Abstract
Context:
The olfactory phenotype in patients with idiopathic hypogonadotropic hypogonadism (IHH) ranges from complete anosmia (Kallmann syndrome) to normosmia (normosmic IHH). However, the true prevalence of intermediary olfactory phenotypes (hyposmia) in IHH patients has not yet been assessed, and systematic correlations with anatomical and genetic abnormalities have not been reported.
Objective:
The objective of this study was to evaluate olfactory function in a large IHH cohort and correlate these findings with olfactory magnetic resonance imaging (MRI) and underlying genetic etiology.
Design and Setting:
We conducted a cross-sectional case-control study at an academic referral center.
Patients:
A total of 286 IHH patients (201 males and 85 females) and 2183 healthy historic controls (1011 males and 1172 females) were studied.
Main Outcome Measures:
We measured olfactory function using the University of Pennsylvania Smell Identification Test; in 208 subjects, the genetic etiology of IHH was ascertained by DNA sequencing; in a minor subset [39 of 286 subjects (13%)], olfactory structures were determined by MRI.
Results:
In the IHH cohort, 31.5% were anosmic, 33.6% were hyposmic, and 34.9% were normosmic. Most hyposmic (seven of 11) subjects with MRI data exhibited olfactory structure abnormalities. Of hyposmic subjects, 39.5% harbored mutations in genes involved in either GnRH neuronal migration or GnRH secretion.
Conclusions:
IHH subjects display a broad spectrum of olfactory function, with a significant hyposmic phenotype in nearly one third of subjects. The hyposmic subjects harbor mutations in genes affecting GnRH neuronal migration and its secretion, suggesting a pathophysiological overlap between Kallmann syndrome and normosmic IHH. Accurate olfactory phenotyping in IHH subjects will inform the pathophysiology of this condition and guide genetic testing.
Pulsatile secretion of GnRH from the hypothalamus initiates the release of the two pituitary gonadotropins, LH and FSH. Idiopathic hypogonadotropic hypogonadism (IHH) is a rare but prismatic human disease that is characterized by frank hypogonadism with low or inappropriately normal gonadotropin levels in the absence of any anatomic abnormality of their hypothalamo-pituitary-gonadal axis. The pathophysiological defect in these patients is an isolated defect in GnRH secretion/action, and the hypothalamo-pituitary-gonadal axis can be restored in most cases by a physiological regimen of exogenous GnRH (1).
Kallmann syndrome (KS) refers to the congenital form of IHH that is characteristically associated with anosmia (absence of sense of smell) (2). This unique pathophysiological association between IHH and impaired olfaction results from a defect in the shared developmental origins of both the GnRH and olfactory neurons (3). Both neurons originate from a hitherto undefined stem cell population within the embryonic olfactory placode and upon fate specification, the resulting GnRH neurons migrate on a structural scaffold provided by the olfactory axons before entering the central nervous system through the cribriform plate. Upon entering the central nervous system, while the olfactory axons synapse in the olfactory bulb, the GnRH neurons migrate further to reach the mediobasal hypothalamus where they form a functional network to initiate pulsatile GnRH secretion.
Developmental abnormalities in this migratory journey like those demonstrated with the deletion of the KAL1 gene result in KS (4, 5). Recently, the fibroblast growth factor (FGF) signaling pathway genes (FGF8 and FGFR1) (6–8), PROK2 signaling pathway genes (PROK2 and PROKR2) (9–13), and CHD7 (14) have joined the KAL1 gene as genetic pathways that have an identical neurodevelopmental function and cause KS in humans. In contrast, IHH also occurs in subjects with a normal sense of smell [normosmic IHH (nIHH)] wherein their GnRH deficiency is secondary to impaired function of genes that govern the neuroendocrine function of GnRH neurons (15–20). Intriguingly, mutations in some genes (e.g. FGF8/FGFR1/PROK2/PROKR2/CHD7) have an overlapping function and can be seen in both KS and nIHH subjects (8, 11, 14, 21).
Olfactory function is a unique prism that can be used to understand the pathophysiological basis of both KS and nIHH. Previously, quantitative olfactory function testing and/or detailed radiological evaluation of the olfactory bulb/tract have been used to assess the olfactory phenotypic spectrum in IHH subjects (22–27). Some studies have also specifically examined the olfactory phenotypes in genetically well-characterized subjects (13, 22, 28, 29). In some of these studies, olfactory function was found to be binary: either anosmic or normosmic, suggesting an “all-or-none” phenomenon (23, 24). In other studies, a broader spectrum including an intermediary hyposmic phenotype (reduced but not complete absence sense of smell) was recognized (6, 22, 25, 27, 29). However, the true prevalence of the intermediary hyposmic phenotype remains unknown. Moreover, many of the above studies involved small cohorts of IHH subjects, and their full genetic spectrum was not analyzed. Hence, by accurate and quantifiable olfactory phenotyping of a large cohort of IHH subjects and by relating olfactory function with anatomic studies of the olfactory system by magnetic resonance imaging (MRI) and with the underlying genetic etiologies, the pathophysiological role of the genes/pathways that underlie GnRH neuronal ontogeny can be ascertained. Therefore, by using a standardized and well-validated 40-item University of Pennsylvania Smell Identification Test (UPSIT) (30), this study determined: 1) the spectrum of olfactory function in a large cohort of IHH subjects; 2) the correlation between these abnormalities with high resolution MRI in a subset; and 3) the genetic etiology across the full spectrum of olfactory function.
