Abstract
Cysteine proteases (CPs) accumulate to high concentration in many fruit, where they are believed to play a role in fungal and insect defense. The fruit of Actinidia species (kiwifruit) exhibit a range of CP activities (e.g. the Actinidia chinensis variety YellowA shows less than 2% of the activity of Actinidia deliciosa variety Hayward). A major quantitative trait locus for CP activity was mapped to linkage group 16 in a segregating population of A. chinensis. This quantitative trait locus colocated with the gene encoding actinidin, the major acidic CP in ripe Hayward fruit encoded by the ACT1A-1 allele. Sequence analysis indicated that the ACT1A locus in the segregating A. chinensis population contained one functional allele (A-2) and three nonfunctional alleles (a-3, a-4, and a-5) each containing a unique frameshift mutation. YellowA kiwifruit contained two further alleles: a-6, which was nonfunctional because of a large insertion, and a-7, which produced an inactive enzyme. Site-directed mutagenesis of the act1a-7 protein revealed a residue that restored CP activity. Expression of the functional ACT1A-1 cDNA in transgenic plants complemented the natural YellowA mutations and partially restored CP activity in fruit. Two consequences of the increase in CP activity were enhanced degradation of gelatin-based jellies in vitro and an increase in the processing of a class IV chitinase in planta. These results provide new insight into key residues required for CP activity and the in vivo protein targets of actinidin.
Cys proteases (CPs) are ubiquitous enzymes that participate in key plant cellular functions, including removing abnormal/misfolded proteins, supplying amino acids needed to make new proteins, regulating the abundance of key enzymes and regulatory proteins, and assisting in the maturation of proenzymes and peptide hormones by specific cleavage (van der Hoorn, 2008). They have also been implicated as catalysts of protein remobilization during seed germination (e.g. aleurain; Wang et al., 2007a) and organ senescence (Hayashi et al., 2001; Wan et al., 2002). CPs may also be important in fungal and insect defense. Tomato (Solanum lycopersicum) CPs, Required for Cladosporium Resistance3 and Phytophthora Inhibited Protease1 are produced upon pathogen attack and are inhibited by pathogen-derived protease inhibitors (Krüger et al., 2002; Tian et al., 2007). Maize Inbred Resistance1 from maize (Zea mays) and papain from papaya (Carica papaya) have both been shown to act against insect larvae (Pechan et al., 2000; Konno et al., 2004). However, the physiological functions of most other CPs in planta are unknown.
Plant CPs are typically between 20 and 30 kD in size and use a catalytic Cys residue to cleave peptide bonds in protein substrates. The catalytic Cys is situated in a broad fold between two domains, one domain being mainly α-helical and the other rich in antiparallel β-sheets (Drenth et al., 1968). The C1A papain-like proteases (Beers et al., 2004) are characterized by the presence of multiple disulfide bridges and accumulate in the vacuole, apoplast, or specific vesicles (van der Hoorn, 2008). The enzymes are synthesized as proproteins and require extensive posttranslational modification to be processed into an active form. The autoinhibitory N-terminal prodomain folds back into the catalytic site cleft and is removed upon activation of the protease (Taylor et al., 1995). Some C1A proteases also contain Cys-rich C-terminal extensions that may participate in the regulation of protease solubility and activation (Yamada et al., 2001).
Kiwifruit (Actinidia) species contain a large number of CPs, including a group of 10 genes that form a distinct kiwifruit-specific clade designated actinidin (Nieuwenhuizen et al., 2007). The actinidin clade belongs to the C1A papain-like subfamily, which includes enzymes such as caricain, bromelain, ficin, and phytolacain (Beers et al., 2004) that accumulate to high concentrations in fruit of other species. Most genetic and biochemical characterization of actinidin has been carried out in the major commercial green-fleshed kiwifruit (Actinidia deliciosa var Hayward), with multiple cDNA sequences (Praekelt et al., 1988; Podivinsky et al., 1989), genomic/promoter sequences (Keeling et al., 1990; Snowden and Gardner, 1990), and protein isoforms (Tello-Solis et al., 1995; Sugiyama et al., 1996, 1997) being reported. Varietal differences in CP activity have been reported in the fruit of four A. deliciosa varieties (Prestamo, 1995), in a range of noncommercial Actinidia genotypes (Boyes et al., 1997), as well as in fruit juices extracted from Actinidia varieties and genotypes (Nishiyama, 2007). Most recently, Nieuwenhuizen et al. (2007) showed that a major commercial yellow-fleshed kiwifruit variety (Actinidia chinensis YellowA) contained less than 2% of the CP activity of A. deliciosa variety Hayward. However, the molecular basis for this difference in CP activity was not clear.
High levels of CP activity are responsible for the meat-tenderizing properties of kiwifruit, pineapple (Ananas comosus), and papaya (Lewis and Luh, 1988; Ashie et al., 2002) and preclude the use of these fruit in processing applications where gelatin is used (Arcus, 1959). CP enzymes may be beneficial to human health, with actinidin being shown to enhance the digestion of a range of common food proteins under simulated gastric conditions (Kaur et al., 2010a, 2010b; Rutherfurd et al., 2011). CP enzymes have also received scientific attention in the medical field for their role in allergenicity. Clinical studies have suggested that actinidin is the major kiwifruit protein responsible for allergic reactions in some European populations (Pastorello et al., 1998; Gavrovic-Jankulovic et al., 2005; Lucas et al., 2007; Palacin et al., 2008). For these reasons, together with the desire to understand the function of actinidin in planta, we have targeted the nature of CP variation in kiwifruit for study. In this paper, we utilize a gene-rich linkage map in A. chinensis (Fraser et al., 2009) to identify a major quantitative trait locus (QTL) for CP activity and show that it colocates with the gene encoding the major acidic isoform of actinidin. Complementation of ACT1A mutations in transgenic YellowA fruit allowed the physiological consequences of increased CP activity in fruit to be investigated.
RESULTS
QTL Mapping of Fruit CP Activity in an A. chinensis Mapping Population
CP activity was studied in an F1 population of 272 individuals obtained from an intraspecific cross (female parent, CK51_05, designated mpM; male parent, CK15_02, designated mpD) within the diploid kiwifruit species A. chinensis. This population was used in the recently developed Actinidia linkage map (Fraser et al., 2009), and preliminary data indicated that CP activity segregated within this population (data not shown). As all Actinidia species are functionally dioecious, CP activity measurements could only be made on fruit from the females (n = 133) in the mapping population. A high-throughput fluorescence assay was used to measure CP activity in individual plants.
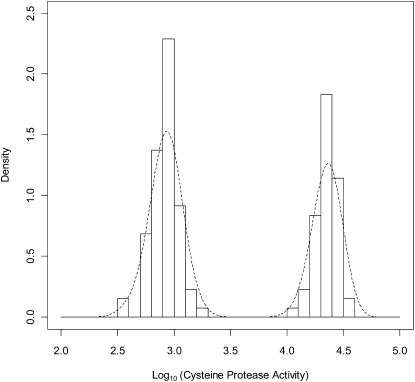
The results (Fig. 1) showed clear evidence of a bimodal distribution: a population with high CP activity (76 plants, at an activity level comparable to that found in green-fleshed Hayward) and a population with low activity (57 plants, at a level comparable to that found in the commercial yellow-fleshed YellowA). Each distribution appeared to be symmetrical and with similar variances. The vines were classified as either “low” CP (log10 [CP activity] ≤ 3.7) or “high” CP (>3.7), and the goodness of fit of the observed counts to a 1:1 ratio was measured by Pearson χ2 (P = 0.097) and by the binomial exact test (P = 0.155). The female parent of the mapping population (mpM) also fell within the low CP activity class. CP activity scores for all individual members of the mapping population and other selected genotypes are given in Supplemental Table S1.
Figure 1.
Distribution of CP activities in female members of the A. chinensis mapping population. Activities were measured with a high-throughput fluorescence assay with Z-Phe-Arg-7-amido-4-methyl coumarin as substrate according to Rassam and Laing (2004). A smoothed density plot is overlaid on the histogram of log10 (CP activity). The activities show a distribution into two CP activity classes: low CP = log10 (CP activity) ≤ 3.7; high CP = log10 (CP activity) > 3.7.
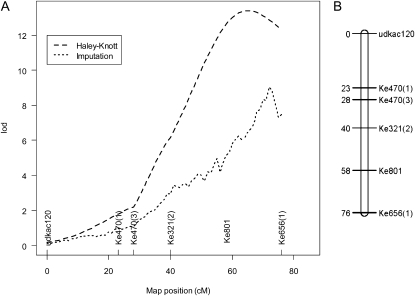
The log10 (CP activity) data were used to map putative QTLs onto the A. chinensis linkage map (Fraser et al., 2009). Single-marker analysis indicated significant associations only with markers on linkage group (LG) 16; hence, further detection of QTLs via interval mapping was restricted to this group. Single QTL scans of LG16 were performed by interval mapping using two different methods: Haley-Knott (Haley and Knott, 1992) and imputation (Sen and Churchill, 2001). The log of the odds (LOD) curves are shown in Figure 2A. Each method produced similar results, placing the QTL at approximately 65 centimorgan (cM; LOD = 13.4) and at approximately 72 cM (LOD = 9.0) for the Haley-Knott and imputation methods, respectively. The LOD thresholds calculated by permutation tests were 3.3 and 2.4, respectively, at a type 1 error rate of 0.1%, providing strong evidence for a putative QTL at this position. Estimates of 95% confidence limits of the putative QTL position determined by the 1-LOD dropoff method were 57 to 76 cM (Haley-Knott) and 69 to 75 cM (imputation). The single putative QTL explained 38% and 27% of the variation in log10 (CP activity) by the two methods, respectively.
