Abstract
Plastids contain multiple copies of the plastid chromosome, folded together with proteins and RNA into nucleoids. The degree to which components of the plastid gene expression and protein biogenesis machineries are nucleoid associated, and the factors involved in plastid DNA organization, repair, and replication, are poorly understood. To provide a conceptual framework for nucleoid function, we characterized the proteomes of highly enriched nucleoid fractions of proplastids and mature chloroplasts isolated from the maize (Zea mays) leaf base and tip, respectively, using mass spectrometry. Quantitative comparisons with proteomes of unfractionated proplastids and chloroplasts facilitated the determination of nucleoid-enriched proteins. This nucleoid-enriched proteome included proteins involved in DNA replication, organization, and repair as well as transcription, mRNA processing, splicing, and editing. Many proteins of unknown function, including pentatricopeptide repeat (PPR), tetratricopeptide repeat (TPR), DnaJ, and mitochondrial transcription factor (mTERF) domain proteins, were identified. Strikingly, 70S ribosome and ribosome assembly factors were strongly overrepresented in nucleoid fractions, but protein chaperones were not. Our analysis strongly suggests that mRNA processing, splicing, and editing, as well as ribosome assembly, take place in association with the nucleoid, suggesting that these processes occur cotranscriptionally. The plastid developmental state did not dramatically change the nucleoid-enriched proteome but did quantitatively shift the predominating function from RNA metabolism in undeveloped plastids to translation and homeostasis in chloroplasts. This study extends the known maize plastid proteome by hundreds of proteins, including more than 40 PPR and mTERF domain proteins, and provides a resource for targeted studies on plastid gene expression. Details of protein identification and annotation are provided in the Plant Proteome Database.
Mitochondria and plastids are descendents of endosymbiotic prokaryotes (Douglas, 1998; Gray, 1999). The genomes of these organelles are organized as DNA-protein complexes named “organelle nucleoids” (Sakai et al., 2004), similar to the prokaryotic nucleoid (Robinow and Kellenberger, 1994). Multiple studies in plants, yeast, and other nonphotosynthetic eukaryotes demonstrated that organelle nucleoids are the site of DNA replication (Kuroiwa, 1973; Kuroiwa et al., 1992; Suzuki et al., 1992; Nerozzi and Coleman, 1997) and transcription (Sakai et al., 1991, 1999; Sasaki et al., 1998) and are the segregation unit of the organelle genome (Kuroiwa et al., 1982, 1994; Lockshon et al., 1995; Nagata et al., 1999). However, it is less clear to what extent posttranscriptional steps of gene expression, such as RNA processing, ribosome assembly, and translation, are associated with nucleoids. Moreover, whereas transcription and translation are coupled in prokaryotes, that is, ribosomes initiate translation on nascent mRNA that is still in the process of synthesis (Gowrishankar and Harinarayanan, 2004; Burmann et al., 2010; Epshtein et al., 2010; Proshkin et al., 2010), there are few data addressing whether a similar coupling occurs in plastids. The mechanisms through which plastid nucleoids participate in plastid gene expression, plastid development, and biogenesis are poorly understood. Moreover, relatively little is known about the proteins involved in the control of plastid DNA quality and copy number. The functions and organization of nucleoids are also likely to change in response to the plastid developmental stage, but little information is available that addresses this possibility. Through extensive proteome analyses of highly enriched nucleoid fractions from undeveloped and mature plastids, this study provides a framework for studying the organization and function of plastid nucleoid proteomes in higher plants.
The plastid genomes of higher plants contain about 100 genes encoding for (1) components of the chloroplast gene expression machinery (RNA polymerase, ribosomal proteins, tRNAs, and rRNAs), (2) subunits of six photosynthetic complexes (Rubisco, PSII, the cytochrome b6f complex, PSI, NAPH dehydrogenase, and ATP synthase), and (3) a few proteins involved in other processes (e.g. ClpP1 and YCF3). The localization and morphology of plastid nucleoids differ between species and also change in response to developmental stage (Kuroiwa, 1991). During the transition from proplastid to chloroplast, nucleoids relocate from the envelope to the thylakoid membranes in some species (tobacco [Nicotiana tabacum] and spinach [Spinacia oleracea]; Selldén and Leech, 1981; Miyamura et al., 1986; Sodmergen et al., 1991) but not in others (e.g. wheat [Triticum aestivum]; Selldén and Leech, 1981). It is not known how this relocation and developmental state affects nucleoid composition and function.
Several procedures have been developed for the isolation of plastid DNA-protein complexes (for review, see Sakai et al., 2004). A procedure for obtaining a plastid transcriptionally active chromosome (pTAC) was originally developed by Hallick et al. (1976) and further modified by others (Krause and Krupinska, 2000). Although isolated pTAC has a high transcriptional activity and retained chromatin-like beaded structures, the native morphological structure of the nucleoid was not preserved (Sato et al., 2003; Sakai et al., 2004). A second procedure for the isolation of plastid DNA-protein complexes aimed at preservation of the morphological integrity of the nucleoid (Cannon et al., 1999). Using 4′,6-diamidino-2-phenylindole fluorescence microscopy, it was shown that such isolated nucleoids were indistinguishable in size and fluorescence intensity from nucleoids in intact plastids (Cannon et al., 1999).
Using protein mass spectrometry (MS) or Edman sequencing, several studies have identified proteins associated with pTACs or nucleoids (Sato et al., 2003; Sakai et al., 2004; Suzuki et al., 2004; Pfalz et al., 2006; Schröter et al., 2010). So far, the richest protein data set resulted from MS analysis of isolated pTACs from Arabidopsis (Arabidopsis thaliana) and mustard (Sinapis alba), in which 35 proteins were identified (Pfalz et al., 2006). Eighteen of these proteins were denoted pTAC proteins, and three of them (pTAC2, -6, and -12) were shown to be required for plastid gene expression (Pfalz et al., 2006). Proteome analysis of an affinity-purified stromal plastid-encoded RNA polymerase (PEP) complex of more than 900 kD from tobacco identified the four PEP subunits as well as seven additional proteins (Suzuki et al., 2004). These PEP subunits and the seven PEP-associated proteins (pTAC2, -3, -6, -10, -17, MurE-like, and pfkB-type carbohydrate kinase FLN1) were also all found in the TAC study (Pfalz et al., 2006). MS analysis of a Triton-insoluble fraction from pea chloroplasts identified several known nucleoid or pTAC proteins as well as other proteins involved in plastid gene expression and multiple proteins with unknown functions (Phinney and Thelen, 2005). This fraction included nucleoids, along with other large plastid complexes (e.g. acetyl-CoA carboxylase, pyruvate dehydrogenase, and ribosomes), but an absence of quantitative information made it difficult to evaluate which proteins should be considered to be nucleoid associated.
Given the observed morphological differences between isolated pTAC and nucleoid complexes, as well as dramatic improvements in protein identification capacity by MS, it is likely that many additional proteins involved in plastid gene expression, DNA replication, recombination, repair, and inheritance could be discovered through an in-depth proteome analysis of intact nucleoids. In fact, proteins involved in plastid gene expression are strongly underrepresented among the approximately 1,000 proteins identified by proteome analyses of chloroplast stromal, envelope, and thylakoid fractions (Zybailov et al., 2008; van Wijk and Baginsky, 2011). We speculated that plastid nucleoids are likely to contain many of these “missing” proteins, a possibility that was already supported by our MS/MS analysis of FPLC-separated Arabidopsis stromal complexes larger than 1 MD (Olinares et al., 2010)
To provide a comprehensive resource for the analysis of nucleoid functions in higher plants, we characterized nucleoid-enriched proteomes of developing maize (Zea mays) proplastids and of mature maize chloroplasts using high-resolution and high-accuracy MS. For this purpose, we took advantage of the natural developmental gradient of the young maize seedling leaf, in which cells are arranged in a linear developmental array, with the youngest cells, containing proplastids, at the base and the oldest cells, containing mature chloroplasts, at the tip (Leech et al., 1973; Baker and Leech, 1977). We used material from the same developmental stages as used in our previous study describing structural and proteome transitions along the developing maize leaf gradient (Majeran et al., 2010): a basal leaf section containing nonphotosynthetic plastids with a minimal thylakoid membrane system (referred to hereafter as proplastids), and an apical leaf section with differentiated photosynthetic chloroplasts characteristic of either bundle sheath or mesophyll cells (Majeran et al., 2010). Quantitative comparisons with proteomes of isolated proplastids (this paper) and mature chloroplasts (Friso et al., 2010), as well as nucleoid fractions that were further purified by immunoprecipitation with antiserum to Whirly1 (WHY1), an abundant DNA-associated chloroplast protein (Prikryl et al., 2008), provided additional criteria for defining an authentic nucleoid-associated proteome. Collectively, our analysis strongly suggests that transcription, mRNA processing, splicing, and editing, as well as ribosome assembly, take place in association with the nucleoid. This study extends the known higher plant plastid proteome by several hundreds of proteins, including more than 50 pentatricopeptide repeat (PPR) and mitochondrial transcription factor domain (mTERF) (for mitochondrial transcription termination factor) proteins and others with domains suggestive of roles in nucleic acid transactions, and thus provides a comprehensive resource for targeted studies on plastid gene expression and DNA metabolism.
RESULTS
Nucleoid-Enriched Proteomes from Nonphotosynthetic Plastids and Chloroplasts
Nucleoids were prepared by nonionic detergent solubilization of plastid preparations, followed by differential centrifugation steps and various washes, essentially as described (Cannon et al., 1999). In the remainder of this paper, we will refer to these fractions as “nucleoids,” even though some nonnucleoid proteins copurify, as will be discussed in detail below. Nucleoids were extracted from proplastids from the yellow to pale-green immature base (just above the ligule) and from combined mesophyll and bundle sheath chloroplasts isolated from the tip of the third leaf of 9- to 10-d-old seedlings (Fig. 1A). In an earlier study, we showed by electron microcopy and proteomics that this leaf base section contained proplastids and developing nonphotosynthetic plastids, whereas the leaf tip section contains mature chloroplasts (Majeran et al., 2010). In addition, nucleoids were purified from plastids isolated from younger 7- to 8-d-old seedlings using the blades of all four leaves (in three biological replicates; Fig. 1B). The average yield for the nucleoid-enriched proteome was around 1 μg of nucleoid protein per 100 μg of chloroplast protein.
Figure 1.
Plant material and selection of maize nucleoid preparations. A, Seedlings used for the isolation of nucleoids from the nonphotosynthetic leaf base (above the ligule) and the photosynthetic tip of the third leaf of 9- to 10-d-old seedlings. B, Seedlings used for the isolation of nucleoids from all leaf blades of 7- to 8-d-old seedlings (assigned as young nucleoids). C, SDS-PAGE protein profile of nucleoids from young leaves. The gel (10.5%–14% acrylamide gradient gel) was loaded with 18 μg of protein and was stained with Sypro Ruby. D, Average abundance (as NadjSPC) of the four subunits of the PEP complex in independent nucleoid preparations from base, tip, and young leaf. sd values are indicated (n = 3). [See online article for color version of this figure.]
Proteins in each of these nucleoid samples were separated by SDS-PAGE (a representative preparation is shown in Fig. 1C). Each gel lane was cut in slices, proteins were in-gel digested with trypsin, and extracted peptides were analyzed by electrospray-MS/MS employing a high-accuracy LTQ-Orbitrap mass spectrometer and an established bioinformatics work flow searching against the maize genome (Friso et al., 2010; Majeran et al., 2010; see “Materials and Methods”). Recent improvements in the sensitivity, mass accuracy, and speed of mass spectrometers (Bantscheff et al., 2007; Mann and Kelleher, 2008; Domon and Aebersold, 2010) have enabled large-scale MS-based label-free proteome quantifications using spectral counting. This approach is based on the observation that the number of MS/MS acquisitions of peptides coming from a protein shows a positive correlation to the relative concentration of this protein in the sample (Liu et al., 2004; Old et al., 2005; Zybailov et al., 2005; Sandhu et al., 2008). Spectral counting is particularly effective to detect large quantitative differences when comparing cellular fractions that are very different in function and composition, as expected in our study. We previously optimized the spectral count (SPC) work flow and tested it for Arabidopsis and maize organelles, cell types, and complexes (Friso et al., 2010; Majeran et al., 2010; Olinares et al., 2011). The relative normalized abundance (relative mass contribution) of each protein within each sample (NadjSPC) was calculated from the number of adjusted matched MS/MS spectra (adjSPC) normalized to the total adjSPC per sample, as defined previously (Friso et al., 2010). A protein with NadjSPC = 0.01 contributes approximately 1% of the protein mass of the analyzed sample. As a general rule, the accuracy of quantification improves with the number of adjSPC per protein; consequently, we have set minimal abundance thresholds when we try to identify (putative) nucleoid-associated proteins.
The PEP complex is an excellent marker for the nucleoid because it interacts directly with plastid DNA and because it has four subunits (Suzuki et al., 2004), allowing for more accurate quantification than would be obtained with a single polypeptide. We observed a positive, linear correlation between PEP abundance and the abundance of pTAC proteins (Supplemental Fig. S1A), as expected if these proteins are generally found in complex with one another. In contrast, the correlation between abundant thylakoid proteins and pTAC proteins was negative (data not shown). For each nucleoid sample type (leaf base, leaf tip, and young leaf), we selected the three best biological replicates based on relative abundance of the PEP subunits Rpo-A, -B, -C1, and -C2 (Supplemental Fig. S1B). The average values for each of the PEP subunits are shown in Figure 1D. Thus, the total PEP complex represented roughly 5%, 3%, and 5% of the protein mass in nucleoid fractions of the base, tip and young leaf, respectively; this corresponded to an average of 500 adjSPC (matched MS/MS spectra) for each of the PEP subunits in each sample type. Within each sample type (base, tip, and young leaf), the variability was relatively small; the sd values translate into coefficients of variation (CVs) between 0.16 and 0.55 (average CV = 0.33). Such CVs are low when compared with other quantitative proteomics studies, in particular considering that we are measuring the protein abundance of isolated dynamic structures. Strikingly, we never observed any nucleus-encoded plastid RNA polymerase (NEP) in the nucleoid fractions, nor did we observe it in proplastids, chloroplasts, or total leaf sections in maize or Arabidopsis, indicating that NEP protein accumulation levels are much lower than PEP levels in these samples.
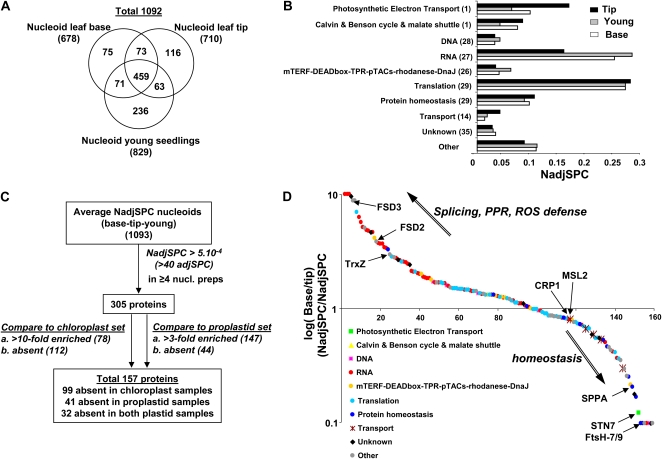
Across the nine selected preparations, we identified 1,092 proteins, counting only the highest scoring protein model per gene (Supplemental Table S1). Where possible, proteins were manually annotated with a putative function and/or functional domains and assigned subcellular localization, based on our experimental MS-based identification in maize proteome analyses to date (Sun et al., 2009; Friso et al., 2010; Majeran et al., 2010), as well as extensive screening of the published literature. Out of the 1,092 identified proteins, we assigned 750 to the plastid (69%) and 235 to nonplastid localizations (21%). A total of 107 proteins (10%) were without clear subcellular localization, using conservative criteria to avoid false-positive plastid assignment (i.e. several unassigned proteins are likely plastid localized; Supplemental Table S1). A total of 67% of the plastid-assigned proteins were predicted by TargetP to localize to plastids. This is lower than we typically observe for Arabidopsis plastid-assigned proteins (Zybailov et al., 2008) and likely resulted from incorrect maize gene models and the bias toward proteins from dicot species in the TargetP training set. Indeed, the Arabidopsis best homologs of these 750 maize proteins showed a chloroplast transit peptide prediction rate of 84%, very close to the previously reported true positive rate of 86% (Zybailov et al., 2008). The nonplastid proteins and proteins without clear location together represented only approximately 2% of the protein mass in the nucleoid preparations; thus, at least 98% of the protein mass was from bona fide plastid proteins. The main nonplastid contaminations were histones, 80S ribosomal subunits, and cytosolic translation factors, a few abundant metabolic enzymes such as phosphoenolpyruvate carboxylase, glycolytic glyceraldehyde 3-phosphate dehydrogenase C, NAD-malate dehydrogenase, cytoskeleton components (actins and tubulins), as well as several nonplastid protein chaperones (Supplemental Table S1). Although we cannot fully eliminate the possibility that some of these proteins are authentic nucleoid components, their low abundance in our preparations and the fact that they are known to be highly abundant outside of the plastid argue strongly that they are contaminants.
To help determine proteins that functionally interact with the nucleoid or that are intrinsic components of the nucleoid, we compared the nucleoid proteome with the previously determined chloroplast stromal and membrane proteomes purified from developed maize leaf tips (1,428 identified proteins; Friso et al., 2010). In addition, we purified proplastids from the yellow base of the leaf blade (Fig. 1A) in triplicate, determined their proteome, and analyzed their functions and subcellular localization using the same methods and criteria as used for the nucleoid preparations. The proplastid analysis identified 1,717 proteins, counting only one model per gene (Supplemental Table S2). Quantitative comparison (including hierarchical clustering) of the nucleoid-enriched data sets with the proplastid and combined thylakoid and stromal data sets allowed us to identify proteins that are substantially enriched in the nucleoid fraction, as will be detailed further below.
Comparison of Maize Nucleoid Proteins with Previously Reported pTAC, PEP, and Nucleoid Proteins
Several previous studies have identified proteins associated with structures related to plastid nucleoids, such as the pTAC (Pfalz et al., 2006) and an affinity-purified PEP preparation (Suzuki et al., 2004; Steiner et al., 2011). These studies were performed with various dicot plants (Arabidopsis, tobacco, and mustard). Except for one pTAC protein (pTAC15), predicted maize orthologs of each of these proteins were detected in our nucleoid samples (Supplemental Table S3). pTAC8 is now known to be a PSI subunit (Khrouchtchova et al., 2005) and was thus a contaminant in the pTAC fraction; pTAC8 was not enriched in our nucleoid fraction in comparison with its abundance in the proplastid or chloroplast. In addition, pTAC16 was also not enriched in nucleoids as compared with chloroplasts, but it could be a candidate for a nucleoid anchor (see below). Collectively, the remaining pTAC proteins represented between 1.5% and 4% of the protein mass in the nucleoid fractions (Supplemental Fig. S1A), and several pTAC proteins were among the most abundant proteins in the nucleoid preparations.
In addition to the 35 pTAC proteins, various studies in angiosperms reported nine additional nucleoid-associated proteins (Supplemental Table S3, bottom section). These proteins were proposed to be involved with DNA repair (ARP1 and NTH2), membrane anchoring of the nucleoids (PEND, TCP34, MFP1, and CND41), DNA packing (sulfite reductase [SiR]), phosphorylation (CK2), nucleoid distribution (YLMG1), translational control (NARA5), or transcription (ETCHED1; for references, see Supplemental Table S3). We identified maize homologs for TCP34, MFP1, and YLMG1 in the nucleoid fractions. However, PEND and SiR were found in unfractionated plastids but not in nucleoids (Supplemental Table S2), suggesting that they are either loosely associated with the nucleoid or are not true nucleoid components. We never detected ETCHED1 or maize homologs for ARP1, NTH2, CK2, or NARA5 in any of our previous or current maize sample analyses, indicating their low abundance in maize leaves.
Comparison of the Nucleoid-Enriched Proteome with Proteomes of Chloroplasts and Proplastids
To understand the range of functions of the nucleoid in proplastids and chloroplasts, it was important to distinguish between proteins that are highly specific for nucleoids and proteins that are also found in other plastid compartments (i.e. stroma, thylakoids, or envelopes). The second category could include proteins that cycle on and off nucleoids, proteins that anchor the DNA to membranes, and proteins that are contaminants. Therefore, we qualitatively and quantitatively compared the proteins detected in our nucleoid fractions with the proteins identified in chloroplast stroma and membranes and in unfractionated proplastids. We made inferences about protein functions based on functional domains predicted by PFAM (Finn et al., 2010) as well as based on homology between maize proteins and proteins analyzed in other species, in particular Arabidopsis. We used the MapMan bin system (Thimm et al., 2004) as the basis to organize the protein functions; we introduced several new bins to accommodate more narrowly defined functions (Supplemental Tables S1 and S2). The protein annotations can also be found through our Plant Proteomics DataBase (PPDB; http://ppdb.tc.cornell.edu/; Sun et al., 2009).
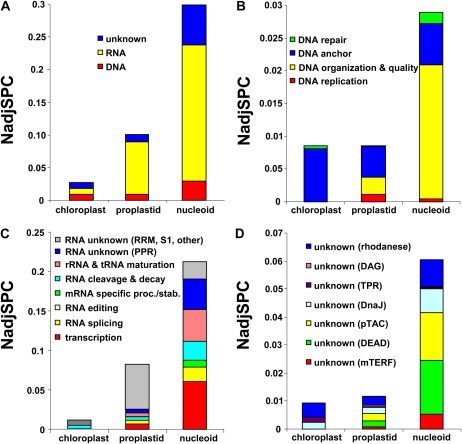
The Venn diagram in Figure 2A shows the overlap between the proteins detected in all nucleoid samples, in chloroplast stroma and membranes, and in proplastids; in total, 2,460 proteins were identified across all samples. The protein mass investments (based on NadjSPC) for these three data sets in 10 different cellular functions is illustrated in Figure 2B. This showed that thylakoid proteins involved in photosynthetic electron transport represented approximately 37% of the protein mass in chloroplasts but approximately 13% in proplastids and approximately 11% in nucleoids. Proteins of the Calvin-Benson cycle and the C4-malate cycle represented approximately 27% of the mass in chloroplasts but only approximately 6% in nucleoids and proplastids. These results are consistent with the nonphotosynthetic nature of proplastids and the successful reduction of photosynthetic contaminants in the nucleoid preparations.
Figure 2.
Comparison of proteomes of chloroplasts, proplastids, and nucleoids. A, Venn diagram showing the overlap between the identified nucleoid, chloroplast, and proplastid proteomes; in total, 2,460 proteins were identified. B, Comparison of protein abundance (based on NadjSPC) of the different functional groups. Ten different functional groups are defined, namely the photosynthetic electron transport chain (bin 1), the Calvin-Benson cycle and malate shuttle (bin 1), DNA (bin 28), RNA (bin 27), proteins with unknown function and with mTERF, DEAD box, TPR, rhodanese, or DnaJ(-like) domains (bin 26) or pTAC proteins with unknown functions, translation (in bin 29), protein homeostasis (bin 29), transport (bin 34), unknown function (bin 35), other functions (all other bins). C, Dendrogram of the distribution pattern of 771 proteins across the three sample types (chloroplasts, proplastids, average nucleoids) obtained by hierarchical clustering. The 771 proteins were observed in nucleoid fractions, they each had at least an average NadjSPC of 1.10−5 across the three sample types (chloroplast, proplastid, and nucleoid), and they were not considered extraplastidic (for location assignments or PPDB, see Supplemental Table S1). Red represents values above the mean, black represents the mean, and green represents values below the mean abundance of a protein across the three sample types. D, Protein mass investments in the clusters for chloroplast, proplastid, and nucleoid samples.
