Abstract
Peptidylarginine deiminase (PAD) catalyzes the posttranslational citrullination of selected proteins in a calcium dependent manner. The PAD4 isoform has been implicated in multiple sclerosis, Rheumatoid arthritis, some types of cancer, and plays a role in gene regulation. However, the substrate selectivity of PAD4 is not well defined, nor is the impact of citrullination on many other pathways. Here, a high-density protein array is used as a primary screen to identify 40 previously unreported PAD4 substrates, 10 of which are selected and verified in a cell lysate-based secondary assay. One of the most prominent hits, human 40S ribosomal protein S2 (RPS2), is characterized in detail. PAD4 citrullinates the Arg-Gly repeat region of RPS2, which is also an established site for Arg methylation by protein arginine methyltransferase 3 (PRMT3). As in other systems, crosstalk is observed; citrullination and methylation modifications are found to be antagonistic to each other, suggesting a conserved posttranslational regulatory strategy. Both PAD4 and PRMT3 are found to co-sediment with the free 40S ribosomal subunit fraction from cell extracts. These findings are consistent with participation of citrullination in the regulation of RPS2 and ribosome assembly. This application of protein arrays to reveal new PAD4 substrates suggests a role for citrullination in a number of different cellular pathways.
Introduction
Mapping the impact of posttranslational modifications on cellular pathways is a daunting task. This challenge is even more difficult when modifications are small and difficult to characterize. One such example is the conversion of protein arginine residues to citrulline. Posttranslational citrullination is catalyzed by a family of enzymes called protein arginine deiminases (PADs). These enzymes convert peptidylarginine residues to peptidylcitrulline in a calcium dependent manner1, 2 and show mechanistic similarity to other enzymes in the pentein superfamily, including the nitric oxide regulating enzyme dimethylarginine dimethylaminohydrolase.3 There are five human PAD isoforms (PAD1-4 and PAD6),4–8 which share 50–55 % amino acid identity but differ in tissue distribution9. The PAD4 isoform has been suggested as a possible target for new therapeutic agents because of its role in several human diseases, including Rheumatoid arthritis (RA),10–12 multiple sclerosis (MS),13 colitis,14 and several types of cancer.15 Additionally, PAD4 is the only isoform known to translocate to the nucleus16 and has been shown to regulate gene expression via its interaction with p5317, 18 and its citrullination of p300,19 the tumor suppressor ING4,20 and histones H321 and H4.22
To further elucidate the biological functions of PAD4 and to identify other pathways putatively affected by citrullination, we sought to use protein arrays to identify previously unknown substrates. Protein arrays are of increasing utility23, 24 and there are many examples that identify new protein-protein interactions as well as other biochemical activities.25–28 One advantage of this approach is the ability to easily identify the positive hits in a defined array and to rapidly progress to validation studies. Also, protein arrays can facilitate the use of transmembrane proteins and low abundance proteins, avoiding the bias against such proteins that is inherent in some complimentary technologies.29
Herein, we describe the use of a protein array to identify a number of previously unknown PAD4 substrates. To maximize library size, we selected a commercially available, high-density, 38,016 spot, redundant array,25, 29 and performed batch wise citrullination by PAD4. Analysis for newly citrullinated spots identified 40 new putative substrates, 10 of which were verified by a secondary cell lysate-based assay. One prominent hit was the human ribosomal protein S2 (RPS2), which immediately was of interest because of its roles in cancer biology,18, 30–33 its importance to pre-40S export competence,34 its accessibility when incorporated into the 40S ribosome small subunit,35, 36 and also, because its Arg residues are known to be targeted for posttranslational methylation by protein arginine methyltransferase-3 (PRMT3).37–39 Antagonistic methylation and citrullination of Arg residues in histones and non-histone proteins is emerging as a conserved regulatory strategy for posttranslational modification of protein function,40 and was considered to be a likely possibility for RPS2. Therefore, the functional impact of citrullination and antagonistic arginine methylation on human RPS2 was explored in more depth.
Materials and methods
Antibodies and plasmids
The expression plasmids pGEX6p1-mouse PRMT3, pGEX6p1-mouse RPS2 and its deletion mutants RPS2ΔRG, RPS2ΔRGGF and RPS2ΔGAR were from Dr. Mark T. Bedford (University of Texas M.D. Anderson Cancer Center).39 pGEX6p1-human RPS2 was generated by PCR using an RPS2-containing plasmid purchased from American Type Culture Collection (ATCC, Manassas, VA) as the template. For maltose binding protein (MBP) fusion constructs, the corresponding GST-RPS2 fusion constructs were digested and the inserts were sub-cloned into the pMal-c2x vector for expression in Escherichia coli strain Rosetta (details below). The coding sequences for Flag-RPS2, PAD4 and PAD4-C645A were cloned into pcDNA-3.1(+) for protein expression in human embryonic kidney epithelial cells (HEK 293T). To generate the Flag-RPS2 (8RQ) mutant, all eight Arg residues in the RPS2 GAR motif were mutated to Gln by PCR-based mutagenesis. The coding sequence for all GST-PAD4 fusion proteins were cloned in pEGX6p1 (see below). The anti-PRMT3 and anti-RPS2 antibodies were from Dr. Mark T. Bedford. The anti-modified citrulline antibody was purchased from Millipore (Catalog number 17-347, Upstate, Billerica, MA), anti-MBP from New England Biolabs (Catalog number E8032S, Ipswich, MA), anti-PAD4 from Abcam (Catalog number ab38772, Cambridge, MA), anti-Flag antibody M2 and the anti-β-tubulin from Sigma-Aldrich (Catalog numbers F1804 and T8203, respectively, St. Louis, MO).
Cell culture and transfection
HEK 293T cells were purchased from ATCC and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10 % fetal bovine serum in a humidified 5 % CO2 incubator at 37 °C. This cell line is a highly transfectable derivative of the human embryonic kidney cell line 293 due to the insertion of the SV40 T-antigen gene. HEK 293T cells express low levels of endeogenous PAD4, which can be detected in nuclear extracts by Western blot assays. DNA transfections were performed using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA).
Screening protein arrays for citrullination
The protein arrays (imaGenes; Berlin, Germany) were originally derived from clones of a human brain cDNA library and contain 38,016 spots spread over two 22 cm × 22 cm PVDF membranes.25 Of these clones, 34,635 have a valid insert, 25,575 have homology to known human proteins, and 6,885 represent different genes. In general, the clones also include partial or frame-shifted sequences (imaGenes).41 The membranes were first blocked by 100 mL TBST (25 mM Tris-HCl, 137 mM NaCl, 2.7 mM KCl, 0.05 % Tween-20, pH 7.4) containing 5 % ECL blocking agent (GE Healthcare, Pittsburgh, PA ) for 1 h at room temperature. They were subsequently rinsed three times with TBST and incubated in 50 mL Screening Buffer (TBST containing 10 mM CaCl2, 5 mM DTT and 0.75 mg purified PAD4) at 4 °C overnight with constant shaking. These reaction conditions were empirically determined to maximize the resulting citrullination signal and likely represent an optimization of the stability of enzyme and substrates, despite a reduced enzymatic activity at low temperature. Citrullinated spots were visualized using an anti-citrulline (modified) detection kit (Millipore, Upstate) according to the manufacturer’s instructions. Arrays were developed by ECL plus Western blotting detection reagents (GE Healthcare) and scanned by a Storm 860 Phosphorimager (GE Electronics). A negative control screen was performed in parallel on a second set of membranes, but using buffers that omit the CaCl2 that is normally required to activate PAD4.
