Abstract
Background:
Preclinical studies in non-small cell lung cancer (NSCLC) suggest the interaction of PTEN and PI3K affects sensitivity to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs). We investigated outcomes in relation to PTEN, PIK3CA and EGFR gene copy number, and chromosome 7 (CEN7) polysomy in NSCLC patients treated with gefitinib.
Methods:
Fluorescent in situ hybridisation analyses of PTEN, PIK3CA, EGFR and CEN7 were performed on tumour specimens from patients treated on the expanded access gefitinib trial. Progression-free survival (PFS) and overall survival (OS) were correlated with outcomes in all patients and EGFR wild-type patients.
Results:
Progression-free survival (hazard ratio=2.54, P<0.001) and OS (hazard ratio=4.04, P<0.001) were significantly shorter in patients whose tumours had all of the following molecular patterns: CEN7 <4 copies per cell, PTEN loss (<2 copies in at least 20% of cells), and PIK3CA gain (>2 copies in at least 40% of cells) both in all and EGFR wild-type only patients.
Conclusion:
The combination of low CEN7 copy number, PTEN loss, and PI3KCA gain may be useful for identifying NSCLC patients unlikely to benefit from treatment with EGFR (TKIs), specifically in wild-type EGFR cases.
Keywords: EGFR, PTEN, PI3KCA, lung cancer, gefitinib
Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) have provided a novel way to treat advanced non-small cell lung cancer (NSCLC; Shepherd et al, 2005; Thatcher Lancet et al, 2005). Non-small cell lung cancers containing activating EGFR mutations in exons 19 and 21 are associated with significantly higher response rates and superior progression-free survival (PFS) in patients treated with gefitinib compared with first-line platinum doublet regimens and to second-line single-agent docetaxel (Mok et al, 2009; Douillard et al, 2010; Maemondo et al, 2010; Mitsudomi et al, 2010). Though EGFR TKIs produce their most pronounced effects in patients with EGFR mutations, there is evidence that patients with wild-type EGFR also benefit from EGFR TKI treatment with significantly longer survival being observed with maintenance erlotinib in patients who had wild-type tumours (Cappuzzo et al, 2010). Similarly, there was a trend for longer survival in patients with wild-type tumours treated with erlotinib as second- or third-line treatment (Zhu et al, 2008).
The Identification of additional molecular markers predictive of clinical benefit with EGFR TKIs in wild-type tumours would have therapeutic and economic implications for NSCLC patients. In addition, molecular markers may identify rationale strategies for combining other targeted agents with EGFR TKIs.
PI3K and PTEN are key downstream components of the EGFR pathway and have significant roles in cell survival, proliferation, and growth (Cantley, 2002; Lou et al, 2003; Bianco et al, 2003a; Engelman, 2009). The expression of these enzymes has also been related to EGFR TKI resistance in preclinical models (Janmaat et al, 2003; Engelman et al, 2005; Yamasaki et al, 2007). Earlier work suggested an association between patient outcomes and the combination of high chromosome 7 copy number (CEN7; a surrogate for EGFR copy number gain due to polysomy) and PTEN expression in advanced NSCLC patients treated with gefitinib (Buckingham et al, 2007). The objective of this study is to evaluate potential relationships between gene copy numbers for PIK3CA (the catalytic subunit of PI3K), PTEN and EGFR, and outcomes for NSCLC patients treated with gefitinib.
Materials and methods
This was a retrospective analysis of specimens from 82 patients in the gefitinib Expanded Access Trial, treated for >1 week, and collected as described previously. (Buckingham et al, 2007) Eligibility criteria and methods for establishing clinical data were also described. Chart review and this study's analyses were approved by the Rush University Medical Center Institutional Review Board. In general, patients were previously treated with at least one chemotherapy regimen or were considered ineligible for chemotherapy.