Figure 2.
QTL mapping of fruit CP activity and the Ke801 marker in the A. chinensis mapping population. A, LOD score curves for single QTL mapping of the log10 (CP activity) phenotype on LG16 calculated using Haley-Knott regression (Haley and Knott, 1992) and the multiple imputation methods of Sen and Churchill (2001). n = 133. QTL mapping was conducted leaving out the Ke801 candidate gene marker. B, Map position of the Ke801 polymorphism in the ACT1A locus on LG16. The Ke801 primer set was developed to the ACT1A-1 cDNA encoding the major acidic actinidin isoform in Hayward. A single-base insertion/deletion identified between the male and female parents of the A. chinensis mapping population was used to place the ACT1A locus on LG16.
The Gene Encoding the ACT1A-1 cDNA Colocates with the CP Activity QTL
The 1:1 segregation pattern of CP activity in the mapping population suggested that a single gene might account for the major QTL on LG16. Previously, Nieuwenhuizen et al. (2007) showed that two actinidin mRNAs (ACT1A and ACT2A) were expressed at similar levels in the fruit of both Hayward and YellowA varieties. The acidic protein isoform encoded by ACT1A was detected at high levels in Hayward but was absent in YellowA. Therefore, ACT1A was targeted as a leading candidate gene to explain the CP segregation pattern in the A. chinensis mapping population. Primers were designed (Ke801; Supplemental Table S1) to cover part of the 5′ promoter region and the first exon of the ACT1A-1 genomic sequence in Hayward (Snowden and Gardner, 1990; Lin et al., 1993). Screening of the parents of the mapping population and a small number of progeny plants indicated that the Ke801 marker was polymorphic and appeared to segregate with the CP phenotype.
The Ke801 primers amplified a single band of 355 bp in the male parent (mpD) of the mapping population and a 356-bp band in the female parent (mpM). The segregation of these bands in the progeny indicated that mpD and mpM each carried a second nonamplifying (or “null”) allele (designated D0 and M0). The segregation pattern of the 355- and 356-bp bands and the D0 and M0 null alleles in the 272 progeny of the mapping population is given in Table I. The Ke801 marker was mapped using JoinMap software to LG16 on the A. chinensis linkage map between Ke321(2) and Ke656(1) at approximately 58 cM (Fig. 2B). The Ke801 marker mapped within the 95% confidence interval of the putative log10 (CP activity) QTL (57–76 cM) estimated by the Haley-Knott method but slightly outside the confidence interval estimated by the imputation method (69–75 cM). Given the level of genotype information in the vicinity, the QTL scanning methods tested spanned the position of the Ke801 marker reasonably well.
Table I. Bands amplified from ACT1A alleles in the A. chinensis mapping population (mp) using Ke801 primers.
Nonamplifying (null) alleles from the male (mpD) and female (mpM) parents were designated D0 and M0, respectively.
| Plant | Band | Band | No. Male | No. Female | No. Total |
| bp | |||||
| mpD | 355 | D0 | |||
| mpM | 356 | M0 | |||
| mp, genotype A | 356 | 355 | 29 | 42 | 71 |
| mp, genotype B | 355 | M0 | 39 | 31 | 70 |
| mp, genotype C | 356 | D0 | 35 | 35 | 70 |
| mp, genotype D | D0 | M0 | 36 | 25 | 61 |
| 272 | |||||
A comparison of the CP activity phenotype and the Ke801 genotyping data for the 133 female members of the mapping population is shown in Table II. Only plants carrying the D0 null allele inherited from the male parent (mpD) showed high CP activity. All plants inheriting the D0/M0 null combination showed high CP activity, while 32 of 35 plants carrying the D0/356-bp band combination showed high CP activity. All plants inheriting the paternal 355-bp band (in combination with the maternal 356-bp band or M0 null) showed low CP activity.
Table II. Bands amplified from ACT1A alleles in females from the A. chinensis mapping population (mp) using Ke801 primers compared with the CP activity in fruit.
Low CP = log10 (CP activity) ≤ 3.7; high CP = log10 (CP activity) > 3.7. Nonamplifying (null) alleles from the male (mpD) and female (mpM) parents were designated D0 and M0, respectively.
| Plant | Band | Band | No. High CP Activity | No. Low CP Activity | No. Total |
| bp | |||||
| mp, genotype A | 356 | 355 | 0 | 42 | 42 |
| mp, genotype B | 355 | M0 | 0 | 31 | 31 |
| mp, genotype C | 356 | D0 | 32 | 3 | 35 |
| mp, genotype D | D0 | M0 | 25 | 0 | 25 |
| 57 | 76 | 133 | |||
As the D0 and M0 null alleles did not amplify a PCR product with the Ke801 primers, a new set of primers (KeAct1) was designed to discriminate between all four possible allelic combinations. Two bands of 321 and 323 bp were amplified in mpD, while from mpM bands of 323 and 325 bp were amplified (Table III). Thirty-two representative plants from the mapping population containing the four possible combinations of ACT1A alleles (eight plants per combination) were also tested with the new primer set (Table III, genotypes A–D). These results allowed a characteristic set of bands to be assigned to each of the alleles identified in mpD and mpM (Table IV).
Table III. Bands amplified from ACT1A alleles using the Ke801 and KeAct1 primer sets in the A. chinensis mapping population (mp) and selected kiwifruit genotypes.
Low CP = log10 (CP activity) ≤ 3.7; high CP = log10 (CP activity) > 3.7. Nonamplifying (null) alleles from the male (mpD) and female (mpM) parents were designated D0 and M0, respectively. nd, No data obtained from male plants. CK01_01_01_01 and CK15_01 are the female and male parents of YellowA, respectively.
| Plant | CP Activity | Ke801 |
KeAct1 |
||
| Band | Band | Band | Band | ||
| bp | |||||
| mpD | nd | 355 | D0 | 323 | 321 |
| mpM | Low | 356 | M0 | 325 | 323 |
| mp, genotype A | Low | 356 | 355 | 323 | 321 |
| mp, genotype B | Low | 355 | M0 | 325 | 321 |
| mp, genotype C | High | D0 | M0 | 325 | 323 |
| mp, genotype D | High | 356 | D0 | 323 | |
| mp210 | Low | 356 | D0 | 325 | 323 |
| mp218 | Low | 356 | D0 | 323 | |
| mp266 | Low | 356 | D0 | 323 | |
| YellowA | Low | 356 | 347 | 325 | 323 |
| CK01_01_01_01 | High | 347 | 325 | 323 | |
| CK15_01 | nd | 356 | 323 | ||
| Hayward | High | 356 | 323 | ||
Table IV. Characteristic bands expected from sequencing data to amplify for each ACT1A allele using the Ke801 and KeAct1 primer sets.
Nonamplifying (null) alleles from the male (mpD) and female (mpM) parents were designated D0 and M0, respectively.
| Allele | Ke801 Band | KeAct1 Band | Functional Gene | Source |
| bp | ||||
| ACT1A-1 | 356 | 323 | Yes | Hayward |
| ACT1A-2 | D0 | 323 | Yes | mpD |
| act1a-3 | 355 | 321 | No | mpD |
| act1a-4 | M0 | 325 | No | mpM |
| act1a-5 | 356 | 323 | No | mpM |
| act1a-6 | 356 | 323 | No | YellowA |
| act1a-7 | 347 | 323 | No | YellowA |
| act1a-8 | 356 | 323 | No | mp218, mp210 |
Sequence Analysis of ACT1A Alleles in the A. chinensis Mapping Population
The four alleles at the ACT1A locus present in the A. chinensis mapping population were amplified from the male parent (mpD) and the female parent (mpM), cloned, and sequenced (for a DNA sequence alignment, see Supplemental Fig. S1). The two alleles identified in mpD were designated ACT1A-2 (A-2) and act1a-3 (a-3), and the two from mpM were designated act1a-4 (a-4) and act1a-5 (a-5). The PCR product corresponding to the male-derived A-2 allele was 2,783 bp and appeared to encode a functional protein. The second male allele (a-3) was 3,190 bp and contained a 1-bp deletion at position 1,285 in exon 2 relative to the consensus ACT1A sequence. From the female parent, the two alleles were 2,968 bp and 3,095 bp, respectively. The a-4 allele contained a 2-bp insertion in exon 3 (position 1,509), while a-5 contained a 2-bp deletion in exon 1 (position 459) and a splice site mutation (position 2,906) at the intron 3/exon 4 boundary. Neither allele (a-4 or a-5) isolated from the mpM would be able to encode a functional protein because of a frameshift (consistent with low CP activity observed in mpM fruit). The a-3 allele from the male parent would also be inactive because of a frameshift.