Preparation of recombinant proteins
Expression of GST-RPS2 was induced by 0.4 mM IPTG (isopropyl β-D-thiogalactopyranside) at 37 °C for 1 h in mid-log phase E. coli Rosetta cells. After induction, cells were harvested (from 500 mL LB) and resuspended in 20 mL PBS containing one complete, mini, EDTA-free protease inhibitor cocktail tablet (Roche Applied Science, Indianapolis, IN). Cells were lysed by sonication on ice and the cell lysate was centrifuged at 4 °C for 30 min at 25,000 × g. GST-RPS2 in the supernatant was bound to glutathione-sepharose 4 Fast Flow resin (GE Healthcare) followed by washing with PBS. GST-RPS2 was then cleaved on-column using PreScission Protease (GE Healthcare). The untagged RPS2 was collected in the flow-through fraction and stored at −80 °C.
Expression of MBP-RPS2 and each of its mutants was induced at 37 °C for 1 h in E. coli Rosetta cells. All MBP fusion proteins were batch-purified using Amylose Resin (New England Biolabs) according to the manufacturer’s instructions. GST-fused PRMT3, PAD4 and PAD4 truncation mutants were prepared as reported previously.39, 42, 43
In vitro and in vivo citrullination assay
For in vitro citrullination assays, purified PAD4 or GST-PAD4 was incubated with substrate proteins in Assay Buffer (100 mM Tris-HCl, pH 7.4, containing 5 mM DTT and 2 mM CaCl2) for 30 min at room temperature. The reactions were stopped by the addition of 5 × SDS-PAGE sample loading buffer. Citrulline was detected using an anti-citrulline (modified) detection kit (Upstate, Millipore).
For the in vivo assay, PAD4 and Flag-RPS2 plasmid were co-transfected into HEK 293T cells. At 36 h post-transfection, cells were treated with 5 μM calcium ionophore A23187 in Locke’s solution for 30 min at 37 °C. Cells were then harvested and lysed in 1 × Cell Lysis Buffer (Cell Signaling, Danvers, MA). Flag-RPS2 was then immunoprecipitated by anti-Flag M2 agarose affinity gel (Sigma Aldrich), eluted by TBS containing 200 μg/mL FLAG peptide and subjected to SDS-PAGE and Western-blot analysis as described above.
In vitro antagonism assay
Purified MBP-GAR was used as a substrate in the following assays. To citrullinate MBP-GAR, 0.1 mg/mL MBP-GAR was incubated with 0.17 μg/mL PAD4 in 40 μL assay buffer (100 mM Tris-HCl, pH 7.4, 50 mM NaCl, 5 mM DTT and 2 mM CaCl2) at room temperature for 1 h. To methylate MBP-GAR, 0.1 mg/mL MBP-GAR was incubated with 0.22 mg/mL GST-PRMT3 under the same conditions as above except that the assay buffer contained 20 μM AdoMet instead of DTT and CaCl2. For antagonism assays, MBP-GAR was 1) first citrullinated by PAD4 and then incubated with PRMT3; 2) first methylated by PRMT3 and then incubated with PAD4, or 3) incubated with PAD4 and PRMT3 at the same time. Reactions were stopped upon the addition of 5 × SDS-PAGE loading buffer. The citrulline and methylarginine content of MBP-GAR were analyzed by Western blots using anti-modified citrulline antibody and anti-mono- and anti-dimethylarginine antibodies, respectively.
Co-immunoprecipitation and GST pull-down assay
HEK 293T cells were transfected with pcDNA3.1(+)-pad4. After 36 h, cells were lysed in 1 × Cell Lysis Buffer (Cell Signaling) and incubated with anti-RPS2 antibody at 4 °C for 1 h. Normal rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA) was used as a control. Protein A/G plus-agarose (Santa Cruz Biotechnology) was added to the mixture and incubated at 4 °C for an additional 1 h with constant shaking. After four washes with TBS, the beads and any protein still bound to them were boiled in 2 × SDS PAGE loading buffer at 95 °C for 10 min and subjected to immunoblotting analysis using anti-RPS2 and anti-PAD4 antibodies. Reciprocally, Flag-PAD4 was expressed in HEK 293T cells, immunoprecipitated by anti-Flag M2 agarose affinity gel and eluted by the Flag peptide. Flag-PAD4 and co-immunoprecipitated RPS2 were visualized by their corresponding antibodies.
For the GST pull-down assay, purified GST, GST-PAD4-N1, GST-PAD4-N1N2 and GST-PAD4 were each bound to the glutathione-Sepharose 4 Fast Flow beads separately. After that, the bead-bound proteins were incubated with MBP or MBP-RPS2 at 4 °C for 1 h with constant shaking. After three washes with TBS, any proteins that remained on the beads were eluted with 40 mM glutathione in TBS and analyzed by SDS-PAGE and Western blot.
Polysome profiling
Ribosome profiles were performed as previously described.44 Briefly, HEK 293T cells from one 100 mm dish were used for each sucrose gradient. Before harvesting, cells were incubated in fresh complete DMEM medium containing 0.1 mg/mL cycloheximide at 37 °C for 30 min and then washed with ice-cold PBS in the presence of 0.1 mg/mL cycloheximide. Cell pellets were then lysed in 300 μL Polysome Lysis Buffer (0.5% Triton X-100, 10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1 mg/ml cycloheximide, 100U/ml RNasin and protease inhibitors) on ice for 10 min and centrifuged at 13,800 × g for 10 min at 4 °C. After that, the supernatant was layered onto 5–46 % sucrose gradients prepared in polysome lysis buffer and centrifuged at 209,700 × g for 3 h at 4 °C. Sucrose gradients were subsequently fractionated by upward displacement with 55 % (W/W) sucrose using a gradient fractionator connected to a UV monitor for continuous measurement of the absorbance at 254 nm. After that, eighteen 0.6 mL fractions were collected. The proteins in each fraction were precipitated with trichloroacetic acid (20 % final), twice washed with cold acetone, vacuum-dried and subjected to Western blotting.
Results
Protein array screening for novel PAD4 substrates
The protein array consists of 38,016 spots spread over two large PVDF membranes. One set of membranes were incubated with purified recombinant PAD4 in the presence of activating concentrations of CaCl2 and served as the variable. A second set of membranes were incubated with PAD4 in the absence of CaCl2 and served as the negative control. After incubation of both sets of membranes, each was treated with an anti-modified citrulline antibody detection kit and scanned for spots that reacted for citrulline content. Hits were identified by appearance of a duplicate spotting pattern within each block matching those defined by the manufacturer, and by their difference from the negative control (Fig. 1A and B). There were 123 duplicately-spotted hits that met these criteria, and the corresponding protein identities were obtained from imaGenes. Because of the redundant nature of the array, a total of 40 different proteins were identified from these 123 hits, some appearing more than once on the array (Table 1). Twenty-two of the 40 citrullinated proteins are ribosomal proteins. Thirteen of the remaining 18 non-ribosomal proteins are nuclear proteins or shuttle between the nucleus and cytoplasm.
Figure 1. Protein array screen for PAD4 substrates.

