Abstract
Phytochromes are the red/far-red photoreceptors in higher plants. Among them, phytochrome A (PHYA) is responsible for the far-red high-irradiance response and for the perception of very low amounts of light, initiating the very-low-fluence response. Here, we report a detailed physiological and molecular characterization of the phyA-5 mutant of Arabidopsis (Arabidopsis thaliana), which displays hyposensitivity to continuous low-intensity far-red light and shows reduced very-low-fluence response and high-irradiance response. Red light-induced degradation of the mutant phyA-5 protein appears to be normal, yet higher residual amounts of phyA-5 are detected in seedlings grown under low-intensity far-red light. We show that (1) the phyA-5 mutant harbors a new missense mutation in the PHYA amino-terminal extension domain and that (2) the complex phenotype of the mutant is caused by reduced nuclear import of phyA-5 under low fluences of far-red light. We also demonstrate that impaired nuclear import of phyA-5 is brought about by weakened binding affinity of the mutant photoreceptor to nuclear import facilitators FHY1 (for FAR-RED ELONGATED HYPOCOTYL1) and FHL (for FHY1-LIKE). Finally, we provide evidence that the signaling and degradation kinetics of constitutively nuclear-localized phyA-5 and phyA are identical. Taken together, our data show that aberrant nucleo/cytoplasmic distribution impairs light-induced degradation of this photoreceptor and that the amino-terminal extension domain mediates the formation of the FHY1/FHL/PHYA far-red-absorbing form complex, whereby it plays a role in regulating the nuclear import of phyA.
Plants, as sessile organisms, have to adapt to the ambient environment. Light is one of the most important environmental factors, because it is not only the energy source for photosynthesis but also a signal regulating a wide range of physiological and developmental processes from seed germination to flowering, affecting almost every aspect of plant life (Chen et al., 2004). Plants can sense the presence or absence, intensity (quantity), wavelength (quality), direction, duration, and diurnal rhythm of light by specialized photoreceptor molecules (Sullivan and Deng, 2003). Plant photoreceptors are categorized by the wavelength of light that they perceive. Receptors of red (R) and far-red (FR) light (approximately 620–750 nm) are phytochromes. Phytochromes in most of the examined species form small gene families (Mathews and Sharrock, 1997). The phytochrome gene family has five members, named phytochrome A (PHYA) through PHYE in Arabidopsis (Arabidopsis thaliana; Sharrock and Quail, 1989; Clack et al., 1994). Phytochrome proteins are synthesized in their Pr form, which can be converted by R light (absorption maximum approximately 665 nm) to the Pfr form, the physiologically active conformer of phytochromes. Upon exposure to FR light, Pfr (absorption maximum approximately 730 nm) can be reversibly converted back to Pr (Schäfer and Bowler, 2002).
According to the classical categorization, phytochromes can be “light-labile” type I (phyA) or “light-stable” type II (phyB–phyE; Sharrock and Quail, 1989). The dominant phytochrome of etiolated plants is phyA, which is quickly degraded upon R light irradiation, and thereby phyB becomes the dominant PHY of light-grown plants (Sharrock and Clack, 2002). The dynamic properties of the light-induced degradation of phyA Pfr has already been described (Hennig et al., 1999; Eichenberg et al., 2000), although the molecular mechanism underlying this phenomenon is only partially revealed (Clough and Vierstra, 1997; Seo et al., 2004; Saijo et al., 2008; Debrieux and Fankhauser, 2010; Toledo-Ortiz et al., 2010).
PhyA-controlled responses are divided to two categories. High levels of phyA in the etiolated seedlings are responsible for the very-low-fluence response (VLFR), which is triggered by extremely low amounts of light. PhyA also controls the FR high-irradiance response (HIR), which can be generated by continuous high-fluence FR light (Schäfer and Bowler, 2002).
The structural motifs of different phytochrome photoreceptors are highly similar (Nagatani, 2010). Functional phytochrome A molecules are dimers of 125-kD monomers, which can be divided into N- and C-terminal halves, connected by a proteolytically vulnerable hinge region (Quail, 1997). The N-terminal half is responsible for defining the functional characteristics of the photoreceptor (Wagner et al., 1996; Mateos et al., 2006). The N-terminal extension domain (NTE; Neff et al., 2000) contains several Ser residues in phyA, which are subject to phosphorylation (Lapko et al., 1997, 1999). Experiments performed on modified phyA carrying Ser/Ala substitutions or deletions in the NTE proved that this region is necessary for correct intracellular localization, biological activity, and signal attenuation (Cherry et al., 1992; Stockhaus et al., 1992; Jordan et al., 1996, 1997; Casal et al., 2002; Trupkin et al., 2007).
The C-terminal half of phyA is involved in the dimerization of phyA monomers (Edgerton and Jones, 1992), presumably via two PER/ARNT/SIM domains. Missense mutations located in these motifs cause impaired light responses (Xu et al., 1995; Yanovsky et al., 2002). Similar to other PHYs, the distant C-terminal part of the PHYA molecule contains the His kinase-related domain, which shows homology to bacterial His kinases (Schneider-Poetsch et al., 1991; Yeh and Lagarias, 1998; Montgomery and Lagarias, 2002). The C-terminal domains of PHYA and PHYB are involved in mediating interaction with several proteins (Ni et al., 1998; Choi et al., 1999; Fankhauser et al., 1999) and in phytochrome nuclear import (Müller et al., 2009). Interestingly, its presence is not essential for PHYB-directed photomorphogenesis (Krall and Reed, 2000; Matsushita et al., 2003; Oka et al., 2008; Palágyi et al., 2010), whereas it seems to be required for phyA-controlled HIR signaling (Cherry et al., 1993; Wolf et al., 2011).
The intracellular distribution of PHYA is tightly controlled by light. The vast majority of the Pr form is localized to the cytoplasm, whereas photoconversion to the Pfr form results in PHYA nuclear import and PHY accumulation in small subnuclear speckles (Sakamoto and Nagatani, 1996; Kircher et al., 1999, 2002; Bauer et al., 2004; Fankhauser and Chen, 2008). PhyA Pfr can also form cytoplasmic bodies (sequestered areas of phytochrome), whereas other PHYs cannot. It was shown nearly a decade ago that in Arabidopsis, phyA signaling is mediated by two small proteins, named FHY1 (for FAR-RED ELONGATED HYPOCOTYL) and FHL (for FHY1-LIKE; Desnos et al., 2001; Zeidler et al., 2001; Zhou et al., 2005). More recently, it has been reported that phyA Pfr interacts with the FHY1 and FHL proteins in yeast and that this interaction is an absolute prerequisite for light-induced nuclear import of phyA in planta (Hiltbrunner et al., 2005, 2006). The fact that constitutively nuclear-localized phyA restores wild-type-like FR signaling in the fhy1 mutant background suggested that the only function of the FHY1 protein in FR-induced signaling is to facilitate the nuclear import of phyA Pfr (Genoud et al., 2008). Saijo et al. (2008), however, reported that phyA Pr-FHY1/FHL, and not the phyA Pfr-FHY1/FHL complex, is readily detectable by coimmunoprecipitation assays in vivo. Additional reports by Shen et al. (2009) and Yang et al. (2009) confirmed this observation. These observations were then interpreted such that the Pr conformer of phyA also plays a role in regulating the nuclear import of phyA. The apparent contradiction between these reports has been elegantly solved by Rausenberger et al. (2011) by demonstrating that the low-abundance phyA Pfr-FHY1/FHL complexes are significantly more stable than the more numerous phyA Pr-FHY1/FHL complexes, which because of their high dissociation rate are dismissible for regulating the nuclear import of phyA. Plants lacking FHY1 and FHL show no phyA nuclear import, and detailed characterization of fhy1/fhl double mutants revealed the cytoplasmic functions of PHYA (Rösler et al., 2007, 2010; Debrieux and Fankhauser, 2010).
We have characterized the phyA-5 mutant bearing a missense mutation in the NTE domain of phyA. This mutation replaces a conserved Ala with Val at position 30. Examination of this mutant allowed us to define the role of the NTE domain in phyA-controlled HIR and VLFR and in nuclear import of the photoreceptor. Data obtained by yeast two-hybrid studies and in vivo by analyzing FR responsiveness of the phyA-5 mutant, wild-type, and transgenic lines suggest that impaired interaction of the mutant protein with the nuclear transport facilitator proteins FHY1/FHL is responsible for the complex phenotype of the phyA-5 mutant.
