Abstract
Described is a method that relies on subtractive tissue-directed shot-gun proteomics to identify tumor proteins in the blood of a patient newly diagnosed with cancer. To avoid analytical and statistical biases caused by physiologic variability of protein expression in the human population, this method was applied on clinical specimens obtained from a single patient diagnosed with non-metastatic renal cell carcinoma (RCC). The proteomes extracted from tumor, normal adjacent tissue and pre-operative plasma were analyzed using 2D-LC-MS. The lists of identified proteins were filtered to discover proteins that i) were found in tumor but not normal tissue, ii) were identified in matching plasma, and iii) whose spectral count was higher in tumor tissue than plasma. These filtering criteria resulted in identification of eight tumor proteins in the blood. Subsequent Western-blot analysis confirmed the presence of cadherin-5, cadherin-11, DEAD-box protein-23, and pyruvate kinase) in the blood of the patient under the study, as well as in the blood of four other patients diagnosed with RCC. These results demonstrate the utility of a combined blood/tissue analysis strategy that permits the detection of tumor proteins in the blood of a patient diagnosed with RCC.
Mass spectrometry (MS) methods allowing for the identification of tumor proteins in blood may enable cancer biomarker. Urgent needs in this domain include assays for: cancer diagnosis, therapy selection, prognosis, and monitoring.1 Both cancer biology and clinical oncology are undergoing rapid transformations, specifically, from an organ-centric to molecular pathways focused disciplines. Therefore, methods allowing for an improved molecular characterization of a patient’s actual malignant process/tumor may facilitate the development of advanced assays for personalized cancer diagnosis and management.2 Molecular profiling of a patient’s tumor may provide better insights into the cancer-induced derangements relevant to the malignancy under study, with the eventual and ultimate hope of benefits to patient outcome.2 MS-based proteomics may play an important role in characterizations of proteins within clinical samples. Therefore, innovative approaches focused on method development for proteomic profiling of clinically relevant specimens are critically needed.3
Despite advances in cancer biomarker research, the translation of proteomic methods and findings to applicable clinical assays has been disappointing.4 Principal factors that hinder mass spectrometry (MS)-based biomarker research using clinical samples include: (i) significant heterogeneity of solid tumors,5 (ii) formidable variability of protein expression in the human population proper, serving as a potential source of analytical/statistical bias,4 (iii) significant mismatches between the dynamic range of MS instrumentation and the protein content of clinical specimens,3 and (iv) the majority of proteomics-derived “potential” cancer biomarkers were not germane to the tumor in question.4 Many of these putative cancer biomarkers fall into the categories of acute-phase reactants and likely lack specificity to the pathologic process under study.4
Identifying relevant differences within the blood proteome from healthy and cancer patients is difficult due to the common lack of specificity of the findings. This may be influenced by a plethora of physiological and analytical factors. Additionally, numerous differences can be detected when comparing such cohorts. The major obstacle is proving which differences are dependent on the presence of the cancer and which result from physiological bias or analytical variability. While blood-based biomarkers would revolutionize cancer management, the commonly followed strategy of only analyzing serum or plasma from patients makes it very difficult to trace the origin of proteomic differences back to a tumor.
In this study, we present the results from a combined tumor/plasma proteome analysis of samples acquired from a single patient. This strategy aims to recognize tumor proteins within the blood and may possess a higher probability of surviving the rigors of verification and validation necessary for generating useful clinical biomarker candidates. The objective of this investigation was to develop a proteomic method capable of reliably profiling the proteome of a solid tumor, and determine whether any of the identified proteins in the tumor proper are detectable in the blood of a patient newly diagnosed with a non-metastatic cancer.
EXPERIMENTAL SECTION
Supporting Information (SI), accessible at http://pubs.acs.org, contains detailed experimental procedures. Briefly, all clinical specimens were procured by the National Cancer Institute (NCI) Cooperative Human Tissue Network (CHTN) in accordance with current regulations and guidance issued by the NCI Office of Human Subjects Review (OHSR). Fresh frozen tumor tissue (pathology report: RCC of a clear cell histology), adjacent non-tumor tissue (kidney stroma and parenchyma), and plasma were collected from a single patient using standard clinical procedure and stored at − 80 °C. Control plasma from matched healthy donor was obtained using an identical procedure. Additional RCC plasma samples were also collected in accordance with NCI OHSR guidelines and approval.