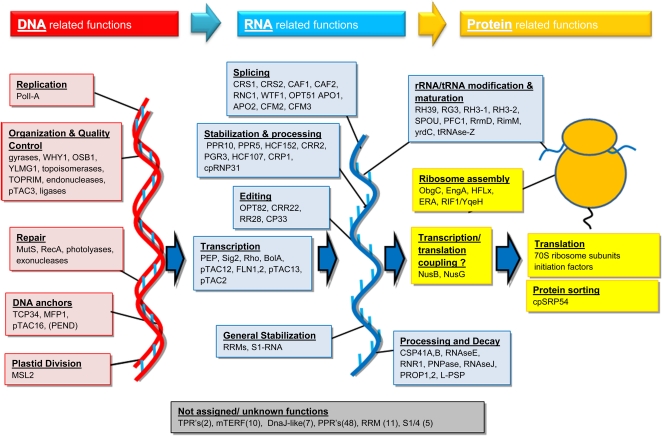
All other identified proteins were placed in the following functional categories (Fig. 2B): (1) DNA-related functions (replication, repair, anchoring, organization); (2) RNA-related functions (RNA polymerases, transcription factors/regulators, splicing, processing, editing, stabilization, degradation, rRNA/tRNA-modifying enzymes, and all PPR proteins without known functions); (3) translation-related functions (ribosome assembly factors, tRNA ligases, ribosomal proteins, initiation, elongation, release factors); (4) protein homeostasis (folding, processing, targeting, posttranslational modifications, assembly, proteolysis); (5) uncharacterized proteins with domains relevant for plastid gene expression and biogenesis (mTERF, DEAD box helicases, rhodanese, DnaJ domain, tetratricopeptide repeat [TPR]) as well as pTAC proteins with unknown functions; (6) transport (bin 34); (7) “unknown” other proteins, which are uncharacterized and also lack predicted functional domains (bin 35); and (8) all other functions (all remaining bins). Explanations for the placements of proteins in the functional categories will follow below.
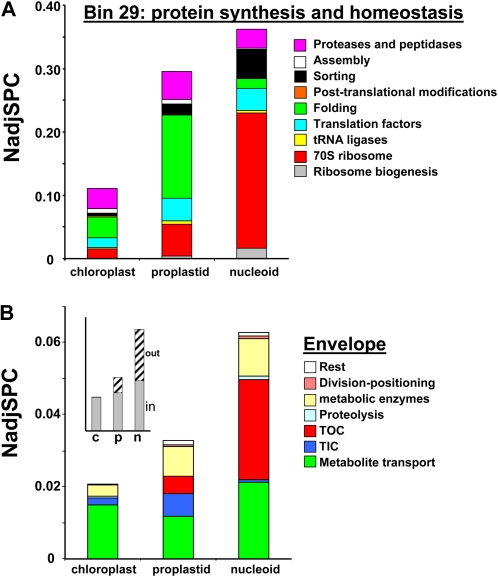
The DNA- and RNA-related functions were approximately 4-fold and approximately 23-fold enriched, respectively, in nucleoids when compared with chloroplasts and approximately 3-fold enriched when compared with proplastids. The dramatic enrichment of RNA-related functions in nucleoids and in proplastids (Fig. 2B) is an important observation, since these functions are strongly underrepresented in chloroplast proteomics studies (Zybailov et al., 2008; van Wijk and Baginsky, 2011). These results show immediately that the nucleoid is a major location of plastid gene expression and RNA metabolism. Compared with chloroplasts and proplastids, nucleoids were also substantially enriched in ribosomal proteins and other proteins involved in translation (approximately 8-fold and approximately 3-fold, respectively), but not in proteins involved in protein homeostasis (sorting, assembly, chaperones, proteases, etc.) that can serve plastid-encoded and/or nucleus-encoded proteins (Fig. 2B). Investments in the plastid translation machinery represented approximately 27% of the protein mass of nucleoids, compared with just a few percent in chloroplasts. Investments in protein homeostasis represented approximately 10% of the mass in nucleoids but more than 20% in proplastids, consistent with the specialized function of nucleoids in the expression of plastid genes (Fig. 2B). Nucleoids were also enriched in a group of proteins with unknown functions that contain functional domains important in different aspects of plastid biogenesis, such as mTERF, TPR, and DEAD box helicases (for details, see below). Finally, large differences were observed among these samples in proteins harboring “other” functions, a class that is particularly high in proplastids (Fig. 2B). As expected from proteome analysis of the maize leaf developmental gradient (Majeran et al., 2010), these other metabolic functions, such as amino acid and fatty acid metabolism, make a large contribution to the proplastid proteome (33%); these functions were 2-fold reduced in chloroplasts and even more reduced in nucleoids. Proteins with entirely unknown functions and without predicted PFAM domains (bin 35) made up about 3% of the protein mass in chloroplasts, proplastids, and nucleoids (Fig. 2B).
Coexpression Analysis to Recognize Proteins Enriched in Nucleoids
Genes or proteins involved in related biological pathways or complexes often accumulate simultaneously, and information on their coexpression is key to understanding biological systems. Conversely, coexpression in many cases implies the presence of functional linkages between genes or proteins, allowing for the identification of new components of processes or protein complexes. Cluster analysis has been used extensively for transcripts (Eisen et al., 1998; Belacel et al., 2006; Long et al., 2008) and more recently for proteomics (Dong et al., 2008; Huang et al., 2009; Pontén et al., 2009; Quintana et al., 2009; Majeran et al., 2010; Olinares et al., 2010). Cluster analysis is based on the notion of unsupervised learning in which data objects within the same cluster are similar to one another and dissimilar to the objects in other clusters. Whereas many clustering algorithms have been developed, hierarchical clustering is most appropriate for the analysis of the proteomics data set used in this study, because no prior assumptions about the number of clusters have to be made (Belacel et al., 2006). The hierarchical clustering algorithms also provide a natural means of graphical representation of the data, in the form of a dendrogram in which each branch forms a group of genes or proteins that share similar behavior.
To help identify proteins enriched in nucleoid fractions, we carried out a hierarchical cluster analysis based on standardized NadjSPC for the chloroplast, proplastid, and nucleoid samples, resulting in a dendrogram (Fig. 2C). We considered only those 771 proteins observed in nucleoid fractions that each had at least an average NadjSPC of 1.10−5 across the three sample types (chloroplast, proplastid, and nucleoid) and that were not considered extraplastidic (for location assignment, see Supplemental Table S1 or the PPDB). This minimal abundance threshold ensured more meaningful quantifications and clustering, as we discussed when analyzing leaf development and C4 differentiation of the vascular bundle and total leaf proteome (Majeran et al., 2010). The dendrogram (Fig. 2C) clearly showed five clusters: clusters 1 and 5 mostly represented contaminating proteins with high accumulation in chloroplasts, whereas clusters 2 and 3 mostly represented proteins with high accumulation in proplastids. Cluster 4, with 374 proteins, represented nucleoid-enriched proteins. Cluster numbers for each protein can be found in Supplemental Table S1 and will be used for more detailed analyses below. Figure 2D shows that proteins in cluster 4 make up approximately 70% of the protein mass in the nucleoid-enriched sample but far less in chloroplasts and proplastids.
In subsequent sections, we discuss these functional classes and individual proteins in more detail. We will first focus on the DNA- and RNA-related functions and on proteins with mTERF, DEAD box, rhodanese, DnaJ, or TPR domains as well as pTAC proteins with unknown functions. A total of 214 of the proteins within these functional classes have a (likely) plastid location, and all are listed in Table I. (Proteins that are likely nonplastid contaminants are not included in Table I and are also not further discussed in this paper.) Figure 3A shows that the 214 proteins within these functions together represent approximately 30% of the protein mass in nucleoids, approximately 10% in proplastids, but only approximately 3% in chloroplasts. For each of these proteins, their relative mass contribution in nucleoids, chloroplasts, and proplastids (based on NadjSPC), nucleoid-chloroplast or nucleoid-proplastid abundance ratios, as well as cluster numbers are provided (Table I). Strong candidates for nucleoid association or nucleoid components are marked (boldface and underlined) in Table I. These proteins fulfill three criteria: (1) they are in cluster 4 (Fig. 2C); (2) they have a nucleoid-proplastid ratio of more than 3 (or were not detected in proplastids); and (3) they have a nucleoid-chloroplast ratio of more than 10 (or were not detected in chloroplasts). In total, 127 marked proteins (or small sets of close homologs [e.g. SIG2]) in Table I should be considered strong candidates for nucleoid localization.
Table I. Abundance, distribution, and (putative) functions of nucleic acid-binding and pTAC proteins as well as TPR, PPR, mTERF, DEAD, rhodanese, and DnaJ domain proteins without known functions in nucleoids, proplastids, and chloroplasts.
| Accession No.a | CRGb | Name | Putative Functionc | Cluster No.(Fig. 2C)d | Cluster No.(Supplemental Fig. S5)e | Abundance Nucleoidf | Abundance Proplastidf | Abundance Chloroplastf | Best AT Hiti | Best Rice Hiti |
| Proteins with (predicted) functions in DNA replication, organization, distribution, membrane anchoring, quality control, and repair | ||||||||||
| GRMZM2G480171_P01 | DNA polymerase PolI-A | DNA replication | 2 | 0.290 | 1.087 | AT1G50840.1 | Os08g07840.1 | |||
| GRMZM2G081519_P01GRMZM2G097898_P01 | DNA exonuclease | DNA organization and quality control | 4 | 0.146 | 0.051 | AT1G34380.2 AT3G52050.3 | Os01g65400.2 Os03g12610.1 | |||
| GRMZM2G033896_P01 | DNA polymerase I | DNA organization and quality control | 2 | 0.012 | 0.051 | AT1G34380.2 | Os01g65400.1 | |||
| GRMZM2G162764_P01 | DNA endonuclease | DNA organization and quality control | 4 | 0.030 | AT3G47490.1 | Os03g07370.1 | ||||
| GRMZM2G095865_P02 | DNA gyrase A | DNA organization and quality control | 4 | 1c | 6.502 | 0.325 | AT3G10690.1 | Os03g59750.1 | ||
| GRMZM2G111014_P01 | DNA gyrase B1/2 | DNA organization and quality control | 4 | 1c | 1.007 | 0.382 | AT5G04130.1 | Os01g16290.1 | ||
| GRMZM2G071304_P01 | DNA ligase 1 (LIG1) | DNA organization and quality control | 4 | 0.100 | AT1G08130.1 | Os10g34750.2 | ||||
| GRMZM2G128432_P01 | OSB1, single-stranded DNA-binding protein | DNA organization and quality control | 3 | 2 | 0.714 | 0.862 | AT1G47720.1 | Os03g43420.1 | ||
| GRMZM2G116526_P01 | pTAC3 (SAP domain) | DNA organization and quality control | 4 | 1c | 4.812 | AT3G04260.1 | Os10g32540.1 | |||
| GRMZM2G313351_P01 | pTAC3-like (SAP domain) | DNA organization and quality control | 4 | 1c | 2.779 | 0.015 | AT3G04260.1 | Os10g32540.1 | ||
| GRMZM2G084928_P02 | Putative homing DNA endonuclease | DNA organization and quality control | 4 | 0.081 | AT1G53800.2 | Os10g12354.1 | ||||
| AC203761.3_FGP002 | Topoisomerase | DNA organization and quality control | 4 | 1c | 0.519 | AT4G31210.1 | Os06g16980.2 | |||
| GRMZM2G164835_P02 | Toprim domain protein (topisomerase?) | DNA organization and quality control | 4 | 0.262 | 0.029 | AT1G30680.1 | Os06g45980.1 | |||
| GRMZM2G155662_P01 | WHY1- pTAC1 | DNA organization and quality control | 4 | 1c | 3.696 | 0.975 | 0.022 | AT2G02740.2 | Os06g05350.1 | |
| GRMZM2G093815_P01 | YlmG1-2 | DNA organization and quality control | 2 | 0.006 | 0.059 | AT4G27990.1 | Os07g08770.1 | |||
| GRMZM2G064663_P01 | YlmG1-2 | DNA organization and quality control | 0.033 | AT5G21920.1 | Os03g08080.1 | |||||
| GRMZM2G142413_P01 | MFP1-1 | DNA anchor | 5 | 1c | 5.463 | 1.101 | 2.383 | AT3G16000.1 | Os01g08510.1 | |
| GRMZM2G106233_P01 | MFP1-2 | DNA anchor | 2 | 0.031 | 0.088 | AT3G16000.1 | Os05g08790.1 | |||
| GRMZM2G034453_P01 | PEND-1 | DNA anchor | 2.163 | AT3G52170.1 | Os12g34350.2 | |||||
| GRMZM2G171006_P02 | PEND-2 | DNA anchor | 0.185 | AT3G52170.1 | Os12g34350.2 | |||||
| GRMZM2G449496_P01 | pTAC16 | DNA anchor | 1 | 3 | 0.828 | 1.001 | 5.448 | AT3G46780.1 | Os05g22614.1 | |
| GRMZM2G098545_P01 | TCP34, TPR protein | DNA anchor | 1 | 0.013 | 0.044 | 0.188 | AT3G26580.1 | Os03g31490.1 | ||
| GRMZM2G306062_P01 | DNA-repair exonuclease II | DNA repair | 0.022 | AT3G63240.1 AT1G05470.1 | Os03g57950.1 Os03g57950.1 | |||||
| GRMZM2G371210_P01 | ||||||||||
| GRMZM2G104608_P03 | FAD photolyase | DNA repair | 0.309 | AT4G25290.1 | Os09g36240.2 | |||||
| GRMZM2G029519_P01 | FAD photolyase | DNA repair | 0.026 | AT1G12370.2 | Os10g08580.1 | |||||
| GRMZM2G060349_P01 | MutS | DNA repair | 4 | 1b | 0.565 | AT5G54090.1 | Os10g36530.1 | |||
| GRMZM2G348956_P01 | MutS-1 | DNA repair | 4 | 1b | 0.870 | AT1G65070.1 | Os04g58410.1 | |||
| AC193754.3_FGP008 | MutS-2 | DNA repair | 0.004 | AT3G18524.1 | Os05g19270.1 | |||||
| GRMZM2G103897_P01 | Photolyase | DNA repair | 4 | 0.084 | AT5G24850.1 | Os06g45100.1 | ||||
| GRMZM2G016602_P01 | RecA | DNA repair | 3 | 0.123 | 0.103 | AT1G79050.1 | Os03g43850.1 | |||
| GRMZM2G124321_P01 | uvrB/uvrC motif protein | DNA repair | 0.241 | AT2G03390.1 | Os08g07540.1 | |||||
| Proteins predicted or known to be involved in transcription, RNA splicing and processing, editing, stabilization, and turnover | ||||||||||
| GRMZM2G134398_P02 | 7 | Antitermination NusB domain protein | Transcription | 4 | 1d | 0.718 | 0.151 | AT4G26370.1 | Os03g45400.1 | |
| GRMZM2G161668_P02 | 7 | Antitermination NusB domain protein | Transcription | 4 | 0.267 | 0.113 | AT4G26370.1 | Os03g45400.1 | ||
| GRMZM2G161244_P01 | BolA-like protein | Transcription | 0.029 | 0.061 | AT5G17560.1 | Os06g28970.1 | ||||
| AC198353.5_FGP004 | NOL1/NOP2/NusB domain protein | Transcription | 4 | 1d | 0.635 | 0.059 | AT3G13180.1 | Os09g30100.2 | ||
| GRMZM2G156937_P01 | pTAC13 – KOW and NusG domain | Transcription | 4 | 1d | 0.907 | 0.044 | AT3G09210.1 | Os03g61030.1 | ||
| GRMZM2G168335_P01 | ρ transcription termination protein | Transcription | 4 | 1c | 2.083 | 0.132 | AT1G06190.1 | Os03g08480.2 | ||
| NP_043055 | RpoA, PEPα | Transcription | 4 | 1c | 9.791 | 2.652 | ATCG00740.1 | Osp1g00660.1 | ||
| NP_043015 | RpoB, PEPβ | Transcription | 4 | 1c | 14.221 | 0.925 | ATCG00190.1 | Osp1g00240.1 | ||
| NP_043017 | RpoC1, PEPβ′-1 | Transcription | 4 | 1c | 16.882 | 1.145 | ATCG00170.1 | Osp1g00260.1 | ||
| NP_043016 | 4 | RpoC1, PEPβ′-2 | Transcription | 4 | 1c | 5.666 | 0.338 | 0.003 | ATCG00180.1 | Osp1g00250.1 |
| GRMZM2G055567_P01 | 4 | RpoC1, PEPβ′-3 | Transcription | 4 | 0.162 | ATCG00180.1 | Osp1g00250.1 | |||
| GRMZM2G143392_P01 | 3 | SIG2 Sigma factor (Sig2-1) | Transcription | 4 | 0.419 | 0.015 | AT1G08540.1 | Os11g26160.1 | ||
| GRMZM2G003182_P01 | 3 | SIG2 Sigma factor (Sig2-2, -3, -4) | Transcription | 0.019 | AT1G08540.1 | Os11g26160.1 | ||||
| GRMZM2G100086_P01 | ||||||||||
| GRMZM2G164084_P01 | ||||||||||
| GRMZM2G053953_P01 | tRNA methyltransferase | Transcription | 4 | 0.035 | AT3G56330.1 | Os05g25880.1 | ||||
| GRMZM2G086277_P01 | Zinc-finger C2H2-type protein | Transcription | 4 | 1c | 0.557 | AT5G52010.1 | Os06g07020.1 | |||
| GRMZM2G042879_P01 | pTAC12 (HEMERA) | Transcriptional regulation | 4 | 1c | 2.343 | 0.353 | AT2G34640.1 | Os01g56350.3 | ||
| GRMZM2G466780_P01 | FLN1, pfkB-type carbohydrate kinase | Transcription (regulation PEP) | 4 | 1c | 1.564 | 0.162 | AT3G54090.1 | Os01g63220.2 | ||
| GRMZM2G103843_P01 | FLN2, pfkB-type carbohydrate kinase | Transcription (regulation PEP) | 4 | 1c | 4.488 | 0.996 | AT1G69200.1 | Os03g40550.1 | ||
| GRMZM2G007453_P01 | APO1, model 0.4 correct | RNA splicing | 0.012 | AT1G64810.2 | Os04g52520.2 | |||||
| GRMZM2G063473_P01 | APO2 | RNA splicing | 4 | 0.307 | 0.059 | AT5G57930.2 | Os02g50010.1 | |||
| AC199526.5_FGP003 | CAF2 (CRM domain) | RNA splicing | 4,4 | 0.650 | AT1G23400.1 | Os01g21990.1 | ||||
| AC199526.5_FGP004 | ||||||||||
| GRMZM2G094072_P01 | CFM2 (CRM domain) | RNA splicing | 4 | 1d | 2.653 | AT3G01370.1 | Os04g39060.1 | |||
| GRMZM2G436001_P02 | CFM3 (CRM domain) | RNA splicing | 4 | 1d | 0.720 | 0.118 | AT4G36390.1 | Os11g37990.1 | ||
| GRMZM2G142740_P01 | CFM3-like (CRM domain) | RNA splicing | 4 | 1d | 0.806 | AT3G23070.1 | Os09g19850.1 | |||
| GRMZM2G161506_P01 | CRS1 (CRM domain) | RNA splicing | 4 | 1d | 0.533 | 0.103 | AT2G21350.1 | Os10g36860.1 | ||
| GRMZM2G025329_P01 | CRS1/YhbY and CRM domain protein | RNA splicing | 4 | 1d | 3.461 | AT3G18390.1 | Os05g47850.1 | |||
| GRMZM2G132021_P03 | CRS2 | RNA splicing | 4 | 1d | 1.026 | 0.088 | AT5G38290.2 | Os03g22610.2 | ||
| NP_043005 | matK maturase | RNA splicing | 4 | 0.327 | ATCG00040.1 | Osp1g00120.1 | ||||
| GRMZM2G325019_P01 | OTP51 (Ath) ycf3 splicing (PPR) | RNA splicing | 4 | 0.351 | 0.029 | AT2G15820.1 | Os02g47360.1 | |||
| GRMZM2G025409_P01 | PPR5, trnG RNA stability and splicing | RNA splicing | 4 | 1d | 0.922 | 0.088 | AT4G39620.1 | Os02g51480.1 | ||
| GRMZM2G035820_P01 | RNC1 | RNA splicing | 4 | 1d | 3.972 | 1.330 | AT4G37510.1 | Os01g59510.1 | ||
| GRMZM2G403797_P01 | WTF1 | RNA splicing | 3 | 1d | 2.359 | 2.428 | AT4G01037.1 | Os05g49610.1 | ||
| GRMZM2G131332_P01 | CRR22, PPR protein (DYW in Ath) | RNA editing | 4 | 0.389 | AT1G11290.1 | Os11g29230.1 | ||||
| GRMZM2G103078_P01 | CRR22 (Ath), DYW-containing PPR protein | RNA editing | 0.012 | AT1G11290.1 | Os06g02200.1 | |||||
| GRMZM2G144843_P01 | CRR28 (Ath), DYW-containing PPR protein | RNA editing | 0.026 | AT1G59720.1 | Os07g09370.1 | |||||
| GRMZM2G426888_P01 | OPT82 (Ath; PPR domain) | RNA editing | 0.023 | AT1G08070.1 | Os06g02400.1 | |||||
| GRMZM2G026614_P01 | CP33, RNA-binding protein (RRM) | Stabilization and editing | 4 | 1c | 1.198 | 0.814 | 0.358 | AT3G52380.1 | Os07g06450.1 | |
| GRMZM2G090271_P01 | CP33, RNA-binding protein (RRM) | Stabilization and editing | 2 | 0.141 | 0.557 | 0.119 | AT2G35410.1 | Os04g50110.1 | ||
| GRMZM2G050697_P01 | HCF152 (Ath) psbH, petB RNA stability (PPR) | mRNA stability | 4 | 0.283 | AT3G09650.1 | Os12g01210.1 | ||||
| GRMZM2G380195_P06 | CRR2, ndhB RNA processing (PPR) | mRNA processing | 0.019 | AT3G46790.1 | Os01g62910.1 | |||||
