In eukaryotes, small noncoding RNAs of approximately 21 to 24 nucleotides function as guide molecules in many biological processes, including genome organization and stability, developmental timing and patterning, and antibacterial and antiviral defense (Carrington and Ambros, 2003; Poethig, 2009; Simon and Meyers, 2011). The small RNAs regulate the functions of target DNA or RNA in a sequence-specific manner at either the transcriptional or posttranscriptional level through an RNA-silencing mechanism (Hammond, 2005; Czech and Hannon, 2011). Based on whether RNase III family proteins participate in the biogenesis, the small RNAs are divided into at least two classes: RNase III family protein-dependent small RNAs, including microRNAs (miRNAs) and many small interfering RNAs (siRNAs); and RNase III family protein-independent small RNAs, including Piwi-RNAs and secondary siRNAs that are processed from single-stranded precursors in worms (Czech and Hannon, 2011). The mature miRNAs and siRNAs are sorted and loaded specifically with Argonaute (AGO) subfamily proteins, forming the RNA-induced silencing complexes (RISCs) that undergo a specific RNA-silencing mechanism (Ender and Meister, 2010; Fabian et al., 2010). Here, we briefly summarize the molecular basis of miRNA biogenesis pathways and provide an update on nuclear dicing bodies (D-bodies), structures involved in miRNA processing in plant cells. For an overview on siRNAs and other small RNAs, readers are referred to recent excellent articles (Li et al., 2006; Pontes et al., 2006; Ahmad et al., 2010; Chen, 2010a; Law and Jacobsen, 2010; Czech and Hannon, 2011; Simon and Meyers, 2011; Zhang and Zhu, 2011).
THE MIRNA BIOGENESIS PATHWAYS IN ANIMALS AND PLANTS
MiRNAs (approximately 21–22 nucleotides) are a class of small, regulatory RNAs that are found in almost all of the eukaryotes (Reinhart et al., 2000; Lau et al., 2001; Llave et al., 2002; Molnár et al., 2007; Zhao et al., 2007; Chen, 2010b). Like protein-coding genes, miRNA genes in both plants and animals are transcribed by RNA polymerase II into primary transcripts, known as pri-miRNAs. Animal miRNAs are often clustered on the same precursor. These pri-miRNAs are subject to 5′ capping, 3′ polyadenylation, and splicing, as some of the pri-miRNAs may contain introns. A pri-miRNA contains a stem-loop structure: an imperfect double-stranded (ds) RNA hairpin that harbors the mature miRNA (Bartel, 2004; Cai et al., 2004; Lee et al., 2004; Xie et al., 2005; Kim and Nam, 2006; Laubinger et al., 2008; Chen, 2010b). The pri-miRNAs are then processed by two substantial site-specific endonucleolytic events and eventually turned into miRNA duplexes. In animals, the initial step is converting the pri-miRNAs into precursor miRNAs (pre-miRNAs) in the nucleus through the microprocessor, a complex that contains the RNase III family protein Drosha and its partner Pasha/DiGeorge syndrome critical region gene 8, a dsRNA-binding (dsRBD) protein (Denli et al., 2004; Han et al., 2004a; Landthaler et al., 2004; Zeng et al., 2005). The specific sites of the pri-miRNA are recognized by the microprocessor and cleaved by Drosha to generate an approximately 60- to 70-nucleotide, folded pre-miRNA with a two-nucleotide overhang at the 3′ end (Han et al., 2006; Chen, 2010a). Mitrons, some intronic pri-miRNAs, are converted into pre-miRNAs by the RNA-splicing machinery rather than the microprocessor complex (Berezikov et al., 2007; Okamura et al., 2007; Ruby et al., 2007; Flynt et al., 2010). Recently, additional factors, including the nuclear export receptor Exportin1, the cap-binding complex, and ARSENITE-RESISITANCE PROTEIN2, were found to participate in this process (Laubinger et al., 2008; Gruber et al., 2009; Sabin et al., 2009; Buessing et al., 2010). The pre-miRNAs are then recognized by the nuclear export protein Exportin5 and transported to the cytoplasm in a Ran-GTP-dependent manner (Yi et al., 2003; Bohnsack et al., 2004; Lund et al., 2004). The secondary step is to generate the approximately 22- to 23-nucleotide miRNA/miRNA* duplex from the pre-miRNA through a cytoplasmic RNase III-like enzyme and its specific dsRBD partner (Bernstein et al., 2001; Hutvágner et al., 2001). In mammals, the RNase III enzyme Dicer functions in association with the cytoplasmic dsRBD protein TAR RNA-binding protein 2, whereas in Drosophila melanogaster, Dicer1 with its cofactor Loquacious plays a role in this transition (Chendrimada et al., 2005; Jiang et al., 2005; Saito et al., 2005; Park et al., 2007). The mature RNAs are sorted and loaded with specific AGO proteins to assemble RISCs. Sorting may depend on the interaction between components of biogenesis and effectors, 5′ end nucleotide and thermodynamics of the small RNA duplexes, and/or the structure biases of the AGO family (Khvorova et al., 2003; Schwarz et al., 2003; Gregory et al., 2005; Miyoshi et al., 2005; Rand et al., 2005; Mi et al., 2008; Okamura et al., 2009; Frank et al., 2010).