Tissue homogenates and plasma were reduced and alkylated prior to high abundant protein depletion using antibody-based multiple affinity removal system (MARS-6) cartridges (Agilent Technologies, Palo Alto, CA). A total of 200 μg of depleted protein sample from each patient’s specimen was digested in buffered methanol and fractionated by strong cation exchange chromatography (SCX).6 Each SCX peptide fraction was analyzed twice using nano-flow reversed phase liquid chromatography (nfRPLC) coupled to a hybrid linear ion trap – Fourier transform ion cyclotron resonance (LIT-FTICR) – MS. All MS/MS spectra were searched against a normal and reversed human protein database (UniProt Human, release 09/2007), using SEQUEST (Thermo-Finnegan, San Jose, CA). The search against the sequence-reversed human protein database was used to assess the peptide false discovery rate (FDR) and establish threshold criteria that permitted a maximum estimated peptide FDR of 1%.
RESULTS AND DISCUSSION
Solid tumors are heterogeneous. They consist of tumor cells, stroma and vasculature elements, forming an essential microenvironment nurturing the malignant process. Interactions/signaling between different cell types, pathways and networks constitute critical aspects of tumor biology and molecular oncology.7 Improved characterization of these molecular-processes should provide better insights to tumor growth/progression and metastasis.7 Hence, the ability to directly and effectively profile clinical specimens is essential, since proteomes of cultured cancer cells (which lack a formal tumor microenvironment) do not accurately resemble those in vivo.8
RCC is known as a silent cancer because it is usually detected at an advanced stage (40% of presentations have metastasis).9 Currently, the standard of care for non-metastatic RCC is surgical resection, but many cases will recur with metastatic disease, an event that usually leads to death. Nonexistent are biomarkers for RCC. Such molecular markers are especially needed for: diagnosis, follow-up, and assessment of treatment response .9 For these reasons, RCC was selected as a cancer model for method development.
Based on recent guidelines published by the St. Gallen International Expert Consensus, effective tumor profiling to better characterize the molecular phenotype is pivotal for targeted cancer treatment.10 The goal of this study was to develop a method to both enable and determine, whether salient proteins identified in a patient’s tumor, are detectable in the blood. This particular patient was newly diagnosed with a localized kidney cancer. Additionally, we sought to explore the utility of a putative tumor marker as a potential drug target for the patient under the study.
The choice of specimens may be critical for cancer biomarker discovery.4 Tumor-derived biomarker proteins are anticipated to be at higher concentration in tumor than in plasma and therefore more easily identifiable by MS.4 However, biofluids are easily accessible and therefore have greater potential for facile diagnostics and/or therapeutic monitoring.4 Previously, we hypothesized that concurrent analysis of tissues and peripheral blood specimens should help to optimize for cancer biomarker identification in blood.11 In accordance with this hypothesis we developed a method that relies on set-based analysis of tissues and plasma for identification of tumor proteins in the blood of a patient newly diagnosed with non-metastatic cancer.