Based on previous work showing higher survival with PTEN expression by immunohistochemistry (IHC), an exploratory analysis was undertaken to study the influence of PTEN copy number loss and PIK3CA copy number gains in conjunction with EGFR alterations by fluorescent in situ hybridisation (FISH) on the same gefitinib cohort previously analysed. As no prospectively validated EGFR FISH analysis has been published to date, EGFR gene copy number status was assessed using multiple measures: the average number of EGFR copies per cell (EGFR/cell), the average number of chromosome 7 copies per cell (CEN7/cell), the average number of EGFR copies per chromosome 7 copies (EGFR/CEN7), the percentage of cells with more copies of EGFR than chromosome 7 (EGFR/CEP7 gain), the percentage of cells with >2 copies of EGFR (EGFR gain), and the percentage of cells with >2 copies of chromosome 7 (CEN7 gain). Fluorescence in situ hybridisation analyses were carried out as follows. The formalin-fixed paraffin-embedded lung tumour tissues and cell pellets were analysed with a dual colour probe set (Abbott Molecular Inc, Desplains, IL, USA) comprising SpectrumOrange LSI EGFR and SpectrumGreen CEP 7 and a four-colour probe set comprising a probe spanning PTEN (LSI PTEN) labelled with SpectrumRed, a probe spanning PIK3CA labelled with SpectrumGold, and probes containing peri-centromeric repeat sequences specific for chromosomes 3 (CEN 3) and 10 (CEN 10) (SpectrumGreen CEP 3 and SpectrumAqua CEP 10). Probes with CEP and LSI designations were obtained from Abbott Molecular Inc., and the four-colour probe set has been previously described. (Morrison et al, 2007) Fluorescence in situ hybridisation signals were enumerated in ⩾40 cells per specimen to obtain copy numbers for each locus, and 72 specimens yielded results for all six probes. Gene copy number gain for EGFR, PTEN, and PIK3CA was defined as >2 gene copies per cell. Conversely, gene loss was defined as <2 copies per cell. Epidermal growth factor receptor/CEN7 was deemed high if the ratio was >1. The response variables considered include, PFS and overall survival (OS). The status of EGFR mutation (exons 19 and 21) was obtained for 55 of the specimens (as described in Buckingham et al, 2007). Statistical analyses were carried out on the total population of patients with FISH analyses and repeated for the EGFR wild-type and mutant populations. Descriptive statistics were obtained and Fisher's exact test was used to measure the association among recurrence, survival, and categorised covariates. For purposes of tabular and time-to-even analyses, the biomarker measurements were divided into two classes (high/low) using optimally chosen marker specific thresholds in the absence of prior published cutoffs for PTEN and PI3KCA FISH testing. The differences in OS and PFS between the low and high biomarker groups were assessed by the log-rank test and Kaplan–Meier method was used to obtain estimates of OS and PFS curves. Predictors that were statistically significant or marginally significant in univariate analyses or were deemed to be clinically or biologically important were included as candidate covariates in multivariate Cox proportional hazards (PHs) regression models. Statistical analyses were performed using Version 9.2 of the SAS software, Version 7.0 of the JMP software (SAS Institute, Cary, NC, USA) and the statistical software R. All reported P-values are two sided and P-values between (0.05–0.10), (0.01–0.05), and (<0.01) are reported as marginally significant, significant, and strongly significant, respectively.
Results
The majority of patients included in this study were over the age of 60 (77%). In all, 54% were female, 85% had a smoking history, 69% had adenocarcinoma, 57% had Eastern Oncology Cooperative Group performance status of 0–1, and 83% had received previous chemotherapy as detailed in Table 1. The gain of PIK3CA is marked as >2 PIK3CA copies and high PIK3CA gain is measured as the percentage of cells with PIK3CA gain being ⩾40%. Similarly, a cutoff of 20% is used for PTEN loss (percentage of cells with <2 PTEN copies). The associations of PIK3CA gain, PTEN loss and the six EGFR-related parameters with PFS and OS were assessed by univariate analyses in Table 2. Chromosome 3 and CEN10 copy numbers did not provide useful associations with outcomes and were not included in the table. Of the six EGFR-related parameters examined, only CEN7/cell, cutoff=4.0, provided statistically significant classification with respect to OS (P=0.02). Epidermal growth factor receptor/CEN7 gain, cutoff=34% (P=0.02), and EGFR/CEN7, cutoff >1.0 (P=0.04), were the only EGFR-related parameters to provide statistically significant classification with respect to PFS. Individually, PIK3CA did not show a statistically significant relationship with the above endpoints, whereas high PTEN loss did correlate with worse OS (P=0.01).
Table 1. Patient characteristics.
| Characteristic | Number of patients (%) | Objective response (%) |
|---|---|---|
| Total | 82 (100) | 12 (15) |
| Age (years) | ||
| ⩾60 | 62 (77) | 8 (13) |
| <60 | 20 (23) | 4 (20) |
| Gender | ||
| Male | 37 (46) | 5 (14) |
| Female | 44 (54) | 7 (16) |
| Smoking status | ||
| Yes | 70 (85) | 5 (7) |
| Never smoked | 12 (15) | 7 (58) |
| Histopathological subtype | ||
| Adenocarcinoma | 56 (69) | 10 (18) |
| Other | 26 (32) | 2 (8) |
| Performance status | ||
| (0–1) | 46 (57) | 6 (13) |
| (2–4) | 34 (43) | 6 (17) |
| Previous chemotherapy | ||
| None | 14 (17) | 2 (14) |
| One | 39 (49) | 7 (18) |
| Two or more | 28 (34) | 3 (11) |
Table 2. Univariate analyses.
