncations that influence Dot/Icm-mediated secretion. J Bacteriol. 2009;191:4232–4242. doi: 10.1128/JB.01656-08. Using Legionella pneumophila as surrogate host, the authors identified 10 Anks from Coxiella burnetii Dugway and one Ank specific to the G and K endocarditis isolates are translocated into the host cytosol in a Dot/Icm-dependent fashion A 10 amino acid C-terminal region is necessary for translocation with some Anks also requiring the chaperone IcmS for secretion Ectopically expressed Anks localized to a variety of subcellular regions in mammalian cells including microtubules, mitochondria, and the PV membrane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vergunst AC, van Lier MCM, den Dulk-Ras A, Grosse Stuve TA, Ouwehand A, Hooykaas PJJ. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci USA. 2005;102:832–837. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rikihisa Y, Lin M, Niu H, Cheng Z. Type IV secretion system of Anaplasma phagocytophilum and Ehrlichia chaffeensis. Ann N Y Acad Sci. 2009;1166:106–111. doi: 10.1111/j.1749-6632.2009.04527.x. [DOI] [PubMed] [Google Scholar]
- 14.Rikihisa Y. Ehrlichia subversion of host innate responses. Curr Opin Microbiol. 2006;9:95–101. doi: 10.1016/j.mib.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Ge Y, Rikihisa Y. Anaplasma phagocytophilum delays spontaneous human neutrophil apoptosis by modulation of multiple apoptotic pathways. Cell Microbiol. 2006;8:1406–1416. doi: 10.1111/j.1462-5822.2006.00720.x. [DOI] [PubMed] [Google Scholar]
- 16.Ge Y, Yoshiie K, Kuribayashi F, Lin M, Rikihisa Y. Anaplasma phagocytophilum inhibits human neutrophil apoptosis via upregulation of bfl-1, maintenance of mitochondrial membrane potential and prevention of caspase 3 activation. Cell Microbiol. 2005;7:29–38. doi: 10.1111/j.1462-5822.2004.00427.x. [DOI] [PubMed] [Google Scholar]
- 17.Xiong Q, Wang X, Rikihisa Y. High-cholesterol diet facilitates Anaplasma phagocytophilum infection and up-regulates macrophage inflammatory protein-2 and CXCR2 expression in apolipoprotein E-deficient mice. J Infect Dis. 2007;195 doi: 10.1086/514819. [DOI] [PubMed] [Google Scholar]
- 18.Rikihisa Y. Intracellular Niches of Ehrlichia and Anaplasma. In: Schaible Ulrich, Haas Albert., editors. Intracellular Niches of Microbes. A pathogens guide through the host cell. Wiley-VCH Verlag GmbH & Co., KGaA; Weinheim: 2009. pp. 301–314. [Google Scholar]
- 19.Niu H, Yamaguchi M, Rikihisa Y. Subversion of cellular autophagy by Anaplasma phagocytophilum. Cell Microbiol. 2008;10:593–605. doi: 10.1111/j.1462-5822.2007.01068.x. [DOI] [PubMed] [Google Scholar]
- 20.Lin M, Rikihisa Y. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect Immun. 2003;71:5324–5331. doi: 10.1128/IAI.71.9.5324-5331.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiong Q, Lin M, Rikihisa Y. Cholesterol-dependent Anaplasma phagocytophilum exploits the low-density lipoprotein uptake pathway. PLoS Pathog. 2009;5:e1000329. doi: 10.1371/journal.ppat.1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Cheng Z, Wang X, Rikihisa Y. Regulation of type IV secretion apparatus genes during Ehrlichia chaffeensis intracellular development by a previously unidentified protein. J Bacteriol. 2008;190:2096–2105. doi: 10.1128/JB.01813-07. This study showed transcription of all five virB/D loci is downregulated prior to the release of E. chaffeensis from host THP-1 cells and is upregulated at the initiation of exponential growth. An E. chaffeensis 12.3-kDa hypothetical protein (EcxR) binds to the promoter regions of these individual virB/D loci, and activates transcription of all five virB/D loci in LacZ reporter constructs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu H, Rikihisa Y, Yamaguchi M, Ohashi N. Differential expression of VirB9 and VirB6 during the life cycle of Anaplasma phagocytophilum in human leucocytes is associated with differential binding and avoidance of lysosome pathway. Cell Microbiol. 2006;8:523–534. doi: 10.1111/j.1462-5822.2005.00643.x. [DOI] [PubMed] [Google Scholar]