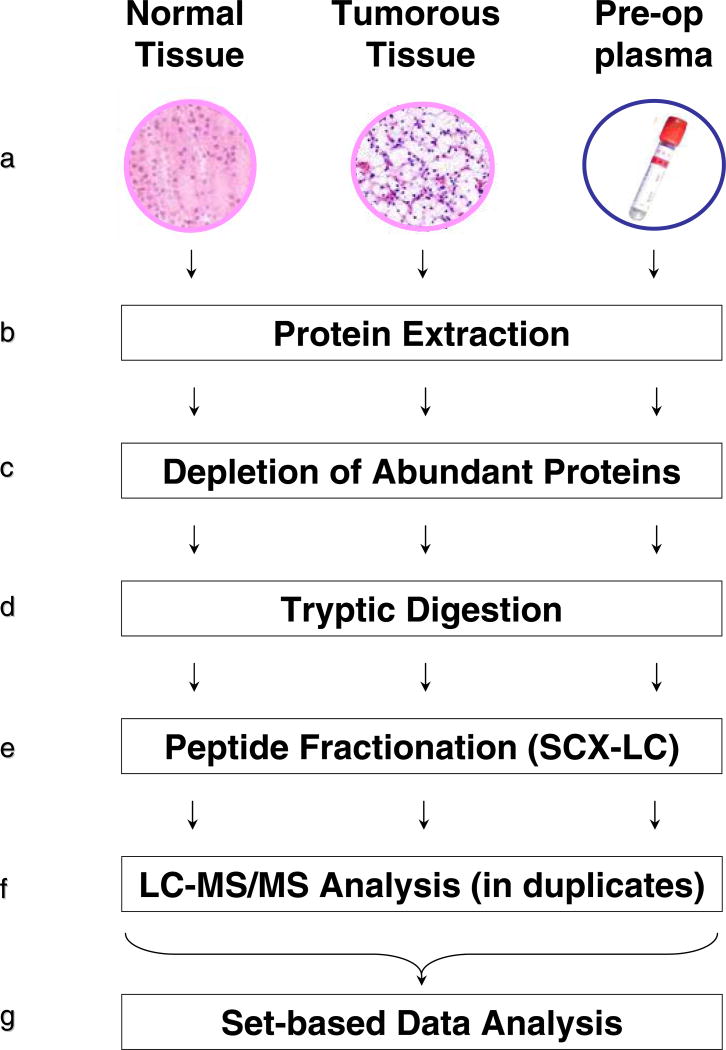
The experimental design is summarized in the Figure 1 (a-g). The workflow encompasses seven basic steps. Specimen collection: tumor, normal adjacent kidney tissue and plasma were prospectively collected from a patient diagnosed with RCC (Figure 1a). To eliminate potential biases caused by leaking of tissue proteins into the vascular system during the surgical procedure, blood was collected prior to surgery. To minimize analytical biases resulting from natural proteome variabilities present within diverse human populations, all specimens were obtained from a single patient. Protein extraction: tissue samples were thawed in lysis buffer followed by protein extraction using homogenization and sonication (Figure 1b). Affinity depletion: since the plasma content of kidney tissue represents up to 22% of its weight,12 depletion of abundant proteins was applied to both, tissue and plasma specimens (Figure 1c). Tryptic digestion: tissue lysates and plasma were tryptically digested in buffered methanol (Figure 1d). The use of methanol provided efficient solubilization and digestion, while the absence of detergents and chaotropes facilitated optimal peptide separation and ionization.13 Fractionation: to achieve optimal protein coverage all digests were fractionated by off-line strong cation exchange (SCX)-LC (Figure 1e). Liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis: a nano-flow RPLC system coupled to a hybrid LIT-FTICR-MS provides an in-depth analysis of tissue and plasma specimens (Figure 1f).14 Data processing: the final list of protein identifications for each specimen was created by including only protein-specific peptides that resulted in the elimination of ambiguous protein identification caused by inclusion of peptides identified in multiple protein species. The identities of proteins identified exclusively in tumor were elicited by subtractive proteomics, which relies on set-based analysis of proteins from tumor and normal kidney (Figure 1g).15 Tumor-only identified proteins were then intersected with those identified in plasma to isolate overlapping protein species. A simple subtractive comparison of overlapping protein species revealed the identities of tumor proteins in plasma as proteins identified by a higher spectral count in tumor versus plasma.16
Figure 1.
a – g. The outline of methodology for identification of tumor proteins in peripheral blood of a patient diagnosed with non-metastatic RCC.
A total of 0.57 g of tumor and 0.56 g of normal adjacent kidney tissue, were acquired from a patient newly diagnosed with RCC. A total of 8,183 unique protein-specific peptides were identified in tumor (SI Table S-1a) with 1,275 proteins being identified by at least two unique peptides (SI Table S-1b). The same analysis resulted in the identification of a total of 8,194 unique peptides in normal kidney (SI Table S-2a) with the identification of 1,281 proteins identified by at least two unique peptides (SI Table S-2b).