Patients and Methods
Patient population
The IHH population was comprised of 286 consented subjects (201 males and 85 females; age range, 18–54 yr) studied at the Reproductive Endocrine Unit of Massachusetts General Hospital (Boston, MA). Criteria for diagnosis of IHH included: 1) current age of at least 18 yr; 2) clinical signs or symptoms of hypogonadism; 3) in men, serum testosterone levels below 100 ng/dl; and in women, primary amenorrhea and estradiol levels below 20 pg/ml in the presence of low or normal gonadotropins; 4) otherwise normal biochemical tests of anterior pituitary function and ferritin concentrations; and 5) normal imaging (MRI or computed tomography scan) of the hypothalamic and pituitary area. Patients were compared with an age- and gender-matched control population (2183 subjects; 1011 males and 1172 females) comprised of healthy subjects who had previously been administered the 40-item UPSIT test at the Smell and Taste Center of the University of Pennsylvania (30). Data from this population forms the basis of the UPSIT normative ranges that are used in the assessment and classification of olfactory function as described in the UPSIT manual (31).
Olfactory function assessment
Before quantitative smell test evaluation, all IHH subjects were asked to self-report their own olfactory function as good, reduced, or no sense of smell. Subjects were asked to refrain from performing the test if they had an acute nasal infection. The olfactory function of both the IHH cohort and the age- and gender-matched controls was then evaluated using the 40-item UPSIT smell identification test. Subjects in the IHH cohort were administered the UPSIT between the years 1995 and 2010, whereas the control groups consisted of historic controls used to validate the UPSIT. The UPSIT is a microencapsulated odor “scratch and sniff” test that is extensively validated, easy to use, and a highly reproducible test of olfactory function that correlates highly with other olfactory tests including odor detection thresholds (30). Each subject's UPSIT score was then interpreted using the age- and sex-related normative classification system described in the UPSIT manual: anosmia (scores ≤18 for both sexes); hyposmia (scores of 19–33 for males and 19–34 for females); or normosmia (scores of 34–40 for males and 35–40 for females) (31). Because smoking history was not uniformly obtained in all subjects, both current and ex-smokers were included in the analysis. Olfactory function of the IHH subjects was compared with the age- and gender-matched cohort of normative subjects using raw UPSIT scores and percentiles. To control for cross-cultural variation of smell identification, every 40-item UPSIT was rescored using the 12-item cross-cultural test (30). When the 12- and 40-item scores were discordant, individual medical records were reexamined, and subjects with potential cultural bias were excluded before inclusion into the study. Of the 286 IHH subjects, a subset of 39 subjects had detailed MRI of their brain that allowed optimal evaluation of the olfactory system as described previously (32). The presence of at least one of the following abnormalities in the neuroradiology report was considered as abnormal olfactory structural development: 1) absent or hypoplastic olfactory bulbs; 2) absent or hypoplastic olfactory sulci; or 3) absent or hypoplastic olfactory tracts.
Genetic analysis
In 208 of the 286 participating IHH subjects, using genomic DNA, the exonic and proximal intronic (at least 15 bp from splice sites) DNA sequences of 12 genes implicated in the etiology of isolated GnRH deficiency were amplified by PCR and determined by direct sequencing as described previously (33). These genes are KAL1 (anosmin-1, MIM: 308700), GNRH1 (GnRH 1, MIM: 152760), GNRHR (GnRH receptor, MIM: 138850), KISS1 (KISS-1 metastin, MIM: 603286), KISS1R (KISS1 receptor, MIM: 604161), NELF (nasal embryonic LHRH factor, MIM: 608137), FGF8 (fibroblast growth factor 8, MIM: 600483), FGFR1 (FGF receptor 1, MIM: 136350), PROK2 (prokineticin 2, MIM: 607002), PROKR2 (prokineticin receptor 2, MIM: 607212), TAC3 (tachykinin 3, MIM: 162330), and TAC3R (tachykinin receptor 3, MIM: 162332). All sequence variations were found on both DNA strands and were confirmed in a separate PCR. The significance of each rare sequence variant (minor allele frequency <1% in control population) was ascertained either based on previously published in vitro functional studies (7, 8, 11, 17, 33–36) or, if no functional study existed for a specific gene (e.g. KAL1), in silico prediction methods (PolyPhen-2 and SIFT) were used to assess functionality as described previously (33). Only functionally detrimental variants (including splice variants) confirmed in either in vitro functional studies or in both in silico methods were included in the analysis. The genetic results of some of the patients in this study have been reported previously (33), whereas sequencing of the additional genes not reported previously were specifically performed for this study in these subjects.
Statistical analysis
A χ2 test was used to determine statistical significance of comparisons with P values < 0.05 considered to be significant.
Results
IHH subjects exhibit a bimodal spectrum of olfactory deficits including a previously underappreciated intermediary hyposmic phenotype
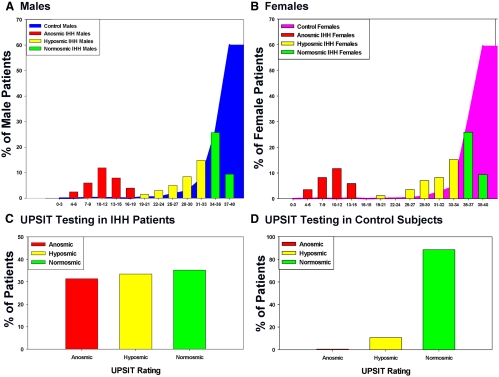
Both male and female IHH subjects showed a bimodal spectrum of distribution of their 40-item UPSIT scores, whereas the control population showed a unimodal score distribution, skewed heavily toward normosmia (Fig. 1, A and B). IHH patients were found to differ significantly in their olfactory function when compared with the control population [anosmia, 90 of 286 (31.5%) vs. 11 of 2183 (0.5%) subjects; hyposmia, 96 of 286 (33.6%) vs. 234 of 2183 (10.7%) subjects; and normosmia, 100 of 286 (34.9%) vs. 1938 of 2183 (88.8%) subjects; P < 0.01; data are shown as IHH vs. control subjects, respectively] (Fig. 1, C and D).