When compared with the phenotyping results, and based on a genetic model with one functional allele (A-2) derived from mpD, three plants gave anomalous genotypes when scored with the Ke801 primers (mp210, mp218, and mp266; Table III). All three plants showed a D0 null/356-bp genotype (A-2/a-5 alleles), which should give high CP activity; instead though, the fruit showed low CP activity. The ACT1A alleles present in mp210, mp218, and mp266 were PCR amplified, cloned, and sequenced. The results indicated that the functional A-2 allele was not present in mp210, mp218, or mp266, which is consistent with the low CP activities found in these plants. Mp266 contained nonfunctional paternal a-3 and maternal a-5 alleles; mp218 contained a nonfunctional maternal a-5 allele and a novel allele designated act1a-8 (a-8; for a DNA alignment, see Supplemental Fig. S1); and mp210 contained a nonfunctional maternal a-4 allele and the a-8 allele. The a-8 allele contained the same 2-bp deletion in exon 1 (position 459) found in a-5, indicating that this allele would not produce a functional protein.
Analysis of ACT1A Alleles in YellowA Kiwifruit
The YellowA kiwifruit variety has been shown to contain low actinidin protein levels and CP activity (Nieuwenhuizen et al., 2007). The male parent of YellowA (CK15_01) and the male parent of the A. chinensis mapping population (mpD; CK15_02) are full siblings. YellowA and both its parents were genotyped using the Ke801 and KeAct1 primer sets to determine if ACT1A alleles were shared with mpD. YellowA amplified 356- and 347-bp bands with Ke801 primers and bands of 323 and 325 bp with KeAct1 primers (Table III). The 347-bp band was different from any band amplified from the mapping population and was inherited from the female parent of YellowA (CK01_01_01_01), which was shown to contain high CP activity. The 356-bp band was inherited from the male parent of YellowA (CK15_01), which only amplifies the 356-bp band.
Although YellowA and the mapping population both amplify a 356-bp band, sequence results indicated that YellowA contained two unique alleles (designated act1a-6 [a-6] and act1a-7 [a-7]; for a DNA alignment, see Supplemental Fig. S1) that were not present in the A. chinensis mapping population. The a-6 allele was 3,478 bp and contained a 383-bp insertion at position 1,585 in exon 3 relative to the consensus ACT1A sequence. This large insertion would disrupt the formation of a functional protein. The a-6 allele is expressed in fruit, as a partial EST was identified in an A. chinensis young YellowA fruit library (GenBank accession nos. FG514838 and GU201527). This EST was polyadenylated 305 bp into the insertion. Interestingly, the a-8 allele found in mp210 and mp218 also contains the 383-bp insertion and appears to be a chimera of the a-5 and a-6 alleles.
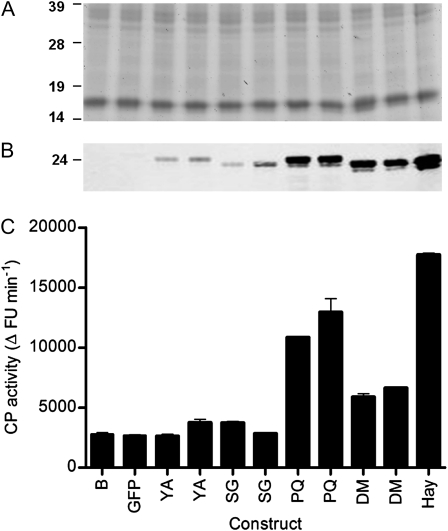
The a-7 allele in YellowA was 2,975 bp and appeared to encode a functional enzyme. The predicted open reading frame encoded a protein that showed only four amino acid differences compared with the functional A-2 allele found in the A. chinensis mapping population. Although the YellowA a-7 allele encodes a complete open reading frame, YellowA fruit shows low CP activity. We hypothesized that one or more amino acid substitutions in the a-7 allele may have led to the production of an inactive enzyme. To test this hypothesis, the complete act1a-7 cDNA (GenBank accession no. GU201528) was amplified from mature YellowA fruit and transiently expressed in Nicotiana benthamiana leaves. The functional ACT1A-1 cDNA from Hayward was used as a positive control. In leaves infiltrated with the binary vector constructs pAct1a-7 and pAct1A-1, actinidin protein was observed by western analysis, albeit at lower amounts in plants inoculated with pAct1a-7 (Fig. 3, A and B). However, the CP activity of leaves inoculated with pAct1a-7 was low and similar to that from control inoculations with buffer or a pGFP control construct, while high CP activity was observed for the pAct1A-1 construct (Fig. 3C). These results indicate that the a-7 allele produces an mRNA that is translated and that accumulates in N. benthamiana, but the protein is inactive. In YellowA fruit, the acidic actinidin isoform does not naturally accumulate (Nieuwenhuizen et al., 2007), suggesting that the inactive act1a-7 protein is subject to degradation in mature kiwifruit.
Figure 3.
Transient expression of act1a-7 and site-directed mutants in N. benthamiana leaves. A, Proteins were separated by 1D SDS-PAGE (20 μg per lane). Molecular mass standards (in kD) were SeeBluePlus2 (Invitrogen). B, Western analysis of actinidin levels in N. benthamiana leaves using an actinidin-specific polyclonal antibody. C, CP activities. Activities were measured in quadruplicate for each sample using the fluorescence assay of Rassam and Laing (2004). Data are normalized to 0.5 mg mL−1 protein per assay. Two clones were independently generated and assayed for each mutant construct. Samples are as follows: B, buffer control; GFP, GFP control; YA, act1a-7 from YellowA; SG, act1a-7 S68G site-directed single mutant; PQ, act1a-7 P75Q site-directed single mutant; DM, act1a-7 S68G/P75Q site-directed double mutant; Hay, ACT1A-1 from Hayward. FU, Fluorescence units.
Structural Basis for the Lack of CP Activity in the act1a-7 Protein
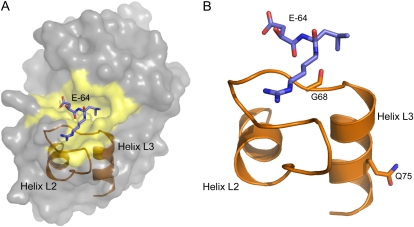
To investigate the structural basis for the lack of CP activity in the act1a-7 protein, a homology model of the active ACT1A-2 enzyme (Fig. 4A) was generated using the crystal structure of ACT1A-1 from Hayward complexed with E-64, a potent and highly selective irreversible inhibitor of CPs, as the template (Varughese et al., 1992). The model was used to identify the topological positioning of amino acids that differed between ACT1A-2 and act1a-7 and, hence, the rational design of mutants, to investigate the absence of CP activity in act1a-7. Of the four amino acid differences between act1a-7 and ACT1A-2, two in particular were identified as mutagenesis candidates (Fig. 4B): an active site Ser (Ser-68) residue and a more distantly located Pro (Pro-75) residue in act1a-7 (corresponding to Gly-68 and Gln-75 residues in ACT1A-2). These residues were targeted in act1a-7 to construct separate single site S68G and P75Q mutants and the double S68G/P75Q mutant.
Figure 4.
ACT1A-2 protein modeling. A, A representation of the ACT1A-2 protein structure based on the ACT1A-1 crystal structure (Varughese et al., 1992). The E-64 inhibitor (purple), active site region (yellow), and the L2 and L3 helices, which help to form part of its topology, are also indicated. B, The positioning of the active-site Gly (G68) and more distal Gln (Q75) residues implicated in ACT1A-2 activity. Helices are named according to the nomenclature of Kamphuis et al. (1984), and the E-64 inhibitor is positioned according to that in the ACT1A-1 crystal structure (Protein Data Bank code 1AEC).
The mutated site act1a-7 cDNAs were generated by overlap extension PCR (Warrens et al., 1997), cloned into binary vectors, and transiently expressed in N. benthamiana. The CP activities of the respective mutant enzymes were measured, and the accumulation of actinidin protein was determined by western analysis. The S68G and S68G/P75Q mutants produced proteins of identical size to the Hayward ACT1A-1 protein but slightly smaller than the YellowA act1a-7 protein. The P75Q mutant produced a predominant protein of equivalent size to the YellowA act1a-7 protein (Fig. 3B). CP activity assays indicated that the S68G mutant was inactive, while the P75Q and S68G/P75Q mutants were now active (Fig. 3C). CP activities for the P75Q and S68G/P75Q mutants were lower than those for the Hayward ACT1A-1 enzyme when expressed on a CP activity per fresh weight basis, possibly because of the different binary vectors used (pART27 for ACT1A-1 versus pGreenII for act1a-7). However, when correcting for the difference in protein accumulation, the S68G/P75Q double mutant showed similar specific activity to the Hayward ACT1A-1 enzyme, while the P75Q single mutant was about 2.5 times more active.
These data show that the P75Q mutation is sufficient to restore CP activity. The double mutant shows a protein of the correct size and with specific activity restored to the ACT1A-1 level.