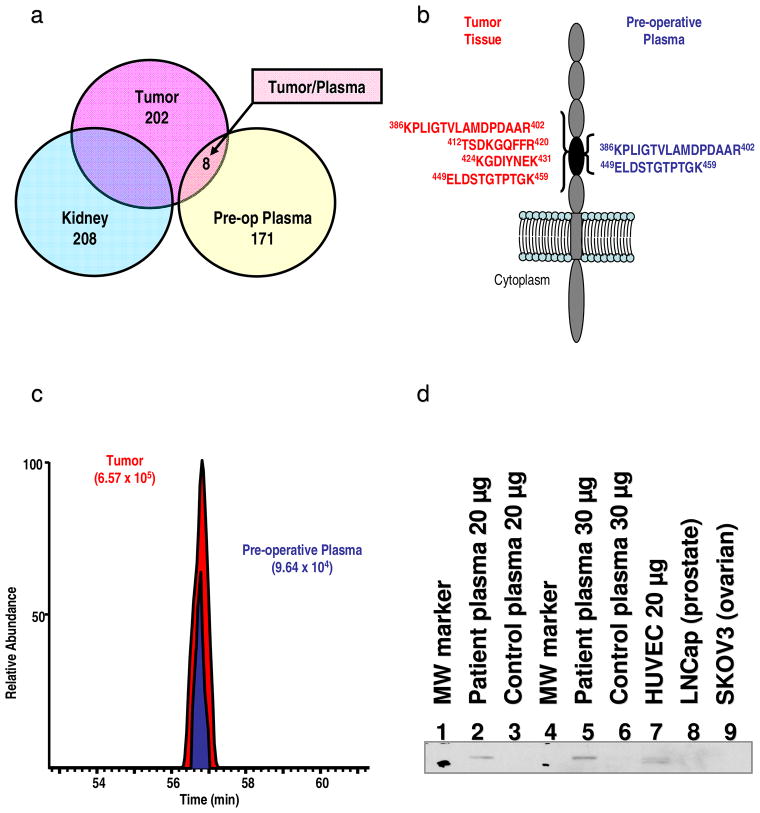
A subtractive proteomic analysis was used to identify proteins detected exclusively in tumor but not in normal-adjacent tissue.15 By using a criterion of two unique peptides per protein, a subset of 202 proteins was identified only in the tumor (SI Figure S-1a). Using the same criteria, a subset of 209 proteins was identified exclusively in normal-adjacent tissue (SI Figure S-2a). Analysis of the same patient’s plasma specimen yielded identification of 2,486 unique peptides (SI Table S-3a) including 179 proteins that where identified by at least 2 unique protein-specific peptides (SI Table S-3b). The 202 proteins identified exclusively in tumor were then compared with these 179 plasma proteins. This comparative list was further culled to find proteins that exhibited a higher spectral count in tumor compared to plasma (Figure S-1b).
This stringent filtering revealed a panel of eight proteins (Figure 2a) meeting the four criteria: i) identified in tumor, ii) not identified in normal-adjacent tissue, iii) identified in plasma, and iv) their spectral count value was higher in tumor versus plasma (SI Table S-4). Three of the identified proteins: cadherin-11 17, pyruvate kinase 18, and vascular cell adhesion molecule-1 19 were previously shown to be implicated in RCC tumorogenesis, while the other five: cadherin-5 20, DEAD box protein 23 21, kidney and brain protein 22, Mi2-beta 23, and nuclear receptor coactivator 6 24, have been proposed to be significant effectors in the biology of other human cancers. Ingenuity Pathway Analysis revealed that all of eight identified proteins were found to be expressed in kidney while three of them (cadherin 11, cadherin 5, and vascular cell adhesion molecule-1) were recognized as membrane proteins (SI Table S-4). Overall, these findings allowed us to hypothesize that the panel of identified proteins is a genuine tumor-set, representing potential biomarkers specific to the RCC molecular-phenotype of the patient under study. Also, these results allowed us to hypothesize that the identification of these putative marker proteins in plasma was not incidental, but a direct outcome of the present method, due to the prior association of these proteins to RCC and cancer in general.
Figure 2.
a – d. Venn diagrams depicting subtractive proteomic analysis employed to reveal identities of tumor-residing proteins. A total of 202 proteins were identified by at least two protein specific peptides in any of the peptide fractions from tumorous tissue but not in any of the technical replicates from normal adjacent tissue (kidney). Subsequent analysis revealed identities of 8 tumor-residing proteins in the plasma by comparing/subtracting proteins identified exclusively in the tumor (202) and those identified in plasma (179). These proteins exhibited higher total peptide count in tumor vs. plasma and are considered as genuine tumor proteins (a). Secondary structure of cadherin-5 depicting the location of identified peptides. Peptides in red font were identified in tumor while peptides depicted by blue font were identified in plasma. All identified peptides in tumor and plasma reside in extracellular domain of this integral plasma membrane protein (b). Extracted ion chromatograms of the KPLIGTVLAMDPDAAR peptide identifying cadherin-5 in tumor (red font) and peripheral plasma (blue font) indicating higher concentration level of this peptide/protein in tumor (c). Western blot analysis of cadherin-5. A total of 20 μg of depleted plasma protein from the patient under study (2) and a total of 20 μg depleted plasma protein from matched healthy donor (3) along with 30 μg of depleted plasma protein from the same patient (5) and 30 μg depleted plasma protein from matched healthy donor (6) were separated on 4-20% Tris-Glycine gradient gels. Also, a total of 20 μg of cellular lysates: HUVEC (7), LNCaP (8) and SKOV3 (9) were separated using the same gradient gel and transferred to Immuno-Blot PVDF membranes. The membranes were blocked by 3% bovine serum albumin and then probed overnight at 4 °C with anti-cadherin-5 MAb followed by peroxidase conjugated goat anti-mouse IgG secondary antibody (d).