Fig. 1.
UPSIT scores in IHH and control subjects. A, The distribution of raw UPSIT scores of male subjects (IHH and controls); B, distribution of raw UPSIT scores of female subjects (IHH and controls); C, UPSIT ratings of all IHH subjects (males and females); D, UPSIT ratings of all control subjects (males and females).
Anosmic and hyposmic subjects display neuroanatomical abnormalities of the olfactory structures
Of the 39 patients in whom MRI of the olfactory system were available, 14 were anosmic, 11 were hyposmic, and 14 were normosmic. All anosmic subjects showed one or more abnormalities in their olfactory structures, and seven of 11 hyposmic subjects showed at least one abnormality in their olfactory structures. Of the normosmic subjects, 13 of 14 had completely normal structures, whereas one normosmic subject showed a hypoplastic left olfactory bulb. These results are summarized in Table 1.
Table 1.
UPSIT ratings stratified by MRI appearances of olfactory structures
| UPSIT rating | Anosmia (n = 14) | Hyposmia (n = 11) | Normosmia (n = 14) |
|---|---|---|---|
| No. of subjects with one or more olfactory structure abnormalities | 14 | 7 | 1 (hypoplastic left bulb) |
| No. of subjects with normal olfactory bulbs and sulci | 0 | 4 | 13 |
Self-reported olfactory assessment grossly underestimates the true olfactory deficit in IHH subjects
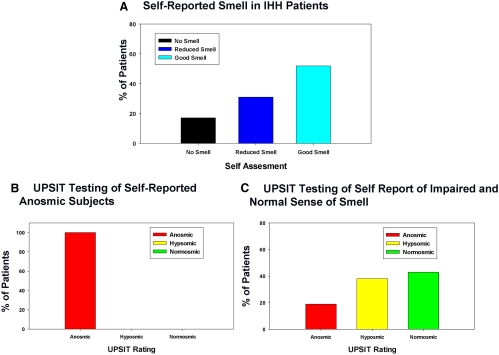
Of the 286 IHH subjects, 285 subjects made self-assessments of their olfactory function before taking the 40-item UPSIT. Of these, 48 of 285 (16.8%) reported having no sense of smell, 87 of 285 (30.5%) claimed a reduced sense of smell, and 150 of 285 (52.7%) reported a good sense of smell (Fig. 2A). Of the 48 subjects that reported to have no smell, all tested as anosmic on the 40-item UPSIT (Fig. 2B). Of the subjects who reported to have some preserved sense of smell on their self-assessments (i.e. good or reduced smell), 43 of 237 (18.1%) were actually anosmic, 93 of 237 (39.2%) were hyposmic, and 101 of 237 (42.7%) proved to be normosmic (Fig. 2C).
Fig. 2.
Correlation between olfactory self-assessment and UPSIT. A, Self-assessment reports of olfactory function in patients with IHH. B, UPSIT rating distributions of IHH patients who report “no smell” during self-assessment of olfactory function. C, UPSIT rating distribution of IHH patients who report to have “reduced” or “good smell” during self-assessment of olfactory function.
Olfactory percentile scores: majority of IHH subjects show olfactory deficit
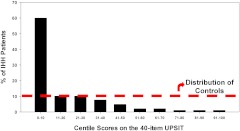
Age- and gender-adjusted percentile values derived from normative control data (30) showed that the olfactory function of IHH subjects was significantly skewed toward the lower centile ranges (47.6% ≤5th centile; 45.1% > 5th < 50th centile; 7.3% > 50th centile) compared with the age- and sex-matched normal population, whose data were used to define the expected normal centiles (Fig. 3).
Fig. 3.
IHH centile-derived data as mapped vs. age- and gender-matched normative population. Histogram showing percentage of all IHH subjects (black bars) lying within respective age- and gender-matched centiles of olfactory function (centile values calculated from Ref. 30). The percentage of control subjects lying within each defined centile range is shown by the red dotted line.
IHH subjects show overlap of genetic defects across the entire olfactory spectrum
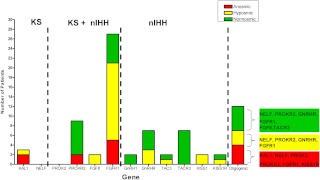
Of the 208 subjects tested for mutations in 12 genes, rare protein-altering variants in a single gene (monogenic variants) were found in 30.3% of the patients tested. Of the monogenic variants, 82.5% were heterozygous (monoallelic), and 17.5% were homozygous or compound heterozygous (biallelic, including KAL1 hemizygous patients). Oligogenic changes (mutations in two or more genes) were seen in 5.8%, whereas the remaining 63.9% did not harbor mutations in any of the 12 genes tested. Rare sequence variants were found in comparable frequencies across the olfactory function spectrum (anosmic subjects, 26.5%; hyposmic subjects, 39.5%; normosmic subjects, 37.3%; P = 0.25). The olfactory phenotypes of the patients harboring monogenic and oligogenic mutations are shown in Fig. 4.