| GRMZM2G083950_P01 | CRP1, mRNA stability and translation (petA, psaC, petB, petD; PPR domain) | mRNA stability (PPR) | 4 | 1c | 3.754 | 0.059 | AT5G42310.1 | Os07g36390.1 | ||
| GRMZM2G018929_P01 | MRL1 (PPR protein), rbcL mRNA stabilization | mRNA stability (PPR) | 4 | 0.285 | 0.003 | AT4G34830.1 | Os10g10170.1 | |||
| GRMZM2G177169_P01 | PPR10, mRNA stability and translation (atpH, psaJ) | mRNA stability (PPR) | 4 | 0.447 | AT2G18940.1 | Os05g19380.1 | ||||
| GRMZM2G121960_P01 | HCF107 (Ath; HAT repeat; psbH) | mRNA stability (TPR) | 4 | 1c | 2.425 | 0.029 | AT3G17040.1 | Os03g19560.1 | ||
| GRMZM2G372632_P01 | PGR3 (Ath) petL RNA stability (PPR domain) | mRNA stability (PPR) | 4 | 1d | 1.384 | AT4G31850.1 | Os10g28600.1 | |||
| GRMZM2G418206_P01 | PRORP2, RNaseP (PPR domain) | RNA cleavage and decay | 4 | 0.084 | AT2G32230.1 | Os02g17360.1 | ||||
| GRMZM2G111216_P01 | Rap41/CSP41A | RNA cleavage and decay | 1 | 0.171 | 0.212 | AT3G63140.1 | Os07g11110.1 | |||
| GRMZM2G002642_P01 | PRORP1, RNaseP (PPR domain) | RNA cleavage and decay | 4 | 0.160 | 0.088 | AT2G32230.1 | Os04g59600.1 | |||
| GRMZM2G103315_P01 | 15 | RNaseJ, RNase | RNA cleavage and decay | 4 | 1c | 7.963 | 0.793 | AT5G63420.1 | Os02g33610.1 | |
| GRMZM2G134439_P01 | 15 | RNaseJ, RNase | RNA cleavage and decay | 4 | 1c | 2.224 | AT5G63420.1 | Os02g33610.1 | ||
| GRMZM2G080546_P02 | Rap38/CSP41B-2 | RNA cleavage and decay | 4,4 | 5.662 | 0.029 | 1.178 | AT1G09340.1 | Os12g23180.1 | ||
| GRMZM2G165655_P01 | ||||||||||
| GRMZM2G328309_P01 | RNaseE/G-type endoribonuclease | RNA cleavage and decay | 4 | 1c | 1.493 | AT2G04270.2 | Os08g23430.1 | |||
| GRMZM2G082260_P01 | 3′ to 5′ exoribonuclease, RNR1 | RNA cleavage and decay | 0.044 | 0.007 | AT5G02250.1 | Os01g62440.1 | ||||
| GRMZM2G377761_P01 | RIF10/PNPase, 3′ exoribonuclease/polyadenylase | RNA cleavage and decay | 4 | 1c | 5.654 | 0.295 | 0.136 | AT3G03710.1 | Os07g07310.1 | |
| GRMZM2G117642_P02 | Endoribonuclease l-PSP | RNA cleavage and decay | 2.007 | 0.177 | AT3G20390.1 | Os07g33240.1 | ||||
| GRMZM2G097282_P01 | tRNA/rRNA methyltransferase (SpoU) | tRNA/rRNA maturation | 4 | 1c | 3.015 | 0.132 | AT2G19870.1 | Os01g53890.1 | ||
| GRMZM2G165694_P01 | 16S rRNA-processing RimM protein | rRNA processing | 4 | 1c | 1.327 | 0.329 | AT5G46420.1 | Os05g49130.2 | ||
| GRMZM2G060070_P01 | spoU rRNA methylase protein | rRNA methylation | 4 | 0.276 | 0.029 | AT4G38020.1 | Os03g48050.1 | |||
| GRMZM2G010801_P01 | RsmD, 16SrRNA methyltransferase | rRNA methylation | 4 | 0.256 | 0.103 | AT3G28460.1 | Os03g52640.3 | |||
| GRMZM2G175867_P02 | RH39, DEAD/DEAH box helicase (23S mat., AthNARA12) | rRNA maturation | 4 | 1c | 4.790 | 0.784 | AT4G09730.1 | Os01g08930.1 | ||
| GRMZM2G094526_P01 | yrdC domain protein (ribosome biogenesis/rRNA maturation) | rRNA maturation | 4 | 0.031 | AT3G01920.1 | Os08g41910.1 | ||||
| GRMZM2G163072_P01 | 6 | RH3-1, DEAD box RNA helicase | rRNA maturation and splicing | 4 | 1c | 23.685 | 2.411 | 0.039 | AT5G26742.2 | Os03g61220.2 |
| GRMZM2G415491_P01 | 6 | RH3-2, DEAD box RNA helicase | rRNA maturation and splicing | 4 | 1d | 7.572 | 1.991 | AT5G26742.2 | Os03g61220.1 | |
| GRMZM2G174669_P01 | rRNA methylase (PFC1) | rRNA methylation | 0.020 | AT1G01860.1 | Os02g03050.1 | |||||
| GRMZM2G125844_P01 | tRNase-Z (3′ end processing) | tRNA cleavage | 4 | 0.235 | AT2G04530.1 | Os09g30466.1 | ||||
| GRMZM2G050845_P01 | RAP domain protein | Unknown (RAP) | 4 | 1c | 1.213 | 0.176 | AT2G31890.1 | Os03g31150.3 | ||
| GRMZM2G078362_P01 | CP29, RNA-binding protein (RRM) | Unknown (RRM) | 0.017 | AT1G60000.1 | Os02g57010.1 | |||||
| GRMZM2G158835_P01 | 8 | CP29B′-1, RNA-binding protein (RRM) | Unknown (RRM) | 2 | 2 | 1.929 | 4.741 | 0.536 | AT2G37220.1 | Os07g43810.1 |
| GRMZM2G068715_P01 | 8 | CP29B′-2, RNA-binding protein (RRM) | Unknown (RRM) | 2 | 2 | 2.771 | 38.888 | 0.029 | AT2G37220.1 | Os07g43810.1 |
| GRMZM2G042683_P01 | CP29B′-3, RNA-binding protein (RRM) | Unknown (RRM) | 2 | 2 | 0.645 | 2.525 | AT2G37220.1 | Os03g25960.1 | ||
| GRMZM2G011129_P01 | CP31A, RNA-binding protein (RRM) | Unknown (RRM) | 4 | 1b | 4.373 | 2.962 | 3.159 | AT4G24770.1 | Os09g39180.1 | |
| GRMZM2G123234_P01 | CP31A, RNA-binding protein (RRM) | Unknown (RRM) | 2 | 2 | 1.846 | 3.220 | 2.167 | AT4G24770.1 | Os08g44290.1 | |
| GRMZM2G158462_P01 | CP31A, RNA-binding protein (RRM) | Unknown (RRM) | 2 | 2 | 0.979 | 1.483 | 0.607 | AT4G24770.1 | Os08g44290.1 | |
| GRMZM2G023591_P01 | RNA-binding protein (RRM) | Unknown (RRM) | 3 | 1c | 1.425 | 0.957 | 0.169 | AT4G09040.1 | Os08g37700.3 | |
| GRMZM2G044422_P02 | RNA-binding protein (RRM) | Unknown (RRM) | 4 | 0.136 | 0.044 | AT3G20930.1 | Os02g49820.1 | |||
| GRMZM2G070015_P01 | RNA-binding protein (RRM) | Unknown (RRM) | 4 | 1d | 0.902 | 0.162 | AT1G70200.1 | Os02g22070.1 | ||
| AC205677.3_FGP005 | RRM domain and kinesin domain | Unknown (RRM) | 4 | 1c | 1.834 | 0.535 | 0.104 | AT3G63480.2 | Os08g02380.1 | |
| GRMZM2G016084_P01 | S1 RNA-binding domain protein | Unknown (S1) | 0.044 | 0.189 | AT3G23700.1 | Os03g62780.1 | ||||
| GRMZM2G087628_P01 | S1 RNA-binding domain protein | Unknown (S1) | 4 | 1c | 0.781 | 0.059 | AT1G71720.1 | Os01g40640.1 | ||
| GRMZM2G088114_P01 | S1 RNA-binding domain protein | Unknown (S1) | 4 | 1b | 0.974 | AT1G12800.1 | Os02g02390.1 | |||
| GRMZM2G546254_P01 | S1 RNA-binding domain protein | Unknown (S1) | 4 | 1c | 1.080 | AT1G71720.1 | Os05g45920.1 | |||
| AC205564.3_FGP001 | S4 RNA-binding domain protein | Unknown (S4) | 4 | 0.333 | 0.029 | AT1G53120.1 | Os01g54390.2 | |||
| GRMZM2G122116_P01 | pTAC2, PPR protein | Unknown (PPR) | 4 | 1c | 7.492 | 1.418 | AT1G74850.1 | Os03g60910.1 | ||
| GRMZM2G150030_P01 | PPR protein | Unknown (PPR) | 4 | 1c | 7.318 | 0.544 | AT3G53700.1 | Os03g40020.1 | ||
| GRMZM2G438524_P01 | PPR protein | Unknown (PPR) | 4 | 1d | 5.037 | 0.706 | AT5G46580.1 | Os03g63910.1 | ||
| GRMZM2G341621_P01 | PPR2 protein | Unknown (PPR) | 4 | 1d | 3.387 | 0.294 | AT3G06430.1 | Os03g19650.3 | ||
| GRMZM2G005938_P01 | PPR protein | Unknown (PPR) | 4 | 1d | 3.023 | 0.162 | AT3G59040.2 | Os08g09270.1 | ||
| GRMZM2G404043_P01 | PPR protein | Unknown (PPR) | 4 | 1b | 1.008 | AT1G09900.1 | Os01g57410.2 | |||
| GRMZM2G059449_P01 | PPR protein | Unknown (PPR) | 4 | 1d | 0.868 | AT1G30610.1 | Os01g48380.1 | |||
| GRMZM2G071162_P01 | PPR protein | Unknown (PPR) | 4 | 1d | 0.859 | AT5G48730.1 | Os06g07550.1 | |||
| GRMZM2G105542_P01 | 9 | PPR protein | Unknown (PPR) | 4 | 1a | 0.826 | AT1G09900.1 | Os02g35750.2 | ||
| GRMZM2G127963_P01 | 9 | PPR protein | Unknown (PPR) | 4 | 0.412 | AT1G09900.1 | Os02g35750.2 | |||
| GRMZM2G076257_P01 | PPR protein | Unknown (PPR) | 4 | 1d | 0.820 | AT4G18750.1 | Os01g10800.1 | |||
| GRMZM2G053196_P01 | PPR protein | Unknown (PPR) | 4 | 1d | 0.818 | AT3G18110.1 | Os06g09880.1 | |||
| GRMZM2G128665_P01 | P67, PPR protein | Unknown (PPR) | 4 | 1c | 0.625 | 0.103 | AT4G16390.1 | Os03g11670.1 | ||
| GRMZM2G353195_P01 | PPR protein | Unknown (PPR) | 4 | 1c | 0.588 | 0.074 | AT1G09900.1 | Os03g21780.1 | ||
| GRMZM2G076950_P01 | PPR protein | Unknown (PPR) | 3 | 1d | 0.518 | 0.368 | 0.006 | AT1G02150.1 | Os04g46010.1 | |
| GRMZM2G475743_P01 | PPR protein | Unknown (PPR) | 4 | 0.449 | AT5G39980.1 | Os01g61630.1 | ||||
| GRMZM2G003869_P02 | PPR protein with Arc domain | Unknown (PPR) | 4 | 0.432 | 0.059 | AT5G25630.1 | Os01g12810.4 | |||
| GRMZM2G076771_P01 | PPR protein | Unknown (PPR) | 4 | 0.416 | AT5G13770.1 | Os04g49350.1 | ||||
| GRMZM2G132140_P01 | PPR protein | Unknown (PPR) | 4 | 0.371 | AT1G71460.1 | Os12g10184.1 | ||||
| GRMZM2G176523_P01 | PPR protein | Unknown (PPR) | 4 | 0.303 | AT4G35130.1 | Os03g04390.1 | ||||
| GRMZM2G176419_P01 | PPR protein | Unknown (PPR) | 4 | 0.282 | 0.029 | AT1G10910.1 | Os01g37870.4 | |||
| GRMZM2G103250_P01 | PPR protein | Unknown (PPR) | 4 | 0.263 | AT4G30825.1 | Os09g25550.1 | ||||
| GRMZM2G164202_P01 | PPR protein | Unknown (PPR) | 4 | 0.244 | AT2G17033.2 | Os02g02770.1 | ||||
| GRMZM2G143646_P01 | PPR protein | Unknown (PPR) | 4 | 0.240 | AT1G19720.1 | Os03g59264.1 | ||||
| GRMZM2G440537_P02 | PPR protein | Unknown (PPR) | 4 | 0.183 | AT3G53170.1 | Os03g02430.1 | ||||
| GRMZM2G170896_P01 | PPR protein | Unknown (PPR) | 4 | 0.174 | AT5G03800.1 | Os07g07620.1 | ||||
| GRMZM2G440349_P01 | PPR protein | Unknown (PPR) | 4 | 0.172 | AT4G01030.1 | Os05g50950.1 | ||||
| GRMZM2G092739_P01 | PPR protein with BTAD domain | Unknown (PPR) | 4 | 0.142 | 0.029 | AT5G02860.1 | Os07g40120.1 | |||
| GRMZM2G056116_P01 | PPR protein | Unknown (PPR) | 4 | 0.139 | AT3G46610.1 | Os12g18640.1 | ||||
| GRMZM2G093291_P01 | PPR protein with BTAD domain | Unknown (PPR) | 4 | 0.128 | AT5G27270.1 | Os06g02120.1 | ||||
| GRMZM2G007372_P01 | PPR protein | Unknown (PPR) | 4 | 0.090 | AT2G35130.1 | Os02g07360.1 | ||||
| GRMZM2G421231_P01 | PPR protein | Unknown (PPR) | 4 | 0.089 | AT1G09900.1 | Os01g57410.2 | ||||
| GRMZM2G019901_P01 | 1 | PPR and CBS domain protein | Unknown (PPR) | 4 | 0.037 | AT5G10690.1 | Os09g26190.1 | |||
| AC206761.3_FGP002 | 1 | PPR and CBS domain protein | Unknown (PPR) | 4 | 0.075 | AT5G10690.1 | Os09g26190.1 | |||
| GRMZM2G105827_P01 | 11 | PPR protein | Unknown (PPR) | 4 | 0.068 | AT4G21190.1 | Os01g55290.2 | |||
| GRMZM2G040970_P02 | 11 | PPR protein | Unknown (PPR) | 4 | 0.042 | AT4G21190.1 | Os01g55290.2 | |||
| GRMZM2G074599_P01 | PPR protein | Unknown (PPR) | 4 | 0.068 | AT3G49240.1 | Os11g24570.1 | ||||
| GRMZM2G090576_P01 | PPR protein | Unknown (PPR) | 4 | 0.063 | 0.022 | AT1G09900.1 | Os03g37260.1 | |||
| GRMZM2G092123_P01 | PPR protein | Unknown (PPR) | 4 | 0.062 | AT5G02830.1 | Os03g17634.1 | ||||
| GRMZM2G012174_P01 | PPR protein | Unknown (PPR) | 4 | 0.055 | AT5G50280.1 | Os05g22870.1 | ||||
| GRMZM2G156472_P01 | PPR protein | Unknown (PPR) | 4 | 0.043 | AT1G31920.1 | Os02g46980.1 | ||||
| GRMZM2G465087_P01 | PPR protein | Unknown (PPR) | 0.025 | AT4G21300.1 | Os03g56850.1 | |||||
| GRMZM2G039016_P01 | PPR protein | Unknown (PPR) | 0.023 | AT1G09900.1 | Os01g57410.2 | |||||
| GRMZM2G017821_P01 | 5 | PPR protein | Unknown (PPR) | 0.021 | AT2G15690.1 | Os04g09530.1 | ||||
| GRMZM2G073551_P01 | 5 | PPR protein | Unknown (PPR) | 0.021 | AT2G15690.1 | Os04g09530.1 | ||||
| GRMZM2G409997_P01 | 5 | PPR protein | Unknown (PPR) | 0.006 | AT2G15690.1 | Os04g09530.1 | ||||
| AC185612.3_FGP001 | DG1, PPR (delayed greening 1; EMB1408) | Unknown (PPR) | 0.019 | AT5G67570.1 | Os05g25060.1 | |||||
| GRMZM2G333142_P01 | PPR protein | Unknown (PPR) | 0.015 | AT4G18520.1 | Os07g14100.1 | |||||
| GRMZM2G166407_P01 | PPR protein | Unknown (PPR) | 0.012 | AT3G42630.1 | Os08g40870.1 | |||||
| Proteins with unknown functions but with specific prediction domains (TPR, DEAD, DnaJ, mTERF, rhodanese) and pTACs with unassigned functions | ||||||||||
| GRMZM2G029933_P01 | mTERF protein | Unknown (mTERF) | 4 | 1d | 3.702 | 0.217 | AT4G02990.1 | Os05g33500.1 | ||
| GRMZM2G017355_P01 | mTERF protein | Unknown (mTERF) | 4 | 0.491 | 0.029 | AT1G21150.1 | Os02g51450.2 | |||
| GRMZM2G024550_P01 | mTERF protein | Unknown (mTERF) | 4 | 0.335 | 0.029 | AT4G14605.1 | Os02g39040.1 | |||
| GRMZM2G017429_P01 | mTERF protein | Unknown (mTERF) | 2 | 0.259 | 0.403 | AT5G07900.1 | Os02g51450.2 | |||
| GRMZM2G426154_P01 | mTERF protein | Unknown (mTERF) | 4 | 0.165 | AT2G21710.1 | Os07g04230.1 | ||||
| GRMZM2G157716_P01 | mTERF protein | Unknown (mTERF) | 4 | 0.099 | AT2G36000.1 | Os03g24590.1 | ||||
| GRMZM2G142150_P01 | mTERF protein | Unknown (mTERF) | 4 | 0.078 | AT1G78930.1 | Os09g38720.1 | ||||
| GRMZM2G130773_P01 | mTERF protein | Unknown (mTERF) | 4 | 0.030 | AT5G55580.1 | Os07g39430.1 | ||||
| GRMZM2G168665_P01 | mTERF protein | Unknown (mTERF) | 0.025 | AT2G36000.1 | Os03g24590.1 | |||||
| GRMZM2G060114_P01 | mTERF protein | Unknown (mTERF) | 0.013 | AT5G07900.1 | Os06g12100.1 | |||||
| GRMZM2G128434_P01 | DEAD box helicase | Unknown (DEAD) | 4 | 1b | 0.661 | AT5G35970.1 | Os09g04440.1 | |||
| GRMZM2G399212_P01 | DEAD box helicase | Unknown (DEAD) | 4 | 0.139 | AT2G35920.1 | Os03g53760.1 | ||||
| AC198169.4_FGP004 | DEAD box helicase | Unknown (DEAD) | 4 | 1c | 2.843 | 0.118 | AT1G12770.1 | Os02g42406.1 | ||
| GRMZM2G113267_P02 | DEAD/DEAH box helicase | Unknown (DEAD) | 4 | 1c | 4.360 | 0.530 | AT3G06980.1 | Os03g01830.1 | ||
| GRMZM2G319509_P01 | DEAD/DEAH box helicase | Unknown (DEAD) | 4 | 1c | 2.450 | AT1G70070.1 | Os02g50560.1 | |||
| GRMZM2G100043_P01 | DEAD/DEAH box helicase | Unknown (DEAD) | 2 | 0.047 | 0.113 | AT1G59990.1 | Os09g21520.2 | |||
| GRMZM2G373175_P01 | DRSM and NACHT domain protein | Unknown (DEAD) | 4 | 0.447 | AT1G48650.1 | Os01g02884.1 | ||||
| GRMZM2G026371_P01 | RH26, DEAD box RNA helicase | Unknown (DEAD) | 4 | 1c | 8.409 | 1.445 | AT5G08610.1 | Os01g43120.1 | ||
| GRMZM2G009070_P01 | MurE-like | Unknown (pTAC) | 4 | 1c | 7.491 | 0.369 | AT1G63680.1 | Os10g40130.1 | ||
| GRMZM2G091419_P01 | pTAC10 (L30 N-terminal domain) | Unknown (pTAC) | 4 | 1c | 4.434 | 0.485 | AT3G48500.1 | Os01g67570.1 | ||
| GRMZM2G111965_P01 | pTAC14 (SET domain) | Unknown (pTAC) | 4 | 1c | 2.378 | 0.147 | AT4G20130.1 | Os05g50980.1 | ||
| GRMZM2G180418_P01 | pTAC17 (various nucleotide BD) | Unknown (pTAC) | 2 | 0.306 | 0.906 | 0.081 | AT1G80480.1 | Os02g55630.1 | ||
| GRMZM2G306104_P01 | pTAC18, Cupin domain | Unknown (pTAC) | 2 | 0.052 | 0.118 | AT2G32650.2 | Os02g54360.1 | |||
| GRMZM2G440746_P01 | pTAC6 | Unknown (pTAC) | 4 | 1c | 1.974 | 0.649 | AT1G21600.2 | Os11g01890.1 | ||
| GRMZM2G020785_P01 | pTAC7 | Unknown (pTAC) | 4 | 0.257 | 0.118 | AT5G24314.1 | Os01g50930.1 | |||
| GRMZM2G159924_P01 | pTAC7 | Unknown (pTAC) | 4 | 0.069 | 0.015 | AT5G24314.1 | Os01g50930.1 | |||
| GRMZM2G071996_P01 | DnaJ domain protein | Unknown (DnaJ) | 1 | 0.013 | 0.048 | AT1G75690.1 | Os03g14040.3 | |||
| GRMZM2G055178_P01 | DnaJ domain protein | Unknown (DnaJ) | 0.015 | AT4G13670.1 | Os04g43420.1 | |||||
| GRMZM2G057091_P01 | DnaJ domain protein | Unknown (DnaJ) | 3 | 0.214 | 0.159 | AT1G08640.1 | Os08g03380.1 | |||
| GRMZM2G054076_P02 | DnaJ domain protein | Unknown (DnaJ) | 4 | 1c | 0.519 | 0.029 | 0.041 | AT1G80030.3 | Os02g56040.1 | |
| GRMZM2G125304_P03 | 2 | DnaJ domain protein (completely unrelated to gene models 1, 2, and 4–6) | Unknown (DnaJ) | 0.027 | AT2G22360.1 | Os05g26914.2 | ||||
| GRMZM2G010389_P01 | DnaJ domain protein (maybe envelope) | Unknown (DnaJ) | 4 | 0.048 | 0.006 | AT5G23040.1 | Os09g21250.1 | |||
| GRMZM2G091811_P01 | 2 | DnaJ domain protein (DnaJ5) | Unknown (DnaJ) | 4 | 1c | 6.110 | 0.676 | 0.021 | AT2G22360.1 | Os05g26914.2 |
| GRMZM2G031721_P01 | pTAC5, DnaJ and PG domains | Unknown (pTAC) | 3 | 1c | 1.550 | 1.085 | 0.076 | AT4G13670.1 | Os04g43420.1 | |
| GRMZM2G068316_P01 | DnaJ domain protein | Unknown (DnaJ) | 0.088 | 1.746 | AT5G21430.1 | Os12g31460.1 | ||||
| GRMZM2G050118_P01 | DnaJ domain protein | Unknown (DnaJ) | 0.018 | AT1G08640.1 | Os08g03380.1 | |||||
| GRMZM2G124644_P01 | DnaJ domain protein | Unknown (DnaJ) | 0.015 | 0.200 | AT4G09350.1 | Os11g10990.1 | ||||
| GRMZM2G113340_P01 | DnaJ domain protein | Unknown (DnaJ) | 0.124 | none | Os05g01950.1 | |||||
| GRMZM2G029698_P02 | TPR protein | TPR protein | 4 | 1c | 0.667 | 0.015 | 0.007 | AT1G78915.1 | Os07g18720.1 | |
| GRMZM2G089075_P01 | TPR protein | TPR protein | 2 | 0.019 | 0.029 | 0.006 | AT4G39470.1 | Os05g08930.1 | ||
| GRMZM2G010973_P01 | TPR protein | TPR protein | 0.103 | 0.121 | AT2G37400.1 | Os02g45880.1 | ||||
| GRMZM2G371670_P01 | TPR protein | TPR protein | 0.031 | AT3G05625.1 | Os01g24090.1 | |||||
| GRMZM2G312910_P02 | TPR protein Shoot1 | TPR protein | 0.103 | 1.642 | AT1G55480.1 | Os07g07540.1 | ||||
| GRMZM2G129080_P01 | 10 | DAG protein | DAG | 2 | 0.204 | 0.333 | 0.001 | AT2G33430.1 | Os04g51280.1 | |
| GRMZM2G175447_P01 | 10 | DAG protein | DAG | 2 | 0.034 | 0.075 | 0.001 | AT2G33430.1 | Os04g51280.1 | |
| GRMZM2G096391_P01 | Rhodanese-like domain protein | Unknown (rhodanese) | 1 | 0.260 | 0.059 | 2.240 | AT3G59780.1 | Os01g67120.1 | ||
| GRMZM2G122715_P02 | Thylakoid Ca2+ sensing receptor, phosphoprotein and rhodanese domain | Unknown (rhodanese) | 4 | 1b | 9.333 | 0.423 | 2.037 | AT5G23060.1 | Os02g49680.1 | |
| GRMZM2G011520_P01 | Rhodanese-like domain protein | Unknown (rhodanese) | 0.011 | AT5G03455.1 | Os10g39860.2 | |||||
| GRMZM2G048085_P01 | Rhodanese-like domain protein | Unknown (rhodanese) | 0.094 | AT4G35770.1 | Os06g50930.1 | |||||
| GRMZM2G052610_P01 | Rhodanese-like domain protein | Unknown (rhodanese) | 0.630 | 0.136 | AT4G27700.1 | Os09g36040.1 | ||||
| GRMZM2G087041_P01 | Rhodanese-like domain protein (one TMD predicted) | Unknown (rhodanese) | 2.256 | 0.605 | AT2G42220.1 | Os09g10750.1 | ||||
| GRMZM2G377487_P01 | Rhodanese-like domain protein | Unknown (rhodanese) | 0.015 | AT1G17850.1 | Os03g64350.1 | |||||
Protein accession number. Accession numbers in boldface and underlined are assigned to the nucleoid based on fulfilling three criteria: (1) they are in cluster 4 (Fig 2C); (2) they have a nucleoid-proplastid ratio of more than 3 (or were not detected in proplastids); and (3) they have a nucleoid-chloroplast ratio of more than 10 (or were not detected in chloroplasts).