RESULTS
Identification of the phyA-5 Mutation
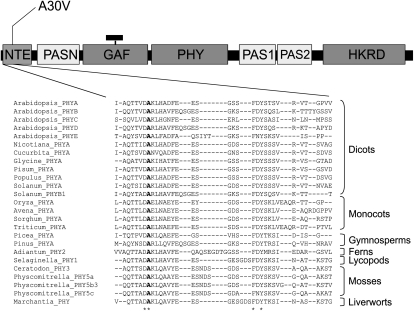
An ethyl methanesulfonate-treated mutant Arabidopsis line in the Wassilewskija (Ws) background showing hyposensitivity in FR light was isolated in the laboratory of Prof. Garry Whitelam. The name phyA-5 follows the guidelines described by Quail et al. (1994). We have previously shown that this mutant can be complemented by overexpression of the PHYA:GFP fusion protein (Kim et al., 2000). This observation suggested that the mutation causing the FR hyposensitivity is intragenic. To define the precise position and nature of this intragenic mutation, we have sequenced the wild-type PHYA and the phyA-5 mutant genes. This effort revealed that a single C/T nucleotide substitution causes an Ala/Val exchange in the NTE domain of the mutant photoreceptor at amino acid position 30 (A30V; Fig. 1). Sequence alignment of different phytochromes revealed that the mutated Ala is highly conserved among not only PHYA homologs from diverse taxa (etc. dicots, monocots, ferns, mosses) but also among Arabidopsis phytochromes (Fig. 1).
Figure 1.
Location of the phyA-5 mutation. Domains of phytochromes are shown (not to scale) together with the sequence alignment of the corresponding region. The position of the phyA-5 missense mutation is indicated (boldface letter). For database accession numbers, see “Materials and Methods.” PASN, N-terminal PER/ARNT/SIM domain; GAF, cGMP-specific phosphodiesterases, adenylyl cyclases, and FhlA domain; PHY, phytochrome domain; PAS1 and PAS2, two additional PER/ARNT/SIM domains; HKRD, His kinase-related domain. The small black rectangle attached to GAF represents the chromophore.
Physiological Characterization of phyA-5
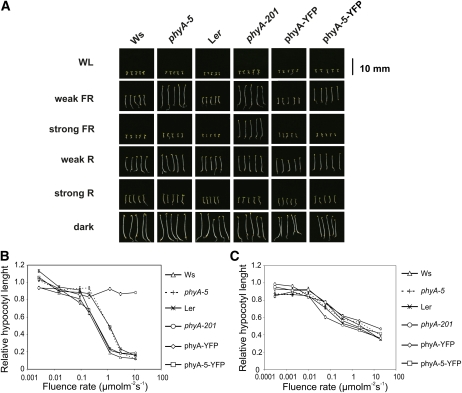
The phyA-5 mutant shows a hyposensitive hypocotyl elongation inhibition response under constant weak FR irradiation but is indistinguishable from the wild type under strong FR light (Fig. 2, A and B). This observation indicates that phyA-5 is a functional photoreceptor, but it does not regulate the inhibition of hypocotyl elongation properly. This finding was supported by cotyledon angle measurements, which also demonstrate the hyposensitivity of phyA-5 under weak FR fluence (Supplemental Fig. S1). The phyA-5 mutation, however, does not affect the inhibition of hypocotyl elongation in seedlings grown under constant R light (Fig. 2C).
Figure 2.
The effect of phyA-5 mutation on the light-dependent phenotype of 4-d-old seedlings. A, Images of seedlings grown under constant irradiation for 4 d. WL, 100 μmol m−2 s−1 fluorescent white light; weak FR, 1 μmol m−2 s−1 FR light; strong FR, 10 μmol m−2 s−1 FR light; weak R, 0.002 μmol m−2 s−1 R light; strong R, 20 μmol m−2 s−1 R light; dark, etiolated seedlings. B, Fluence rate-dependent inhibition of hypocotyl elongation, measured on 4-d-old seedlings grown in FR light. The obtained values were normalized to the hypocotyl length of the corresponding dark-grown seedlings. Error bars indicate se. C, Fluence rate-dependent inhibition of hypocotyl elongation, measured on 4-d-old seedlings grown in R light. The obtained values were normalized to the hypocotyl length of the corresponding dark-grown seedlings. Error bars indicate se. Analyzed genotypes are as follows: Ws; phyA-5 mutant (ecotype Ws); Ler; phyA-201 (ecotype Ler); phyA-YFP, PHYA:PHYA-YFP in the phyA-201 background; phyA-5-YFP, PHYA:PHYA-5-YFP in the phyA-201 background. [See online article for color version of this figure.]
To confirm that the phenotype of the phyA-5 mutant is caused by the missense mutation described, we expressed phyA-5 fused to the yellow fluorescent protein (YFP) under the control of the PHYA promoter in the phyA-201 background (Landsberg erecta [Ler] ecotype). The PHYA-5-YFP fusion protein reestablished the phenotype of the phyA-5 mutant (Fig. 2, A and B). This experiment demonstrates that the A30V mutation in the phyA molecule is indeed fully responsible for the observed phenotype.
The phyA-5 Mutation Causes a Spectral Shift in Light Sensitivity and Impaired VLFR
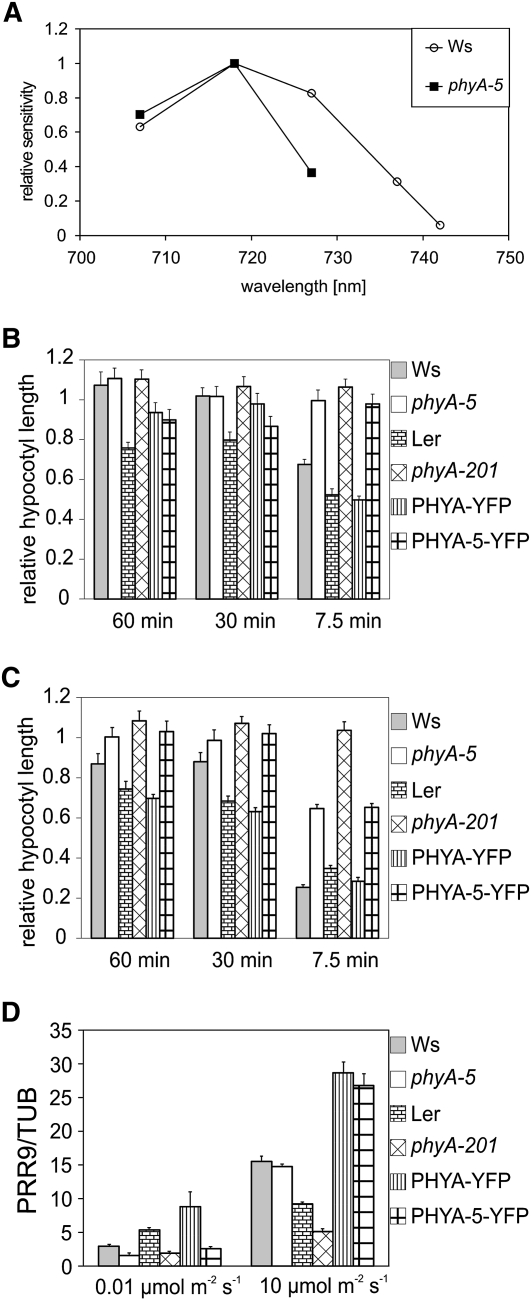
We constructed additional fluence rate curves of hypocotyl elongation using different narrow-band interference filters in order to obtain an action spectrum for hypocotyl elongation (Supplemental Fig. S2). The phyA-5 mutation causes a strong reduction in light sensitivity at every examined wavelength, especially at longer wavelengths (Fig. 3A). The most pronounced reduction was observed at 742 nm, resulting in complete insensitivity (Supplemental Fig. S2). To examine phyA-5 VLFR, we raised seedlings in darkness supplied with regular FR pulses and measured their hypocotyl lengths. Figure 3B shows that the applied weak (0.6 μmol m−2 s−1) FR pulses can induce the inhibition of hypocotyl elongation only in genotypes in which wild-type phyA is present (e.g. Ws, Ler, and PHYA:PHYA-YFP in phyA-201). phyA-5, like the phyA-201 null mutant, cannot initiate the response. Frequent stronger (6 μmol m−2 s−1) FR pulses, however, can induce a phyA-5-driven response, which is less pronounced than in the case of phyA. phyA-201 showed no response at this intensity either (Fig. 3C).