| Biomarkers | Patients (%) | Median PFS months | Hazard ratio | P-value (log-rank test) | Median survival months | Hazard ratio | P-value (log-rank test) | |
|---|---|---|---|---|---|---|---|---|
| EGFR/cell | ||||||||
| Low | <4.5 | 59 (73) | 2.53 | 7.27 | ||||
| High | ⩾4.5 | 22 (27) | 4.31 | 0.78 | 0.32 | 9.44 | 0.74 | 0.24 |
| CEN7/cell | ||||||||
| Low | <4 | 67 (83) | 2.86 | 6.9 | ||||
| High | ⩾4 | 14 (17) | 4.04 | 0.78 | 0.42 | 17.2 | 0.49 | 0.02 |
| EGFR/CEN7 | ||||||||
| Low | ⩽1 | 24 (27) | 2.04 | 5.95 | ||||
| High | >1 | 57 (73) | 4.04 | 0.54 | 0.02 | 8,78 | 0.66 | 0.11 |
| EGFR/CEN7 gain | ||||||||
| Low | <34% | 41 (51) | 2.47 | 6.61 | ||||
| High | ⩾34% | 40 (49) | 4.32 | 0.58 | 0.02 | 11.2 | 0.68 | 0.1 |
| EGFR- gain | ||||||||
| Low | <70% | 52 (64) | 2.53 | 7.27 | ||||
| High | ⩾70% | 29 (36) | 4.31 | 0.82 | 0.43 | 11.5 | 0.73 | 0.2 |
| CEN7 gain | ||||||||
| Low | <80% | 68 (84) | 3.22 | 7.27 | ||||
| high | ⩾80% | 13 (16) | 4.32 | 0.73 | 0.31 | 17.2 | 0.62 | 0.12 |
| PTEN loss | ||||||||
| Low | <20% | 15 (21) | 5.92 | 20 | ||||
| High | ⩾20% | 58 (79) | 3.25 | 1.47 | 0.19 | 6.9 | 2.13 | 0.01 |
| PI3KCA gain | ||||||||
| Low | <40% | 33 (46) | 3.52 | 9.6 | ||||
| High | ⩾40% | 39 (54) | 3.61 | 1.46 | 0.13 | 6.64 | 1.31 | 0.27 |
Abbreviations: CEN7=chromosome 7; EGFR=epidermal growth factor receptor; PFS=progression-free survival; PI3KCA=phosphatidylinositol 3-kinase catalytic subunit alpha; PTEN=phosphatase and tensin homologue.
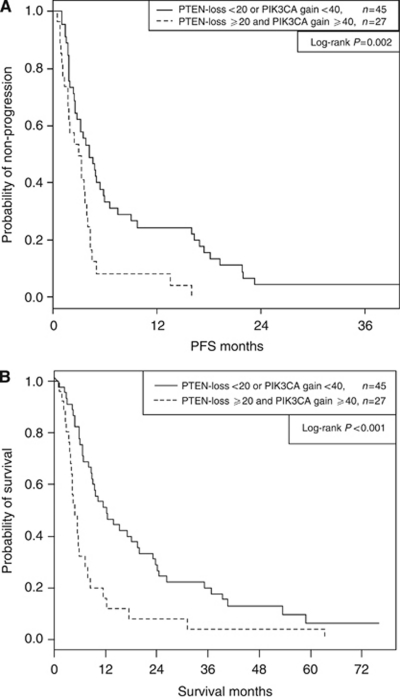
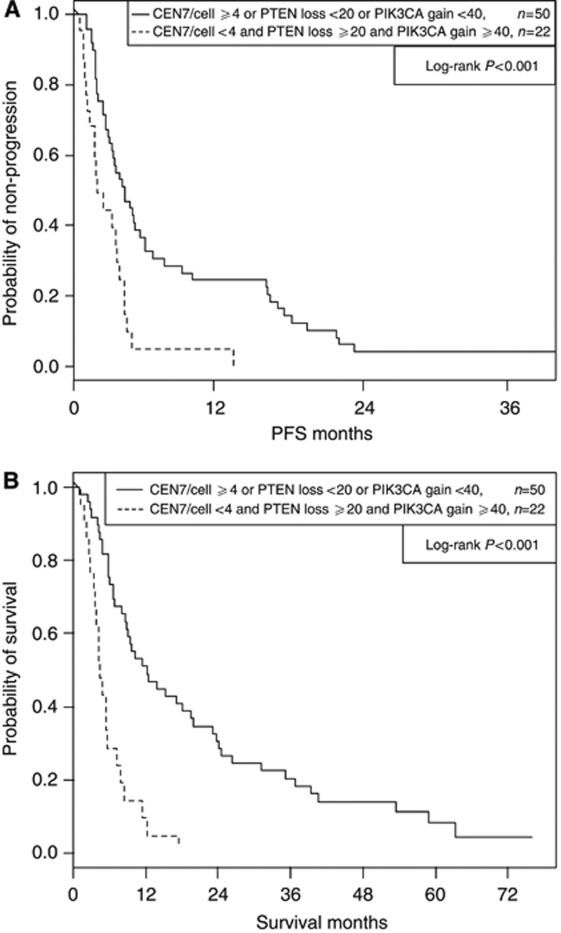
Thirty-eight percent of 72 patients had the double combination of high levels of PIK3CA gain (cutoff=40%) and high levels of PTEN loss (cutoff=20%) and had strongly significantly shorter PFS (P=0.002) and OS (P<0.001) than the remaining patients (3.02 vs 4.27 months median PFS, 4.93 vs 12.3 months median OS). Corresponding PFS and OS curve estimates are plotted in (Figure 1 A and B), respectively. Thirty-one percent of the 72 patients had the triple combination of low CEN7/cell (cutoff=4), high PIK3CA gain (cutoff=40%) and high PTEN loss (cutoff=20%), and experienced further shortened PFS (P<0.001) and OS (P<0.001) than the remaining patients (2.04 vs 4.21 months median PFS, 4.34 vs 12.3 months median OS). Corresponding PFS and OS curve estimates are plotted in (Figure 2A and B), respectively, for this three variate combination. Several EGFR-related parameters other than CEN7/cell provided highly significant associations in combination with PTEN loss and PIK3CA gain. Low EGFR gain (cutoff=75%), high PTEN loss, and high PIK3CA gain provided highly significant associations with PFS (P<0.001) and OS (P<0.001), and grouping low EGFR/CEN7 gain (cutoff=34%) with the same PTEN and PIK3CA parameters also provided high association with PFS (P=0.002) and OS (P<0.001). See Table 3 for double and triple covariate analyses.
