Summary
Hepatocyte nuclear factor 4α (HNF4α) is essential for liver development and hepatocyte function. Here, we show that transient inhibition of HNF4α initiates hepatocellular transformation through a microRNA-inflammatory feedback loop circuit consisting of miR-124, IL6R, STAT3, miR-24 and miR-629. Moreover, we show that once this circuit is activated, it maintains suppression of HNF4a and sustains oncogenesis. Systemic administration of miR-124, which modulates inflammatory signaling, prevents and suppresses hepatocellular carcinogenesis by inducing tumor-specific apoptosis without toxic side-effects. As we also show that this HNF4α circuit is perturbed in human hepatocellular carcinomas, our data raise the possibility that manipulation of this microRNA feedback-inflammatory loop has therapeutic potential for treating liver cancer.
Keywords: HNF4α, microRNA, inflammation, feedback loop, liver cancer
Introduction
Hepatocellular carcinoma (HCC) is the main type of liver cancer and the third most common cause of cancer mortality worldwide. The major risk factor for HCC is chronic hepatitis, due to hepatotropic viruses (HBV, HCV) (El-Serag and Rudolph, 2007), but the molecular mechanisms leading to HCC have not been well characterized. Hepatocellular carcinogenesis involves many genetic and epigenetic alterations and is influenced by environmental factors. Genes such as c-myc, cyclin D1, p53, p16, E-cadherin and PTEN have been linked to hepatocarcinogenesis (Villanueva et al., 2007). Persistent inflammation also impacts the course of liver tumor development (Coussens and Werb, 2002), and chronic inflammatory stimuli and increased STAT3 activation recapitulate hepatic oncogenesis in various animal models (He et al., 2010). In addition, the inflammatory responses induced by obesity or administration of the diethylnitrosamine (DEN) are known to promote HCC in mice promote HCC (Park et al., 2010; Maeda et al., 2005).
HNF4α is a member of the nuclear receptor superfamily of ligand-dependent transcription factors (NR2A1) this is enriched in liver tissue (Zhong et al., 1993). HNF4α is indispensable for development and maintenance of the hepatic epithelium (Parviz et al., 2003) and also has links to a variety of human diseases including diabetes, colitis, and cancer. A number of mutations within the HNF4A gene are considered to contribute to several forms of maturity-onset diabetes in children (Gupta and Kaestner, 2004). Suggesting a potential link between HNF4α and inflammation, genome-wide association studies have identified HNF4A as a susceptibility locus for ulcerative colitis (Barrett et al., 2009), and recent evidence supports an oncogenic role for HNF4α in intestinal cancer (Darsigny et al., 2010), but conflicting reports have assigned HNF4α both tumor promoting and tumor suppressing roles in liver cancer (Xu et al., 2001; Yin et al., 2008).
Here we show that HNF4α is a key regulator of the hepatocellular carcinogenesis. During hepatocellular transformation, transient inhibition of HNF4α becomes a stable event, with a feedback loop consisting of miR-124, IL6R, STAT3, miR-24 and miR-629 maintaining of the hepatocyte transformed phenotype in vitro and in vivo. Perturbation of this network, through miR-124 systemic administration, prevents and suppresses HCC development in a murine liver cancer model. Components of the HNF4α feedback loop circuit are differentially expressed in human hepatocellular carcinomas relative to normal liver tissues. While the epigenetic switch described here resembles an epigenetic switch that converts a non-transformed breast cell line into a stably transformed line in that relies on an inflammatory feedback look involving STAT3 (Iliopoulos et al., 2009a; Iliopoulos et al., 2010), the microRNA, transcription factors, and target genes mediating these epigenetic switches differ considerably in the breast and liver contexts. Overall, our data suggest that epigenetic switches are regulatory events essential for cancer initiation and maintenance in addition to mutational events.
Results
Transient inhibition of HNF4α induces hepatocellular oncogenesis
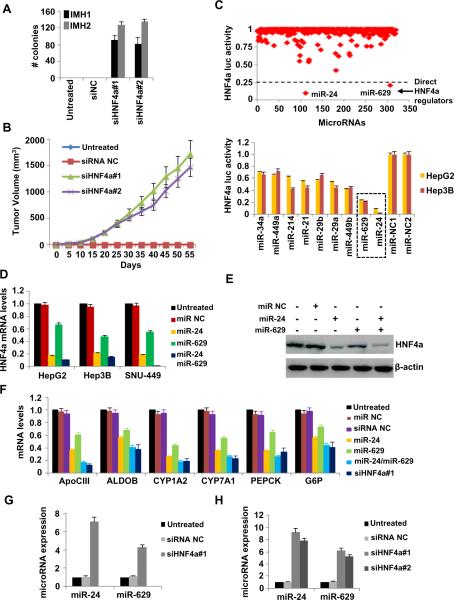
To elucidate the role and function of HNF4α in liver cancer initiation, we modulated its expression in non-transformed immortalized human hepatocytes (IMH). We found that HNF4α inhibition transformed IMH cells and increased their invasiveness (Figures 1A–B, and S1A–C). Strikingly, transient inhibition of HNF4α was sufficient to induce transformation of IMH cells and promote tumor formation in immunodeficient mice (Figures 1A, B). In these tumors (day 55), HNF4α mRNA expression was still suppressed (Figure S1D), suggesting that inhibition of HNF4α initiates a feedback loop that continuously suppresses HNF4α expression and induces a stable transformed phenotype. In accordance with the data from our primary IMH cells, transient inhibition of HNF4α increased colony formation and invasiveness of HepG2 and SNU-449 cancer cells (Figure S1E, F) and decreased expression levels of HNF4a direct metabolic target genes (Figure S1G). Overall, these data suggest that HNF4α inhibition induces transformation of immortalized hepatocytes through a feedback regulatory mechanism.
Figure 1.
HNF4α suppression through miR-24 and miR-629 induce hepatocellular transformation. (A) Soft-agar colony assay of non transformed immortalized hepatocytes (IMH1, IMH2) treated for 48h with siRNA negative control (siNC) or two different siRNAs against HNF4α (siHNF4α#1, siHNF4α#2). Colonies (mean ± SD) 50 μm were counted using a microscope 20 days later. (B) Tumor volume (mean ± SD) in mice injected with IMH1 cells untreated or treated for 48h with siRNA NC, siHNF4α#1 or siHNF4α#2. (C) Effects of microRNAs (primary screen) on HNF4α luciferase activity in HepG2 cells (top panel). The top 9 hits identified from the primary microRNA library screen, were tested in secondary screen in HepG2 and Hep3B cells (bottom panel). (D) HNF4α mRNA levels (mean ± SD of three independent experiments) assessed by real-time RT-PCR analysis in HepG2, Hep3B and SNU-449 cells untreated or treated with 100nM miR NC or miR-24 and/or miR-629 for 48h. (E) HNF4α protein levels in HepG2 cells untreated or treated with 100nM miR NC or miR-24 and/or miR-629 for 48h. (F) mRNA levels of HNF4α direct targets (mean ± SD of three independent experiments) assessed by real-time RT-PCR analysis in HepG2 cells untreated or treated with 100nM miR NC or miR-24 and/or miR-629 or siRNA NC or siRNA or siHNF4α#1 for 48h. (G) miR-24 and miR-629 expression levels (mean ± SD of three independent experiments) assessed by real-time RT-PCR analysis in IMH1 cells that were untreated or treated for 48h with siRNA NC or siHNF4α#1. (H) miR-24 and miR-629 expression levels (mean ± SD of three independent experiments) assessed by real-time RT-PCR analysis in tumors derived from injected IMH1 cells that were untreated or treated for 48h with siRNA NC or siHNF4α#1 or siHNF4α#2.