a) Image of citrullinated proteins on the array. Only a section of the large array is shown (X and Y axis coordinates are marked). Each 5 × 5 block of spots contains one reference ink spot (center) and twelve proteins spotted in duplicate patterns. Selected blocks containing positive hits are outlined and marked with the GenBank accession number corresponding to the citrullinated protein (see Table I). For example, P15880 corresponds to human RPS2. b) Selected blocks (bottom row) containing independent hits for citrullinated RPS2 are shown in detail. The white dot in the center is the reference spot. The black dots are citrullinated RPS2 present in various duplicate spotting patterns matching those described by the manufacturer. Blocks shown in the top row are the corresponding blocks from the negative control array. c) Secondary screen for selected hits using a cell-lysate based assay. Along with PAD4, 12 selected protein hits were co-expressed in HEK 293T cells with addition of a C-terminal Flag tag to each hit. The corresponding cell lysates were incubated with CaCl2 (5 mM) to activate PAD4. Each target protein was immunoprecipitated, separated by SDS-PAGE, transferred to a PVDF membrane, and probed by an anti-Flag antibody to assess expression (left panel) and by an anti-modified citrulline antibody to assess total citrulline content (right panel). The expected position of each target protein as predicted by molecular weight is marked by an asterisk (*).
Table 1.
Putative PAD4 Substrates Identified by the Primary Array Screen
| Substrates | No. of clonesa | GenBank accession No. | Subcellular locationb |
|---|---|---|---|
| Ribosomal proteins | |||
| 40S subunit components | |||
| ribosomal protein S2 (RPS2) | 41 | P15880 | N/C |
| ribosomal protein S8 (RPS8) | 13 | P62241 | N/C |
| ribosomal protein S10 (RPS10) | 3 | P46783 | N/C |
| ribosomal protein S13 (RPS13) | 3 | P62277 | N/C |
| ribosomal protein S14 (RPS14) | 2 | P62263 | N/C |
| ribosomal protein S15a (RPS15A) | 2 | P62244 | N/C |
| ribosomal protein S18 (RPS18) | 2 | P62269 | N/C |
| ribosomal protein S3A (RPS3A) | 1 | P61247 | N/C |
| 60S subunit components | |||
| ribosomal protein L7 (RPL7) | 8 | P18124 | N/C |
| ribosomal protein L18a (RPL18A) | 6 | Q02543 | N/C |
| ribosomal protein L15 (RPL15) | 4 | P61313 | N/C |
| ribosomal protein L10 (RPL10) | 3 | P27635 | N/C |
| ribosomal protein L18 (RPL18) | 3 | Q07020 | N/C |
| ribosomal protein L3 (RPL3) | 2 | P39023 | N/C |
| ribosomal protein L9 (RPL9) | 2 | P32969 | N/C |
| ribosomal protein L7a (RPL7A) | 1 | P62424 | N/C |
| ribosomal protein L14 (RPL14) | 1 | P50914 | N/C |
| ribosomal protein L17 (RPL17) | 1 | NM_001035006 | N/C |
| ribosomal protein L26-like 1 (RPL26L1) | 1 | Q9UNX3 | N/C |
| ribosomal protein L35 (RPL35) | 1 | P42766 | N/C |
| mitochondrial ribosomal proteins | |||
| mitochondrial ribosomal protein L27 (MRPL27) | 2 | Q9P0M9 | Mt. |
| mitochondrial ribosomal protein S18A (MRPS18A) | 1 | Q9NVS2 | Mt. |
| Non-ribosomal proteins | |||
| Putative RNA-binding protein 15 | 2 | Q96T37 | N |
| Probable rRNA-processing protein EBP2 | 1 | Q99848 | N |
| Heterogeneous nuclear ribonucleoprotein A1 | 1 | P09651 | N/C |
| Cold-inducible RNA-binding protein | 1 | Q14011 | N |
| RNA-binding protein 5 | 1 | P52756 | N |
| Nuclease sensitive element-binding protein1 | 1 | P67809 | N/C |
| Proteasome subunit beta type 4 | 1 | P28070 | N/C |
| Ufm-1-conjugating enzyme 1 | 1 | Q9Y3C8 | N/C |
| Protein arginine N-methyltransferase 1 | 1 | Q99873 | N |
| Inhibitor of growth protein 4 | 1 | Q9UNL4 | N |
| Ubiquitin-like protein FUBI | 1 | P35544 | N/C |
| Fasciculation and elongation protein zeta 1 | 1 | Q99689 | N/C/M |
| Nuclear mitotic apparatus protein 1 | 2 | Q14980 | N |
| 1-Phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma1 | 1 | P19174 | C/M |
| Spectrin beta chain, brain 2 | 1 | O15020 | C |
| Adenylyl cyclase-associated protein 1 (CAP1) | 1 | Q01518 | M |
| Cytohesin-2 | 1 | Q99418 | C/M |
| PDZ domain-containing protein GIPC1 | 1 | O14908 | C/M |
No. of clones indicates the number of times the protein was identified as a duplicately spotted hit on the redundant array.
N, Nucleus; C, Cytoplasm; Mt., Mitochondrion; M, Membrane.
To verify a portion of the primary screen’s results, 12 proteins (RPS2 along with 11 nuclear proteins) were selected and the coding sequence for each was cloned into a pFlag-CMV™-5a expression vector which incorporates a Flag epitope. The resulting plasmids were each co-transfected into HEK 293T cells along with an expression vector for PAD4. As a negative control, one protein that was not identified as a PAD4 substrate in the primary screen, lysine acetyltransferase-5 (KAT5), was selected to include in the secondary validation screen. (KAT5 is also known as TIP 60 (60 kDa Tat-interactive protein)). After transfection, cells were lysed and PAD4 was activated by the addition of CaCl2 to the lysates. Each targeted protein was then immunoprecipitated using an anti Flag antibody and probed for citrulline content using an anti-modified citrulline antibody. Eleven of the of the 12 selected proteins were solubly expressed in HEK 293T cells and were subsequently confirmed to be substrates of PAD4 in fresh cell lysates (Fig. 1C). Some additional citrullinated bands are observed and may represent degradation products (of lower apparent molecular weight) or proteins that co-immunoprecipitate with the target substrate (or bind nonspecifically to the resin) that are either PAD4 substrates, or nonspecifically cross react with the first or second antibody. One protein, nuclease-sensitive element-binding protein-1 (YBP) did not express well in our hands and so was not evaluated as a PAD4 substrate. Based on the ratio of the signal intensities derived from the Flag and modified citrulline epitopes, the ten citrullinated proteins were further categorized into three groups: high, medium and low total citrulline content (Table 2). RPS2, along with six nuclear proteins, showed high citrulline content and is predicted to be a good substrate of PAD4.
Table 2.
Relative Ranking of Total Citrulline Content in Selected Hits
| Substrate | Expression a | Citrullination b | Citrulline content c |
|---|---|---|---|
| Lysine acetyltransferase 5 (KAT5) – Negative control | ++ | - | No |
| Cold-inducible RNA-binding protein (CIRP) | ++ | +++ | high |
| Probable rRNA-processing protein (EBP2) | +++ | +++ | high |
| Fasciculation and elongation protein zeta 1 (FEZ ) | + | + | medium |
| Ubiquitin-like protein FUBI (Fau) | +++ | +++ | high |
| Heterogeneous nuclear ribonucleoprotein A1 (HNRPA1) | +++ | +++ | high |
| Inhibitor of growth protein 4 (ING4) | +++ | +++ | high |
| Protein arginine N-methyltransferase 1 (PRMT1) | +++ | +++ | high |
| Proteasome subunit beta type 4 (PSBT4) | ++ | + | low |
| ribosomal protein S2 (RPS2) | +++ | ++ | high |
| Ufm-1-conjugating enzyme 1 (UFC) | +++ | + | low |
| Nuclease sensitive element-binding protein 1 (YBP) | - | - | - |
Recombinant target proteins were expressed in HEK 293T cells and ranked by relative response to an anti-flag antibody.
Citrullination of target proteins was visualized using an anti modified citrulline antibody.
+++, strong signal; ++, medium signal; +, weak signal; -, no signal.
Citrulline content is based on the relative ratio of signal intensities for modified-citrulline and Flag epitopes.