Figure 1.
Progression free survival (PFS) and OS by PTEN loss and PIK3CA gain. (A) Progression free survival in all patients. (B) Overall survival in all patients.
Figure 2.
Progression-free survival(PFS) and OS by PTEN loss, PIK3CA gain and chromosome 7 (CEN7) polysomy. (A) Progression free survival in all patients. (B) Overall survival in all patients.
Table 3. Two and three markers analyses.
| Biomarkers | Patients | % | Median PFS months | Hazard ratio | P-value (log-rank test) | Median survival months | Hazard ratio | P-value (log-rank test) |
|---|---|---|---|---|---|---|---|---|
| High PTEN loss and high PIK3CA gain | ||||||||
| No | 45 | 62 | 4.27 | 12.3 | ||||
| Yes | 27 | 38 | 3.02 | 2.23 | 0.002 | 4.93 | 2.38 | <0.001 |
| Low CEN7/cell, high PTEN loss, and high PIK3CA gain | ||||||||
| No | 50 | 69 | 4.21 | 12.26 | ||||
| Yes | 22 | 31 | 2.04 | 2.54 | <0.001 | 4.34 | 4.04 | <0.001 |
| Low EGFR/CEN7 gain, high PTEN loss, and high PIK3CA gain | ||||||||
| No | 56 | 78 | 4.04 | 10.4 | ||||
| Yes | 16 | 22 | 2.04 | 2.62 | 0.002 | 4.93 | 3.37 | <0.001 |
| Low EGFR gain, high PTEN loss, and high PIK3CA gain | ||||||||
| No | 55 | 76 | 4.04 | 10.36 | ||||
| Yes | 17 | 24 | 2.04 | 2.52 | 0.003 | 4.93 | 3.35 | <0.001 |
| Wild Type (n=37) | ||||||||
| High PTEN loss and high PIK3CA gain | ||||||||
| No | 20 | 54 | 3.55 | 12.3 | ||||
| Yes | 17 | 46 | 3.02 | 1.84 | 0.09 | 5.49 | 2.41 | 0.012 |
| Low CEN7/cell, high PTEN loss, and high PIK3CA gain | ||||||||
| No | 24 | 65 | 3.35 | 12.5 | ||||
| Yes | 13 | 35 | 2.04 | 2.41 | 0.02 | 4.18 | 6.77 | <0.001 |
| Low EGFR/CEN7 gain, high PTEN loss, and high PIK3CA gain | ||||||||
| No | 27 | 73 | 3.35 | 10.6 | ||||
| Yes | 10 | 27 | 2.04 | 1.94 | 0.098 | 5.49 | 3.97 | 0.001 |
| Low EGFR gain, high PTEN loss, and high PIK3CA gain | ||||||||
| No | 26 | 70 | 3.25 | 10.64 | ||||
| Yes | 11 | 30 | 2.04 | 1.84 | 0.13 | 5.49 | 3.97 | 0.001 |
| EGFR Mutant (n=15) | ||||||||
| High PTEN loss, high PIK3CA gain yes/no | ||||||||
| No | 12 | 80 | 16.31 | 26.4 | ||||
| Yes | 3 | 20 | 4.54 | 5.01 | 0.02* | 5.57 | 8.14 | 0.002 |
| Low CEN7/cell, high PTEN loss, and high PIK3CA gain | ||||||||
| No | 13 | 87 | 16.31 | 26.4 | ||||
| Yes | 2 | 13 | 9.07 | 3.07 | 0.2* | 11.5 | 4.35* | 0.07 |
Abbreviations: CEN7=chromosome 7; EGFR=epidermal growth factor receptor; PFS=progression-free survival; PI3KCA=phosphatidylinositol 3-kinase catalytic subunit alpha; PTEN=phosphatase and tensin homologue.