Figure 3.
The phyA-5 mutation affects spectral sensitivity and the VLFR. A, Action spectra for hypocotyl elongation in wild-type (Ws) and phyA-5 seedlings. The reciprocal value of the fluence rate that results in 60% inhibition of hypocotyl elongation compared with the corresponding dark controls was determined from the analysis of fluence rate response curves determined at different wavelengths. For better comparison, the highest value obtained in each line was set to 1, and all corresponding data were normalized to this value. B, Seedlings grown for 4 d in the dark were irradiated with 150-s FR (DAL715 filter) pulses of 0.6 μmol m−2 s−1 once every 60, 30, or 7.5 min. The hypocotyl lengths were measured after 4 d of growth, and each value obtained was normalized to the corresponding etiolated control. Error bars indicate se. C, The treatment and the analyzed lines were exactly as presented in B, with the exception of the intensity of the applied FR light pulse, which was 6 μmol m−2 s−1. Error bars indicate se. D, Four-day-old etiolated seedlings were irradiated with 0.01 or 10 μmol m−2 s−1 R light pulse for 1 min and were subsequently incubated for 60 min in the dark before sample collection. PRR9 mRNA level was determined by qRT-PCR. Data normalized to TUBULIN2/3 levels are shown. Error bars indicate se. Analyzed genotypes are as in Figure 2.
The transcript level of PRR9 (for PSEUDORESPONSE REGULATOR9) is up-regulated by light, and this sensitive marker can be used to examine VLFR (Khanna et al., 2006). The results obtained show that PRR9 mRNA induction by very-low-intensity R light is impaired in phyA-5, whereas no significant reduction compared with the wild type can be observed after a strong R pulse (Fig. 3D).
These observations are supported by studies on transgenic seedlings expressing phyA-YFP and phyA-5-YFP (Fig. 3, B–D).
The phyA-5 Mutant Has Altered Protein Stability in Weak FR
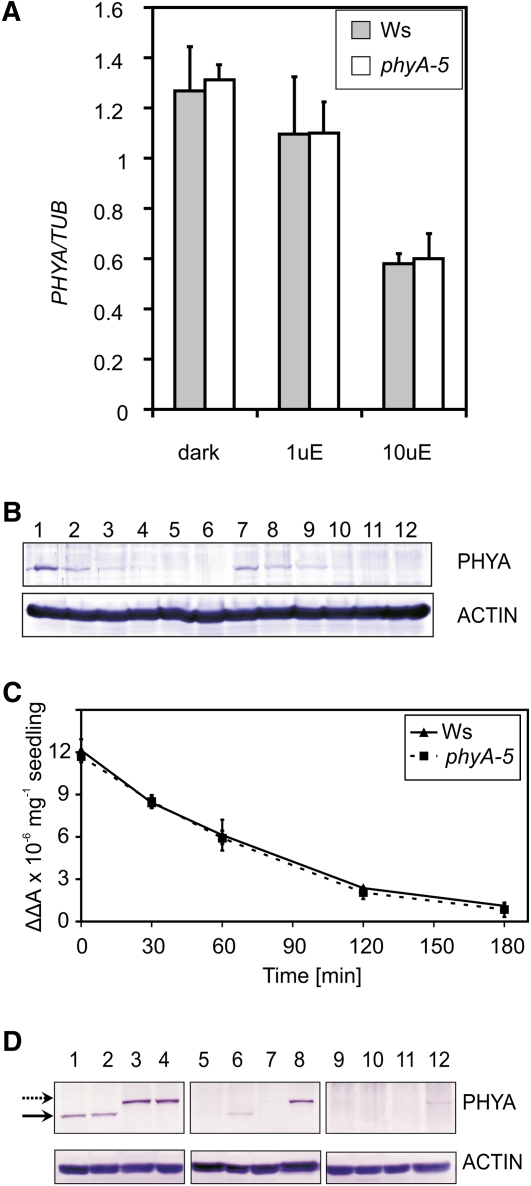
The observed hyposensitivity in FR light can be explained by changes in (1) the amount of PHYA transcript or (2) phyA protein level in the phyA-5 mutant. The activity of the PHYA promoter is down-regulated by light (Hennig et al., 1999). Under FR irradiation, light responses are only controlled by PHYA; thus, mutations in phyA can affect PHYA promoter control. This possibility, however, is not supported by quantitative real-time (qRT)-PCR analysis of the PHYA transcript (Fig. 4A). No detectable difference can be observed between PHYA and PHYA-5 transcript levels in FR-grown seedlings.
Figure 4.
PHYA mRNA and protein levels in phyA-5 and wild-type seedlings. A, Ws and phyA-5 seedlings were grown in darkness (dark), 1 μmol m−2 s−1 (1uE), or 10 μmol m−2 s−1 (10uE) FR light for 4 d. After performing RNA extraction, PHYA mRNA levels were determined by qRT-PCR. Data normalized to TUBULIN2/3 levels are shown. Error bars indicate se. B, Four-day-old Ws (lanes 1–6) or phyA-5 (lanes 7–12) etiolated seedlings irradiated with 25 μmol m−2 s−1 R light were subjected to total protein isolation and western-blot analysis using PHYA (top panel) or ACTIN-specific (bottom panel) antiserum. The lengths of the R light treatments were 0 h (lanes 1 and 7), 1 h (lanes 2 and 8), 2 h (lanes 3 and 9), 3 h (lanes 4 and 10), 4 h (lanes 5 and 11), and 6 h (lanes 6 and 12). C, Four-day-old etiolated seedlings were irradiated with continuous R light (25 μmol m−2 s−1). The amount of total phytochrome was measured by in vivo spectrophotometry. Error bars indicate se. D, Seedlings were grown for 4 d in darkness (lanes 1–4), 1 μmol m−2 s−1 (lanes 5–8), and 10 μmol m−2 s−1 (lanes 9–12) FR light and were subjected to total protein isolation and subsequent western-blot analysis using PHYA (top panels) or ACTIN-specific (bottom panels) antiserum. The examined genotypes are as follows: Ws (lanes 1, 5, and 9); phyA-5 (lanes 2, 6, and 10); PHYA:PHYA-YFP in phyA-201 (lanes 3, 7, and 11); and PHYA:PHYA-5-YFP in phyA-201 (lanes 4, 8, and 12). The solid arrow marks the bands corresponding to endogenous PHYA, whereas the dashed arrow marks the PHYA-YFP-specific bands. [See online article for color version of this figure.]
In order to examine the second possibility (i.e. altered phyA-5 protein stability), levels of phyA and phyA-5 in dark-grown seedlings were determined. It is known that the highest levels of phyA can be measured in etiolated seedlings, and the light-induced degradation of phyA is triggered by the Pr-Pfr transition of the photoreceptor (Hennig et al., 1999). Both western-blot analysis (Fig. 4B) and in vivo spectroscopy (Fig. 4C) show that the dark level of phyA-5 does not differ from that of wild-type phyA. These figures also demonstrate that no significant difference can be observed between the degradation of phyA and phyA-5 under continuous R irradiation. Additionally, we could not detect any difference between the steady-state levels of phyA and phyA-5 in seedlings grown under strong FR light. However, phyA-5 reaches easily observable levels, whereas the wild-type phyA photoreceptor remains below the detection limit in 4-d-old weak FR-grown seedlings (Fig. 4D). Figure 4D also shows that the transgenic PHYA-YFP fusion proteins (PHYA and PHYA-5) showed levels comparable to their endogenous counterparts under each type of irradiation. Careful examination of the western blots also revealed that the YFP tag only slightly increases the stability of the photoreceptor, if at all, resulting in higher steady-state levels as compared with the corresponding nontagged PHYAs.