- 24.Felek S, Huang H, Rikihisa Y. Sequence and expression analysis of virB9 of the type IV secretion system of Ehrlichia canis strains in ticks, dogs, and cultured cells. Infect Immun. 2003;71:6063–6067. doi: 10.1128/IAI.71.10.6063-6067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Bao W, Kumagai Y, Niu H, Yamaguchi M, Miura K, Rikihisa Y. Four VirB6 paralogs and VirB9 are expressed and interact in Ehrlichia chaffeensis-containing vacuoles. J Bacteriol. 2009;191:278–286. doi: 10.1128/JB.01031-08. This study showed that all four virB6 paralogs (virB6-1, -2, -3, and -4) that are 3- to 10-fold larger than A. tumefaciens virB6 are cotranscribed in THP-1 human leukemia and ISE6 tick cell cultures. VirB9 interacts with VirB6-1 and VirB6-2; VirB6-4 interacts with VirB6-1, VirB6-2, and VirB6-3; and VirB6-2 80-kDa fragment interacts with VirB6-3 and VirB6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson CM, Herron MJ, Felsheim RF, Schloeder BR, Grindle SM, Chavez AO, Kurtti TJ, Munderloh UG. Whole genome transcription profiling of Anaplasma phagocytophilum in human and tick host cells by tiling array analysis. BMC Genomics. 2008;9:364. doi: 10.1186/1471-2164-9-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin M, Zhang C, Gibson K, Rikihisa Y. Analysis of complete genome sequence of Neorickettsia risticii: causative agent of Potomac horse fever. Nucleic Acids Res. 2009;37:6076–6091. doi: 10.1093/nar/gkp642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge Y, Rikihisa Y. Surface-exposed proteins of Ehrlichia chaffeensis. Infect Immun. 2007;75:3833–3841. doi: 10.1128/IAI.00188-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franco IS, Shuman HA, Charpentier X. The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell Microbiol. 2009;11:1435–1443. doi: 10.1111/j.1462-5822.2009.01351.x. [DOI] [PubMed] [Google Scholar]
- 30.Scheidegger F, Ellner Y, Guye P, Rhomberg TA, Weber H, Augustin HG, Dehio C. Distinct activities of Bartonella henselae type IV secretion effector proteins modulate capillary-like sprout formation. Cell Microbiol. 2009;11:1088–1101. doi: 10.1111/j.1462-5822.2009.01313.x. [DOI] [PubMed] [Google Scholar]
- 31.Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 32.Kotob SI, Hausman SZ, Burns DL. Localization of the promoter for the ptl genes of Bordetella pertussis, which encode proteins essential for secretion of pertussis toxin. Infect Immun. 1995;63:3227–3230. doi: 10.1128/iai.63.8.3227-3230.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu J, den Dulk-Ras A, Hooykaas PJ, Glover JN. Agrobacterium tumefaciens VirC2 enhances T-DNA transfer and virulence through its C-terminal ribbon-helix-helix DNA-binding fold. Proc Natl Acad Sci U S A. 2009;106:9643–9648. doi: 10.1073/pnas.0812199106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol. 2006;9:207–217. doi: 10.1016/j.mib.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Ninio S, Roy CR. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 2007;15:372–380. doi: 10.1016/j.tim.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Schulein R, Guye P, Rhomberg TA, Schmid MC, Schroder G, Vergunst AC, Carena I, Dehio C. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc Natl Acad Sci U S A. 2005;102:856–861. doi: 10.1073/pnas.0406796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci U S A. 2005;102:826–831. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massung RF, Owens JH, Ross D, Reed KD, Petrovec M, Bjoersdorff A, Coughlin RT, Beltz GA, Murphy CI. Sequence analysis of the ank gene of granulocytic ehrlichiae. J Clin Microbiol. 2000;38:2917–2922. doi: 10.1128/jcm.38.8.2917-2922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Loewenich FD, Baumgarten BU, Schroppel K, Geissdorfer W, Rollinghoff M, Bogdan C. High diversity of ankA sequences of Anaplasma phagocytophilum among Ixodes ricinus ticks in Germany. J Clin Microbiol. 