In contrast to the nuclear cropping and cytoplasmic dicing of pri-miRNAs in animals, both of the steps in pri-miRNA processing occur in the nucleus of plant cells. In addition, a single plant RNase III family protein, DICER-LIKE1 (DCL1), plays similar roles to both nuclear Drosha and cytoplasmic Dicer in animals (Park et al., 2002; Reinhart et al., 2002; Schauer et al., 2002; Kurihara and Watanabe, 2004). Plant miRNA genes are also transcribed by RNA polymerase II, and the primary transcripts are 5′ capped and 3′ polyadenylated (Xie et al., 2005; Kim and Nam, 2006; Chen, 2010a). The pri-miRNAs are cropped into pre-miRNAs with a shorter stem-loop structure, which are further cut into the miRNA/miRNA* duplex. This process requires the interaction of DCL1 with its dsRBD protein partner HYPONSTIC LEAVES1 (HYL1), similar to Drosha and Dicer in animals, which are assisted by a specific dsRBD protein partner (Han et al., 2004a, 2004b; Vazquez et al., 2004; Kurihara et al., 2006; Yang et al., 2010). The C2H2 zinc finger protein SERRATE (SE) also interacts with DCL1 and HYL1 and participates in the transition process (Yang et al., 2006a; Laubinger et al., 2008; Montgomery and Carrington, 2008). HYL1 together with SE promotes the accuracy of miRNA processing (Dong et al., 2008). Many miRNAs are reduced in abundance, while their corresponding pri-miRNAs accumulate in the dcl1, hyl1, or se mutant (Han et al., 2004b; Kurihara and Watanabe, 2004; Vazquez et al., 2004; Yang et al., 2006a). Other proteins such as DAWDLE (an RNA-binding protein) and the nuclear cap-binding complex also act in this process, probably by facilitating DCL1 to access or recognize pri-miRNAs or the loading of miRNA processing factors onto pri-miRNAs (Laubinger et al., 2008; Yu et al., 2008). Recently, plant Mediator was found to participate in miRNA biogenesis by recruiting RNA polymerase II to MIR gene promoters and then promoting their transcription (Kim et al., 2011). After the miRNA/miRNA* duplex is released from the precursor, each strand of the duplex is methylated by the small RNA methyltransferase HUA ENHANCER1 (HEN1) in the 2′-OH of the 3′ terminal nucleotide (Li et al., 2005; Yu et al., 2005; Yang et al., 2006b). The methylation is a protection from further polyuridylation and from the subsequent degradation by the exonucleases of the Small RNA-Degrading Nuclease family (Li et al., 2005; Yu et al., 2005; Ramachandran and Chen, 2008).
The 3′ methylated miRNA/miRNA* duplex may be transported by a plant Exportin5 homolog HASTY (HST) or through HST-independent mechanisms to the cytoplasm (Park et al., 2005; Eamens et al., 2009), where RISC can be assembled as in animals. However, the exact form of the exported miRNA and the subcellular localization of plant RISC loading and maturation are still not clear (Voinnet, 2009). Plant RISC may also be assembled in the nucleus. In this scenario, only mature AGO1 with a single-stranded miRNA can be exported to the cytoplasm (Eamens et al., 2009). Plant AGO1 is the major part of RISC and has an endonucleolytic cleavage activity that cleaves complementary mRNAs in the center of the miRNA-mRNA paired region (Vaucheret et al., 2004; Baumberger and Baulcombe, 2005; Qi et al., 2005).
