Abstract
Abscission in plants is a crucial process used to shed organs such as leaves, flowers, and fruits when they are senescent, damaged, or mature. Abscission occurs at predetermined positions called abscission zones (AZs). Although the regulation of fruit abscission is essential for agriculture, the developmental mechanisms remain unclear. Here, we describe a novel transcription factor regulating the development of tomato (Solanum lycopersicum) pedicel AZs. We found that the development of tomato pedicel AZs requires the gene MACROCALYX (MC), which was previously identified as a sepal size regulator and encodes a MADS-box transcription factor. MC has significant sequence similarity to Arabidopsis (Arabidopsis thaliana) FRUITFULL, which is involved in the regulation of fruit dehiscent zone development. The MC protein interacted physically with another MADS-box protein, JOINTLESS, which is known as a regulator of fruit abscission; the resulting heterodimer acquired a specific DNA-binding activity. Transcriptome analyses of pedicels at the preabscission stage revealed that the expression of the genes involved in phytohormone-related functions, cell wall modifications, fatty acid metabolism, and transcription factors is regulated by MC and JOINTLESS. The regulated genes include homologs of Arabidopsis WUSCHEL, REGULATOR OF AXILLARY MERISTEMS, CUP-SHAPED COTYLEDON, and LATERAL SUPPRESSOR. These Arabidopsis genes encode well-characterized transcription factors regulating meristem maintenance, axillary meristem development, and boundary formation in plant tissues. The tomato homologs were specifically expressed in AZs but not in other pedicel tissues, suggesting that these transcription factors may play key roles in pedicel AZ development.
Abscission is a cell separation process by which plants shed senescing leaves, flower organs, mature fruits, or seeds. This separation occurs within a specific tissue, the abscission zone (AZ), which has several layers of small, densely cytoplasmic cells at the junction of the organ and the main body of the plant (Patterson, 2001). Control of abscission in fruit and grain crops is a key agricultural concern. For example, during cereal crop domestication, mutants that reduce seed shattering have been preferentially selected for, because shattering is a major limiting factor for yield (Li et al., 2006). Many fruit tree species shed some of their young fruits because the trees usually bear too many fruit to support; too much or too little abscission of young fruit can affect the yield and quality of fruit crops (Bangerth, 2000). In tomato (Solanum lycopersicum), a mutant phenotype called jointless, which is characterized by defective AZs in fruit pedicels, is an important trait for tomatoes that are processed into commercial tomato products. This phenotype facilitates large-scale harvesting of the tomatoes by saving time removing the calices, because when the jointless fruit is harvested, the calyx remains attached to the plant, not to the fruit (Zahara and Scheuerman, 1988).
Abscission can be divided into four major steps (Patterson, 2001): (1) development of the AZ tissue; (2) acquisition of competence to respond to abscission signals; (3) activation of abscission; and (4) differentiation of a protective layer on the main body side of the AZ. Regulation of steps 2 and 3 has been extensively investigated by transcriptome analyses of shedding organs (Roberts et al., 2002; Agustí et al., 2008; Cai and Lashbrook, 2008; Meir et al., 2010; Botton et al., 2011). The genes involved in these steps are highly similar among different plant species and in different plant organs. The genes commonly found in these steps, for instance, are involved in ethylene and auxin biosynthesis and signal transduction, cell wall modification, and various stress responses. For step 1, in contrast, no commonalities have yet been found between different plant species and between different organs, even though a number of the genes regulating the development of AZs and other cell separation tissues have been identified. In tomato, the mutants jointless (j), jointless2 (j2), and lateral suppressor (ls) suppress the development of pedicel AZs (Roberts et al., 2002). JOINTLESS encodes a Short Vegetative Phase group MADS-box transcription factor (Mao et al., 2000), and Ls encodes a GRAS family transcription factor (Schumacher et al., 1999). Research is ongoing to identify the j2 locus (Yang et al., 2005). In rice (Oryza sativa), genes involved in the development of abscission layers for seed shattering include three loci, sh4, qSH1, and OsCPL1, that encode a transcription factor with a Myb3 DNA-binding domain, a BEL1-type homeobox transcription factor, and a protein with a C-terminal phosphatase domain, respectively (Konishi et al., 2006; Li et al., 2006; Ji et al., 2010). In Arabidopsis (Arabidopsis thaliana), SEEDSTICK (STK), which encodes an AGAMOUS (AG) group MADS-box transcription factor, is involved in the regulation of seed AZ development (Pinyopich et al., 2003). The regulation of Arabidopsis pod dehiscence, which is another typical cell separation process, has been extensively investigated, and a number of key genes involved in fruit dehiscence zone (DZ) development have been identified. Two AG group MADS-box genes, SHATTERPROOF1 (SHP1) and SHP2, and two basic helix-loop-helix-type transcription factor genes, INDEHISCENT (IND) and ALCATRAZ (ALC), are required for the development of DZs, and another MADS-box gene, FRUITFULL (FUL), which is a homolog of APETALA1 (AP1), and the homeodomain transcription factor REPLUMLESS are also involved in DZ development via restricting the expression of IND, SHP, and ALC to the valve margin (Lewis et al., 2006). Anther dehiscence is another type of cell separation process, and the Arabidopsis wuschel (wus) mutant, which was originally identified as affecting stem cell maintenance, was also shown to have nonopening anthers (Mayer et al., 1998; Deyhle et al., 2007). Although the diverse genes involved in the development of these cell separation tissues have been identified, no homologous genes, except the AG homologs STK and SHP, have been found between these cell separation processes; thus, it remains unknown whether a conserved system regulates these cell separation processes.
To clarify the mechanism underlying AZ development, we focused on tomato pedicels of the fruit-ripening mutant ripening-inhibitor (rin). Tomato plants with the rin mutation develop incomplete pedicel AZ structures that show a knuckle region on the pedicels similar to wild-type plants, but frequently they show insufficient fruit abscission (Fig. 1, A and B). The rin mutation was previously shown to be a deletion that affects two tandemly arranged genes, RIN and MACROCALYX (MC), both of which encode MADS-box transcription factors; RIN controls fruit ripening and MC regulates sepal size and inflorescence determinacy (Vrebalov et al., 2002). Phylogenetic analyses revealed that MC belongs to the MADS-box family of the AP1/FUL group (Hileman et al., 2006). The mc mutant, which was isolated independently of the rin mutant, reportedly develops incomplete pedicel AZs (Butler, 1952). Although there is no molecular evidence that the mc locus is allelic to the rin locus, linkage analysis revealed that these two loci could not be separated (Robinson and Tomes, 1968; Rick, 1980).
Figure 1.
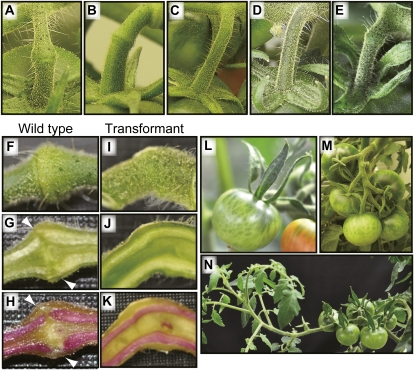
Suppression of MC resulted in the inhibition of pedicel AZ development in tomato. A to E, Fruit pedicels of the wild type (A), rin mutant (B), j mutant (C), and MC-suppressed transformants (D and E). F to K, A pedicel AZ of the wild type and a vestigial organ of the transformant harvested at the fruit-ripening stage. External appearance (F and I), longitudinal sections (G and J), and lignified cells within pedicels stained with phloroglucinol-HCl (H and K) are shown. Arrowheads indicate the interruption of lignified cells in the wild type. L, Large sepal phenotype of the transformant. M, Normal inflorescence of the wild type. N, Indeterminate inflorescence of the transformant.