2003;41:5033–5040. doi: 10.1128/JCM.41.11.5033-5040.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.IJdo J, Carlson AC, Kennedy EL. Anaplasma phagocytophilum AnkA is tyrosine-phosphorylated at EPIYA motifs and recruits SHP-1 during early infection. Cell Microbiol. 2007;9:1284–1296. doi: 10.1111/j.1462-5822.2006.00871.x. The authors showed that a 190-kDa protein, AnkA, is increasingly tyrosine-phosphorylated infected host cell. Recombinant AnkA can be phosphorylated by Src in vitro and AnkA expressed in COS-7 cells is tyrosine phosphorylation by Src in EPIYA motifs and AnkA binds the host cell phosphatase SHP-1 during early infection. [DOI] [PubMed] [Google Scholar]
- 41.Park J, Kim KJ, Choi KS, Grab DJ, Dumler JS. Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins. Cell Microbiol. 2004;6:743–751. doi: 10.1111/j.1462-5822.2004.00400.x. [DOI] [PubMed] [Google Scholar]
- 42••.Garcia-Garcia JC, Rennoll-Bankert KE, Pelly S, Milstone AM, Dumler JS. Silencing of host cell CYBB gene expression by the nuclear effector AnkA of the intracellular pathogen Anaplasma phagocytophilum. Infect Immun. 2009;77:2385–2391. doi: 10.1128/IAI.00023-09. The authors reported that AnkA accumulates in nuclei of infected cells and interacts with transcriptional regulatory regions of the CYBB locus. AnkA binds to regions with high AT content. Histone H3 acetylation decreased at the CYBB locus during A. phagocytophilum infection. Transcription of CYBB and other defense genes is significantly decreased in AnkA-transfected HL-60 cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Felipe KS, Pampou S, Jovanovic OS, Pericone CD, Ye SF, Kalachikov S, Shuman HA. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol. 2005;187:7716–7726. doi: 10.1128/JB.187.22.7716-7726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho NH, Kim HR, Lee JH, Kim SY, Kim J, Cha S, Darby AC, Fuxelius HH, Yin J, Kim JH, et al. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc Natl Acad Sci U S A. 2007;104:7981–7986. doi: 10.1073/pnas.0611553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iturbe-Ormaetxe I, Burke GR, Riegler M, O’Neill SL. Distribution, expression, and motif variability of ankyrin domain genes in Wolbachia pipientis. J Bacteriol. 2005;187:5136–5145. doi: 10.1128/JB.187.15.5136-5145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker T, Klasson L, Sebaihia M, Sanders MJ, Thomson NR, Parkhill J, Sinkins SP. Ankyrin repeat domain-encoding genes in the wPip strain of Wolbachia from the Culex pipiens group. BMC Biol. 2007;5:39. doi: 10.1186/1741-7007-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, Podowski RM, Naslund AK, Eriksson AS, Winkler HH, Kurland CG. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 49••.Pan X, Luhrmann A, Satoh A, Laskowski-Arce MA, Roy CR. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science. 2008;320:1651–1654. doi: 10.1126/science.1158160. Authors showed Legionella pneumophila AnkX protein is secreted by T4S system and prevents microtubule-dependent vesicular transport to interfere with fusion of the L. pneumophila-containing vacuole with late endosomes after infection of macrophages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu B, Nethery KA, Kuriakose JA, Wakeel A, Zhang X, McBride JW. Nuclear translocated Ehrlichia chaffeensis ankyrin protein interacts with a specific adenine-rich motif of host promoter and intronic Alu elements. Infect Immun. 2009;77:4243, 4255. doi: 10.1128/IAI.00376-09. This study reported that a 200-kDa AnkA of E. chaffeensis is detected in the nuclei of Ehrlichia-infected THP-1 cells. AnkA interacts host promoter and intronic Alu-Sx elements; a specific adenine-rich (mid A-stretch) motif. Genes (n=456) with promoter Alu elements primarily related to transcription, apoptosis, ATPase activity and structural proteins associated with the nucleus and membrane-bound organelles are the targets of p200. [DOI] [PMC free article] [PubMed] [Google Scholar]


