Abstract
A large number of plants accumulate N-acylated polyamines (phenolamides [PAs]) in response to biotic and/or abiotic stress conditions. In the native tobacco (Nicotiana attenuata), the accumulation of two major PAs, caffeoylputrescine and dicaffeoylspermidine (DCS), after herbivore attack is known to be controlled by a key transcription factor, MYB8. Using a broadly targeted metabolomics approach, we show that a much larger spectrum of PAs composed of hydroxycinnamic acids and two polyamines, putrescine and spermidine, is regulated by this transcription factor. We cloned several novel MYB8-regulated genes, annotated as putative acyltransferases, and analyzed their function. One of the novel acyltransferases (AT1) is shown to encode a hydroxycinnamoyl-coenzyme A:putrescine acyltransferase responsible for caffeoylputrescine biosynthesis in tobacco. Another gene (acyltransferase DH29), specific for spermidine conjugation, mediates the initial acylation step in DCS formation. Although this enzyme was not able to perform the second acylation toward DCS biosynthesis, another acyltransferase gene, CV86, proposed to act on monoacylated spermidines, was isolated and partially characterized. The activation of MYB8 in response to herbivore attack and associated signals required the activity of LIPOXYGENASE3, a gene involved in jasmonic acid (JA) biosynthesis in N. attenuata. These new results allow us to reconstruct a complete branch in JA signaling that defends N. attenuata plants against herbivores: JA via MYB8’s transcriptional control of AT1 and DH29 genes controls the entire branch of PA biosynthesis, which allows N. attenuata to mount a chemically diverse (and likely efficient) defense shield against herbivores.
Phenolamides (PAs) are a diverse group of plant secondary metabolites found in many dicotyledonous plants as well as in monocots, including wheat (Triticum aestivum), barley (Hordeum vulgare), rice (Oryza sativa), and maize (Zea mays; Martin-Tanguy et al., 1978; Martin-Tanguy, 1985; Facchini et al., 2002; Edreva et al., 2007; Bassard et al., 2010). Although the chemical structures of PAs are well studied, the regulation of their biosynthetic pathways, patterns of accumulation, and roles of PAs in plants remain largely unknown. PAs often accumulate in reproductive organs and developing tissues, suggesting their role in plant growth and development (Martin-Tanguy, 1985; Facchini et al., 2002; Edreva et al., 2007; Grienenberger et al., 2009; Luo et al., 2009). In addition, the involvement of PAs in plant defense against abiotic (mineral deficiency, drought and salt stress, and UV irradiation) and biotic (pathogen infection and tissue-chewing herbivores) stresses has been proposed (Stoessl and Unwin, 1970; Martin-Tanguy, 1985; Back et al., 2001; Tanaka et al., 2003; Camacho-Cristóbal et al., 2005; Izaguirre et al., 2007; Kaur et al., 2010).
Jasmonates are important plant hormones that mediate plant responses to attack from herbivores and necrotrophic pathogens (Howe and Jander, 2008; Bari and Jones, 2009). Rapid accumulation of jasmonic acid (JA) after herbivore attack (Halitschke and Baldwin, 2003) results in altered secondary metabolite profiles in tobacco (Nicotiana attenuata) plants, as exemplified by the accumulation of alkaloids and phenylpropanoids (Baldwin, 1996; Shoji et al., 2000; Keinänen et al., 2001; Bernards and Båstrup-Spohr, 2008; Gaquerel et al., 2010). Goossens et al. (2003) demonstrated that the application of methyl jasmonate (MeJA) strongly induces the expression of genes involved in the alkaloid and phenylpropanoid biosynthetic pathways in Nicotiana tabacum cell cultures. Because JA-mediated responses are controlled by the activation of transcription factors (TFs) and synchronized expression of defense-related genes (Eulgem et al., 2000; Sugimoto et al., 2000; Singh et al., 2002; Vom Endt et al., 2002; Zheng et al., 2007; Woldemariam et al., 2011), the discovery of regulatory TFs greatly facilitates the identification of novel biosynthetic enzymes that can potentially serve as important tools for the efficient regulation of metabolic pathways through the manipulation of a single gene in the plant genome.
In PA biosynthesis, the expression of methyl jasmonate-induced MYB-related TF (NtMYBJS1) was rapidly elicited by MeJA in BY-2 cell suspension cultures, and the overexpression of this gene resulted in a strong ectopic accumulation of caffeoylputrescine (CP) in N. tabacum cells (Galis et al., 2006). When Kaur et al. (2010) targeted a homolog of the NtMYBJS1 gene in N. attenuata (NaMYB8) to down-regulate the levels of CP and dicaffeoylspermidine (DCS), both metabolites appeared to be essential for N. attenuata’s defense against chewing herbivores. While it highlighted the role of PAs in plant defense, the identity of the specific CP and DCS biosynthetic genes controlled by NaMYB8 TF remained unknown.
It has been shown that the N-coupling reaction of polyamines to phenolic acids (such as cinnamic, p-coumaric, caffeic, ferulic, and sinapic acids) in plants is catalyzed by a specific class of acyltransferase enzymes. A significant number of these proteins have been shown to belong to the previously characterized BAHD protein superfamily (for review, see D’Auria, 2006). However, the conventional use of sequence homology to identify substrates and enzyme activities of BAHD proteins often fails, because many BAHD enzymes with similar substrate specificities have evolved independently through a process of convergent evolution (Pichersky and Lewinsohn, 2011). This may include the case of CP and DCS secondary metabolites: although they are very abundant in plants, their biosynthetic genes remained unresolved. In addition, proteins from other groups can also be involved in the biosynthesis of PAs in plants, such as structurally divergent tyramine hydroxycinnamoyltransferases (Bassard et al., 2010) that contain a GCN5-related N-acetyltransferase-like domain in their structure.
Due to difficulties associated with the identification of PA biosynthetic enzymes, the development of novel approaches, such as the correlation analysis of metabolite accumulation and gene expression used in this work, is essential to find and characterize novel enzymes (Luo et al., 2007). In the initial analysis, using a metabolomics approach, we show that NaMYB8 (referred to as MYB8 in this paper) TF controls a surprisingly wide variety of PAs in N. attenuata plants. Next, we used data mining and microarray analysis to identify several novel PA biosynthetic candidate genes controlled by the activity of the MYB8 gene. Finally, silencing the expression of the three most promising candidate genes by virus-induced gene silencing (VIGS) combined with metabolomics analysis revealed that MYB8-controlled AT1, DH29, and CV86 genes are directly responsible for PA biosynthesis in N. attenuata plants.
RESULTS
MYB8 Controls a Large Spectrum of PAs in N. attenuata Plants
In our previous study, the silencing of the MYB8 TF resulted in a strong suppression of CP and DCS in N. attenuata (Kaur et al., 2010). To examine whether MYB8 also controls other less abundant PAs that occur in tobacco plants (Bassard et al., 2010), we used a broadly targeted metabolomics approach to compare the herbivory-elicited metabolic profiles of wild-type and MYB8-silenced (irMYB8) leaves. The young rosette leaves of wild-type and irMYB8 plants were elicited by treating puncture wounds (W) with diluted oral secretions (OS) from Manduca sexta larvae (W+OS), which strongly elicits plant defenses and the accumulation of CP and DCS in N. attenuata. The extracts from leaves collected 24 h after elicitation were subjected to ultraperformance liquid chromatography coupled to electrospray ionization time-of-flight mass spectrometry (UPLC-ESI-TOF-MS; Gaquerel et al., 2010). Peak matrices obtained from the analysis of wild-type and irMYB8 leaf extracts (for data processing, see “Materials and Methods”) were first subjected to a principal component analysis (PCA) to examine the variance in our data set without referring to the class labels.
Metabolic profiles from both irMYB8 constitutive and induced leaf samples strongly separated from those of wild-type leaves in the PCA projection plots (Supplemental Fig. S1), suggesting the existence of major metabolic differences between the two genotypes (the cumulative variance of PC1 and PC2 accounted for 53.3% of the total variance between wild-type and irMYB8 samples). Mass-to-charge ratio (m/z) features (parameters defined as discrete m/z values at defined retention times) that were most affected by OS elicitation exerted strong loading values on PC1, as observed by comparing the rankings of the loadings on the two first PCs and their relative fold changes (wild type/W+OS versus wild type/control in Supplemental Fig. S2). The m/z patterns controlled by MYB8 (revealed from the comparison irMYB8/W+OS versus wild type/W+OS in Supplemental Fig. S2) affected the orientation of both PCs, indicating that other signals that are less affected by the W+OS, and likely developmentally controlled, could also be regulated through MYB8 signaling.
When we annotated the pseudospectra corresponding to the m/z signals exhibiting the strongest signals on these two components, ions characteristic caffeic acid-containing metabolites showed the highest contributions to the PCA separations (Supplemental Fig. S1; Supplemental Table S1); however, metabolites predicted to contain coumaric and ferulic acid residues also contributed strongly to the separation. This reinforced our initial idea that the influence of MYB8 silencing on N. attenuata metabolism greatly surpasses its previously reported role in CP and DCS biosynthesis.
Annotations of Novel MYB8-Dependent Metabolites
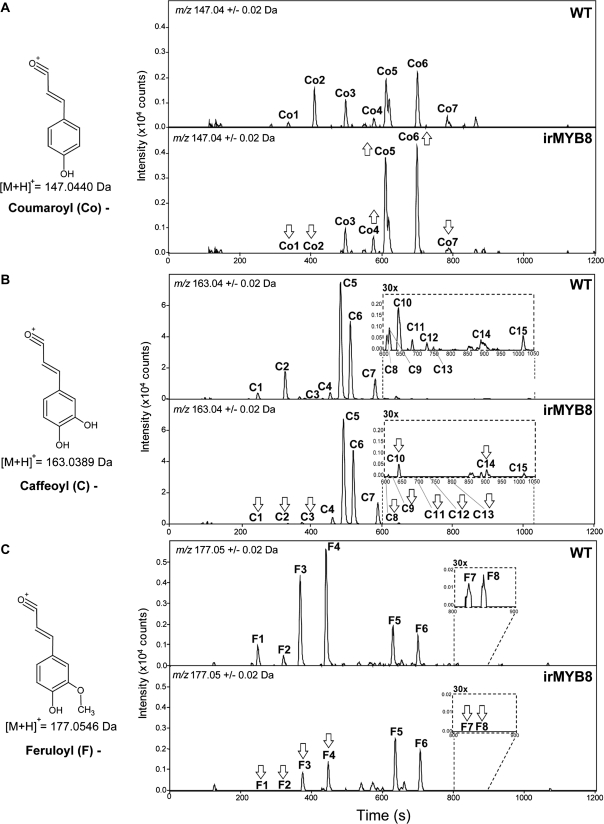
We next performed a direct pairwise statistical analysis of irMYB8 versus wild-type metabolic profiles (Supplemental Table S2). In total, 387 m/z signals (5.6% of the normalized m/z profile) were up-regulated (2-fold change; unpaired t test, P < 0.05) and 707 (10.3%) were down-regulated in irMYB8 compared with wild-type leaves at 24 h after W+OS elicitation. The m/z signal behavior was again strongly influenced by OS elicitation treatment (Supplemental Fig. S3). To directly visualize the phenolic constituents controlled by MYB8 TF, we computed extracted ion traces corresponding to the typical signatures of coumaroyl ([M+H]+ m/z 147.04 ± 0.02), caffeoyl ([M+H]+ m/z 163.04 ± 0.02), and feruloyl ([M+H]+ m/z 177.05 ± 0.02), moieties after in-source fragmentation of molecular ions and cleavage of the core molecules (e.g. sugars, polyamines, and small acids). The overlay and visual inspection of extracted ion traces in Figure 1 revealed that the MYB8 TF silencing affects a number of coumaric acid-, caffeic acid-, and ferulic acid-containing metabolites with distinct retention times. In contrast, ion traces of cinnamic and sinapic acid moieties showed only minor differences between wild-type and irMYB8 leaf samples (data not shown). When the predicted parent ions of the most abundant PAs composed of coumaric, caffeic, and ferulic acids coupled to putrescine or spermidine were extracted, even more striking differences in the accumulation of these metabolites were observed, further supporting a central role of MYB8 in PA biosynthesis (Supplemental Fig. S4). From this analysis, MYB8 TF can regulate at least 29 different (includes isomeric forms) hydroxycinnamoylputrescine/spermidine conjugates (Table I; Supplemental Fig. S4) in N. attenuata. Given the complexity of the observed regulated pool of metabolites, we predicted the existence of multiple MYB8 TF-controlled hydroxycinnamoyl-CoA:putrescine/spermidine transferases and pursued this prediction in the following text.