In this report, we provide direct evidence that MC regulates tomato pedicel AZ development and also show the molecular function of the MC protein. The biochemical and genetic experiments shown here suggest that the products of MC and JOINTLESS form a transcription factor complex regulating the expression of genes involved in abscission. In addition, we discuss the similarity of transcriptional regulation between tomato fruit abscission and another cell separation processes (i.e. Arabidopsis fruit dehiscence).
RESULTS
MC, a MADS-Box Transcription Factor Gene, Regulates Tomato Pedicel AZ Formation
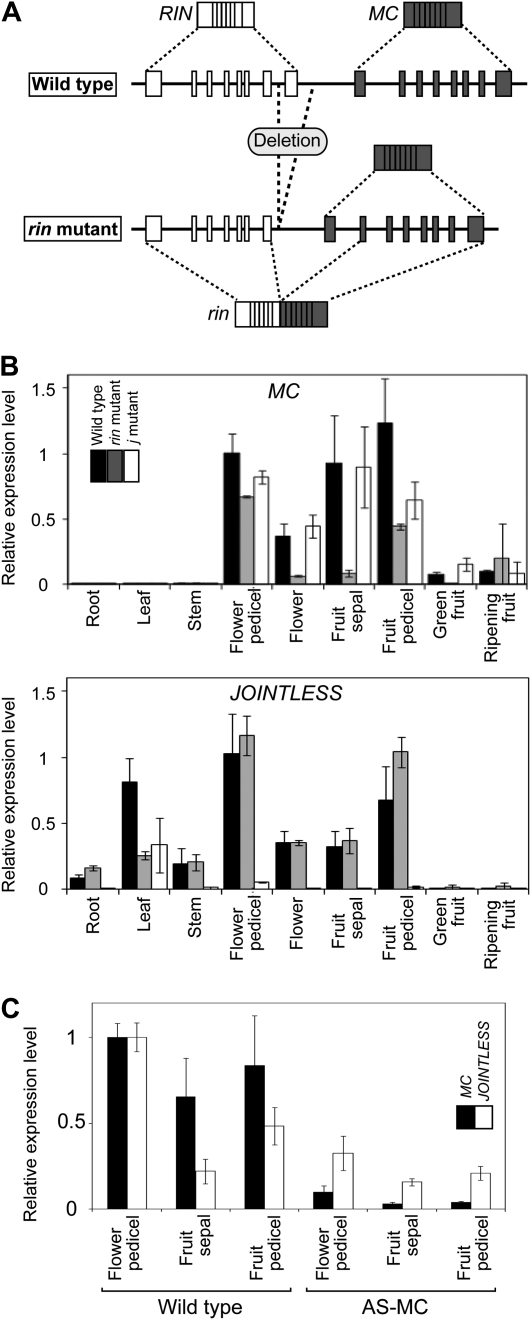
As described above, the rin mutation causes incomplete pedicel AZ development but does not show the complete jointless phenotype as observed in the j mutant (Fig. 1, A–C). The genomic region deleted by the rin mutation includes both the last exon of RIN and part of a putative cis-acting regulatory region of MC (Giovannoni, 2004; Fig. 2A). Therefore, we hypothesized that this partial deletion of the putative cis-acting sequence may cause a reduction of MC expression and, consequently, could cause imperfect pedicel AZ development in the rin mutant. Expression analyses revealed that in the rin mutants, MC was actually expressed, but at significantly lower levels in most organs, including flower and fruit pedicels, and the tissue specificity of MC expression during reproductive growth was maintained (Fig. 2B). This result supports our hypothesis that the decreased expression of MC may cause incomplete AZ development in rin mutant pedicels.
Figure 2.
Expression analysis of MC and JOINTLESS. A, Structures of genomic DNA and mRNA of the rin locus in the wild type and the rin mutant. Exons of RIN and MC are indicated by white and gray boxes, respectively. From the wild-type genome, two normal mRNAs (RIN and MC) are transcribed, whereas from the rin mutant genome, the normal mRNA of MC and the mutant mRNA, which is a chimera of RIN and MC, are transcribed (Vrebalov et al., 2002; Kitagawa et al., 2005). B, MC and JOINTLESS expression in various organs detected by qRT-PCR analysis. Levels of transcripts of MC and JOINTLESS are shown as fold change values compared with flower pedicels of the wild type. The results are means of three biological replicates of all samples, except for the ripening fruits of the j mutant and the rin mutant, whose results are means of two biological replicates. Error bars indicate sd. SlACTIN51 (SGN-E1300032) was used as the reference. mRNA of flowers and flower pedicels was sampled at anthesis, and mRNA of fruit sepals and fruit pedicels was sampled at preripening stage. C, MC and JOINTLESS expression in a transformant expressing the MC antisense gene detected by qRT-PCR. Levels of transcripts are shown as fold change values compared with flower pedicels of the wild type.
To directly test this hypothesis, we next used an antisense gene to specifically reduce MC expression. MC-suppressed transformants were produced by the introduction of an MC antisense construct (Fig. 2C), resulting in distinct inhibition of abscission at the pedicels in 12 of the 21 transformants. The degree of inhibition of AZ formation varied among transformants: some produced pedicels that completely lacked any AZ structures, whereas others produced pedicels with the knuckle-like structure in which a groove was barely visible (Fig. 1, D and E), but neither of these types of pedicels exhibited abscission of their fruits or abscission of unfertilized flowers. Sections of AZ tissue revealed that the development of AZ cells at the fruit-ripening stage resulted in the separation of a lignified cell layer in the wild type but not in the transformants (Fig. 1, F–K), indicating that MC suppression causes the inhibition of AZ development. In the transformants and their T1 progeny, inhibited abscission was always associated with the transgene (Supplemental Fig. S1), with the phenotype of the sepal shape modification (Fig. 1L), and with the leafy inflorescence that exhibits a reversion of inflorescence meristems to vegetative growth after forming flowers (Fig. 1, M and N), as reported previously (Vrebalov et al., 2002). From these observations, we concluded that MC is required for pedicel AZ formation as well as for sepal development and inflorescence determination.
MC Directly Interacts with JOINTLESS, a MADS-Box Transcription Factor That Regulates Tomato Pedicel AZ Formation
The jointless and leafy phenotypes shown in the MC-suppressed transformants are clearly similar to the j mutant phenotype (Fig. 1C; Mao et al., 2000), suggesting a possible interaction between MC and JOINTLESS. Expression analysis revealed that MC mRNA accumulation was slightly reduced in flower and fruit pedicels of j mutants (Fig. 2B). JOINTLESS mRNA accumulation was not affected in rin mutant organs except leaves but was reduced by one-third in flower pedicels of the MC-suppressed transformant (Fig. 2, B and C). These results suggest that MC and JOINTLESS may play roles in modulating the expression of each other in pedicel cells.