CRG, Closely related group; shared peptides, but also unique peptides.
Function based on experimental information in maize or plant homologs or based on predicted PFAM domains.
Cluster number from the dendrogram in Figure 2C. Clusters 1 and 5 mostly represented contaminating proteins with high accumulation in chloroplasts, whereas clusters 2 and 3 mostly represented contaminating proteins with high accumulation in proplastids. Cluster 4 (in boldface) should be considered nucleoid-enriched proteins.
Cluster number from the dendrogram in Supplemental Figure S5, with the purpose to determine developmental effects on nucleoid composition. Cluster 1 (in boldface), with proteins that should be mostly considered nucleoid associated, is split into four subclusters, a to d. Subclusters 1b and 1d are base and tip enriched, respectively; subclusters 1a and 1c do not show developmental effects. Clusters 2 and 3 represent proteins with high accumulation in chloroplasts and/or proplastids.
Protein abundance based on NadjSPC × 1,000.
Figure 3.
Comparison of protein investments of chloroplasts, proplastids, and nucleoids for the proteins listed in Table I. A, Relative abundance of the proteins in the three main functions as in Table I, namely DNA ([putative] DNA-related functions: DNA replication, DNA organization and quality control, anchoring, and DNA repair), RNA (putative RNA-related functions: transcription, RNA splicing, editing, cleavage, stability, rRNA and tRNA maturation, and other predicted RNA-binding proteins with unknown functions), and Unknown (proteins with unknown functions with TPR, mTERF, DEAD, rhodanese, or DnaJ domains or pTAC proteins with unknown functions). B, Relative mass distribution of proteins within the function DNA. C, Relative abundance of proteins within the function RNA. D, Relative abundance of proteins within the function Unknown. Details are provided in Table I.
Proteins Predicted To Be Involved in Plastid DNA Replication, Repair, or Organization (Bin 28)
This functional category included 33 proteins; 25 of these were detected in nucleoids and only eight in chloroplasts and/or proplastids (Table I; top section). Most proteins were detected in both nucleoids and proplastids, but several were many-fold enriched in the nucleoid. Together, this suggests that many of these proteins were specifically located in the nucleoid. The most abundant (NadjSPC > 0.002, normalized spectral abundance factor > 0.001) and highly nucleoid-enriched proteins (high nucleoid-plastid ratio) with DNA-related functions were DNA gyrase A, two pTAC3 co-orthologs with SAP domains (a DNA-binding motif; Aravind and Koonin, 2000), the coiled-coil protein MFP1-1 (Meier et al., 1996; Jeong et al., 2003; Samaniego et al., 2006), and the nucleid acid-binding protein WHY1 (also known as pTAC1) involved in genome stability and RNA splicing (Prikryl et al., 2008; Maréchal et al., 2009; Table I).
These 33 proteins were divided into four putative functional groups: DNA anchoring, DNA organization and quality control, DNA replication, and DNA repair (Table I; Fig. 3B). The relative investments across these four functions differed strongly between chloroplasts, proplastids, and nucleoids (Fig. 3B). Proteins involved in DNA organization and quality control were particularly overrepresented in nucleoids, with DNA gyrase A, WHY1/pTAC1, and two pTAC3 homologs being the most abundant (Table I). The polymerases involved in DNA replication were of relatively low abundance in nucleoids and proplastids and were not detected in chloroplasts (Fig. 3B). DNA repair enzymes were enriched in nucleoids, but a few DNA repair enzymes were not detected in proplastids or nucleoids (a FAD photolyase and a uvrB/C motif protein), suggesting that they cycle on/off the nucleoid or that their interactions were disrupted during nucleoid purification (Fig. 3B). Three proteins (MFP1-1, pTAC16, and TCP34) were unusual in that they were abundant in nucleoids and proplastids but were also found at high or even higher relative concentrations in chloroplasts (Table I). Homologs of MFP1 and TCP34 have been shown to interact with plastid DNA and were proposed to anchor the DNA to the thylakoid membrane (Jeong et al., 2003; Weber et al., 2006). The quantitative distribution of the proteins observed here is consistent with such an anchoring function. The function of pTAC16 (GRMZM2G449496_P01) is unknown, but the protein is very abundant in chloroplast membranes (Majeran et al., 2008) but not in stroma (Friso et al., 2010). It is interesting that pTAC16 was observed in complexes of up to approximately 700 kD (Majeran et al., 2008) and showed strong induction along the developmental leaf gradient from base to tip (Majeran et al., 2010; Supplemental Fig. S2A; see also the expression viewer in PPDB [http://ppdb.tc.cornell.edu/dbsearch/plotgradient.aspx]). The distribution of pTAC16 between membranes and nucleoids is compatible with an anchoring function to chloroplast membranes, similar to TCP34 and MFP1.
We identified two PEND homologs (PEND-1 and PEND-2) in proplastids, neither of which was detected in chloroplasts or nucleoids. Within those proteins with DNA-related functions, PEND-1 is the most abundant protein in proplastids (Table I). PEND homologs in Arabidopsis and other species bind to plastid nucleoids in GFP fusion visualization experiments, and it has been proposed that PEND anchors the nucleoid to the inner envelope membrane (Sato et al., 1998; Terasawa and Sato, 2005a, 2005b). Our data support the notion that PEND is most important in nonphotosynthetic, developing plastids, but the interaction between PEND and nucleoid must be weak, as it does not withstand our nucleoid purification procedure. Interestingly and consistently, WHY1 (one of the most abundant nucleoid proteins) does not strictly colocalize with PEND (Melonek et al., 2010), while we find very high accumulation of WHY1 in nucleoids. This further supports the idea that PEND is not a strict nucleoid protein, at least not in maize. PEND was also not observed in pTAC complexes isolated from Arabidopsis (Pfalz et al., 2006).
Proteins Involved in Transcription and RNA Metabolism
We identified 131 proteins that are known or predicted to be involved in plastid transcription or RNA metabolism (Table I, middle section); these represent only 1% mass in chloroplasts but 8% in proplastids and 22% in nucleoids (Fig. 3A). The most abundant RNA-related proteins in the nucleoid fractions were the subunits of the PEP complex, several DEAD box RNA helicases (RH3 homologs and RH39), two nucleases (RNaseJ and polynucleotide phosphorylase RIF10/PNPase; Li et al., 1998; Baginsky et al., 2001), a few PPR proteins with unknown function (pTAC2, GRMZM2G150030_P01, and GRMZM2G438524_P01), FLN2, a protein kinase likely involved in the regulation of PEP activity (Arsova et al., 2010), and many splicing factors (e.g. RNC1, WTF1, APO1, and CRS1; Till et al., 2001; Asakura and Barkan, 2006; Watkins et al., 2007, 2011; Kroeger et al., 2009). Importantly, these proteins were not detected or were at very low levels in chloroplasts and were enriched (more than 3-fold) in nucleoids as compared with proplastids (Table I). This strongly suggests that most RNA processing occurs in association with the nucleoid (Fig. 3A).
To obtain a general overview of the functions of these proteins and their quantitative distribution across nucleoids, proplastids, and chloroplasts, we divided them into eight more narrowly defined functional groups: transcription and transcriptional regulation (e.g. PEP subunits, SIG2, NUSB, NUSG, ρ, BolA, pTAC12, FLN1/2), RNA splicing (e.g. PPR5, OTP51, RNC1, CRS1, CRS2, CFM2, CFM3, CAF2, APO1, APO2), RNA editing (e.g. OPT82, CRR22, CRR28), processing/stabilization of specific mRNAs (HCF152, CRR2, MRL1, CRP1, HCF107, PPR10, PGR3), RNA cleavage and decay (PROP/RNaseP, Rap41/CSP41A, RNaseE/G, RNaseJ, RNR1, RIF10/PNPase, l-PSP), rRNA and tRNA maturation and methylation (RH39, RG3, RH3-1/2 SPOU, PFC1, SrmD, RimM, tRNA-Z), PPR proteins with unknown functions (48 in total), and uncharacterized proteins with other RNA-binding domains (e.g. with RNA recognition motif [RRM], S1, and S4 domains). Figure 3C shows the quantitative distribution of these nine functional groups in nucleoids, proplastids, and chloroplasts.
In the cases of chloroplasts and proplastids, but not nucleoids, most (more than 70%) of the investments in RNA-related functions were in the abundant RRM proteins CP29 and CP31, here assigned as RNA-related proteins with unknown function, as well as RRM protein CP33, involved in editing and stabilization (Tillich et al., 2009). These proteins have been proposed to have general functions in RNA stabilization, RNA editing, and as RNA chaperones under cold stress (Ruwe et al., 2011). Proteins involved in transcription were strongly overrepresented in nucleoids (18 proteins) and virtually undetectable in chloroplasts, with low levels in proplastids; they included the highly abundant PEP subunits, four co-orthologs of Sigma2 (but no other Sigma factors), and various proteins that may function in the regulation of transcription (FLN1/2 and BolA homologs) or that have domains found in bacterial proteins involved in transcriptional termination (Rho protein) and coupling between transcription and translation (Nus proteins; Burmann et al., 2010; Proshkin et al., 2010). Proteins involved in tRNA and rRNA maturation and methylation were also overrepresented in nucleoids; by far, the most abundant proteins in this group were the orthologs of the DEAD box helicases RH39 and RH3 (Table I), which are discussed below.
Ribonucleases involved in either RNA cleavage or RNA decay showed a much more varied distribution between chloroplasts, proplastids, and nucleoids (Table I). We identified a total of 10 ribonucleases in the nucleoid and plastid preparations, including the well-studied proteins RNR1, PNPase, RNaseE/G, CSP41a/RAP41, and CSP41b/RAP38 (for review, see Stern et al., 2010). Only two of these nucleases (l-PSP and RNR1) were not detected in nucleoids, but they were detected in both chloroplasts and proplastids (Table I). l-PSP endonuclease was very abundant in chloroplasts and proplastids; its function has not been studied in plants. The nucleoid proteome further included four PPR proteins (MRL1, PPR10, CRP1, PGR3) and a “HAT” type of TPR protein (HCF107) known to be involved in the stabilization and/or translation of specific mRNA species. HCF107 and CRP1 were among the most abundant nucleoid proteins but were not detected in chloroplasts and were more than 50-fold enriched in nucleoids in comparison with unfractionated proplastids (Table I). These results suggest that these RNA-stabilizing proteins bind cotranscriptionally to their RNA ligands (see “Discussion”).
We also identified 48 PPR proteins with unknown function; they were all detected in nucleoids, 12 were also detected in proplastids and only one in chloroplasts, with PPR proteins consistently most abundant in nucleoids (Table I). Most of these can be anticipated to function in plastid RNA metabolism, based on the functions assigned to characterized PPR proteins (O’Toole et al., 2008; Schmitz-Linneweber and Small, 2008; Barkan, 2011). pTAC2 was the most abundant of the nucleoid-associated PPR proteins and was 1,200-fold more abundant than the least abundant PPR protein in the nucleoid. The strong enrichment of PPR proteins in nucleoids (Fig. 3C) further supports our conclusion that nucleoids are a key site of RNA metabolism.
Abundance and Distribution of TPR, mTERF, DEAD Box, Rhodanese, and DnaJ(-Like) Domain Proteins and pTAC Protein with Unknown Functions
Figure 3D shows the abundance of 52 proteins without known function and with predicted domains (mTERF, DEAD box, TPR, rhodanese, DnaJ) that play important roles in diverse aspects of plastid biogenesis. In addition, it shows the abundance of nine pTAC proteins without known functions. A total of 40, 34, and 22 of these proteins were found in nucleoids, proplastids, and chloroplasts, respectively. DEAD box proteins and pTAC proteins were the most abundant group in this set of proteins in nucleoids but were insignificant in unfractionated chloroplast samples (Fig. 3D).
mTERF proteins contain repeats of approximately 30 amino acids, the mTERF motif, that fold into helical hairpins resembling TPR and PPR motifs (Roberti et al., 2009). mTERF proteins in animal mitochondria have a range of functions, including transcription, DNA replication, and ribosome biogenesis (Pellegrini et al., 2009; Yakubovskaya et al., 2010; Cámara et al., 2011). The Arabidopsis genome encodes 35 mTERF proteins, and GFP fusion studies suggested that 11 of these are plastid localized, 17 are localized to mitochondria, while the localization of the remaining seven has not been determined (Babiychuk et al., 2011). Several plastid-localized mTERF proteins (At4g02990, BSM; At2g03050, SOLDAT10; At2g21710, EMB2219; At4g02990, RUGOSA2) have been genetically characterized in Arabidopsis; the null mutants have pale-green or embryo-lethal phenotypes with defects in plastid gene expression, but the precise molecular functions are unclear (Tzafrir et al., 2004; Meskauskiene et al., 2009; Babiychuk et al., 2011; Quesada et al., 2011). We identified 10 mTERF proteins in the maize nucleoid samples; eight of these have Arabidopsis orthologs based on reciprocal BLAST analysis, all of which were shown to be chloroplast localized in GFP fusion assays (Babiychuk et al., 2011). These 10 maize mTERF proteins are highly enriched in nucleoid samples in comparison with their concentrations in chloroplasts (seven passed the criteria that we used for nucleoid assignment; boldface in Table I). Thus, mTERF protein abundance generally parallels that of PPR proteins and correlates with early chloroplast development. The identification of so many nucleoid-localized mTERF proteins is intriguing and raises questions about the roles of these proteins in plastid gene expression and nucleoid function.
DEAD box proteins catalyze the ATP-dependent unwinding of double-stranded nucleic acids and/or remodel protein/nucleic acid complexes (Rocak and Linder, 2004; Linder, 2006; Hilbert et al., 2009). Three chloroplast DEAD box proteins have been studied in plastids: VDL (Wang et al., 2000), RH3 (Y. Asakura, E. Galarneau, K.P. Watkins, R.E. Williams-Carrier, A. Vichas, G. Friso, A. Barkan, and K.J. van Wijk, unpublished data), and RH39 (Nishimura et al., 2010): the precise function of VDL is unknown, whereas both RH3 and RH39 are involved in rRNA maturation; RH3, in addition, promotes the splicing of some group II introns (Y. Asakura, E. Galarneau, K.P. Watkins, R.E. Williams-Carrier, A. Vichas, G. Friso, A. Barkan, and K.J. van Wijk, unpublished data). Both of these were enriched in nucleoids, as were eight uncharacterized DEAD box proteins. None of these eight proteins were detected in the chloroplast samples, and only four of them were detected in proplastid samples, indicating that they were highly enriched in nucleoids (seven passed the criteria that we used for nucleoid assignment; boldface in Table I). Thus, these DEAD box proteins are likely to function in association with the nucleoid, for instance in ribosome biogenesis or splicing (Table I).
Except for pTAC17 and pTAC5, none of the nine pTAC proteins without known function were detected in chloroplast samples. The homolog of a bacterial MurE-like ligase (GRMZM2G009070_P01) had highest relative concentration among these pTAC proteins and was 20-fold enriched in nucleoids in comparison with proplastids (Table I). Mur ligases in bacteria are involved in peptidoglycan synthesis for cell walls (Smith, 2006), but homologs in plants are not involved in cell wall biogenesis (Takano and Takechi, 2010). MurE in the moss Physcomitrella patens is localized to the chloroplast, and MurE gene disruption prevented chloroplast division (Machida et al., 2006). However, inactivation of the MurE homolog in Arabidopsis (At1g63680) did not affect plastid division, but it did inhibit chloroplast biogenesis and reduce the abundance of RNA from PEP-dependent genes (Garcia et al., 2008). MurE was also found in the Arabidopsis pTAC preparation (Pfalz et al., 2006) and in the tobacco PEP complex (Suzuki et al., 2004; Supplemental Table S3). We thus suggest that maize MurE directly or indirectly influences PEP activity; this could be through an influence on DNA packaging or a more direct effect on PEP itself. The next most abundant pTAC proteins were pTAC10, pTAC14, and pTAC6 (cluster 4); these were between 3- and 16-fold enriched in comparison with unfractionated proplastids. pTAC17 with CobW nucleotide-binding domains and pTAC18 with a predicted Cupin domain were more abundant in the proplastid samples than in nucleoids (and part of cluster 2), suggesting that they cycle on and off the nucleoids (Table I).
Plastid DnaJ(-like) proteins play diverse roles in plastid biogenesis, including plastid division and protein assembly and disassembly, and the DnaJ domain is best known as a nucleotide-exchange factor of ATP-dependent chaperones such as HSP70 (Albrecht et al., 2008; Chen et al., 2010). Other well-studied plastid DnaJ domain proteins include Rubisco assembly factor BSD2 (Brutnell et al., 1999) and ARC6, involved in plastid division (Glynn et al., 2009). Recently, several plastid DnaJ domain proteins have been studied in Arabidopsis (Albrecht et al., 2008; Chiu et al., 2010; Chen et al., 2011), but none of them appeared to be localized to nucleoids. Of the 12 maize DnaJ proteins with unknown functions, eight were found in nucleoids; two of them (GRMZM2G091811_P01 and GRMZM2G054076_P02) stand out for their strong enrichment in the nucleoid fraction (Table I). The DnaJ domain protein pTAC5 (protein GRMZM2G031721_P0) was abundant in nucleoids, but it did not pass our criteria to be a candidate nucleoid protein. Escherichia coli nucleoids contain a DnaJ-related protein, CbpA, with DNA-binding activity involved in DNA aggregation that protects DNA from degradation by nucleases (Cosgriff et al., 2010). Therefore, it seems possible that some of the nucleoid-localized DnaJ domain proteins interact directly with DNA.
TPR proteins generally bind protein ligands and function in various protein targeting, scaffolding, and assembly processes (D’Andrea and Regan, 2003). Examples of chloroplast TPR proteins include the PSII assembly protein LPA1 (Peng et al., 2006) and the FLU protein involved in the regulation of tetrapyrrole synthesis (Peng et al., 2006). Two TPR proteins of unknown function were detected in nucleoids; in particular, GRMZM2G029698_P02 (TPR) was highly enriched in nucleoids compared with chloroplasts (95-fold) and proplastids (45-fold) and indeed passed our three criteria for nucleoid association.
We identified two maize homologs of the DAL or DAG protein, which are essential for early steps in chloroplast development in Arabidopsis and Antirrhinum (Chatterjee et al., 1996; Bisanz et al., 2003). The Arabidopsis DAG protein is required for the maturation of the plastid ribosomal RNAs, but the molecular mechanism is unclear. Interestingly, the maize homologs were strongly (more than 50 fold) enriched in nucleoids compared with chloroplasts but not when compared with proplastids. This suggests that these proteins can cycle on/off the nucleoid, similar to pTAC17 and pTAC18.
The rhodanese domain is a ubiquitous fold found in proteins that function in sulfur metabolism and other processes (Bordo and Bork, 2002; Cipollone et al., 2007). Among the nucleoid, chloroplast, and proplastid maize proteomes, we identified seven proteins of unknown functions with rhodanese domains, two of which were detected in nucleoids (GRMZM2G122715_P02 and GRMZM2G096391_P01; Table I). The rhodanese protein GRMZM2G122715_P02 was one of the most abundant proteins in nucleoids and is the maize homolog of the Arabidopsis thylakoid protein CaS (At5g23060.1). CaS is phosphorylated and has been suggested to be involved in stress responses and signaling (Vainonen et al., 2008). Although maize CaS with the nucleoid did not pass our stringent criteria to be firmly assigned as a nucleoid protein, its possible association with the nucleoid is very interesting, because this could provide a link between the redox/excitation state of the thylakoid and plastid gene regulation.