*P-values, though statistically significant, are reflective of a small sample size.
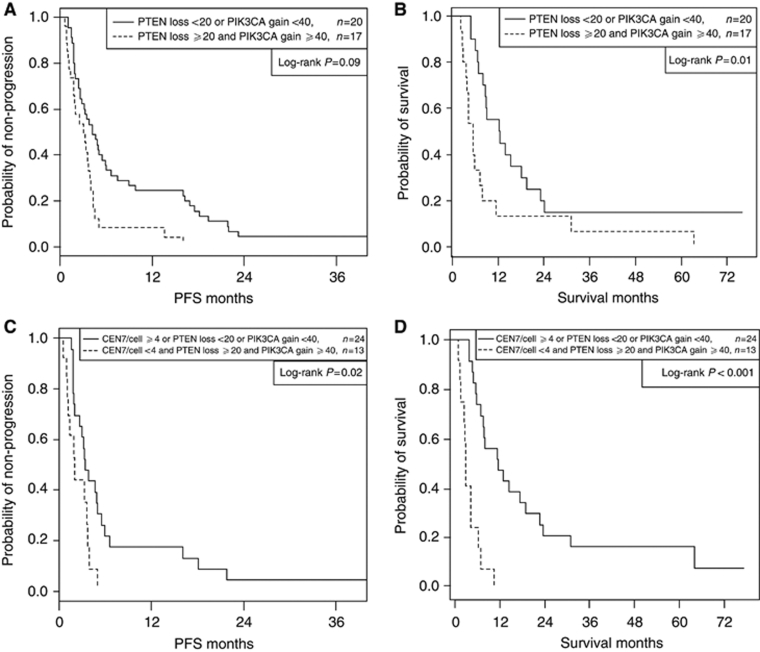
Seventeen of 55 patients tested had an EGFR activating gene mutation in exon 19 or exon 21. Patients with EGFR gene mutation had superior PFS compared with EGFR wild-type patients (13.6 vs 3.25 months median PFS, P=.003) but the association with OS was not significant (23.8 vs 7.98 months OS, P=0.18). In the 37 patients with wild-type EGFR and complete FISH data, patients with both high PIK3CA gain (cutoff=40%) and high PTEN loss (cutoff=20%) showed marginally significant shortening of PFS (3.02 vs 3.55 months median PFS, P=0.09) and significant shortening of OS (5.49 vs 12.3 months OS, P=0.01). Corresponding PFS and OS curve estimates are plotted in (Figure 3A and B), respectively. Wild-type patients with the triple combination of low CEN7/cell (cutoff=4), high PIK3CA gain (cutoff=40%), and high PTEN loss (cutoff=20%) showed significant shortening of PFS (2.04 vs 3.35 months, P=0.02) and strongly significant shortening of OS (4.18 vs 12.5 months, P<0.001). Corresponding PFS and OS curve estimates for patients with and without this triple combination are plotted in Figure 3C and D, respectively. Triple combinations using EGFR/CEN7 gain and EGFR gain were also significant and are presented in Table 3. Of note, ∼30% of wild-type patients presented with an unfavourable triple combination and had shortened PFS and OS. In the small group of 15 patients with activating EGFR mutations and complete FISH data, the combination of low CEN7/cell (cutoff=4), high PIK3CA gain (cutoff=40%), and high PTEN loss (cutoff=20%) showed marginally significant shortening of OS (P=0.07).
Figure 3.
(A) Progression-free survival in EGFR wild-type patients (two covariates). (B) Overall survival in EGFR wild-type patients (two covariates). (C) Progression-free survival in EGFR wild-type patients (three covariates). (D) Overall survival in EGFR wild-type patients (three covariates).
The results of multivariate Cox PHs regression analysis are listed in Table 4 for the subset of patients tested for EGFR mutations with full FISH data (N=52). Chromosome 7/cell (cutoff=4), PTEN loss (cutoff=20%) and PIK3CA gain (cutoff=40%) continued to have strongly significant association with OS after adjusting for gender, smoking status and histology, and PIK3CA gain was marginally associated with PFS. Epidermal growth factor receptor mutation did not show a significant association with OS, but had strongly significant association with PFS.