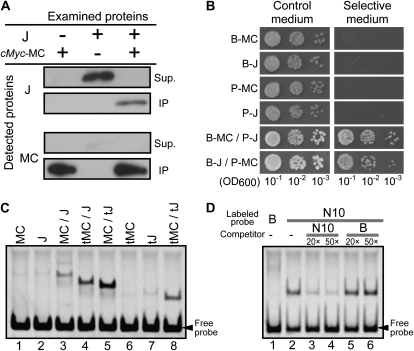
Because several MADS-box proteins reportedly function as heterodimers (Schwarz-Sommer et al., 1992; Goto and Meyerowitz, 1994), we hypothesized that the products of MC and JOINTLESS might form a transcription factor complex. To test this hypothesis, first we used an immunoprecipitation assay on in vitro-synthesized MC and JOINTLESS proteins. As shown in Figure 3A, JOINTLESS did not precipitate with the cMyc antibody but did coprecipitate with added cMyc-tagged MC, demonstrating a direct interaction between MC and JOINTLESS. Second, we confirmed this interaction by yeast two-hybrid assays. MC and JOINTLESS were expressed in yeast cells as bait and prey, respectively, or vice versa. As shown in Figure 3B, the yeast grew on selective medium when both proteins were expressed together, in either bait/prey combination, but did not grow when only MC or JOINTLESS was expressed, also demonstrating the direct interaction between MC and JOINTLESS. Thus, our data shown here, including immunoprecipitation and yeast two-hybrid assays, show that the MADS-domain proteins MC and JOINTLESS can directly interact.
Figure 3.
MC and JOINTLESS form a heterodimer with DNA-binding ability. A, Coimmunoprecipitation of MC and JOINTLESS. The cMyc-tagged MC (cMyc-MC) and JOINTLESS (J) were cotranslated or translated individually and then precipitated using anti-cMyc antibody. Supernatants (Sup.) and immunoprecipitates (IP) were analyzed using immunoblotting with anti-JOINTLESS or anti-MC antiserum as indicated. B, Yeast two-hybrid assays of MC and JOINTLESS. Yeast expressing MC or JOINTLESS with the GAL4 DNA-binding domain as a bait protein (B) or with the GAL4 activation domain as a prey protein (P) were examined on nonselective (left panel) or selective (right panel) medium. OD600, Optical density at 600 nm. C, Gel retardation assay for MC and JOINTLESS. Full-length proteins (MC and J) or truncated proteins with C-terminal domain deletions (tMC and tJ) were produced independently or simultaneously and then assayed with a labeled DNA that included a CArG motif (N10 [West et al., 1998]; CTATTTATAG). The C-terminal domain is not responsible for the DNA-binding ability; thus, proteins lacking the domain still show DNA binding, but with different electrophoretic mobility from the full-length proteins. D, The MC-JOINTLESS heterodimer binds to the specific DNA sequence known as the CArG motif. Competition assays were performed to examine the specific DNA-binding properties of the MC-JOINTLESS heterodimer. The C-terminal domain-deleted versions of MC and JOINTLESS were synthesized simultaneously and assayed by a gel retardation assay with the CArG motif used in A (N10) and a negative control probe (probe B; GGATGCATCC; Huang et al., 1993). Probe B did not bind to MADS-box proteins (lane 1). A binding competition assay was performed using the CArG probe and 20- or 50-fold concentrations of either the nonlabeled CArG sequence or probe B.
MADS domains bind to the CArG-box DNA motif (Messenguy and Dubois, 2003), and in dimers of MADS-box proteins, both of the MADS domains take part in binding to a single DNA site (Pellegrini et al., 1995; Santelli and Richmond, 2000). We examined the DNA-binding ability of MC and JOINTLESS using gel retardation assays with in vitro DNA-protein-binding reactions. When a CArG-box DNA motif (CTATTTATAG) was used for the reaction, a more intense binding signal was generated by the cotranslated products of MC and JOINTLESS than by either gene product alone (Fig. 3C, lanes 1–3). The analysis using the full-length MC and the C-domain-truncated JOINTLESS protein (tJ) showed an intense binding signal with intermediate migration between the weaker signals of the MC homodimer and the tJ homodimer (Fig. 3C, lanes 1, 5, and 7), indicating that the intense signal was likely the heterodimer of MC and tJ. The experiment using truncated MC and full-length JOINTLESS showed similar results (Fig. 3C, lanes 2, 4, and 6), indicating that the heterodimerization of MC and JOINTLESS confers strong DNA-binding ability. The DNA-binding abilities of these proteins were also compared by using two types of CArG motifs [CC(A/T)6GG and CTA(A/T)4TAG]. The heterodimer of MC and JOINTLESS showed more binding to the CTA(A/T)4TAG-type motifs than to the CC(A/T)6GG type, whereas JOINTLESS alone showed more binding to the CC(A/T)6GG-type motifs (Supplemental Fig. S2). MC showed weak binding to the CTA(A/T)4TAG-type motifs, and we detected no binding between MC and the CC(A/T)6GG-type motifs. The DNA competition assay shown in Figure 3D showed that the heterodimer exhibited specific binding to the CArG-box motif. These results demonstrate that MC and JOINTLESS form a heterodimer that can bind to the CArG-box DNA motif. Moreover, the DNA-binding specificities of the heterodimer and the homodimers of MC and JOINTLESS may be different.
Global Transcriptional Regulation of MC and JOINTLESS
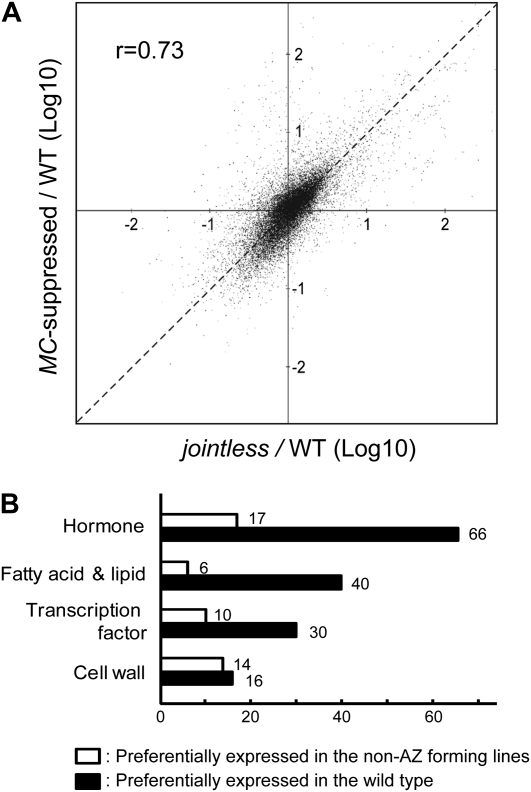
To elucidate the transcriptional regulation by MC and JOINTLESS, we conducted transcriptome assays using whole flower pedicels at anthesis stage and compared the expression pattern of the wild type with that of non-AZ-forming lines, namely, a j mutant and a MC-suppressed transformant. The transcription patterns in the non-AZ-forming lines clearly differed from the wild type. The j mutant and the MC-suppressed transformant produced very similar changes in total expression patterns compared with the wild type (Fig. 4A).
Figure 4.