PLANT MIRNA PROCESSING PROTEINS CONCENTRATE IN DISCRETE D-BODIES
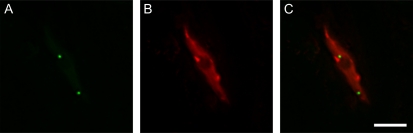
Recent progress in live-cell imaging proposed that nuclear chromatin is packaged into a higher order three-dimensional structure that may correlate to the regulation of the genes (Hübner and Spector, 2010; Misteli, 2010). In addition, the interchromatin region in the cell nucleus is highly heterogeneous and contains various nuclear domains or bodies, for example, nuclear speckle, paraspeckle, nucleolus, perinucleolar compartment, Cajal body (CB), cleavage body, gemini of coiled bodies, OPT (for Oct1/PTF/transcription) domain, SAM68 (for Src associated in mitosis of 68 kD) nuclear body, polymorphic interphase karyosomal association, polycomb body, promyelocytic leukemia body (Mao et al., 2011), and plant-specific nuclear bodies, such as cyclophilin, phytochrome, or abscisic acid-activated protein kinase-containing nuclear bodies (Shaw and Brown, 2004; Chen et al., 2010). These bodies are present in the nucleus at steady state and dynamically respond to basic cellular processes as well as to diverse metabolic conditions, alterations in cellular signaling, and various forms of stress (Dundr and Misteli, 2010; Mao et al., 2011). Live-cell imaging of plant miRNA processing proteins DCL1 and HYL1, which were fused to fluorescent proteins and expressed in transgenic Arabidopsis (Arabidopsis thaliana) plants under the control of their endogenous promoters, revealed that DCL1 was enriched in round nuclear bodies measuring 0.2 to 0.8 μm in diameter as well as being diffusely distributed throughout the nucleoplasm, predominantly excluded from nucleoli (Fig. 1; Fang and Spector, 2007). The number of nuclear bodies present in each nucleus ranged from zero to four, with the majority of the nuclei having one nuclear body. A population of DCL1 bodies (approximately 60%) localize in close proximity to nucleoli in projection images, but three-dimensional deconvolution analysis revealed that they are not within nucleoli. The DCL1 partner protein HYL1 displays a similar localization pattern of nuclear bodies to DCL1 bodies. Colocalization analysis revealed that DCL1 bodies and HYL1 bodies are the same structures as they colocalize (Fang and Spector, 2007; Song et al., 2007). Unlike DCL1 and HYL1, SE was distributed in nuclear speckles or interchromatin granule clusters containing the Ser/Arg (SR) splicing factor SR33 (Fang et al., 2004; Fang and Spector, 2007). In a small population of cells, the SE signal was also present in HYL1 bodies in addition to its nucleoplasmic distribution. The dual localization patterns of SE both in nuclear speckles and DCL1/HYL1 nuclear bodies may correlate with its dual roles in both splicing and miRNA processing (Fang and Spector, 2007; Laubinger et al., 2008).
Figure 1.
Arabidopsis nuclear D-bodies. A, HYL1-YFP signals in the nucleus of a leaf epidermal cell. B, 4′,6-Diamidino-2-phenylindole staining of the nucleus in A. C, Overlay of A and B. Two D-bodies are observed in the image, one of them close to the nucleolus as shown in C. Bar = 10 μm.
The DCL1/HYL1-containing bodies are different from most known nuclear bodies, due to their round shape, size, and average number per nucleus (Shaw and Brown, 2004), but are similar to CBs, as they are round in shape and their distribution is frequently perinucleolar (Nizami et al., 2010). CBs contain components involved in the processing/assembly of small nuclear RNAs, small nucleolar RNAs, and possibly siRNAs (Li et al., 2006; Pontes et al., 2006; Nizami et al., 2010). However, colocalization analysis indicated that DCL1/HYL1-containing bodies are different from CBs (Fang and Spector, 2007; Song et al., 2007), since they show no overlay with the AtCoilin signal, a signature marker of CBs. DCL1/HYL1-containing bodies are called dicing bodies or D-bodies (Fang and Spector, 2007; Fig. 1). In living cells of Arabidopsis plants, D-bodies move in the nuclei in a constrained manner.
HOW ARE D-BODIES FORMED?