Mir-24 and miR-629 suppress directly HNF4α expression during hepatocellular transformation
How is HNF4α suppression triggered and maintained during hepatocellular transformation? Recently, we and others have described the existence of dynamic microRNA-transcription factor networks in a variety of cancers (Iliopoulos et al., 2009a; Iliopoulos et al., 2010; Kent et al., 2010). To identify microRNAs that regulate directly HNF4α expression we performed a microRNA-based genetic screen (Figure 1C, top panel). MicroRNAs that inhibited HNF4α 3'UTR luciferase activity by more than 75% were scored as positive hits. These were further validated in HepG2 and Hep3B cells, seeded in 6-well plates, according to the same criteria (Figure 1C, bottom panel). Our approach resulted in the identification of two microRNAs, miR-24 and miR-629 as direct regulators of HNF4α expression.
Several lines of evidence indicate that miR-24 and miR-629 target HNF4α directly, binding to its 3'UTR. Sequence complementarity analysis revealed that HNF4α is a gene target of miR-24 and miR-629, and upon overexpression of miR-24 or miR-629, HNF4α mRNA levels are reduced 5-fold and 2-fold, respectively (Figure 1D). In addition, HNF4α protein levels drop (Figure 1E), and the direct downstream targets are down-regulated by miR-24 and miR-629 (Figure 1F). In addition, combined expression of these two miRNAs resembles the effects of HNF4α knockdown (Figure 1F).
Transient inhibition of HNF4α by siRNA resulted in up-regulation of both miR-24 and miR-629 in IMH cells (Figure 1G). We also identified increased expression of miR-24 and miR-629 in tumors derived from IMH cells treated with two different siRNAs against HNF4α (Figure 1H). Taken together, these data suggest that both microRNAs regulate directly HNF4α expression and are part of the feedback loop circuit.
Mir-24 and miR-629 play a key role in hepatocellular cancer initiation and growth
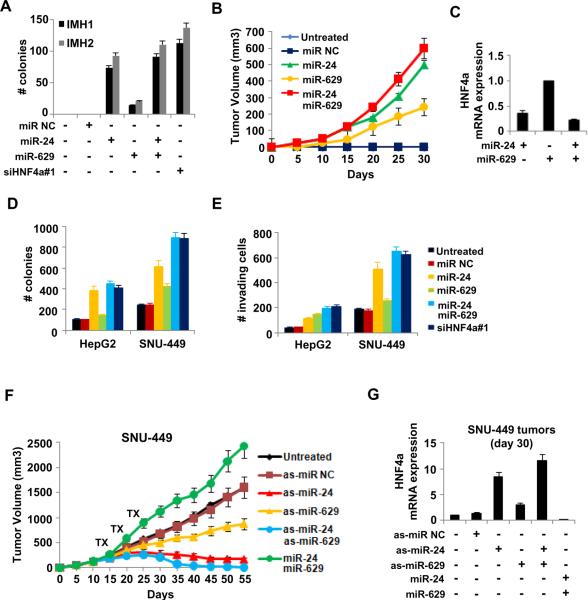
To assess the functional role of miR-24 and miR-629 in tumorigenicity we tested whether their overexpression can transform two distinct immortalized hepatocyte cell lines. Expression of miR-24 and/or miR-629 is sufficient for hepatocellular transformation and colony formation in soft agar (Figure 2A). While miR-24 has a stronger effect than miR-629, the combination of the two microRNAs closely resembles HNF4α knockdown. The ability of miR-24 or miR-629 to induce transformation in vitro, led us to extend our results and examine their ability to regulate tumor initiation in vivo. Overexpression of miR-24 or miR-629, to a lesser extent, was sufficient for the induction of tumor initiation and growth (Figure 2B). These observations indicate that transient expression of either miR-24 or miR-629 is sufficient to induce stable transformation of hepatocytes in vitro and in vivo. Reduced HNF4α expression in miR-24/miR-629 treated tumors (Figure 2C) also indicates that both microRNAs cooperatively suppress HNF4α expression inducing a stable transformed state.
Figure 2.
MiR-24 and miR-629 regulate the induction and stability of the hepatocellular transformed phenotype. (A) Number of colonies (>50 μm) (mean ± SD) of IMH1 and IMH2 cells treated with 100nM miR NC, miR-24 and/or miR-629 or siHNF4α#1 for 48h. (B) Tumor volume (mean ± SD) in mice injected with IMH1 cells untreated or treated for 48h with 100nM miR NC or miR-24 and/or miR-629. (C) HNF4α mRNA levels assessed by real-time RT-PCR analysis in tumors (day 30) derived from IMH1 cells untreated or treated for 48h with 100nM miR-24 and/or miR-629. (D) Soft-agar colony assay (mean ± SD) and (E) invasion assay (mean ± SD) of HepG2 and SNU-449 cells treated with 100nM miR NC, miR-24, miR-629 or siHNF4α#1 for 24h. (F) Tumor volume (mean ± SD) in mice injected with SNU-449 cells and treated with as-miR NC or as-miR-24 and/or as-miR-629, or miR-24 and miR-629. (G) HNF4α mRNA levels (mean ± SD) in tumors (day 30) derived from mice treated with as-miR NC or as-miR-24 and/or as-miR-629 or miR-24 and miR-629.
To address the functional role of miR-24 and miR-629 in the maintenance of the transformed phenotype, we tested the effects of their up-regulation on the tumorigenicity of hepatocellular cancer cells. Overexpression of miR-24 or miR-629 in HepG2 and SNU-449 cells increased their ability to form colonies (Figure 2D) and their invasive capacity (Figure 2E). As expected, combination of the two microRNAs exhibited the same effects with HNF4α inhibition. To delineate the role of miR-24 and miR-629 in HCC growth in vivo, we performed xenograft experiments in which SNU-449 cells were injected subcutaneously in immunodeficient mice (Figure 2F). We found that overexpression of miR-24 and miR-629 increased the growth of SNU-449 xenograft tumors (Figure 2F), while simultaneous inhibition of both microRNAs completely suppressed tumor growth. Are the effects of miR-24 and miR-629 on tumor growth related to HNF4α expression? We tested HNF4α mRNA levels in xenograft tumors (day 30) from the same mice, as described above. Tumors treated with the antisense microRNAs are smaller, contain many apoptotic cells (Figure S2) and exhibit elevated HNF4α mRNA levels (Figure 2G).
STAT3 is a direct regulator of miR-24 and miR-629 expression
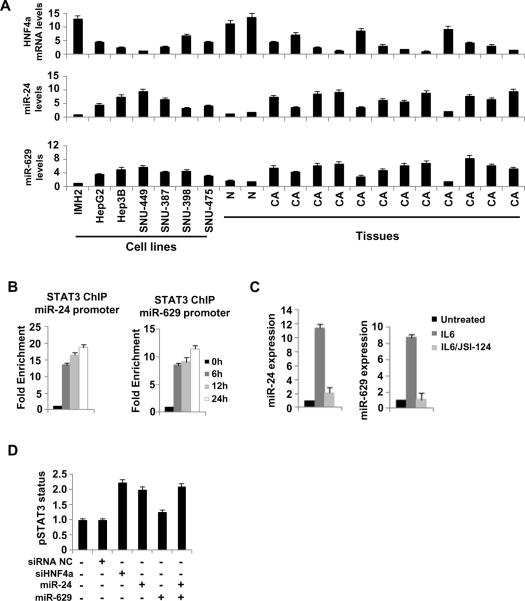
According to our data, both miR-24 and miR-629 directly suppress HNF4α expression and they are activated by inhibition of HNF4α expression in hepatocytes as part of the feedback loop circuit. We found that miR-24 and miR-629 are coordinately up-regulated in both hepatocellular cell lines and human tumors (Figure 3A). Examination of potential common transcription factor binding sites in miR-24 and miR-629 promoter areas revealed a highly conserved STAT3 binding motif in miR-24 promoter and a moderately conserved STAT3 motif in miR-629 promoter (Table S1). Chromatin immunoprecipitation (ChIP) analysis in SNU-449 cells revealed that upon IL6 stimulation, STAT3 binds in miR-24 and miR-629 promoter regions, with binding to the highly conserved miR-24 site being stronger (Figure 3B). STAT3 activation by IL6 treatment resulted in up-regulation of both miR-24 and miR-629 levels, while pharmacological inhibition of STAT3 (JSI-124) strongly reduced miR-24 and miR-629 expression levels (Figure 3C).