Gene expression profiles in tomato pedicels. A, Effects of reduced activities of MC and JOINTLESS on gene expression in pedicels at anthesis. The scatterplot shows fold changes from the wild type (WT) for either the MC-suppressed transformant or the j mutant on the 20,915 probes that exhibited reliable signal intensity (see “Materials and Methods”). The horizontal axis shows the gene expression signal ratio of the j mutant to the wild type, and the vertical axis shows the signal ratio of the MC-suppressed transformant to the wild type. If the expression signal ratio of a gene in the wild type to the same gene in the j mutant is similar to that in the MC-suppressed transformant, the dot for that gene is plotted close to the dashed line with a slope of 1.0. B, Distribution of differentially expressed transcripts in pedicels between the wild type and non-AZ-forming lines. Genes exhibiting significantly different expression between the wild type and non-AZ-forming lines were selected from the results of two independent microarray analyses according to the method described in the text. The selected genes were then grouped into functional categories. Some genes were categorized redundantly; for example, AK322587, a tomato EST showing high homology to the Arabidopsis gene CYTOKININ-RESPONSIVE GATA FACTOR1, was categorized into both the transcription factor group and the hormone group.
To find genes regulated by MC and JOINTLESS, we selected ESTs whose array probes exhibited at least a 2.0-fold change in signal intensity between the wild type and both the non-AZ-forming lines in two biological replicates. The ESTs preferentially expressed in the wild type (743 independent ESTs) are listed in Supplemental Table S1. We inferred the function of each EST from the function of its Arabidopsis homolog. Based on the putative functions of these genes, we found four major functional groups: phytohormone-associated functions, cell wall modification, metabolism of fatty acids and lipids, and transcription factors (Fig. 4B). We also list the ESTs preferentially expressed in the non-AZ-forming lines (163 independent ESTs) in Supplemental Table S2. Of the genes identified as differentially expressed by microarray analysis (Supplemental Table S1), we selected four genes and used quantitative reverse transcription (qRT)-PCR to confirm that their expression was altered in the mutants (Supplemental Fig. S3).
The timing of abscission is mainly regulated by the phytohormones auxin and ethylene: auxin inhibits abscission and ethylene promotes it (Roberts et al., 2002). A number of ESTs showing similarity to auxin-related Arabidopsis genes were down-regulated in the non-AZ-forming lines (Supplemental Table S1). These include homologs of the indole-3-acetic acid (IAA) amido synthetase DWARF IN LIGHT1 (TA45520_4081), methyl IAA esterase METHYL ESTERASE3 (AK323887), IAA inducible transcription factor IAA19 (AF022022), auxin efflux transmembrane transporter ATP-BINDING CASETTE SUBFAMILY B1 (BI924211), auxin efflux carrier PIN-FORMED4 (CK714987), and two other auxin efflux carrier family protein genes (GO375493 and AI488645). We found a few ESTs associated with ethylene-related functions among the genes down-regulated in the non-AZ-forming lines. These genes were homologs of the ethylene signaling transcription factor ETHYLENE-RESPONSIVE ELEMENT BINDING PROTEIN (DV105056 and AK327476) and EIN3-BINDING F-BOX PROTEIN1 (DQ307489), which inhibits ethylene signaling in the absence of ethylene by reducing the accumulation of the ethylene-associated transcription factor ETHYLENE-INSENSITIVE3 (EIN3; Binder et al., 2007). Although the homolog of ACC SYNTHASE (AK326692), which is part of the gene family that encodes a key enzyme for ethylene synthesis, is expressed higher in the wild type, the homologous protein (ACS10) in Arabidopsis has no 1-aminocyclopropane-1-carboxylic acid synthase activity (Yamagami et al., 2003). In the non-AZ-forming lines, only a few auxin- or ethylene-related genes were up-regulated in comparison with the wild type (Supplemental Table S2), suggesting that the activities related to both phytohormones may be low in the pedicels of the non-AZ-forming lines.
The activation of cell wall modification and degradation at the AZ is required for cell separation and abscission. Although the flower pedicels examined in this study are likely to be at a preabscission stage, many genes encoding cell wall modification-related proteins showed differences in expression between the wild type and the non-AZ-forming lines. Of the genes previously identified to be up-regulated during abscission (Agustí et al., 2008, 2009; Cai and Lashbrook, 2008; Meir et al., 2010), homologs that show similarity to an endo-β-1,4-glucanase (Cel; U13054), xyloglucan endohydrolase endotransglycosylase (XET; AI485569), and expansins (EXP; BT014091, BP902391, and AI772225) were down-regulated in the pedicels of the non-AZ-forming lines. In addition, several putative cellulose synthase genes were also down-regulated in the non-AZ-forming lines (TA55960_4081, AK320713, and AK320411).
We found many ESTs possibly involved in lipid metabolism among the down-regulated genes in the non-AZ-forming lines (Supplemental Table S1). This group includes a number of ESTs showing similarity to the genes involved in cutin and cuticular wax biosynthesis and transport: ECERIFERUM1 (CER1; BM535116, TA36741_4081, TA37792_4081, BE354679, and TA36739_4081), CER3 (AK320525), CER4 (AK321666), ACETYL-COENZYME A CARBOXYLASE (BG124815), WHITE-BROWN COMPLEX HOMOLOG PROTEIN11 (BI929069), HOTHEAD (AK321597), GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE4 (GPAT4; BI924670 and TA42756_4081), GPAT6 (AK324536), CYTOCHROME P450 86 A2 (AK324391), and BODYGUARD1 (AK321255).
Genes in diverse transcription factor gene families were down-regulated in the non-AZ-forming lines. Among these genes, BT013459 showed similarity to the Arabidopsis BEL1-LIKE HOMEODOMAIN1, whose rice homolog qSH1 regulates seed shattering (Konishi et al., 2006). AK324219, which shows similarity to the Arabidopsis basic helix-loop-helix-type transcription factor UNE10 and the pod-DZ regulator ALCATRAZ, were also down-regulated. Interestingly, we also detected the down-regulation of several transcription factor genes that were previously identified as regulators of meristem cell fate. For example, Blind (Bl; AF426174) and GOBLET (GOB; FJ435163) are homologs of REGULATOR OF AXILLARY MERISTEMS (RAX) and CUP-SHAPED COTYLEDON (CUC), respectively, which are the transcription factor genes involved in axillary meristem initiation in Arabidopsis (Raman et al., 2008). Also, LeWUS (AJ538329) is a homolog of the meristem-regulating transcription factor WUS.
The Ls gene, which is known to regulate axillary meristem initiation and AZ development (Schumacher et al., 1999; Roberts et al., 2002), was not selected in the set of genes preferentially expressed in the wild type because it was eliminated for technical reasons: one of the replicate microarray experiments in MC-suppressed transformant lines showed a fold change value above the threshold (greater than 0.5; Supplemental Table S1). However, because of the very low fold change values (less than 0.1; Supplemental Table S1) in other microarray experiments (another experiment with MC-suppressed transformants and duplicate experiments with j mutants) and our knowledge of its biological function, we further investigated the Ls gene in addition to the other transcription factor genes that we identified.
