Plants are sessile organisms living in an environment rich in microbes that are potentially able to cause disease. As a result, plant survival depends on the ability to couple rapid pathogen detection to an efficient defense response. In contrast to the somatic adaptive immune system of mammals involving mobile defender cells, plant immune responses rely on the ability of each cell to recognize and respond to pathogen invasion and on systemic signals originating from infection sites. A first line of plant defense is activated after recognition of pathogen- or microbe-associated molecular patterns (PAMPs or MAMPs), which are highly conserved among microbes. PAMP recognition by specific pattern recognition receptors leads to PAMP-triggered immunity (PTI), which induces basal defenses thereby preventing further pathogen ingress (Jones and Dangl, 2006). To counteract PTI, thriving pathogens gained the ability to secrete virulence effectors able to target crucial PTI regulators. In turn, plants evolved to produce resistance (R) proteins that recognize pathogen-produced effectors or their action on host components, leading to effector-triggered immunity (ETI). ETI provides a second layer of plant defense that is frequently associated to the development of a form of programmed cell death, the so-called hypersensitive response (HR; Jones and Dangl, 2006).
Plant immunity against microbial infection engages highly dynamic responses that involve multiple organelles during the recognition and signaling mechanisms associated to defense. A particularly important role in plant defense responses has been attributed to nuclear dynamics, since a growing number of reports revealed that nuclear localization of pathogen effectors, R proteins, and key host components, including transcription factors (TFs) and regulators, is essential for plant immunity (Deslandes and Rivas, 2011). Spatial restriction of defense regulators by the nuclear envelope as well as their stimulus-induced nuclear translocation provide an important mechanism for defense regulation, as their level of nuclear accumulation determines the output of the defense response. Interestingly, mutations in cellular factors involved in the transport of macromolecules through the nuclear envelope, compromise plant resistance signaling, underlining the importance of nucleocytoplasmic trafficking during plant innate immunity (Deslandes and Rivas, 2011).
Together, these findings situate the nucleus at the forefront of the mutual recognition between plants and pathogens. Here, I provide a summary of our current knowledge about nuclear dynamics during plant immune responses.
THE DOORWAY IN AND OUT THE NUCLEUS: HOW TO CROSS THE NUCLEAR ENVELOPE
Communication between the cytoplasm and the nucleus is a fundamental feature conserved among eukaryotic systems. Transport of macromolecules across the nuclear envelope occurs through nuclear pore complexes (NPCs), which are composed of nucleoporins, and depends on import and export receptors, importins and exportins that respectively recognize nuclear localization signals (NLSs) and nuclear export signals on cargo proteins (Meier and Brkljacic, 2009). The Ras-related nuclear (Ran) protein provides the directionality of transport through its binding to GDP (cytoplasmic side) or GTP (nuclear side). For a more detailed account on dynamics at the nuclear envelope, refer to Meier (2007).
Several cellular factors involved in the transport of macromolecules through the nuclear envelope, including nucleoporins, importins, and Ran-GTP-related components, are essential to mount an efficient immune response in response to different pathogens (Palma et al., 2005; Zhang and Li, 2005; Tameling and Baulcombe, 2007; Cheng et al., 2009). In addition, several reports strongly suggest that components of the NPC specifically mediate the transport of R proteins, immunity components, as well as TFs and regulators that are necessary for activation of disease resistance (García and Parker, 2009). These and other findings support the notion that specific modulation of the nuclear concentration of a set of defense regulators is crucial for the fine tuning of plant immunity.
PATHOGEN EFFECTORS AND PLANT RESISTANCE PROTEINS
Following delivery into the plant cell, microbial effectors may be targeted to different cellular compartments where they may manipulate a variety of host cellular functions. Importantly, a significant number of effector proteins from different pathogens, including nematodes, fungi, viruses, bacteria, and oomycetes, are targeted to the nucleus by co-opting the host nuclear import machinery (Deslandes and Rivas, 2011). This observation suggests that effectors may manipulate host transcription or directly target nuclear essential host components for the benefit of the pathogen. It has been additionally proposed that some effectors may affect histone modification and chromatin remodelling. Alternatively, nuclear translocation of effectors may affect subcellular localization of their cognate R proteins in a process that is essential for R-protein-mediated plant immunity.