Figure 1.
MYB8 silencing induces specific alterations in the accumulation of phenylpropanoid-containing metabolites. A to C show representative ion chromatograms (n = 5) calculated for coumaroyl (A; m/z 147.04; peaks Co1–Co7), caffeoyl (B; m/z 163.04; peaks C1–C15), and feruloyl (C; m/z 177.05; peaks F1–F8) ion moieties generated upon in-source fragmentation during the analysis by UPLC-TOF-MS in positive mode of methanol-water extracts of 24-h W+OS-elicited leaves from untransformed wild-type (WT) and MYB8-silenced (irMYB8) plants. Raw data were deconvoluted with the R package XCMS prior to statistical analysis. Up and down arrows indicate significant (unpaired t test, P < 0.05) increases and decreases, respectively, in the accumulation of coumaroyl-, caffeoyl-, and feruloyl-containing metabolites in irMYB8 compared with wild-type leaves. C, Co, and F numbers refer to the major phenylpropanoid derivatives present in N. attenuata leaves as summarized in Table I.
Table I. Major coumaroyl-, caffeoyl- and feruloyl-containing metabolites measured by UPLC-TOF-MS in methanol-water leaf extracts of N. attenuata wild-type and irMYB8 plants.
Rosette leaves of wild-type and irMYB8 plants were wounded (W) with a fabric pattern wheel, immediately treated with M. sexta oral secretions (OS), and harvested 24 h later. Elemental formulas and relative mass errors (in ppm) were calculated using Smart Formula from the UPLC-TOF-MS operating software (see “Materials and Methods”). Candidate formulas were ranked according to both mass deviation and isotope pattern accuracy reflected in the σ value. MS/MS+ spectra for some of the reported parent ions have been published by our group (Gaquerel et al., 2010), and the strategy used for compound annotation is summarized into Supplemental Text S1. Asterisks indicate significant differences (unpaired t test, performed on log2-transformed data) in the relative intensity of hydroxycinnamic acid-containing metabolites between W+OS-elicited wild-type leaves and untreated wild-type controls (24 h; WT W+OS versus WT Ctrl) and between irMYB8 leaves induced by W+OS compared with identically treated wild-type leaves (24 h; irMYB8 W+OS versus WT W+OS): * P < 0.05, ** P < 0.001, *** P < 0.0001. Different fonts indicate either significant increases (boldface) or decreases (italics) due to W+OS elicitation or MYB8 silencing. Numbers in the compound name column refer to the different annotation levels using the four levels of the metabolite annotation nomenclature proposed by the Metabolome Standard Initiative. Indexes after nitrogen atoms indicate that structural rearrangements during in-source or collision-induced dissociation-MS/MS fragmentation did not allow the unequivocal assignment of the phenylpropanoid residues to the N1, N5, or N10 position of spermidine. N.V., Not visible in Figure 1 but presented in Supplemental Figure S4; NS, not significant.
|
P (Unpaired t Test) |
Fold Change |
||||||||||
| Label in Figure 1 | WT W+OS Versus WT Ctrl | irMYB8 W+OS Versus WT W+OS | WT W+OS > WT Ctrl | irMYB8 W+OS > WT W+OS | Retention Time | Precursor m/z | Ion Type | Elemental Formula | Error | Abbreviated Name | Annotation |
| s | ppm | ||||||||||
| Co1 | * | ** | 3.98 | 0.00 | 337 | m/z 235.143 | [M+H]+ | C13H19N2O2+ | 4.1 | CoP | 2N -Coumaroylputrescine isomer 1 |
| Co2 | *** | *** | 8.48 | 0.00 | 410 | m/z 235.143 | [M+H]+ | C13H19N2O2+ | 4.1 | CoP | 2N -Coumaroylputrescine isomer 2 |
| Co3 | NS | NS | – | – | – | – | – | – | Unk | 4Unknown | |
| Co4 | * | ** | 9.58 | 2.13 | 575 | m/z 361.097 | [M+H]+ | C11H21O13+ | 0.9 | Unk | 4Unknown |
| Co5 | * | * | 1.48 | 1.71 | 612 | m/z 339.106 | [M+H]+ | C16H19O8+ | 1.3 | CoQ | 3O -Coumaroylquinic acid isomer 1 |
| Co6 | NS | ** | 1.11 | 1.87 | 700 | m/z 339.110 | [M+H]+ | C16H19O8+ | 2.8 | CoQ | 3O -Coumaroylquinic acid isomer 2 |
| Co7 | *** | *** | 0.05 | 0.25 | 780 | m/z 498.261 | [M+H]+ | C27H36N3O6+ | 0.8 | CoSiS | N′,N″-Coumaroyl, sinapoyl spermidine isomer 1 |
| C1 | ** | ** | 2.72 | 0.01 | 254 | m/z 251.138 | [M+H]+ | C13H19N2O3+ | 3.6 | CP | 1N -Caffeoylputrescine isomer 1 |
| C2 | ** | *** | 5.74 | 0.00 | 333 | m/z 251.139 | [M+H]+ | C13H19N2O3+ | 3.6 | CP | 1N -Caffeoylputrescine isomer 2 |
| C3 | NS | ** | 0.90 | 1.12 | 373 | m/z 401.122 | [M+H]+ | C13H19N3O7+ | 0.3 | Unk | 4Unknown |
| C4 | NS | NS | 1.41 | 0.93 | 460 | m/z 355.103 | [M+H]+ | C16H19O9+ | 2.2 | CQ | 3O -Caffeoylquinic acid isomer 1 |
| C5 | NS | NS | 0.98 | 0.93 | 490 | m/z 355.109 | [M+H]+ | C16H19O9+ | 1.2 | CGA | 13-O -Caffeoylquinic acid isomer 2 (CGA) |
| C6 | NS | NS | 0.96 | 0.92 | 517 | m/z 355.104 | [M+H]+ | C16H19O9+ | 1.4 | CQ | 3O -Caffeoylquinic acid isomer 3 |
| C7 | * | NS | 0.83 | 0.94 | 586 | m/z 355.104 | [M+H]+ | C16H19O9+ | 1.4 | CQ | 3O -Caffeoylquinic acid isomer 4 |
| C8 & C9 | NS | ** | 0.80 | 0.06 | 612 | m/z 470.229 | [M+H]+ | C25H32N3O6+ | 2 | DCS | 2N′,N″-Dicaffeoylspermidine isomers 2 and 3 |
| C10 | * | *** | 1.77 | 0.08 | 646 | m/z 470.230 | [M+H]+ | C25H32N3O6+ | 2 | DCS | 2N′,N″-Dicaffeoylspermidine isomer 4 |
| C11 | ** | ** | 5.68 | 0.01 | 687 | m/z 347.196 | [M+H]+ | C19H27N2O4+ | 0.3 | Unk | 3Unknown putrescine metabolite isomer 1 |
| C11 | ** | ** | 1.80 | 0.03 | 687 | m/z 484.244 | [M+H]+ | C26H34N3O6+ | 0.3 | CFS | 2N′,N″-Caffeoyl, feruloyl spermidine isomer 1 |
| C12 | ** | ** | 4.22 | 0.00 | 731 | m/z 347.198 | [M+H]+ | C19H27N2O4+ | 3.3 | Unk | 3Unknown putrescine metabolite isomer 2 |
| C12 | *** | ** | 2.38 | 0.03 | 730 | m/z 484.245 | [M+H]+ | C26H34N3O6+ | 1.2 | CFS | 2N′,N″-Caffeoyl, feruloyl spermidine isomer 2 |
| C13 | *** | ** | 4.88 | 0.05 | 751 | m/z 484.245 | [M+H]+ | C26H34N3O6+ | 1.2 | CFS | 2N′,N″-Caffeoyl, feruloyl spermidine isomer 3 |
| C14 | NS | ** | 1.07 | 0.19 | 899 | m/z 566.286 | [M+H]+ | C31H40N3O7+ | 0.1 | Unk | 3Unknown spermidine metabolite |
| C15 | * | * | 2.91 | 0.13 | 1,019 | m/z 580.300 | [M+H]+ | C32H42N3O7+ | 0.3 | Unk | 3Unknown spermidine metabolite |
| F1 | *** | *** | 4.38 | 0.01 | 263 | m/z 322.212 | [M+H]+ | C17H28N3O3+ | 0.7 | MFS | 2N -Feruloylspermidine isomer 1 |
| F2 | *** | *** | 13.92 | 0.01 | 338 | m/z 322.212 | [M+H]+ | C17H28N3O3+ | 0.7 | MFS | 2N -Feruloylspermidine isomer 2 |
| F3 | ** | *** | 17.51 | 0.17 | 388 | m/z 265.153 | [M+H]+ | C14H21N2O3+ | 4.5 | FP | 2N -Feruloylputrescine isomer 1 |
| F4 | *** | *** | 25.71 | 0.19 | 464 | m/z 265.153 | [M+H]+ | C14H21N2O3+ | 3.8 | FP | 2N -Feruloylputrescine isomer 2 |
| F5 | * | NS | 1.71 | 1.22 | 661 | m/z 369.119 | [M+H]+ | C17H21O9+ | 0.6 | FQ | 3O -Feruloylquinic acid isomer 1 |
| F6 | * | NS | 1.74 | 1.31 | 731 | m/z 369.121 | [M+H]+ | C17H21O9+ | 3.6 | FQ | 3O -Feruloylquinic acid isomer 2 |
| F7 | NS | NS | 1.93 | 0.04 | 823 | m/z 498.260 | [M+H]+ | C27H36N3O6+ | 0.7 | DFS | 2N′,N″-Di-feruloyl-spermidine isomers 1 and 2 |
| F8 | ** | * | 2.43 | 0.03 | 847 | m/z 498.259 | [M+H]+ | C27H36N3O6+ | 0.7 | DFS | 2N′,N″-Di-feruloyl-spermidine isomer 3 |
| N.V. | *** | ** | 0.16 | 0.00 | 218 | m/z 292.202 | [M+H]+ | C16H26N3O2+ | 0.4 | MCoS | 2N -Coumarylspermidine isomer 1 |
| N.V. | NS | ** | 0.06 | 0.00 | 289 | m/z 292.201 | [M+H]+ | C16H26N3O2+ | 0.4 | MCoS | 2N -Coumarylspermidine isomer 2 |
| N.V. | NS | ** | 1.20 | 0.00 | 175 | m/z 308.195 | [M+H]+ | C16H26N3O3+ | 0.9 | MCS | 2N Caffeoylspermidine isomer 1 |
| N.V. | *** | ** | 11.08 | 0.00 | 229 | m/z 308.197 | [M+H]+ | C16H26N3O3+ | 0.9 | MCS | 2N Caffeoylspermidine isomer 2 |
| N.V. | * | ** | 0.31 | 0.03 | 572 | m/z 470.229 | [M+H]+ | C25H32N3O6+ | 0.9 | DCS | 2N′,N″-Dicaffeoylspermidine isomer 1 |
| N.V. | NS | *** | 0.80 | 0.00 | 658 | m/z 454.233 | [M+H]+ | C25H32N3O5+ | 1.7 | CoCS | 2N -Coumaryl, caffeoyl spermidine isomer 1 |
| N.V. | NS | ** | 0.30 | 0.00 | 702 | m/z 454.233 | [M+H]+ | C25H32N3O5+ | 1.7 | CoCS | 2N -Coumaryl, caffeoyl spermidine isomers 2 and 3 |
| N.V. | * | *** | 0.10 | 0.01 | 725 | m/z 454.235 | [M+H]+ | C25H32N3O5+ | 2.5 | CoCS | 2N -Coumaryl, caffeoyl spermidine isomer 4 |
Identification of Novel MYB8-Regulated Acyltransferases
In the previous report, a custom oligo-DNA microarray was used to identify differentially regulated genes in irMYB8 plants after W+OS elicitation (Kaur et al., 2010). Although the microarray contained a relatively small number of oligoprobes (1,421), two cDNA fragments from N. attenuata encoding potential acyltransferase genes controlled by MYB8 TF were found. The DH29 and CV86.1 (referred to as CV86 in this paper) gene fragments, originally cloned as herbivory-regulated ESTs (DH29 [Hui et al., 2003] and CV86.1 [Voelckel and Baldwin, 2003]), showed 3.8- and 3.7-fold down-regulation in irMYB8 compared with wild-type leaves at 90 min after W+OS elicitation, respectively (Kaur et al., 2010). First, we tried to extend the partial cDNA fragments of DH29 (CA591847.1) and CV86 (BU494535.1) genes by repeatedly BLASTing the fragments against public ESTs libraries from N. tabacum available in the National Center for Biotechnology Information (NCBI) database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). When the putative full-length size of N. tabacum DH29 and CV86 genes was reached (i.e. no additional overlapping ESTs were found), primers outside the longest open reading frame were designed (Supplemental Table S3) and full-length coding sequence of N. attenuata DH29 and CV86 were cloned, sequenced, and deposited in the database under accession numbers JN390824 (N. attenuata DH29) and JN390825 (N. attenuata CV86).