LeWUS, Bl, GOB, and Ls Are Specifically Expressed in Pedicel AZs
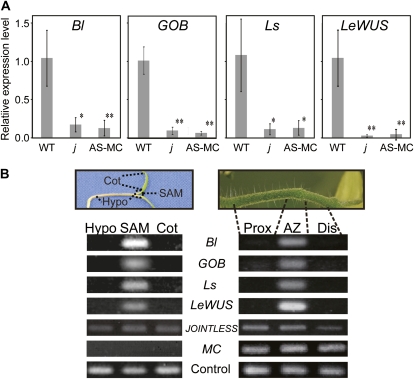
Genes that were transcribed more abundantly in the wild type than in the non-AZ-forming lines may be involved in abscission processes. Among the genes that were down-regulated in the non-AZ-forming lines, we focused on the transcription factor genes involved in the maintenance of shoot apical meristems (SAMs) and the initiation of axillary meristems: LeWUS, Bl, GOB, and Ls. The expression specificities of these genes were also examined by qRT-PCR using RNAs extracted from whole flower pedicels at anthesis. These analyses indicated that the expression of these genes in the pedicel of the j mutant and the MC-suppressed transformant were 6.2- to 25.4-fold less than in the wild type (Fig. 5A), confirming the results of the microarray analyses. Next, we analyzed local expression of these genes by dividing flower pedicels into three parts: the AZ, the region between the flower and the AZ (distal region), and the region between the AZ and the main stem of the inflorescence (proximal region; Fig. 5B). RT-PCR analyses revealed that the expression of these genes was detected specifically in the AZ, although MC and JOINTLESS were expressed throughout pedicels (Fig. 5B). The genes specifically detected in the AZ, LeWUS, Bl, GOB, and Ls, coincided with the genes specifically expressed in the SAMs of seedlings (Fig. 5B).
Figure 5.
AZ-specific expression of genes involved in meristem cell regulation. A, Expression in tomato flower pedicels of transcription factor genes affected by MC and JOINTLESS. Whole flower pedicels of the wild type (WT), the j mutant (j), and the MC-suppressed transformant (AS-MC) were examined by qRT-PCR. Levels of transcripts of LeWUS, Bl, GOB, and Ls are shown as fold change values compared with the wild type. Error bars indicate sd (n = 3). A SAND family protein gene (SGN-U316474) was used as the reference for tomato pedicels, and CAC (SGN-U314153) was used as the reference for seedlings (Expósito-Rodríguez et al., 2008). * P < 0.05, ** P < 0.01. B, Expression specificities of LeWUS, Bl, GOB, and Ls in tomato pedicels and seedlings as shown by RT-PCR. mRNAs were sampled from AZs, proximal (Prox) and distal (Dis) regions within tomato flower pedicels, hypocotyls (Hypo), SAM, and cotyledons (Cot) of tomato seedlings. [See online article for color version of this figure.]
DISCUSSION
MC Is Likely to Regulate Tomato Pedicel AZ Development as a Heterodimer with JOINTLESS
In this study, we demonstrated that the MC gene is involved in the regulation of tomato pedicel AZ development. This indicates that in the rin mutant, incomplete pedicel AZ development is caused by the modified expression of MC. Although no molecular analysis of the mc mutant that develops incomplete AZ structures has been reported, the results of this study and the previous genetic linkage analyses strongly suggest that MC in the rin locus is allelic to the gene affected in the mc mutant (Butler, 1952; Robinson and Tomes, 1968; Vrebalov et al., 2002). MC is not an allele of j2, because the two loci map to different chromosomes (Vrebalov et al., 2002; Yang et al., 2005). Phylogenetic analysis of MADS-box proteins has revealed that MC belongs to the Arabidopsis AP1/FUL clade, which is separate from the Short Vegetative Phase clade to which JOINTLESS belongs (Hileman et al., 2006). In addition, MC is involved in sepal size determination, whereas the j mutation does not affect sepal development. These observations indicate that MC function is not redundant with that of JOINTLESS. Therefore, to our knowledge, MC is a newly identified member of the set of genes regulating pedicel AZ development. MC shows significant sequence similarity to FUL, which is required to develop pod DZs in Arabidopsis (Liljegren et al., 2004; Hileman et al., 2006), suggesting that MC and FUL may play homologous roles in these distinct cell separation processes.
Our biochemical analyses reported here showed that MC and JOINTLESS form a heterodimer with specific DNA-binding ability. Leseberg et al. (2008) have also described the possibility of an interaction between MC and JOINTLESS based on the results of yeast two-hybrid assays (Leseberg et al., 2008). MC suppression produced similar phenotypes to j mutants, including jointless pedicels and leafy inflorescences. The gene expression profile of the MC-suppressed transformant was highly similar to that of the j mutant (Fig. 4A), consistent with the model that MC and JOINTLESS cooperate by forming a heterodimer. These observations suggest that the MC-JOINTLESS heterodimer most likely acts as a regulator of pedicel AZ development. However, although MC suppression and the j mutation had similar phenotypic effects, they also showed a different effect on sepal size determination (Fig. 1). In addition, MC and JOINTLESS homodimers can bind to CArG-box sequences, and the binding specificity of the JOINTLESS homodimer was different from the specificity of the heterodimer (Fig. 3C; Supplemental Fig. S2). Thus, we cannot exclude the possibility that both MC and JOINTLESS may have additional, independent functions.
MC and JOINTLESS Regulate Key Factors in Abscission, Such as Phytohormones, Cell Wall Modifications, and Lipid Metabolism
We conducted transcriptome assays on pedicels of the MC-suppressed transformant and j mutants at anthesis. At this stage, these pedicels had already acquired competence to respond to the abscission signal, but abscission was not yet initiated. Meanwhile, growth of the pedicel including AZ tissue is ongoing to develop thick fruit pedicels. Thus, the genes showing different expression between the wild type and the non-AZ-forming lines may include genes involved in these stage-specific activities. The transcriptome assay results indicate that both MC and JOINTLESS regulate the activities of key factors for abscission, including phytohormone metabolism and transport, cell wall modification, and lipid metabolism and transport.
The phytohormone auxin prevents the AZ from activating abscission, whereas ethylene acts as an auxin antagonist that promotes abscission in various tissues, including tomato pedicels (Agustí et al., 2008; Meir et al., 2010). Our results showed the expression of many auxin-related genes in wild-type pedicels, suggesting that MC and JOINTLESS activate the auxin-mediated pathway to hold abscission in check and prevent the abscission of anthesis-stage flowers. Meanwhile, ethylene-mediating pathways for abscission were not up-regulated in the non-AZ-forming lines, although these lines were defective in auxin regulation, which is an antagonist of ethylene (Supplemental Table S2). These results suggest that MC and JOINTLESS are required to confer the pedicel-specific auxin/ethylene regulation that is the most critical factor in determining the timing of abscission of the pedicel. Meanwhile, the down-regulation of the genes related to auxin in the non-AZ-forming lines suggests the possibility that auxin may play a role in the regulation of AZ development.
Previous studies indicated that the expression of many genes encoding cell wall modification-related proteins, such as Cel, polygalacturonase (PG), XET, and EXP, are drastically up-regulated at the onset of abscission (Agustí et al., 2008, 2009; Cai and Lashbrook, 2008; Meir et al., 2010). The expression of these genes, however, is low before the onset of abscission (Meir et al., 2010), which was the stage analyzed in this study. Here, at the preonset stage, we found that, except for PG, the low expression of these genes decreased further in the MC-suppressed lines or j mutants (Supplemental Table S1), suggesting that MC and JOINTLESS are involved in the transcriptional regulation of these genes at the preabscission stage. Meanwhile, ethylene is required to increase the expression of these cell wall modification genes with the onset of abscission (Meir et al., 2010). These observations suggest that the MADS-box transcription factors MC and JOINTLESS may be involved in initiating the expression of these cell wall modification genes and that ethylene may play a role in enhancement of the expression. Similar divisional cooperation is proposed in tomato fruit ripening, where LeACS2, which is a rate-limiting gene of ethylene synthesis, would be initiated by a developmental factor, after which robust expression would be caused by ethylene in an autocatalytic manner (Yokotani et al., 2009); indeed, the MADS-box transcription factor RIN is a promising candidate for the developmental factor (Ito et al., 2008).