Figure 3.
STAT3 regulates miR-24 and miR-629 during hepatocellular transformation. (A) HNF4α, miR-24 and miR-629 levels in non-transformed immortalized hepatocytes (IMH2), different HCC lines, 2 normal liver tissues (N) and 12 hepatocellular cancer tissues (CA). (B) STAT3 occupancy (fold enrichment) at the miR-24 and miR-629 loci as determined by chromatin immunoprecipitation of cross-linked SNU-449 cells treated with IL6 (20ng/ml) for 6, 12 or 24h. (C) miR-24 and miR-629 expression levels (mean ± SD) in SNU-449 cells treated with IL6 (10ng/ml) for 24h or JSI-124 (5μg/ml) for 24h and then IL6 for 24h. (D) STAT3 phosphorylation status (Tyr 705) assessed by ELISA in SNU-449 cells treated with 1nM siRNA NC, siHNF4α#1, miR-24 and/or miR-629 for 24h. The data are presented as mean ± SD of three independent experiments.
To determine whether STAT3 is a member of the HNF4α feedback loop circuit we measured STAT3 phosphorylation levels upon overexpression of miR-24 and/or miR-629 or inhibition of HNF4α in SNU-449 cells (Figure 3D). Strikingly, all treatments significantly induced STAT3 phosphorylation when compared to the negative control samples. In accordance with our data above, miR-24 had a more pronounced effect (compared to miR-629) similar with that of HNF4α knockdown and the combinatorial expression of the two microRNAs. These results strongly suggest that these microRNAs, STAT3 and HNF4α are part of an inflammatory feedback loop and not simply downstream effectors of IL6.
MiR-124 is a direct downstream effector of HNF4α activity and part of the feedback loop network
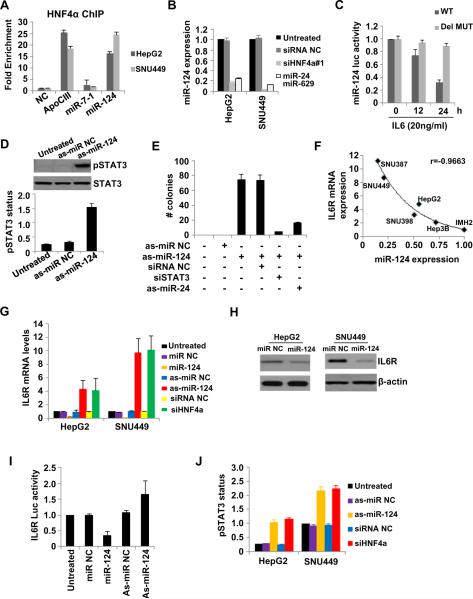
Recent studies have identified microRNA-transcription factor feedback loops in cancer cells (Fabbri et al., 2011; Iliopoulos et al., 2010). To further unravel the mechanism by which inhibition of HNF4α expression induces hepatocellular transformation through a feedback loop, we looked for HNF4a binding sites in miRNA promoters. Lever algorithm analysis revealed HNF4α binding sites in eight microRNA promoter areas (Table S2). ChIP analysis showed that HNF4α binds strongly (15- to 25-fold enrichment) to miR-124 promoter in HepG2 and SNU-449 cells (Figure 4A), and inhibition of HNF4α expression resulted in significant reduction of miR-124 levels (~5 fold) (Figure 4B). Similarly, miR-124 expression is significantly inhibited upon the combined overexpression of miR-24 and miR-629 and this inhibition is comparable with the one caused by HNF4α knockdown. Based on our observation of an inverse correlation between STAT3 activation and HNF4α expression, we examined how IL6 treatment influenced activity of a luciferase reporter construct containing the miR-124 promoter (Figure 4C). Treatment of HepG2 cells with IL6 significantly inhibited the activity of the miR-124 luciferase reporter, while there was not effect when the HNF4α site was mutated.
Figure 4.
HNF4α binds and regulates miR-124 which controls directly IL6R expression in hepatocytes. (A) HNF4α occupancy (fold enrichment) in ApoCIII, miR-7-1 and miR-124 promoter areas. (B) miR-124 levels (mean ± SD) in HepG2 and SNU-449 treated with siRNA NC, siHNF4α#1, miR-24 and miR-629 for 24h. (C) Luciferase activity (mean ± SD) of a reporter construct harboring miR-124 promoter (wild type or deletion mutant in the HNF4α binding site) 12 and 24h post treatment with IL6 (10ng/ml) in HepG2 cells. (D) STAT3 phosphorylation status (Tyr 705) (mean ± SD) evaluated by ELISA and western blot analyses after treatment with as-miR-NC or as-miR-124 for 24h in HepG2 cells. (E) Number of colonies (mean ± SD) of non-transformed immortalized hepatocytes (IMH1) treated with as-miR NC or as-miR-124 together with siRNA NC or siRNA against STAT3 (siSTAT3) or as-miR-24 for 24h. The data are presented as mean ± SD of three independent experiments. (F) miR-124 and IL6R levels in the indicated cell lines and a correlation coefficient (r) is shown. (G) IL6R mRNA levels (mean ± SD) in HepG2 and SNU-449 cells treated for 24h with miR-NC, miR-124, as-miR NC, as-miR-124, siRNA NC, siHNF4α. (H) IL6R protein levels in HepG2 and SNU-449 cells treated for 24h with miR-NC or miR-124. (I) Luciferase assay using a reporter construct containing the 3'UTR of IL6R, 24h after transfection with miR-NC, miR-124, as-miR NC, as-miR-124. The data are presented as (mean ± SD). (J) STAT3 phosphorylation status (Tyr 705) evaluated by ELISA in HepG2 and SNU-449 cells treated for 24h with as-miR NC, as-miR-124, siRNA NC and siHNF4α. The data are presented as mean ± SD of three independent experiments.
As HNF4α directly regulates miR-124 expression in HCC lines, we tested the possibility that miR-124 may mediate the HNF4α-regulated inhibition of STAT3. Interestingly, STAT3 activation was induced upon miR-124 suppression when compared to the respective negative controls (Figure 4D). The above experiments suggest that miR-124 participates also in the HNF4α feedback loop. To further show that miR-124 is a member of this loop, we examined IMH1 transformation efficiency upon inhibition of miR-124 expression. As expected, inhibition of miR-124 expression strongly induces colony formation and this effect is reversed by STAT3 knockdown or combined suppression of miR-24 expression (Figure 4E). Likewise, suppression of miR-124 or knockdown of HNF4α induces colony formation and invasiveness of HepG2 and SNU-449 cells, while overexpression of miR-124 in these cell lines reverses the phenotype (Figure S3). Taken together, these observations are consistent with a pathway in which STAT3 activation inhibits HNF4α expression which leads to suppressed expression of miR-124 and establishes an inflammatory feedback loop that is necessary and sufficient for human hepatocyte transformation.