We then used the N. attenuata DH29 and CV86 sequences as protein queries to search for additional acyltransferase-like enzymes in tobacco, exploring the N. tabacum EST libraries in the NCBI (for details, see “Materials and Methods”). In total, we obtained 32 potential acyltransferase-like sequences that could be examined for their expression using our currently established herbivory-regulated microarray data set. In this experiment, time kinetics of labeled copy RNA probes from W+OS, W+W, and control leaves treated for 1, 5, 9, 13, 17, and 21 h were hybridized to a N. attenuata-specific Agilent microarray platform (GPL13527). Raw and normalized data from the triplicate experiment showing gene expression in locally treated leaves, systemic leaves, and roots were already deposited in the public Gene Expression Omnibus (GEO) microarray repository under sample numbers GSM750588 to GSM750721. Searching the expression patterns of our 32 putative acyltransferases, we found 15 novel herbivory-regulated transcripts, using a strong elicitation by W and/or W+OS and systemic induction in untreated leaves as the main criteria for herbivory-regulated genes (Supplemental Table S4, worksheet C).
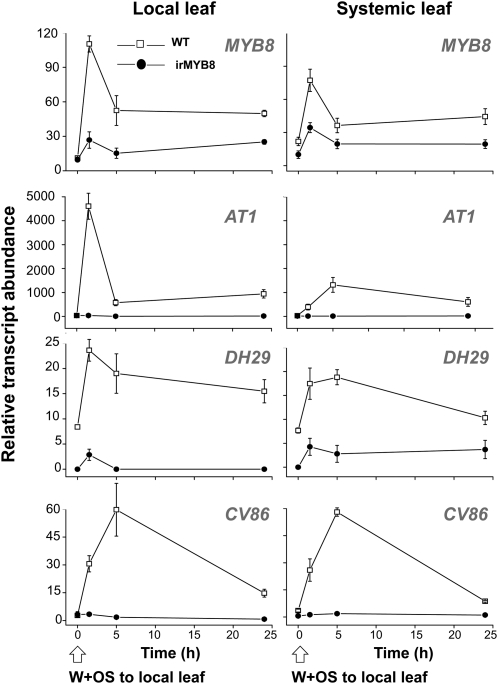
Next, we performed a visual inspection of all expression patterns, searching for those that matched the expression of MYB8, DH29, and CV86 genes, assuming that the use of a common TF will result in significant coregulation of downstream genes. The main criteria were (1) strong W+OS elicitation in local leaves; (2) strong W+OS elicitation in systemic leaves; and (3) low expression of genes in the roots (Supplemental Table S4, worksheet C; Supplemental Fig. S5). After screening, the AT1 gene represented by the CUST_51472_PI422650789 oligoprobe on the microarray was selected; this acyltransferase-like gene was strongly induced and closely coregulated with DH29 and CV86 genes. After cloning the entire N. attenuata AT1 gene (deposited in the database as JN390826), the AT1 expression in W+OS-elicited wild-type and irMYB8 leaves was examined by reverse transcription-quantitative (RT-q)PCR. The AT1 gene was strongly down-regulated in irMYB8 OS-elicited leaves compared with wild-type plants, suggesting that it is indeed located downstream of the MYB8 control element (Fig. 2). At the same time, MYB8-dependent expression of DH29 and CV86 in OS locally induced and systemic unwounded leaves was confirmed by RT-qPCR (Fig. 2). In summary, our results indicated that MYB8 TF acts upstream of AT1, DH29, and CV86 putative acyltransferases and that MYB8 controls the expression of these genes in N. attenuata both locally and systemically during herbivory.
Figure 2.
AT1, DH29, and CV86 transcript levels are dependent on MYB8 transcriptional activity in local and systemic tissues of W+OS-elicited N. attenuata plants. RT-qPCR analysis was performed with cDNA samples prepared from local (wounded) and systemic young (unwounded) leaves before and 1.5, 5, and 24 h after W+OS elicitation. Values are means of three replicate measurements ± se. WT, Wild type.
Silencing of Acyltransferases Impairs PA Accumulation
When we compared our novel acyltransferases against the set of known BAHD proteins (Luo et al., 2009), adding several other characterized hydroxycinnamoyl-CoA:polyamine transferases (Bassard et al., 2010; Supplemental Fig. S6), the AT1 protein clustered with a barley ACT gene known to utilize a short-chain polyamine agmatine as its acyl acceptor (Burhenne et al., 2003). In contrast, DH29 and CV86 clustered with proteins that contained the acetyl-CoA:benzylalcohol acetyltransferase protein from Clarkia breweri (Dudareva et al., 1998). Neither of the proteins closely grouped with the Arabidopsis (Arabidopsis thaliana) spermidine utilizing spermidine dicoumaroyltransferase, spermidine disinapoyltransferase, or spermidine-hydroxycinnamoyltransferase N-hydroxycinnamoyltransferases (Grienenberger et al., 2009; Luo et al., 2009), as would be expected from our metabolic profiles shown in Figure 1. Therefore, we proceeded to the direct functional characterization of AT1, DH29, and CV86 proteins.
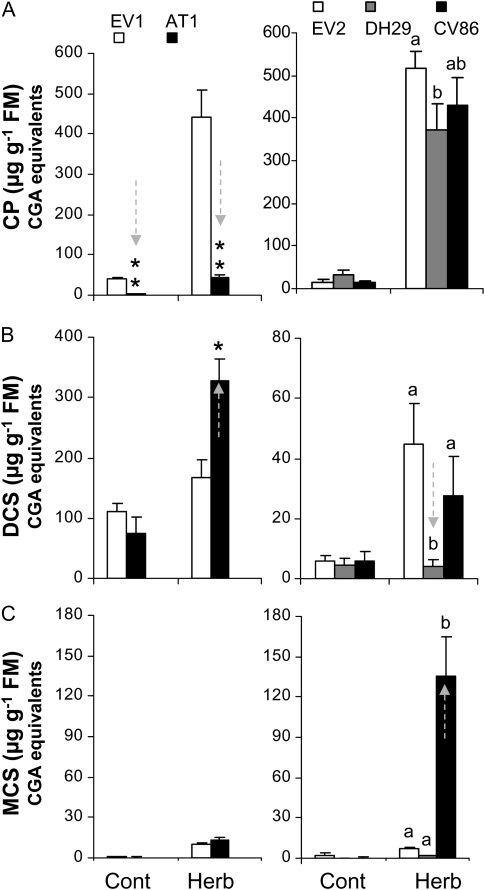
When the expression of AT1, DH29, and CV86 was separately silenced by VIGS, all three genes showed reduced transcript levels in their respective VIGS plants (Supplemental Fig. S7). Next, the PAs in an empty vector (EV) control and AT1-, DH29-, and CV86-VIGS plants that had their leaves fed on by M. sexta neonates for 4 d were examined. Because of the large number of plants involved in our functional screens, we performed our experiments in two groups, each time using a new set of control EV-inoculated plants (group 1, AT1-EV1; group 2, DH29-CV86-EV2). Herbivore-fed and control leaves were extracted and subjected to HPLC coupled to photodiode array detection (HPLC-PDA). An abundant UV-absorbing peak corresponding to CP was strongly suppressed in AT1-VIGS plants (Fig. 3A), while the peak corresponding to pooled DCS increased. In contrast, pooled DCS content was strongly reduced in DH29 leaves (Fig. 3B). Only small amounts of monocaffeoylspermidine (MCS) were typically detected in the samples, suggesting a rapid conversion of monoacylated spermidines into PAs like DCS; however, CV86-VIGS plants accumulated abnormally high levels of MCS during herbivory (Fig. 3C).
Figure 3.
HPLC-PDA determination of differentially accumulated metabolites in herbivory-elicited leaves of AT1-, DH29-, and CV86-VIGS plants. Caffeoylputrescine (A), dicaffeoylspermidine (B), and monocaffeoylspermidine (C) contents, expressed as CGA equivalents, were determined in control (Cont) and 4-d continuously M. sexta neonate-fed leaves (Herb). VIGS plants treated with an EV construct were used as controls in all experiments; EV1 and EV2 show controls in two VIGS experiments conducted with AT1 (EV1) and DH29 + CV86 (EV2), respectively. Asterisks or different letters indicate significantly down- and up-regulated metabolites in response to silencing of the respective acyltransferase gene activity as follows: AT1-EV1, Student’s t test, * P < 0.05 (*), ** P < 0.05 (n = 5); DH29-CV86-EV2, one-way ANOVA (n = 5). FM, Fresh mass.
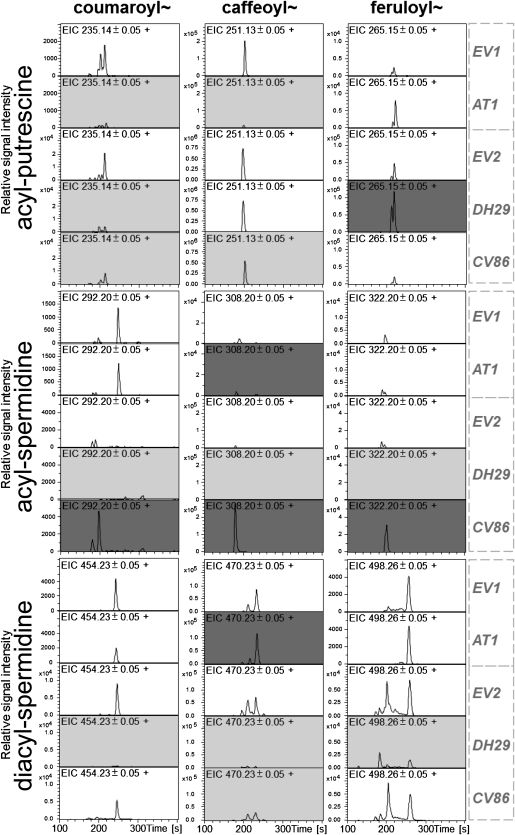
Because HPLC-PDA only detects the most dominant metabolites in the otherwise complex plant extracts, we utilized the more sensitive UPLC-ESI-TOF-MS to examine the metabolic changes in our herbivore-fed leaves from VIGS plants. As expected from HPLC-PDA profiles, CP isomeric peaks at m/z 251.13 ± 0.05 were strongly reduced in AT1-VIGS (Fig. 4; Supplemental Table S5). In addition, a putative p-coumaroylputrescine peak (CoP; m/z 235.14 ± 0.05) was reduced in AT1-VIGS, but surprisingly, putative feruloylputrescine (FP; m/z 265.15 ± 0.05) content did not change after silencing of the AT1 enzyme, suggesting that FP might be produced by another yet unknown MYB8-dependent enzyme. The isomeric peaks belonging to DCS (m/z 470.23 ± 0.05) were strongly and partially reduced in DH29- and CV86-VIGS leaf extracts, respectively. Silencing the expression of DH29 affected the entire spectrum of acylated spermidines (Fig. 4), suggesting a broad donor substrate specificity of this enzyme. The overaccumulation of MCS dominated the UPLC-ESI-TOF-MS traces in CV86-VIGS leaves, but this accumulation pattern now extended to include putative mono-p-coumaroylspermidine and monoferuloylspermidine metabolites (Fig. 4).