Proteins Involved in Plastid Protein Synthesis, Folding, Assembly, and Posttranslational Modifications
Thirty percent and 36% of the protein mass of proplastids and nucleoids, respectively, was invested in functions related to protein translation and homeostasis, but only approximately 11% of the protein mass of chloroplasts was (Fig. 4A). A total of 320 proteins were assigned to these functions (Supplemental Table S4). Figure 4A shows a more detailed functional breakdown of these functions for identified nucleoid, proplastid, and chloroplast proteomes. The largest investment in nucleoids was in ribosomal proteins, with a much smaller contribution from translation factors (initiation, elongation, and tRNA and peptide-release factors; Fig. 4A). The abundance of ribosomal proteins in the nucleoid fraction suggests that translation can initiate cotranscriptionally in chloroplasts, as it does in bacteria. However, the inclusion of EDTA during nucleoid preparation would be expected to dissociate polysomes and 70S ribosomes, complicating the interpretation of these findings. We identified at least a dozen nucleoid-enriched proteins that are related to known ribosome biogenesis factors in bacteria (Shajani et al., 2011) or that have been shown to influence ribosome assembly in chloroplasts, supporting a functional linkage between nucleoids and ribosome biogenesis. Predicted ribosome assembly factors in the nucleoid were 3.5 and more than 200 times more abundant than in proplastids and chloroplasts, respectively (Fig. 4A). These include several GTPases (ObgC, EngA, HFLx, ERA, RIF1/YqeH) and others (e.g. SVR1 protein). We note that additional proteins that directly or indirectly affect rRNA processing are also listed under RNA-related functions (Table I; Fig. 3C); most of these are likely involved with ribosome assembly. In addition, several of the DEAD box proteins with unknown function, discussed previously, are also likely involved in rRNA maturation and ribosome assembly (Fig. 3D). The nucleoid enrichment for ribosome assembly factors is also highly consistent with our recent analysis of FPLC-separated stromal megadalton complexes in Arabidopsis; here, we identified 14 ribosome biogenesis factors, most of which are homologs of those identified in the maize nucleoids (Olinares et al., 2010). Arabidopsis homologs for a few of these proteins were recently identified in plastid mutant screens. Examples included pseudouridine synthase SVR1 (At2g39140.1), which was discovered as a suppressor of the var2 mutant, devoid of thylakoid protease FTSH2 (Yu et al., 2008). Furthermore, RIF1 (At3g47450), a homolog of the Bacillus subtilis YqeH protein GTPase required for proper ribosome assembly, was discovered as a mutant with modified expression of enzymes of the plastid-localized methylerythritol phosphate pathway (Flores-Pérez et al., 2008). This combined information strongly suggests that ribosome assembly occurs in association with the nucleoid.
Figure 4.
Comparison of the abundance of proteins involved in protein synthesis and homeostasis (bin 29) and of envelope-localized proteins in maize chloroplasts, proplastids, and nucleoids. A, Distribution of different functions within bin 29, namely ribosome biogenesis, 70S ribosomes, tRNA ligases, “synthesis” (translation, elongation, and termination factors), protein folding, posttranslational modifications, protein sorting and translocation, protein assembly, and proteases. B, Distribution of different functions of envelope proteins, namely metabolite transporters, protein translocon of the inner envelope membrane (TIC) and outer envelope membrane (TOC), proteases, metabolic enzymes (mostly fatty acid/lipid synthesis), plastid division and positioning, and other proteins. The inset in B shows the abundance distribution between detected inner and outer envelope membrane proteins.
Chaperones involved in protein folding (e.g. CPN60/20, HSP70/GRPE, protein isomerases, etc.) were strongly underrepresented in nucleoids as compared with chloroplasts and, in particular, proplastids (Fig. 4A). This can be explained in part by the fact that most plastid proteins are nucleus encoded and thus posttranslationally folded within the plastid and, therefore, do not function in the context of chloroplast protein synthesis.
Protein sorting factors in the nucleoid were overrepresented as compared with chloroplasts and proplastids (Fig. 4A). The biggest contributions were from the envelope TIC/TOC protein translocon as well as the 54-kD subunit of the signal recognition particle (cpSRP54). cpSRP54 is involved in both posttranslational targeting of chlorophyll a/b-binding proteins and cotranslational sorting of plastid-encoded proteins (Richter et al., 2010). cpSRP54 was highly abundant and highly enriched in nucleoids (25- and 110-fold compared with proplastids and chloroplasts). Its high abundance in the nucleoid is most likely through the association with the subunits of the plastid ribosome and possibly nascent peptides (Franklin and Hoffman, 1993; Nilsson et al., 1999). Importantly, cpSRP43, only involved in posttranslational targeting of nucleus-encoded proteins, was not at all enriched in nucleoids (Supplemental Table S4), supporting our suggestion that cpSRP54 was nucleoid enriched due to its involvement in the cotranslational targeting of plastid-encoded proteins. Finally, thylakoid protein sorting components of the SEC, TAT, and ALB3 translocon were underrepresented in the nucleoids; this is consistent with the role of the nucleoid in plastid gene expression but not in posttranslational protein biogenesis steps.
Surprisingly the ratio between TIC (TIC110, TIC55, TIC40, and TIC22) and TOC (TOC34, TOC35, TOC75, and TOC159) proteins was much lower in nucleoids (0.02) than in proplastids (1.4) and chloroplasts (250). To understand the significance, we carefully annotated all 2,460 identified proteins for envelope location based on known function or location in other plant species, and we found more than 100 proteins (Supplemental Table S2). They could be separated into six main categories, namely metabolite transport, TIC, TOC, proteolysis, metabolic enzymes (mostly lipid, FA, and tetrapyroles), and plastid division positioning. The distribution of these categories across chloroplasts, proplastids, and nucleoids in shown in Figure 4B. This showed a 2- to 3-fold enrichment of envelope-associated proteins in nucleoids compared with proplastids and chloroplasts. Outer envelope proteins were underrepresented in chloroplasts and proplastids as compared with nucleoids (Fig. 4B, inset), mainly due to the strong enrichment of TOC proteins in nucleoids (Fig. 4B). We speculate that this could relate to Nonidet P-40 detergent-resistant domains in the outer envelope and subsequent copurification with the nucleoid particles.
Across the chloroplast, proplastid, and nucleoid samples, we identified 28 proteins involved in protein complex assembly (Supplemental Table S4), three of which were enriched in the nucleoid fractions; these are homologs of two Arabidopsis PSII assembly factors, LPA1 (Peng et al., 2006) and LPA2 (Ma et al., 2007), as well as protein ENH1, with rubredoxin and PDZ domains and a predicted transmembrane domain (Zhu et al., 2007). Like that observed for the thylakoid sorting components, the underrepresentation of protein assembly factors is consistent with the role of the nucleoid in plastid gene expression but not in posttranslational protein biogenesis steps.
Finally, we identified 64 proteases and processing peptidases in the chloroplast, proplastid, and nucleoid samples, 31 of which were also identified in the nucleoids. Proteases SPPA, FtsHi3, and FtsH7/9 stood out as abundant proteases that were enriched in nucleoids compared with proplastids and chloroplasts; they each passed our three criteria for nucleoid association (Supplemental Table S4). Other proteases detected in nucleoids were of low abundance and/or were not enriched compared with chloroplasts or proplastids (e.g. FtsH2, DegP2). It seems logical that proteases might be required for the removal of components of plastid gene expression localized to the nucleoid. SPPA is a thylakoid-associated protease that is up-regulated upon light stress (Lensch et al., 2001). It has no known substrates, and the null mutant phenotype is very weak and only observed upon light stress (Wetzel et al., 2009). SPPA seems a possible candidate for a protease involved in the degradation of nucleoid proteins. FtsHi3 and FtsH7/9 are members of the chloroplast protease family that have not been studied, in contrast to the four abundant FtsH proteins (FtsH1, -2, -5, and -8) that form a complex in the thylakoid membrane (Kato and Sakamoto, 2010; Liu et al., 2010a). FtsHi3 lacks the zinc-binding motif required for proteolytic activity (Sokolenko et al., 2002). The role of FtsHi proteins is not known, but the proteins might be involved in chaperone functions or play a structural role, comparable to the essential inactive members of the plastid Clp protease family (Olinares et al., 2011).
Comparison of Nucleoids from Base, Tip, and Young Leaf
To discover so far unrecognized nucleoid proteins and determine possible developmental effects on nucleoid proteome composition, we compared the nucleoids from base, tip, and young leaf. The Venn diagram in Figure 5A shows the overlap between the nucleoid-enriched proteomes of these three sample types. Proteins identified in all three sample types represented 92% to 95% of the mass for each sample type, indicating that the three types of nucleoid samples were generally similar, even if they were derived from different leaf development stages or leaf sections. The proteins uniquely identified in only one of the tissue types were mostly low-abundance proteins, including proteins considered as contaminants. The protein mass investments for these three nucleoid proteomes in different cellular functions are illustrated in Figure 5B. This showed that thylakoid proteins involved in photosynthetic electron transport represented 16% of the protein mass in the tip, 10% in the base, and 6% in the nucleoids from young seedlings. Proteins of the Calvin-Benson cycle and the C4-malate cycle represented 8% in the tip, 7% in the base, and 4% in nucleoid from young leaf. Thus, nucleoids from the fully photosynthetic cells at the leaf tip contained a larger contribution from photosynthetic and transport proteins, mostly at the expense of proteins with RNA-related functions (Fig. 5B). Investment in proteins involved in RNA-related functions or in translation was by far the most abundant irrespective of whether the nucleoids were isolated from base, tip, or young leaf (Fig. 5B; Supplemental Fig. S3).
Figure 5.
Comparison between nucleoid preparations isolated from the leaf base, tip, or young seedlings. A, Venn diagram showing the overlap between the identified nucleoid proteomes; in total, 1,092 proteins were identified. B, Comparison of protein abundance (based on NadjSPC) of the 10 different functional groups defined as for Figure 2B. C, Work flow for determination of the nucleoid core proteome. D, Base-tip ratio plot for the 157 proteins defined in the work flow in C. Data points are color coded based on the 10 functions defined for Figure 2. Identities of a few proteins are indicated. Details for all 157 proteins are provided in Supplemental Table S5.
The Core Nucleoid Proteome
The similarity between the three sets of nucleoids suggested that there is a set of proteins that could be considered the “core” nucleoid proteome (i.e. proteins that specifically locate to the nucleoid). Defining a core nucleoid proteome is challenging, since the nucleoid is not an organelle (i.e. it lacks a membrane separating it from the rest of the plastid); therefore, it is unlikely that a strict nucleoid proteome can be defined, because many proteins cycle on and off the nucleoid structure. Moreover, few other large plastid complexes (e.g. PDH and starch particles) partly copurified with nucleoids, making it difficult to use strictly quantitative methods to assign proteins to the nucleoid. However, a working model of core plastid nucleoid proteome will help to (1) find putative nucleoid proteins without bias based on (postulated) function, (2) obtain insights on the main functions of the nucleoid, and (3) determine developmental effects on nucleoid proteome and function.
To define the core nucleoid proteome, we applied four types of thresholds to the identified proteins in the combined nucleoid samples from base, tip, and young leaf (Fig. 5C): (1) each core protein should represent at least 0.05% of the nucleoid protein mass, as calculated from the NadjSPC (NadjSPC > 5.10−4; this corresponds to approximately 40 adjSPC); (2) each core protein must be observed in at least four of the nine nucleoid preparations; (3) more than 10-fold enrichment in nucleoids (based on NadjSPC) as compared with chloroplasts or not detected in the chloroplast; and (4) more than 3-fold enrichment as compared with the proplastid. The first and second thresholds were chosen to select against proteins with low abundance that are more likely to represent contaminations or that are infrequently associated with the nucleoid; however, this relatively high threshold was conservative and removed several bona fide nucleoid proteins, such the DNA polymerases, most mTERFs, and others. Nevertheless, we felt that a conservative threshold was beneficial to discover nucleoid proteins that had not previously been suspected to have nucleoid-related functions and to determine developmental differences. The third and fourth thresholds removed proteins not specifically enriched in the nucleoid of each plastid type, even if a subset of such proteins do associate transiently with the nucleoid. We note that the criteria for the core nucleoid were more stringent than those applied for the cluster analysis in Figure 2, C and D, and when discussing Table I, since we also included a specific threshold for average nucleoid abundance.
The work flow resulted in 157 proteins, accounting for 50% of the mass in the nucleoid but only 6% of proplastids and just 1% of the chloroplast, demonstrating their enrichment. Proteins involved in bins 26 (mostly PPRs), 27 (DNA), 28 (RNA), 29 (protein), and 35 (unknown functions) dominated the mass of this nucleoid core. All except two agglutinin proteins had an assigned plastid location (Supplemental Table S5). These two agglutinin proteins have no homologs in Arabidopsis or rice (Oryza sativa) and were nearly exclusively detected in nucleoids of young leaves. Our previous leaf proteome analysis (Majeran et al., 2010) showed that they have peak expression around 3.5 to 4 cm from the base and are very low in the base, explaining their enrichment in nucleoids of “young” leaves (Supplemental Fig. S2B).
Thirty-six proteins of the core nucleoid were so far not considered, as they were not assigned to RNA, DNA, or “protein”-related functions, and were manually evaluated for functional significance. Several proteins were likely contaminants in starch (putative glycogen synthase and an α-amylase) and fatty acid (PDH) metabolism and were likely cofractionated because of their association with high-molecular-mass particles; these proteins should not be considered nucleoid proteins (Supplemental Table S5). A number of proteins were likely localized to the inner (e.g. a Mg2+ channel, various unknowns) or outer (e.g. OEP37, CHUP1) envelope and may have been enriched due to the anchoring of nucleoids to chloroplast membranes. Some have no known functions but have interesting predicted functional domains, such as the structural maintenance of chromosomes domain, a combined zinc-finger and SRR domain, whereas other proteins with no obvious domains were found specifically in nucleoids with very high spectral counts (e.g. GRMZM2G388855_P01; Supplemental Table S5). The identification of a maize homolog of Arabidopsis MSL2 was very interesting, since members of this small gene family colocalize with the plastid division protein MinE and influence plastid size, shape, and perhaps division during normal plant development (Haswell and Meyerowitz, 2006; Wilson et al., 2011). This work flow also identified two iron superoxide dismutases (FSD2 and -3); Arabidopsis homologs were also found in nucleoids and proposed to play a role in protecting plastid DNA from oxygen radical damage (Myouga et al., 2008). The localization of low-abundance glutamyl-tRNA reductase involved in early steps of tetrapyrrole synthesis is intriguing, given the postulated roles of tetrapyrrole precursors in plastid-nucleus signaling. Moreover, this enzyme is the target of negative feedback inhibition by the FLU protein (Meskauskiene et al., 2001). FLU was also observed in the nucleoids but was not dramatically enriched compared with proplastids and chloroplasts. Perhaps most exciting was the identification of the maize homolog of the state transition kinase STN7. Arabidopsis STN7 targets several thylakoid proteins and is particularly important in balancing photosynthetic electron transfer rates for optimal growth and development of the plant (Rochaix, 2011; Tikkanen and Aro, 2011). These and other proteins listed in Supplemental Table S5 provide new avenues to study the regulation of plastid gene expression.
Developmental Effects on the Core Nucleoid Proteome
To determine possible developmental effects, we plotted the base-tip ratio of this core proteome (Fig. 5D). Proteins with high base-tip ratios (greater than 3) were strongly enriched in PPR proteins, splicing factors (CRS, CFM2, CFM3), ribosome biogenesis (SVR1), and both SODs (FSD2 and FSD3). This high base-tip ratio is consistent with high plastid gene expression rates in the undeveloped part of the leaf. The high base-tip ratio of the SODs is also consistent with the proposal that they act as reaction oxygen species (ROS) scavengers in the maintenance of early chloroplast development by protecting the chloroplast nucleoids from ROS (Myouga et al., 2008). Proteins with a base-tip ratio of less than 0.33 (14 proteins) were enriched for proteases (FtsH7/9 and SPPA), DNA repair (both MutS homologs), a few unknown proteins, and a few metabolic enzymes as well as proteins involved with the biogenesis of photosystems (maize homologs of HCF173 [Schult et al., 2007] and LPA1; Fig. 5D; Supplemental Table S5). This suggests that plastid gene expression in the photosynthetic tip is oriented toward maintaining the photosynthetic apparatus. Because of the high expression threshold used on the filter procedure (Fig. 5C), several low-abundance authentic nucleoid proteins (e.g. DNA polymerase) were not included in the base-tip analysis. Therefore, we also manually evaluated the nucleoid proteins in Table I for base-tip ratio (Supplemental Table S1). This confirmed the base-tip core analysis in that proteins with high base-tip accumulation ratios were predominantly involved with transcription, RNA metabolism, and DNA replication.
An alternative work flow to determine the core nucleoid and possible developmental effects was also tested (Supplemental Fig. S4A). This work flow first separately identified proteins enriched in the base nucleoid (148 proteins) or in the tip nucleoid (117 proteins; Supplemental Table S5). Ninety proteins were found enriched in both base and tip and were considered an alternative core nucleoid proteome (Supplemental Fig. S4A). Eighty-four out of these 90 proteins were also found in the core proteome of 157 proteins (Supplemental Fig. S4B; Supplemental Table S5). Supplemental Figure S4C shows the base-tip ratio for the sets of core and base- and tip-enriched proteins. The base-tip distribution for the 90 core proteins generally shows that high base-tip proteins are enriched in RNA metabolism, whereas those with low base-tip ratios are enriched for envelope proteins and later steps in protein biogenesis and assembly. This is consistent with the work flow analysis presented above (Fig. 5).
As an alternative approach to determine the nucleoid core and the developmental effect on the nucleoid composition, we carried out a hierarchical cluster analysis based on standardized NadjSPC for the chloroplast, proplastid, and specific nucleoid samples from base, tip, or young leaf. Starting with the 771 proteins used for coexpression analysis above (Fig. 5C), we applied an additional, more stringent abundance threshold just for the nucleoids. This ensured that there were enough observations to recognize developmental effects (Supplemental Fig. S5). We thus considered only those 305 proteins that each had at least an average NadjSPC of 5.10−4 across the three nucleoid types and that were observed in at least four of the nine nucleoid preparations; this was the same abundance threshold as applied in the work flow of Figure 5C. The dendrogram (Supplemental Fig. S5) showed three main clusters. Clusters 2 and 3 mostly represented proteins with highest accumulation in proplastids and chloroplasts, respectively. Proteins in cluster 1 (217 proteins) should be considered nucleoid-enriched proteins and could be divided into four subclusters (a–d). Subclusters 1b (33 proteins) and 1d (49 proteins) represented tip- and base-enriched proteins, respectively. An excellent overlap was observed between proteins in cluster 1 and the analyses from Figure 5C and Supplemental Figure S4A, as can be observed in Supplemental Figure S5.
Coimmunoprecipitation with anti-WHY1 and RNase Treatment
To determine if the purification of the nucleoids included systematic contaminations, we also analyzed isolated nucleoids that were coimmunoprecipitated (co-IP) with antiserum against the maize DNA-binding protein WHY1 (Prikryl et al., 2008). Since the nucleoid samples are tightly packed particles that are prone to aggregation (even in the presence of nonionic detergents), centrifugation could result in pelleting of such aggregated nucleoids or other particles. Therefore, we used magnetic beads for the co-IPs, since this allowed us to avoid collecting the co-IP proteome as a pellet. Because we observed so many proteins involved in RNA metabolism or 70S ribosomes in nucleoids, we also tested the consequence of RNase treatment on the nucleoid proteome.
A nucleoid preparation, purified from young 7- to 8-d-old seedlings, was split into three equal fractions. One fraction was pretreated with RNase and subsequently coimmunoprecipitated by anti-WHY1, in parallel with a mock-treated fraction. The two co-IP fractions were then run side by side on an SDS-PAGE gel with the starting material (Fig. 6A), and proteins were analyzed by MS/MS. In total, 888 proteins were identified (Supplemental Table S6). Assigned non-plastid-localized proteins represented 2.1% of the total protein mass (percentage NadjSPC) for the starting material and 2.1% and 1.6% for the co-IPs; as before, this showed that the nucleoids were not systematically contaminated with large amounts of nonplastid proteins. Figure 6B shows the mass distribution of all proteins in the starting material and in the two co-IPs according to functional group, following the same assignments as before. In general, the co-IPs only modestly affected the distribution across protein functions. The contribution of thylakoid or “Calvin cycle and malate shuttle” proteins was around 3%, before or after co-IPs, indicating that the residual (likely) contaminants are quite tightly associated with the nucleoid particles (Fig. 6B). Furthermore, small decreases in the categories “protein homeostasis” and “RNA” and an increase in the category “translation” were observed due to the co-IPs.
Figure 6.
Co-IP of nucleoids by anti-Why1 serum and effect of RNase treatment. A, One-dimensional SDS-PAGE gel of the nucleoids prior to co-IP, and nucleoid co-IP by anti-WHY1 with or without RNase treatment. B, Quantitative distribution of protein functions across the three nucleoid samples. The functional groups are as defined in Figure 2. C, Quantitative effect of RNase treatment on the co-IP nucleoids using anti-WHY1 serum. Ribosomal proteins decreased in abundance by the RNase treatment are circled (in red). D, Effect of co-IP and RNase treatment of the quantitative presence of protein in the nucleoid. Cross-correlation of nucleoid protein abundance before and after co-IP and RNase treatment is shown. Proteins quantitatively reduced by RNase are numbered as follows: 1, NP_043018, 30S ribosomal protein S2; 2, GRMZM2G005973_P01, 50S ribosomal protein L1; 3, GRMZM2G377761_P01, 3′ to 5′ PNPase exoribonuclease; 4, GRMZM2G170870_P01, 50S ribosomal protein L6-2; 5, NP_043046, 30S ribosomal protein S18; 6, GRMZM2G105570_P01, ABC and structural maintenance of chromosomes domain protein; 7, GRMZM2G165694_P01, 16S rRNA-processing protein RimM family; 8, GRMZM2G028216_P01, 50S ribosomal protein L29. ^ Proteins only found in co-IPs; * proteins lost by co-IP. Detailed information is provided in Supplemental Table S6.
The RNase treatment (compare the co-IPs with or without RNase treatment) showed generally little effect and did not remove significant amounts of proteins involved in RNA metabolism or components of the translational apparatus (Fig. 6B). This indicates that the RNA molecules associated with the nucleoid are generally protected from degradation, most likely by associated proteins. However, we did observe an approximately 20% decrease in 70S ribosomal subunits after RNase treatment (Supplemental Fig. S6). Quantitative cross-correlation at the individual protein level of the nucleoid proteome with or without RNase treatment showed that proteins reduced after the RNase treatment were indeed primarily 70S ribosomal proteins (Fig. 6C, circled data points). The cross-correlation further showed that RH3-1, RpoB, RpoC, CaS, ribosomal protein L4, pTAC2, and MurE are the most abundant proteins in the immunoprecipitated nucleoid fractions, irrespective of the RNase treatment (Fig. 6C) and consistent with earlier observations (Table I; Supplemental Table S1).