Our results also revealed that MC and JOINTLESS play an important role in the transcriptional regulation of the genes for fatty acid metabolism and transport. This group includes genes involved in cutin or cuticular wax metabolism and transport (Kurdyukov et al., 2006; Li-Beisson et al., 2010), and in pedicels, the products are expected to protect the AZ from microbial attack (Wu and Burns, 2003). A previous observation revealed that the cuticle is continuous within an indentation of the AZ in the flower pedicel (Jensen and Valdovinos, 1967).
MC and JOINTLESS Regulate Transcription Factor Genes Involved in Meristem Identity
Our transcriptome analyses also revealed that a number of transcription factor genes showed altered expression patterns caused by MC suppression or j mutation. Among these, we found four transcription factor genes (LeWUS, Bl, GOB, and Ls) whose homologs have been shown to regulate cell fate at the shoot apex. The expression of these genes in tomato pedicel AZs was highly specific to pedicel AZs (Fig. 5B). These results suggest that these transcription factors may play an important role in abscission processes, such as the regulation of pedicel AZ development, the maintenance of the AZ cells in an undifferentiated state, or the acquisition of competence to respond to abscission signals. Mutations in Ls and Bl reportedly affect the jointless phenotype (Roberts et al., 2002; Shalit et al., 2009; Supplemental Fig. S4), supporting the possibility that these two genes act in AZ development. In addition, the CUC gene family, of which GOB is a member, reportedly plays a crucial role in several boundary-formation processes, regulating separation of cotyledons, serration of leaf margins, dissection of leaflet margins, and separation of sepal margins (Aida et al., 1997; Blein et al., 2008; Berger et al., 2009). In tomato, pedicels can be divided into a proximal region and a distal region; the groove in the AZ is likely to be the boundary between the two regions. The AZ-specific expression of GOB may be involved in the regulation of boundary formation at pedicel AZs. In axillary meristem formation in Arabidopsis, RAX, which is a homolog of Bl, appears to act upstream of CUC, whereas LAS acts downstream (Keller et al., 2006; Raman et al., 2008). RAX, CUC, and LAS are expressed in the boundaries established between a leaf primordium and the adjacent meristem (Greb et al., 2003; Vroemen et al., 2003; Keller et al., 2006). The AZ-specific expression of these homologs suggests that the transcriptional cascade found in axillary meristem initiation is also conserved in AZ tissues. Tomato pedicel AZs have the potential to develop adventitious shoots, although the event is rare (Supplemental Fig. S5), supporting the possibility that pedicel AZs and leaf axils share some regulatory mechanisms. In the Arabidopsis shoot apex, WUS is expressed in the cells underneath the stem cells and plays a central role in the maintenance of stem cells in an undifferentiated state (Mayer et al., 1998). In the development of AZs, small undifferentiated cells are arranged transversely to the axis of the pedicels, even though pedicel cells outside of the AZ have already differentiated (van Nocker, 2009). Analogous to the role of WUS in the shoot apex, LeWUS might be required for the maintenance of AZ cells in an undifferentiated state. The wus mutant of Arabidopsis shows a suggestive phenotype in which the anthers do not open and pollen is not released due to the improper development of the cells for dehiscence in the anther (Deyhle et al., 2007). LeWUS expressed in pedicel AZs may take part in the regulation of AZ development, similar to the role of WUS in anther dehiscence.
Our results show that MC and JOINTLESS are necessary to induce the expression of four transcription factor genes (LeWUS, Bl, GOB, and Ls). However, these four genes are expressed specifically at AZs but MC and JOINTLESS are expressed throughout pedicels (Fig. 5B), indicating that MC and JOINTLESS may be necessary but not sufficient to induce these four transcription factor genes. To account for this discrepancy, certain additional factors may be required to induce the specific expression of the four transcription factors in the AZ. Local expression of other MADS-box proteins may determine the AZ-specific expression of the four genes via the region-specific formation of MADS-box protein multimers with the MC-JOINTLESS heterodimer. The model of region-specific MADS-box protein combinations is well established for the quartet model of floral organ development (Coen and Meyerowitz, 1991). Comprehensive expression analyses between AZs and other pedicel parts may provide meaningful information on additional key factor(s) that induce specific gene expression in AZs and cause AZ tissue differentiation.
The non-AZ-forming tomato lines exhibited normal SAM development, suggesting that the expression of GOB, Bl, Ls, and LeWUS at AZs is regulated by MC and JOINTLESS, but at shoot apices, these genes are likely regulated by other factor(s). Although further studies are necessary to confirm the possibility that these transcription factors are required for abscission, elucidation of the functional roles of these transcription factors in the AZ may provide a significant breakthrough in our understanding of abscission.
Regulation of AZ Development May Be Conserved among a Wide Variety of Cell Separation Processes
Cell separation is a common plant activity in various plant tissues, although little is known about similarities in the regulation among the different types of the process. Here, we found AZ-specific expression of several transcription factor genes that are key regulators in meristem cell differentiation. Interestingly, a common transcriptional regulation between apical meristem cell differentiation and pod dehiscence has been proven in Arabidopsis by examination of the roles of YABBY and JAGGED (Dinneny et al., 2005). The functions of the tomato homologs MC and FUL suggest the possibility of similarities between AZ development and DZ development. These similarities suggest that the regulation of abscission, dehiscence, and meristem cell differentiation may derive from a common ancestral transcriptional pathway.
MATERIALS AND METHODS
Plasmid Construction
Primers for gene amplification are listed in Supplemental Table S3. The plasmid expressing the antisense MC gene was constructed as follows. A 473-nucleotide fragment of the MC cDNA (accession no. AF448521) was amplified with the primer pair AntiMC-F and AntiMC-R and then inserted into SacI-BamHI-cleaved pBI121 (Clontech); the resulting plasmid was designated pBI-ASMC. The plasmids expressing full-length MC, truncated MC (amino acids 1–160), JOINTLESS, and truncated JOINTLESS (amino acids 1–161) via an in vitro translation system were constructed as follows. Each gene fragment was amplified by using the following primer pairs: MC-F1 and MC-R1 for MC, MC-F1 and MC-R2 for truncated MC, J1-F1 and J1-R1 for JOINTLESS (accession no. AX403041), and J1-F1 and J1-R2 for truncated JOINTLESS. The amplified fragments were digested with NcoI and SacI and inserted into pEU-3a (Ito et al., 2002). The resulting plasmids were designated pEU-MC, pEU-MCMIK, pEU-J1, and pEU-J1MIK. The plasmid pEU-cM-MC, expressing cMyc epitope-tagged MC, was constructed by inserting the cMyc epitope tag sequence into the NcoI site of pEU-MC. The plasmids for the yeast two-hybrid assays were constructed as follows. The plasmids pEU-MC and pEU-J1 were digested with NcoI and EcoRI, and each fragment was inserted into pGBKT7 (Clontech; http://www.clontech.com/), resulting in the plasmids pGBK-MC and pGBK-J1, respectively. The plasmids pEU-MC and pEU-J1 were digested with NcoI and SacI, and each fragment was inserted into pGADT7 (Clontech), resulting in the plasmids pGAD-MC and pGAD-J1, respectively.