Nuclear bodies are membraneless subnuclear organelles. A specific nuclear body is formed in a stochastic or ordered assembly manner (Dundr and Misteli, 2010). In addition, a seeding mechanism has been proposed to assemble, maintain, and regulate particular nuclear bodies (Mao et al., 2011). DCL1 contains two C-terminal dsRBDs. The mutant DCL1-6, with truncation of its two dsRBDs, is embryo lethal, while DCL1-9, a mutant with truncation of only the second dsRBD, results in infertility and severe defects in the biogenesis of most miRNAs, suggesting an important role of these dsRBDs in miRNA processing (Schauer et al., 2002). Live-cell imaging revealed that DCL1-9 failed to localize to D-bodies but instead distributed diffusely in the nucleoplasm, demonstrating that the dsRBD of DCL1 is critical for its localization to D-bodies. HYL1 contains two N-terminal dsRBDs, and these two dsRBDs are sufficient for pre-miRNA processing and localization to D-bodies (Wu et al., 2007). These results suggested that the dsRBDs in these miRNA processing proteins are essential for their targeting to D-bodies, possibly forming the seed for the assembly of D-bodies.
WHAT ARE THE FUNCTIONS OF D-BODIES?
In vivo tracking of a pri-miRNA using 24 tandem MS2 translational operators (MS2 repeats) and the MS2 coat protein-yellow fluorescent protein (MS2-YFP) system demonstrated that an introduced pri-RNA concentrates in DCL1-containing D-bodies in addition to being present in a diffuse distribution in the nucleoplasm, indicating that the pri-miRNAs can be recruited to D-bodies, where the machinery for their processing is enriched (Fang and Spector, 2007).
The precise and efficient pri-miRNA processing requires protein-protein interactions between the miRNA processing proteins (Kurihara et al., 2006). Using bimolecular fluorescence complementation, Fang and Spector (2007) found that DCL1, HYL1, and SE interact in the nuclear D-bodies in vivo, while the bimolecular fluorescence complementation signal in the surrounding nucleoplasm is very weak. In addition, DCL1 and HYL1 self-interact in the D-bodies. By contrast, DCL1-9 showed no interaction with SE, HYL1, or DCL1. Together, these results suggested a role of D-bodies in the dicing reaction of pri-miRNAs mediated by DCL1 and its interacting partner HYL1 (Fig. 2).
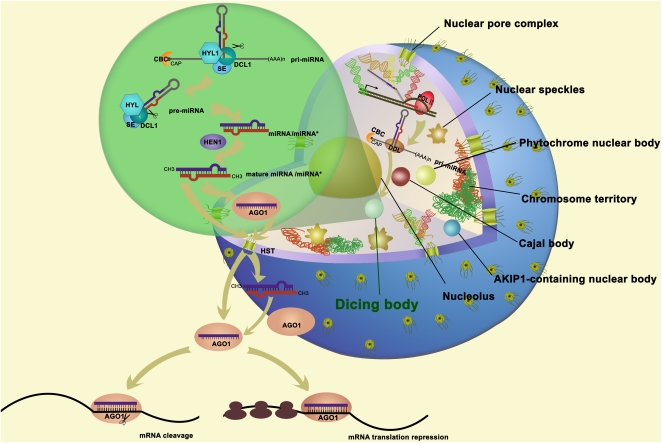
Figure 2.
Plant nuclear D-bodies, which contain proteins for miRNA processing and are involved in miRNA biogenesis. In eukaryotic cells, the nucleus is encapsulated in two layers of membranes in which nuclear pore complexes are embedded for transport between the nucleus and the cytoplasm. Chromosomes in the nucleus are organized into chromosome territories. The interchromatin region of the cell nucleus is highly heterogeneous, containing various nuclear domains or bodies. In a plant cell, these nuclear bodies include nucleolus, CB, nuclear speckles, phytochrome nuclear body, AAPK-Interacting Protein1 (AKIP1)-containing nuclear body, and D-body. These bodies have different sizes, shapes, components, dynamics, and functions. D-bodies play a role in the biogenesis of miRNAs. Plant miRNA genes are transcribed by RNA polymerase II to generate pri-miRNAs. The RNA-binding protein DAWDLE presumably stabilizes pri-miRNAs and facilitates DCL1 to access or recognize pri-miRNAs. The nuclear cap-binding complex (CBC) likely facilitates the loading of miRNA processing factors onto pri-miRNAs. The pri-miRNAs are then recruited to D-bodies, which contain DCL1, the dsRBD protein HYL1, and the C2H2 zinc finger protein se. These pri-miRNAs are then processed into a shorter stem-loop structure called pre-miRNAs and then further into the miRNA/miRNA* duplex. The miRNA/miRNA* duplex is methylated by the small RNA methyltransferase HEN1 in the 2′-OH of the 3′ terminal nucleotide. The mature miRNA/miRNA* may be transported in an HST-dependent or -independent manner through the nuclear pore complex, or the guide strand of mature miRNA/miRNA* is probably selectively loaded into AGO1-RISC in the nucleus and the miRISC is transported into the cytoplasm. The miRISC carries out the silencing reactions through translation repression or mRNA cleavage in the cytoplasm.