MiR-124 targets IL6R and consequently modulates IL6R/STAT3 pathway during hepatocellular transformation
Because STAT3 activation is suppressed by miR-124, we hypothesized that miR-124 might target one of the components of the IL6-STAT3 pathway. In support of this hypothesis, sequence complementarity and conservation analysis revealed that interleukin 6 receptor (IL6R) is a potential direct gene target of miR-124. Furthermore, miR-124 and IL6R expression levels are inversely correlated in IMH1 cells and five hepatocellular cancer cell lines (Figure 4F). In addition, suppression of miR-124 expression, either directly by antisense miR-124 or indirectly by knockdown of HNF4α, leads to induced expression of IL6R (Figure 4G). Conversely, overexpression of miR-124 significantly reduced IL6R mRNA and protein levels (Figure 4G, H). Also, miR-124 overexpression inhibits the activity of a luciferase reporter construct containing the IL6R 3'UTR and vice versa (Figure 4I). Next, phosphorylation of STAT3, a downstream target of IL6R, is induced by inhibition of miR-124 expression or knockdown of HNF4α (Figure 4J). In addition to IL6R, we found that inhibition of miR-124 expression results in increased IL6 production (Figure S4A), suggesting that miR-124 regulates STAT3 activity by affecting the IL6-IL6R levels and pathway. Similar effects were identified when HNF4α was suppressed. Specifically, HNF4α inhibition resulted in increased levels of soluble IL6 and IL6R (Figure S4B, C), which, in turn, increased liver tumorigenicity (Figure S4D). These experiments support a central role for HNF4a in regulating the IL6-STAT3 inflammatory response.
The feedback loop involving HNF4α, miR-124, IL6R, STAT3, miR-24 and miR-629 is required for the induction and maintenance of the transformed phenotype in hepatocytes
To examine the dynamics of this circuit during the transformation of hepatocytes, IMH1 cells were transiently transfected with the respective microRNAs or siRNAs and 96–480h post-transfection were plated in soft agar and injected in mice (Figure S5A, B). Suppression of miR-124 or HNF4α or overexpression of miR-24 or miR-629 induced hepatocellular transformation. We also find that the kinetics of STAT3 activation along with expression levels of miR-124, miR-24 miR-629 and HNF4α demonstrate the establishment and maintenance of the regulatory loop even 480 h after transfection (Figure S5C–G). In addition to transcriptional activation, we show that suppression of HNF4α led to increased soluble IL6 and IL6R levels (Figure S5H), hepatocyte hyperproliferation and decreased apoptosis (Figure S6). On the other hand, breaking the regulatory circuit by manipulation of different members of the loop blocked the stable transformed phenotype of human hepatocytes (Figure S5I, J). Overall, these data indicate that HNF4α is a central regulator of hepatocyte growth and transformation.
HNF4α-miRNA inflammatory circuit is perturbed during HCC development in mice
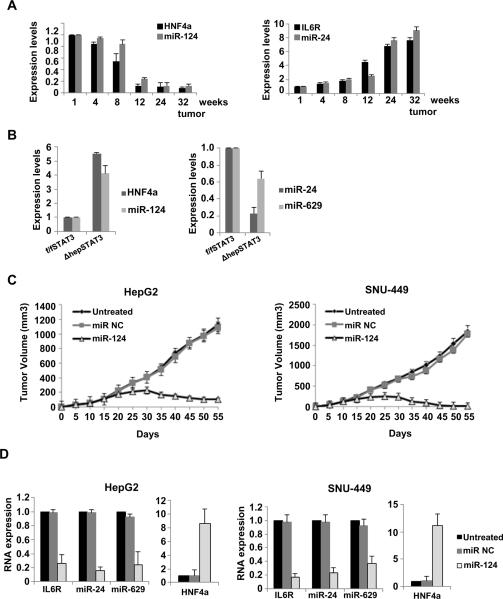
Building on our in vitro findings, we asked whether the HNF4α circuit is perturbed during development of chemical-induced hepatocellular carcinogenesis in vivo. To exclude the possibility that IL6/STAT3 pathway is activated by Kupffer cells, we examined the expression levels of HNF4α, miR-124, IL6R and miR-24 in purified hepatocytes derived from DEN-treated mice (Figure 5A). In accordance with our in vitro data, we identified that the HNF4α-miRNA circuit is perturbed in hepatocytes during HCC development in mice. Interestingly, HNF4α suppression started on week 4, while on the other hand miR-24 was up-regulated on week 24, when the tumors have already been formed. These data are consistent with the idea that early suppression of HNF4α leads to activation of the miRNA-inflammatory circuit during HCC development.
Figure 5.
The HNF4α circuit is perturbed during HCC development. (A) Assessment of HNF4α, miR-124, IL6R and miR-24 levels (mean ± SD) in purified hepatocytes during DEN-induced liver carcinogenesis in mice. (B) Evaluation of HNF4α mRNA levels and miR-124, miR-24 and miR-629 levels derived from DEN-treated male STAT3f/f and STAT3Δhep mice. The experiments have been performed in triplicate and data shown mean ± SD. (C) Tumor volume (mean ± SD) in mice injected with HepG2 and SNU-449 cells, treated with miR NC or miR-124. Treatments were repeated every five days and tumor volume was monitored every five days for 55 days. (D) IL6R, miR-24, miR-629 and HNF4α levels (mean ± SD) assessed by real-time RT-PCR analysis, in tumors (day 30) derived from mice treated with miR NC or miR-124.
In addition we tested if the HNF4α-miRNA circuit is perturbed in hepatocyte specific STAT3 deficient mice (STAT3f/f/Alb-Cre = STAT3Δhep). It is known that the DEN-treated STAT3Δhep mice develop less and much smaller tumors in comparison to the DEN-treated STAT3f/f mice (He et al., 2010). Consistent with our hypothesis, we identified that tumors derived from DEN-treated STAT3Δhep mice had increased HNF4α and miR-124 levels and decreased miR-24 and miR-629 levels in comparison to DEN-treated STAT3f/f mice (Figure 5B). These data show that suppression of the inflammatory response in vivo perturbs the HNF4α circuit, suggesting that this circuit can be affected at any step.
Perturbation of the HNF4α circuit has therapeutic and preventive effects in different murine liver cancer models
To further validate the in vivo significance of the HNF4α circuit, we asked how perturbation of this circuit would affect tumor growth in different HCC mouse models. Specifically, the inhibitory role of miR-124 on hepatocellular neoplastic transformation, suggested the possibility that HCC-derived tumors could be eradicated efficiently by interference with the feedback loop on the level of miR-124. We found that miR-124 treatment suppressed HepG2 and SNU-449 xenograft tumor growth (Figure 5C) by reducing IL6R, miR-24 and miR-629 expression levels and significantly increasing HNF4α expression (Figure 5D).