The Nuclear Localization of phyA-5 Is Impaired in Low-Fluence FR Light
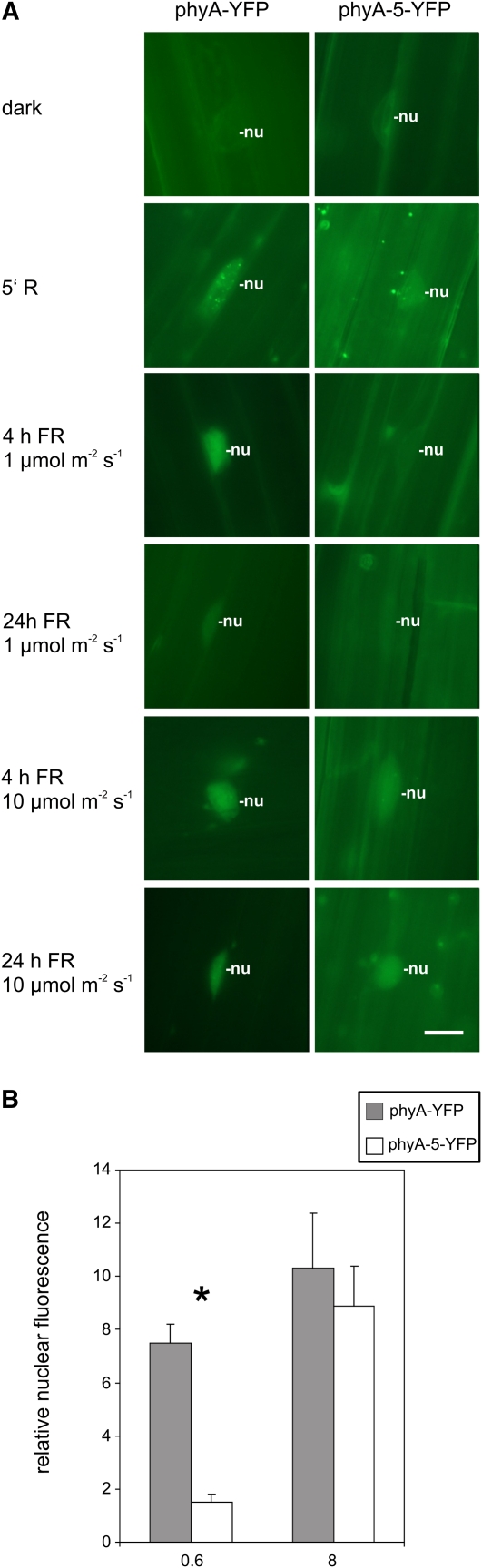
In order to study comparatively the nuclear localization of the mutant phyA-5 and wild-type phyA, we expressed these proteins as PHYA:YFP fusions under the control of the PHYA promoter in phyA-201 plants (PHYA:PHYA-5-YFP). Phenotypic analysis of these transgenic plants shows that both phyA-YFP and phyA-5-YFP are functional photoreceptors under HIR conditions (Fig. 2) but display distinct sensitivities to constant FR. Microscopic data revealed that the nuclear import of phyA-5-YFP but not that of phyA-YFP is decreased below the detection level in weak FR light irrespective of the duration of the irradiation. In contrast, strong FR light does not lead to different localization patterns of phyA-5 compared with wild-type phyA (Fig. 5A).
Figure 5.
Intracellular dynamics of phyA and phyA-5. A, Transgenic Arabidopsis seedlings expressing the phyA-YFP or phyA-5-YFP fusion protein under the control of the PHYA promoter were grown for 4 d in darkness. Subsequently, the plantlets were directly analyzed (dark) or irradiated for 5 min with R light or FR light of 1 or 10 μmol m−2 s−1 prior to epifluorescence microscopy. The various FR light treatments were given for 4 and 24 h. nu, Nucleus. Bar = 10 μm. B, Etiolated seedlings expressing PHYA:PHYA-YFP or PHYA:PHYA-5-YFP in phyA-201 were used to quantify the nuclear import of phyA-YFP and phyA-5-YFP. The fluorescent signal detected from nuclei irradiated with a 1-h FR light pulse (0.6 or 8 μmol m−2 s−1) was background corrected and normalized to the corresponding averaged dark control. Error bars indicate se. Dark mean values were as follows: phyA-YFP, 11.5 ± 2.11; phyA-5-YFP, 10.2 ± 1.75. Statistically significant differences between phyA-YFP and phyA-5-YFP signals were determined by Student’s two-tailed heteroscedastic t test. The asterisk indicates sample sets where P < 0.001.
The efficiency of the photoreceptor import induced by brief pulses of different intensities of FR light was also determined. Figure 5B and Supplemental Figure S3 show that a short, strong FR light pulse can clearly induce the nuclear accumulation of phyA and phyA-5 to the same extent. A weak pulse, however, was significantly less effective in promoting phyA-5 nuclear accumulation compared with the wild type.
The Binding of PHYA-5 to FHY1 and FHL Is Weaker Than That of the Wild Type
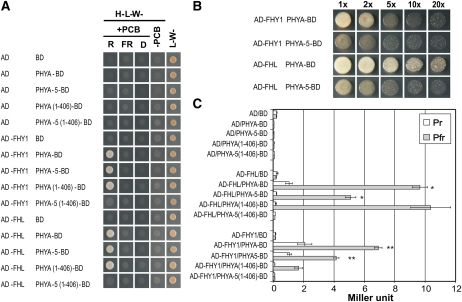
Yeast two-hybrid assays were used to study the binding of nuclear import facilitators FHY1 and FHL to phyA (Hiltbrunner et al., 2005, 2006). We employed the same experimental approach to compare the binding of phyA-5 and wild-type phyA to these proteins. To this end, the PHYA-5 coding sequence was fused to the GAL4 DNA-binding domain (BD), whereas the FHY1 and FHL coding sequences were fused to the GAL4 activation domain (AD). These fusion proteins were coexpressed in yeast cells growing on solid medium (Fig. 6A). Two different plates were used in this assay: the nonselective plates that allow the growth of yeast containing both AD and BD plasmids; and selective plates that only allow the growth of cells that express interacting proteins tagged with AD and BD. To show that the interaction of phyA and phyA-5 with FHY1/FHL is Pfr specific, the phycocyanobilin chromophore was added to the medium. This allows phyA to undergo Pr-Pfr transition after light treatments, rendering the experimental system suitable to examine the different interaction properties of Pr and Pfr forms of phyA. To reveal the possible differences between phyA Pfr and phyA-5 Pfr in the interaction with nuclear import facilitators, a dilution series was made from the overnight cultures before plating. Figure 6B clearly shows that the A30V mutation weakens phyA-5 binding to FHY1 and FHL compared with wild-type phyA. This difference is even more pronounced if only the N-terminal 1 to 406 amino acids of the phyAs are expressed. While phyA(1-406) shows normal growth as described by Hiltbrunner et al., (2006), phyA-5(1-406) shows no detectable interaction with FHL and FHY1. These results are further supported by the β-galactosidase activity assay, which allows the quantification of protein interactions. These data clearly confirm the difference between phyA and phyA-5 in the FHY1/FHL interaction, since the mutated photoreceptor shows no detectable binding to either FHY1 or FHL (Fig. 6C).
Figure 6.
The binding affinity of phyA-5 protein to the nuclear import machinery is impaired. A, Yeast strain AH-109 was cotransformed with the indicated plasmids. Five microliters of overnight cultures grown in liquid L-W- medium was dropped on nonselective (L-W-) or selective (H-L-W-; containing 1 mm 3-aminotriazole) synthetic dropout plates. The selective plates also contained 10 μm phycocyanobilin chromophore (PCB), except for plates marked −PCB. After dropping, the plates were incubated at 28°C for 2 d under 1 μmol m−2 s−1 R light or 10 μmol m−2 s−1 FR light or in darkness (D). B, Cotransformed overnight-grown yeast cultures as indicated in A were diluted to the same optical density (OD600 = 1 [1×]), and sets of dilutions (2×–20×) were made. Five microliters from each dilution were dropped on a H-L-W- plate supplied with 1 mm 3-aminotriazole and 10 μm PCB. The plate was incubated for 2 d at 28°C under 1 μmol m−2 s−1 R light. C, Yeast strain Y187 was cotransformed with the indicated plasmids. Liquid cultures at 0.5 mL were propagated in nonselective medium (L-W-) supplied with 20 μm PCB overnight. Before a further 4-h propagation in the dark, cultures were irradiated either with 30 μmol m−2 s−1 R light (Pfr) for 5 min or with the R pulse followed by 5 min of 20 μmol m−2 s−1 FR light (Pr). β-Galactosidase activity was measured using orthonitrophenyl-β-galactoside substrate. Triplicate assays were performed, and mean values are plotted. Error bars indicate se. Student’s two-tailed heteroscedastic t test was used to determine the statistical significance of differences between values indicated by one or two asterisks (each bar represents 20 replicates; P < 0.001). [See online article for color version of this figure.]