Table 4. Multivariate Cox proportional hazards regression of patients with EGFR mutation status.
| PFS HR | P-value | OS | P-value | |
|---|---|---|---|---|
| Female | 1.19 | 0.6 | 0.77 | 0.45 |
| Smoker | 1.48 | 0.35 | 1.32 | 0.53 |
| Adenocarcinoma | 1.49 | 0.35 | 1.06 | 0.9 |
| EGFR mutation | 0.32 | 0.006 | 0.74 | 0.47 |
| CEN7/cell <4 | 0.99 | 0.97 | 3.36 | 0.006 |
| PTEN loss ⩾20% | 1.62 | 0.26 | 4.06 | 0.002 |
| PIK3CA gain ⩾40% | 2 | 0.06* | 3.02 | 0.003 |
| n=52 |
Abbreviations: CEN7=chromosome 7; EGFR=epidermal growth factor receptor; HR=hazard ratio; PFS=progression-free survival; PI3KCA=phosphatidylinositol 3-kinase catalytic subunit alpha; PTEN=phosphatase and tensin homologue.
Discussion
Patients with EGFR mutations in exons 19 and 21 have been shown to have significantly higher response rates and improved PFS when treated with frontline gefitinib, and EGFR mutation has become an established criterion for selecting an EGFR TKI as first-line therapy in stage IV NSCLC patients. (Mok et al, 2009; Maemondo et al, 2010; Mitsudomi et al, 2010). In the absence of an EGFR-activating gene mutation, it seems likely that a functional EGFR pathway is necessary for EGFR TKIs to be effective. Much work has been done that correlates retrospectively applied EGFR gene copy number with outcomes in NSCLC patients treated with EGFR TKIs (Cappuzzo et al, 2005; Tsao et al, 2005; Zhu et al, 2008). A recent meta-analysis showed that high EGFR gene copy number was associated with longer survival in NSCLC patients treated with an EGFR TKI (Dahebreh et al, 2011). The most commonly applied criteria for FISH positivity is complex, has yet to be validated when prospectively applied to a clinical trial and seems to have less importance when comparing outcomes in patients treated with TKIs vs second-line chemotherapy (Douillard et al, 2010). In this study, an exploratory analysis was conducted measuring EGFR copy number in several different ways. Both high chromosome 7 copy number and a high ratio of EGFR to chromosome 7 copy numbers were associated with either prolonged OS or PFS.
Preclinical work suggests that the striking benefit of EGFR TKI therapies in mutation-positive tumours is related to massive apoptosis (Sordella et al, 2004). involving PI3K and PTEN. Recently, PTEN loss has been suggested as a potential mechanism of EGFR TKI resistance in NSCLC, which contain activating EGFR mutations (Sos et al, 2009). In addition, multivariate analysis in our initial studies showed that PTEN expression, detected by IHC, was significantly related to OS in gefitinib-treated patients (Buckingham et al, 2007). These considerations suggested PTEN might also be a determinant of the efficacy of EGFR TKIs in EGFR wild-type tumours and prompted us to evaluate PTEN and PIK3CA gene copy number in our gefitinib-treated patients. Although PTEN gene copy number alone was significantly related to OS, this was not the case for PIK3CA. However, the combination of gene copy data for PTEN and PIK3CA was strongly associated with both PFS and OS, and may be a more useful stratification than PTEN alone (Table 3).
The most powerful correlate of improved survival was the combination of EGFR or chromosome 7 copy number with PIK3CA and PTEN copy numbers. Although our results were obtained in a relatively small, single arm study, gefitinib-treated patients whose tumours contained low CEN7/cell (or low EGFR/CEN7 gain or low EGFR gain), high PTEN loss, and high PIK3CA gain had significantly shorter PFS and OS than other patients. The poor outcome with this molecular signature was seen both in the entire group and in the wild-type EGFR subset. The subset of patients with EGFR mutations and complete FISH data (n=15) was too small to support strong conclusions in this study. We are currently evaluating this set of markers in a larger group of patients treated with erlotinib. If our ongoing study shows similar results, this molecular profile, which was found in 30% of our patients, may identify a significant subset of NSCLC patients who derive minimal or no benefit from treatment with EGFR TKI.
In addition to the clinical implications of excluding patients who are unlikely to benefit from treatment with EGFR inhibitors, this selection strategy might have significant economic impact. Bradbury et al (2010) recently reported that the cost–benefit ratio for erlotinib was marginal. Subset analysis showed that the cost–benefit ratio was more favourable in never smokers and in patients with high EGFR gene copy numbers. The authors recommended increasing efforts to identify the most cost-effective way to use EGFR TKIs.
Our observations might also be useful in designing combination regimens targeting both the EGFR TKIs and downstream pathways. Preclinical studies have shown that reduced PTEN expression increases cancer cell survival and proliferation, and has been associated with resistance to EGFR inhibitors in NSCLC and colon cancers, and resistance to trastuzumab in breast cancer (Bianco et al, 2003b; Fujita et al, 2006; Berns et al, 2007; Sierra et al, 2010). It was somewhat surprising to find that a relatively high percentage of tumours (80%) had PTEN gene loss (defined as ⩾20% of cells with <2 copies of PTEN), and that relatively subtle PTEN loss was associated with significantly shorter PFS and OS. If additional studies yield similar results, relatively minor alterations in wild-type PTEN gene copy number might have prognostic and therapeutic implications for NSCLC patients in identifying a practical patient group likely to benefit from multi-targeted therapy.