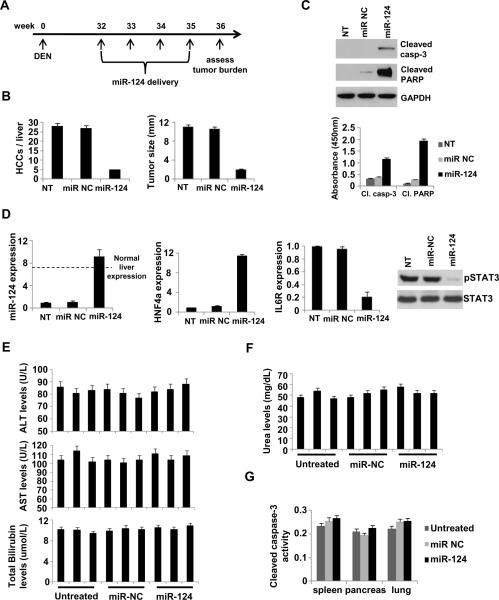
In addition to the subcutaneous HCC mouse model, we tested if systemic administration of miR-124 is able to suppress HCC tumor growth in DEN-treated mice. According to our treatment protocol, miR-NC or miR-124 was systemically administered in DEN-treated mice in a weekly basis (first day of the week) for 4 cycles (week 32, 33, 34, 35) (Figure 6A). On week 36 the mice were sacrificed and we assessed the tumor burden. We found that miR-124 suppressed >80% HCC tumor growth and size (Figure 6B) through induction of apoptosis (Figure 6C) and actually, miR-124 administration resulted in restoration of physiological miR-124 expression, while miR-NC administration did not have any effect (Figure 6D). In addition miR-124 delivery perturbed the HNF4α circuit, through up-regulation of HNF4α mRNA levels, IL6R suppression and inhibition of STAT3 activation (phosphorylation). Importantly, we found that systemic delivery of miR-NC or miR-124 did not affect liver and kidney function (Figure 6E, F) and did not have any toxicity effects on essential organs (Figure 6G). These data demonstrate that miR-124 administration does not affect the physiology of mice through induction of cytotoxic effects.
Figure 6.
Modulation of the HNF4α circuit prevents and suppresses HCC development in mice. (A) Timeline of miR-124 therapeutic delivery experiment. (B) Number of HCC tumors/liver and tumor size (mm3) in non-treated (NT), miR-NC and miR-124 treated mice (week 36). (C) Levels of cleaved PARP and caspase-3 in untreated, miR NC and miR-124 treated mice (week 36) assessed by ELISA and western blot analyses. (D) Evaluation of miR-124 levels, HNF4α levels, IL6R levels by real-time PCR and STAT3 phosphorylation status (Tyr705) by western blot, in tumors derived from non-treated (NT), miR-NC and miR-124 treated mice (week 36). The data are shown as mean ± SD. (E) Levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bilirubin and (F) of urea were assessed in the serum of mice treated with 10mg/kg miR-NC or miR-124 for 48h. Each bar represents a different mouse. The experiment was performed in triplicate and the data show mean ± SD. (G) Cleaved Caspase-3 activity (mean ± SD) assessed by ELISA assays in tissues (spleen, pancreas, heart) derived from untreated, miR-NC or miR-124 treated mice.
In addition to therapeutic effects, we examined whether perturbation of the HNF4α circuit can prevent HCC development in mice. We identified that miR-124 delivery restored the physiological levels of this microRNA in liver tumors, even two weeks post injection (Figure S7A). According to these data, miR-124 was administered systemically on week 12, every two weeks until week 30 and at week 32, we assessed tumor burden (Figure S7B). We found that miR-124 delivery prevented efficiently HCC tumor growth in DEN-treated mice (Figure S7C), suggesting that the HNF4α-miRNA inflammatory circuit is essential for HCC development in vivo.
The HNF4α regulatory circuit is perturbed in human HCC tissues
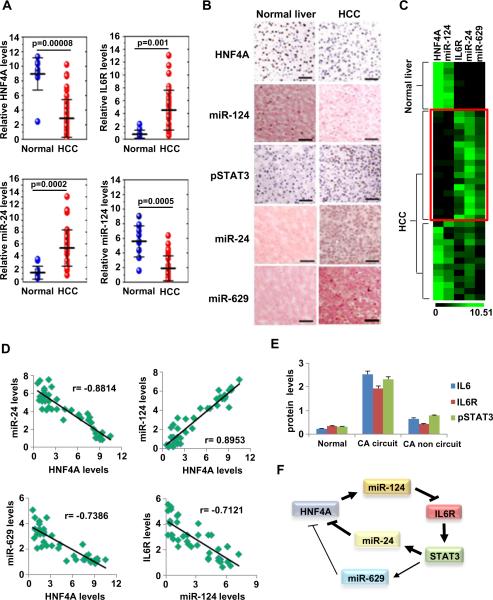
We examined the expression levels of miR-24, IL6R, miR-124 and HNF4α in total RNA extracted from 12 normal liver tissues and 45 hepatocellular carcinomas (HCCs). We found that HNF4α and miR-124 were down-regulated, while miR-24 and IL6R mRNA levels were increased in liver cancers relative to normal tissues (Figure 7A). In addition, immunohistochemical (IHC) analysis for HNF4α and phosphorylated STAT3 and in situ hybridization for miR-124, miR-24 and miR-629 revealed that in 13/30 (43.3%) of HCC tumors the circuit is perturbed (Figure 7B).
Figure 7.
HNF4α circuit is perturbed in human hepatocellular carcinomas. (A) Assessment of HNF4α, IL6R, miR-24 and miR-124 levels (mean ± SD) by real-time PCR analysis in total RNAs derived from 12 normal liver tissues and 45 hepatocellular carcinomas. (B) Immunohistochemistry for HNF4α, pSTAT3 and in situ hybridization for miR-124 and miR-24 in FFPE sections of hepatocellular carcinomas and normal liver tissues. Sections were subjected to immunohistochemistry for HNF4α (DAB staining, brown color) and phospho-STAT3 (Tyr705) (DAB staining) and counterstained with haematoxylin (blue color) and in situ hybridization for miR-124, miR-24, and miR-629 and counterstained with nuclear fast red. Bar, 100 μm. (C) Heatmap representation of HNF4α, IL6R, miR-24 and miR-124 levels assessed by real-time PCR (mean ± SD) in tissue-microdissected FFPE sections of 8 normal liver tissues and 31 hepatocellular carcinomas. (D) Correlation between the expression levels of different members of the HNF4α circuit (same samples as in figure 7C). Each data point represents an individual liver tissue sample and a correlation coefficient (r) is shown. (E) Levels of IL6, IL6R and pSTAT3 (Tyr705) assessed by ELISA, in 8 normal liver tissues, 31 hepatocellular carcinomas [18 tissues with activation of the HNF4α circuit (CA circuit) and 13 liver cancer tissues without activation of the HNF4α circuit (CA non circuit)]. The data are presented as mean ± SD of three independent experiments. (F) Schematic representation of the proposed HNF4α feedback circuit in hepatocellular oncogenesis.
Due to the fact that our in vitro data suggest that activation of an inflammatory response through suppression of HNF4α levels is cell autonomous, we examined the activation of the inflammatory circuit in the absence of Kuppfer cells. We tested expression levels of each member of the HNF4α circuit in RNA samples derived from laser capture microdissected hepatocytes, which were negative for CD45 expression. Specifically, in all (8/8) human normal liver tissues we found high HNF4α and miR-124 levels and low IL6R, miR-24 and miR-629 levels. On the other hand, we identified that the HNF4α circuit is perturbed (HNF4α and miR-124 low levels; IL6R, miR-24 and miR-629) in 18/31 of human hepatocellular carcinomas (Figure 7C). Furthermore, in the same samples we tested if there is any correlation between the RNA expression levels of the different members of this circuit. We found an inverse correlation between HNF4α and miR-24 or miR-629 levels, an inverse correlation between miR-124 and IL6R levels and a positive correlation between HNF4α and miR-124 levels (Figure 7D). Also, in the same human tissue samples, we examined IL6 and IL6R protein levels and STAT3 phosphorylation status and identified that the HCC samples (n=18) with perturbed HNF4α circuit have higher levels in comparison to the HCC samples (n=13) with non perturbed HNF4α circuit or normal liver tissues (n=8) (Figure 7E).