Fig. 4.
Olfactory phenotype of IHH subjects harboring monogenic and oligogenic mutations. Number of IHH subjects with protein altering mutations in each gene are shown with their respective olfactory function as determined by UPSIT (anosmic, red; hyposmic, yellow; normosmic, green). The genes are arranged on the x-axis and are divided using vertical dotted lines to represent genes previously linked only to KS (KAL1, NELF); genes previously linked to both KS and nIHH (PROK2, PROKR2, FGF8, FGFR1); and genes previously linked only to nIHH (GNRH1, GNRHR, TAC3, TACR3, KISS1, KISS1R). The last column represents the number of subjects with oligogenic changes and the individual genes with protein-altering variants found within each olfactory phenotype.
Genetic mutations and their olfactory phenotypes
The olfactory phenotypes of subjects with genetic mutations are shown in Table 2. Two male subjects with hemizygous KAL1 variants were anosmic, whereas one female IHH subject harboring a heterozygous KAL1 mutation was hyposmic (UPSIT score, 32/50; 6th centile for age). All PROKR2 variants in this study were monoallelic and were associated with normosmia and anosmia as previously published (13, 37). In the FGF signaling pathway, two subjects with monoallelic FGF8 variants were both hyposmic, whereas monoallelic FGFR1 variants were seen across the full olfactory function spectrum. Biallelic FGFR1 variants were associated with hyposmia and normosmia. All GNRH1 and TACR3 variants were monoallelic, and all of them exhibited normosmia. In contrast, monoallelic and biallelic mutations in GNRHR and TAC3 were associated with normosmia and hyposmia (Table 2). All KISS1 and KISS1R variants were monoallelic and were associated with both hyposmia and normosmia. Hyposmia was seen in subjects with monogenic variants in almost all genes studied (KAL1, FGFR1, FGF8, GNRHR, TAC3, KISS1, and KISS1R). The gene-gene interactions in the oligogenic group are shown in Fig. 4. No subjects with monogenic PROK2 or NELF variants were seen in this study.
Table 2.
Olfactory phenotype of monoallelic and biallelic genetic mutations in a single gene (monogenic changes)
| Genesa | Olfactory phenotype (no. of subjects) |
|
|---|---|---|
| Subjects with monoallelic mutations | Subjects with biallelic mutations | |
| KAL1 (n = 3; 1.4%) | Hyposmic (1)b | Anosmic (2)c |
| PROKR2 (n = 9; 4.3%) | Anosmic (2), normosmic (7) | |
| FGF8 (n = 2; 1%) | Hyposmic (2) | |
| FGFR1 (n = 27; 13%) | Anosmic (5), hyposmic (14), normosmic (4) | Hyposmic (2), normosmic (2) |
| GNRHR (n = 7; 3.4%) | Hyposmic (2), normosmic (1) | Hyposmic (1), normosmic (3) |
| GNRH1 (n = 2; 1%) | Normosmic (2) | |
| TAC3 (n = 2; 1%) | Normosmic (1) | Hyposmic (1) |
| TACR3 (n = 7; 3.4%) | Normosmic (7) | |
| KISS1 (n = 2; 1%) | Hyposmic (2) | |
| KISS1R (n = 2; 1%) | Hyposmic (1), normosmic (1) | |
No mutations in PROK2 or NELF were seen in this study.
Monoallelic female subject.
Hemizygous KAL1 changes.
Discussion
Although some studies in IHH subjects have documented a broad spectrum of olfactory deficits including intermediary hyposmic phenotypes (6, 22, 25, 27, 29), a few studies have supported a binary olfactory phenotype [anosmia (KS) or normosmia (nIHH)], with a bimodal distribution of smell test scores (23, 24). The current study confirms a bimodal distribution of UPSIT scores in IHH subjects but, in addition, confirms a broader spectrum of olfactory deficits and reveals a higher prevalence of intermediary hyposmic phenotype than previously appreciated. Nearly one third of IHH subjects were hyposmic, and of the hyposmic subjects in whom olfactory MRI was available, seven of 11 subjects demonstrated abnormal olfactory structures, confirming a GnRH/olfactory neurodevelopmental basis for their smell defect. Mutations in most genes, including genes previously thought to primarily regulate GnRH secretion, were associated with the intermediary hyposmic phenotype. This surprising finding suggests either that these patients harbor a oliogogenic “second hit” (33) that contributes to the hyposmia or that neuroendocrine genes may have hitherto unrecognized neurodevelopmental roles in addition to their known roles in regulating GnRH secretion.
Traditionally, KS and nIHH have been considered to be distinct clinical entities, with KS subjects representing a neurodevelopmental phenotype with a primary defect in GnRH neuronal migration, whereas nIHH subjects represent a pure neuroendocrine defect in GnRH secretion/action. Indeed, most genes identified in subjects with KS have been shown to play a predominant GnRH migratory role (KAL1, NELF, PROK2/PROKR2, FGF8/FGFR1) (38), whereas genes identified in nIHH subjects have been shown to primarily affect neuroendocrine regulation of GnRH secretion/action (GNRH1, GNRHR, TAC3, TACR3, KISS1R) (17, 18, 20, 34). However, over the last decade, mutations in genes such as FGFR1, FGF8, PROK2, and PROKR2 have been shown to be associated with both KS and nIHH subjects (11, 13, 21), suggesting that the pathophysiological basis of KS and nIHH may indeed overlap. The hyposmic subjects in this study harbored mutations in both KS- and nIHH-related genes. This observation strongly supports a shared pathophysiological basis between KS and nIHH phenotypes. This notion is further supported by the observation in this study that when the UPSIT scores in IHH subjects are expressed in age- and gender-adjusted centiles, the centile ranges of olfactory function in the entire IHH cohort were significantly skewed toward the lowest centiles, hinting that olfactory defects may indeed exist in a large majority of these subjects (Fig. 3). This observation also suggests that compared with a simple UPSIT score-based olfactory function assessment, a centile-based evaluation of olfactory function may offer higher statistical certitude for classification of olfactory function and can identify subtle olfactory dysfunction in IHH subjects. These observations are also in keeping with a previous report by Hudson et al. (24), who showed that the olfactory function scores of apparently normosmic subjects lay within the lower end of the normal ranges for control subjects, arguing for a subtle olfactory abnormality within this subset. Likewise, Vogl et al. (26) also demonstrated that a small percentage of apparently nIHH subjects had abnormal olfactory bulbs and their smell tests were in the lower range of the normal distribution.