Our observation that the combination of PTEN and PIK3CA gene copy number more strongly related to outcome than either marker alone is consistent with recent results reported in breast cancer patients treated with trastuzumab Berns et al (2007). observed higher rates of disease progression on trastuzumab in breast cancer patients with either PIK3CA mutations or with low PTEN expression. Further, they suggested that assessing both molecular markers might be required for optimal prediction of disease progression during the treatment with trastuzumab. These preliminary observations might be particularly pertinent xin defining the roles of PI3K inhibitors and mTOR inhibitors in NSCLC.
In summary, if our results with CEN7, EGFR, PIK3CA, and PTEN FISH analyses are confirmed in larger groups of patients, this molecular profile could have clinical and economic implications for patients being considered for EGFR TKI treatment. Similarly, if our ongoing PTEN and PIK3CA gene copy number study in erlotinib-treated patients shows results consistent with our observations in gefitinib-treated patients, evaluation of PTEN and PIK3CA gene copy numbers should be considered in single agent and combination trials testing PI3K and mTOR inhibitors in NSCLC patients.
Acknowledgments
FISH probes and analysis support were provided by Abbott Molecular Inc., Desplains, IL, USA.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM, Stemke-Hale K, Hauptmann M, Beijersbergen RL, Mills GB, van de Vijver MJ, Bernards R (2007) A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell 12: 395–402 [DOI] [PubMed] [Google Scholar]
- Bianco R, Shin I, Ritter CA, Yakes FM, Basso A, Rosen N, Tsurutani J, Dennis PA, Mills GB, Arteaga CL (2003) Loss of PTEN/MMAC1/TEP in EGF receptor-expressing tumor cells counteracts the antitumor action of EGFR tyrosine kinase inhibitors. Oncogene 22: 2812–2822 [DOI] [PubMed] [Google Scholar]
- Bianco R, Shin I, Ritter CA, Yakes FM, Basso A, Rosen N, Tsurutani J, Dennis PA, Mills GB, Arteaga ClL (2003b) Lossof PTEN/MMAC1/TEP in EGF receptor expressing tumor cells counteracts the antitumor effects of EGFR tyrosine kinase imhbitors. Oncogene 22: 2812–2822 [DOI] [PubMed] [Google Scholar]
- Bradbury PA, Tu D, Seymour L, Isogai PK, Zhu L, Ng R, Mittmann N, Tsao MS, Evans WK, Shepherd FA, Leighl NB (2010) Economic analysis: randomized placebo-controlled clinical trial of erlotinib in advanced non-small cell lung cancer. J Nat Cancer Inst 102: 298–306 [DOI] [PubMed] [Google Scholar]
- Buckingham LE, Coon JS, Morrison LE, Jacobson KK, Jewell SS, Kaiser KA, Mauer AM, Muzzafar T, Polowy C, Basu S, Gale M, Villaflor VM, Bonomi P (2007) The prognostic value of chromosome 7 polysomy in non-small cell lung cancer patients treated with gefitinib. J Thorac Oncol 2: 414–422 [DOI] [PubMed] [Google Scholar]
- Cantley LC (2002) The phosphoinositide 3-kinase pathway. Science 296: 1655–1657 [DOI] [PubMed] [Google Scholar]
- Cappuzzo F, Ciuleanu T, Stelmakh L, Cicenas S, Szczesna A, Juhasz E, Esteban E, Molinier O, Brugger W, Melezinek I, Klingelschmitt G, Klughammer B, Giaccone G (2010) Erlotinib as maintenance treatment in advanced non-small-cell lung cancer: a multicentre, randomised, placebo-controlled phase 3 study. Lancet Oncol 11: 521–529 [DOI] [PubMed] [Google Scholar]
- Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, Haney J, Witta S, Danenberg K, Domenichini I, Ludovini V, Magrini E, Gregorc V, Doglioni C, Sidoni A, Tonato M, Franklin WA, Crino L, Bunn Jr PA, Varella-Garcia M (2005) Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst 97: 643–655 [DOI] [PubMed] [Google Scholar]
- Dahebreh IJ, Linardou H, Kosimidis P, Bafaloukos D, Murray S. (2011) EGFR gene copy number as a predictive biomarker for patients receiving tyrosine kinase inhibitor treatment: a systematic review and meta-analysis in non-small cell lung cancer. Ann Oncol 22(3): 545–552 [DOI] [PubMed] [Google Scholar]
- Douillard JY, Shepherd FA, Hirsh V, Mok T, Socinski MA, Gervais R, Liao ML, Bischoff H, Reck M, Sellers MV, Watkins CL, Speake G, Armour AA, Kim ES (2010) Molecular predictors of outcome with gefitinib and docetaxel in previously treated non-small-cell lung cancer: data from the randomized phase III INTEREST trial. J Clin Oncol 28: 744–752 [DOI] [PubMed] [Google Scholar]
- Engelman JA (2009) Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer 9: 550–562 [DOI] [PubMed] [Google Scholar]