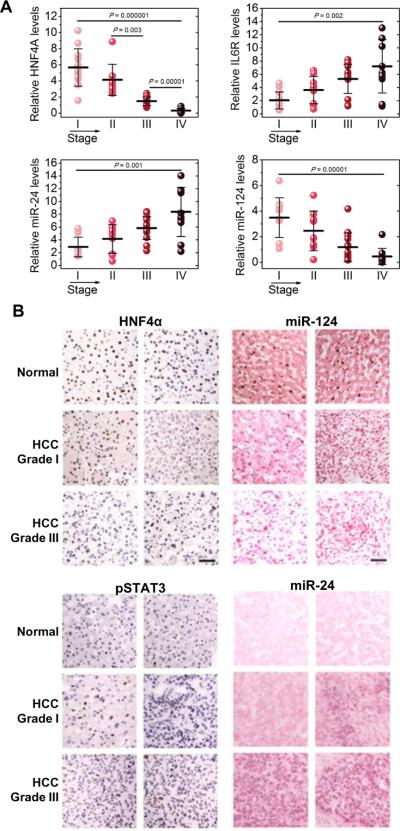
Furthermore, we were interested in identifying if the HNF4α circuit is perturbed not only during liver cancer initiation but also during liver cancer progression. Thus, we examined the mRNA expression levels of the different members of the circuit in different stages of HCC oncogenesis. We found that HNF4α and miR-124 levels were decreased, while IL6R and miR-24 levels were increased during HCC progression (Figure 8A). Interestingly, the activity of this circuit correlated to HCC grade (Figure 8B). Overall, these data strongly suggest that in addition to tumor initiation, the HNF4α-miRNA inflammatory feedback circuit is important for the progression of human cancer.
Figure 8.
HNF4α circuit is perturbed during HCC progression. (A) Assessment of HNF4α, IL6R, miR-24 and miR-124 levels in total RNAs derived from 45 hepatocellular carcinomas, according to their tumor stage. The experiments have been performed in triplicate and data are shown as mean ± SD. (B) HCC sections were subjected to immunohistochemistry for HNF4α and phospho-STAT3 (Tyr705) (DAB staining, brown) and counterstained with haematoxylin (blue) and in situ hybridization for miR-124 and miR-24 and counterstained with nuclear fast red. Representative pictures are shown from normal, HCC grade I and HCC grade III tissues. Bar, 50μm.
Discussion
An HNF4α Circuit is essential for the Transformation of Immortalized Hepatocytes
Our data reveal the dynamics of a complex molecular self-reinforcing circuit that involves HNF4α, miR-124, IL6R, STAT3 and miR-24/miR-629 in the regulation of hepatocellular transformation and liver cancer (Figure 7F). The first component of the circuit links HNF4α to STAT3 activation, with HNF4α controlling IL6R expression through transcriptional regulation of miR-124. Although, miR-124 has been identified as a cancer-associated tumor suppressive microRNA (Lujambio et al., 2007) its regulation and mode of action has been elusive. Here, we show that HNF4α binding and transcriptional regulation of miR-124 is comparable to the bona fide HNF4α target ApoCIII (Kardassis et al., 1997; Ladias et al., 1992). The second component of the circuit connects STAT3 activity to HNF4α expression via regulation of miR-24 and miR-629. Perturbations of the STAT3-HNF4A axis interfere with processes that govern hepatic transformation and oncogenesis, mechanistically linking inflammation and liver cancer.
An Epigenetic Switch regulates Hepatocyte Transformation
The main characteristic of the HNF4α feedback circuit is that it transforms immortalized human hepatocytes by converting a transient signal (e.g., acute HNF4α inhibition) into a stable signal. Overexpression of any positive factor (miR-24, miR-629) or inhibition of any negative factor (HNF4α, miR-124) transforms immortalized hepatocytes, indicating that the loop can be affected at any step. Thus, the initiating event in different HCC mouse models and patients can be different. It is not necessary that the loop begins with reduction of HNF4α. According to our data, suppression of HNF4α expression is the first event in DEN-treated mice, followed by perturbation of the other members of the loop. In other scenarios, the IL6-STAT3 axis may activate the loop by different extracellular stimuli. Specifically, secreted IL6 from different immune cells in the tumor microenvironment, including Kupffer cells, could initiate this axis. For example, recent studies show that IL-22, a cytokine secreted by Th17 cells, controls hepatocellular oncogenesis via up-regulation of STAT3 activity (Jiang et al., 2011) and hepatitis C viral infection can promote STAT3 activation (Tacke et al., 2011). Interestingly, miR-124 has been found epigenetically silenced, through tumor-specific methylation, both in human HCC cell lines and tissues (Furuta et al., 2010), and therefore miR-124 down-regulation may be the first event that triggers hepatic carcinogenesis. Together, all these data suggest that the initial event that activates this circuit could differ from patient to patient.
Due to the fact that the epigenetic switch in immortalized hepatocytes occurs within a few days, it is extremely unlikely to involve changes in the DNA sequence, which is consistent with the definition of a true epigenetic switch (Ptashne, 2009). This notion of a self-reinforcing feedback loop which controls hepatocellular transformation comes in line with our previous observation of an epigenetic switch which mediated transformation of immortalized mammary epithelial (MCF10A) cells to a stably transformed cell (Iliopoulos et al., 2009a; Iliopoulos et al., 2010). Furthermore, the identification of an epigenetic switch in hepatocellular transformation indicates that it is not a rare mechanism involved in cellular transformation and supports the possibility that cancer cells of diverse developmental origin might share a common mechanism for the establishment of the transformed state.
Our data also suggest that microRNA-transcription factor regulatory circuits mediate epigenetic switches that induce transformation of immortalized cells. Recent reports posit that transcriptional (TFs) and non-transcriptional elements (microRNAs) may cooperate to tune gene expression in various biological processes (Chen et al., 1994; Gerstein et al., 2010; Martinez et al., 2008), including oncogenesis (Fabbri et al., 2011; Kent et al., 2010). Various network motifs have been proposed, but miRNA-TF feed-forward and feedback loops predominate. For example, a feed-forward regulatory circuit (KRAS-miR-143/miR-145) plays an essential role in pancreatic tumorigenesis (Kent et al., 2010). Taken together these observations demonstrate that transcription factors participate in similar circuits that regulate induction and maintenance of stable transformation programs, suggesting that use of analogous regulatory loops may be a widespread property of oncogenic processes.
Role of HNF4α and its Downstream Effectors in Hepatocellular Oncogenesis
HNF4α has long been considered a key transcription factor during liver embryonic development (Kyrmizi et al., 2006; Parviz et al., 2003). In the adult liver, HNF4α is expressed at high levels and binds to the promoter of 12% of genes expressed (Odom et al., 2004). Nevertheless, HNF4α role in hepatocellular cancer and the mechanisms involved are far from clear. It has been shown that HNF4α is up-regulated in human hepatocellular carcinoma (Xu et al., 2001) and on the other hand impedes the formation of liver tumors in mice by inducing differentiation of malignant cells-including cancer stem cells-into mature hepatocytes (Yin et al., 2008). Recent findings that the Wnt/β-catenin pathway interacts with HNF4α in intestinal epithelial cells (Cattin et al., 2009) and hepatocytes (Colletti et al., 2009) strengthens the notion that HNF4α acts as a tumor-suppressor gene in both cancer types. Our study refines the repressive role of HNF4α in hepatic neoplasia, suggesting that HNF4α inhibition mediates an epigenetic switch essential for the transformation of immortalized hepatocytes.