The 40S RPS2 is citrullinated by PAD4
The most frequent hit in the redundant array, RPS2, was identified in 41 separate, duplicate spotting patterns (Table 1) and was verified as a good substrate of PAD4 in cell lysates (Table 2). Therefore, citrullination of RPS2 was investigated in more detail. Recombinant human RPS2 tagged with an N-terminal maltose binding protein (MBP) was purified and incubated with PAD4 in the presence or absence of CaCl2. Citrullination of RPS2 was only observed in the presence of CaCl2 (Fig. 2A). PAD4 was not active with CaCl2 concentration lower than 0.4 mM and reached its maximum activity at 0.8 mM CaCl2 (Figure 2B). The calcium concentration required for PAD4 to citrullinate RPS2 in vitro is considerably higher than most basal intracellular physiological calcium concentrations, which are typically near 100 nM.45 However, this result is very consistent with those reported for other bona fide PAD4 substrates including F-actin capping protein α-1 subunit.46 When visualized on an SDS-PAGE gel, citrullination of RPS2 induced a progressive upward shift in its apparent molecular mass (Figure 2a, lower panel). This observation is also seen with other PAD4 substrates and is likely due to the decreased ability of citrullinated proteins to bind SDS in their denatured state.47
Figure 2. PAD4 citrullinates RPS2 in a CaCl2-dependent manner.
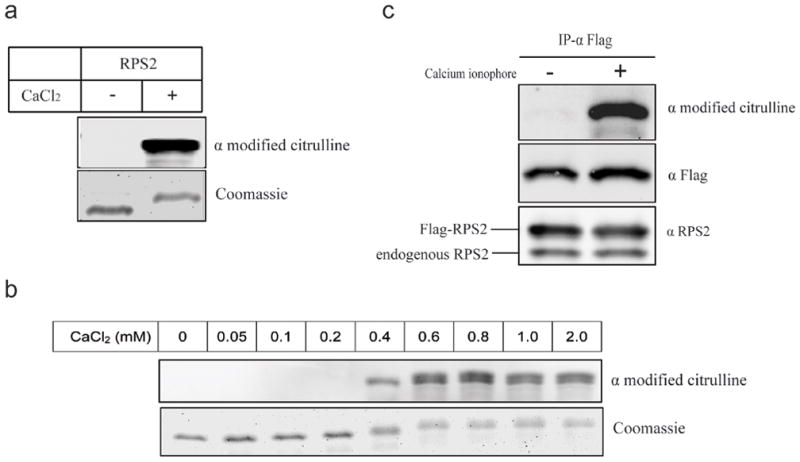
a) Purified human RPS2 was incubated with PAD4 in the absence (left lane) or presence (right lane) of CaCl2 (2 mM) for 30 min at 25 °C. Proteins were resolved by SDS-PAGE and citrullination of RPS2 was detected using an anti-modified citrulline antibody (upper panel). A duplicate gel stained with Coomassie Blue indicates loading amounts (lower panel). b) Citrullination of RPS2 by PAD4 is calcium dependent. Samples are prepared as in (a) except that the concentration of CaCl2 used is indicated for each lane. c) Flag-tagged RPS2 is citrullinated in vivo. HEK 293T cells were transiently co-transfected with Flag-RPS2 and PAD4 expression plasmids. After transfection, cells were treated with or without calcium ionophore A23187 as described in Methods. Immunoprecipitations (IP) were performed with anti-Flag M2 affinity gel. Citrullinated RPS2 was detected in cells treated with calcium ionophore (top panel). Equal loading of RPS2 was demonstrated by response to anti-Flag (middle panel) and anti-RPS2 (bottom panel) antibodies.
To test whether RPS2 is citrullinated in cells, a construct encoding Flag-tagged RPS2 was generated and co-transfected with an expression vector for PAD4 into HEK 293T cells. After transfection, cells were treated with the calcium ionophore A23187 to increase the intracellular calcium concentration. RPS2 was subsequently immunoprecipitated and its citrulline content determined as described above. In cells treated with A23187, the citrullination of RPS2 by PAD4 is observed (Fig. 2C, upper panel). Citrullination of RPS2 by endogenous PAD4 could not detected, regardless of treatment with the calcium ionophore, and may reflect the low expression levels of PAD4 and the detection limits of the antibodies used here.
Mapping of the citrullinated region in RPS2
RPS2 contains an extensive glycine arginine rich (GAR) motif at its N-terminus. This particular GAR motif is composed of several distinct repeating patterns: two Arg-Gly-Gly-Phe (RGGF) repeats and eight Arg-Gly (RG) repeats. To map the sites of citrullination in RPS2, we generated three deletion mutants of mouse RPS2 (mRPS2) (Figure 3a): MBP-mRPS2ΔRGGF (the two RGGF repeats are deleted), MBP-mRPS2ΔRG (the 8 RG repeats are deleted) and MBP-mRPS2ΔGAR (all the repeats described above are deleted). Mouse RPS2 has 98% amino acid sequence identity to human RPS2 and can also be citrullinated by PAD4 (Figure S1). Deletion of the RG repeats (ΔRG) was observed to drastically inhibit citrullination of RPS2 in vitro. This contrasts with deletion of the RGGF repeats, which did not inhibit citrullination, suggesting that the RG repeat region is the primary site of citrullination on RPS2 (Figure 3b).
Figure 3. Mapping the citrullination sites of RPS2.

a) Diagram of RPS2 deletion mutants. Deletion mutants were expressed as maltose binding protein (MBP) fusions. Mouse RPS2 (98% AA identity to human RPS2) was used in these experiments. b) Purified deletion mutant proteins were incubated with PAD4 in the presence of CaCl2 and separated by SDS-PAGE. Citrulline content was detected by western blot (upper panel). An anti-MBP antibody was used to indicate equal loading (lower panel).
Citrullination of RPS2 antagonizes its methylation
Notably, the same RG repeat region of RPS2 that is citrullinated by PAD4 is also known to be specifically methylated by protein arginine methyltransferase-3 (PRMT3), and this posttranslational methylation affects the balance of the ribosomal free 40S:60S subunit ratio, with possible implications for regulation of ribosome biogenesis.38, 39 Therefore, we tested whether posttranslational citrullination can antagonize methylation of the same RG repeats in RPS2. For these experiments, we used a truncated MBP-GAR motif that is more stable than full-length RPS2. Seen in Figure 4a (lane 8), the pre-citrullination of MBP-GAR inhibited its methylation by PRMT3. Use of a higher PAD4 concentration or a longer incubation time completely blocked MBP-GAR methylation. Reciprocally, pre-methylation of the GAR motif partly inhibited citrullination (Figure 4a, lane 9) but could not block it completely, even with an increased PRMT3 concentration or a longer incubation time.
Figure 4. Antagonism between posttranslational Arg methylation and citrullination of RPS2.