Figure 4.
Representative (n = 4) extracted ion chromatograms (EIC) for monoacylated putrescines and monoacylated and diacylated spermidines in 4-d herbivory-elicited leaves of EV plants and AT1-, DH29-, and CV86-silenced plants. Ion types selected for the calculation of EIC traces correspond to the protonated [M+H]+ m/z signals of the different isomers as follows: (first row, left to right), N-coumaroylputrescine, N-caffeoylputrescine, N-feruloylputrescine; (second row), N-coumaroylspermidine, N-caffeoylspermidine, N-feruloylspermidine; (third row), N′,N″-coumaroyl,caffeoylputrescine, N′,N″-dicaffeoylspermidine, N′,N″-diferuloylspermidine. Indexes after nitrogen atoms indicate that structural rearrangements during in-source or collision-induced dissociation-MS/MS fragmentation did not allow the unequivocal assignment of the phenylpropanoid residues to the N1, N5, or N10 position of spermidine. Metabolites showing at least 1.5-fold reduced and increased content are highlighted using light gray and dark gray background, respectively (Supplemental Table S5). EV1 and EV2 show control treatments with an EV infiltration in two independent VIGS experiments conducted separately for AT1 (EV1) and DH29 + CV86 (EV2).
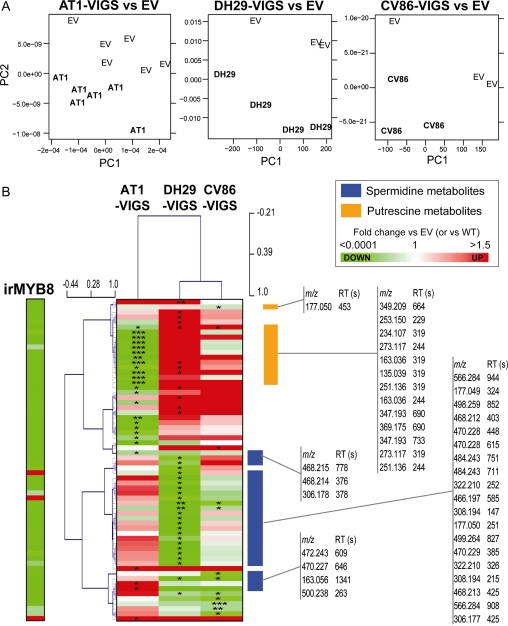
Even after a single W+OS elicitation treatment, similar metabolic patterns could be observed in the leaves from VIGS plants (Supplemental Fig. S8; Supplemental Table S6); however, they were much less pronounced compared with herbivore-fed leaves. When W+OS-elicited samples were subjected to PCA analysis, the metabolic profiles of AT1-, DH29-, and CV86-VIGS-silenced plants strongly separated from their respective EV-VIGS controls (Fig. 5A). Interestingly, while the majority of metabolites in irMYB8 leaves were down-regulated (Table I; Supplemental Fig. S3), the regulation of the same set of metabolites in the individual AT1-, DH29-, and CV86-silenced plants was more complex, showing the highly interconnected character of PA biosynthesis (Fig. 5B). Typically, silencing of one acyltransferase enzyme impaired the accumulation of several compounds while increasing another set of metabolites, most likely a result of silencing-mediated “metabolic tension” in the PA biosynthetic pathway. This was particularly discernible in the case of AT1- and DH29-regulated putrescine- and spermidine-containing metabolites, respectively (Fig. 5B; follow the contrasting red/green colors in the heat maps).
Figure 5.
AT1, DH29, and CV86 contribute to distinct, but interconnected, MYB8-controlled hydroxycinnamoylputrescine and hydroxycinnamoylspermidine pools. A, PCA showing a separation of herbivory-elicited large-scale metabolic profiles in AT1-, DH29-, and CV86-VIGS leaves compared with an EV-treated leaf profile. PCA analysis was performed after preprocessing using the R package XCMS and log2 transformation of the raw UPLC-TOF-MS files obtained from the measurement of the leaf tissues harvested 24 h after W+OS treatment. B, Hierarchical clustering analysis, using Pearson correlation as a distance metric, and the heat map of MYB8-dependent m/z signals from UPLC-TOF-MS analysis (log2-scaled intensities are shown in Supplemental Table S2) that were differentially regulated in AT1-, DH29-, and CV86-VIGS plants (fold change versus EV < 0.5 or EV > 2, unpaired t test on log2-transformed data: * P < 0.05, ** P < 0.001, *** P < 0.0001) compared with EV plants. The hierarchical clustering analysis separated AT1-dependent putrescine-based PA metabolic clusters from DH29- and putatively CV86-dependent spermidine-based PAs.
In Vitro Activity of Recombinant Acyltransferase Enzymes
Our in vivo metabolic profiles of VIGS plants suggested that AT1 encodes a hydroxycinnamoyl-CoA:putrescine transferase, DH29 showed properties predicted for a hydroxycinnamoyl-CoA:spermidine transferase, and the role of CV86 was suggestive of hydroxycinnamoyl-CoA:hydroxycinnamoylspermidine-conjugating activity. AT1, DH29, and CV86, therefore, were expressed and purified (using 6×His tag sequence attached to the C terminus of each of the proteins; Supplemental Fig. S9A) to examine their biochemical function in vitro. While the proteins accumulated in Escherichia coli cells, native CV86 protein was recovered exclusively as inactive protein in pellets during extraction. Therefore, we purified soluble CV86 protein under denaturing conditions and subsequently renatured the protein by dialysis. Despite numerous attempts under numerous conditions, the CV86 protein showed no activity with substrates predicted from our previous metabolic studies.
Therefore, we focused on the AT1 (50.8-kD) and DH29 (52-kD) proteins and successfully determined their enzymatic activities. Notably, recombinant DH29 protein showed a significant tendency for precipitation, especially when frozen in the elution buffer with imidazole, and it was unstable during storage at −80°C. The protein precipitation could be efficiently avoided by directly desalting the protein after elution from the nickel column, and frozen protein aliquots were used immediately after defrosting on ice to minimize the unavoidable loss of enzyme activity. Compared with DH29, the AT1 recombinant enzyme was fully soluble and more resistant to temperature changes during handling.
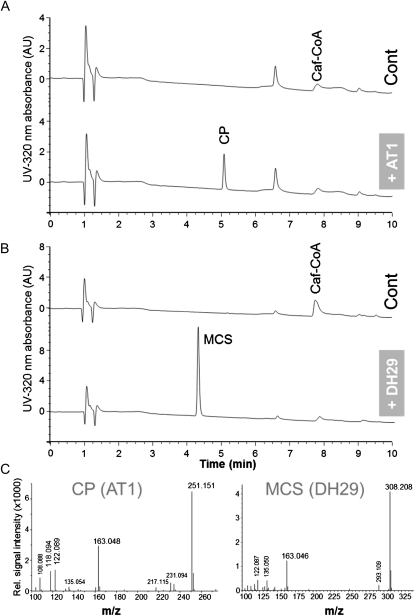
Enzyme activity assays with recombinant AT1 and DH29 proteins, informed by close examination of our VIGS metabolomics data, were performed with enzymatically prepared p-coumaroyl-, caffeoyl-, and feruloyl-CoA esters as acyl donors and the polyamines putrescine, spermidine, and spermine as acyl acceptors (Supplemental Fig. S9, B and C). When using caffeoyl-CoA as acyl donor, AT1 was only active with putrescine (Fig. 6A; Table II), while DH29 required spermidine for its activity (Fig. 6B; Table II). Neither of the proteins was active with spermine. These data were fully consistent with the metabolic changes observed in our VIGS-silenced plants (Fig. 3–5). When using fixed concentrations of caffeoyl-CoA as an acyl donor and variable spermidine levels, a low apparent Km value (27.2 ± 5.3 μm) of DH29 for spermidine was observed. In contrast, an apparent Km value of AT1 for putrescine was significantly higher (664 ± 56 μm) in a similar experiment with putrescine (variable) and caffeoyl-CoA (fixed). In a relative comparison, caffeoyl-CoA was the best substrate for both AT1 (Km = 24.0 ± 4.9 μm) and DH29 (Km = 325 ± 12 μm) when their obligatory polyamines were used (Table II). DH29 also metabolized feruloyl-CoA (70% relative activity compared with caffeoyl-CoA) and showed minor activity with p-coumaroyl-CoA (8% relative activity of caffeoyl-CoA). AT1 showed low but detectable activity with feruloyl-CoA (10% relative activity of caffeoyl-CoA), while it was almost inactive in vitro with p-coumaroyl-CoA as a substrate (Table II). AT1 and DH29 were found to be most active at pH 8 and 7.5, respectively (Supplemental Fig. S10). Finally, the main reaction product of AT1, determined by UPLC-ESI-TOF, matched the structure of synthetic CP standard (Fig. 6C, left), while the product of DH29 showed the expected spectral properties of MCS (Fig. 6C, right). In our experiments, we did not detect any reaction products corresponding to higher acylated (di or tri) forms of polyamines.
Figure 6.
Biochemical characterization of AT1 and DH29 using recombinant proteins and in vitro enzymatic assays. A and B, HPLC chromatograms (signal detected at 320 nm) show reaction products without and after the addition of AT1 (A) and DH29 (B) recombinant enzymes to reaction mixtures containing caffeoyl-CoA and the polyamines putrescine (A) or spermidine (B). The bottom chromatograms show the formation of CP at a retention time of 5.1 min (A) and of MCS at a retention time of 4.3 min (B) in samples supplied with recombinant AT1 and DH29 enzymes, respectively. Putrescine and spermidine that do not absorb UV are not visible in the chromatograms. C, MS spectra of the reaction products of AT1 (CP) and DH29 (MCS) recombinant enzymes determined with high-resolution UPLC-TOF-MS.
Table II. Summary of determined biochemical properties of the recombinant proteins AT1 and DH29 in vitro.
Recombinant His tag-containing AT1 and DH29 enzymes were examined for substrate specificity based on the observed spectra of naturally occurring metabolites found in N. attenuata. Km values were determined with caffeoyl-CoA (0–750 μm) as the best substrate and 0 to 2 mm putrescine (AT1) and spermidine (DH29).
| Sample | Relative Activity |
Km |
||
| AT1 | DH29 | AT1put | DH29spd | |
| % | μm | |||
| Acyl donor | ||||
| Caffeoyl-CoA | 100 | 100 | 24.0 ± 4.9 | 325 ± 12 |
| Feruloyl-CoA | 10 | 70 | ND | ND |
| Coumaroyl-CoA | <1 | 8 | ND | ND |
| AT1CafCoA | DH29CafCoA | |||
| Acyl acceptor | ||||
| Putrescine (put) | 100 | 0 | 664 ± 56 | ND |
| Spermidine (spd) | 0 | 100 | ND | 27.2 ± 5.3 |
| Spermine | 0 | 0 | ND | ND |
In conclusion, we annotated AT1 as an authentic CP synthase that correlates with the accumulation of CP but not dicaffeoylputrescine in N. attenuata. The function of DH29 was tentatively assigned to the biosynthesis of MCS (as well as coumaroylspermidine and feruloylspermidine). However, DCS in N. attenuata occurs in at least four isomeric forms (Supplemental Fig. S4). The silencing of the CV86 gene did not provide sufficiently conclusive information, as only some DCS isomers were significantly reduced in CV86-VIGS leaves (Supplemental Fig. S8; Supplemental Table S6). Therefore, the identification of an enzyme(s) involved in the conversion of MCS to DCS, which partially overlapped with the experimentally determined function of the CV86 enzyme, still remains to be resolved.
Accumulation of DCS during Ontogeny
While the pattern of CP accumulation during ontogeny was published earlier by our group (Kaur et al., 2010), a comparably detailed accumulation profile of DCS was not reported. Returning to the original data set used by Kaur et al. (2010) for CP quantification, we now determined the pooled DCS content in the W+OS-elicited N. attenuata plants at five distinct developmental stages (analyzed by HPLC-PDA in all available tissues). The highest concentration of DCS was found in the young rosette leaves, which was further enhanced by W+OS elicitation of the young leaves. Basal stems of early elongated plants also contained very high induced levels of DCS (Fig. 7). The overall accumulation patterns of DCS were similar to those of CP (Kaur et al., 2010), where younger tissues contained significantly more DCS (or CP) compared with the older leaves and roots. The levels of DCS in the vegetative parts of the flowering and mature plants were low and less inducible by W+OS treatment (Fig. 7). However, DCS was still detected in the capsules of mature N. attenuata plants (Fig. 7). When the accumulation of two other phenolic metabolites, rutin and chlorogenic acid (CGA), was determined, these metabolites lacked the herbivory-inducible character (Supplemental Fig. S11). Indeed, the accumulation of rutin and CGA does not depend on MYB8 TF, which specifically regulates the accumulation of direct defenses in N. attenuata plants against herbivores.