Complementation of ACT1A Mutations in Transgenic Fruit
If the low-CP phenotype in YellowA is caused by the defective ACT1A alleles, high CP levels should be restored by complementation with a functional actinidin gene copy. To test this hypothesis, transgenic YellowA plants were produced that overexpressed the Hayward ACT1A-1 cDNA under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter. Multiple transgenic lines were generated by Agrobacterium tumefaciens-mediated transformation, and fruit were obtained from four transgenic plants. Under containment greenhouse conditions, no differences in vegetative growth and the physical appearance of flowers or fruit were observed between transgenic and control plants.
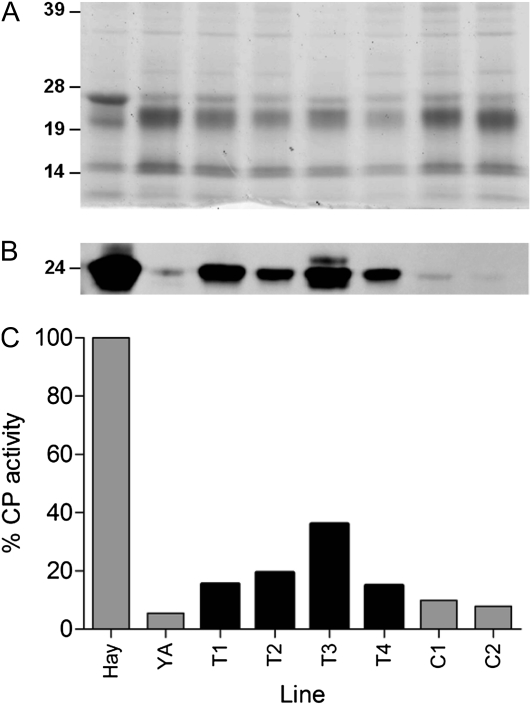
To confirm the transgenic nature of the transgenic plants and to determine the number of T-DNA inserts in the four fruiting lines (designated T1–T4), quantitative PCR was employed using primers specific to the Hayward ACT1A-1 transgene. After comparison with amplification from a single-copy reference gene, the copy number of the transgenic lines was estimated to range between one and four copies per line (Supplemental Table S2). Line T3 appeared to contain four T-DNA copies, while line T4 appeared to contain a single copy insert.
Transgenic fruit were analyzed for the accumulation of actinidin protein by western analysis (Fig. 5, A and B). All four transgenic YellowA plants accumulated actinidin protein at much higher levels than control plants and compared with wild-type YellowA but less than the very high levels observed in control Hayward fruit. Fruit from all transgenic lines exhibited increases in CP activity (Fig. 5C) compared with fruit from YellowA and control transgenic lines. The best transgenic line, T3, showed at least four times the activity of transgenic controls and restored CP activity to approximately 40% that of Hayward. These results indicate that the Hayward ACT1A-1 cDNA can partially complement the mutations observed in the YellowA a-6 and a-7 alleles.
Figure 5.
Molecular characterization of transgenic YellowA fruit overexpressing the ACT1A-1 cDNA from Hayward. A, Proteins were extracted from mature fruit and separated by 1D SDS-PAGE (2 μg per lane). Molecular mass standards (in kD) were SeeBluePlus2 (Invitrogen). B, Western analysis of actinidin levels in wild-type, control, and transgenic fruit. Actinidin was detected using an actinidin-specific polyclonal antibody. C, CP activities were measured using the fluorescence assay of Rassam and Laing (2004). Data were normalized to 0.1 mg mL−1 protein per assay and are shown with the activity of Hayward set at 100%. Samples are as follows: Hay, Hayward wild type; YA, YellowA wild type; T1 to T4, transgenic YellowA lines overexpressing the ACT1A-1 cDNA from Hayward; C1 and C2, control YellowA lines.
Consequences of Increased CP Activity in Transgenic Fruit
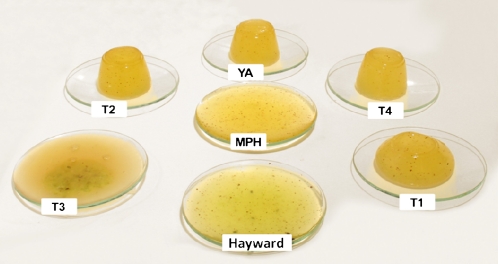
The consequences of complementing the defective ACT1A alleles and restoring high levels of CP activity in the YellowA background were initially investigated in vitro using gel-setting assays. Homogenized fruit extracts were mixed with gelatin, poured into plastic molds, and allowed to set at 4°C. The jellies were then inverted from the molds and incubated at 24°C for 24 h (Fig. 6). Fruit extracts with high CP activity (e.g. Hayward) rapidly degraded the gelatin, and the jellies collapsed. Jellies containing YellowA fruit extracts with low CP activity retained their shape. The fruit extract from the T3 transgenic line with the highest CP activity degraded the gelatin fully within 24 h (Fig. 6). T1 fruit extracts partially degraded the gelatin in 24 h, while longer incubations were required to observe degradation by extracts from the T2 and T4 lines (data not shown).
Figure 6.
Complementation of ACT1A mutations in transgenic YellowA fruit leads to an increase in the degradation of gelatin-based jellies in vitro. Homogenized fruit extracts and a gelatin solution were mixed, poured into plastic molds, and set at 4°C. Jellies were then inverted onto watch glasses, transferred to an incubator at 24°C, and monitored over 24 h. Samples are as follows: Hayward wild type; YA, YellowA wild type; T1 to T4, transgenic YellowA lines overexpressing the ACT1A-1 cDNA from Hayward; MPH, mp181 from the A. chinensis mapping population with high CP activity. [See online article for color version of this figure.]
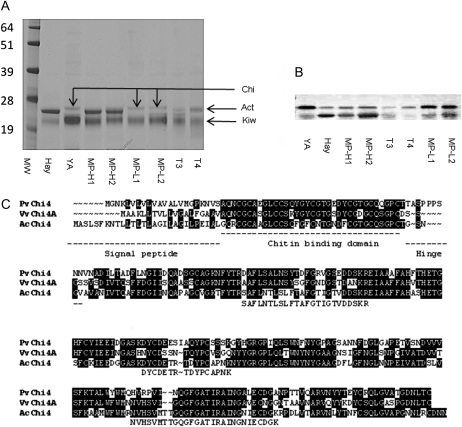
To identify protein targets for the CP activity of actinidin in planta, one-dimensional (1D) SDS-PAGE gels were used to separate proteins from the fruit of transgenic lines T3 and T4, YellowA, Hayward, and members of the A. chinensis mapping population with either high or low CP activity (Fig. 7A). All extractions were performed in the presence of protease inhibitors to protect proteins from the CP activity of actinidin during the extraction process. The three samples that contained low levels of actinidin (YellowA, MP-L1, and MP-L2) shared a common band of approximately 27 kD that was absent from all the other samples that contained actinidin (including the transgenic lines T3 and T4). The band was excised from YellowA and subjected to liquid chromatography-mass spectrometry (LC-MS) analysis, which revealed the presence of multiple peptides that showed strong homology to class IV chitinase proteins. The peptide fragments also matched the deduced amino acid sequence of a contig of ESTs from A. chinensis (Fig. 7C). One full-length EST from this contig was sequenced and designated AcChi4.
Figure 7.
Complementation of ACT1A mutations in transgenic YellowA fruit leads to an increase in processing of chitinase in planta. A, 1D SDS-PAGE of proteins extracted from mature fruit (2 μg per lane). The band at approximately 27 kD (arrow Chi) in YA, MP-L1, and MP-L2 was identified by LC-MS from YellowA as showing homology to class IV chitinases. The bands at approximately 24 kD (arrow Act) and approximately 22 kD (arrow Kiw) correspond to actinidin (Nieuwenhuizen et al., 2007) and kiwellin (Tamburrini et al., 2005) proteins, respectively. B, Western analysis using a chitinase-specific polyclonal antibody. C, Comparison of the deduced amino acid sequences of class IV chitinases from kiwifruit (AcChi4; GenBank accession no. JN701537), bean (PvChi4; X57187), and grape (VvChi4A; U97522). Residues highlighted in black were conserved in at least two of the sequences. The tryptic peptide fragments obtained from LC-MS analysis are aligned below the AcChi4 sequence. The signal peptide, chitin-binding domain, and hinge region reported for the grape and bean proteins are underlined (Lange et al., 1996; Robinson et al., 1997). Samples are as follows: MW, molecular mass standards (in kD; SeeBluePlus2 [Invitrogen]); Hay, Hayward wild type; YA, YellowA wild type; MP-H1 and MP-H2, mp181 and mp175 from the A. chinensis mapping population with high CP activity; MP-L1 and MP-L2, mp105 and mp98 from the A. chinensis mapping population with low CP activity; T3 and T4, transgenic YellowA lines overexpressing the ACT1A-1 cDNA from Hayward.