Finally we explored which proteins were absent in both co-IPs but present in the starting material. Proteins identified by ≤2 adjSPC across the three data sets were removed from consideration, since proteins identified with significant numbers of spectra (i.e. more than five to 10) are needed to make quantitative statements (Zybailov et al., 2008). The resulting filtered data set contained 611 proteins; Figure 6D shows the quantitative differences as a result of the co-IPs with and without RNase treatment for these proteins. Thirty-four proteins were only detected in the starting material (i.e. lost in both co-IP samples), were a mixture of proteins of diverse functions, and had on average 4.6 adjSPC in the starting material. The three most abundant proteins that were lost were stromal ATP phosphoribosyl transferase involved in His synthesis (GRMZM2G068862_P01; 15 adjSPC), a PPR protein without known function (GRMZM2G074599_P01; 11 adjSPC), and a pseudouridylate synthase TruB family protein involved in nucleotide metabolism (GRMZM2G174716_P01; 7 adjSPC). None of them were known pTAC proteins or proteins assigned as DNA-binding proteins. There were 24 proteins that were identified in the co-IP without RNase treatment but not in the starting material; they were identified with on average 3 adjSPC, consistent with the variability of observation for low-abundance proteins. These included four cytosol 80S ribosomal proteins, several likely plastid contaminants such as psaB, the very abundant acetyl-coA carboxylase ACC1A, SPP, and three thylakoid proteins of the NDH and PSI complex. The RNase treatment did result in additional loss of proteins, as compared with the co-IP effect alone, as we identified in this fraction 473 proteins compared with 546 in the co-IP sample without RNase treatment (Supplemental Table S6). The 10 most abundant proteins lost after RNase treatment were PSRP-2, known to be associated with 30S ribosome particles (Yamaguchi and Subramanian, 2003), two homologs of Rap38/CSP41B-2 (Yang et al., 1996; Bollenbach et al., 2009), 50S ribosomal protein L9, an RRM protein, iron superoxide dismutase (Fe-SOD or FSD3), an agglutinin domain protein, RNA-binding protein cp33 (RRM), DNA polymerase I, and another RRM-containing protein (Supplemental Table S6).
DISCUSSION
A New Conceptual Framework for Nucleoid Functions through Proteome Analysis
In this study, we provide an in-depth proteome analysis of nucleoids of mature and immature chloroplasts in maize. Our study provides, by far, the most comprehensive description to date of the functions that are tethered to the nucleoid structure. Our findings for nucleoid function are summarized in Figure 7; here, we distinguish between proteins likely to be bound directly to DNA (e.g. gyrases, DNA and RNA polymerases, DNA anchors), those likely to be tethered via nascent RNA transcripts (e.g. PPRs and splicing factors), and those likely to be tethered via ribosomes (e.g. translation factors). The three-dimensional organization of the plastid nucleoid, including the folding state of the DNA, the localization of ribosomes, and the possible presence of transcription foci (areas with enhanced transcriptional activities within the nucleoid), is poorly understood; in the case of bacterial nucleoids, it has been shown that this organization is dynamic, with observed changes dependent on growth phase and transcription rates and a peripheral localization of ribosomes (Dillon and Dorman, 2010). In Figure 7, we have not speculated about the three-dimensional organization of the plastid nucleoid; however, we believe that this will need to be addressed in future experimental studies to truly understand plastid gene expression.
Figure 7.
Summarizing schematic view of nucleoid functions. [See online article for color version of this figure.]
The Nucleoid-Enriched Proteome: Significance and Extending Plastid Proteome Coverage
The challenges to determine proteins that are intrinsic to the nucleoid or that have functional associations with the nucleoid are several-fold: (1) many proteins with nucleoid-associated functions likely cycle on and off the dynamic nucleoid structure, depending on the developmental and/or metabolic state of the plastid and specific needs, such as DNA repair or translation initiation; (2) the nucleoid is not an organelle (i.e. it lacks a membrane that separates it from the rest of the plastid) and therefore it is impossible to make claims of strict and exclusive nucleoid localization; (3) nucleoids are membrane associated and need to be released through the use of (nonionic) detergents, which can result in the loss of nucleoid-associated proteins that interact via hydrophobic interactions; and (4) other large plastid complexes, such as PDH and starch particles (Phinney and Thelen, 2005) and possibly TOC complexes (this study), appear to copurify with the large nucleoid structures, thus resulting in a systematic contamination.
Through quantitative proteome comparisons (based on matched MS/MS spectra) of nucleoid-enriched fractions isolated from different developmental stages of the plastid with chloroplasts and unfractionated proplastids (each with multiple biological replicates), we were able to determine whether proteins were enriched in the nucleoid as compared with unfractionated plastids. Even if we could not assign P values to nucleoid enrichment, the combination of minimal abundance thresholds, abundance ratios, and hierarchical clustering allowed us to identify proteins that likely functionally contribute to nucleoid structure and function.
The comparison of known nucleoid proteins (Sato et al., 2003; Sakai et al., 2004) and previous analysis of PEP and pTAC complexes (Suzuki et al., 2004; Pfalz et al., 2006) with our quantitative proteome information (including the reproducibility of independent replicates) showed that the proteomics information did identify and recognize most known nucleoid proteins and provided solid assignments for nucleoid enrichment. We have provided several types of tables (Table I; Supplemental Tables S1, S2, and S5) from which the reader can determine (1) if proteins were observed in the nucleoid, (2) if proteins were enriched compared with proplastids and/or chloroplasts based on abundance ratios, and (3) if proteins clustered with known nucleoid proteins. We also explicitly listed previously reported nucleoid proteins that were not observed in our study. Comparison with previously published information on pTAC or nucleoid proteins showed that we greatly expanded the identification of proteins highly enriched in the nucleoid. Moreover, compared with our previous extensive proteome analysis of chloroplast stroma and membranes (Majeran et al., 2005, 2008; Friso et al., 2010), we identified several hundred new proteins; about 50% of these were also identified in our “proplastid fraction.” In many cases, the homologs have neither been identified nor studied in Arabidopsis or other plant species.
Finally, we did not attach P values (through statistical tests) to nucleoid enrichment or nucleoid assignments because of the nature of the comparisons and data sets. This mostly relates to the “missing observation problem” in the case of comparison of nucleoids with proplastids or chloroplasts; simply put, we identified many proteins in the nucleoids with relatively high abundances (and in several of the nine independent nucleoid preparations) that were not or only infrequently detected in the proplastid or chloroplasts. It is possible to equate such lack of detection with zero values, but this leads to artificially low sd values. We also considered using modeled sd values for such cases, but we believe that because there are so many proteins to which this applies, we would lose statistical rigor.
The identified nucleoid proteins included proteins involved in DNA replication, quality control, organization, and repair as well as transcription, mRNA processing, splicing, and editing. In particular, proteins involved in RNA metabolism were highly enriched, indicating that RNA can be cotranscriptionally processed. Proteins affecting ribosome biogenesis, including rRNA maturation, were also highly enriched in the nucleoid, as were ribosomal subunits, again suggesting cotranscriptional assembly as possibly coupled transcription-translation. In addition, many nucleoid-enriched proteins of unknown function, including PPR, TPR, DnaJ, and mTERF domain proteins, were identified. Many of these proteins have not been studied and not previously been identified in earlier plant (plastid) proteomics studies, indicative of their low abundance. Estimating that the nucleoid proteome represents approximately 1% protein mass in chloroplasts, the isolated nucleoids provide an approximately 100-fold enrichment, explaining why we were able to discover so many previously unobserved proteins. The identification of 62 PPR proteins, of which 48 have no known function, and 10 mTERF proteins was very rewarding; these PPRs and mTERFs were without exception enriched in nucleoids as compared with proplastids and chloroplasts. This study thus extends the known plastid proteome by several hundred proteins, in particular those involved in DNA and RNA metabolism, and provides a resource for targeted studies on plastid gene expression.
A small number of previously assigned nucleoid proteins (mostly in species other than maize) were not observed in our maize nucleoid preparations. In particular, several that were discovered through genetics, rather than through proteomics-type experiments, likely have too low expression levels to be identified by MS. For a few others, in particular PEND, SiR, and CND41, for which substantial biochemical evidence was obtained for other plant species, the proteins were not found in nucleoids but were found at significant levels in unfractionated proplastids or chloroplasts, suggesting that they are not nucleoid enriched. Alternatively, their interaction with the nucleoid was particularly sensitive to detergent. To our initial surprise, we never observed any NEP in the nucleoids, believed to be important in early stages of plastid development (Liere et al., 2011), nor did we observe it in proplastids, chloroplasts, or basal leaf sections in maize, indicating that NEP levels are much lower than PEP levels in leaves, regardless of developmental stage. Indeed, NEP has never been identified in any plant proteomics studies, except for the MS analysis of Arabidopsis cell cultures (http://fgcz-atproteome.unizh.ch/), which did identify two of the three NEP isoforms (RpoT-1/2).
DNA Replication, Quality Control, and DNA Repair
Our understanding of DNA replication, quality control, and DNA repair of the plastid chromosome is limited, and many of the genes have not been identified, despite their importance for plant and plastid viability (Day and Madesis, 2007). Our nucleoid analysis identified more than 25 proteins that could fulfill such functions; these genes provide an excellent starting point for systematic studies on plastid DNA replication, quality control, and DNA repair (Fig. 7).
Replication of plastid DNA occurs through the activity of two PolI-type polymerases (PoII-A/B), as has been shown for Arabidopsis (Mori et al., 2005; Parent et al., 2011). In the case of Arabidopsis, PolI-B is also involved in DNA repair (Mori et al., 2005) and prevents, together with WHY1 and WHY3, deleterious rearrangements of plastid DNA through unwanted recombination, safeguarding plastid genome integrity (Maréchal et al., 2009; Parent et al., 2011). During DNA replication and recombination, erroneous insertions, deletions, and misincorporation of bases can occur. To repair these mistakes, a DNA mismatch repair system must be in place. In prokaryotes, this involves MutS, which recognizes the mismatched base, DNA exonucleases that excise the incorrect base, followed by the activity of DNA polymerase and DNA ligases. Very little is known about this mismatch repair system in plastids, but we identified nucleoid-enriched candidates for each of these repair enzymes, including MutS homologs, several exonucleases, DNA polymerases, and DNA ligase, as well as a RecA homolog. Recently, another MutS homolog (At3g24320) was found to be dual targeted to mitochondria and plastids; in plastids, this MutS homolog influenced genome stability and plastid development (Xu et al., 2011). Surprisingly, this homolog has never been identified by proteomics and was not found in our nucleoid fractions.
DNA can also be altered by oxidative damage, in particular when high levels of ROS are produced in the plastid; this is also a topic that has received very little attention. A recent study identified several enzymes in Arabidopsis with DNA glycolylase-lyase/endonuclease activities involved in DNA base excision/repair (Gutman and Niyogi, 2009). This study also suggested that there might be additional pathways for DNA repair in chloroplasts. We identified several proteins, such as FAD photolyases, that are likely involved in DNA quality control and repair, but we did not detect the maize homologs of these Arabidopsis DNA glycolylase-lyase/endonucleases. However, we did identify a putative DNA endonuclease that could function in the DNA repair pathway.
DNA Organization, Packaging, Distribution, and Anchoring of Nucleoids
Little is known about how the plastid chromosome is compacted into the nucleoid, what determines the DNA copy number per nucleoid, and what regulates nucleoid distribution throughout the plastid. Nevertheless, this packaging and distribution are likely important in maintaining DNA quality, inheritance, copy number, and transcription. Our nucleoid proteome analysis identified numerous new candidate proteins that could play a role in the nucleoid organization.
Based on experimental information for prokaryotic nucleoids, it is likely that the structural organization of plastid DNA is determined by supercoiling through the activity of topoisomerases and DNA folding involving DNA-protein-DNA bridges. Importantly, we identified nucleoid-enriched topoisomerase and a Toprim domain protein involved in supercoiling and several gyrases (A and B type) involved in unwinding of DNA to facilitate replication.
No plastid homolog or functional equivalent of histones has been found in higher plant plastids, whereas nucleoids in algae have Hu-like proteins that appear to fulfill the packing function (Kobayashi et al., 2002; Karcher et al., 2009). Based on its association with plastid DNA and in vitro DNA packaging activity, it has been suggested that SiR functions in plastid DNA condensation in soybean (Glycine max) and pea (Pisum sativum) chloroplasts, in addition to its established function in sulfur assimilation (Cannon et al., 1999; Sato et al., 2001; Chi-Ham et al., 2002). Silencing of the SiR gene in Nicotiana benthamiana resulted in “abnormal” chloroplasts and differentially reduced expression of plastid-encoded genes and resulted in larger nucleoids with increased amounts of DNA (Kang et al., 2010). Surprisingly, whereas maize SiR (GRMZM2G090338_P01) was very abundant in both proplastids and chloroplasts, we did not find it in isolated nucleoids, consistent with an earlier observation for maize (Sekine et al., 2007), suggesting (at least in maize) that it is not a functional equivalent of Hu/HP proteins or histones. SiR was not identified in TAC preparations either (Pfalz et al., 2006). In contrast, we suggest that pTAC3 is an excellent candidate as a DNA packing protein, because pTAC3 with a SAP domain, a conserved DNA-binding motif involved in chromosomal organization, was highly abundant and nearly exclusively observed in nucleoids from base, tip, and young leaf. We note that pTAC3 was part of cluster 4 (corresponding to nucleoid-enriched proteins; Fig. 2C) as well as the core proteome defined by both work flows (Fig. 5C; Supplemental Fig. 4A).
In developing plastids of dicots, nucleoids exist in the periphery of plastids in close contact to the envelope, but they appear to relocate to thylakoids and stroma in mature chloroplasts. However, in the case of the monocotyledons maize and wheat, no relocation from envelope to thylakoid has (yet) been observed. The purpose of anchoring nucleoids to chloroplast membranes is not clear, but it could possibly facilitate the insertion of plastid-encoded membrane proteins and help to provide feedback from the thylakoid redox state to plastid gene expression. Our nucleoid analysis does support maize TCP34 and MFP1 as putative membrane anchors for nucleoids but suggests that PEND may not serve this function. In addition, the abundance, shared membrane, and nucleoid location of pTAC16 make it another candidate for a nucleoid anchor protein.
One of the most significant findings regarding regulators of nucleoid distribution and its relation to plastid division has been the Arabidopsis nucleoid-associated YLMG1-1 protein (At3g07430; Kabeya et al., 2010). Loss of YLMG1-1 resulted in the concentration of nucleoids in a few large structures, but it did not affect plastid division (Kabeya et al., 2010). In contrast, overexpression of YLMG1 impaired chloroplast division, indicating that proper distribution of the nucleoid is needed to allow division to occur, even if the division machinery does not interact directly with the nucleoid; this mechanism is conserved between cyanobacteria and chloroplasts. We identified maize homolog GRMZM2G093815_P01, with high scores in maize proplastids and in nucleoids isolated from young leaves; in contrast, the other, slightly more distant maize homolog, GRMZM2G064663_P01, was only detected in thylakoids (Majeran et al., 2008). Another YGGT family member, CCB3 (At5g36120.1), is involved in heme biogenesis of the cytochrome b6f complex (Lezhneva et al., 2008); consistently, the rather abundant maize homolog (GRMZM2G167766_P01) was detected in thylakoids of chloroplasts and in proplastids but not in nucleoids. Thus, additional studies on YLMG1 and its interactors in Arabidopsis and maize could further elucidate how nucleoid distribution is regulated. We found little evidence for physical connections between the plastid division machinery and the nucleoid, but there appears to be a connection between nucleoids and the mechanosensitive ion channel protein MSL2 (Wilson et al., 2011).
Transcription and Transcriptional Regulation
Plastid transcription is likely regulated by (1) the structural organization of DNA, affecting the access of RNA polymerases and other proteins to the DNA, and (2) the copy number and activity of the NEP and PEP polymerases. In bacteria, Rho transcription termination factors (Epshtein et al., 2010) and members of the antitermination complex (Nus proteins) also contribute to transcriptional regulation and coupling to translation (Burmann et al., 2010; Proshkin et al., 2010), but it is not clear if this is relevant for plant plastids. Historically, most attention has been focused on the plastid RNA polymerases, possibly because little is known about the structural organization of plastid nucleoid DNA. In prokaryotes, it has been postulated that the nucleoid forms so-called transcription foci, which are regions of the chromosome that show particularly high transcriptional activities and possibly also coupled posttranscriptional steps of gene expression such as translation initiation (Dillon and Dorman, 2010). It is quite possible that such transcription foci also exist in plastids, and this should be experimentally tested.
The activity and regulation of the NEP and PEP RNA polymerases have received a great deal of attention over the last 20 to 30 years (Liere et al., 2011). Depending on their promoter, plastid genes are either primarily transcribed by NEP or by PEP, although there appears to be less of a strict division than initially suggested (for review, see Liere et al., 2011). Moreover, NEP activity is particularly important in developing tissues, whereas PEP activity is clearly the dominant polymerase is more developed, photosynthetic tissue. Promoter recognition by PEP requires Sigma factors: Arabidopsis has six Sigma factors that have been studied (Schweer et al., 2010). NEP protein levels appeared to be below detection by MS in leaves, plastids, and even nucleoids. We did identify four homologs of SIG2, but not of any other Sigma factor. Whereas SIG3, SIG4, and SIG5 each targets single or lower numbers of genes, SIG2 appears to play a more general role in transcription, in particular during early leaf development; therefore, it is perhaps not surprising that SIG2 is by far the most abundant Sigma factor. The four SIG2 proteins that we observed were a subfamily of three homologs with relatively low expression and the far more abundant protein GRMZM2G143392_P01, detected in nucleoids as well as unfractionated proplastids. The significance of the maize homologs is not yet clear. PEP activity is also believed to be regulated by the plastid redox state, likely through the activity of the thioredoxin system involving TrxZ and (de)phosphorylation, possibly involving the fructokinase-like FLN1/2 and casein kinases (e.g. CK2; Steiner et al., 2009; Bräutigam et al., 2010). FLN and TrxZ are abundant proteins in the nucleoids, in contrast to the CK-type kinases, which we did not detect in nucleoids (or in maize or Arabidopsis plastids). We identified the thylakoid phosphoprotein CaS as well as STN7 as potential nucleoid-associated factors, with low base-tip ratios, suggesting that they may provide feedback to plastid gene expression in photosynthetic plastids. Functional connections between STN7, CaS, and the FLN kinases remain to be determined.
RNA Splicing, Processing, Editing, Methylation, Stabilization, and Decay
Proteins involved in various aspects of RNA metabolism were overrepresented in the nucleoids. These include most known splicing factors, RNA nucleases, and other RNA-modifying enzymes (for recent review, see Stern et al., 2010; Barkan, 2011), as well as some 100 proteins with unknown functions but with predicted RNA interaction based on functional domains, such as PPR, RRM, S1, and S4 domains. This shows that the nucleoid is intimately involved with RNA metabolism and that many of these RNA-processing steps occur cotranscriptionally. A few of the identified proteins have been shown to target specific transcripts, such as PPR4, PPR5, PPR10, CRS1, and CRP1 (in maize) and CRR2, MRL1, and HCF107 (in Arabidopsis), whereas others likely serve all or most transcripts. The large set of earlier unidentified proteins provides an excellent resource for targeted studies on RNA metabolism.
Ribosome Biogenesis, Translation Initiation, and Coupled Transcription-Translation in Nucleoids
Ribosomes are highly complex ribonucleoprotein particles with approximately 65 protein subunits. The maize plastid genome encodes for 12 and nine proteins of the 30S and 50S particles, respectively, in addition to the 16S rRNA (30S) and the 4.5S, 5S, and 23S rRNA (50S), with the remaining ribosomal proteins encoded by the nuclear genome. Ribosomal proteins represented about 20% of the nucleoid protein mass but only 5% and 2% of the proplastid and chloroplast, respectively. We detected an approximately 18% to 20% higher ratio between plastid- and nucleus-encoded ribosomal proteins in nucleoids as compared with unfractionated proplastids or chloroplasts, indicating a modest overrepresentation of plastid-encoded ribosomal proteins in nucleoids. This perhaps suggests that a minority of ribosomal subunits is undergoing assembly, whereas the majority of detected ribosomal subunits in the nucleoids were assembled and engaged in translation. Estimating that the nucleoid proteome represents approximately 1% of the chloroplast protein mass, then approximately 10% of ribosomal proteins were located in nucleoids (20%/2% × 1% = 10%). We did observe a strong overrepresentation of rRNA-modifying enzymes and (predicted) ribosome biogenesis factors in nucleoids, supporting the interpretation that ribosomes are assembled in association with the nucleoid.
Support for nucleoid-associated translating ribosomes comes from the highly enriched translation initiation factors in nucleoids (IF1/2/3). Given that the plastid gene expression machinery still shares many features of prokaryotes and that no membrane separates nascent mRNA from ribosomes, it is quite possible that translation can initiate cotranscriptionally in plastids. The RNase treatment released only a very small fraction of nucleoid-associated proteins; however, those released were predominantly ribosomal subunits, which is consistent with the tethering of translating ribosomes via the nascent mRNA.
We considered whether the ribosome enrichment could have been due to a systematic contamination. Indeed, ribosomes, and in particular polysomes, are large structures that could copurify with the nucleoids; however, when we analyzed megadalton-soluble complexes from Arabidopsis chloroplast stroma by gel filtration and MS/MS, we were able to mostly separate the pTAC complex (more than 3 MD) from monosomes (Olinares et al., 2010). Moreover, in this study, we did not use ribosome-stabilizing conditions (such as high MgCl2); therefore, it is likely that free polysome and monosomes mostly destabilized into 30S and 50S complexes, resulting in decreased copurification with nucleoids. Finally, it is formally possible that the (mostly basic) ribosomal proteins nonspecifically interacted with negatively charged RNA or DNA. However, the systematic and significant copurification through this mechanism seems unlikely to explain the strong enrichment of ribosomes (and their assembly factors) in nucleoid fractions. Thus, a significant nucleoid-ribosome interaction does appear to exist in vivo.
Developmental Effects on Nucleoid Proteome and Function
Earlier experimental studies suggested that nucleoid protein composition was different between chromoplasts, nongreen plastids from cell cultures, and chloroplasts (for review, see Sato et al., 1999, 2003; Sakai et al., 2004). Moreover, nucleoid morphology and localization changed during leaf development, with large but few nucleoids in proplastids and smaller but a higher number of nucleoids in mature chloroplasts. Using modern MS techniques, our quantitative comparisons (including hierarchical clustering; Supplemental Fig. S5) of nucleoids from proplastids and chloroplasts showed both quantitative and qualitative effects in proteome composition, even if the majority were identified in both types of nucleoid. Indeed, the total abundance of designated pTAC protein linearly correlated with PEP abundance in the isolated nucleoids, independently of developmental state. Nucleoid function shifted from RNA metabolism and ROS defense in the undeveloped tissues to protein homeostasis and DNA repair. Furthermore, comparing total (unfractionated) proplastid and chloroplast proteomes showed clearly that the concentration of many nucleoid-associated proteins decreased strongly, to the point that many nucleoid proteins could not be detected in chloroplast. This is in agreement with older observations that transcription and DNA replication decrease with development and that nucleoid content per plastid is low in mature tissue (for review, see Sato et al., 1999, 2003; Sakai et al., 2004). It is not yet clear if this is simply due to dilution, with photosynthetic proteins synthesized at high rates during greening, or to the reduced expression of genes encoding nucleoid proteins as chloroplast development proceeds, or to an active degradation. We did identify several plastid proteases (SPPA, FtsH7/9, Ftshi) that showed enrichment in the nucleoid fraction from mature tissue; these are candidates for turnover of nucleoid proteins.