PERSPECTIVES
Apart from DCL1, HYL1, and SE, which localize predominantly or transiently to D-bodies, the miRNA/miRNA* duplex, the methyltransferase HEN1, and the slicer AGO1 also exhibited some localization to D-bodies in addition to their nucleoplasmic and cytoplasmic distribution patterns when examined by colocalization analysis with HYL1 (Fang and Spector, 2007). Therefore, it is of interest to investigate if HEN1 and AGO1 are recruited to D-bodies to methylate the miRNA/miRNA* duplex and load mature miRNAs to AGO1 to assemble the RISC complex in D-bodies. In this case, only mature AGO1 and miRNA containing RISC can be exported to the cytoplasm through nuclear pore complexes (Eamens et al., 2009; Fig. 2). In addition, HYL1 was observed to colocalize with its homolog DRB4 in D-bodies (Y. Fang and D.L. Spector, unpublished data). It was known that DRB4 interacts with DCL4 in vivo and is involved in the biogenesis of siRNAs (Fukudome et al., 2011). Therefore, more extensive studies are needed to learn about the potential roles of D-bodies in orchestrating the processing, sorting, RISC assembly, and functioning of small RNAs.
Acknowledgments
We thank Dr. David L. Spector for critical reading of the manuscript and members of Y.F.’s laboratory for insightful discussions. We apologize to all colleagues whose relevant work could not be cited due to space limitations.
References
- Ahmad A, Zhang Y, Cao XF. (2010) Decoding the epigenetic language of plant development. Mol Plant 3: 719–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC. (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung W-J, Willis J, Cuppen E, Lai EC. (2007) Mammalian mirtron genes. Mol Cell 28: 328–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D. (2004) Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10: 185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buessing I, Yang J-S, Lai EC, Grosshans H. (2010) The nuclear export receptor XPO-1 supports primary miRNA processing in C. elegans and Drosophila. EMBO J 29: 1830–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. (2004) Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10: 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. (2003) Role of microRNAs in plant and animal development. Science 301: 336–338 [DOI] [PubMed] [Google Scholar]
- Chen M, Galvão RM, Li M, Burger B, Bugea J, Bolado J, Chory J. (2010) Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141: 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2010a) Small RNAs: secrets and surprises of the genome. Plant J 61: 941–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2010b) Plant microRNAs at a glance. Semin Cell Dev Biol 21: 781. [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. (2005) TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. (2011) Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet 12: 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BBJ, Plasterk RHA, Ketting RF, Hannon GJ. (2004) Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235 [DOI] [PubMed] [Google Scholar]
- Dong Z, Han M-H, Fedoroff N. (2008) The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci USA 105: 9970–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Misteli T. (2010) Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol 2: a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamens AL, Smith NA, Curtin SJ, Wang M-B, Waterhouse PM. (2009) The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 15: 2219–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Meister G. (2010) Argonaute proteins at a glance. J Cell Sci 123: 1819–1823 [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. (2010) Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79: 351-379 [DOI] [PubMed] [Google Scholar]
- Fang Y, Hearn S, Spector DL. (2004) Tissue-specific expression and dynamic organization of SR splicing factors in Arabidopsis. Mol Biol Cell 15: 2664–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Spector DL. (2007) Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol 17: 818–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Greimann JC, Chung W-J, Lima CD, Lai EC. (2010) MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol Cell 38: 900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, Nagar B. (2010) Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465: 818–822 [DOI] [PubMed] [Google Scholar]
- Fukudome A, Kanaya A, Egami M, Nakazawa Y, Hiraguri A, Moriyama H, Fukuhara T. (2011) Specific requirement of DRB4, a dsRNA-binding protein, for the in vitro dsRNA-cleaving activity of Arabidopsis Dicer-like 4. RNA 17: 750–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. (2005) Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123: 631–640 [DOI] [PubMed] [Google Scholar]
- Gruber JJ, Zatechka DS, Sabin LR, Yong J, Lum JJ, Kong M, Zong W-X, Zhang Z, Lau C-K, Rawlings J, et al. (2009) Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell 138: 328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM. (2005) Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett 579: 5822–5829 [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom K-H, Kim Y-K, Jin H, Kim VN. (2004a) The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom K-H, Nam J-W, Heo I, Rhee J-K, Sohn SY, Cho Y, Zhang B-T, Kim VN. (2006) Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125: 887–901 [DOI] [PubMed] [Google Scholar]
- Han M-H, Goud S, Song L, Fedoroff N. (2004b) The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA 101: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner MR, Spector DL. (2010) Chromatin dynamics. Annu Rev Biophys 39: 471–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. (2005) Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev 19: 1674–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Kim VN, Nam J-W. (2006) Genomics of microRNA. Trends Genet 22: 165–173 [DOI] [PubMed] [Google Scholar]