The sequence of AcChi4 was compared with class IV chitinases from grape (Vitis vinifera) and bean (Phaseolus vulgaris; Lange et al., 1996; Robinson et al., 1997). These alignments (Fig. 7C) suggested that the AcChi4 protein was likely to be synthesized as an approximately 30-kD protein that would be cleaved initially to remove a signal peptide. Further processing at a hinge region would be expected to remove a chitin-binding domain of approximately 4 kD, leaving a catalytic domain of approximately 23 kD (Fig. 7C). Western analysis using a polyclonal chitinase antibody (Fig. 7B) showed that the major band for chitinase in fruit of YellowA and members of the A. chinensis mapping population with low actinidin levels was larger (by approximately 4 kD) than the major isoform present in Hayward and members of the A. chinensis mapping population with high actinidin levels. The banding pattern of transgenic plant T4 (with low levels of actindin and CP activity) resembled YellowA, while the transgenic line T3 (with higher levels of CP activity) showed a reduction in the upper band and greater accumulation of the lower band. The intensity of the lower band was not as strong as observed in Hayward, perhaps reflecting that the T3 line is only partially complemented for CP activity.
DISCUSSION
In the past 5 years, significant new tools have become available to understand the genetic control of key consumer and production traits in fruit and to facilitate fast, accurate, and efficient development of new varieties. These tools include full genome sequences for grape (Jaillon et al., 2007) and papaya (Ming et al., 2008) and large EST collections for apple (Malus domestica; Newcomb et al., 2006), tomato (Fei et al., 2004), and grape (Peng et al., 2007). In kiwifruit, a large database of EST sequences has also become available (Crowhurst et al., 2008), along with efficient transformation systems for assessing gene function (Wang et al., 2006, 2007b) and a gene-rich linkage map in A. chinensis (Fraser et al., 2009). In this paper, we utilize all these genomic tools to gain insight into the genetic control of CP activity in kiwifruit. In contrast to most other fruit quality traits in Actinidia, CP activity is shown to be controlled by a single major QTL located on LG16 that colocates with the gene encoding the major acidic CP actinidin.
Mapping QTLs and genes in an obligate outbreeding species, such as A. chinensis, is a more challenging task than mapping in species using inbred or homozygous parents. Two maps are used, one for the male parent and one for the female parent, and these maps are integrated using markers that are heterozygous in both parents (Fraser et al., 2009). The major QTL for fruit CP activity was located on the male map at position approximately 65 cM on LG16. The accuracy of this map position was restricted by the low density of markers in the region and the relatively small number of female plants (n = 133) available in the cross. We have subsequently performed two-dimensional scanning for putative QTLs of the log10 (CP activity) phenotype on LG16 by the imputation method (Sen and Churchill, 2001). The only high LOD scores were achieved at the very distal end of LG16, and within a narrow range, providing insufficient evidence for distinctively positioned additive QTLs (Supplemental Fig. S2A, below the diagonal plot). LOD scores were also not significant for a model based on two interacting QTLs (Supplemental Fig. S2A, above the diagonal plot). We also found no evidence of a second QTL on LG16 using the imputation method, where we assumed that 58 cM was the true position of the putative CP activity QTL (Supplemental Fig. S2B).
These mapping data do not preclude the possibility that there are multiple actinidin genes at (or near) the ACT1A locus on LG16. Two plants in the mapping population (mp210 and mp218) were identified that contained a novel a-8 allele that appeared to be a chimera of the a-5 allele sequenced from the mapping population and the a-6 allele sequenced from YellowA. Although it is possible that these two plants are derived from stray pollen in the original cross to generate the mapping population, or that the sequences were derived from DNA contamination, we have no evidence to indicate that this is the case, using other markers or sequencing information. The possibility of more than one actinidin gene copy is also suggested by the genotyping data with the KeAct1a primer set, where YellowA amplifies bands of 325 and 323 bp, even though sequencing data indicate that the a-6 and a-7 alleles will only amplify a band of 323 bp with KeAct1a primers. The nature and complexity of the ACT1A locus will be better understood once an Actinidia genome sequence is completed.
Homology modeling in conjunction with site-directed mutagenesis revealed the structural basis for the lack of CP activity in the act1a-7 protein. In act1a-7, a single nonsynonymous nucleotide substitution (CAG to CCG) results in the conversion of the wild-type Gln (Gln-75) residue to a Pro. A P75Q “reverse” mutation was sufficient to restore CP activity to the act1a-7 protein. Although the Pro-75 in act1a-7 is removed from the active site, it is located in a conserved L3 helix. The presence of a conformationally rigid Pro within the L3 helix has the potential to destabilize this helix and hence alter the positioning of a catalytic Gly (Gly-68 in ACT1A-2), which is not only conserved in many other CPs (Berti and Storer, 1995) but also directly implicated in substrate H-bond interactions (Patel et al., 1992). The single-site S68G reverse mutation was not sufficient to restore activity to the act1a-7 protein. However, the S68G mutant, the S68G/P75Q double mutant, and Hayward ACT1A-1 proteins all ran at a slightly lower molecular mass to the act1a-7 protein and the P75Q mutant. The reason for the difference (approximately 3 kD) in molecular mass is unclear but appears to be due to a change in protein folding, as LC-MS analysis of the recombinant S68G mutant did not reveal any differences in protein processing (data not shown). Actinidin contains a number of Cys bonds, and even under fully denaturing conditions, it is difficult to get actinidin to resolve at the predicted size (Larocca et al., 2010).
In the A. chinensis mapping population, high CP activity segregates as a single dominant Mendelian trait that is associated with the presence of the ACT1A-2 allele. Three ACT1A alleles in the mapping population and two further alleles present in the commercial variety YellowA were shown to be inactive, either by insertion/deletion, leading to protein frameshifts, or by a point mutation in the CP active site, leading to the formation of an inactive protein. Rapid evolution through mutation and gain/loss of activities is a characteristic of genes involved in pathogen defense. This has been shown for chitinases (Bishop et al., 2000), β-1-3-endoglucanases (Bishop et al., 2005), polygalacturonases (Stotz et al., 2000), protease inhibitors, and other CPs (Moeller and Tiffin, 2005; Tiffin and Moeller, 2006). Actinidin may also be directly involved in pathogen defense, as it has been shown to have a small detrimental effect on the growth of neonate leaf worm larvae (Malone et al., 2005).
Our results indicate that actinidin may also play a role in pathogen defense through the processing of a class IV chitinase in planta. Chitinases are well-known pathogenesis proteins that can be induced by a range of stress conditions, both biotic and abiotic, and by plant growth regulators such as ethylene and salicylic acid (Kasprzewska, 2003). Some chitinases have been shown to have direct antifungal activity in vitro (Schlumbaum et al., 1986; Mauch et al., 1988). In green-fleshed kiwifruit, chitinase activity has been shown to significantly increase in response to prolonged fruit storage and the pathogen Botrytis cinerea (Wurms et al., 1997). In yellow-fleshed kiwifruit, chitinase activity was higher than in green-fleshed kiwifruit irrespective of fruit maturity, and this correlated with an increase in resistance to B. cinerea (Wurms, 2004). It is tempting to think that the increase in resistance to B. cinerea in yellow-fleshed kiwifruit might be associated with the reduction in processing of AcChi4 by actinidin. Future experiments using the actinidin-overexpressing YellowA plants may help resolve this hypothesis and address what other direct or indirect roles actinidin plays in pathogen defense in kiwifruit.
MATERIALS AND METHODS
Plant Material
Actinidia deliciosa var. deliciosa ‘Hayward’, the Actinidia chinensis var. chinensis mapping population, A. chinensis ‘Hort16A’ (designated here YellowA), and other A. chinensis genotypes were grown in the New Zealand Institute for Plant and Food Research orchard in Te Puke, New Zealand. The male parent (mpD; CK15_02) of the mapping population is a sibling of the male parent (CK15_01) of YellowA. The female parent of the mapping population (mpM; CK51-05) and the female parent of YellowA (CK01_01_01_01) are not closely related.
CP Activity Assays
For fluorescence assays, fruit were collected at approximately 18 weeks after anthesis, when fruit were mature but unripe. Whole fruit were also frozen at −80°C until required. A longitudinal representative wedge of about 2.5 g was cut from each fruit, mixed with 2 volumes of ice-cold extraction buffer (Wein et al., 2002), and homogenized using an Ultra-Turrax T25 Basic homogenizer (IKA Works GmBH & Co.). The supernatant was cleared by centrifugation and assayed immediately on 96-well assay plates using 10 μL of extract in 120 μL of MOPS buffer with Z-Phe-Arg-7-amido-4-methyl coumarin as substrate according to Rassam and Laing (2004). A subset of samples (n = 29) was analyzed in duplicate to confirm the assay reproducibility.
For gel-setting assays, ripe fruit tissue (without peel) was homogenized and mixed 1:1 (w/v) with buffer (0.5 m Tris-HCl, 8 mm EDTA, and 8 mm dithiothreitol, pH 7). A 10% clear gelatin (Davis Gelatin NZ) solution was made up in the same buffer and cooled to 45°C. The fruit slurry (7.5 mL) and gelatin solution (22.5 mL) were mixed and poured into 30-mL plastic molds. Jellies were set at 4°C for 2 h, inverted onto watch glasses, and transferred to an incubator at 24°C.