Assorted Effector Stratagems to Manipulate Host Transcription
Most of the molecular mechanisms by which nuclear localization of microbial effectors contributes to promotion of virulence remain to be elucidated. However, some well-characterized examples, mainly from the bacterial field, have been seminal to illustrate pathogen strategies hidden behind effector nuclear targeting.
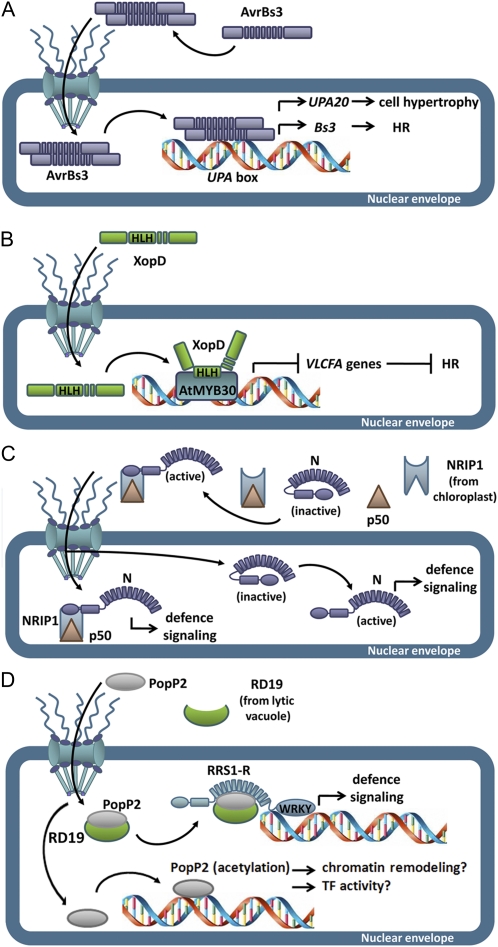
To promote virulence, some nuclear effectors are able to induce transcriptional reprogramming in host cells. For example, the Xanthomonas campestris pv vesicatoria effector AvrBs3 and likely other members of the transcription activator-like effector family provoke developmental reprogramming of host cells by mimicking eukaryotic TFs (Kay and Bonas, 2009). AvrBs3 dimerizes in the plant cell cytoplasm and is translocated to the nucleus via the specific recognition of its NLSs by importin α. In the nucleus, AvrBs3 is able to bind to a specific DNA sequence, the Up-regulated by AvrBs3 (UPA) box, and induce the expression of the so-called UPA genes, via its acidic transcriptional activation domain. Among the UPA genes, UPA20, which encodes a TF containing a basic helix-loop-helix domain, has been identified as a master regulator of plant cell hypertrophy (Kay et al., 2007; Fig. 1A). Plants have evolved a sophisticated strategy to recognize AvrBs3 using the promoter of the R gene Bs3 as a molecular trap. In resistant pepper (Capsicum annuum) plants activation of Bs3 leads to HR development (Römer et al., 2007; Fig. 1A).
Figure 1.
Nuclear translocation of bacterial effectors and plant resistance proteins. A, After dimerizing in the plant cell cytoplasm, AvrBs3 is translocated into the nucleus, where it is able to bind to the UPA box and act as a TF. Transcriptional activation of UPA20 induces plant cell hypertrophy, whereas in resistant pepper, activation of the R gene Bs3 leads to HR. B, XopD nuclear interaction with the Arabidopsis TF AtMYB30 is mediated by the HLH domain of XopD, which is necessary and sufficient to repress AtMYB30 transcriptional activation and thereby the plant defense response. C, In resting cells, the N immune receptor presents a dual cytoplasmic and nuclear localization. Upon TMV infection, the viral replicase p50 recruits to the cytoplasm the tobacco NRIP1 protein, which is localized in the chloroplast of uninfected cells. The NRIP1/p50 prerecognition complex interacts with and activates cytoplasmic N, which is then able to cross the nuclear envelope and/or send a signal to activate nuclear N, thereby activating defense signaling. D, The PopP2 effector induces nuclear accumulation of its cognate R protein RRS1-R, which acts as a transcriptional repressor in resting cells, and of the vacuolar protease RD19. The nuclear interaction between the PopP2/RD19 complex and RRS1-R leads to activation of plant defense, perhaps through modification of the transcriptional activity of RRS1-R WRKY domain or by activation of additional TFs. It has been additionally proposed that PopP2 acetyltransferase activity may disrupt higher-order packaging of the chromatin or alter DNA-binding activities and interaction properties of host TFs, leading to modification of their activity.