The presence of a hyposmic phenotype in subjects harboring mutations in the KAL1/PROK2/FGF signaling pathway is not unexpected, given their known effects on GnRH neuronal migration. However, the precise mechanism(s) of hyposmia occurring in subjects carrying mutations in nIHH-associated genes in this study (GNRHR, KISS1, KISS1R, and TAC3) remains unclear. It is possible that these hyposmic subjects may harbor additional unidentified neurodevelopmental gene mutations that confer the impaired olfactory phenotype. Alternatively, it is possible that a hyposmic UPSIT test per se may not necessarily indicate an underlying neurodevelopmental defect and that hyposmia may be related to local nasal factors. However, subjects with active nasal infection were asked to refrain from testing, and in those hyposmic subjects on whom we obtained additional olfactory assessment with MRI, the majority (seven of 11) showed neuroanatomical abnormalities in the olfactory system arguing for an intrinsic neurodevelopmental abnormality. Similarly, in a recent study (27), olfactory structural abnormalities were present in KS subjects who were hyposmic (by UPSIT), confirming that hyposmia per se is indicative of a pathophysiological defect in GnRH neuronal migration. Finally, it can be hypothesized that the nIHH-related neuroendocrine genes may directly impair olfactory function. However, studies of murine knockouts of these genes have not revealed any gross olfactory defects in these animals. So far, quantitative olfactory function has not been reported in these knockout animals. Of the hyposmic subjects who harbored mutation in the nIHH-related genes, only a minority (two of seven subjects) carried biallelic changes (one each in GNRHR and TAC3) (Table 2). Although it is intriguing that hyposmia was seen in these two subjects, a direct neurodevelopmental role for these genes cannot be implied unless more subjects with biallelic changes in the nIHH-related genes are shown to have impaired olfactory function. Taken together, oligogenicity is the most plausible reason for the hyposmic phenotype seen in IHH subjects harboring nIHH-gene mutations, and any direct neurodevelopmental role for the nIHH-related genes requires further evaluation.
Most clinicians frequently rely on patients' self-assessment to determine whether an IHH subject has KS or nIHH. In the current study, self-assessment is shown to be an inaccurate measure of olfactory function because nearly 50% of subjects who self-reported to have some preserved sense of smell were found to be either anosmic or hyposmic upon formal UPSIT evaluation. In contrast, when GnRH-deficient subjects self-reported no sense of smell, a subsequent 40-item UPSIT was uniformly abnormal (UPSIT score ≤18). Thus, a self-report of anosmia is a reliable estimate of olfactory function, but in those IHH subjects reporting preserved sense of smell, a standardized method for assessing olfactory function should be administered for accurate olfactory phenotypic ascertainment. Similar lack of accuracy of self-reported olfaction has been shown in other clinical syndromes (39) as well as in the normal population (40).
In the last two decades, our understanding of the molecular basis of IHH has been greatly advanced by genetic studies in IHH subjects. However, with the increasing number of genes linked to IHH, targeted genetic testing will be needed to minimize cost and improve efficiency. It is already known that nonreproductive phenotyping of IHH subjects can guide appropriate genetic testing (e.g. renal agenesis and KAL1 gene screening) (38). Likewise, the findings in this study indicate that accurate olfactory phenotyping may also guide targeted genetic screening in IHH subjects. Specifically, in subjects with either self-reported anosmia or anosmia by UPSIT, genetic screening can be targeted primarily to traditional neurodevelopmental genes. However, in both hyposmic and normosmic subjects, a more comprehensive genetic screening will be required. The presence of nIHH-related genes in hyposmic subjects indicates that more neurodevelopmental genes remain to be discovered, and given that oligogenicity is highly likely in these subjects, appropriate genetic counseling will also be required for such subjects.
A few limitations of this study need to be considered. The 40-item UPSIT is an odor identification test and does not test olfactory threshold. However, the 40-item UPSIT is highly correlated with traditional odor detection threshold tests and hence reflects the true olfactory ability (30). The olfactory function of the control subjects in this study was obtained from UPSIT tests done several years ago, and hence subtle changes in UPSIT odors and their recognition may have changed since that time. For example, some persons today are unfamiliar with previously used odors such as turpentine or motor oil. However, even if such effects entered into the present data, our finding of a sizable hyposmic group would not be negated given the number and distribution of the involved cases. High-resolution MRI images of the olfactory bulbs and tracts were only available in a limited subset of the IHH cohort. Nonetheless, in the limited number of MRI images that were available, the presence of olfactory abnormalities in the hyposmic subjects confirmed the neuroanatomical defect in these subjects. Mutations in the gene CHD7 have recently been implicated in IHH and are a relatively common genetic cause of IHH (14). However, this gene was not sequenced in this study due to high sequencing costs given its large size. Because CHD7 is thought to play a key neurodevelopmental role in olfactory/GnRH neuronal development (14), the clinical spectrum olfactory phenotypes of subjects with CHD7 mutations will certainly need evaluation in future studies. Finally, the genetic etiology of nearly 60% of IHH subjects is still unknown, and hence an oligogenic interaction may partly or fully explain the genetic findings seen in the hyposmic subjects. With the falling sequencing costs, future studies will be able to address the role of oligogenicity on the various olfactory phenotypes.