QTL and Gene-Mapping Analysis
CP activity phenotypic data were log10 transformed to meet the assumptions of homogeneity of variance and normal distribution. An initial genome-wide scan for putative QTLs was carried out by single-marker analysis using a t test. Separate analyses were done with the male and female marker maps, and corresponding P values were determined. Testing of large numbers of hypotheses warranted control of the error rate, and both the Bonferroni correction and false discovery rate (Benjamini and Hochberg, 1995) methods were considered. Any LGs that were indicative of a marker association with the phenotype were further analyzed by interval mapping using the R/qtl package (Broman et al., 2003), which is an add-on to the R statistical software (http://www.r-project.org). Both the single and pairwise genome scan functions were used. Two separate methods for interval mapping were used: Haley-Knott regression (Haley and Knott, 1992) and the multiple imputation method of Sen and Churchill (2001). Again, multiple testing was an issue, and this was handled by computing LOD thresholds as percentiles of maximum LOD scores resulting from analyses of permuted trait data (5,000 permutations) while the marker data remained fixed (Churchill and Doerge, 1994). The 95% confidence intervals of the discovered QTL position were estimated by the 1 − LOD dropoff and bootstrap methods. The proportion of variation explained by the QTL was estimated as 1 − 10−2LOD/n. For relevant LGs, the LOD curves were presented graphically.
Genotyping
Leaf tissue for DNA extractions was taken at bud break, held at 4°C for 24 h, then stored at −80°C until required. DNA was extracted from tissue that was ground to powder in liquid nitrogen before being processed through a DNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. The final eluate was 200 μL in volume, and 5 μL of a 1:10 dilution of this eluate was used in each PCR.
Genotypes were scored using actinidin-specific primer pairs (Supplemental Table S3) in PCR mixes of 15 μL containing 1× PCR buffer (20 mm Tris-HCl and 50 mm KCl), 5 mm MgCl2, 0.2 mm of each deoxyribonucleotide triphosphate, 4.5 pmol of each primer, and 1.25 units of Platinum Taq polymerase (Invitrogen). About 12.5 ng of genomic DNA was added in 5 μL to bring the total PCR volume to 20 μL. PCR was performed in a Techne TC-412 thermal cycler with a single cycle of 94°C for 3 min preceding 35 cycles of denaturing at 94°C for 30 s, annealing for 30 s (at 57°C for Ke801 primers and 58°C for KeAct1 primers), and elongation at 72°C for 1 min. PCR was carried out individually before three color multiplexes of products labeled with the dyes 6FAM, VIC, or NED (filter set D) were prepared for analysis. The allelic content of each plant was determined by capillary electrophoresis in an ABI Prism 3100 Genetic Analyzer (filter set D; ROX size standard) and analyzed with GeneMapper Software version 3.7 (Applied Biosystems). All alleles were independently verified by at least two people.
Structural Modeling
Three-dimensional structure predictions were carried out using SWISS-MODEL (Arnold et al., 2006), available on the Expasy Web site (http://www.expasy.org). The program was run using project mode, and the template selected was the crystal structure of the A. deliciosa actinidin-E-64 complex (Protein Data Bank code 1AEC; Varughese et al., 1992). Template and target sequences were between 98.4% and 98.7% identical. The semirefined model was then sent to the SWISS-MODEL server for final refinement. Images were generated in the molecular graphics and modeling package Pymol (http://www.pymol.org/).
Binary Vector Construction and Site-Directed Mutagenesis
The actinidin cDNA clone pKIWI450 (Podivinsky et al., 1989) isolated from A. deliciosa Hayward (corresponding to ACT1A-1) was digested with XbaI and EcoRI and cloned into the corresponding sites of pART7 (Gleave, 1992). This construct was digested with NotI, and the insert was cloned into the binary vector pART27 (Gleave, 1992) to form pAct1A-1. The full-length act1a-7 cDNA was amplified from mRNA isolated from YellowA fruit using primers Actc1F1 (5′-CCCAATCTTTTTCTTCTAAAATTCA-3′) and Actc1R2 (5′-GGACAAACAAATCCCCTGAA-3′) and cloned into pCR-Blunt II-TOPO (Invitrogen) to generate pCRII-Act1a-7. The insert was then PCR amplified using the universal M13F and M13R vector primers, digested with PstI and HindIII, and cloned into the binary vector pGreenII 62-SK (Hellens et al., 2005) to generate pAct1a-7. pGFP was created by recombining the pENTR-GFP entry vector into the pHEX2 destination binary vector according to the manufacturer’s instructions (Invitrogen). pART27 and pHEX2 contain the CaMV 35S promoter and octopine synthase terminator (Gleave, 1992), while pGreenII contains the CaMV 35S promoter and terminator sequences (Hellens et al., 2005).
Act1a-7 site-directed mutants were generated from pCRII-Act1a-7 by overlap extension PCR (Warrens et al., 1997). The universal vector primers M13F and M13R were used in combination with the forward and reverse complement mutation primers 16A_GF (5′-ACCAGGGGCTGCAATGGCGGTTACATAACCGACG-3′), 16A_QF (5′-ATAACCGACGGGTTTCAGTTCATCATCAACAAC-3′), and 16A_G/QF (5′-AATGGCGGTTACATAACCGACGGGTTTCAGTTCATC-3′), to generate two overlapping mutated gene fragments. In the second stage, the two overlapping fragments were mixed and PCR amplified with primers M13F and M13R. The full-length mutated gene products were digested with PstI and HindIII, gel purified, cloned into the pGreenII 62-SK binary vector, and sequence verified.
Transient Expression, Stable Plant Transformation, and Growth
For transient expression in Nicotiana benthamiana, 6-week-old greenhouse-grown seedlings were infiltrated with Agrobacterium tumefaciens strain GV3101 harboring the binary vector of choice with the helper plasmid pSOUP vector as described previously (Nieuwenhuizen et al., 2007). After 14 d, the leaves were detached, and protein was extracted for western analysis and to determine CP activity.
Transgenic A. chinensis YellowA shoots were produced using the method of Fraser et al. (1995) using the pAct1A-1 binary vector after electroporation into the A. tumefaciens strain A281. Transgenic and control lines were maintained in a containment greenhouse under ambient light and temperature conditions. Plants received 8 weeks of winter chilling at 7°C each year. Flowers were hand pollinated each spring, and mature unripe fruit were harvested at approximately 18 weeks post anthesis.
Allele Sequencing and Copy Number Estimation
Actinidin alleles for sequencing were amplified with the primers given in Supplemental Table S3 using Pwo SuperYield polymerase (Roche Applied Science) and transferred by blunt TOPO cloning into pCR-Blunt II-TOPO (Invitrogen) according to the manufacturer’s instructions. Multiple clones (n = 3–8) of all alleles were verified by full-length DNA sequencing.
Transgene copy numbers were determined using the LightCycler 480 Real-Time PCR System (Roche) according to Nieuwenhuizen et al. (2009) using 1 ng of genomic DNA per reaction. Genomic DNA was isolated from transgenic leaves using the DNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. The transgene was amplified with two independent primer sets specific for the Hayward actinidin ACT1A-1 cDNA in the pAct1A-1 binary vector (Actc1RT_F/R and Actc1RT_F2/R2; Supplemental Table S3) and compared with the amplification from a reference kiwifruit gene amplified with AlfyRT_F/R primers (Supplemental Table S3). Transgene copy numbers were determined using the LightCycler 480 software with the presumption that diploid A. chinensis contains two copies of the reference gene.
Western Analysis and LC-MS
Protein was extracted from fruit tissues ground to powder with mortar and pestle under liquid nitrogen. Powder (500 mg) was mixed with polyvinylpolypyrrolidone (50 mg) and extracted with 500 μL of extraction buffer (Wein et al., 2002) with reducing agent. All extractions were performed in the presence of Complete Protease Inhibitor Cocktail Tablets (Roche) to protect proteins from the CP activity of actinidin during the extraction process. 1D SDS-PAGE, electroblotting, and membrane blocking for western analysis were performed as described by Nieuwenhuizen et al. (2007). Proteins were immunolocalized with polyclonal antibodies (diluted 1:200 [w/v] for kiwifruit actinidin and 1:100 [w/v] for chitinase) in Tris-buffered saline buffer containing 5% nonfat milk powder and 0.05% (v/v) Tween 20. For detection, the polyvinylidene difluoride blots were incubated with a Qdot 655 goat F(ab′)2 anti-rabbit IgG conjugate according to the manufacturer’s instructions (Invitrogen) in the same buffer, and binding was visualized using a Typhoon 9400 Variable Mode Imager and quantified using ImageQuant TL software (both GE Healthcare Life Sciences).