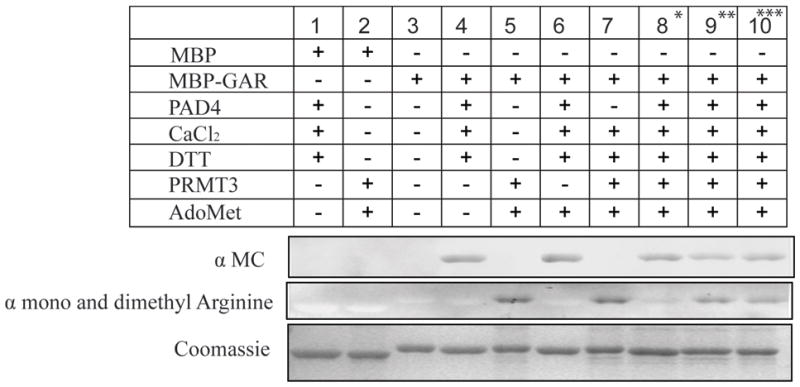
The purified MBP-GAR domain of RPS2 was treated under conditions indicated by the top grid. An anti-modified citrulline antibody was used to detect the citrullinated MBP-GAR (top panel), see Methods. A monoclonal antibody specific to mono and dimethylarginine was used to detect methylated MBP-GAR (middle panel). A duplicate gel was stained with Coomassie Blue to demonstrate equal loading (bottom panel). * In sample 8, MBP-GAR was first incubated with PAD4 (in the presence of CaCl2 and DTT) and subsequently with GST-PRMT3 (in the presence of AdoMet). ** In sample 9, MBP-GAR was first incubated with GST-PRMT3, and subsequently with PAD4 (with addition of cofactors, as above). *** In sample 10, MBP-GAR was treated with PAD4 and GST-PRMT3 at the same time (with addition of cofactors, as above).
One of several possible mechanisms leading to partial citrullination of pre-methylated sequences is that monomethyl (but not dimethyl) arginine residues might be targeted for citrullination. This would be consistent with previous in vivo21, 22 (but not in vitro40, 48) reports of demethyliminase activity of PAD4. Therefore, we monitored changes in the monomethylarginine content of MBP-GAR upon treatment by PAD4 by using an antibody that detects monomethyl arginine in a context independent manner.21 However, no changes in the monomethyl- or dimethylarginine content of pre-methylated MBP-GAR was observed upon treatment with calcium-activated PAD4 (Figure S2).
Protein-protein interaction of RPS2 and PAD4
To test for binding interactions between RPS2 and PAD4, PAD4 was transiently expressed in HEK 293T cells and used for the following experiments: PAD4 could be co-immunoprecipitated with endogenous RPS2 by using the anti-RPS2 antibody, but not by using the normal rabbit IgG (Figure 5a). Reciprocally, RPS2 could be co-immunoprecipitated with Flag-PAD4 using the anti-Flag antibody. Together, these co-immunoprecipitation experiments suggest that a stable binding interaction occurs between RPS2 and PAD4, even in the absence of activating concentrations of CaCl2.
Figure 5. Mapping the interacting domains of PAD4 and RPS2.
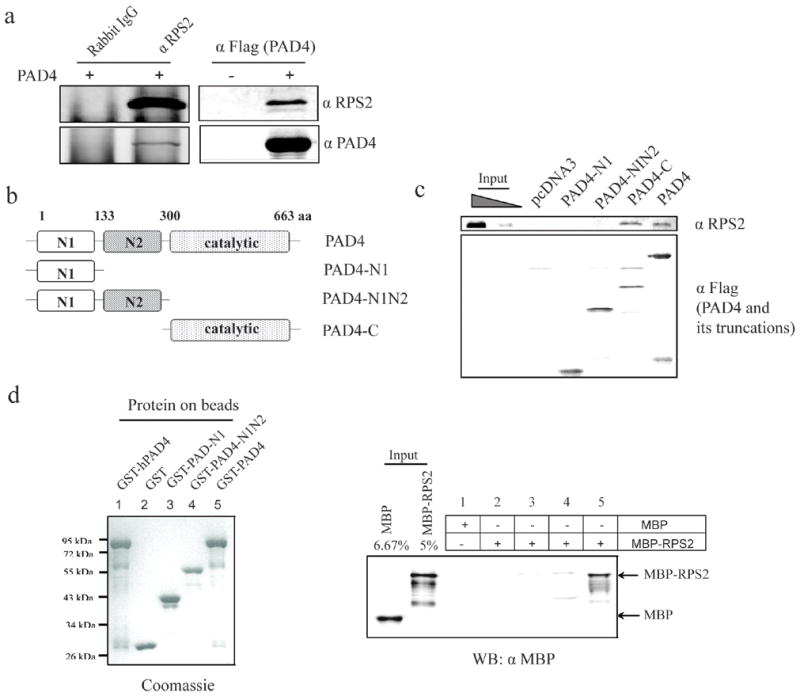
a) Recombinant PAD4 was expressed in HEK 293T cells and coimmunoprecipitated with endogenous RPS2 using an anti-RPS2 antibody (left panels). Reciprocally, endogenous RPS2 was coimmunoprecipitated by the M2 agarose beads from the Flag-PAD4-expressing HEK 293T cells (right panels). Each was detected by an anti-RPS2 antibody (top panel) and an anti-PAD4 antibody (bottom panel). b) Diagram of PAD4 and its deletion mutants. c) The catalytic domain of PAD4 interacts with RPS2. PAD4 and its deletion mutants were expressed with N-terminal flag tags in HEK 293T cells and immunoprecipitated. Any co-precipitated endogenous RPS2 was visualized using an anti RPS2 antibody (top panel). The loading amounts of PAD4 and its deletion mutants were indicated by Western blot using an anti-Flag antibody (bottom panel). C645A is a catalytically inactive point mutant of full-length PAD4. The left two lanes were loaded with 5 % and 1.25 % of the input, respectively. d) Direct binding interaction between PAD4 and RPS2. Right panel: Purified GST (lane 2), GST-PAD4-N1 (lane 3), GST-PAD4-N1N2 (lane 4) and GST-PAD4 (lane 1 and 5) were bound to glutathione-sepharose beads and incubated with purified MBP or MBP-RPS2. MBP-RPS2 pull downs were visualized using an anti-MBP antibody. The leftmost two lanes with MBP and MBP-RPS2 were loaded with 6.67 % and 5 % of the input, respectively. Left panel: A duplicate gel was stained by coomassie blue to indicate loading amounts.
The PAD4 structure can be divided into two N-terminal immunoglobulin-like (IgL) domains and one C-terminal catalytic domain.49 To map which domains interact with RPS2, Flag-tagged PAD4 truncation mutants were generated (PAD4-N1 (amino acids 1-133), PAD4-N1N2 (amino acids 1-299) and PAD4-C (amino acids 300-663)) (Figure 5b). Each PAD4 truncation mutant was expressed and immunoprecipitated from HEK 293T cell lysates and the presence of co-immunoprecipitated endogenous RPS2 was assayed using an anti RPS2 antibody. RPS2 only co-immunoprecipitated when the PAD4 catalytic domain was present. To further verify the direct interaction between PAD4 and RPS2, PAD4 and its truncation mutants were purified as GST fusion proteins and used for pull-down assays (Figure 5d). Unfortunately, the recombinant PAD4 catalytic domain fragment was not soluble when expressed in E. coli and could not be used directly. However, we observed that the immobilized full length PAD4, but not its N-terminal IgL domains, was able to pull down MBP-RPS2 (Figure 5d), consistent with direct binding of the C-terminal domain of PAD4 to RPS2.
RPS2, which is homologous to RPS5 in E. coli, is found on the surface of the small ribosomal subunit and is accessible for posttranslational modification.35, 36, 50, 51 The RPS2-modifying PRMT3 enzyme is known to bind free 40S ribosomal subunits.38, 39 Because PAD4 can bind RPS2 and because citrullination can inhibit methylation by PRMT3, we investigated whether PAD4 also associates with ribosomal particles. Lysates from HEK 293T cells co-expressing PAD4 and PRMT3 were subjected to sucrose gradient sedimentation. Fractions collected from the gradient elutions were analyzed by Western blot (Figure 6a). The majority of ectopically-expressed PAD4 was detected in fractions containing 40S small ribosomal subunits, 80S monosomes and polysomes. A trace amount of PAD4 was observed in the 60S large subunits fractions, and no PAD4 was observed in the low-density fractions. This clearly contrasts with the distribution of PRMT3, which was previously detected in low-density and free 40S subunits fractions.38, 39 RPS2 was observed in monosome (40S and 60S) and polysome fractions. Similar results were obtained when endogenous PAD4 and PRMT3 were probed in non-transfected HEK 293T cells (Figure 6b). Both PAD4 and PRMT3 cosedimented with RPS2 in the 40S subunit fractions, thereby further validating their interactions with RPS2.