Inflammation is one of the downstream mechanisms linking HNF4α to hepatocellular carcinogenesis. The protective action of HNF4α against inflammatory bowel diseases (Ahn et al., 2008; Darsigny et al., 2010) and the potential associations between the HNF4A locus and ulcerative colitis (Barrett et al., 2009) raise the possibility that this multifaceted transcription factor is a potent mediator of inflammatory responses (Marcil et al., 2010). Several studies have identified STAT3 as an oncogenic transcription factor activated by inflammatory responses (Bromberg et al., 1999; Grivennikov et al., 2009; Iliopoulos et al., 2009a; Iliopoulos et al., 2010) and IL6 is known to directly activate STAT3 (Zhong et al., 1994). STAT3 activity has been correlated with poor prognosis in HCC patients (Calvisi et al., 2006), and STAT3 inhibitors inhibit the growth of several human cancers (Hedvat et al., 2009), including HCC development and growth in mice (He et al., 2010).
As genetic alterations that result in constitutive STAT3 activation in hepatocytes only cause benign hepatic adenomas, unless combined with oncogenic mutations (Rebouissou et al., 2009), it will be important to discover the parameters that distinguish primary hepatocytes from non-transformed immortalized cells. Although a transient inflammatory signal is insufficient to trigger such an epigenetic switch in normal hepatocytes, our in vivo data suggest that the epigenetic switch described here is relevant to human cancer. The epigenetic switch requires that a transient inflammatory response is converted to a chronic inflammatory response, with no resolution phase but continuous enhancement of the inflammatory signal. Thus, the results presented here provide a paradigm in which a key step in transformation involves an epigenetic switch in response to an inflammatory signal as opposed to a mutational change in a tumor suppressor or oncogene. In support of this idea, recent data suggest that chronic activation of the IL6-STAT3 axis contributes to the transformation of hepatocytes that have acquired oncogenic mutations upon exposure to environmental and dietary carcinogens (Park et al., 2010).
Therapeutic and Preventive Effects of MiR-124 Delivery in Hepatocellular Carcinogenesis
We show that miR-124 administration restored miR-124 expression to physiological levels in the liver, inhibiting and preventing DEN-induced hepatocellular carcinogenesis in mice. Previous studies have aimed to suppress microRNA expression in animal models, through delivery of antagomiRs or locked nucleic acid (LNA) oligomers (Elmen et al., 2008; Esau et al., 2006). Few studies have investigated the therapeutic delivery of microRNAs in vivo. A recent study has shown that restoration of miR-26a expression levels by an adeno-associated virus (AAV) suppresses liver tumorigenesis in liver-specific MYC transgenic mice (Kota et al., 2009) without any cytotoxic effects.
Our data suggest that systemic delivery of miR-124 may be a clinically viable anticancer therapeutic approach for liver cancer. Delivery of microRNAs in the liver is more efficient in comparison to other tissues and a recent study revealed that delivery of the antisense-microRNA-122 suppressed hepatitis C viremia in primates, with no evidence of viral resistance or side effects (Lanford et al., 2010), leading to the initiation of phase I clinical trials in HCV-infected patients. This work lays the ground work for testing whether miR-124 can also exert tumor suppressive effects in human liver cancers. Together, our findings elucidate a molecular mechanism responsible for the initiation and maintenance of the hepatocyte transformed phenotype which enhances our understanding of liver cancer pathogenesis and provides a microRNA therapeutic strategy for prevention and treatment of liver cancer. While we have identified a novel molecular circuit that is essential for the transformation of hepatocytes and is found to be perturbed in different HCC models and in human hepatocellular carcinomas, significant work remains to identify the driver signaling pathways involved in hepatocellular carcinogenesis.
Experimental Procedures
Cell Culture
Human non-transformed immortalized hepatocytes (IMH1, IMH2) were purchased from ATCC (cat no. CRL-4020) and from Xenotech LLC (cat. no. IFH15), respectively. Detailed description of the origin of these immortalized hepatocytes and their culture conditions could be found in the supplementary experimental procedures. In addition, human liver cancer cell lines (HepG2, Hep3B, SNU-449, SNU-398 and SNU-387) were purchased from ATCC. Human liver cancer cell lines HepG2 and Hep3B were maintained in DMEM medium (Gibco) supplemented with 10% FBS and 10 units/ml penicillin, and 100 μg/ml streptomycin. SNU-449, SNU-398 and SNU-387 were maintained in RPMI-1640 medium (Gibco) supplemented with 10% FBS and 10 units/ml penicillin, and 100 μg/ml streptomycin.
MicroRNA Library HNF4α Screening
A microRNA library, consisting of 317 microRNA mimics and 2 microRNA negative control mimics (100nM) (Dharmacon Inc) was transfected in HepG2 cells plated in 96-well plates. 24h post-transfection, the cells were transfected with a firefly luciferase vector harboring the 3'UTR of HNF4α for 24h and the luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega, WI, USA). MicroRNAs that inhibited >75% the luciferase activity of HNF4α were considered as positive hits. Detailed experimental description can be found in the supplementary experimental procedures.
TUNEL assay
Apoptosis was determined using the DeadEnd Fluorometric TUNEL System (G3250, Promega), as previously described (Polytarchou et al., 2008).
Real-time PCR analysis
RNA purified from IMH, HepG2, Hep3B, SNU-449, SNU-398, SNU-387 and SNU-475 cells under different transfection conditions with siRNAs or microRNAs was reverse-transcribed to form cDNA, which was subjected to SYBR Green based real-time PCR analysis. MicroRNA expression levels were tested using the Exiqon PCR Primer Sets, according to the manufacturer's instructions (Exiqon Inc, Denmark). Primer sequences can be found in the supplementary experimental procedures.
Identification of transcription factor sites in microRNA regulatory areas
The Lever and PhylCRM algorithms have been used to identify STAT3 and HNF4α binding motifs in an area of 5kb upstream and 2kb downstream of microRNAs. A detailed description of this method has been included in the supplementary experimental procedures.
Mouse experiments
All the experiments in xenografts and DEN-treated mice are described analytically in the supplementary experimental procedures.
RNA Expression Studies from Patient Samples
RNAs from 45 hepatocellular carcinomas and 12 normal tissues were purchased from Biochain (Hayward, CA) and Origene (Rockville, MD). The expression levels of miR-24, IL6R, miR-124 and HNF4α were analyzed by real-time RT-PCR in all the tissue described above. Each sample was run in triplicate and the data represent the mean ± SD.
Supplementary Material
Highlights
-
•
HNF4α transient inhibition induces hepatocellular transformation.
-
•
The HNF4α feedback loop circuit links inflammation to liver cancer.
-
•
MiR-124 regulates IL6-STAT3 signaling pathway.
-
•
Systemic delivery of miR-124 inhibits HCC development.
Acknowledgements
This work was supported by Dana-Farber start-up funds to D.I. and research grants to M.K from the National Institutes of Health (RO1-CA118165 and P42-ES0100337). We would like to thank the Nikon Imaging Center at Harvard Medical School for their help with light microscopy. Also, we would like to thank Hye-Jung Kim for her assistance with the flow cytometry analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Extensive details for all experimental procedures are provided in the Supplemental Information.