For LC-MS analysis, bands were excised from Coomassie blue-stained gels, and the protein was digested with 0.65 μg of trypsin (modified sequencing grade; Roche) in 0.2 m ammonium bicarbonate and 4.3% acetonitrile. Tryptic peptides were separated and analyzed using an Ettan multidimensional liquid chromatograph (GE Healthcare Bio-Sciences) coupled to an LTQ linear ion-trap mass spectrometer with a nanospray electrospray ionization interface (Thermo Fisher Scientific) as described (Nieuwenhuizen et al., 2007). MS/MS data were analyzed using the TurboSEQUEST protein identification software (Eng et al., 1994), and proteins were identified by searching spectra against an in house Actinidia EST database (Crowhurst et al., 2008).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers GU201520 to GU201526 (alleles ACT1A-2 through act1a-8), GU201527 and GU201528 (cDNA sequences corresponding to the act1a-6 and act1a-7 alleles), and JN701537 (AcChi4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence alignment of ACT1A alleles from the A. chinensis mapping population and YellowA.
Supplemental Figure S2. Scanning for a second QTL on LG16 using the imputation method.
Supplemental Table S1. Genotypes and CP activities of individual plants.
Supplemental Table S2. Copy number estimation in YellowA transgenic plants complemented with the ACT1A cDNA.
Supplemental Table S3. Primers used for genotyping, allele amplification, and determining transgene copy number.
Acknowledgments
We thank Julie Nichols and Gnanaseela Wadasinghe for plant care, Elspeth MacRae and Sarah Thromber for preliminary characterization of CP activity in the mapping population, Tim Holmes for photography, Ramon Lopez Perez and Simona Nardozza for help with fruit collection, and Mark McNeilage, David Brummell, Andrew Allen, and Bryan Parkes for their useful edits in the preparation of the manuscript.
References
- Arcus AC. (1959) Proteolytic enzyme of Actinidia chinensis. Biochim Biophys Acta 33: 242–244 [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. (2006) The SWISS-MODEL workspace: a Web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Ashie INA, Sorensen TL, Nielsen PM. (2002) Effects of papain and a microbial enzyme on meat proteins and beef tenderness. J Food Sci 67: 2138–2142 [Google Scholar]
- Beers EP, Jones AM, Dickerman AW. (2004) The S8 serine, C1A cysteine and A1 aspartic protease families in Arabidopsis. Phytochemistry 65: 43–58 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Berti PJ, Storer AC. (1995) Alignment/phylogeny of the papain superfamily of cysteine proteases. J Mol Biol 246: 273–283 [DOI] [PubMed] [Google Scholar]
- Bishop JG, Dean AM, Mitchell-Olds T. (2000) Rapid evolution in plant chitinases: molecular targets of selection in plant-pathogen coevolution. Proc Natl Acad Sci USA 97: 5322–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JG, Ripoll DR, Bashir S, Damasceno CMB, Seeds JD, Rose JKC. (2005) Selection on glycine beta-1,3-endoglucanase genes differentially inhibited by a Phytophthora glucanase inhibitor protein. Genetics 169: 1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes S, Strubi P, Marsh H. (1997) Actinidin levels in fruit of Actinidia species and some Actinidia arguta rootstock scion combinations. Lebensm Wiss Technol 30: 379–389 [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890 [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. (1994) Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowhurst RN, Gleave AP, MacRae EA, Ampomah-Dwamena C, Atkinson RG, Beuning LL, Bulley SM, Chagne D, Marsh KB, Matich AJ, et al. (2008) Analysis of expressed sequence tags from Actinidia: applications of a cross species EST database for gene discovery in the areas of flavor, health, color and ripening. BMC Genomics 9: 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenth J, Jansonius JN, Koekoek R, Swen HM, Wolthers BG. (1968) Structure of papain. Nature 218: 929–932 [DOI] [PubMed] [Google Scholar]
- Eng JK, Mccormack AL, Yates JR. (1994) An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J Am Soc Mass Spectrom 5: 976–989 [DOI] [PubMed] [Google Scholar]
- Fei ZJ, Tang X, Alba RM, White JA, Ronning CM, Martin GB, Tanksley SD, Giovannoni JJ. (2004) Comprehensive EST analysis of tomato and comparative genomics of fruit ripening. Plant J 40: 47–59 [DOI] [PubMed] [Google Scholar]
- Fraser LG, Kent J, Harvey CF. (1995) Transformation studies of Actinidia chinensis Planch. N Z J Crop Hortic Sci 23: 407–413 [Google Scholar]
- Fraser LG, Tsang GK, Datson PM, De Silva HN, Harvey CF, Gill GP, Crowhurst RN, McNeilage MA. (2009) A gene-rich linkage map in the dioecious species Actinidia chinensis (kiwifruit) reveals putative X/Y sex-determining chromosomes. BMC Genomics 10: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrovic-Jankulovic M, Polovic N, Prisic S, Jankov RM, Atanaskovic-Markovic M, Vuckovic O, Cirkovic Velickovic T. (2005) Allergenic potency of kiwi fruit during fruit development. Food Agric Immunol 16: 117–128 [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Haley CS, Knott SA. (1992) A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324 [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Yamada K, Shimada T, Matsushima R, Nishizawa NK, Nishimura M, Hara-Nishimura I. (2001) A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol 42: 894–899 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Kamphuis IG, Kalk KH, Swarte MB, Drenth J. (1984) Structure of papain refined at 1.65 A resolution. J Mol Biol 179: 233–256 [DOI] [PubMed] [Google Scholar]
- Kasprzewska A. (2003) Plant chitinases: regulation and function. Cell Mol Biol Lett 8: 809–824 [PubMed] [Google Scholar]
- Kaur L, Rutherfurd SM, Moughan PJ, Drummond L, Boland MJ. (2010a) Actinidin enhances gastric protein digestion as assessed using an in vitro gastric digestion model. J Agric Food Chem 58: 5068–5073 [DOI] [PubMed] [Google Scholar]
- Kaur L, Rutherfurd SM, Moughan PJ, Drummond L, Boland MJ. (2010b) Actinidin enhances protein digestion in the small intestine as assessed using an in vitro digestion model. J Agric Food Chem 58: 5074–5080 [DOI] [PubMed] [Google Scholar]
- Keeling J, Maxwell P, Gardner RC. (1990) Nucleotide sequence of the promoter region from kiwifruit actinidin genes. Plant Mol Biol 15: 787–788 [DOI] [PubMed] [Google Scholar]
- Konno K, Hirayama C, Nakamura M, Tateishi K, Tamura Y, Hattori M, Kohno K. (2004) Papain protects papaya trees from herbivorous insects: role of cysteine proteases in latex. Plant J 37: 370–378 [DOI] [PubMed] [Google Scholar]
- Krüger J, Thomas CM, Golstein C, Dixon MS, Smoker M, Tang SK, Mulder L, Jones JDG. (2002) A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296: 744–747 [DOI] [PubMed] [Google Scholar]
- Lange J, Mohr U, Wiemken A, Boller T, Vögeli-Lange R. (1996) Proteolytic processing of class IV chitinase in the compatible interaction of bean roots with Fusarium solani. Plant Physiol 111: 1135–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocca M, Rossano R, Riccio P. (2010) Analysis of green kiwi fruit (Actinidia deliciosa cv. Hayward) proteinases by two-dimensional zymography and direct identification of zymographic spots by mass spectrometry. J Sci Food Agric 90: 2411–2418 [DOI] [PubMed] [Google Scholar]
- Lewis DA, Luh BS. (1988) Application of actinidin from kiwifruit to meat tenderization and characterization of beef muscle protein hydrolysis. J Food Biochem 12: 147–158 [Google Scholar]
- Lin E, Burns DJW, Gardner RC. (1993) Fruit developmental regulation of the kiwifruit actinidin promoter is conserved in transgenic petunia plants. Plant Mol Biol 23: 489–499 [DOI] [PubMed] [Google Scholar]
- Lucas JSA, Nieuwenhuizen NJ, Atkinson RG, Macrae EA, Cochrane SA, Warner JO, Hourihane JOB. (2007) Kiwifruit allergy: actinidin is not a major allergen in the United Kingdom. Clin Exp Allergy 37: 1340–1348 [DOI] [PubMed] [Google Scholar]
- Malone LA, Todd JH, Burgess EPJ, Philip BA, Christeller JT. (2005) Effects of kiwifruit (Actinidia deliciosa) cysteine protease on growth and survival of Spodoptera litura larvae (Lepidoptera: Noctuidae) fed with control or transgenic avidin-expressing tobacco. N Z J Crop Hortic Sci 33: 99–105 [Google Scholar]