Figure 7.
Time- and tissue-scale-resolved accumulation of DCS in N. attenuata plants. Wild-type (WT) plants were grown in sand; at each developmental stage, a single fully expanded rosette leaf at the +1 position was OS elicited, and 3 d later, samples from representative plant parts were collected and analyzed by HPLC-PDA. The dotted arrows show the position of the locally W+OS-elicited rosette leaf at different stages of development. DH29 transcripts, shown in the gray inset, were analyzed by RT-qPCR from tissues at the elongated stage of development. R, Root; SB, stem basal; SU, stem upper; SeL, senescent leaf; OL, old leaf; LL, locally W+OS-induced rosette leaf; SL, systemic rosette leaf; 1SL, first stem leaf; YL, young stem leaf; B, flower buds; EB, elongating flower buds; F, open flowers; GC, green capsules with seeds; RC, ripe capsules with seeds; FM, fresh mass; nd, metabolite not detected in the sample. Only samples with the abbreviated label on x axes in each graph and developmental stage were available for HPLC-PDA. The distribution and accumulation of two herbivory-noninducible phenylpropanoid metabolites, CGA and rutin, from an identical set of samples are shown for comparison in Supplemental Figure S11.
MYB8 Connects JA to Metabolic Genes and PAs
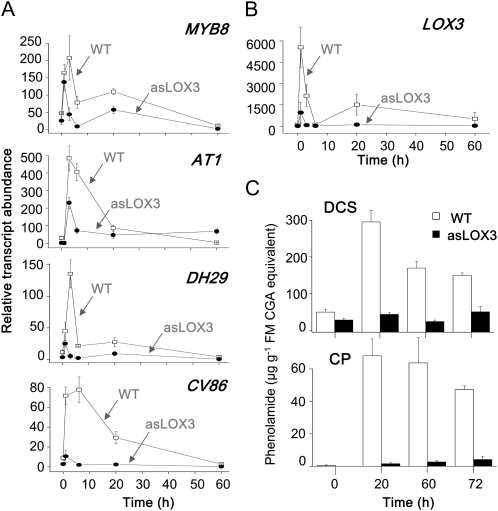
Previously, low levels of CP and DCS were reported in both JA biosynthesis- and JA perception-deficient N. attenuata plants (Paschold et al., 2008; Demkura et al., 2010). To determine if MYB8 directly links JA to downstream metabolic genes, we examined the accumulation patterns of MYB8, AT1, DH29, and CV86 transcripts (Fig. 8A) in plants silenced in the expression of the LOX3 gene (asLOX3; Fig. 8B) and therefore having very low accumulated levels of JA (Halitschke and Baldwin, 2003; Onkokesung et al., 2010). Relative transcript levels of all studied genes were coordinately reduced in asLOX3 compared with wild-type plants before and 1, 6, 20, and 60 h after W+OS elicitation (Fig. 8A). The levels of CP and DCS determined in the same samples confirmed the previously demonstrated reduction of CP and DCS in the asLOX3 genetic background (Fig. 8C), establishing the important role of JA, MYB8, and the MYB8-controlled acyltransferase gene network in the PA accumulation process in response to herbivory in N. attenuata (Fig. 9).
Figure 8.
Silencing of jasmonate biosynthesis in N. attenuata impairs CP and DCS accumulation that is mediated by the MYB8 regulatory network. A, The transcript abundance of MYB8 and MYB8-dependent genes AT1, DH29, and CV86 was determined by RT-qPCR in W+OS-elicited leaves from wild-type (WT) and JA biosynthesis-deficient (asLOX3) plants. B, Silencing of the LOX3 gene in asLOX3 plants. C, Strongly reduced contents of CP and DCS in the leaves of asLOX3 plants. FM, Fresh mass. Values are means of three replicate measurements with se indicated.
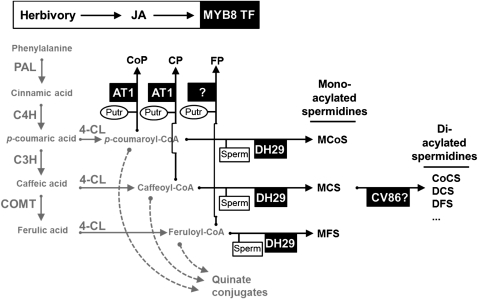
Figure 9.
Schematic summary of AT1, DH29, and CV86 action in herbivore-attacked N. attenuata plants. Metabolic function of AT1 and DH29 and putative function of CV86 are placed in the context of phenylpropanoid metabolism regulated by MYB8 transcriptional activity after herbivore attack. AT1 is directly involved in the biosynthesis of CoP and CP, while FP biosynthesis involves another unknown acyltransferase enzyme. In another branch of PA metabolism, DH29 mediates the biosynthesis of monoacylated spermidines such as monocoumaroylspermidine (MCoS), MCS, and monoferuloylspermidine (MFS). CV86 is proposed to encode the second acylation of monoacylated spermidines to DCS and related metabolites in N. attenuata. PAL, Phenylalanine ammonia-lyase; C4H, cinnamate-4-hydroxylase, C3H, coumarate-3-hydroxylase; COMT, caffeate O-methyltransferase; 4CL, 4-coumarate:coenzyme A ligase; CoCS, p-coumaroylcaffeoylspermidine; DFS, diferuloylspermidine; Putr, putrescine; Sperm, spermidine.
DISCUSSION
Although the biochemical activity leading to CP and DCS biosynthesis in N. tabacum was demonstrated nearly 20 years ago (Negrel, 1989; Negrel et al., 1991, 1992), cloning and characterization of the respective acyltransferase genes have not been reported. At present, the combination of gene and metabolite profiling methods in N. attenuata provide a new powerful molecular tool for the functional genetics in this established ecological plant model. Taking advantage of this novel platform, we show that JA-regulated AT1, DH29, and CV86 genes encode three essential acyltransferases in PA biosynthesis, including the two most abundant PAs, CP and DCS, in tobacco plants.
Distribution and Role of PAs in N. attenuata
PAs, such as CP, di-p-coumaroylspermidine, and p-coumaroyltyramine, are abundant floral constituents in several Solanaceae plant species. Therefore, it was proposed that PAs should play important role(s) in plant development, and the occurrence of PAs in plants was intensively examined (Cabanne at al., 1981; Martin-Tanguy, 1997, 1985). In N. attenuata, CP and DCS can be readily determined by HPLC in all aboveground tissues (CP [Kaur et al., 2010]; DCS [Fig. 7]). In addition, both CP and DCS are more abundant in young vegetative tissues and reproductive organs, and their accumulation is strongly amplified by the attack of chewing herbivores in N. attenuata plants (CP [Kaur et al., 2010]; DCS [Fig. 7]). These accumulation patterns, therefore, strongly support a defense-related function of PAs against herbivores (Kaur et al., 2010). Notably, the plants that were completely deprived of CP and DCS by MYB8 silencing showed no developmental abnormalities and produced normal flowers and seeds (Kaur et al., 2010). Consistent with stably silenced irMYB8 plants, VIGS plants silenced separately in CP (AT1-VIGS) and DCS (DH29-VIGS) biosynthesis showed completely normal morphology, providing another argument against the proposed developmental roles of PAs.
While the regulatory role of MYB8 in Kaur et al. (2010) was limited to CP and DCS, Gaquerel et al. (2010) demonstrated that a much broader range of PAs can be elicited by simulated herbivory treatment (W+OS) in N. attenuata leaves. Indeed, we found that the majority (if not all) of putrescine- and spermidine-containing PAs in N. attenuata accumulate in a MYB8-dependent manner (Fig. 1; Table I; Supplemental Figs. S1–S4). Although a direct application of CP to N. attenuata leaves retarded the growth of M. sexta larvae (Kaur et al., 2010), the exact mode of toxicity of PAs in insects remains unknown. A broad spectrum of PAs regulated during herbivory, as shown in this paper, suggests that a mixture of various PAs may be required to exert the maximal efficiency of PAs against insects. In addition, as the application of synthetic PAs to insect artificial diet usually did not affect the growth of insects (Bassard et al., 2010), the presence of plant factor(s), such as low molecular mass compounds or enzymes, may be required for the transformation of PAs to toxic metabolites, most likely during the digestion of plant material in insect guts.
The Limited Functionality of DH29 Suggests a Two-Step Biosynthesis of DCS
Previously published data in Arabidopsis suggested that acyltransferases are most likely multifunctional enzymes. For example, Luo et al. (2009) reported two novel spermidine-specific acyltransferases, spermidine dicoumaroyltransferase and spermidine disinapoyltransferase, in Arabidopsis, which mediated the diacylation of spermidine in vitro. The Arabidopsis spermidine-hydroxycinnamoyltransferase recombinant enzyme was even able to produce triferuloylspermidine in vitro (Grienenberger et al., 2009). In contrast, the DH29 enzyme reported here, which readily transferred one acyl group to spermidine, failed to produce DCS or tricaffeoylspermidine from the caffeoyl-CoA and spermidine in vitro. While this could be an artifact of our in vitro reaction conditions and/or the recombinant origin of the protein, we obtained a potential explanation of our data by partially solving the function of the CV86 protein. This enzyme, while being structurally very similar to the DH29 protein, was not involved in the first acylation of spermidine (shown in vivo by silencing CV86). Instead, the inhibition of CV86 activity by VIGS resulted in a strong accumulation of monoacylated spermidines (products of DH29) in herbivore-attacked N. attenuata leaves, strongly suggesting that the CV86 protein might be utilizing monoacylated spermidines as its acyl acceptor substrates.
According to our high-resolution MS data and MS2 experiments, at least four isomeric peaks of DCS exist in N. attenuata (Table I; Supplemental Fig. S4). Interestingly, even when the CV86 gene was strongly silenced by VIGS (Supplemental Fig. S7), the accumulation of most of the DCS isomers was only partially affected (Figs. 3B and 4; Supplemental Fig. S8; Supplemental Tables S5 and S6). This suggests that CV86 might not be the only acyltransferase involved in DCS biosynthesis; therefore, the existence of additional stereospecific acyltransferases is predicted by our data. Perhaps, as already proposed by Bassard et al. (2010), the ability to separately control the accumulation of the different DCS isomers became important if the different isomers acquired specific functions in plant defense and/or development. Functional analysis of the CV86 enzyme and/or identification of the additional CV86-like acyltransferases in tobacco will allow this hypothesis to be directly falsified.
Evidence for a Metabolic Exchange in the PA Pathway
Previously, it was shown that silencing MYB8 TF results in the complete shutdown of PA biosynthesis without substantially affecting fluxes into other potentially interconnected metabolic pathways (nicotine, CGA, and rutin; Kaur et al., 2010). However, working with the singly silenced acyltransferase plants allowed us to appreciate the interconnections and substantial plasticity of the PA biosynthetic pathway. For example, silencing of AT1 (and therefore CP biosynthesis) increased the accumulation of DCS in the herbivore-attacked leaves (Fig. 3, A and B; Supplemental Table S5). In contrast, plants silenced in DH29 expression accumulated significantly more FP (Fig. 4; Supplemental Table S5). This shows that in PA biosynthesis, hydroxycinnamoyl-CoA substrates can be diverted into the production of other PAs, as long as the enzymatic activity of AT1 or DH29 is not sufficient. In addition to limiting acyltransferase activity, similar substrate-switch scenarios, although not demonstrated here, can be predicted if one or more of the polyamine substrates become limiting in the biosynthesis.