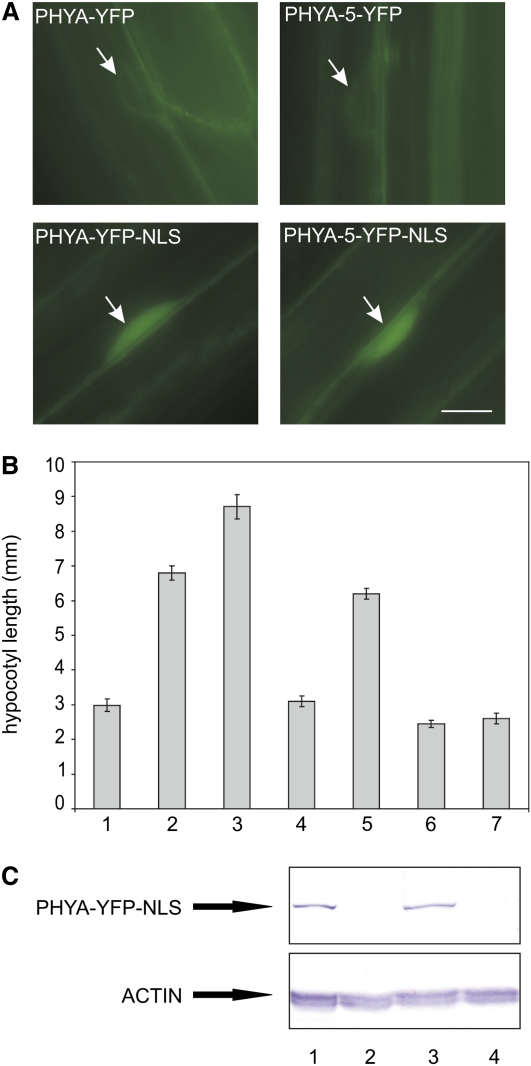
phyA-5-YFP-NLS and phyA-YFP-NLS Fusion Proteins Are Equally Effective to Complement the phyA-201 Mutant
The observed phenotype of the phyA-5 mutant can be explained by insufficient nuclear import and/or modified nuclear signaling. To answer this question, we generated transgenic seedlings expressing the phyA-5 and wild-type phyA proteins fused to YFP and the nuclear localization signal (NLS; Kalderon et al., 1984) under the control of the PHYA promoter (PHYA:PHYA-5-YFP-NLS and PHYA:PHYA-YFP-NLS, respectively). As expected, phyA-YFP-NLS and phyA-5-YFP-NLS proteins were constitutively localized in the nucleus (Fig. 7A). Analysis of FR light-induced hypocotyl growth inhibition showed that both phyA-5-YFP-NLS and phyA-YFP-NLS fusion proteins complemented the phyA-201 mutant in a similar fashion (Fig. 7B). These data clearly demonstrate that constitutively nuclear-localized phyA-5 signals properly and that FHY1/FHL is required only to facilitate the nuclear import of these photoreceptors. Additionally, we show that the kinetics of light-induced degradation of the phyA-5-YFP-NLS and phyA-YFP-NLS fusion proteins is identical (Figs. 4D and 7C). These findings indicate that the aberrant phyA levels detected in the mutant exposed to FR light are due to the compromised nuclear import of phyA-5 Pfr.
Figure 7.
Expression of PHYA-5-YFP-NLS or PHYA-YFP-NLS equally complements the phyA-201 mutant. A, Transgenic Arabidopsis seedlings expressing the indicated chimeric proteins under the control of the PHYA promoter in the phyA-201 background were grown for 4 d in darkness prior to epifluorescence microscopy. Arrows point to nuclei. Bar = 10 μm. B, Hypocotyl lengths of seedlings grown under 1 μmol m−2 s−1 constant FR light for 4 d were measured. Analyzed genotypes are as follows: bar 1, Ws; bar 2, phyA-5; bar 3, phyA-201; bar 4, PHYA:PHYA-YFP in phyA-201; bar 5, PHYA:PHYA-5-YFP in phyA-201; bar 6, PHYA:PHYA-YFP-NLS in phyA-201; bar 7, PHYA:PHYA-5-YFP-NLS in phyA-201. Error bars indicate se. C, Seedlings grown for 4 d in the dark (lanes 1 and 3) or under 1 μmol m−2 s−1 FR light (lanes 2 and 4) were subjected to total protein extraction and western-blot hybridization applying PHYA (top panel) or ACTIN-specific antiserum (bottom panel). The analyzed genotypes are PHYA:PHYA-YFP-NLS in phyA-201 (lanes 1 and 2) and PHYA:PHYA-5-YFP-NLS in phyA-201 (lanes 3 and 4).
DISCUSSION
This study describes the characterization of phyA-5, isolated in the laboratory of the late Garry Whitelam. phyA-5 is a loss-of-function PHYA allele whose complex phenotype is caused by a single amino acid exchange (A30V) at a highly conserved position in the NTE domain of the phyA photoreceptor. The sequence alignment presented in Figure 1 shows that the mutated Ala residue is conserved in phytochromes isolated from a wide variety of evolutionally different taxa and also among all type II Arabidopsis phytochromes. Our knowledge about the function of the NTE domain in phyA-controlled signaling is rather limited. So far, it has been reported that the integrity of this domain is necessary for full biological activity of oat (Avena sativa) PHYA in tobacco (Nicotiana tabacum; Cherry et al., 1992), whereas Casal et al. (2002) demonstrated that the expression of the oat phyA molecule lacking amino acid residues 6 to 12 (Δ6-12) interferes with VLFR or HIR in Arabidopsis and tobacco. More recently Trupkin et al. (2007) used the Arabidopsis homologous system and found that Δ6-12 phyA signaling is reduced under continuous FR light and not changed under FR pulses. In addition, they reported that truncated phyA displayed normal binding to FHY1 and FHL, yet the mutated photoreceptor is less stable under constant R or FR irradiation compared with wild-type phyA. Some other studies concentrated on the examination of Ser residues located in the NTE of monocot phyA (Stockhaus et al., 1992; Lapko et al., 1997, 1999). These studies showed that the presence of Ser residues in the NTE domain (6–12) is not essential for PHYA activity, but this domain negatively regulates VLFR and continuous FR-driven inhibition of hypocotyl elongation (HIR).
The Ser residues are not altered in the NTE domain of the phyA-5 mutant, and its phenotype is fairly complex. First, FR-grown phyA-5 seedlings show pronounced hyposensitivity in the inhibition of hypocotyl elongation and cotyledon opening (Fig. 2, A and B; Supplemental Fig. S2). Second, the action spectrum for hypocotyl elongation shows that the phyA-5 mutation results in a reduction in HIR, especially at higher FR wavelengths (Fig. 3A). This decrease of HIR of hypocotyl growth is so pronounced at 742 nm that phyA-5 shows no detectable response (Supplemental Fig. S2). Finally, phyA-5 shows impaired VLFR (Fig. 3, B–D). Thus, we can conclude that the conserved Ala residue at position 30 plays a role in mediating both VLFR and HIR, independently of the Ser residues located between 6 and 12 in the NTE of phyA.
To provide a mechanistic explanation for the observed hyposensitive phenotype, we performed a series of experiments. First, we examined whether the negative feedback loop resulting in the down-regulation of the phyA promoter in constant FR is intact in the phyA-5 mutant. Results of qRT-PCR analysis show that the PHYA transcript level is not altered in phyA-5 seedlings (Fig. 4A). This observation proves that the phyA-5 photoreceptor, like wild-type phyA, can down-regulate its own expression under FR irradiation. To test whether the accumulation/degradation of phyA-5 and phyA is different, we measured protein levels by western-blot analysis in seedlings grown under various light conditions. Our data show that the phyA-5 level in etiolated seedlings and the R light-induced degradation of the mutant photoreceptor were unaltered compared with phyA (Fig. 4, B and C). Surprisingly, we measured higher than wild-type phyA-5 levels in seedlings grown under constant weak FR fluences, although no difference was observed in seedlings grown under strong FR (Figs. 2, A and B, and 4D). This finding may indicate that the degradation machinery has limited access to the phyA-5 Pfr molecules under low FR, whereas high FR or saturating R light can maintain wild-type-like phyA-5 levels via more effective degradation.