In summary, this study demonstrates that olfactory function in IHH subjects encompasses a broad spectrum ranging from anosmia to normosmia with a significant intermediary hyposmic phenotype. Hyposmic subjects display olfactory structural abnormalities and harbor mutations in both neurodevelopmental and neuroendocrine genes, suggesting a pathophysiological overlap between the KS and nIHH phenotypes. Correlation of olfactory function with the genetic etiology in subjects with isolated GnRH deficiency will improve our understanding of the GnRH neuronal ontogeny and potentially guide targeted genetic testing in this condition.
Acknowledgments
The authors thank Douglas Hayden of the Biostatistics Center at the Massachusetts General Hospital for statistical analysis of the data. We also thank all physicians who referred valuable patients from across the world who contributed valuable patients for this study.
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development/National Institutes of Health, through cooperative agreement U54 HD028138-21 as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research. R.L.D. is supported, in part, by U.S. Army Medical Research Acquisition Activity Grant W81XWH-09-1-0467.
Disclosure Summary: R.L.D. is the president and major shareholder of Sensonics, Inc., the manufacturer of the olfactory test employed in this study. The other authors have nothing to declare.
Footnotes
- FGF
- Fibroblast growth factor
- IHH
- idiopathic hypogonadotropic hypogonadism
- KS
- Kallmann syndrome
- MRI
- magnetic resonance imaging
- nIHH
- normosmic IHH.
References
- 1. Hoffman AR, Crowley WF., Jr 1982. Induction of puberty in men by long-term pulsatile administration of low-dose gonadotropin-releasing hormone. N Engl J Med 307:1237–1241 [DOI] [PubMed] [Google Scholar]
- 2. Kallmann F, Schoenfeld W, Barrera S. 1944. The genetic aspects of primary eunuchoidism. Am J Ment Defic 48:203–236 [Google Scholar]
- 3. Schwanzel-Fukuda M, Pfaff DW. 1989. Origin of luteinizing hormone-releasing hormone neurons. Nature 338:161–164 [DOI] [PubMed] [Google Scholar]
- 4. Legouis R, Hardelin JP, Levilliers J, Claverie JM, Compain S, Wunderle V, Millasseau P, Le Paslier D, Cohen D, Caterina D, Bougueleret L, Delemarre-Van de Waal H, Lutfalla G, Weissenbach J, Petit C. 1991. The candidate gene for the X-linked Kallmann syndrome encodes a protein related to adhesion molecules. Cell 67:423–435 [DOI] [PubMed] [Google Scholar]
- 5. Franco B, Guioli S, Pragliola A, Incerti B, Bardoni B, Tonlorenzi R, Carrozzo R, Maestrini E, Pieretti M, Taillon-Miller P, Brown CJ, Willard HF, Lawrence C, Graziella Persico M, Camerino G, Ballabio A. 1991. A gene deleted in Kallmann's syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature 353:529–536 [DOI] [PubMed] [Google Scholar]
- 6. Dodé C, Levilliers J, Dupont JM, De Paepe A, Le Dû N, Soussi-Yanicostas N, Coimbra RS, Delmaghani S, Compain-Nouaille S, Baverel F, Pêcheux C, Le Tessier D, Cruaud C, Delpech M, Speleman F, Vermeulen S, Amalfitano A, Bachelot Y, Bouchard P, Cabrol S, Carel JC, Delemarre-van de Waal H, Goulet-Salmon B, Kottler ML, Richard O, Sanchez-Franco F, Saura R, Young J, Petit C, Hardelin JP. 2003. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet 33:463–465 [DOI] [PubMed] [Google Scholar]
- 7. Pitteloud N, Acierno JS, Jr, Meysing A, Eliseenkova AV, Ma J, Ibrahimi OA, Metzger DL, Hayes FJ, Dwyer AA, Hughes VA, Yialamas M, Hall JE, Grant E, Mohammadi M, Crowley WF., Jr 2006. Mutations in fibroblast growth factor receptor 1 cause both Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA 103:6281–6286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Falardeau J, Chung WC, Beenken A, Raivio T, Plummer L, Sidis Y, Jacobson-Dickman EE, Eliseenkova AV, Ma J, Dwyer A, Quinton R, Na S, Hall JE, Huot C, Alois N, Pearce SH, Cole LW, Hughes V, Mohammadi M, Tsai P, Pitteloud N. 2008. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Invest 118:2822–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dodé C, Teixeira L, Levilliers J, Fouveaut C, Bouchard P, Kottler ML, Lespinasse J, Lienhardt-Roussie A, Mathieu M, Moerman A, Morgan G, Murat A, Toublanc JE, Wolczynski S, Delpech M, Petit C, Young J, Hardelin JP. 2006. Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet 2:e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pitteloud N, Zhang C, Pignatelli D, Li JD, Raivio T, Cole LW, Plummer L, Jacobson-Dickman EE, Mellon PL, Zhou QY, Crowley WF., Jr 2007. Loss-of-function mutation in the prokineticin 2 gene causes Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA 104:17447–17452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cole LW, Sidis Y, Zhang C, Quinton R, Plummer L, Pignatelli D, Hughes VA, Dwyer AA, Raivio T, Hayes FJ, Seminara SB, Huot C, Alos N, Speiser P, Takeshita A, Van Vliet G, Pearce S, Crowley WF, Jr, Zhou QY, Pitteloud N. 2008. Mutations in prokineticin 2 and prokineticin receptor 2 genes in human gonadotrophin-releasing hormone deficiency: molecular genetics and clinical spectrum. J Clin Endocrinol Metab 93:3551–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abreu AP, Trarbach EB, de Castro M, Frade Costa EM, Versiani B, Matias Baptista MT, Garmes HM, Mendonca BB, Latronico AC. 2008. Loss-of-function mutations in the genes encoding prokineticin-2 or prokineticin receptor-2 cause autosomal recessive Kallmann syndrome. J Clin Endocrinol Metab 93:4113–4118 [DOI] [PubMed] [Google Scholar]
- 13. Sarfati J, Guiochon-Mantel A, Rondard P, Arnulf I, Garcia-Piñero A, Wolczynski S, Brailly-Tabard S, Bidet M, Ramos-Arroyo M, Mathieu M, Lienhardt-Roussie A, Morgan G, Turki Z, Bremont C, Lespinasse J, Du Boullay H, Chabbert-Buffet N, Jacquemont S, Reach G, De Talence N, Tonella P, Conrad B, Despert F, Delobel B, Brue T, Bouvattier C, Cabrol S, Pugeat M, Murat A, Bouchard P, Hardelin JP, Dodé C, Young J. 2010. A comparative phenotypic study of Kallmann syndrome patients carrying monoallelic and biallelic mutations in the prokineticin 2 or prokineticin receptor 2 genes. J Clin Endocrinol Metab 95:659–669 [DOI] [PubMed] [Google Scholar]
- 14. Kim HG, Kurth I, Lan F, Meliciani I, Wenzel W, Eom SH, Kang GB, Rosenberger G, Tekin M, Ozata M, Bick DP, Sherins RJ, Walker SL, Shi Y, Gusella JF, Layman LC. 2008. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet 83:511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Roux N, Young J, Misrahi M, Genet R, Chanson P, Schaison G, Milgrom E. 1997. A family with hypogonadotropic hypogonadism and mutations in the gonadotropin-releasing hormone receptor. N Engl J Med 337:1597–1602 [DOI] [PubMed] [Google Scholar]
- 16. Layman LC, Cohen DP, Jin M, Xie J, Li Z, Reindollar RH, Bolbolan S, Bick DP, Sherins RR, Duck LW, Musgrove LC, Sellers JC, Neill JD. 1998. Mutations in gonadotropin-releasing hormone receptor gene cause hypogonadotropic hypogonadism. Nat Genet 18:14–15 [DOI] [PubMed] [Google Scholar]
- 17. Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O'Rahilly S, Carlton MB, Crowley WF, Jr, Aparicio SA, Colledge WH. 2003. The GPR54 gene as a regulator of puberty. N Engl J Med 349:1614–1627 [DOI] [PubMed] [Google Scholar]
- 18. Topaloglu AK, Reimann F, Guclu M, Yalin AS, Kotan LD, Porter KM, Serin A, Mungan NO, Cook JR, Ozbek MN, Imamoglu S, Akalin NS, Yuksel B, O'Rahilly S, Semple RK. 2009. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for neurokinin B in the central control of reproduction. Nat Genet 41:354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. 2003. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci USA 100:10972–10976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bouligand J, Ghervan C, Tello JA, Brailly-Tabard S, Salenave S, Chanson P, Lombès M, Millar RP, Guiochon-Mantel A, Young J. 2009. Isolated familial hypogonadotropic hypogonadism and a GNRH1 mutation. N Engl J Med 360:2742–2748 [DOI] [PubMed] [Google Scholar]
- 21. Pitteloud N, Meysing A, Quinton R, Acierno JS, Jr, Dwyer AA, Plummer L, Fliers E, Boepple P, Hayes F, Seminara S, Hughes VA, Ma J, Bouloux P, Mohammadi M, Crowley WF., Jr 2006. Mutations in fibroblast growth factor receptor 1 cause Kallmann syndrome with a wide spectrum of reproductive phenotypes. Mol Cell Endocrinol 254–255:60–69 [DOI] [PubMed] [Google Scholar]
- 22. Quinton R, Duke VM, de Zoysa PA, Platts AD, Valentine A, Kendall B, Pickman S, Kirk JM, Besser GM, Jacobs HS, Bouloux PM. 1996. The neuroradiology of Kallmann's syndrome: a genotypic and phenotypic analysis. J Clin Endocrinol Metab 81:3010–3017 [DOI] [PubMed] [Google Scholar]
- 23. Quinton R, Duke VM, Robertson A, Kirk JM, Matfin G, de Zoysa PA, Azcona C, MacColl GS, Jacobs HS, Conway GS, Besser M, Stanhope RG, Bouloux PM. 2001. Idiopathic gonadotrophin deficiency: genetic questions addressed through phenotypic characterization. Clin Endocrinol (Oxf) 55:163–174 [DOI] [PubMed] [Google Scholar]