Figure 6. PAD4 co-sediments with PRMT3 and 40S ribosomal subunits.
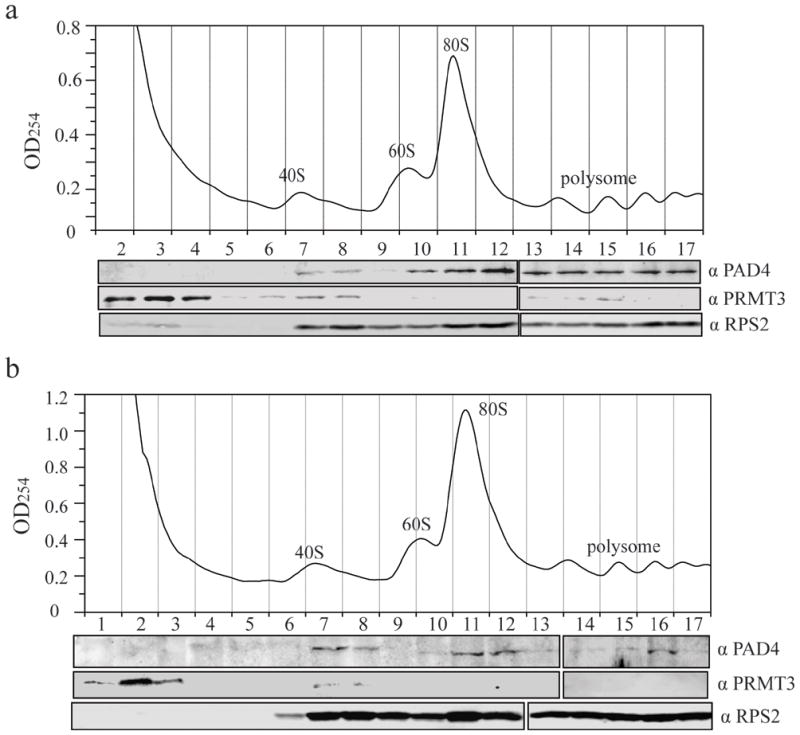
a) Recombinant PAD4 and PRMT3 were expressed in HEK 293T cells and cell lysates were subjected to a polysome profiling analysis. The profile was performed using a 5–46 % sucrose density gradient and recorded by continuous measurement of absorbance at 254 nm. The positions of 40S, 60S subunits, monosomes (80S) and polysomes are indicated. Fractions were collected, precipitated by tricholroacetic acid and used for Western blots with anti-PAD4 (top panel), anti-PRMT3 (middle panel) and anti-RPS2 (bottom panel) antibodies. b) Polysome profiling of endogenous PAD4, PRMT3 and RPS2 was performed as described in (a).
Discussion
The impact of protein citrullination on cellular processes is an area of increasing interest. Some of the most intensely studied areas include the contribution of this posttranslational modification to disease states. In multiple sclerosis, inappropriate citrullination of myelin basic protein adversely effects the stability of myelin sheaths and is thought to contribute to the etiology of the disease.45 In Rheumatoid arthritis, citrullinated proteins act as autoantigens to induce autoimmune responses in the synovium.45, 52 In cancer biology, there is a growing interest in the role of PAD4 in gene regulation with the discovery of the functional impact of citrullination on histones ING4 and p300.19–22 These, and a number of other proteins have been identified as substrates of PAD4 and other PAD isoforms.
However, to our knowledge, there have only been a limited number of proteomic approaches applied to identify novel PAD4 substrates, and many of these have been focused on particular pathophysiological states such as Rheumatoid arthritis46, 53–55 or glaucoma.56 Immunoscreens of human chondrocyte and synoviocyte cDNA expression libraries have also been performed to identify potentially citrullinated proteins.57, 58 However, these screens are limited in a number of ways including their focus on the synovium, the optic nerve, or diseased tissues. In an attempt to conduct a relatively unbiased screen, we used a high-density protein array to query a large number of proteins as potential PAD4 substrates.
A 38,016 spot array was treated batch wise with PAD4 and analyzed to identify any newly citrullinated proteins. This primary screen revealed 40 putative protein substrates for PAD4. These hits were initially categorized into two groups, the largest of which consists of 22 ribosomal proteins. Because of the relative abundance of ribosomal proteins and the derivation of the arrays from a cDNA library, ribosomal proteins are overrepresented by approximately 7-fold (imaGenes). This is consistent with the appearance of a larger proportion of ribosomal proteins in our hits, and the greater number of times that each was detected as a citrullinated product. The most prominent hit, identified 41 separate times, is the ribosomal protein RPS2, which is characterized in more detail (see below). As part of the ribosome’s assembly process, many of the ribosomal proteins spend some time in the nucleus (see below). Of the remaining 18 non-ribosomal protein hits, 13 are also reported to have nuclear localization. These results are consistent with this PAD4 isoform’s ability to translocate into the nucleus.
In order to validate the primary screen, a selection of the hits including RPS2 and 11 more nuclear proteins were subjected to further testing. Each of these 12 proteins were co-expressed with PAD4 and assayed for citrullination in the resulting cell lystates. One protein, YBP, did not express well and therefore could not be verified as a PAD4 substrate. However, each of the remaining 11 proteins was expressed solubly. One protein of many that were not identified as PAD4 substrates in the primary screen, KAT5, was also not a PAD4 substrate in the secondary cell lysate based assay. The remaining 10 proteins could all be citrullinated by PAD4, corroborating the primary screening results. Notably, each substrate was not citrullinated to the same degree, with two proteins, PSBT4 and UFC, showing only a weak citrullination signal. At this time, it is not clear whether the weaker intensity of these citrullination signals corresponds to low catalytic efficiency of citrullination by PAD4, or to a limited number of total citrullination sites on each target protein. However, 8 of the remaining proteins showed a moderate or strong citrullination signal. Three of these proteins, CIRP, EBP2 and HNRPA1, are RNA binding proteins. Two are involved in cancer biology and cell cycle regulation, Fau and ING4. One protein, PRMT1, functions in gene regulation as a histone methyltransferase and interestingly, also targets selected Arg residues for posttranslational modification. The remaining PAD4-substrate, RPS2, is a ribosomal protein and is discussed below. Taken together, these results represent a number of different pathways in which posttranslational citrullination may have a functional impact.
Because of its prominence in the primary screen and its importance in cancer biology,18, 30–33 RPS2 was selected for a more detailed characterization with respect to its interactions with PAD4 in vitro and in cells. In eukaryotes, the ribosome is composed of 40S and 60S ribonucleoprotein subunits, and RPS2 is a component of the 40S subunit. Construction of the ribosome is a complicated process wherein the component ribosomal proteins, together with pre-rRNA, assemble and become modified in the nucleolus to form a pre-ribosome that is subsequently exported to nucleoplasm for further maturation before its final destination in the cytoplasm.59 The shuttling of RPS2 through a number of subcellular compartments in consistent with its ability to be citrullinated by PAD4.