It was shown earlier that phyA, like other phytochromes, translocates to the nucleus after Pfr formation (Kircher et al., 1999, 2002; Kim et al., 2000). It was also published that phyA degradation, although not limited to the nucleus (Dieterle et al., 2005), occurs faster after nuclear translocation (Debrieux and Fankhauser, 2010; Toledo-Ortiz et al., 2010). In order to examine the possibility that phyA-5 mislocalization can be the reason for the observed phenotype, we examined the intracellular localization of phyA-5-YFP, a protein perfectly reconstituting the phyA-5 phenotype (Fig. 2). Our data show that the nuclear import of phyA-5-YFP is seriously reduced under low FR light, whereas strong FR pulses initiate normal nuclear import (Fig. 5; Supplemental Fig. S3). Thus, we conclude that a malfunction of phyA-5 nuclear import could be responsible for the higher phyA-5 level under these conditions.
FHY1 and FHL proteins can bind directly to the Pfr form of phyA and specifically manage its import to the nucleus (Hiltbrunner et al., 2005, 2006). This binding is an essential step in phyA nuclear import, which triggers PHYA-dependent nuclear signaling (Genoud et al., 2008). Our data obtained by yeast two-hybrid assays show that the binding of phyA-5 Pfr to FHY1 and FHL proteins is detectable but produces significantly lower values than the binding of phyA (Fig. 6). This observation underlines the importance of the NTE domain of PHYA in the establishment of the interaction with the FHY1/FHL system, but we have to point out that the NTE domain alone is not sufficient for the interaction with the nuclear import machinery (Hiltbrunner et al., 2006).
The reduced affinity of phyA-5 to transport facilitators provides a reasonable explanation for the impaired nuclear import observed under weak FR irradiation. Under saturating R light irradiation, a high Pfr/Pr ratio is established (Mancinelli, 1994), which makes feasible sufficient nuclear import of phyA-5 for proper signaling despite its lower affinity to the FHY1/FHL system. High-intensity FR irradiation, although resulting in much lower Pfr/Pr ratios than R light, still produces enough Pfr to induce nuclear import and signaling, whereas low-fluence FR results in a lower Pfr/Pr ratio. Under these conditions, the available limited amount of phyA-5 Pfr is unable to signal due to its much reduced nuclear import caused by the impaired binding of the phyA-5 protein to the nuclear import machinery. The same situation occurs under weak FR or R pulses (VLFR), whereas strong light pulses reduce the difference between the effects of phyA and phyA-5.
It had been shown by Genoud et al. (2008) that the phyA-NLS fusion protein is constitutively nucleus localized in the fhy1/phyA mutant and restores responsiveness to FR in the absence of FHY1. To test whether the impaired binding of phyA-5 Pfr to FHY1 and FHL proteins affects only the nuclear localization of the photoreceptor and/or nuclear signaling, we produced transgenic lines in the phyA-201 background expressing the phyA-YFP-NLS and phyA-5-YFP-NLS fusion proteins. Analysis of transgenic seedlings indicates that nuclear-localized phyA-5 and phyA launched signaling cascades that are equally efficient (Fig. 7B). Expression of phyA-5-YFP-NLS and phyA-YFP-NLS resulted in full complementation of the phyA-201 mutant, and both fusion proteins displayed proper light-induced degradation (Fig. 7C). Our conclusions are validated by a very recent report by Rausenberger et al. (2011). Based on experimental findings and mathematical modeling, those authors concluded that the stability of the phyA Pfr-FHY/FHL complex is a critical factor in regulating phyA nuclear import and signaling. Our data support the conclusion by Rausenberger et al. (2011) and show that a subtle change that affects the stability of phyA Pfr-FHY/FHL can significantly modulate phyA signaling. On the other hand, our results also eliminate the apparent contradiction between high phyA-5 levels and a hyposensitive phenotype under low-Pfr conditions. We show that (1) under low Pfr, the nuclear import of phyA-5 Pfr is much reduced; (2) the suboptimal level of phyA-5 Pfr in the nucleus manifests as ineffective signaling; and (3) under these conditions, phyA-5 Pfr is stranded in the cytoplasm, where it degrades slower than its nuclear counterpart (Debrieux and Fankhauser, 2010). Thus, a subtle change in the nucleo/cytoplasmic distribution of phyA-5 Pfr manifests itself in higher phyA-5 protein levels under low Pfr. These observations underline the biological importance of phyA nuclear import in mediating phyA signaling and degradation of the photoreceptor.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Light Sources
The phyA-5 mutant of Arabidopsis (Arabidopsis thaliana Ws ecotype) was a generous gift of Prof. Garry Whitelam. The phyA-201 (Nagatani et al., 1993) mutant is in the Ler ecotype. White light illumination was provided by cool-white fluorescent tubes or monochromatic light-emitting diode light sources (R, λmax = 667 nm; FR, λmax = 730 nm). Seedling growth was performed as described previously by Bauer et al. (2004). Seedlings for hypocotyl length and cotyledon angle measurements were placed on 1% agar plates and scanned with a flatbed scanner (Canon). Hypocotyl length and cotyledon angle values were measured with ImageJ software, and calculations were performed in Microsoft Excel 2003. Each plotted data point represents at least 20 to 25 seedlings. Experiments were repeated three times, and a representative data set is shown here.
Cloning of Plasmid Constructs and Generation of Transgenic Plants
The full-length genomic DNA fragment, including the 1,555-bp promoter sequence of the Arabidopsis PHYA gene (At1g09570) from Ws and phyA-5, was amplified using PCR with Pfu Ultra polymerase (Stratagene) according to the manufacturer’s instructions using the following primers: PHYA-F (5′-AAACTCGAGGAGAAGAAGAAAGAGATAAC-3′) and PHYA-R (5′-CGGAACTCGCTGCAGCAAACAAGCCCGGG-3′). The PCR products were inserted into pBluescript II SK+ (pBSK) vector (Stratagene) after digestion with XhoI and SmaI restriction enzymes to obtain PHYA pBSK and PHYA-5 pBSK, respectively. pPCVB812 including the coding sequence of the YFP and nopaline synthase terminator (Bauer et al., 2004) was digested with SalI and SmaI restriction endonucleases. XhoI-SmaI fragments of PHYA pBSK or PHYA-5 pBSK were inserted into this vector, resulting in PHYA:PHYA-YFP pPCVB or PHYA:PHYA-5-YFP pPCVB, respectively.
Cloning of the SV40 NLS sequence is described in detail by Wolf et al. (2011). PHYA promoter, PHYA, and PHYA-5 coding sequences were cloned to the YFP-NLS pPCV vector using the restriction sites indicated above.
These binary constructs were transformed into the phyA-201 mutant applying the Agrobacterium tumefaciens-mediated floral dip transformation method (Clough and Bent, 1998). Several transformed T1 lines were tested, and lines containing a single-copy transgene were bred to homozygosity. Next, the abundance of PHYA-YFP fusion proteins was determined, and selected lines were chosen for further studies.
The following yeast two-hybrid plasmid constructs were already described previously: PHYA pD153 (Shimizu-Sato et al., 2002), PHYA(1-406) pD153, FHY1 pGADT7, and FHL pGADT7 (Hiltbrunner et al., 2006). PHYA-5 pD153 was created as follows: PHYA-5 pBSK was used as a template in a PCR performed using PHYA-F (5′-TTTGGATCCATATGTCAGGCTCTAGGCCGAC-3′) and PHYA406-R (5′-TTTCCCGGGGTGTTATCGAGTTCCACCTCC-3′) primers. The resulting product was digested with BamHI-HindIII and inserted into PHYA pD153 to BamHI-HindIII sites. The same PCR product was cloned as a BamHI-SmaI fragment into PHYA(1-406) pD153 to obtain PHYA-5(1-406) pD153. Every construct containing a PCR product was verified by automated sequencing.