Plant Transformation
The tomato (Solanum lycopersicum) cv Ailsa Craig and a breeding line of Kagome Co., PK331, were transformed with pBI-ASMC via the Agrobacterium tumefaciens-mediated method, as described previously (Sun et al., 2006).
Immunoprecipitation Assays
For immunoprecipitation experiments, cMyc-tagged MC and JOINTLESS were synthesized using TnT Quick Coupled Transcription/Translation Systems (Promega; http://www.promega.com/) with the plasmids pEU-cM-MC and pEU-J1. Immunoprecipitation reactions with anti-cMyc monoclonal antibody were carried out by using the BD Matchmaker Co-IP Kit (Clontech). Precipitated proteins were separated on 10% (w/v) SDS-polyacrylamide gels and then detected by immunoblot analysis with an antiserum against MC or JOINTLESS.
Yeast Two-Hybrid Assays
The bait vector pGBK-MC or pGBK-J1 and the prey vector pGAD-MC or pGAD-J1 were cotransformed into the yeast strain AH109 (Clontech) and selected on synthetic dropout (SD) plates lacking Leu and Trp. Five independent transformants for each combination were incubated in 1 mL of liquid SD medium lacking Leu and Trp at 30°C overnight with shaking, and cells were harvested by centrifugation and suspended in water. Five microliters of cell suspension was spotted on SD medium lacking His, adenine (Ade), Leu, and Trp or on the control medium lacking Leu and Trp. As negative controls, the transformants with just pGBK-MC or pGBK-J1 were incubated on SD medium lacking His, Ade, and Trp or on the control medium lacking Trp, and transformants with just pGAD-MC or pGAD-J1 were incubated on the medium lacking His, Ade, and Leu or on the control medium lacking Leu.
Gel Retardation Assays
Fluorescein isothiocyanate-labeled probes were produced using the method described previously (Ito et al., 2008). For labeling the target DNA fragments, the plasmid for the target sequence, pGEM-N10 (CTATTTATAG), and that for negative control sequences, pGEM-B (GGATGCATCC), were used. Protein-DNA-binding reactions were done according to the method described previously (Riechmann et al., 1996). Fluorescence images of the fluorescein isothiocyanate-labeled probe were detected by using Typhoon 8600 (GE Healthcare Bioscience; http://www.gehealthcare.com/worldwide.html).
Phloroglucinol Lignin Staining
Phloroglucinol-HCl reagent was prepared by mixing 2 volumes of 2% (w/v) phloroglucinol in 95% (v/v) ethanol with 1 volume of concentrated HCl. The flower pedicel was cut longitudinally, and a solution of phloroglucinol-HCl was applied directly to fresh cut tissue.
Real-Time and Semiquantitative RT-PCR Analyses
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen; http://www.qiagen.com/). One microgram of total RNA was reverse transcribed into cDNA using the PrimeScript II First-Strand cDNA Synthesis Kit (Takara; www.takara-bio.co.jp). PCR amplification was carried out using ExTaq (Takara; www.takara-bio.co.jp). Real-time PCR amplification was carried out with the 7300 Real-Time PCR System (Applied Biosystems; www.appliedbiosystems.com). Relative quantification of the expression of each gene was performed using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Primer sequences are listed in Supplemental Table S3.
Microarray Experiments
The first cDNA strands and Cy3-labeled complementary RNA were synthesized from 0.5 μg of total RNA using the Quick Amp Labeling Kit (Agilent; http://www.chem.agilent.com). Agilent Tomato Oligo Microarray 44K slides were used for hybridization. Microarray slides were scanned using the Agilent Technology Microarray Scanner. Spot signal intensities were acquired using feature extraction software version 10.5.1.1 (Agilent). To compare the results between the different experiments, signal intensities were normalized by the per chip normalization method to the 75th percentile using Genespring version 10.0 software (Agilent). Data from outlier probes were removed according to the following criteria: if the feature was signal saturated, if the signal was a nonuniform outlier, or if the signal was a population outlier. The complete data set has been deposited at the Gene Expression Omnibus with accession number GSE22682 (National Center for Biotechnology Information). For scatterplot drawing, microarray data from probes with less reliable signal intensity were further removed if the background-subtracted signal was less than the background signal plus 13-fold of the sd of the background signal. The description of each tomato EST on the microarray was determined using similarity with Arabidopsis (Arabidopsis thaliana) sequences; to do this, a BLASTX search was performed against the National Center for Biotechnology Information Arabidopsis Refseq protein database (http://www.ncbi.nlm.nih.gov/genome/seq/BlastGen/BlastGen.cgi?taxid=3702). For the functional classification of each sequence, the Gene Ontology annotation search was performed at The Arabidopsis Information Resource (http://arabidopsis.org/), and the obtained results were confirmed and modified manually.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Segregation of the transgene and the jointless phenotype in T1 progeny.
Supplemental Figure S2. Gel retardation assay of two types of CArG-box motifs.
Supplemental Figure S3. Expression analyses of the gene down-regulated in the non-AZ-forming lines by qRT-PCR.
Supplemental Figure S4. Pedicel of the ls mutant.
Supplemental Figure S5. Development of adventitious shoots from a tomato pedicel AZ.
Supplemental Table S1. Tomato ESTs for which expression in flower pedicels is down-regulated by the j mutation and MC suppression.
Supplemental Table S2. Tomato ESTs for which expression in flower pedicels is up-regulated by the j mutation and MC suppression.
Supplemental Table S3. Primers used for PCR.