Of the eight putative tumor proteins identified in plasma, cadherin-5 was selected for cross validation based on the observation that all peptides identifying this integral membrane protein in the tumor (SI Table S-4) and plasma (SI Table S-5) originate from its extracellular domain (Figure 2b). The MS/MS spectrum of the KPLIGTVLAMDPDAAR peptide identifying cadherin-5 in tumor is shown in Figure S-2a. The MS/MS spectrum of the same peptide identified in plasma (SI Figure S-2b) exhibits higher background and lower normalized intensity of fragment ions. Taken together, the spectral counts shown in Tables S-5 and S-6, MS/MS spectra shown in Figure S-2 (a-b), and respective extracted ion-chromatogram of the peptide identified in the tumor and plasma (Figure 2c) illustrate the existing gradient and higher abundance of cadherin-5 in the tumor.
Subsequent Western blot analysis confirmed the presence of cadherin-5 in the preoperative plasma specimen while it was not detected in the plasma of a matched healthy donor (Figure 2d). Further analyses revealed the absence of cadherin-5 in ovarian (SKOV-3) and prostate cancer (LNCaP) cell lysates, but confirmed its presence in HUVEC lysate (positive control) (Figure 2d). Altogether, these findings suggest an active role of cadherin-5 in the RCC molecular cancer biology of our patient under study. Interestingly, cadherin-5 has been previously described as potential anti-angiogenic drug target.25 Thus, cadherin-5 may be rationally selected as a potential drug target in a hypothetical personalized adjuvant therapy plan, for this individual diagnosed with localized RCC. Although this study investigated only a single RCC tumor/patient, a panel of putative protein markers identified using this method may be further investigated using high-throughput techniques employing suitable immunoassays (i.e. ELISA) or MS-assays (i.e. MRM) to test the general applicability in larger RCC patient cohorts. To test this hypothesis, cadherin-5, cadherin-11, DEAD box protein, 23 and pyruvate kinase were selected for cross-validation by Western-blot analysis in the blood of the patient under the study and in the blood of four additional patients diagnosed with RCC. The analysis confirmed the presence of these proteins in analyzed blood specimens (Figure S-3) indicating further confirmation of our initial findings. Additionally, these results may serve as sufficient evidence to warrant further scientific and clinical study via a clinical trial, where a much larger number of patients with RCC would now be evaluated and followed regarding the reported putative biomarkers from this pilot study.
In summary, this method represents a small but significant step toward improved proteomic analysis of solid tumors. Demonstrated was the detection of tumor-residing proteins in the blood of a patient newly diagnosed with RCC. This method enabled an in-depth proteomic profiling of a single patient’s tumor for further elucidation of the molecular phenotype. Despite the fact that in this investigation only a single RCC tumor/patient was analyzed the totality of findings has identified a panel of putative protein markers, which now may be further investigated with high-throughput techniques employing suitable immunoassays (i.e. ELISA) or MS-assays (i.e. MRM) to test thir general applicability in larger RCC patient cohorts. Finally, the concept contained within this method may accelerate the transformation of oncology from the current paradigm of categorically assigned cancer treatments derived from population-based statistics (survival functions) to an evidence-based, personalized individually-targeted treatments derived by the rational molecular profiling of an actual patient’s tumor.