Plastid Mutants and New Entry Points for the Study of Plastid Gene Expression
Arabidopsis and maize mutant analysis has shown that many nuclear genes encoding factors in chloroplast gene expression are critical for plastid development and biogenesis. Such mutants show often embryo-lethal phenotypes in Arabidopsis and pale-green or albino seedling-lethal phenotypes in maize (Stern et al., 2004; Bryant et al., 2011). The identification of several nucleoid-localized suppressors for FtsH thylakoid proteases with a role in plastid gene expression, such as the PPR protein At4g16390 (SVR7; Liu et al., 2010b) and pseudouridine synthase (SVR1; Yu et al., 2008), underscores that posttranslational plastid protein homeostasis is well integrated with plastid gene expression. Through a combination of the nucleoid-enriched proteome in our study and the use of large collections of nonphotosynthetic mutants in Arabidopsis (http://rarge.psc.riken.jp/chloroplast/) and maize (http://pml.uoregon.edu/photosyntheticml.html), it will be possibly to accelerate our understanding of the gene network controlling plastid gene expression and protein homeostasis.
CONCLUSION
Collectively, our analysis strongly suggests that nucleoids encompass a continuum of functions ranging from DNA replication, organization and quality control, transcription, splicing and processing, ribosome assembly, and translation initiation. This study provides a comprehensive resource for targeted studies on all aspects of plastid gene expression and DNA metabolism and extends the known higher plant plastid proteome by several hundred proteins.
MATERIALS AND METHODS
Plant Material, Chloroplast, and Nonphotosynthetic Plastid Isolation
For purification of nucleoids along the leaf developmental gradient, maize (Zea mays) inbred line W22-T43 was grown in a growth chamber for 9 to 10 d (16 h of light/8 h of dark, 400 μmol photons m–2 s–1, constant 28°C). Alternatively, maize inbred line B73 was grown for 7.5 to 9 d in a growth chamber (12 h of light/12 h of dark, 400 μmol photons m−2 s−1, 31°C day and 22°C night). Mixed mesophyll and bundle sheath chloroplast plastids were isolated from the base of the third leaf (the first 3 cm from the ligule), the middle 3 cm of the leaf, and the tip of the leaf (last 4 cm) as follows. Maize leaves (80–140 g fresh weight) were cut to approximately 5 mm in length, homogenized in grinding medium (50 mm HEPES-KOH, pH 8.0, 330 mm sorbitol, 2 mm EDTA, 8 mm ascorbic acid, and 5 mm l-Cys) with four bursts of 2- to 3-s high-speed (one-third the maximum speed) and 3-s low-speed pulses by a modified Waring blender in which the original blades were replaced by a razor blade, and filtered through two to four layers of 20-μm Miracloth. The leaf residues on nylon membranes were returned to the blender, and the homogenization step was repeated one to three times. The crude chloroplasts were overlaid onto a 35%/80% (v/v) PF Percoll (Percoll contains 0.98% [w/v] Ficoll PM 400 and 2.9% [w/v] polyethylene glycol 4000) step gradient that was osmotically adjusted (50 mm HEPES-KOH, pH 8.0, 330 mm sorbitol, and 2 mm EDTA) and spun in a swinging-bucket rotor at 4,740g for 15 min. The broken chloroplasts of the upper band were removed, the intact chloroplasts from the interface of the 35% and 80% Percoll layers were collected and washed with approximately 5 volumes of the wash buffer, and the chloroplasts were collected by centrifugation at 1,940g for 4 min. All plastid isolations were carried out under low-intensity green light at 4°C, and all buffers were chilled on ice. Separate mesophyll and bundle sheath chloroplasts were purified from the top 4-cm section of the third leaf and harvested about 2 h after the onset of the light period, using about 200 leaf tips (about 80 g of fresh tissue) as described (Majeran et al., 2005).
For anti-WHY1 co-IP experiments, maize plants (B73) were grown for 7.5 d in a growth chamber (12 h of light/12 h of dark, 400 μmol photons m−2 s−1, 31°C day and 22°C night). All three leaves were harvested and separated from the plant at its ligule 1 h after the onset of the light period. Intact plastids were isolated essentially as described above.
For proteome analysis of proplastids, we used the first 2 cm (counting from the ligule) of each third leaf blade of 9- to 10-d-old B73 seedlings, in total collecting 8 to 10 g fresh weight per plastid isolation. The plastid isolation protocol described above was slightly modified to increase yield as follows: (1) leaf sections were ground in a Waring blender at half maximum speed with five pulses of approximately 5 s; (2) the Percoll cushion was 30%/80% instead of 35%/80% for 10 min instead of 15 min; and (3) centrifugations steps to collect crude and pure protoplastids were at higher g values and longer times than for chloroplasts, typically 10 min at 6,000 rpm in a JS13.1 rotor (average 3,649g).
Isolation of Nucleoids
Nucleoids were isolated from intact isolated maize chloroplasts essentially as described by Cannon et al. (1999) with minor modifications, as follows. Half of the chloroplast pellets were suspended in 8 mL of nucleoid isolation buffer (20 mm Tris-HCl, pH 7.0, 0.5 mm EDTA, 1.5 mm spermidine, 7 mm 2-mercaptohethanol, and 40 μg mL−1 Pefabloc SC) in 10-mL beakers, and 2 mL of 10% (v/v) Igepal CA-630 (instead of Nonidet P-40) was slowly added while stirring to a final concentration to 2% on ice and left stirring for 30 min. The lysate was spun in a Ti50.2 rotor (Beckman) at 148,000g for 30 min at 4°C. The resulting nucleoid pellets were resuspended with 1 mL of nucleoid isolation buffer with 2% Igepal CA-630 and was brought up to 20 mL of the buffer, homogenized, and spun again at 148,000g for 30 min at 4°C. (In an attempt to increase the purity of the isolated nucleoids, we resuspended the nucleoid pellet in 2 mL of nucleoid resuspension buffer and loaded it onto a 30-mL 40%/80% step Suc gradient. The gradient was spun at 8,000 rpm for 15 min [Beckman SW-13.1 rotor], and the nucleoid particles were harvested from the 40% to 80% Suc interface. The harvested nucleoid particles were diluted at least 10 times in nucleoid resuspension buffer and collected by centrifugation at 48,000g for 30 min. However, we found that this procedure greatly reduced the yield, and we did not further employ this extra step.) The pellets were washed and spun again with nucleoid isolation buffer with 2% Igepal CA-630. The final nucleoid pellets were resuspended in 440 μL of nucleoid isolation buffer with 0.5% Igepal CA-630 and protease inhibitors (50 μg mL−1 antipain, 40 μg mL−1 bestatin, 20 μg mL−1 chymostein, 10 μg mL−1 E64, 10 μg mL−1 phosphamidon, 2 μg mL−1 aprotinin, and 250 μg mL−1 Pefabloc SC) and stored at −80°C until use. Protein concentrations in nucleoids were estimated from the gel stain pattern, since the high levels of nucleic acid prevented accurate protein determination using the Bradford or bicinchoninic acid assays.
Anti-Why1 Co-IP of Nucleoids and RNaseA Treatment
The generation and purification of antiserum to maize Why1 were described previously (Prikryl et al., 2008). Anti-Why1 immunoprecipitation was performed as described previously for chloroplast stroma with modifications (Watkins et al., 2007). Four hundred microliters of Dynabeads protein A (Invitrogen) was washed three times with phosphate-buffered saline (PBS), pH 8.0, and 0.1% Igepal CA-630. Washed beads were split to 120 μL in two tubes, and the rest (160 μL) was stored at 4°C to preclear the nucleoids (see below). Fifty microliters of anti-Why1 antibody or a mock solution (without antibody) was incubated with washed 120-μL beads for 2 h at 4°C, then the beads were washed with PBS, pH 8.0, and 0.1% Igepal CA-630 and three times with 0.2 m triethanolamine, pH 8.2, and 0.1% Igepal CA-630. Anti-Why1-Dynabeads complex or Dynabeads (mock) were cross-linked by 500 μL of 25 mm dimethyl pimelimidate (Pierce) in 0.2 m triethanolamine, pH 8.2, and 0.1% Igepal CA-630 for 1 h at room temperature, washed with triethanolamine, pH 8.2, and 0.1% Igepal CA-630, and the cross-linking reaction was quenched by washing three times with 0.1 m ethanolamine, pH 8.2, and 0.1% Igepal CA-630. For the last wash, beads were incubated for 30 min at room temperature. After five washes with PBS, pH 8.0, and 0.1% Igepal CA-630, beads were stored in PBS, pH 8.0, 0.1% Igepal CA-630, and 0.1% sodium azide at 4°C until use.
Four hundred microliters of nucleoids was sonicated three times for 30 s with a 1-min interval in an ice-cold bath sonicator. In the first experiment, the nucleoids were clarified by centrifugation at 4,000g for 10 min at 4°C. In the second trial, the centrifugation step was omitted due to the loss of nucleoids. To preclear nucleoids, 160 μL of washed Dynabeads was added in the nucleoids and incubated for 30 min at 4°C, and the beads were removed by DynaMag (Invitrogen). Precleared nucleoids were split into four tubes to approximately 110 μL, and 40 μL was saved as a total sample.
Beads were divided into two 60-μL aliquots for RNaseA +/− treatments and washed once with nucleoid co-IP buffer (20 mm Tris-HCl, pH 7.0, 0.5 mm EDTA, 1.5 mm spermidine, 7 mm 2-mercaptoethanol, 0.5% Igepal CA-630, and 5 μg mL−1 aprotinin). Nucleoids (approximately 110 μL) were incubated with anti-Why1-coupled-Dynabeads or Dynabeads in 0.4 mL of nucleoid co-IP buffer. For the RNaseA treatment, 3 μL of 1 μg μL−1 RNaseA was added to the reactions. Samples were incubated for more than 90 min at 4°C and then washed six times with nucleoid co-IP buffer. Bound proteins were eluted with 40 μL of 1.5× Laemmli buffer without 2-mercaptoethanol for 5 min at 95°C, and the beads were removed by DynaMag. Before running on SDS-PAGE gels, 4 μL of 2-mercaptoethanol was added to the samples, heated for 10 min at 75°C, and microfuged for 5 min to remove insoluble materials. For MS, 30- and 40-μL total and eluted samples were separated, respectively, by the Criterion 10.5% to 14% Tris-HCl gel (Bio-Rad) and stained with MS-compatible silver stain (Shevchenko et al., 1996).
Proteome Analysis
After protein separation by one-dimensional SDS-PAGE, each gel lane was cut into six to 10 slices. Proteins were digested with trypsin, and the extracted peptides were analyzed by nano-LC LTQ-Orbitrap MS using data-dependent acquisition and dynamic exclusion, as described (Friso et al., 2010). Peak lists (.mgf format) were generated using DTA supercharge (version 1.19) software (http://msquant.sourceforge.net/) and searched with Mascot version 2.2 (Matrix Science) against maize genome release 4a.53 (with 53,764 models) from http://www.maizesequence.org/, supplemented with the plastid-encoded proteins (111 protein models) and mitochondria-encoded proteins (165 protein models). Details for the calibration and control of false positive rate are described by Friso et al. (2010). MS-based information for all identified proteins was extracted from the Mascot search pages and filtered for significance (e.g. minimum ion scores, etc.), ambiguities, and shared spectra as described (Friso et al., 2010). All filtered results were uploaded into the PPDB (http://ppdb.tc.cornell.edu/; Sun et al., 2009).
Hierarchical Clustering Analysis
To group proteins with similar distribution patterns across the sample types, hierarchical clustering was employed using the Statistics toolbox of MATLAB version 7 (Mathworks). The linear correlation (ρ) between every pair of proteins with NadjSPC distribution across the samples X1…Xn and Y1…Yn, where n = 6,
 |
was derived. This was then converted into a distance measure: ΔXY = 1 − ρXY. Protein pairs with similar distribution patterns have higher correlations and, in turn, have smaller distance values. A linkage map based on the average distance among protein pairs was then constructed to yield a hierarchical cluster tree (dendrogram).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Quality of nucleoid preparations.
Supplemental Figure S2. Protein accumulation profiles for pTAC16 (A) and for three agglutinin domain homologs (B) along the gradient of a maize leaf.
Supplemental Figure S3. Comparison between nucleoids isolated from base, tip, and young seedlings for proteins in bin 29.
Supplemental Figure S4. Alternative work flow to detect the developmental accumulation of core nucleoid proteins.
Supplemental Figure S5. Hierarchical cluster analysis of 305 proteins to detect the developmental accumulation of nucleoid-enriched proteins.
Supplemental Figure S6. Effect of co-IPs and RNase treatment on proteins annotated to bin 29.
Supplemental Table S1. Detailed quantitative and qualitative information about the proteins identified in maize nucleoids from base or tip from the third leaf of 10-d-old seedlings or from young leaves (all four leaves of 7-d-old seedlings).
Supplemental Table S2. Proteins identified in maize nucleoids, chloroplasts, and proplastids. with summarized quantitative information based on average NadjSPC and normalized spectral abundance factor.
Supplemental Table S3. Nucleoid or pTAC proteins (46 in total) identified in Arabidopsis or other higher plant species as reported in the literature, and information about predicted maize homologs.
Supplemental Table S4. Proteins involved in protein synthesis, folding, assembly, and posttranslational modifications (bin 29) and identified in chloroplasts, proplastids, or nucleoids, their putative functions, and their relative abundances.
Supplemental Table S5. The unfiltered core nucleoid proteomes based on the quantitative work flows shown in Figure 5 (157 proteins) and Supplemental Figure S4A (175 proteins) .
Supplemental Table S6. Co-IP of nucleoids with anti-maize WHY1 serum with or with RNase treatment.
References
- Albrecht V, Ingenfeld A, Apel K. (2008) Snowy cotyledon 2: the identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves. Plant Mol Biol 66: 599–608 [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. (2000) SAP: a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci 25: 112–114 [DOI] [PubMed] [Google Scholar]
- Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustun S, Melzer M, Petersen K, Lein W, Bornke F. (2010) Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 22: 1498–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Barkan A. (2006) Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol 142: 1656–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiychuk E, Vandepoele K, Wissing J, Garcia-Diaz M, De Rycke R, Akbari H, Joubes J, Beeckman T, Jansch L, Frentzen M, et al. (2011) Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc Natl Acad Sci USA 108: 6674–6679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky S, Shteiman-Kotler A, Liveanu V, Yehudai-Resheff S, Bellaoui M, Settlage RE, Shabanowitz J, Hunt DF, Schuster G, Gruissem W. (2001) Chloroplast PNPase exists as a homo-multimer enzyme complex that is distinct from the Escherichia coli degradosome. RNA 7: 1464–1475 [PMC free article] [PubMed] [Google Scholar]
- Baker NR, Leech RM. (1977) Development of photosystem I and photosystem II activities in leaves of light-grown maize (Zea mays). Plant Physiol 60: 640–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem 389: 1017–1031 [DOI] [PubMed] [Google Scholar]
- Barkan A. (2011) Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol 155: 1520–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belacel N, Wang Q, Cuperlovic-Culf M. (2006) Clustering methods for microarray gene expression data. OMICS 10: 507–531 [DOI] [PubMed] [Google Scholar]
- Bisanz C, Bégot L, Carol P, Perez P, Bligny M, Pesey H, Gallois JL, Lerbs-Mache S, Mache R. (2003) The Arabidopsis nuclear DAL gene encodes a chloroplast protein which is required for the maturation of the plastid ribosomal RNAs and is essential for chloroplast differentiation. Plant Mol Biol 51: 651–663 [DOI] [PubMed] [Google Scholar]
- Bollenbach TJ, Sharwood RE, Gutierrez R, Lerbs-Mache S, Stern DB. (2009) The RNA-binding proteins CSP41a and CSP41b may regulate transcription and translation of chloroplast-encoded RNAs in Arabidopsis. Plant Mol Biol 69: 541–552 [DOI] [PubMed] [Google Scholar]
- Bordo D, Bork P. (2002) The rhodanese/Cdc25 phosphatase superfamily: sequence-structure-function relations. EMBO Rep 3: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam K, Dietzel L, Pfannschmidt T. (2010) Hypothesis: a binary redox control mode as universal regulator of photosynthetic light acclimation. Plant Signal Behav 5: 81–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell TP, Sawers RJ, Mant A, Langdale JA. (1999) BUNDLE SHEATH DEFECTIVE2, a novel protein required for post-translational regulation of the rbcL gene of maize. Plant Cell 11: 849–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant N, Lloyd J, Sweeney C, Myouga F, Meinke D. (2011) Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol 155: 1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, Rösch P. (2010) A NusE:NusG complex links transcription and translation. Science 328: 501–504 [DOI] [PubMed] [Google Scholar]
- Cámara Y, Asin-Cayuela J, Park CB, Metodiev MD, Shi Y, Ruzzenente B, Kukat C, Habermann B, Wibom R, Hultenby K, et al. (2011) MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab 13: 527–539 [DOI] [PubMed] [Google Scholar]
- Cannon GC, Ward LN, Case CI, Heinhorst S. (1999) The 68 kDa DNA compacting nucleoid protein from soybean chloroplasts inhibits DNA synthesis in vitro. Plant Mol Biol 39: 835–845 [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Sparvoli S, Edmunds C, Garosi P, Findlay K, Martin C. (1996) DAG, a gene required for chloroplast differentiation and palisade development in Antirrhinum majus. EMBO J 15: 4194–4207 [PMC free article] [PubMed] [Google Scholar]
- Chen KM, Holmström M, Raksajit W, Suorsa M, Piippo M, Aro EM. (2010) Small chloroplast-targeted DnaJ proteins are involved in optimization of photosynthetic reactions in Arabidopsis thaliana. BMC Plant Biol 10: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KM, Piippo M, Holmstrom M, Nurmi M, Pakula E, Suorsa M, Aro EM. (2011) A chloroplast-targeted DnaJ protein AtJ8 is negatively regulated by light and has rapid turnover in darkness. J Plant Physiol 168: 1780–1783 [DOI] [PubMed] [Google Scholar]
- Chi-Ham CL, Keaton MA, Cannon GC, Heinhorst S. (2002) The DNA-compacting protein DCP68 from soybean chloroplasts is ferredoxin:sulfite reductase and co-localizes with the organellar nucleoid. Plant Mol Biol 49: 621–631 [DOI] [PubMed] [Google Scholar]
- Chiu CC, Chen LJ, Li HM. (2010) Pea chloroplast DnaJ-J8 and Toc12 are encoded by the same gene and localized in the stroma. Plant Physiol 154: 1172–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipollone R, Ascenzi P, Visca P. (2007) Common themes and variations in the rhodanese superfamily. IUBMB Life 59: 51–59 [DOI] [PubMed] [Google Scholar]
- Cosgriff S, Chintakayala K, Chim YT, Chen X, Allen S, Lovering AL, Grainger DC. (2010) Dimerization and DNA-dependent aggregation of the Escherichia coli nucleoid protein and chaperone CbpA. Mol Microbiol 77: 1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea LD, Regan L. (2003) TPR proteins: the versatile helix. Trends Biochem Sci 28: 655–662 [DOI] [PubMed] [Google Scholar]
- Day A, Madesis P. (2007) DNA replication, recombination, and repair in plastids. Bock R, , Cell and Molecular Biology of Plastids, Vol 19. Springer-Verlag, Berlin, pp 65–119 [Google Scholar]
- Dillon SC, Dorman CJ. (2010) Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol 8: 185–195 [DOI] [PubMed] [Google Scholar]
- Domon B, Aebersold R. (2010) Options and considerations when selecting a quantitative proteomics strategy. Nat Biotechnol 28: 710–721 [DOI] [PubMed] [Google Scholar]
- Dong M, Yang LL, Williams K, Fisher SJ, Hall SC, Biggin MD, Jin J, Witkowska HE. (2008) A “tagless” strategy for identification of stable protein complexes genome-wide by multidimensional orthogonal chromatographic separation and iTRAQ reagent tracking. J Proteome Res 7: 1836–1849 [DOI] [PubMed] [Google Scholar]
- Douglas SE. (1998) Plastid evolution: origins, diversity, trends. Curr Opin Genet Dev 8: 655–661 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epshtein V, Dutta D, Wade J, Nudler E. (2010) An allosteric mechanism of Rho-dependent transcription termination. Nature 463: 245–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. (2010) The Pfam protein families database. Nucleic Acids Res 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Pérez U, Sauret-Güeto S, Gas E, Jarvis P, Rodríguez-Concepción M. (2008) A mutant impaired in the production of plastome-encoded proteins uncovers a mechanism for the homeostasis of isoprenoid biosynthetic enzymes in Arabidopsis plastids. Plant Cell 20: 1303–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin AE, Hoffman NE. (1993) Characterization of a chloroplast homologue of the 54-kDa subunit of the signal recognition particle. J Biol Chem 268: 22175–22180 [PubMed] [Google Scholar]
- Friso G, Majeran W, Huang M, Sun Q, van Wijk KJ. (2010) Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol 152: 1219–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M, Myouga F, Takechi K, Sato H, Nabeshima K, Nagata N, Takio S, Shinozaki K, Takano H. (2008) An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development. Plant J 53: 924–934 [DOI] [PubMed] [Google Scholar]
- Glynn JM, Yang Y, Vitha S, Schmitz AJ, Hemmes M, Miyagishima SY, Osteryoung KW. (2009) PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J 59: 700–711 [DOI] [PubMed] [Google Scholar]
- Gowrishankar J, Harinarayanan R. (2004) Why is transcription coupled to translation in bacteria? Mol Microbiol 54: 598–603 [DOI] [PubMed] [Google Scholar]
- Gray MW. (1999) Evolution of organellar genomes. Curr Opin Genet Dev 9: 678–687 [DOI] [PubMed] [Google Scholar]
- Gutman BL, Niyogi KK. (2009) Evidence for base excision repair of oxidative DNA damage in chloroplasts of Arabidopsis thaliana. J Biol Chem 284: 17006–17012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallick RB, Lipper C, Richards OC, Rutter WJ. (1976) Isolation of a transcriptionally active chromosome from chloroplasts of Euglena gracilis. Biochemistry 15: 3039–3045 [DOI] [PubMed] [Google Scholar]
- Haswell ES, Meyerowitz EM. (2006) MscS-like proteins control plastid size and shape in Arabidopsis thaliana. Curr Biol 16: 1–11 [DOI] [PubMed] [Google Scholar]
- Hilbert M, Karow AR, Klostermeier D. (2009) The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol Chem 390: 1237–1250 [DOI] [PubMed] [Google Scholar]