- Kim YJ, Zheng B, Yu Y, Won SY, Mo B, Chen X. (2011) The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J 30: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Takashi Y, Watanabe Y. (2006) The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12: 206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y. (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. (2004) The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14: 2162–2167 [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Rätsch G, Weigel D. (2008) Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 8795–8800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom K-H, Lee S, Baek SH, Kim VN. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. (2006) An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126: 93–106 [DOI] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X. (2005) Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol 15: 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC. (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. (2004) Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- Mao YS, Zhang B, Spector DL. (2011) Biogenesis and function of nuclear bodies. Trends Genet 27: 295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. (2008) Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. (2010) Higher-order genome organization in human disease. Cold Spring Harb Perspect Biol 2: a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. (2005) Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev 19: 2837–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. (2007) miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature 447: 1126–1129 [DOI] [PubMed] [Google Scholar]
- Montgomery TA, Carrington JC. (2008) Splicing and dicing with a SERRATEd edge. Proc Natl Acad Sci USA 105: 8489–8490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizami ZF, Deryusheva S, Gall JG. (2010) Cajal bodies and histone locus bodies in Drosophila and Xenopus. Cold Spring Harb Symp Quant Biol 75: 313–320 [DOI] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. (2007) The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Liu N, Lai EC. (2009) Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell 36: 431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JK, Liu X, Strauss TJ, McKearin DM, Liu Q. (2007) The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr Biol 17: 533–538 [DOI] [PubMed] [Google Scholar]
- Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. (2005) Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci USA 102: 3691–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig RS. (2009) Small RNAs and developmental timing in plants. Curr Opin Genet Dev 19: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Li CF, Costa Nunes P, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. (2006) The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 126: 79–92 [DOI] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ. (2005) Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19: 421–428 [DOI] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. (2008) Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321: 1490–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X. (2005) Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123: 621–629 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. (2002) MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. (2007) Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, Lau C-K, Thompson CB, Cherry S. (2009) Ars2 regulates both miRNA- and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila. Cell 138: 340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Ishizuka A, Siomi H, Siomi MC. (2005) Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol 3: e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A. (2002) DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci 7: 487–491 [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Brown JW. (2004) Plant nuclear bodies. Curr Opin Plant Biol 7: 614–620 [DOI] [PubMed] [Google Scholar]
- Simon SA, Meyers BC. (2011) Small RNA-mediated epigenetic modifications in plants. Curr Opin Plant Biol 14: 148–155 [DOI] [PubMed] [Google Scholar]
- Song L, Han MH, Lesicka J, Fedoroff N. (2007) Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci USA 104: 5437–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crété P, Bartel DP. (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crété P, Vaucheret H. (2004) The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14: 346–351 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wu F, Yu L, Cao W, Mao Y, Liu Z, He Y. (2007) The N-terminal double-stranded RNA binding domains of Arabidopsis HYPONASTIC LEAVES1 are sufficient for pre-microRNA processing. Plant Cell 19: 914–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. (2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138: 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu Z, Lu F, Dong A, Huang H. (2006a) SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J 47: 841–850 [DOI] [PubMed] [Google Scholar]
- Yang SW, Chen H-Y, Yang J, Machida S, Chua N-H, Yuan YA. (2010) Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure 18: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ebright YW, Yu B, Chen X. (2006b) HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res 34: 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Bi L, Zheng B, Ji L, Chevalier D, Agarwal M, Ramachandran V, Li W, Lagrange T, Walker JC, et al. (2008) The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci USA 105: 10073–10078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307: 932–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. (2005) Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J 24: 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhu JK. (2011) RNA-directed DNA methylation. Curr Opin Plant Biol 14: 142–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y. (2007) A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev 21: 1190–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]