- Mauch F, Mauch-Mani B, Boller T. (1988) Antifungal hydrolases in pea tissue. II. Inhibition of fungal growth by combinations of chitinase and β-1,3-glucanase. Plant Physiol 88: 936–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Hou SB, Feng Y, Yu QY, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KLT, et al. (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452: 991–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller DA, Tiffin P. (2005) Genetic diversity and the evolutionary history of plant immunity genes in two species of Zea. Mol Biol Evol 22: 2480–2490 [DOI] [PubMed] [Google Scholar]
- Newcomb RD, Crowhurst RN, Gleave AP, Rikkerink EHA, Allan AC, Beuning LL, Bowen JH, Gera E, Jamieson KR, Janssen BJ, et al. (2006) Analyses of expressed sequence tags from apple. Plant Physiol 141: 147–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuizen NJ, Beuning LL, Sutherland PW, Sharma NN, Cooney JM, Bieleski LRF, Schröder R, MacRae EA, Atkinson RG. (2007) Identification and characterisation of acidic and novel basic forms of actinidin, the highly abundant cysteine protease from kiwifruit. Funct Plant Biol 34: 946–961 [DOI] [PubMed] [Google Scholar]
- Nieuwenhuizen NJ, Wang MY, Matich AJ, Green SA, Chen X, Yauk Y-K, Beuning LL, Nagegowda DA, Dudareva N, Atkinson RG. (2009) Two terpene synthases are responsible for the major sesquiterpenes emitted from the flowers of kiwifruit (Actinidia deliciosa). J Exp Bot 60: 3203–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama I. (2007) Fruits of the Actinidia genus. Adv Food Nutr Res 52: 293–324 [DOI] [PubMed] [Google Scholar]
- Palacin A, Rodriguez J, Blanco C, Lopez-Torrejon G, Sánchez-Monge R, Varela J, Jiménez MA, Cumplido J, Carrillo T, Crespo JF, et al. (2008) Immunoglobulin E recognition patterns to purified kiwifruit (Actinidia deliciosa) allergens in patients sensitized to kiwi with different clinical symptoms. Clin Exp Allergy 38: 1220–1228 [DOI] [PubMed] [Google Scholar]
- Pastorello EA, Conti A, Pravettoni V, Farioli L, Rivolta F, Ansaloni R, Ispano M, Incorvaia C, Giuffrida MG, Ortolani C. (1998) Identification of actinidin as the major allergen of kiwi fruit. J Allergy Clin Immunol 101: 531–537 [DOI] [PubMed] [Google Scholar]
- Patel M, Kayani IS, Mellor GW, Sreedharan S, Templeton W, Thomas EW, Thomas M, Brocklehurst K. (1992) Variation in the P2-S2 stereochemical selectivity towards the enantiomeric N-acetylphenylalanylglycine 4-nitroanilides among the cysteine proteinases papain, ficin and actinidin. Biochem J 281: 553–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechan T, Ye L, Chang Y, Mitra A, Lin L, Davis FM, Williams WP, Luthe DS. (2000) A unique 33-kD cysteine proteinase accumulates in response to larval feeding in maize genotypes resistant to fall armyworm and other Lepidoptera. Plant Cell 12: 1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng FY, Reid KE, Liao N, Schlosser J, Lijavetzky D, Holt R, Martínez Zapater JM, Jones S, Marra M, Bohlmann J, et al. (2007) Generation of ESTs in Vitis vinifera wine grape (Cabernet Sauvignon) and table grape (Muscat Hamburg) and discovery of new candidate genes with potential roles in berry development. Gene 402: 40–50 [DOI] [PubMed] [Google Scholar]
- Podivinsky E, Forster RLS, Gardner RC. (1989) Nucleotide sequence of actinidin, a kiwi fruit protease. Nucleic Acids Res 17: 8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praekelt UM, McKee RA, Smith H. (1988) Molecular analysis of actinidin, the cysteine proteinase of Actinidia chinensis. Plant Mol Biol 10: 193–202 [DOI] [PubMed] [Google Scholar]
- Prestamo G. (1995) Actinidin in kiwifruit cultivars. Z Lebensm Unters Forsch 200: 64–66 [Google Scholar]
- Rassam M, Laing WA. (2004) Purification and characterization of phytocystatins from kiwifruit cortex and seeds. Phytochemistry 65: 19–30 [DOI] [PubMed] [Google Scholar]
- Robinson SP, Jacobs AK, Dry IB. (1997) A class IV chitinase is highly expressed in grape berries during ripening. Plant Physiol 114: 771–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherfurd SM, Montoya CA, Zou ML, Moughan PJ, Drummond LN, Boland MJ. (2011) Effect of actinidin from kiwifruit (Actinidia deliciosa cv. Hayward) on the digestion of food proteins determined in the growing rat. Food Chem 129: 1681–1689 [Google Scholar]
- Schlumbaum A, Mauch F, Vogeli U, Boller T. (1986) Plant chitinases are potent inhibitors of fungal growth. Nature 324: 365–367 [Google Scholar]
- Sen S, Churchill GA. (2001) A statistical framework for quantitative trait mapping. Genetics 159: 371–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden KC, Gardner RC. (1990) Nucleotide sequence of an actinidin genomic clone. Nucleic Acids Res 18: 6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz HU, Bishop JG, Bergmann CW, Koch M, Albersheim P, Darvill AG, Labavitch JM. (2000) Identification of target amino acids that affect interactions of fungal polygalacturonases and their plant inhibitors. Physiol Mol Plant Pathol 56: 117–130 [Google Scholar]
- Sugiyama S, Ohtsuki K, Sato K, Kawabata M. (1996) Purification and characterization of six kiwifruit proteases isolated with two ion-exchange resins, Toyopearl-SuperQ and Bakerbond WP-PEI. Biosci Biotechnol Biochem 60: 1994–2000 [Google Scholar]
- Sugiyama S, Ohtsuki K, Sato K, Kawabata M. (1997) Enzymatic properties, substrate specificities and pH-activity profiles of two kiwifruit proteases. J Nutr Sci Vitaminol (Tokyo) 43: 581–589 [DOI] [PubMed] [Google Scholar]
- Tamburrini M, Cerasuolo I, Carratore V, Stanziola AA, Zofra S, Romano L, Camardella L, Ciardiello MA. (2005) Kiwellin, a novel protein from kiwi fruit: purification, biochemical characterization and identification as an allergen. Protein J 24: 423–429 [DOI] [PubMed] [Google Scholar]
- Taylor MAJ, Baker KC, Briggs GS, Connerton IF, Cummings NJ, Pratt KA, Revell DF, Freedman RB, Goodenough PW. (1995) Recombinant pro-regions from papain and papaya proteinase IV are selective high affinity inhibitors of the mature papaya enzymes. Protein Eng 8: 59–62 [DOI] [PubMed] [Google Scholar]
- Tello-Solis SR, Valle-Guadarrama ME, Hernández-Arana A. (1995) Purification and circular dichroism studies of multiple forms of actinidin from Actinidia chinensis (kiwifruit). Plant Sci 106: 227–232 [Google Scholar]
- Tian MY, Win J, Song J, van der Hoorn R, van der Knaap E, Kamoun S. (2007) A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol 143: 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin P, Moeller DA. (2006) Molecular evolution of plant immune system genes. Trends Genet 22: 662–670 [DOI] [PubMed] [Google Scholar]
- van der Hoorn RAL. (2008) Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol 59: 191–223 [DOI] [PubMed] [Google Scholar]
- Varughese KI, Su Y, Cromwell D, Hasnain S, Xuong NH. (1992) Crystal structure of an actinidin-E-64 complex. Biochemistry 31: 5172–5176 [DOI] [PubMed] [Google Scholar]
- Wan LL, Xia Q, Qiu X, Selvaraj G. (2002) Early stages of seed development in Brassica napus: a seed coat-specific cysteine proteinase associated with programmed cell death of the inner integument. Plant J 30: 1–10 [DOI] [PubMed] [Google Scholar]
- Wang JQ, Li YB, Lo SW, Hillmer S, Sun SSM, Robinson DG, Jiang LW. (2007a) Protein mobilization in germinating mung bean seeds involves vacuolar sorting receptors and multivesicular bodies. Plant Physiol 143: 1628–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Atkinson R, Janssen B. (2007b) The choice of Agrobacterium strain for transformation of kiwifruit. Acta Hortic 753: 227–232 [Google Scholar]
- Wang TC, Ran YD, Atkinson RG, Gleave AP, Cohen D. (2006) Transformation of Actinidia eriantha: a potential species for functional genomics studies in Actinidia. Plant Cell Rep 25: 425–431 [DOI] [PubMed] [Google Scholar]
- Warrens AN, Jones MD, Lechler RI. (1997) Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene 186: 29–35 [DOI] [PubMed] [Google Scholar]
- Wein M, Lavid N, Lunkenbein S, Lewinsohn E, Schwab W, Kaldenhoff R. (2002) Isolation, cloning and expression of a multifunctional O-methyltransferase capable of forming 2,5-dimethyl-4-methoxy-3(2H)-furanone, one of the key aroma compounds in strawberry fruits. Plant J 31: 755–765 [DOI] [PubMed] [Google Scholar]
- Wurms KV. (2004) The incidence of Botrytis cinerea and expression of putative host defences in green- and golden-fleshed kiwfruit of differing harvest maturity. New Zealand Plant Protection 57: 125–129 [Google Scholar]
- Wurms KV, Sharrock KR, Long PG, Greenwood DR, Ganesh S. (1997) Responses of chitinases in kiwifruit to curing and to long-term storage. N Z J Crop Hortic Sci 25: 213–220 [Google Scholar]
- Yamada K, Matsushima R, Nishimura M, Hara-Nishimura I. (2001) A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiol 127: 1626–1634 [PMC free article] [PubMed] [Google Scholar]