- Huang S, Taylor NL, Narsai R, Eubel H, Whelan J, Millar AH. (2009) Experimental analysis of the rice mitochondrial proteome, its biogenesis, and heterogeneity. Plant Physiol 149: 719–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SY, Rose A, Meier I. (2003) MFP1 is a thylakoid-associated, nucleoid-binding protein with a coiled-coil structure. Nucleic Acids Res 31: 5175–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Nakanishi H, Suzuki K, Ichikawa T, Kondou Y, Matsui M, Miyagishima SY. (2010) The YlmG protein has a conserved function related to the distribution of nucleoids in chloroplasts and cyanobacteria. BMC Plant Biol 10: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YW, Lee JY, Jeon Y, Cheong GW, Kim M, Pai HS. (2010) In vivo effects of NbSiR silencing on chloroplast development in Nicotiana benthamiana. Plant Mol Biol 72: 569–583 [DOI] [PubMed] [Google Scholar]
- Karcher D, Köster D, Schadach A, Klevesath A, Bock R. (2009) The Chlamydomonas chloroplast HLP protein is required for nucleoid organization and genome maintenance. Mol Plant 2: 1223–1232 [DOI] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W. (2010) New insights into the types and function of proteases in plastids. Int Rev Cell Mol Biol 280: 185–218 [DOI] [PubMed] [Google Scholar]
- Khrouchtchova A, Hansson M, Paakkarinen V, Vainonen JP, Zhang S, Jensen PE, Scheller HV, Vener AV, Aro EM, Haldrup A. (2005) A previously found thylakoid membrane protein of 14kDa (TMP14) is a novel subunit of plant photosystem I and is designated PSI-P. FEBS Lett 579: 4808–4812 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Takahara M, Miyagishima SY, Kuroiwa H, Sasaki N, Ohta N, Matsuzaki M, Kuroiwa T. (2002) Detection and localization of a chloroplast-encoded HU-like protein that organizes chloroplast nucleoids. Plant Cell 14: 1579–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause K, Krupinska K. (2000) Molecular and functional properties of highly purified transcriptionally active chromosomes from spinach chloroplasts. Physiol Plant 109: 188–195 [Google Scholar]
- Kroeger TS, Watkins KP, Friso G, van Wijk KJ, Barkan A. (2009) A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc Natl Acad Sci USA 106: 4537–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa T. (1973) Studies on mitochondrial structure and function in Physarium polycephalum. I. Fine structure, cytochemistry, and 3H-uridine autoradiography of a central body in mitochondria. Exp Cell Res 78: 351–359 [DOI] [PubMed] [Google Scholar]
- Kuroiwa T. (1991) The replication, differentiation, and inheritance of plastids with emphasis on the concept of organelle nuclei. Int Rev Cytol 128: 1–62 [Google Scholar]
- Kuroiwa T, Fujie M, Kuroiwa H. (1992) Studies on the behavior of mitochondrial DNA: synthesis of mitochondrial DNA occurs actively in a specific region just above the quiescent center in the root apical meristem of Pelargonium zonale. J Cell Sci 101: 483–493 [Google Scholar]
- Kuroiwa T, Kawano S, Nishibayashi S, Sato C. (1982) Epifluorescent microscopic evidence for maternal inheritance of chloroplast DNA. Nature 298: 481–483 [DOI] [PubMed] [Google Scholar]
- Kuroiwa T, Ohta T, Kuroiwa H, Shigeyuki K. (1994) Molecular and cellular mechanisms of mitochondrial nuclear division and mitochondriokinesis. Microsc Res Tech 27: 220–232 [DOI] [PubMed] [Google Scholar]
- Leech RM, Rumsby MG, Thomson WW. (1973) Plastid differentiation, acyl lipid, and fatty acid changes in developing green maize leaves. Plant Physiol 52: 240–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lensch M, Herrmann RG, Sokolenko A. (2001) Identification and characterization of SppA, a novel light-inducible chloroplast protease complex associated with thylakoid membranes. J Biol Chem 276: 33645–33651 [DOI] [PubMed] [Google Scholar]
- Lezhneva L, Kuras R, Ephritikhine G, de Vitry C. (2008) A novel pathway of cytochrome c biogenesis is involved in the assembly of the cytochrome b6f complex in Arabidopsis chloroplasts. J Biol Chem 283: 24608–24616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QS, Gupta JD, Hunt AG. (1998) Polynucleotide phosphorylase is a component of a novel plant poly(A) polymerase. J Biol Chem 273: 17539–17543 [DOI] [PubMed] [Google Scholar]
- Liere K, Weihe A, Börner T. (2011) The transcription machineries of plant mitochondria and chloroplasts: composition, function, and regulation. J Plant Physiol 168: 1345–1360 [DOI] [PubMed] [Google Scholar]
- Linder P. (2006) Dead-box proteins: a family affair. Active and passive players in RNP-remodeling. Nucleic Acids Res 34: 4168–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., III (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76: 4193–4201 [DOI] [PubMed] [Google Scholar]
- Liu X, Yu F, Rodermel S. (2010a) Arabidopsis chloroplast FtsH, var2 and suppressors of var2 leaf variegation: a review. J Integr Plant Biol 52: 750–761 [DOI] [PubMed] [Google Scholar]
- Liu X, Yu F, Rodermel S. (2010b) An Arabidopsis pentatricopeptide repeat protein, SUPPRESSOR OF VARIEGATION7, is required for FtsH-mediated chloroplast biogenesis. Plant Physiol 154: 1588–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshon D, Zweifel SG, Freeman-Cook LL, Lorimer HE, Brewer BJ, Fangman WL. (1995) A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell 81: 947–955 [DOI] [PubMed] [Google Scholar]
- Long TA, Brady SM, Benfey PN. (2008) Systems approaches to identifying gene regulatory networks in plants. Annu Rev Cell Dev Biol 24: 81–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Peng L, Guo J, Lu Q, Lu C, Zhang L. (2007) LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 19: 1980–1993 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Machida M, Takechi K, Sato H, Chung SJ, Kuroiwa H, Takio S, Seki M, Shinozaki K, Fujita T, Hasebe M, et al. (2006) Genes for the peptidoglycan synthesis pathway are essential for chloroplast division in moss. Proc Natl Acad Sci USA 103: 6753–6758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Cai Y, Sun Q, van Wijk KJ. (2005) Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. Plant Cell 17: 3111–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Friso G, Ponnala L, Connolly B, Huang M, Reidel E, Zhang C, Asakura Y, Bhuiyan NH, Sun Q, et al. (2010) Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. Plant Cell 22: 3509–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Zybailov B, Ytterberg AJ, Dunsmore J, Sun Q, van Wijk KJ. (2008) Consequences of C4 differentiation for chloroplast membrane proteomes in maize mesophyll and bundle sheath cells. Mol Cell Proteomics 7: 1609–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M, Kelleher NL. (2008) Precision proteomics: the case for high resolution and high mass accuracy. Proc Natl Acad Sci USA 105: 18132–18138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Parent JS, Véronneau-Lafortune F, Joyeux A, Lang BF, Brisson N. (2009) Whirly proteins maintain plastid genome stability in Arabidopsis. Proc Natl Acad Sci USA 106: 14693–14698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I, Phelan T, Gruissem W, Spiker S, Schneider D. (1996) MFP1, a novel plant filament-like protein with affinity for matrix attachment region DNA. Plant Cell 8: 2105–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melonek J, Mulisch M, Schmitz-Linneweber C, Grabowski E, Hensel G, Krupinska K. (2010) Whirly1 in chloroplasts associates with intron containing RNAs and rarely co-localizes with nucleoids. Planta 232: 471–481 [DOI] [PubMed] [Google Scholar]
- Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K. (2001) FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskiene R, Würsch M, Laloi C, Vidi PA, Coll NS, Kessler F, Baruah A, Kim C, Apel K. (2009) A mutation in the Arabidopsis mTERF-related plastid protein SOLDAT10 activates retrograde signaling and suppresses 1O2-induced cell death. Plant J 60: 399–410 [DOI] [PubMed] [Google Scholar]
- Miyamura S, Nagata T, Kuroiwa T. (1986) Quantitative fluorescence microscopy on dynamic changes of plastid nucleoids during wheat development. Protoplasma 133: 66–72 [Google Scholar]
- Mori Y, Kimura S, Saotome A, Kasai N, Sakaguchi N, Uchiyama Y, Ishibashi T, Yamamoto T, Chiku H, Sakaguchi K. (2005) Plastid DNA polymerases from higher plants, Arabidopsis thaliana. Biochem Biophys Res Commun 334: 43–50 [DOI] [PubMed] [Google Scholar]
- Myouga F, Hosoda C, Umezawa T, Iizumi H, Kuromori T, Motohashi R, Shono Y, Nagata N, Ikeuchi M, Shinozaki K. (2008) A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell 20: 3148–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata N, Saito C, Sakai A, Kuroiwa H, Kuroiwa T. (1999) The selective increase or decrease of organellar DNA in generative cells just after pollen mitosis one controls cytoplasmic inheritance. Planta 209: 53–65 [DOI] [PubMed] [Google Scholar]
- Nerozzi AM, Coleman AW. (1997) Localization of plastid DNA replication on a nucleoid structure. Am J Bot 84: 1028–1041 [PubMed] [Google Scholar]
- Nilsson R, Brunner J, Hoffman NE, van Wijk KJ. (1999) Interactions of ribosome nascent chain complexes of the chloroplast-encoded D1 thylakoid membrane protein with cpSRP54. EMBO J 18: 733–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Ashida H, Ogawa T, Yokota A. (2010) A DEAD box protein is required for formation of a hidden break in Arabidopsis chloroplast 23S rRNA. Plant J 63: 766–777 [DOI] [PubMed] [Google Scholar]
- Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics 4: 1487–1502 [DOI] [PubMed] [Google Scholar]
- Olinares PD, Ponnala L, van Wijk KJ. (2010) Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol Cell Proteomics 9: 1594–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinares PDB, Kim J, Davis JI, van Wijk KJ. (2011) Subunit stoichiometry, evolution, and functional implications of an asymmetric plant plastid ClpP/R protease complex in Arabidopsis. Plant Cell 23: 2348–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, Sugita M, Small I. (2008) On the expansion of the pentatricopeptide repeat gene family in plants. Mol Biol Evol 25: 1120–1128 [DOI] [PubMed] [Google Scholar]
- Parent JS, Lepage E, Brisson N. (2011) Divergent roles for the two PolI-like organelle DNA polymerases of Arabidopsis. Plant Physiol 156: 254–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M, Asin-Cayuela J, Erdjument-Bromage H, Tempst P, Larsson NG, Gustafsson CM. (2009) MTERF2 is a nucleoid component in mammalian mitochondria. Biochim Biophys Acta 1787: 296–302 [DOI] [PubMed] [Google Scholar]
- Peng L, Ma J, Chi W, Guo J, Zhu S, Lu Q, Lu C, Zhang L. (2006) LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R. (2006) pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18: 176–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phinney BS, Thelen JJ. (2005) Proteomic characterization of a Triton-insoluble fraction from chloroplasts defines a novel group of proteins associated with macromolecular structures. J Proteome Res 4: 497–506 [DOI] [PubMed] [Google Scholar]
- Pontén F, Gry M, Fagerberg L, Lundberg E, Asplund A, Berglund L, Oksvold P, Björling E, Hober S, Kampf C, et al. (2009) A global view of protein expression in human cells, tissues, and organs. Mol Syst Biol 5: 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J, Watkins KP, Friso G, van Wijk KJ, Barkan A. (2008) A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res 36: 5152–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proshkin S, Rahmouni AR, Mironov A, Nudler E. (2010) Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328: 504–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Sarmiento-Manus R, Gonzalez-Bayon R, Hricova A, Perez-Marcos R, Gracia-Martinez E, Medina-Ruiz L, Leyva-Diaz E, Ponce MR, Micol JL. (2011) Arabidopsis RUGOSA2 encodes an mTERF family member required for mitochondrion, chloroplast and leaf development. Plant J 68: 738–753 [DOI] [PubMed] [Google Scholar]
- Quintana LF, Campistol JM, Alcolea MP, Bañon-Maneus E, Sol-González A, Cutillas PR. (2009) Application of label-free quantitative peptidomics for the identification of urinary biomarkers of kidney chronic allograft dysfunction. Mol Cell Proteomics 8: 1658–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter CV, Bals T, Schünemann D. (2010) Component interactions, regulation and mechanisms of chloroplast signal recognition particle-dependent protein transport. Eur J Cell Biol 89: 965–973 [DOI] [PubMed] [Google Scholar]
- Roberti M, Polosa PL, Bruni F, Manzari C, Deceglie S, Gadaleta MN, Cantatore P. (2009) The MTERF family proteins: mitochondrial transcription regulators and beyond. Biochim Biophys Acta 1787: 303–311 [DOI] [PubMed] [Google Scholar]
- Robinow C, Kellenberger E. (1994) The bacterial nucleoid revisited. Microbiol Rev 58: 211–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocak S, Linder P. (2004) DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5: 232–241 [DOI] [PubMed] [Google Scholar]
- Rochaix JD. (2011) Regulation of photosynthetic electron transport. Biochim Biophys Acta 1807: 375–383 [DOI] [PubMed] [Google Scholar]
- Ruwe H, Kupsch C, Teubner M, Schmitz-Linneweber C. (2011) The RNA-recognition motif in chloroplasts. J Plant Physiol 168: 1361–1371 [DOI] [PubMed] [Google Scholar]
- Sakai A, Suzuki T, Nagata N, Sasaki N, Miyazawa Y, Saito C, Inada N, Nishimura Y, Kuroiwa T. (1999) Comparative analysis of DNA synthesis activity in plastid-nuclei and mitochondrial-nuclei simultaneously isolated from cultured tobacco cells. Plant Sci 140: 9–19 [Google Scholar]
- Sakai A, Takano H, Kuroiwa T. (2004) Organelle nuclei in higher plants: structure, composition, function, and evolution. Int Rev Cytol 238: 59–118 [DOI] [PubMed] [Google Scholar]
- Sakai A, Yamashita H, Nemoto Y, Kawano S, Kuroiwa T. (1991) Transcriptional activity of morphologically intact proplastid-nuclei (nucleoids) isolated from tobacco cultured cells. Plant Cell Physiol 32: 835–843 [Google Scholar]
- Samaniego R, Jeong SY, Meier I, de la Espina SM. (2006) Dual location of MAR-binding, filament-like protein 1 in Arabidopsis, tobacco, and tomato. Planta 223: 1201–1206 [DOI] [PubMed] [Google Scholar]
- Sandhu C, Hewel JA, Badis G, Talukder S, Liu J, Hughes TR, Emili A. (2008) Evaluation of data-dependent versus targeted shotgun proteomic approaches for monitoring transcription factor expression in breast cancer. J Proteome Res 7: 1529–1541 [DOI] [PubMed] [Google Scholar]
- Sasaki N, Sakai A, Kawano S, Kuroiwa H, Kuroiwa T. (1998) DNA synthesis in isolated mitochondrial nucleoids from plasmodia of Physarium polycephalum. Protoplasma 203: 221–231 [Google Scholar]
- Sato N, Nakayama M, Hase T. (2001) The 70-kDa major DNA-compacting protein of the chloroplast nucleoid is sulfite reductase. FEBS Lett 487: 347–350 [DOI] [PubMed] [Google Scholar]
- Sato N, Ohshima K, Watanabe A, Ohta N, Nishiyama Y, Joyard J, Douce R. (1998) Molecular characterization of the PEND protein, a novel bZIP protein present in the envelope membrane that is the site of nucleoid replication in developing plastids. Plant Cell 10: 859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Rolland N, Block MA, Joyard J. (1999) Do plastid envelope membranes play a role in the expression of the plastid genome? Biochimie 81: 619–629 [DOI] [PubMed] [Google Scholar]
- Sato N, Terasawa K, Miyajima K, Kabeya Y. (2003) Organization, developmental dynamics, and evolution of plastid nucleoids. Int Rev Cytol 232: 217–262 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I. (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Schröter Y, Steiner S, Matthäi K, Pfannschmidt T. (2010) Analysis of oligomeric protein complexes in the chloroplast sub-proteome of nucleic acid-binding proteins from mustard reveals potential redox regulators of plastid gene expression. Proteomics 10: 2191–2204 [DOI] [PubMed] [Google Scholar]
- Schult K, Meierhoff K, Paradies S, Töller T, Wolff P, Westhoff P. (2007) The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell 19: 1329–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweer J, Türkeri H, Kolpack A, Link G. (2010) Role and regulation of plastid sigma factors and their functional interactors during chloroplast transcription: recent lessons from Arabidopsis thaliana. Eur J Cell Biol 89: 940–946 [DOI] [PubMed] [Google Scholar]
- Sekine K, Fujiwara M, Nakayama M, Takao T, Hase T, Sato N. (2007) DNA binding and partial nucleoid localization of the chloroplast stromal enzyme ferredoxin:sulfite reductase. FEBS J 274: 2054–2069 [DOI] [PubMed] [Google Scholar]
- Selldén G, Leech RM. (1981) Localization of DNA in mature and young wheat chloroplasts using the fluorescent probe 4′-6-diamidino-2-phenylindole. Plant Physiol 68: 731–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR. (2011) Assembly of bacterial ribosomes. Annu Rev Biochem 80: 501–526 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Smith CA. (2006) Structure, function and dynamics in the mur family of bacterial cell wall ligases. J Mol Biol 362: 640–655 [DOI] [PubMed] [Google Scholar]
- Sodmergen S, Kawano S, Kuroiwa T. (1991) Degradation of chloroplast DNA in second leaves of rice (Oryza sativa) before leaf yellowing. Protoplasma 160: 89–98 [Google Scholar]
- Sokolenko A, Pojidaeva E, Zinchenko V, Panichkin V, Glaser VM, Herrmann RG, Shestakov SV. (2002) The gene complement for proteolysis in the cyanobacterium Synechocystis sp. PCC 6803 and Arabidopsis thaliana chloroplasts. Curr Genet 41: 291–310 [DOI] [PubMed] [Google Scholar]
- Steiner S, Dietzel L, Schröter Y, Fey V, Wagner R, Pfannschmidt T. (2009) The role of phosphorylation in redox regulation of photosynthesis genes psaA and psbA during photosynthetic acclimation of mustard. Mol Plant 2: 416–429 [DOI] [PubMed] [Google Scholar]
- Steiner S, Schroter Y, Pfalz J, Pfannschmidt T. (2011) Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol 157: 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR. (2010) Chloroplast RNA metabolism. Annu Rev Plant Biol 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Stern DB, Hanson MR, Barkan A. (2004) Genetics and genomics of chloroplast biogenesis: maize as a model system. Trends Plant Sci 9: 293–301 [DOI] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ. (2009) PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res 37: D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki JY, Ytterberg AJ, Beardslee TA, Allison LA, Wijk KJ, Maliga P. (2004) Affinity purification of the tobacco plastid RNA polymerase and in vitro reconstitution of the holoenzyme. Plant J 40: 164–172 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Kawano S, Sakai A, Fujie M, Kuroiwa H, Nakamura H, Kuroiwa T. (1992) Preferential mitochondrial and plastid DNA synthesis before multiple cell divisions in Nicotiana tabacum. J Cell Sci 103: 831–837 [Google Scholar]
- Takano H, Takechi K. (2010) Plastid peptidoglycan. Biochim Biophys Acta 1800: 144–151 [DOI] [PubMed] [Google Scholar]
- Terasawa K, Sato N. (2005a) Occurrence and characterization of PEND proteins in angiosperms. J Plant Res 118: 111–119 [DOI] [PubMed] [Google Scholar]
- Terasawa K, Sato N. (2005b) Visualization of plastid nucleoids in situ using the PEND-GFP fusion protein. Plant Cell Physiol 46: 649–660 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tikkanen M, Aro EM. (2011) Thylakoid protein phosphorylation in dynamic regulation of photosystem II in higher plants. Biochim Biophys Acta (in press) [DOI] [PubMed] [Google Scholar]
- Till B, Schmitz-Linneweber C, Williams-Carrier R, Barkan A. (2001) CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 7: 1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Hardel SL, Kupsch C, Armbruster U, Delannoy E, Gualberto JM, Lehwark P, Leister D, Small ID, Schmitz-Linneweber C. (2009) Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proc Natl Acad Sci USA 106: 6002–6007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainonen JP, Sakuragi Y, Stael S, Tikkanen M, Allahverdiyeva Y, Paakkarinen V, Aro E, Suorsa M, Scheller HV, Vener AV, et al. (2008) Light regulation of CaS, a novel phosphoprotein in the thylakoid membrane of Arabidopsis thaliana. FEBS J 275: 1767–1777 [DOI] [PubMed] [Google Scholar]
- van Wijk KJ, Baginsky S. (2011) Update on plastid proteomics in higher plants: current state and future goals. Plant Physiol 155: 1578–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Duby G, Purnelle B, Boutry M. (2000) Tobacco VDL gene encodes a plastid DEAD box RNA helicase and is involved in chloroplast differentiation and plant morphogenesis. Plant Cell 12: 2129–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins KP, Kroeger TS, Cooke AM, Williams-Carrier RE, Friso G, Belcher SE, van Wijk KJ, Barkan A. (2007) A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell 19: 2606–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins KP, Rojas M, Friso G, van Wijk KJ, Meurer J, Barkan A. (2011) APO1 promotes the splicing of chloroplast group II introns and harbors a plant-specific zinc-dependent RNA binding domain. Plant Cell 23: 1082–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P, Fulgosi H, Piven I, Müller L, Krupinska K, Duong VH, Herrmann RG, Sokolenko A. (2006) TCP34, a nuclear-encoded response regulator-like TPR protein of higher plant chloroplasts. J Mol Biol 357: 535–549 [DOI] [PubMed] [Google Scholar]
- Wetzel CM, Harmacek LD, Yuan LH, Wopereis JL, Chubb R, Turini P. (2009) Loss of chloroplast protease SPPA function alters high light acclimation processes in Arabidopsis thaliana L. (Heynh.). J Exp Bot 60: 1715–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ME, Jensen GS, Haswell ES. (2011) Two mechanosensitive channel homologs influence division ring placement in Arabidopsis chloroplasts. Plant Cell 23: 2939–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YZ, Arrieta-Montiel MP, Virdi KS, de Paula WB, Widhalm JR, Basset GJ, Davila JI, Elthon TE, Elowsky CG, Sato SJ.et al (2011) MutS HOMOLOG1 is a nucleoid protein that alters mitochondrial and plastid properties and plant response to high light. Plant Cell 23: 3428–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakubovskaya E, Mejia E, Byrnes J, Hambardjieva E, Garcia-Diaz M. (2010) Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell 141: 982–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Subramanian AR. (2003) Proteomic identification of all plastid-specific ribosomal proteins in higher plant chloroplast 30S ribosomal subunit. Eur J Biochem 270: 190–205 [DOI] [PubMed] [Google Scholar]
- Yang J, Schuster G, Stern DB. (1996) CSP41, a sequence-specific chloroplast mRNA binding protein, is an endoribonuclease. Plant Cell 8: 1409–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Liu X, Alsheikh M, Park S, Rodermel S. (2008) Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis. Plant Cell 20: 1786–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Fu X, Koo YD, Zhu JK, Jenney FE, Jr, Adams MW, Zhu Y, Shi H, Yun DJ, Hasegawa PM, et al. (2007) An enhancer mutant of Arabidopsis salt overly sensitive 3 mediates both ion homeostasis and the oxidative stress response. Mol Cell Biol 27: 5214–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Coleman MK, Florens L, Washburn MP. (2005) Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal Chem 77: 6218–6224 [DOI] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]