References
- Agustí J, Merelo P, Cercós M, Tadeo FR, Talón M. (2008) Ethylene-induced differential gene expression during abscission of citrus leaves. J Exp Bot 59: 2717–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agustí J, Merelo P, Cercós M, Tadeo FR, Talón M. (2009) Comparative transcriptional survey between laser-microdissected cells from laminar abscission zone and petiolar cortical tissue during ethylene-promoted abscission in citrus leaves. BMC Plant Biol 9: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangerth F. (2000) Abscission and thinning of young fruit and their regulation by plant hormones and bioregulators. Plant Growth Regul 31: 43–59 [Google Scholar]
- Berger Y, Harpaz-Saad S, Brand A, Melnik H, Sirding N, Alvarez JP, Zinder M, Samach A, Eshed Y, Ori N. (2009) The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136: 823–832 [DOI] [PubMed] [Google Scholar]
- Binder BM, Walker JM, Gagne JM, Emborg TJ, Hemmann G, Bleecker AB, Vierstra RD. (2007) The Arabidopsis EIN3 binding F-box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 19: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blein T, Pulido A, Vialette-Guiraud A, Nikovics K, Morin H, Hay A, Johansen IE, Tsiantis M, Laufs P. (2008) A conserved molecular framework for compound leaf development. Science 322: 1835–1839 [DOI] [PubMed] [Google Scholar]
- Botton A, Eccher G, Forcato C, Ferrarini A, Begheldo M, Zermiani M, Moscatello S, Battistelli A, Velasco R, Ruperti B, et al. (2011) Signaling pathways mediating the induction of apple fruitlet abscission. Plant Physiol 155: 185–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler L. (1952) The linkage map of the tomato. J Hered 43: 25–35 [Google Scholar]
- Cai S, Lashbrook CC. (2008) Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2. Plant Physiol 146: 1305–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM. (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Deyhle F, Sarkar AK, Tucker EJ, Laux T. (2007) WUSCHEL regulates cell differentiation during anther development. Dev Biol 302: 154–159 [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Weigel D, Yanofsky MF. (2005) A genetic framework for fruit patterning in Arabidopsis thaliana. Development 132: 4687–4696 [DOI] [PubMed] [Google Scholar]
- Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA. (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. (2004) Genetic regulation of fruit development and ripening. Plant Cell (Suppl) 16: S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. (1994) Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev 8: 1548–1560 [DOI] [PubMed] [Google Scholar]
- Greb T, Clarenz O, Schafer E, Muller D, Herrero R, Schmitz G, Theres K. (2003) Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev 17: 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman LC, Sundstrom JF, Litt A, Chen M, Shumba T, Irish VF. (2006) Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Mol Biol Evol 23: 2245–2258 [DOI] [PubMed] [Google Scholar]
- Huang H, Mizukami Y, Hu Y, Ma H. (1993) Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res 21: 4769–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, Ito H, Inakuma T, Hiroi S, Kasumi T. (2008) DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J 55: 212–223 [DOI] [PubMed] [Google Scholar]
- Ito Y, Ozawa A, Sawasaki T, Endo Y, Ochi K, Tozawa Y. (2002) OsRALyase1, a putative F-box protein identified in rice, Oryza sativa, with enzyme activity identical to that of wheat RALyase. Biosci Biotechnol Biochem 66: 2727–2731 [DOI] [PubMed] [Google Scholar]
- Jensen TE, Valdovinos JG. (1967) Fine structure of abscission zone. Planta 77: 298–318 [DOI] [PubMed] [Google Scholar]
- Ji H, Kim SR, Kim YH, Kim H, Eun MY, Jin ID, Cha YS, Yun DW, Ahn BO, Lee MC, et al. (2010) Inactivation of the CTD phosphatase-like gene OsCPL1 enhances the development of the abscission layer and seed shattering in rice. Plant J 61: 96–106 [DOI] [PubMed] [Google Scholar]
- Keller T, Abbott J, Moritz T, Doerner P. (2006) Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18: 598–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Ito H, Shiina T, Nakamura N, Inakuma T, Kasumi T, Ishiguro Y, Yabe K, Ito Y. (2005) Characterization of tomato fruit ripening and analysis of gene expression in F-1 hybrids of the ripening inhibitor (rin) mutant. Physiol Plant 123: 331–338 [Google Scholar]
- Konishi S, Izawa T, Lin SY, Ebana K, Fukuta Y, Sasaki T, Yano M. (2006) An SNP caused loss of seed shattering during rice domestication. Science 312: 1392–1396 [DOI] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Trenkamp S, Bär S, Franke R, Efremova N, Tietjen K, Schreiber L, Saedler H, Yephremov A. (2006) Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain alpha-,omega-dicarboxylic fatty acids and formation of extracellular matrix. Planta 224: 315–329 [DOI] [PubMed] [Google Scholar]
- Leseberg CH, Eissler CL, Wang X, Johns MA, Duvall MR, Mao L. (2008) Interaction study of MADS-domain proteins in tomato. J Exp Bot 59: 2253–2265 [DOI] [PubMed] [Google Scholar]
- Lewis MW, Leslie ME, Liljegren SJ. (2006) Plant separation: 50 ways to leave your mother. Curr Opin Plant Biol 9: 59–65 [DOI] [PubMed] [Google Scholar]
- Li C, Zhou A, Sang T. (2006) Rice domestication by reducing shattering. Science 311: 1936–1939 [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, DeBono A, Durrett TP, et al. (2010) Acyl-lipid metabolism. The Arabidopsis Book; 8: e0133, doi/10.1199/tab.0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Roeder AH, Kempin SA, Gremski K, Østergaard L, Guimil S, Reyes DK, Yanofsky MF. (2004) Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116: 843–853 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mao L, Begum D, Chuang HW, Budiman MA, Szymkowiak EJ, Irish EE, Wing RA. (2000) JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 406: 910–913 [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Meir S, Philosoph-Hadas S, Sundaresan S, Selvaraj KS, Burd S, Ophir R, Kochanek B, Reid MS, Jiang CZ, Lers A. (2010) Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol 154: 1929–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F, Dubois E. (2003) Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316: 1–21 [DOI] [PubMed] [Google Scholar]
- Patterson SE. (2001) Cutting loose: abscission and dehiscence in Arabidopsis. Plant Physiol 126: 494–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L, Tan S, Richmond TJ. (1995) Structure of serum response factor core bound to DNA. Nature 376: 490–498 [DOI] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF. (2003) Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88 [DOI] [PubMed] [Google Scholar]
- Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K. (2008) Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J 55: 65–76 [DOI] [PubMed] [Google Scholar]
- Rick CM. (1980) Tomato linkage survey. Rep Tomato Genet Coop 30: 2–17 [Google Scholar]
- Riechmann JL, Krizek BA, Meyerowitz EM. (1996) Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci USA 93: 4793–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JA, Elliott KA, Gonzalez-Carranza ZH. (2002) Abscission, dehiscence, and other cell separation processes. Annu Rev Plant Biol 53: 131–158 [DOI] [PubMed] [Google Scholar]
- Robinson RW, Tomes ML. (1968) Ripening-inhibitor: a gene with multiple effects on ripening. Rep Tomato Genet Coop 18: 36–37 [Google Scholar]
- Santelli E, Richmond TJ. (2000) Crystal structure of MEF2A core bound to DNA at 1.5 A resolution. J Mol Biol 297: 437–449 [DOI] [PubMed] [Google Scholar]
- Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K. (1999) The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Natl Acad Sci USA 96: 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lönnig WE, Saedler H, Sommer H. (1992) Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J 11: 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalit A, Rozman A, Goldshmidt A, Alvarez JP, Bowman JL, Eshed Y, Lifschitz E. (2009) The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc Natl Acad Sci USA 106: 8392–8397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HJ, Uchii S, Watanabe S, Ezura H. (2006) A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol 47: 426–431 [DOI] [PubMed] [Google Scholar]
- van Nocker S. (2009) Development of the abscission zone. Stewart Postharvest Review 1: 1–6 [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC. (2003) The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15: 1563–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AG, Causier BE, Davies B, Sharrocks AD. (1998) DNA binding and dimerisation determinants of Antirrhinum majus MADS-box transcription factors. Nucleic Acids Res 26: 5277–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Burns JK. (2003) Isolation and characterization of a cDNA encoding a lipid transfer protein expressed in ‘Valencia’ orange during abscission. J Exp Bot 54: 1183–1191 [DOI] [PubMed] [Google Scholar]
- Yamagami T, Tsuchisaka A, Yamada K, Haddon WF, Harden LA, Theologis A. (2003) Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J Biol Chem 278: 49102–49112 [DOI] [PubMed] [Google Scholar]
- Yang TJ, Lee S, Chang SB, Yu Y, de Jong H, Wing RA. (2005) In-depth sequence analysis of the tomato chromosome 12 centromeric region: identification of a large CAA block and characterization of pericentromere retrotransposons. Chromosoma 114: 103–117 [DOI] [PubMed] [Google Scholar]
- Yokotani N, Nakano R, Imanishi S, Nagata M, Inaba A, Kubo Y. (2009) Ripening-associated ethylene biosynthesis in tomato fruit is autocatalytically and developmentally regulated. J Exp Bot 60: 3433–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahara MB, Scheuerman RW. (1988) Hand-harvesting jointless vs. jointed-stem tomatoes. Calif Agric 42: 14 [Google Scholar]