Similar to transcription activator-like effectors, HsvG and HsvB effectors of gall-forming Pantoea agglomerans, which present two functional NLSs required for their nuclear targeting and pathogenicity, are able to bind DNA and activate transcription (Nissan et al., 2006; Weinthal et al., 2011). Although the mode of action of HsvG and HsvB remains unknown, it has been hypothesized that it involves modulation of host phytohormones associated with gall formation.
The PopP2 effector from Ralstonia solanacearum presents an NLS that is required for its nuclear targeting (Deslandes et al., 2003). It has been reported that PopP2 is able to display acetyltransferase activity, suggesting that PopP2 may directly manipulate host transcription (Tasset et al., 2010). Indeed, acetylation of Lys residues of histone tails facilitates access of TFs to DNA by disrupting higher-order packaging of the chromatin and also by neutralizing the positive charge of histones, which reduces their affinity for DNA. Acetylation also impairs the ability of the Lys side chain to form hydrogen bonds, thereby enhancing specific or inhibiting nonspecific DNA-binding activities of TFs. In addition, acetylation forms docking sites for recruitment of transcriptional coactivators. Therefore, it has been proposed that PopP2 autoacetylation and/or acetylation of its host targets may affect gene transcription in host cells (Tasset et al., 2010; Fig. 1D).
XopD, a modular effector of 760 amino acids from X. campestris pv vesicatoria, displays SUMO protease activity and is targeted to plant cell subnuclear structures named nuclear bodies (or nuclear foci), suggesting that it may target SUMO-conjugated nuclear proteins (Hotson et al., 2003; Canonne et al., 2010). The molecular mechanism allowing XopD nuclear import remains unknown, but a truncated XopD version containing only the helix-loop-helix domain of XopD (amino acids 216–405), and not comprising its putative NLS, is sufficient for XopD nuclear import and subnuclear targeting (Canonne et al., 2011). Expression of XopD appears to induce reorganization of the host nuclear structure that leads to nonspecific relocalization of all tested nuclear proteins into nuclear bodies. In addition, 4′,6-diamino-phenylindole staining showed that DNA accumulation is weaker in nuclear bodies, where XopD is expressed, compared to the nucleoplasm, where DNA distribution remains otherwise unaltered (Canonne et al., 2011). It is thus tempting to speculate that XopD-induced modification of the nuclear structure and protein distribution may be part of a general virulence strategy, which allows Xanthomonas to perturb plant cell responses to bacterial infection. Along these lines, it has been proposed that XopD may affect host transcription by affecting chromatin remodelling (Kay and Bonas, 2009). Significantly, in agreement with the idea that plant TFs and/or regulators might be direct targets of XopD, recent data show that XopD is able to interact with AtMYB30, a previously described Arabidopsis (Arabidopsis thaliana) MYB TF that positively regulates defense and HR responses through the activation of the lipid biosynthesis pathway that leads to the production of very-long-chain fatty acids (VLCFAs; Raffaele et al., 2008). Indeed, XopD-specific interaction with AtMYB30 leads to inhibition of the transcriptional activation of AtMYB30 target genes and suppression of plant defense during Xanthomonas infection (Canonne et al., 2011; Fig. 1B). Importantly, XopD interaction with AtMYB30 appears to be independent of nuclear foci formation.