Interestingly, the AT1 gene product was required for CoP and CP biosynthesis but not for FP formation in vivo (Fig. 4; Supplemental Table S5). Previously, it has been shown that O-methyltransferase enzymes may contribute to the formation of feruloyl-containing PAs in Arabidopsis pollen (Grienenberger et al., 2009), suggesting an alternative pathway and indirect production of FP from CP by O-methylation in N. attenuata plants. In our in vitro recombinant assays, we were still able to detect a limited activity of the AT1 enzyme with feruloyl-CoA as a substrate (Table II), suggesting that the production of FP in N. attenuata plants may be facilitated by two or more functionally overlapping biosynthetic pathways. Notably, FP production was strongly suppressed in irMYB8 plants, predicting the existence of MYB8-regulated O-methyltransferase(s) genes in N. attenuata.
Functional Diversification of PAs
As shown above, tobacco plants are able to accumulate many PAs, and this number currently increases with the analytical ability to resolve isomeric and hydrated forms of PAs. Such a broad spectrum of PAs may reflect the natural variety of stresses, biotic and abiotic, giving rise to the existence of highly diversified chemical defenses in plants. For example, the accumulation of putrescine/spermidine PAs in N. attenuata was strongly induced by herbivory that is efficiently coordinated by MYB8 TF (Kaur et al., 2010; this report); however, we did not find substantial amounts of other polyamine conjugate forms featuring tyramine, agmatine, or serotonin in the N. attenuata plants challenged with herbivores and/or under the regulation of herbivory-related MYB8 protein (Fig. 1; Table I). Interestingly, the contents of feruloyltyramine, coumaroyltyramine, feruloylagmatine, and coumaroylagmatine are known to strongly increase in response to pathogen infection in plants (Stoessl and Unwin, 1970; Fattorusso et al., 1999; Newman et al., 2001; Matsuda et al., 2009; Muroi et al., 2009; Bassard et al., 2010). This observation suggests that the biological activity of PAs may be linked to the polyamine structure present in the particular defense metabolite. To test this hypothesis, the role of MYB8 in defense against pathogens should be examined in order to complement its already demonstrated role in defense against herbivores.
Future Perspectives of MYB8 Research
While we can now fully appreciate the role of MYB8-regulated PAs in defense against herbivores in N. attenuata, the role of PAs in defense against pathogens and/or UV stress requires more attention (Izaguirre et al., 2007; Demkura et al., 2010). The previously found induction of MYB8 by UV irradiation (Pandey and Baldwin, 2008) thus provides another independent chapter in the functional repertoires of MYB8 TF and PAs in plants exposed to a multitude of stress conditions in nature. In our future experiments, by studying plants in their natural environment, we hope to uncover the full cross talk of signaling pathways and understand the complete roles of MYB8 and PAs in the stress resistance of plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Nicotiana attenuata (31st inbred generation) seeds, originally collected from a native population at a field site located in Utah, were used in all experiments. The transformants used were irMYB8 plants previously described by Kaur et al. (2010) and antisense LOX3 plants as described by Halitschke and Baldwin (2003).
Seeds were germinated on Gamborg’s B5 medium (Duchefa) as described previously by Krügel et al. (2002). Seedlings were maintained at 26°C, 16 h of 155 μmol m−2 s−1 light/24°C, 8 h of dark (Percival) for 10 d and then transplanted to soil in Teku plastic pots. After 10 d, early rosette plants were transferred to 1-L pots and grown in the glasshouse with a day/night cycle of 16 h (26°C–28°C)/8 h (22°C–24°C) under supplemental light from Master Sun-T PIA Agro 400 or Master Sun-T PIA Plus 600 high-pressure sodium lamps (Philips Sun-T Agro; http://www.nam.lighting.philips.com). Plants for VIGS experiments were grown in climate chambers with a day/night cycle of 16 h (20°C)/8 h (20°C) under supplemental light from Master Sun-T PIA Agro 400 or 600.
Plant Elicitations
Elicitation by Direct Feeding
One freshly hatched Manduca sexta neonate was placed on stem leaves of AT1-, DH29-, CV86-, and EV-VIGS plants, and neonates were allowed to feed on the plants for 4 d. After removing the larvae, the remaining leaf tissues were collected for metabolite and transcript analyses. The untreated leaf tissues from similarly positioned EV leaves were collected at the same time and used as a control. Leaf tissues were frozen in liquid nitrogen and kept at −80°C until analysis.
Simulated Herbivory Treatment
Transition rosette leaves (stable lines) or stem leaves (VIGS plants) were wounded with a fabric pattern wheel and immediately treated with 20 μL of 1:5 (v/v) water-diluted OS from M. sexta. Control plants remained untreated. Treated and untreated leaf tissues were collected at designated time points, frozen in liquid nitrogen, and kept at −80°C until analysis. M. sexta larval OS were collected after larvae were reared on N. attenuata wild-type plants until the third to fifth instar; OS were collected after regurgitation through a Teflon tube connected to vacuum and stored under argon at −20°C until use.
VIGS
Vector construction, plant growth, and inoculation conditions were as described by Saedler and Baldwin (2004). Briefly, 200- to 300-bp fragments of N. attenuata AT1, DH29, and CV86 were amplified by PCR using specific primer pairs as listed in Supplemental Table S3. Amplified fragments were cloned into pTV00 vector, and plasmids were transformed by electroporation into Agrobacterium tumefaciens strain GV3101. A pTV00 plasmid without insert (EV) was used as a negative control in all experiments. Three leaves of 24- to 25-d-old N. attenuata plants were infiltrated with a 1:1 mixture of A. tumefaciens transformed with pBINTRA and one of the pTVAT1, pTVDH29, pTVCV86, or pTV00 constructs. Phytoene desaturase (pTVPDS) causing bleaching of tobacco leaves due to the depletion of carotenoids was used as a positive control to monitor the progression of VIGS in a separate set of inoculated plants. AT1-, DH29-, CV86-, and EV-VIGS-silenced plants were used for treatment after PDS-VIGS leaves developed a strong bleaching phenotype. Silencing efficiency was verified by RT-qPCR of target gene transcripts after RNA extraction and cDNA synthesis.
RT-qPCR Analysis
Total RNA was extracted by adding Trizol reagent (Invitrogen; http://www.invitrogen.com) to approximately 150 mg of powdered leaf material ground in liquid nitrogen following the manufacturer’s protocol. Crude RNA samples were treated with RQ1 DNase (Promega; http://www.promega.com), followed by phenol:chloroform:isoamyl alcohol (25:24:1) extraction and ethanol precipitation. A total of 500 ng of DNA-free RNA samples was reverse transcribed using oligo(dT)18 primers and SuperScript II enzyme (Invitrogen) following the manufacturer’s recommendations. All RT-qPCR assays were performed with a Stratagene MX3005P instrument (http://www.stratagene.com) as recommended by the manufacturer. For normalization of transcript levels, primers specific for the elongation factor-1α gene from Nicotiana tabacum (EF1-α; accession no. D63396) were used. Specific primers in the 5′ to 3′ direction used for SYBR Green-based analyses are listed in Supplemental Table S3. Typically, data from three to five biological replicates were used for statistical analysis.
Identification of the Novel Acyltransferase Gene AT1
The NCBI “non-human, non-mouse ESTs (est_others)” database limited to “Nicotiana tabacum (taxid:4097)” and the TBLASTN program with default settings were used to identify a set of 500 TBLASTN hits with corresponding e-values of less than 0.059 and less than 0.020 for DH29 and CV86, respectively. All FASTA-formatted EST sequences similar to DH29 and CV86 were uploaded from the database (Supplemental Table S4, worksheet A) and included in the assembly of ESTs using the publicly available CAP3 program with default parameters (http://deepc2.psi.iastate.edu/aat/cap/cap.html). Each assembled contig (Supplemental Text S2) was BLASTed using our local BLASTN engine against the N. attenuata-specific microarray 60-mer Agilent oligoprobe database (all 43,533 oligonucleotide sequences can be downloaded from the GEO microarray repository as part of the N. attenuata Agilent platform GPL13527). All contigs that did not provide a significant hit against the microarray oligonucleotide database were reBLASTed against N. tabacum ESTs using the NCBI BLASTN program, and where possible, the 5′ and 3′ ends of the contigs were extended (Supplemental Text S2). Extended contigs (e.g. contig5-1 and contig5-2 represent the extensions of contig5) were reBLASTed against Agilent 60-mer oligonucleotides to obtain a set of 53 contigs that showed a positive greater than 84% match to the 60-mer Agilent oligonucleotide sequence database, representing 32 unique genes from N. attenuata (Supplemental Table S4, worksheet B).
Microarray Analysis
N. attenuata seedlings were cultivated in 1-L pots with sand to facilitate sampling of the roots. Plants were watered by a flood irrigation system with 200 g of Ca(NO3)2.4H2O and 200 g of Flory B1 fertilizer in 400 L of water and grown in the glasshouse under standard conditions. Three rosette leaves at positions 0, +1, and +2 of early elongated stage plants were elicited with simulated herbivory (W+OS), wounding (W+W), or remained untreated. Locally treated leaves, systemic unwounded leaves (two younger leaves above the treated leaves), and roots were collected at 1, 5, 9, 13, 17, and 21 h after treatment, and tissues were frozen in liquid nitrogen and kept at −80°C until analysis. Microarrays were performed exactly as described by Gilardoni et al. (2011). Raw and normalized data for each triplicate treatment (a total of 134 samples) were deposited in the GEO repository under sample numbers GSM750588 to GSM750721 (Agilent platform identification no. GPL13527). In this report, raw data in the Agilent “gProcessedSignal” column were normalized by dividing each value by the 75th percentile value of each experiment. Expression data for the individual genes shown in Supplemental Figure S5 were manually extracted from the array matrix and plotted in x-y plots using the Excel program (Microsoft Office 2003).
Expression and Purification of Recombinant Proteins
Full-length cDNA of AT1, DH29, and CV86 without natural stop codons were amplified using specific primers as listed in Supplemental Table S3. Sequence-verified PCR products were cloned into pET23a (Novagen; http://www.merck-chemicals.de) as an NdeI-XhoI fragment (AT1 and DH29) or an NdeI-NotI fragment (CV86) to produce His-tagged fusion proteins (C-terminal 6× His tag) in the expression vector (pET23a-AT1, pET23a-DH29, and pET23a-CV86). Expression vectors were transformed into BL21 (DE3) pLysS Escherichia coli host cells (Novagen) by standard electroporation. For each protein, expression conditions were first optimized in 10-mL bacterial cultures. For large-scale protein extractions, 250-mL cultures were allowed to grow at optimal temperature (28°C) until the culture reached an optical density at 600 nm of 1; isopropyl β-d-1-thiogalactopyranoside (Roth; http://www.carlroth.com) was added to a final concentration of 1 mm to induce recombinant genes, and cultivation was continued for 24 h. Bacterial cultures were centrifuged at 2,500g at 4°C for 20 min, and pellets were frozen in liquid nitrogen after completely removing the supernatants and stored at −80°C. Cell pellets were resuspended in lysis buffer (50 mm Na2HPO4/300 mm NaCl, 1 mg mL−1 lysozyme, and 10 mm imidazole) and incubated on ice for 30 min. All buffers were supplemented with 6 m urea in the case of CV86 protein extraction under denaturing conditions. Homogenates were disrupted by 3 min of sonication in a Bandelin UW2070 sonicator (Sonoplus; power at 60%, 4× cycle) and centrifuged at 10,000g, 4°C for 30 min. Supernatants were loaded onto 2-mL Hi-Trap charged nickel chloride (Jena Biosciences) columns in 5-mL plastic reservoirs. Proteins were eluted from the columns with 50 mm Na2HPO4 buffer containing 300 mm NaCl and a gradient of 0 to 0.5 m imidazole. Twenty-microliter eluted protein fractions were subjected to SDS-PAGE to identify recombinant protein-containing fractions. The positive fractions were pooled and desalted with Zeba Spin Desalting Columns (7,000 molecular weight cutoff; 10 mL; Thermo Scientific; http://www.piercenet.com). One hundred-microliter aliquots of purified AT1 and DH29 proteins were snap frozen in liquid nitrogen and stored at −80°C. CV86 protein extracted under denaturing conditions was renatured by dialysis of the protein fraction against 50 mm Tris-HCl buffer, pH 7.5, containing gradually decreasing concentrations of urea at 4°C in a cold room. SDS-PAGE was performed using 8% to 10% acrylamide gels with Tris-Gly SDS running buffer. Protein bands were visualized using bio-safe Coomassie blue stain (Bio-Rad).