- Engelman JA, Janne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, Cichowski K, Johnson BE, Cantley LC (2005) ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc Natl Acad Sci USA 102: 3788–3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Doihara H, Kawasaki K, Takabatake D, Takahashi H, Washio K, Tsukuda K, Ogasawara Y, Shimizu N (2006) PTEN activity could be a predictive marker of trastuzumab efficacy in the treatment of ErbB2-overexpressing breast cancer. Br J Cancer 94: 247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmaat ML, Kruyt FA, Rodriguez JA, Giaccone G (2003) Response to epidermal growth factor receptor inhibitors in non-small cell lung cancer cells: limited antiproliferative effects and absence of apoptosis associated with persistent activity of extracellular signal-regulated kinase or Akt kinase pathways. Clin Cancer Res 9: 2316–2326 [PubMed] [Google Scholar]
- Lou J, Manning BD, Cantley LC (2003) Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell 4: 257. [DOI] [PubMed] [Google Scholar]
- Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, Fujita Y, Okinaga S, Hirano H, Yoshimori K, Harada T, Ogura T, Ando M, Miyazawa H, Tanaka T, Saijo Y, Hagiwara K, Morita S, Nukiwa T (2010) Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 362: 2380–2388 [DOI] [PubMed] [Google Scholar]
- Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, Asami K, Katakami N, Takada M, Yoshioka H, Shibata K, Kudoh S, Shimizu E, Saito H, Toyooka S, Nakagawa K, Fukuoka M (2010) Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 11: 121–128 [DOI] [PubMed] [Google Scholar]
- Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M (2009) Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 361: 947–957 [DOI] [PubMed] [Google Scholar]
- Morrison LE, Jewell SS, Usha L, Blondin BA, Rao RD, Tabesh B, Kemper M, Batus M, Coon J (2007) Effects of ERBB2 amplicon size and genomic alterations of chromosomes 1, 3, and 10 on patient response to trastuzumab in metastatic breast cancer. Genes Chromosomes Cancer 46(4): 397–405 [DOI] [PubMed] [Google Scholar]
- Sierra JR, Cepero V, Giordano S (2010) Molecular mechanisms of acquired resistance to tyrosine kinase targeted therapy. Mol Cancer 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordella R, Bell DW, Haber DA, Settleman J (2004) Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science 305: 1163–1167 [DOI] [PubMed] [Google Scholar]
- Shepherd FA, Pereira JR, Ciuleanu T, Tan EH, Hirsh V, Thongprasent S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R, van Koote M, Dediv M, Findlay B, Tu D, Johnston D, Bezjak A, Clark G, Santabarbara P, Seymour L (2005) Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 353: 123–132 [DOI] [PubMed] [Google Scholar]
- Sos ML, Koker M, Weir B, Heynck S, Rabinovsky R, Zander T, Seeger JN, Weiss J, Fischer F, Frommolt P, Michel K, Peifer M, Mermel C, Girard L, Peyton M, Gazdar AZ, Minna JD, Garraway LA, Kashkar H, Pao W, Meyerson M, Thomas RK. (2009) PTEN loss contributes to erlotinib resistance in egfr - mutant lung cancer by activation of akt and egfr. Cancer Res 69: 32256–32261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher N, Chang A, Parikh P, Pereira JR, Ciuleanu T, von Pawel J, Thongprasert S, Tan EH, Pemberton K, Archer V, Carrol K (2005) Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer). Lancet 366: 1527–1537 [DOI] [PubMed] [Google Scholar]
- Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, Lorimer I, Zhang T, Liu N, Daneshmand M, Marrano P, da Cunha Santos G, Lagarde A, Richardson F, Seymour L, Whitehead M, Ding K, Pater J, Shepherd FA (2005) Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med 353: 133–144 [DOI] [PubMed] [Google Scholar]
- Yamasaki F, Johansen MJ, Zhang D, Krishnamurthy S, Felix E, Bartholomeusz C, Aguilar RJ, Kurisu K, Mills GB, Hortobagyi GN, Ueno NT (2007) Acquired resistance to erlotinib in A-431 epidermoid cancer cells requires down-regulation of MMAC1/PTEN and up-regulation of phosphorylated Akt. Cancer Res 67: 5779–5788 [DOI] [PubMed] [Google Scholar]
- Zhu CQ, da Cunha Santos G, Ding K, Sakurada A, Cutz JC, Liu N, Zhang T, Marrano P, Whitehead M, Squire JA, Kamel-Reid S, Seymour L, Shepherd FA, Tsao MS, National Cancer Institute of Canada Clinical Trials Group Study BR. 21 (2008) Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol 26: 4268–4275 [DOI] [PubMed] [Google Scholar]