Plant R Proteins: Recognition of the Invader May Also Occur in the Nucleus
Different R proteins have been reported to dynamically traffic between the cytoplasm and the nucleus where they act as cotranscriptional regulators to activate innate immune responses. For example, the tobacco (Nicotiana tabacum) immune receptor N, which confers resistance to Tobacco mosaic virus (TMV), has been shown to function in the nucleus. In resting cells, N is found in the cytoplasm and the nucleus. During TMV infection, the viral replicase p50 is delivered into the plant cell cytoplasm where it recruits the tobacco rhodanase sulfurtransferase NRIP1, which otherwise localizes to the stroma of chloroplasts. As part of this cytoplasmic prerecognition complex, NRIP1 interacts with and activates cytoplasmic N (Caplan et al., 2008). Shuttling of p50-activated N from the cytoplasm to the nucleus appears to be required for an efficient defense response, although it is also possible that cytoplasmic N is able to send a signal that activates the N nuclear pool (Fig. 2C). Nuclear relocalization of NRIP1 is required to provide full resistance to TMV infection (Caplan et al., 2008). The molecular mechanism behind p50-mediated NRIP1 nuclear relocalization remains unclear. It has been proposed that p50 may disrupt global chloroplast import by an unknown mechanism that would affect NRIP1 translocation. Otherwise, interaction with p50 may mask NRIP1 chloroplast targeting signal and allow its nuclear recruitment. Alternatively, NRIP1 may be released from chloroplasts into the cytoplasm and the nucleus following p50-induced permeabilization of the outer membrane. Finally, since close contact between stromules and nuclei has been observed, it has been proposed that this close association may enhance the nuclear import of chloroplastic factors, such as NRIP1.
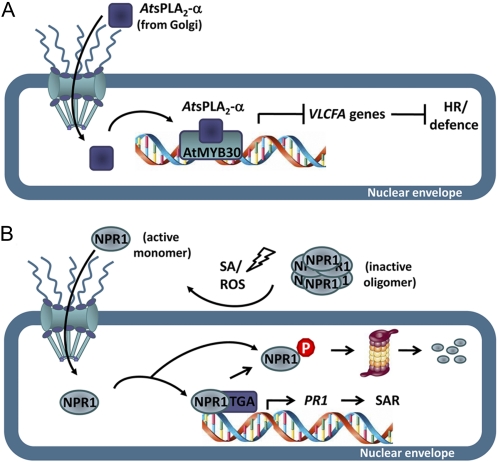
Figure 2.
Nucleo-cytoplasmic trafficking of TFs and regulators during plant defense responses. A, The secreted phospholipase AtsPLA2-α is partially relocalized from Golgi-associated vesicles to the nucleus, where it interacts with the positive defense and HR regulator AtMYB30. This nuclear protein interaction leads to down-regulation of AtMYB30 transcriptional activation of VLCFA-related genes and, therefore, suppression of HR and defense responses. B, Pathogen-induced SA and reactive oxygen species (ROS) production leads to a change in the oligomerization state of the transcriptional coactivator NPR1, which is translocated into the nucleus in its monomeric active form. In the nucleus, active NPR1 interacts with TGA TFs and enhance their DNA-binding properties during the establishment of SAR. NPR1 nuclear accumulation is regulated by the proteasome.
Activation of the immune response mediated by the barley (Hordeum vulgare) MLA10 R protein, which confers resistance to the powdery mildew fungus Blumeria graminis f. sp. hordei, also requires its nuclear accumulation, even though the protein is also found in the cytoplasm (Shen et al., 2007). Indeed, recognition of MLA10 cognate effector AVRA10 induces the nuclear association of MLA10 and barley HvWRKY1 and HvWRKY2, which belong to the WRKY class of zinc finger plant TFs and act as PAMP-inducible repressors of PTI. In this context, MLA appears to interfere with the WRKY repressor function, thereby derepressing PAMP-triggered basal defense responses. In conclusion, activation of MLA would allow rapid derepression of PTI by directly connecting pathogen perception with regulation of defense-related transcriptional reprogramming in the nucleus.