In Vitro Enzyme Assays
Hydroxycinnamoyl-CoAs of p-coumaric, caffeic, and ferulic acids were synthesized using N. tabacum recombinant 4CL enzyme as described by Beuerle and Pichersky (2002). However, this enzyme was found to be completely inactive with cinnamic and sinapic acids under our conditions. All enzymatic assays were performed in a total volume of 100 μL containing available acyl donors (caffeoyl-, feruloyl-, and p-coumaroyl-CoA) and polyamines (putrescine, spermidine, and spermine) as acyl acceptors in 100 mm Tris-HCl buffer, pH 7.5, containing 5 mm EDTA (pH 8). For testing enzyme activity, the reactions were initiated by the addition of purified enzymes, incubated at 30°C for 15 min, and stopped by adding 1 μL of 12 n HCl and 15 μL of acetonitrile. Undiluted samples were injected into the HPLC-PDA device and analyzed for reaction products based on RT and UV absorbance spectra. The identities of the reaction products were confirmed by UPLC-ESI-TOF-MS analysis. The kinetic constants of AT1 and DH29 were determined using 0 to 750 μm caffeoyl-CoA as acyl donor and 0 to 2 mm polyamine as acyl acceptor. Reactions were monitored in an Infinite M200 spectrophotometer (Tecan; http://www.tecan.com) at 1-min intervals by following the decrease of specific UV A365 from caffeoyl-CoA substrate.
Phylogenetic Analysis
BAHD protein sequences described by Luo et al. (2009) and several other functionally characterized hydroxycinnamoyl-CoA:amine N-(hydroxycinnamoyl) transferases (Bassard et al., 2010) were supplemented with AT1, DH29, and CV86 sequences from N. attenuata and used to generate a multiple alignment using the MUSCLE program (http://www.ebi.ac.uk/Tools/msa/muscle/) with default parameters (Supplemental Data Set S1). The protein alignment was trimmed in order to remove highly variable regions of the aligned proteins that contained frequent insertions and deletions. Trimming of the sequence alignment shown in Supplemental Data Set S1 was performed using Gblocks (http://molevol.cmima.csic.es/castresana/Gblocks.html), with the maximum number of contiguous nonconserved positions set to 100 and the minimum length of a block set to two. Gblocks trimming has defined 16 blocks showing sequence conservation, which gave 133-amino acid trimmed sequences, as shown in Supplemental Data Set S2. A tree was built using the neighbor-joining clustering method with the MEGA 4 program. One thousand iterations were used to calculate the bootstrap values assessing the confidence of each tree clade.
Secondary Metabolite Quantification by HPLC-PDA
PAs in VIGS experiments were quantified with a HPLC-PDA detector. Approximately 100 mg of liquid nitrogen-ground leaf powders was extracted by adding 1 mL of acidified 40% methanol prepared with 0.5% acetic acid water to each sample in 2-mL microcentrifuge tubes with metal balls. The samples were homogenized in the ball mill (Genogrinder 2000; SPEX CertiPrep) for 45 s at 1× rate and 250 strokes per min. Homogenized samples were centrifuged at 16,000g, 4°C for 30 min, and supernatants were transferred into 1.5-mL microcentrifuge tubes and recentrifuged as before. Supernatants of 400 μL were transferred to 2-mL glass vials before analyzing on an Agilent HPLC 1100 series device (http://www.chem.agilent.com); 1 μL of the sample was injected in a Chromolith FastGradient RP 18-e column (50 × 2 mm; monolithic silica with bimodal pore structure, macropores with 1.6 μm diameter; Merck) attached to a precolumn (Gemini NX RP18, 2 × 4.6 mm, 3 μm). The mobile phases (0.1% formic acid + 0.1% ammonium water, pH 3.5) as solvent (A) and methanol as solvent (B) were used in a gradient mode with the following conditions: time/concentration (min/%) for B was 0.0/0, 0.5/0, 6.5/80, 9.5/80, and reconditioning for 5 min to 0% B. The flow rate was 0.8 mL min−1, and column oven temperature was set to 40°C. DCS, CGA, and rutin determination in N. attenuata development was performed as described previously by Kaur et al. (2010). Peaks from individual DCS isoforms were pooled and reported as total DCS in Figures 3, 7 and 8. CP and DCS were quantified based on the calibration curves constructed for CGA and expressed as CGA equivalents.
UPLC-ESI-TOF-MS Measurements
Two microliters of the leaf extracts prepared as above was typically separated using a Dionex Rapid Separation LC system equipped with a Dionex Acclaim 2.2-μm 120A, 2.1- × 150-mm column, applying a short separation binary gradient (flow rate of 300 mL min -1) with the following parameters: 0 to 0.5 min, isocratic 80% A (deionized water, 0.1% [v/v] acetonitrile [Baker; HPLC grade], and 0.05% formic acid), 20% B (acetonitrile, 0.05% formic acid); 0.5 to 2 min, linear gradient to 40% B; 2 to 6 min, isocratic 40% B; 6 to 10 min, linear gradient to 80% B. For determination of PAs in Fig. 1 and Supplemental Figs. 4 and 8, an optimized long separation gradient was used (flow rate of 300 μL min–1); 0 to 5 min, isocratic 95% A, 5% B; 5 to 20 min, linear gradient to 32% B; 20 to 22 min, linear gradient to 80% B; isocratic for 6 min. Eluted compounds were detected by a MicroToF mass spectrometer (Bruker Daltonics) equipped with an ESI source in positive ionization mode. Typical instrument settings were as follows: capillary voltage, 4,500 V; capillary exit, 130 V; dry gas temperature, 200°C; dry gas flow of 8 L min–1. Ions were detected from m/z 200 to 1,400 at a repetition rate of 1 Hz. Mass calibration was performed using sodium formate clusters (10 mm solution of NaOH in 50%:50% [v/v] isopropanol:water containing 0.2% formic acid). The level of identification of the metabolites reported in this study is denoted in Table I using the four levels of the metabolite annotation nomenclature proposed by the Metabolome Standard Initiative (Sumner et al., 2007), employed as described by Matsuda et al. (2010).
Processing of UPLC-ESI-TOF-MS Data
Raw data files were converted to netCDF format using the export function of the Data Analysis version 4.0 software (Bruker Daltonics) and processed using the XCMS package (http://metlin.scripps.edu/download/). Peak detection was performed using the “centWave” method and the following parameter settings: ppm = 20, snthresh = 10, peakwidth = c(5,18). Retention time correction was achieved using the following parameter settings: minfrac = 1, bw = 10 s, mzwid = 0.1 D, span = 1, missing = extra = 0. After peak grouping and filling in of missing features using the fillPeaks routine of the XCMS package, the obtained data matrix was imported into Microsoft Excel for statistical analysis. Ion traces were deconvoluted, and putative in-source pseudospectra were reconstructed with the R package CAMERA (http://www.bioconductor.org/packages/release/bioc/html/CAMERA.html) with defaults parameters. Isotopic peaks and multicharged m/z signals detected by CAMERA were excluded to reduce the redundancy within the data matrix. Consistent mass features, which were at least present (for a single factorial group) in four out of the five biological replicates with retention time greater than 1 min, were considered for further analysis. Zero values remaining after applying the “filling in” function in XCMS were replaced by half of the minimum positive value of the row in the original data.
Statistical Analysis
For PCA of UPLC-ESI-TOF-MS profiling data, we first filtered the data in order to identify and remove m/z signals that are unlikely to be of use when modeling the data. No phenotype information was used during the filtering process, so the results can be used with any downstream analysis. Forty percent of the original m/z features were considered as near constant throughout the experiment conditions, based on their coefficient of variation (mean divided by sd), and removed. Normalization was used to make each variable comparable to each other within the same sample. We used the Pareto scaling (mean centered and divided by the square root of the sd of each variable) method for data normalization, as recommended by van den Berg et al. (2006). The PCA analysis was performed using the prcomp package for R via the MetaboAnalyst interface. The calculation is based on singular value decomposition. The HPLC data and RT-qPCR data were analyzed with PASW statistic 18 (SPSS) software.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: DH29 (fragment, CA591847; full-length CDS, JN390824); CV86 (fragment, BU494534; full-length coding sequence, JN390825); AT1 (full-length CDS, JN390826); EF1-α (D63396).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. PCA separates herbivory-induced large-scale metabolic profiles in irMYB8 leaves from those of wild-type leaves.
Supplemental Figure S2. PC ranking analysis, W+OS inducibility, and irMYB8-silencing effects on leaf metabolomics profiles.
Supplemental Figure S3. MYB8 silencing results in large-scale alterations in N. attenuata leaf secondary metabolism.
Supplemental Figure S4. Extracted ion chromatograms for selected monoacylated putrescines and mono- and diacylated spermidines in the 24-h W+OS-elicited leaves of wild-type and irMYB8 plants.
Supplemental Figure S5. Expression profiles of MYB8, AT1, DH29, and CV86 determined by microarray analysis in the control (untreated), W+W-, and W+OS-elicited tobacco tissues.
Supplemental Figure S6. Phylogenetic tree of the BAHD enzymes from Arabidopsis and other PA biosynthetic enzymes identified from other plant species.
Supplemental Figure S7. Silencing efficiency of AT1, DH29, and CV86 genes in the VIGS plants.
Supplemental Figure S8. Representative extracted ion chromatograms for monoacylated putrescines and monoacylated and diacylated spermidines in the 24-h W+OS-elicited leaves of EV plants and AT1-, DH29-, and CV86-silenced plants.
Supplemental Figure S9. Characterization of AT1 and DH29 recombinant enzymes in vitro.
Supplemental Figure S10. Determination of the pH optima of AT1 and DH29 recombinant enzymes in vitro.
Supplemental Figure S11. Time- and tissue-resolved accumulation of CGA and flavonoid rutin in N. attenuata plants.
Supplemental Table S1. Summary of loadings exerted on PC1 and PC2 by 4,128 m/z signals obtained after preprocessing with the R package XCMS, and log2-transformed fold changes.
Supplemental Table S2. Summary of molecular fragment relative intensities and fold changes in irMYB8 compared with wild-type leaves.
Supplemental Table S3. Primer sequences used for RT-qPCR and cloning of VIGS vectors into pTV00 or full-length cDNA sequences into pET23a for protein expression analysis.
Supplemental Table S4. Identification of the AT1 gene as a novel herbivory-regulated acyltransferase in N. attenuata.
Supplemental Table S5. Fold changes and related statistical analysis (n = 4) of the metabolites depicted in Figure 4.
Supplemental Table S6. Summary of up- and down-regulated m/z features and predicted metabolites in the leaves of plants transiently silenced by VIGS for CV86, DH29, and AT1 expression compared with EV-infiltrated plants.
Supplemental Text S1. Description of the method and tandem MS measurements for the precursor ions of selected PAs.
Supplemental Text S2. CAP3-assembled contigs with significant similarity to DH29 and CV86 proteins represent a pool of novel acyltransferase enzymes in tobacco.
Supplemental Data Set S1. BAHD protein alignment used for trimming by Gblocks before generating the phylogenetic tree by MEGA4 displayed in Supplemental Figure S6.
Supplemental Data Set S2. Gblocks-trimmed BAHD protein alignment used for generating the phylogenetic tree by MEGA4 displayed in Supplemental Figure S6.
Acknowledgments
We thank Aura Navarro Quezada for advice and help with the phylogenetic analysis of acyltransferase genes; Klaus Gase and Gustavo Bonaventure for help with the preparation of materials for microarray construction; Wibke Kröber for hybridization of Agilent microarrays; San Gyu Kim for providing plant materials for a time-course microarray experiment; Antje Wissgott for help with VIGS vector constructs; John D’Auria for the donation of cloned 4CL enzyme in an expression vector; Andreas Weber and Andreas Schünzel for growing the plants in the glasshouse; and Aleš Svatoš, Nicolas Heinzel, and Mathias Schöttner for help with the elucidation and consultation of unknown PA structures.