Epifluorescence Microscopy and in Vivo Spectroscopy
Seedling growth, epifluorescence microscopy setup, and observation techniques were described previously (Bauer et al., 2004). Semiquantitative epifluorescence microscopy was performed as follows: 4-d-old etiolated seedlings were irradiated with 1-h FR light (718-nm DEPIL filter) pulses of different light intensities. Twelve-bit TIFF images, not containing any saturated pixels, were taken of the observed nuclei. In order to minimize the effect of the microscopic light, every image was taken within the first 30 s of the onset of excitation light. The same exposure time and excitation light intensity setting were applied throughout the whole experiment. The average intensity of pixels was calculated in the examined nuclei using ImageJ software. After subtraction of the signal from the vacuole background in each image, the mean value of data obtained from at least 15 independent nuclei was normalized to the corresponding dark control.
In vivo spectroscopy was performed according to Dieterle et al. (2005) using 25 μmol m−2 s−1 R light.
Analysis of Transcript Levels
Total RNA extraction, cDNA preparation, and qRT-PCR assays were carried out as described previously (Kevei et al., 2007). All graphs show mRNA levels relative to the TUBULIN2/3 mRNA transcript (Endo et al., 2007). Primers used were the following: for PHYA, PHYA-RT-F (5′-ATCTAGAGATCAGGTTAACGCA-3′) and PHYA-RT-R (5′-CCTTCTTCTGACACATCTTCC-3′); for TUBULIN2/3, TUB2/3-F (5′-CCAGCTTTGGTGATTTGAAC-3′) and TUB2/3-R (5′-CCAGCTTTCGGAGGTCAGAG-3′); and for PRR9, PRR9-F (5′-CCTTCTCAAGATTTGAGGAAAGC-3′) and PRR9-R (5′-TTTGGCTCACCTGAAGTACTCTC-3′). The assays were repeated three times, and representative data sets are shown.
Western-Blot Analysis
Protein extraction and immunoblot analysis were done as described previously (Bauer et al., 2004). The antiserum raised against PHYA was also described earlier (Hiltbrunner et al., 2006). The actin-specific antibody was obtained from Sigma.
Yeast Two-Hybrid Assays
Yeast two-hybrid assays were performed as described previously (Hiltbrunner et al., 2006).
Protein Sequence Alignment
Protein sequence alignment was made using Clone Manager 9 software.
Database accession numbers are as follows: Arabidopsis_PHYA, NM_100828; Arabidopsis_PHYB, NP_179469; Arabidopsis_PHYC, ABG21336; Arabidopsis_PHYD, AAW56595; Arabidopsis_PHYE, CAB53654; Nicotiana_PHYA, CAA47284; Cucurbita_PHYA, P06592; Glycine_PHYA, P42500; Pisum_PHYA, AAT97643; Populus_PHYA, O49934; Solanum_PHYA, P30733; Solanum_PHYB1, CAA05293; Oryza_PHYA, A2XLG5; Avena_PHYA, P06593; Sorghum_PHYA, AAB41397; Triticum_PHYA, CAC85512; Picea_PHYA, Q40762; Pinus_PHYA, CAA65510; Adiantum_PHY2, BAA33775; Selaginella_PHY1, Q01549; Ceratodon_PHY3, AAM94956; Physcomitrella_PHY5a, XP_001761145; Physcomitrella_PHY5b3, XP_001767224; Physcomitrella_PHY5c, XP_001754366; Marchantia_PHY, BAB39687.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Fluence rate dependency of cotyledon angle.
Supplemental Figure S2. Hypocotyl elongation inhibition at different wavelengths.
Supplemental Figure S3. Selection of representative images used for semiquantitative fluorescence microscopy shown in Figure 5B.
Acknowledgments
We are grateful to Erzsébet Fejes for critical reading of the manuscript.
References
- Bauer D, Viczián A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adám E, Fejes E, Schäfer E, et al. (2004) Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Davis SJ, Kirchenbauer D, Viczián A, Yanovsky MJ, Clough RC, Kircher S, Jordan-Beebe ET, Schäfer E, Nagy F, et al. (2002) The serine-rich N-terminal domain of oat phytochrome A helps regulate light responses and subnuclear localization of the photoreceptor. Plant Physiol 129: 1127–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C. (2004) Light signal transduction in higher plants. Annu Rev Genet 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Cherry JR, Hondred D, Walker JM, Keller JM, Hershey HP, Vierstra RD. (1993) Carboxy-terminal deletion analysis of oat phytochrome A reveals the presence of separate domains required for structure and biological activity. Plant Cell 5: 565–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JR, Hondred D, Walker JM, Vierstra RD. (1992) Phytochrome requires the 6-kDa N-terminal domain for full biological activity. Proc Natl Acad Sci USA 89: 5039–5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G, Yi H, Lee J, Kwon YK, Soh MS, Shin B, Luka Z, Hahn TR, Song PS. (1999) Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature 401: 610–613 [DOI] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA. (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol 25: 413–427 [DOI] [PubMed] [Google Scholar]
- Clough RC, Vierstra RD. (1997) Phytochrome degradation. Plant Cell Environ 20: 713–721 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Debrieux D, Fankhauser C. (2010) Light-induced degradation of phyA is promoted by transfer of the photoreceptor into the nucleus. Plant Mol Biol 73: 687–695 [DOI] [PubMed] [Google Scholar]
- Desnos T, Puente P, Whitelam GC, Harberd NP. (2001) FHY1: a phytochrome A-specific signal transducer. Genes Dev 15: 2980–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M, Bauer D, Büche C, Krenz M, Schäfer E, Kretsch T. (2005) A new type of mutation in phytochrome A causes enhanced light sensitivity and alters the degradation and subcellular partitioning of the photoreceptor. Plant J 41: 146–161 [DOI] [PubMed] [Google Scholar]
- Edgerton MD, Jones AM. (1992) Localization of protein-protein interactions between subunits of phytochrome. Plant Cell 4: 161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberg K, Hennig L, Schäfer E. (2000) Variation in dynamics of phytochrome A in Arabidopsis ecotypes and mutants. Plant Cell Environ 23: 311–319 [Google Scholar]
- Endo M, Mochizuki N, Suzuki T, Nagatani A. (2007) CRYPTOCHROME2 in vascular bundles regulates flowering in Arabidopsis. Plant Cell 19: 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chen M. (2008) Transposing phytochrome into the nucleus. Trends Plant Sci 13: 596–601 [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Yeh KC, Lagarias JC, Zhang H, Elich TD, Chory J. (1999) PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284: 1539–1541 [DOI] [PubMed] [Google Scholar]
- Genoud T, Schweizer F, Tscheuschler A, Debrieux D, Casal JJ, Schäfer E, Hiltbrunner A, Fankhauser C. (2008) FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet 4: e1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Büche C, Eichenberg K, Schäfer E. (1999) Dynamic properties of endogenous phytochrome A in Arabidopsis seedlings. Plant Physiol 121: 571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltbrunner A, Tscheuschler A, Viczián A, Kunkel T, Kircher S, Schäfer E. (2006) FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol 47: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A, Viczián A, Bury E, Tscheuschler A, Kircher S, Tóth R, Honsberger A, Nagy F, Fankhauser C, Schäfer E. (2005) Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr Biol 15: 2125–2130 [DOI] [PubMed] [Google Scholar]
- Jordan ET, Cherry JR, Walker JM, Vierstra RD. (1996) The amino-terminus of phytochrome A contains two distinct functional domains. Plant J 9: 243–257 [DOI] [PubMed] [Google Scholar]
- Jordan ET, Marita JM, Clough RC, Vierstra RD. (1997) Characterization of regions within the N-terminal 6-kilodalton domain of phytochrome A that modulate its biological activity. Plant Physiol 115: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. (1984) A short amino acid sequence able to specify nuclear location. Cell 39: 499–509 [DOI] [PubMed] [Google Scholar]
- Kevei E, Schäfer E, Nagy F. (2007) Light-regulated nucleo-cytoplasmic partitioning of phytochromes. J Exp Bot 58: 3113–3124 [DOI] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Toledo-Ortiz G, Kikis EA, Johannesson H, Hwang Y-S, Quail PH. (2006) Functional profiling reveals that only a small number of phytochrome-regulated early-response genes in Arabidopsis are necessary for optimal deetiolation. Plant Cell 18: 2157–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L, Kircher S, Toth R, Adam E, Schäfer E, Nagy F. (2000) Light-induced nuclear import of phytochrome-A:GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis. Plant J 22: 125–133 [DOI] [PubMed] [Google Scholar]