Supplementary Material
Acknowledgments
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the United States Government.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
SUPPORTING INFORMATION AVAILABLE
Supporting Information (SI), as noted in text is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hanash SM, Pitteri SJ, Faca VM. Nature. 2008;452:571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 2.Liotta LA, Kohn EC, Petricoin EF. Jama. 2001;286:2211–2214. doi: 10.1001/jama.286.18.2211. [DOI] [PubMed] [Google Scholar]
- 3.Anderson NL. Mol Cell Proteomics. 2005;4:1441–1444. doi: 10.1074/mcp.I500001-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Rifai N, Gillette MA, Carr SA. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 5.Johann DJ, Rodriguez-Canales J, Mukherjee S, Prieto DA, Hanson JC, Emmert-Buck M, Blonder J. J Proteome Res. 2009;8:2310–2318. doi: 10.1021/pr8009403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blonder J, Chan KC, Issaq HJ. Nat Protoc. 2006;1:2783–2790. doi: 10.1038/nprot.2006.359. [DOI] [PubMed] [Google Scholar]
- 7.Liotta LA, Kohn EC. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 8.Sandberg R, Ernberg I. Proc Natl Acad Sci U S A. 2005;102:2052–2057. doi: 10.1073/pnas.0408105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tunuguntla HS, Jorda M. J Urol. 2008;179:2096–2102. doi: 10.1016/j.juro.2008.01.083. [DOI] [PubMed] [Google Scholar]
- 10.Goldhirsch A, Ingle JN, Gelber RD, Coates AS, Thurlimann B, Senn HJ. Annals of Oncology. 2009 doi: 10.1093/annonc/mdp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johann DJ, Jr, Blonder J. Expert Rev Mol Diagn. 2007;7:473–475. doi: 10.1586/14737159.7.5.473. [DOI] [PubMed] [Google Scholar]
- 12.Akber SF. Am J Clin Oncol. 2000;23:345–348. doi: 10.1097/00000421-200008000-00006. [DOI] [PubMed] [Google Scholar]
- 13.Blonder J, Conrads TP, Yu LR, Terunuma A, Janini GM, Issaq HJ, Vogel JC, Veenstra TD. Proteomics. 2004;4:31–45. doi: 10.1002/pmic.200300543. [DOI] [PubMed] [Google Scholar]
- 14.Mann M, Kelleher NL. Proc Natl Acad Sci U S A. 2008 [Google Scholar]
- 15.Schirmer EC, Florens L, Guan T, Yates JR, 3rd, Gerace L. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- 16.Liu H, Sadygov RG, Yates JR., 3rd Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 17.Shimazui T, Giroldi LA, Bringuier PP, Oosterwijk E, Schalken JA. Cancer Res. 1996;56:3234–3237. [PubMed] [Google Scholar]
- 18.Hegele A, Varga Z, Kosche B, Stief T, Heidenreich A, Hofmann R. Urol Int. 2003;70:55–58. doi: 10.1159/000067707. [DOI] [PubMed] [Google Scholar]
- 19.Vasselli JR, Shih JH, Iyengar SR, Maranchie J, Riss J, Worrell R, Torres-Cabala C, Tabios R, Mariotti A, Stearman R, Merino M, Walther MM, Simon R, Klausner RD, Linehan WM. Proc Natl Acad Sci U S A. 2003;100:6958–6963. doi: 10.1073/pnas.1131754100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin TA, Watkins G, Lane J, Jiang WG. Histopathology. 2005;46:422–430. doi: 10.1111/j.1365-2559.2005.02104.x. [DOI] [PubMed] [Google Scholar]
- 21.Causevic M, Hislop RG, Kernohan NM, Carey FA, Kay RA, Steele RJ, Fuller-Pace FV. Oncogene. 2001;20:7734–7743. doi: 10.1038/sj.onc.1204976. [DOI] [PubMed] [Google Scholar]
- 22.Rayala SK, den Hollander P, Manavathi B, Talukder AH, Song C, Peng S, Barnekow A, Kremerskothen J, Kumar R. J Biol Chem. 2006;281:19092–19099. doi: 10.1074/jbc.M600021200. [DOI] [PubMed] [Google Scholar]
- 23.Denslow SA, Wade PA. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 24.Yang SH, Seo MY, Jeong HJ, Jeung HC, Shin J, Kim SC, Noh SH, Chung HC, Rha SY. Clin Cancer Res. 2005;11:612–620. [PubMed] [Google Scholar]
- 25.Vincent L, Kermani P, Young LM, Cheng J, Zhang F, Shido K, Lam G, Bompais-Vincent H, Zhu Z, Hicklin DJ, Bohlen P, Chaplin DJ, May C, Rafii S. J Clin Invest. 2005;115:2992–3006. doi: 10.1172/JCI24586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.