References
- Back K, Jang SM, Lee BC, Schmidt A, Strack D, Kim KM. (2001) Cloning and characterization of a hydroxycinnamoyl-CoA:tyramine N-(hydroxycinnamoyl)transferase induced in response to UV-C and wounding from Capsicum annuum. Plant Cell Physiol 42: 475–481 [DOI] [PubMed] [Google Scholar]
- Baldwin IT. (1996) Methyl jasmonate-induced nicotine production in Nicotiana attenuata: inducing defenses in the field without wounding. Entomol Exp Appl 80: 213–220 [Google Scholar]
- Bari R, Jones JD. (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Bassard JE, Ullmann P, Bernier F, Werck-Reichhart D. (2010) Phenolamides: bridging polyamines to the phenolic metabolism. Phytochemistry 71: 1808–1824 [DOI] [PubMed] [Google Scholar]
- Bernards MA, Båstrup-Spohr L. (2008) Phenylpropanoid metabolism induced by wounding and insect herbivory. Schaller A, , Induced Plant Resistance to Herbivory. Springer, Dordrecht, The Netherlands, pp 189–211 [Google Scholar]
- Beuerle T, Pichersky E. (2002) Enzymatic synthesis and purification of aromatic coenzyme A esters. Anal Biochem 302: 305–312 [DOI] [PubMed] [Google Scholar]
- Burhenne K, Kristensen BK, Rasmussen SK. (2003) A new class of N-hydroxycinnamoyltransferases: purification, cloning, and expression of a barley agmatine coumaroyltransferase (EC 2.3.1.64). J Biol Chem 278: 13919–13927 [DOI] [PubMed] [Google Scholar]
- Cabanne F, Dalebroux MA, Martin-Tanguy J, Martin C. (1981) Hydroxycinnamic acid-amides and ripening to flower of Nicotiana tabacum var Xanthi nc. Physiol Plant 53: 399–404 [Google Scholar]
- Camacho-Cristóbal JJ, Maldonado JM, Gonzláez-Fontes A. (2005) Boron deficiency increases putrescine levels in tobacco plants. J Plant Physiol 162: 921–928 [DOI] [PubMed] [Google Scholar]
- D’Auria JC. (2006) Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol 9: 331–340 [DOI] [PubMed] [Google Scholar]
- Demkura PV, Abdala G, Baldwin IT, Ballaré CL. (2010) Jasmonate-dependent and -independent pathways mediate specific effects of solar ultraviolet B radiation on leaf phenolics and antiherbivore defense. Plant Physiol 152: 1084–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, D’Auria JC, Nam KH, Raguso RA, Pichersky E. (1998) Acetyl-CoA:benzylalcohol acetyltransferase: an enzyme involved in floral scent production in Clarkia breweri. Plant J 14: 297–304 [DOI] [PubMed] [Google Scholar]
- Edreva AM, Velikova VB, Tsonev TD. (2007) Phenylamides in plants. Russ J Plant Physiol 54: 287–301 [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Hagel J, Zulak KG. (2002) Hydroxycinnamic acid amide metabolism: physiology and biochemistry. Can J Bot 80: 577–589 [Google Scholar]
- Fattorusso E, Lanzetii V, Taglialatela-Scafati O. (1999) Antifungal N-feruloylamides from roots of Allium species. Plant Biosyst 133: 199–203 [Google Scholar]
- Galis I, Simek P, Narisawa T, Sasaki M, Horiguchi T, Fukuda H, Matsuoka K. (2006) A novel R2R3 MYB transcription factor NtMYBJS1 is a methyl jasmonate-dependent regulator of phenylpropanoid-conjugate biosynthesis in tobacco. Plant J 46: 573–592 [DOI] [PubMed] [Google Scholar]
- Gaquerel E, Heiling S, Schoettner M, Zurek G, Baldwin IT. (2010) Development and validation of a liquid chromatography-electrospray ionization-time-of-flight mass spectrometry method for induced changes in Nicotiana attenuata leaves during simulated herbivory. J Agric Food Chem 58: 9418–9427 [DOI] [PubMed] [Google Scholar]
- Gilardoni P, Hettenhausen C, Baldwin IT, Bonaventure G. (2011) The Nicotiana attenuata NaLecRK1 gene suppresses the insect-mediated inhibition of induced defense responses during Manduca sexta herbivory. Plant Cell 23: 3512–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens A, Häkkinen ST, Laakso I, Seppänen-Laakso T, Biondi S, De Sutter V, Lammertyn F, Nuutila AM, Söderlund H, Zabeau M, et al. (2003) A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc Natl Acad Sci USA 100: 8595–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger E, Besseau S, Geoffroy P, Debayle D, Heintz D, Lapierre C, Pollet B, Heitz T, Legrand M. (2009) A BAHD acyltransferase is expressed in the tapetum of Arabidopsis anthers and is involved in the synthesis of hydroxycinnamoyl spermidines. Plant J 58: 246–259 [DOI] [PubMed] [Google Scholar]
- Halitschke R, Baldwin IT. (2003) Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J 36: 794–807 [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G. (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Hui DQ, Iqbal J, Lehmann K, Gase K, Saluz HP, Baldwin IT. (2003) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. V. Microarray analysis and further characterization of large-scale changes in herbivore-induced mRNAs. Plant Physiol 131: 1877–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaguirre MM, Mazza CA, Svatos A, Baldwin IT, Ballaré CL. (2007) Solar ultraviolet-B radiation and insect herbivory trigger partially overlapping phenolic responses in Nicotiana attenuata and Nicotiana longiflora. Ann Bot (Lond) 99: 103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Heinzel N, Schöttner M, Baldwin IT, Gális I. (2010) R2R3-NaMYB8 regulates the accumulation of phenylpropanoid-polyamine conjugates, which are essential for local and systemic defense against insect herbivores in Nicotiana attenuata. Plant Physiol 152: 1731–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinänen M, Oldham NJ, Baldwin IT. (2001) Rapid HPLC screening of jasmonate-induced increases in tobacco alkaloids, phenolics, and diterpene glycosides in Nicotiana attenuata. J Agric Food Chem 49: 3553–3558 [DOI] [PubMed] [Google Scholar]
- Krügel T, Lim M, Gase K, Halitschke R, Baldwin IT. (2002) Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology 12: 177–183 [Google Scholar]
- Luo J, Fuell C, Parr A, Hill L, Bailey P, Elliott K, Fairhurst SA, Martin C, Michael AJ. (2009) A novel polyamine acyltransferase responsible for the accumulation of spermidine conjugates in Arabidopsis seed. Plant Cell 21: 318–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nishiyama Y, Fuell C, Taguchi G, Elliott K, Hill L, Tanaka Y, Kitayama M, Yamazaki M, Bailey P, et al. (2007) Convergent evolution in the BAHD family of acyl transferases: identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J 50: 678–695 [DOI] [PubMed] [Google Scholar]
- Martin-Tanguy J. (1985) The occurrence and possible function of hydroxycinnamoyl acid-amides in plants. Plant Growth Regul 3: 381–399 [Google Scholar]
- Martin-Tanguy J. (1997) Conjugated polyamines and reproductive development: biochemical, molecular and physiological approaches. Physiol Plant 100: 675–688 [Google Scholar]
- Martin-Tanguy J, Cabanne F, Perdrizet E, Martin C. (1978) Distribution of hydroxycinnamic acid-amides in flowering plants. Phytochemistry 17: 1927–1928 [Google Scholar]
- Matsuda F, Hirai MY, Sasaki E, Akiyama K, Yonekura-Sakakibara K, Provart NJ, Sakurai T, Shimada Y, Saito K. (2010) AtMetExpress development: a phytochemical atlas of Arabidopsis development. Plant Physiol 152: 566–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Yonekura-Sakakibara K, Niida R, Kuromori T, Shinozaki K, Saito K. (2009) MS/MS spectral tag-based annotation of non-targeted profile of plant secondary metabolites. Plant J 57: 555–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroi A, Ishihara A, Tanaka C, Ishizuka A, Takabayashi J, Miyoshi H, Nishioka T. (2009) Accumulation of hydroxycinnamic acid amides induced by pathogen infection and identification of agmatine coumaroyltransferase in Arabidopsis thaliana. Planta 230: 517–527 [DOI] [PubMed] [Google Scholar]
- Negrel J. (1989) The biosynthesis of cinnamoylputrescines in callus tissue cultures of Nicotiana tabacum. Phytochemistry 28: 477–481 [Google Scholar]
- Negrel J, Javelle F, Paynot M. (1991) Separation of putrescine and spermidine hydroxycinnamoyltransferases extracted from tobacco callus. Phytochemistry 30: 1089–1092 [Google Scholar]
- Negrel J, Paynot M, Javelle F. (1992) Purification and properties of putrescine hydroxycinnamoyltransferase from tobacco (Nicotiana tabacum) cell suspensions. Plant Physiol 98: 1264–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, von Roepenack-Lahaye E, Parr A, Daniels MJ, Dow JM. (2001) Induction of hydroxycinnamoyl-tyramine conjugates in pepper by Xanthomonas campestris, a plant defense response activated by hrp gene-dependent and hrp gene-independent mechanisms. Mol Plant Microbe Interact 14: 785–792 [DOI] [PubMed] [Google Scholar]
- Onkokesung N, Gális I, von Dahl CC, Matsuoka K, Saluz H-P, Baldwin IT. (2010) Jasmonic acid and ethylene modulate local responses to wounding and simulated herbivory in Nicotiana attenuata leaves. Plant Physiol 153: 785–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SP, Baldwin IT. (2008) Silencing RNA-directed RNA polymerase 2 increases the susceptibility of Nicotiana attenuata to UV in the field and in the glasshouse. Plant J 54: 845–862 [DOI] [PubMed] [Google Scholar]
- Paschold A, Bonaventure G, Kant MR, Baldwin IT. (2008) Jasmonate perception regulates jasmonate biosynthesis and JA-Ile metabolism: the case of COI1 in Nicotiana attenuata. Plant Cell Physiol 49: 1165–1175 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Lewinsohn E. (2011) Convergent evolution in plant specialized metabolism. Annu Rev Plant Biol 62: 549–566 [DOI] [PubMed] [Google Scholar]
- Saedler R, Baldwin IT. (2004) Virus-induced gene silencing of jasmonate-induced direct defences, nicotine and trypsin proteinase-inhibitors in Nicotiana attenuata. J Exp Bot 55: 151–157 [DOI] [PubMed] [Google Scholar]
- Shoji T, Yamada Y, Hashimoto T. (2000) Jasmonate induction of putrescine N-methyltransferase genes in the root of Nicotiana sylvestris. Plant Cell Physiol 41: 831–839 [DOI] [PubMed] [Google Scholar]
- Singh KB, Foley RC, Oñate-Sánchez L. (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5: 430–436 [DOI] [PubMed] [Google Scholar]
- Stoessl A, Unwin CH. (1970) Antifungal factors in barleys. V. Antifungal activity of hordatines. Can J Bot 48: 465–470 [Google Scholar]
- Sugimoto K, Takeda S, Hirochika H. (2000) MYB-related transcription factor NtMYB2 induced by wounding and elicitors is a regulator of the tobacco retrotransposon Tto1 and defense-related genes. Plant Cell 12: 2511–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW, Fiehn O, Goodacre R, Griffin JL, et al. (2007) Proposed minimum reporting standards for chemical analysis. Metabolomics 3: 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka E, Tanaka C, Mori N, Kuwahara Y, Tsuda M. (2003) Phenylpropanoid amides of serotonin accumulate in witches’ broom diseased bamboo. Phytochemistry 64: 965–969 [DOI] [PubMed] [Google Scholar]
- van den Berg RA, Hoefsloot HCJ, Westerhuis JA, Smilde AK, van der Werf MJ. (2006) Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics 7: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelckel C, Baldwin IT. (2003) Detecting herbivore-specific transcriptional responses in plants with multiple DDRT-PCR and subtractive library procedures. Physiol Plant 118: 240–252 [Google Scholar]
- Vom Endt D, Kijne JW, Memelink J. (2002) Transcription factors controlling plant secondary metabolism: what regulates the regulators? Phytochemistry 61: 107–114 [DOI] [PubMed] [Google Scholar]
- Woldemariam MG, Baldwin IT, Galis I. (2011) Transcriptional regulation of plant inducible defenses against herbivores: a mini-review. J Plant Interact 6: 113–119 [Google Scholar]
- Zheng ZY, Mosher SL, Fan BF, Klessig DF, Chen ZX. (2007) Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol 7: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]