- Kircher S, Gil P, Kozma-Bognár L, Fejes E, Speth V, Husselstein-Muller T, Bauer D, Adám E, Schäfer E, Nagy F. (2002) Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schafer E, Nagy F. (1999) Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krall L, Reed JW. (2000) The histidine kinase-related domain participates in phytochrome B function but is dispensable. Proc Natl Acad Sci USA 97: 8169–8174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapko VN, Jiang XY, Smith DL, Song PS. (1997) Posttranslational modification of oat phytochrome A: phosphorylation of a specific serine in a multiple serine cluster. Biochemistry 36: 10595–10599 [DOI] [PubMed] [Google Scholar]
- Lapko VN, Jiang XY, Smith DL, Song PS. (1999) Mass spectrometric characterization of oat phytochrome A: isoforms and posttranslational modifications. Protein Sci 8: 1032–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancinelli AL. (1994) The physiology of phytochrome action. Kendrick RE, Kronenberg GHM, , Photomorphogenesis in Plants, Ed 2. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 51–59 [Google Scholar]
- Mateos JL, Luppi JP, Ogorodnikova OB, Sineshchekov VA, Yanovsky MJ, Braslavsky SE, Gärtner W, Casal JJ. (2006) Functional and biochemical analysis of the N-terminal domain of phytochrome A. J Biol Chem 281: 34421–34429 [DOI] [PubMed] [Google Scholar]
- Mathews S, Sharrock RA. (1997) Phytochrome gene diversity. Plant Cell Environ 20: 666–671 [Google Scholar]
- Matsushita T, Mochizuki N, Nagatani A. (2003) Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424: 571–574 [DOI] [PubMed] [Google Scholar]
- Montgomery BL, Lagarias JC. (2002) Phytochrome ancestry: sensors of bilins and light. Trends Plant Sci 7: 357–366 [DOI] [PubMed] [Google Scholar]
- Müller R, Fernández AP, Hiltbrunner A, Schäfer E, Kretsch T. (2009) The histidine kinase-related domain of Arabidopsis phytochrome A controls the spectral sensitivity and the subcellular distribution of the photoreceptor. Plant Physiol 150: 1297–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A. (2010) Phytochrome: structural basis for its functions. Curr Opin Plant Biol 13: 565–570 [DOI] [PubMed] [Google Scholar]
- Nagatani A, Reed JW, Chory J. (1993) Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol 102: 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J. (2000) Light: an indicator of time and place. Genes Dev 14: 257–271 [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Oka Y, Matsushita T, Mochizuki N, Quail PH, Nagatani A. (2008) Mutant screen distinguishes between residues necessary for light-signal perception and signal transfer by phytochrome B. PLoS Genet 4: e1000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palágyi A, Terecskei K, Adám E, Kevei E, Kircher S, Mérai Z, Schäfer E, Nagy F, Kozma-Bognár L. (2010) Functional analysis of amino-terminal domains of the photoreceptor phytochrome B. Plant Physiol 153: 1834–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH. (1997) An emerging molecular map of the phytochromes. Plant Cell Environ 20: 657–666 [Google Scholar]
- Quail PH, Briggs WR, Chory J, Hangarter RP, Harberd NP, Kendrick RE, Koornneef M, Parks B, Sharrock RA, Schafer E, et al. (1994) Spotlight on phytochrome nomenclature. Plant Cell 6: 468–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausenberger J, Tscheuschler A, Nordmeier W, Wüst F, Timmer J, Schäfer E, Fleck C, Hiltbrunner A. (2011) Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell 146: 813–825 [DOI] [PubMed] [Google Scholar]
- Rösler J, Jaedicke K, Zeidler M. (2010) Cytoplasmic phytochrome action. Plant Cell Physiol 51: 1248–1254 [DOI] [PubMed] [Google Scholar]
- Rösler J, Klein I, Zeidler M. (2007) Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A. Proc Natl Acad Sci USA 104: 10737–10742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Zhu D, Li J, Rubio V, Zhou Z, Shen Y, Hoecker U, Wang H, Deng XW. (2008) Arabidopsis COP1/SPA1 complex and FHY1/FHY3 associate with distinct phosphorylated forms of phytochrome A in balancing light signaling. Mol Cell 31: 607–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Nagatani A. (1996) Nuclear localization activity of phytochrome B. Plant J 10: 859–868 [DOI] [PubMed] [Google Scholar]
- Schäfer E, Bowler C. (2002) Phytochrome-mediated photoperception and signal transduction in higher plants. EMBO Rep 3: 1042–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Poetsch HA, Braun B, Marx S, Schaumburg A. (1991) Phytochromes and bacterial sensor proteins are related by structural and functional homologies: hypothesis on phytochrome-mediated signal-transduction. FEBS Lett 281: 245–249 [DOI] [PubMed] [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH. (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18: 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Clack T. (2002) Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol 130: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Quail PH. (1989) Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev 3: 1745–1757 [DOI] [PubMed] [Google Scholar]
- Shen Y, Zhou Z, Feng S, Li J, Tan-Wilson A, Qu LJ, Wang H, Deng XW. (2009) Phytochrome A mediates rapid red light-induced phosphorylation of Arabidopsis FAR-RED ELONGATED HYPOCOTYL1 in a low fluence response. Plant Cell 21: 494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. (2002) A light-switchable gene promoter system. Nat Biotechnol 20: 1041–1044 [DOI] [PubMed] [Google Scholar]
- Stockhaus J, Nagatani A, Halfter U, Kay S, Furuya M, Chua NH. (1992) Serine-to-alanine substitutions at the amino-terminal region of phytochrome A result in an increase in biological activity. Genes Dev 6: 2364–2372 [DOI] [PubMed] [Google Scholar]
- Sullivan JA, Deng XW. (2003) From seed to seed: the role of photoreceptors in Arabidopsis development. Dev Biol 260: 289–297 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Kiryu Y, Kobayashi J, Oka Y, Kim Y, Nam HG, Mochizuki N, Nagatani A. (2010) Subcellular sites of the signal transduction and degradation of phytochrome A. Plant Cell Physiol 51: 1648–1660 [DOI] [PubMed] [Google Scholar]
- Trupkin SA, Debrieux D, Hiltbrunner A, Fankhauser C, Casal JJ. (2007) The serine-rich N-terminal region of Arabidopsis phytochrome A is required for protein stability. Plant Mol Biol 63: 669–678 [DOI] [PubMed] [Google Scholar]
- Wagner D, Fairchild CD, Kuhn RM, Quail PH. (1996) Chromophore-bearing NH2-terminal domains of phytochromes A and B determine their photosensory specificity and differential light lability. Proc Natl Acad Sci USA 93: 4011–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf I, Kircher S, Fejes E, Kozma-Bognár L, Schäfer E, Nagy F, Adám E. (2011) Light-regulated nuclear import and degradation of Arabidopsis phytochrome-A N-terminal fragments. Plant Cell Physiol 52: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Parks BM, Short TW, Quail PH. (1995) Missense mutations define a restricted segment in the C-terminal domain of phytochrome A critical to its regulatory activity. Plant Cell 7: 1433–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SW, Jang IC, Henriques R, Chua NH. (2009) FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome A signals for inhibition of hypocotyl elongation. Plant Cell 21: 1341–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Luppi JP, Kirchbauer D, Ogorodnikova OB, Sineshchekov VA, Adam E, Kircher S, Staneloni RJ, Schäfer E, Nagy F, et al. (2002) Missense mutation in the PAS2 domain of phytochrome A impairs subnuclear localization and a subset of responses. Plant Cell 14: 1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh KC, Lagarias JC. (1998) Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA 95: 13976–13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler M, Bolle C, Chua NH. (2001) The phytochrome A specific signaling component PAT3 is a positive regulator of Arabidopsis photomorphogenesis. Plant Cell Physiol 42: 1193–1200 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Hare PD, Yang SW, Zeidler M, Huang LF, Chua NH. (2005) FHL is required for full phytochrome A signaling and shares overlapping functions with FHY1. Plant J 43: 356–370 [DOI] [PMC free article] [PubMed] [Google Scholar]