- 24. Hudson R, Laska M, Berger T, Heye B, Schopohl J, Danek A. 1994. Olfactory function in patients with hypogonadotropic hypogonadism: an all-or-none phenomenon? Chem Senses 19:57–69 [DOI] [PubMed] [Google Scholar]
- 25. Bhagavath B, Podolsky RH, Ozata M, Bolu E, Bick DP, Kulharya A, Sherins RJ, Layman LC. 2006. Clinical and molecular characterization of a large sample of patients with hypogonadotropic hypogonadism. Fertil Steril 85:706–713 [DOI] [PubMed] [Google Scholar]
- 26. Vogl TJ, Stemmler J, Heye B, Schopohl J, Danek A, Bergman C, Balzer JO, Felix R. 1994. Kallman syndrome versus idiopathic hypogonadotropic hypogonadism at MR imaging. Radiology 191:53–57 [DOI] [PubMed] [Google Scholar]
- 27. Koenigkam-Santos M, Santos AC, Versiani BR, Diniz PR, Junior JE, de Castro M. 21 May 2011. Quantitative magnetic resonance imaging evaluation of the olfactory system in Kallmann syndrome: correlation with a clinical smell test. Neuroendocrinology doi: 10.1159/000328437 [DOI] [PubMed] [Google Scholar]
- 28. Massin N, Pêcheux C, Eloit C, Bensimon JL, Galey J, Kuttenn F, Hardelin JP, Dodé C, Touraine P. 2003. X chromosome-linked Kallmann syndrome: clinical heterogeneity in three siblings carrying an intragenic deletion of the KAL-1 gene. J Clin Endocrinol Metab 88:2003–2008 [DOI] [PubMed] [Google Scholar]
- 29. Parenti G, Rizzolo MG, Ghezzi M, Di Maio S, Sperandeo MP, Incerti B, Franco B, Ballabio A, Andria G. 1995. Variable penetrance of hypogonadism in a sibship with Kallmann syndrome due to a deletion of the KAL gene. Am J Med Genet 57:476–478 [DOI] [PubMed] [Google Scholar]
- 30. Doty RL, Shaman P, Dann M. 1984. Development of the University of Pennsylvania Smell Identification Test: a standardized microencapsulated test of olfactory function. Physiol Behav 32:489–502 [DOI] [PubMed] [Google Scholar]
- 31. Doty RL. 2001. The Smell Identification Test Administration Manual. Haddon Heights, NJ: Sensonics Inc [Google Scholar]
- 32. Yousem DM, Geckle RJ, Bilker W, McKeown DA, Doty RL. 1996. MR evaluation of patients with congenital hyposmia or anosmia. AJR Am J Roentgenol 166:439–443 [DOI] [PubMed] [Google Scholar]
- 33. Sykiotis GP, Plummer L, Hughes VA, Au M, Durrani S, Nayak-Young S, Dwyer AA, Quinton R, Hall JE, Gusella JF, Seminara SB, Crowley WF, Jr, Pitteloud N. 2010. Oligogenic basis of isolated gonadotropin-releasing hormone deficiency. Proc Natl Acad Sci USA 107:15140–15144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chan YM, de Guillebon A, Lang-Muritano M, Plummer L, Cerrato F, Tsiaras S, Gaspert A, Lavoie HB, Wu CH, Crowley WF, Jr, Amory JK, Pitteloud N, Seminara SB. 2009. GNRH1 mutations in patients with idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA 106:11703–11708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bo-Abbas Y, Acierno JS, Jr, Shagoury JK, Crowley WF, Jr, Seminara SB. 2003. Autosomal recessive idiopathic hypogonadotropic hypogonadism: genetic analysis excludes mutations in the gonadotropin-releasing hormone (GnRH) and GnRH receptor genes. J Clin Endocrinol Metab 88:2730–2737 [DOI] [PubMed] [Google Scholar]
- 36. Gianetti E, Tusset C, Noel SD, Au MG, Dwyer AA, Hughes VA, Abreu AP, Carroll J, Trarbach E, Silveira LF, Costa EM, de Mendonça BB, de Castro M, Lofrano A, Hall JE, Bolu E, Ozata M, Quinton R, Amory JK, Stewart SE, Arlt W, Cole TR, Crowley WF, Kaiser UB, Latronico AC, Seminara SB. 2010. TAC3/TACR3 mutations reveal preferential activation of gonadotropin-releasing hormone release by neurokinin B in neonatal life followed by reversal in adulthood. J Clin Endocrinol Metab 95:2857–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martin C, Balasubramanian R, Dwyer AA, Au MG, Sidis Y, Kaiser UB, Seminara SB, Pitteloud N, Zhou QY, Crowley WF., Jr 2011. The role of the prokineticin 2 pathway in human reproduction: evidence from the study of human and murine gene mutations. Endocr Rev 32:225–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hardelin JP, Dodé C. 2008. The complex genetics of Kallmann syndrome: KAL1, FGFR1, FGF8, PROKR2, PROK2, et al. Sex Dev 2:181–193 [DOI] [PubMed] [Google Scholar]
- 39. Doty RL, Deems DA, Stellar S. 1988. Olfactory dysfunction in Parkinsonism: a general deficit unrelated to neurologic signs, disease stage, or disease duration. Neurology 38:1237–1244 [DOI] [PubMed] [Google Scholar]
- 40. Nordin S, Monsch AU, Murphy C. 1995. Unawareness of smell loss in normal aging and Alzheimer's disease: discrepancy between self-reported and diagnosed smell sensitivity. J Gerontol B Psychol Sci Soc Sci 50:P187–P192 [DOI] [PubMed] [Google Scholar]