Ribosomal proteins carry numerous posttranslational modifications, including phosphorylation, methylation, and acetylation,60 but the crosstalk between these modifications and their impact on ribosome assembly and function is not well characterized. Here, we find a number of ribosomal proteins that can be citrullinated by PAD4. Specifically with RPS2, the same RG repeat regions that are targeted for calcium-dependent citrullination by PAD4 are also targeted for posttranslational methylation by protein arginine methyltransferase 3 (PRMT3).37,39 Since PRMT3 can functionally impact ribosome biogenesis,38, 39 we investigated whether there was any crosstalk between these posttranslational modifications. First, from cell extracts, PRMT3 and PAD4 are found to both cosediment with the 40S ribosomal subunit which contains RPS2. Second, when assayed in vitro, pre-citrullination of RPS2 by PAD4 blocks methylation by PRMT3. In turn, pre-methylation of RPS2 by PRMT3 inhibits citrullination by PAD4, suggesting that these two posttranslational Arg modifications are antagonistic to each other. In general terms, this finding is consistent with other examples of antagonistic Arg methylation and citrullination observed with both peptide and protein substrates, and suggests a conserved regulatory strategy. With regards to this specific system, citrullination of a different protein, nucleophosmin, has also been demonstrated and has a predicted functional impact on ribosome biogenesis.61 These results suggest a multitargeted impact of citrullination on ribosome assembly. Because PRMT3 is predominately located in the cytoplasm62, not the nucleus, the competition between these posttranslational modifications may also have a temporal component. Previous work has shown that RPS2 is methylated in the cytoplasm after assembly of the 40S subunit. Therefore, the timing and cellular conditions conducive for citrullination of RPS2 and the impact of citrullination on ribosome biogenesis are important topics to be addressed in future studies.
In summary, to our knowledge, this is the first use of a protein array to indentify previously unknown substrates of PAD4. Citrullination of 40 protein substrates that are involved in a number of different pathways were identified by array screening and 10 were confirmed by a secondary cell lysate-based screen. The most prominent hit, RPS2, is citrullinated in a calcium-dependent manner at its N-terminal RG repeat region and this modification antagonizes Arg methylation at the same region by PRMT3. Both of these Arg modifying enzymes co-sediment with RPS2 in the 40S ribosomal subunit. These findings contribute to the understanding of citrullination in ribosome biogenesis, of antagonistic Arg methylation, and also suggest a number of different pathways potentially impacted by posttranslational citrullination.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (GM69754 to W.F. and DK62248 to M.T.B.), the Robert A. Welch Foundation (F-1572 to W.F.) and a seed grant to W.F. from the Texas Institute for Drug and Diagnostic Development (Welch Foundation Grant # H-F-0032).
Footnotes
Electronic Supplementary Information (ESI) available: Two figures showing citrullination of mouse RPS2 and methylation status of deiminated RPS2 domains. See DOI: 10.1039/b000000x
References
- 1.Jones JE, Causey CP, Knuckley B, Slack-Noyes JL, Thompson PR. Curr Opin Drug Discov Devel. 2009;12:616–627. [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson PR, Fast W. ACS Chem Biol. 2006;1:433–441. doi: 10.1021/cb6002306. [DOI] [PubMed] [Google Scholar]
- 3.Linsky T, Fast W. Biochim Biophys Acta. 1804:1943–1953. doi: 10.1016/j.bbapap.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chavanas S, Mechin MC, Takahara H, Kawada A, Nachat R, Serre G, Simon M. Gene. 2004;330:19–27. doi: 10.1016/j.gene.2003.12.038. [DOI] [PubMed] [Google Scholar]
- 5.Guerrin M, Ishigami A, Mechin MC, Nachat R, Valmary S, Sebbag M, Simon M, Senshu T, Serre G. Biochem J. 2003;370:167–174. doi: 10.1042/BJ20020870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishigami A, Ohsawa T, Asaga H, Akiyama K, Kuramoto M, Maruyama N. Arch Biochem Biophys. 2002;407:25–31. doi: 10.1016/s0003-9861(02)00516-7. [DOI] [PubMed] [Google Scholar]
- 7.Kanno T, Kawada A, Yamanouchi J, Yosida-Noro C, Yoshiki A, Shiraiwa M, Kusakabe M, Manabe M, Tezuka T, Takahara H. J Invest Dermatol. 2000;115:813–823. doi: 10.1046/j.1523-1747.2000.00131.x. [DOI] [PubMed] [Google Scholar]
- 8.Nakashima K, Hagiwara T, Ishigami A, Nagata S, Asaga H, Kuramoto M, Senshu T, Yamada M. J Biol Chem. 1999;274:27786–27792. doi: 10.1074/jbc.274.39.27786. [DOI] [PubMed] [Google Scholar]
- 9.Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. Bioessays. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 10.Lundberg K, Nijenhuis S, Vossenaar ER, Palmblad K, van Venrooij WJ, Klareskog L, Zendman AJ, Harris HE. Arthritis Res Ther. 2005;7:R458–467. doi: 10.1186/ar1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schellekens GA, de Jong BA, van den Hoogen FH, van de Putte LB, van Venrooij WJ. J Clin Invest. 1998;101:273–281. doi: 10.1172/JCI1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willis VC, Gizinski AM, Banda NK, Causey CP, Knuckley B, Cordova KN, Luo Y, Levitt B, Glogowska M, Chandra P, Kulik L, Robinson WH, Arend WP, Thompson PR, Holers VM. J Immunol. 2011;186:4396–4404. doi: 10.4049/jimmunol.1001620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moscarello MA, Mastronardi FG, Wood DD. Neurochem Res. 2007;32:251–256. doi: 10.1007/s11064-006-9144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chumanevich AA, Causey CP, Knuckley BA, Jones JE, Poudyal D, Chumanevich AP, Davis T, Matesic LE, Thompson PR, Hofseth LJ. Am J Physiol Gastrointest Liver Physiol. 2011 doi: 10.1152/ajpgi.00435.2010. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang X, Han J, Pang L, Zhao Y, Yang Y, Shen Z. BMC Cancer. 2009;9:40. doi: 10.1186/1471-2407-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakashima K, Hagiwara T, Yamada M. J Biol Chem. 2002;277:49562–49568. doi: 10.1074/jbc.M208795200. [DOI] [PubMed] [Google Scholar]
- 17.Li P, Yao H, Zhang Z, Li M, Luo Y, Thompson PR, Gilmour DS, Wang Y. Mol Cell Biol. 2008;28:4745–4758. doi: 10.1128/MCB.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao H, Li P, Venters BJ, Zheng S, Thompson PR, Pugh BF, Wang Y. J Biol Chem. 2008;283:20060–20068. doi: 10.1074/jbc.M802940200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Proc Natl Acad Sci U S A. 2005;102:3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Q, Fast W. J Biol Chem. 2011 doi: 10.1074/jbc.M111.230961. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, Kouzarides T. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 23.Kricka LJ, Master SR, Joos TO, Fortina P. Ann Clin Biochem. 2006;43:457–467. doi: 10.1258/000456306778904731. [DOI] [PubMed] [Google Scholar]