References
- Ahn SH, Shah YM, Inoue J, Morimura K, Kim I, Yim S, Lambert G, Kurotani R, Nagashima K, Gonzalez FJ, et al. Hepatocyte nuclear factor 4alpha in the intestinal epithelial cells protects against inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:908–920. doi: 10.1002/ibd.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, Phillips A, Wesley E, Parnell K, Zhang H, Drummond H, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Calvisi DF, Ladu S, Gorden A, Farina M, Conner EA, Lee JS, Factor VM, Thorgeirsson SS. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006;130:1117–1128. doi: 10.1053/j.gastro.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Cattin AL, Le Beyec J, Barreau F, Saint-Just S, Houllier A, Gonzalez FJ, Robine S, Pincon-Raymond M, Cardot P, Lacasa M, et al. Hepatocyte nuclear factor 4alpha, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol Cell Biol. 2009;29:6294–6308. doi: 10.1128/MCB.00939-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, Darnell JE., Jr. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- Colletti M, Cicchini C, Conigliaro A, Santangelo L, Alonzi T, Pasquini E, Tripodi M, Amicone L. Convergence of Wnt signaling on the HNF4alpha-driven transcription in controlling liver zonation. Gastroenterology. 2009;137:660–672. doi: 10.1053/j.gastro.2009.05.038. [DOI] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsigny M, Babeu JP, Seidman EG, Gendron FP, Levy E, Carrier J, Perreault N, Boudreau F. Hepatocyte nuclear factor-4alpha promotes gut neoplasia in mice and protects against the production of reactive oxygen species. Cancer Res. 2010;70:9423–9433. doi: 10.1158/0008-5472.CAN-10-1697. [DOI] [PubMed] [Google Scholar]
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Bottoni A, Shimizu M, Spizzo R, Nicoloso MS, Rossi S, Barbarotto E, Cimmino A, Adair B, Wojcik SE, et al. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. JAMA. 2011;305:59–67. doi: 10.1001/jama.2010.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulci V, Colombo T, Chiaretti S, Messina M, Citarella F, Tavolaro S, Guarini A, Foa R, Macino G. Characterization of B- and T-lineage acute lymphoblastic leukemia by integrated analysis of MicroRNA and mRNA expression profiles. Genes Chromosomes Cancer. 2009;48:1069–1082. doi: 10.1002/gcc.20709. [DOI] [PubMed] [Google Scholar]
- Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Kaestner KH. HNF-4alpha: from MODY to late-onset type 2 diabetes. Trends Mol Med. 2004;10:521–524. doi: 10.1016/j.molmed.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Hatziapostolou M, Iliopoulos D. Epigenetic aberrations during oncogenesis. Cell Mol Life Sci. 2011;68:1681–1702. doi: 10.1007/s00018-010-0624-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Yu GY, Temkin V, Ogata H, Kuntzen C, Sakurai T, Sieghart W, Peck-Radosavljevic M, Leffert HL, Karin M. Hepatocyte IKKbeta/NF-kappaB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer Cell. 2010;17:286–297. doi: 10.1016/j.ccr.2009.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedvat M, Huszar D, Herrmann A, Gozgit JM, Schroeder A, Sheehy A, Buettner R, Proia D, Kowolik CM, Xin H, et al. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16:487–497. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009a;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Polytarchou C, Hatziapostolou M, Kottakis F, Maroulakou IG, Struhl K, Tsichlis PN. MicroRNAs differentially regulated by Akt isoforms control EMT and stem cell renewal in cancer cells. Sci Signal. 2009b;2:ra62. doi: 10.1126/scisignal.2000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Tan Z, Deng L, Chen Y, Xia Y, Gao Y, Wang X, Sun B. IL-22 promotes human hepatocellular carcinoma by activation of STAT3. Hepatology. 2011 doi: 10.1002/hep.24486. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Kardassis D, Tzameli I, Hadzopoulou-Cladaras M, Talianidis I, Zannis V. Distal apolipoprotein C-III regulatory elements F to J act as a general modular enhancer for proximal promoters that contain hormone response elements. Synergism between hepatic nuclear factor-4 molecules bound to the proximal promoter and distal enhancer sites. Arterioscler Thromb Vasc Biol. 1997;17:222–232. doi: 10.1161/01.atv.17.1.222. [DOI] [PubMed] [Google Scholar]
- Kent OA, Chivukula RR, Mullendore M, Wentzel EA, Feldmann G, Lee KH, Liu S, Leach SD, Maitra A, Mendell JT. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev. 2010;24:2754–2759. doi: 10.1101/gad.1950610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev. 2006;20:2293–2305. doi: 10.1101/gad.390906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladias JA, Hadzopoulou-Cladaras M, Kardassis D, Cardot P, Cheng J, Zannis V, Cladaras C. Transcriptional regulation of human apolipoprotein genes ApoB, ApoCIII, and ApoAII by members of the steroid hormone receptor superfamily HNF-4, ARP-1, EAR-2, and EAR-3. J Biol Chem. 1992;267:15849–15860. [PubMed] [Google Scholar]
- Lanford RE, Hilderbrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Chu IS, Mikaelyan A, Calvisi DF, Heo J, Reddy JK, Thorgeirsson SS. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet. 2004;36:1306–1311. doi: 10.1038/ng1481. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Git A, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Marcil V, Seidman E, Sinnett D, Boudreau F, Gendron FP, Beaulieu JF, Menard D, Precourt LP, Amre D, Levy E. Modification in oxidative stress, inflammation, and lipoprotein assembly in response to hepatocyte nuclear factor 4alpha knockdown in intestinal epithelial cells. J Biol Chem. 2010;285:40448–40460. doi: 10.1074/jbc.M110.155358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros VR, Walhout AJ. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008;22:2535–2549. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG, Osterreicher CH, Takahashi H, Karin M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140:197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- Polytarchou C, Pfau R, Hatziapostolou M, Tsichlis PN. The JmjC domain histone demethylase Ndy1 regulates redox homeostasis and protects cells from oxidative stress. Mol Cell Biol. 2008;28:7451–7464. doi: 10.1128/MCB.00688-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. Binding reactions: epigenetic switches, signal transduction and cancer. Curr Biol. 2009;19:R234–241. doi: 10.1016/j.cub.2009.02.015. [DOI] [PubMed] [Google Scholar]
- Rebouissou S, Amessou M, Couchy G, Poussin K, Imbeaud S, Pilati C, Izard T, Balabaud C, Bioulac-Sage P, Zucman-Rossi J. Frequent in-frame somatic deletions activate gp130 in inflammatory hepatocellular tumours. Nature. 2009;457:200–204. doi: 10.1038/nature07475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke RS, Tosello-Trampont A, Nguyen V, Mullins DW, Hahn YS. Extracellular hepatitis C virus core protein activates STAT3 in human monocytes/macrophages/dendritic cells via an IL-6 autocrine pathway. J Biol Chem. 2011;286:10847–55. doi: 10.1074/jbc.M110.217653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva A, Newell P, Chiang DY, Friedman SL, Llovet JM. Genomics and signaling pathways in hepatocellular carcinoma. Semin Liver Dis. 2007;27:55–76. doi: 10.1055/s-2006-960171. [DOI] [PubMed] [Google Scholar]
- Xu L, Hui L, Wang S, Gong J, Jin Y, Wang Y, Ji Y, Wu X, Han Z, Hu G. Expression profiling suggested a regulatory role of liver-enriched transcription factors in human hepatocellular carcinoma. Cancer Res. 2001;61:3176–3181. [PubMed] [Google Scholar]
- Yin C, Lin Y, Zhang X, Chen YX, Zeng X, Yue HY, Hou JL, Deng X, Zhang JP, Han ZG, et al. Differentiation therapy of hepatocellular carcinoma in mice with recombinant adenovirus carrying hepatocyte nuclear factor-4alpha gene. Hepatology. 2008;48:1528–1539. doi: 10.1002/hep.22510. [DOI] [PubMed] [Google Scholar]
- Zhong W, Sladek FM, Darnell JE., Jr. The expression pattern of a Drosophila homolog to the mouse transcription factor HNF-4 suggests a determinative role in gut formation. EMBO J. 1993;12:537–544. doi: 10.1002/j.1460-2075.1993.tb05685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Wen Z, Darnell JE., Jr. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.