- 24.Spurrier B, Honkanen P, Holway A, Kumamoto K, Terashima M, Takenoshita S, Wakabayashi G, Austin J, Nishizuka S. Biotechnol Adv. 2008;26:361–369. doi: 10.1016/j.biotechadv.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 25.de Graaf K, Hekerman P, Spelten O, Herrmann A, Packman LC, Bussow K, Muller-Newen G, Becker W. J Biol Chem. 2004;279:4612–4624. doi: 10.1074/jbc.M310794200. [DOI] [PubMed] [Google Scholar]
- 26.Grelle G, Kostka S, Otto A, Kersten B, Genser KF, Muller EC, Walter S, Boddrich A, Stelzl U, Hanig C, Volkmer-Engert R, Landgraf C, Alberti S, Hohfeld J, Strodicke M, Wanker EE. Mol Cell Proteomics. 2006;5:234–244. doi: 10.1074/mcp.M500198-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Lee J, Bedford MT. EMBO Rep. 2002;3:268–273. doi: 10.1093/embo-reports/kvf052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahlknecht U, Ottmann OG, Hoelzer D. J Biotechnol. 2001;88:89–94. doi: 10.1016/s0168-1656(01)00271-1. [DOI] [PubMed] [Google Scholar]
- 29.O’Connell DJ, Bauer MC, O’Brien J, Johnson WM, Divizio CA, O’Kane SL, Berggard T, Merino A, Akerfeldt KS, Linse S, Cahill DJ. Mol Cell Proteomics. 9:1118–1132. doi: 10.1074/mcp.M900324-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antoine M, Reimers K, Wirz W, Gressner AM, Muller R, Kiefer P. Biochem Biophys Res Commun. 2005;338:1248–1255. doi: 10.1016/j.bbrc.2005.10.079. [DOI] [PubMed] [Google Scholar]
- 31.Grade M, Hummon AB, Camps J, Emons G, Spitzner M, Gaedcke J, Hoermann P, Ebner R, Becker H, Difilippantonio MJ, Ghadimi BM, Beissbarth T, Caplen NJ, Ried T. Int J Cancer. 128:1069–1079. doi: 10.1002/ijc.25453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loging WT, Reisman D. Cancer Epidemiol Biomarkers Prev. 1999;8:1011–1016. [PubMed] [Google Scholar]
- 33.Wang M, Hu Y, Stearns ME. J Exp Clin Cancer Res. 2009;28:6. doi: 10.1186/1756-9966-28-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perreault A, Bellemer C, Bachand F. Nucleic Acids Res. 2008;36:6132–6142. doi: 10.1093/nar/gkn625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marion MJ, Marion C. FEBS Lett. 1988;232:281–285. doi: 10.1016/0014-5793(88)80753-1. [DOI] [PubMed] [Google Scholar]
- 36.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 37.Swiercz R, Cheng D, Kim D, Bedford MT. J Biol Chem. 2007;282:16917–16923. doi: 10.1074/jbc.M609778200. [DOI] [PubMed] [Google Scholar]
- 38.Bachand F, Silver PA. EMBO J. 2004;23:2641–2650. doi: 10.1038/sj.emboj.7600265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swiercz R, Person MD, Bedford MT. Biochem J. 2005;386:85–91. doi: 10.1042/BJ20041466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raijmakers R, Zendman AJ, Egberts WV, Vossenaar ER, Raats J, Soede-Huijbregts C, Rutjes FP, van Veelen PA, Drijfhout JW, Pruijn GJ. J Mol Biol. 2007;367:1118–1129. doi: 10.1016/j.jmb.2007.01.054. [DOI] [PubMed] [Google Scholar]
- 41.Bussow K, Nordhoff E, Lubbert C, Lehrach H, Walter G. Genomics. 2000;65:1–8. doi: 10.1006/geno.2000.6141. [DOI] [PubMed] [Google Scholar]
- 42.Kearney PL, Bhatia M, Jones NG, Yuan L, Glascock MC, Catchings KL, Yamada M, Thompson PR. Biochemistry. 2005;44:10570–10582. doi: 10.1021/bi050292m. [DOI] [PubMed] [Google Scholar]
- 43.Stone EM, Schaller TH, Bianchi H, Person MD, Fast W. Biochemistry. 2005;44:13744–13752. doi: 10.1021/bi051341y. [DOI] [PubMed] [Google Scholar]
- 44.Shin HS, Jang CY, Kim HD, Kim TS, Kim S, Kim J. Biochem Biophys Res Commun. 2009;385:273–278. doi: 10.1016/j.bbrc.2009.05.055. [DOI] [PubMed] [Google Scholar]
- 45.Gyorgy B, Toth E, Tarcsa E, Falus A, Buzas EI. Int J Biochem Cell Biol. 2006;38:1662–1677. doi: 10.1016/j.biocel.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 46.Matsuo K, Xiang Y, Nakamura H, Masuko K, Yudoh K, Noyori K, Nishioka K, Saito T, Kato T. Arthritis Res Ther. 2006;8:R175. doi: 10.1186/ar2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarcsa E, Marekov LN, Mei G, Melino G, Lee SC, Steinert PM. J Biol Chem. 1996;271:30709–30716. doi: 10.1074/jbc.271.48.30709. [DOI] [PubMed] [Google Scholar]
- 48.Hidaka Y, Hagiwara T, Yamada M. FEBS Lett. 2005;579:4088–4092. doi: 10.1016/j.febslet.2005.06.035. [DOI] [PubMed] [Google Scholar]
- 49.Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. Nat Struct Mol Biol. 2004;11:777–783. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 50.Chandramouli P, Topf M, Menetret JF, Eswar N, Cannone JJ, Gutell RR, Sali A, Akey CW. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 52.Anzilotti C, Pratesi F, Tommasi C, Migliorini P. Autoimmun Rev. 9:158–160. doi: 10.1016/j.autrev.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 53.Goeb V, Thomas-L’Otellier M, Daveau R, Charlionet R, Fardellone P, Le Loet X, Tron F, Gilbert D, Vittecoq O. Arthritis Res Ther. 2009;11:R38. doi: 10.1186/ar2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kinloch A, Tatzer V, Wait R, Peston D, Lundberg K, Donatien P, Moyes D, Taylor PC, Venables PJ. Arthritis Res Ther. 2005;7:R1421–1429. doi: 10.1186/ar1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Masson-Bessiere C, Sebbag M, Girbal-Neuhauser E, Nogueira L, Vincent C, Senshu T, Serre G. J Immunol. 2001;166:4177–4184. doi: 10.4049/jimmunol.166.6.4177. [DOI] [PubMed] [Google Scholar]
- 56.Bhattacharya SK, Crabb JS, Bonilha VL, Gu X, Takahara H, Crabb JW. Invest Ophthalmol Vis Sci. 2006;47:2508–2514. doi: 10.1167/iovs.05-1499. [DOI] [PubMed] [Google Scholar]
- 57.Okazaki Y, Suzuki A, Sawada T, Ohtake-Yamanaka M, Inoue T, Hasebe T, Yamada R, Yamamoto K. Biochem Biophys Res Commun. 2006;341:94–100. doi: 10.1016/j.bbrc.2005.12.160. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki A, Yamada R, Ohtake-Yamanaka M, Okazaki Y, Sawada T, Yamamoto K. Biochem Biophys Res Commun. 2005;333:418–426. doi: 10.1016/j.bbrc.2005.05.137. [DOI] [PubMed] [Google Scholar]
- 59.Dinman JD. J Biol Chem. 2009;284:11761–11765. doi: 10.1074/jbc.R800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee SW, Berger SJ, Martinovic S, Pasa-Tolic L, Anderson GA, Shen Y, Zhao R, Smith RD. Proc Natl Acad Sci U S A. 2002;99:5942–5947. doi: 10.1073/pnas.082119899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tanikawa C, Ueda K, Nakagawa H, Yoshida N, Nakamura Y, Matsuda K. Cancer Res. 2009;69:8761–8769. doi: 10.1158/0008-5472.CAN-09-2280. [DOI] [PubMed] [Google Scholar]
- 62.Tang J, Gary JD, Clarke S, Herschman HR. J Biol Chem. 1998;273:16935–16945. doi: 10.1074/jbc.273.27.16935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