As previously mentioned, the R. solanacearum effector PopP2 is targeted to the nucleus of plant cells. Its cognate Arabidopsis R protein RRS1-R is an atypical R protein that confers resistance to R. solanacearum and presents a C-terminal WRKY motif and a putative bipartite NLS (Deslandes et al., 2003). Intriguingly, PopP2 promotes nuclear accumulation of RRS1-R, possibly by preventing its proteasomal degradation. PopP2 and RRS1-R physically interact in the nucleus, but whether and how this protein interaction affects host transcription remain to be determined. In addition to the previously discussed putative modulation of host transcription that may be mediated through PopP2 acetyltransferase activity, it has been proposed that PopP2 interaction with RRS1-R may lead to regulation of defense-related gene expression either directly via the RRS1-R WKRY domain or through the action of additional plant TFs (Deslandes et al., 2003; Tasset et al., 2010; Fig. 1D). Nuclear recognition of PopP2 by RRS1-R exemplifies an additional molecular shortcut to rapidly connect pathogen perception with defense-related signaling.
Intriguingly, the Arabidopsis RPS4 protein, which confers resistance to Pseudomonas syringae pv tomato DC3000 expressing AvrRps4 (Gassmann et al., 1999), has been shown to function cooperatively with RRS1-R in triggering resistance against a variety of pathogens (Birker et al., 2009; Narusaka et al., 2009). RPS4, which contains a functional NLS, distributes between endomembranes and nuclei. RPS4 nuclear accumulation is necessary for triggering immunity through activation by AvrRps4 (Wirthmueller et al., 2007).
Finally, recognition of the coat protein (CP) of Potato virus X (PVX) by the potato (Solanum tuberosum) immune receptor Rx confers resistance to the virus without triggering HR, in a process that is known as extreme resistance (Bendahmane et al., 1999). The identified protein interaction between Rx and Ran-GTPase-activating protein2 (RanGAP2) suggests the implication of Ran-regulated components of the nuclear pore in the control of plant immune responses (Sacco et al., 2007; Tameling and Baulcombe, 2007). This idea was additionally supported by the finding that silencing of RanGAP2 impairs Rx-mediated resistance (Sacco et al., 2007; Tameling and Baulcombe, 2007). Recent data show that (1) despite the absence of an obvious NLS within its sequence, Rx localizes to both the cytoplasm and the nucleus in Nicotiana benthamiana cells and that (2) this dual localization is required for full functionality (Slootweg et al., 2010; Tameling et al., 2010). The physical interaction between Rx and cytoplasmic RanGAP2 results in sequestration of Rx in the cytoplasm, independently of RanGAP2 GAP activity. This leads to enhanced Rx-mediated immune signaling, which correlates with stronger HR and increased resistance to PVX. In contrast, Rx nuclear hyperaccumulation compromises HR and resistance responses to PVX. This is in agreement with the finding that Rx is activated in the cytoplasm, where recognition of the CP elicitor occurs, and cannot be activated in the nucleus (Slootweg et al., 2010). Together, these data identify RanGAP2 as a cytoplasmic retention factor that regulates nucleocytoplasmic partitioning of Rx in a process that is required for proper regulation of defense signaling. Distinct roles have been suggested for nuclear and cytoplasmic Rx during ETI induction. Nuclear Rx may play a role in transcriptional reprogramming, leading to ETI induction, whereas cytoplasmic Rx may directly activate an antiviral mechanism in the cytoplasm, where PVX replication and detection by Rx occur (Slootweg et al., 2010; Tameling et al., 2010).
IMMUNITY COMPONENTS
In addition to R and effector proteins, essential regulators of the plant immune response shuttle between the cytoplasm and the nucleus, providing a possible framework for defense signal trafficking between the two compartments. One of the best-characterized examples of this dynamic traffic of immunity components is provided by the plant immune regulatory complex formed by ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1), PHYTOALEXIN DEFICIENT4 (PAD4), and SENESCENCE-ASSOCIATED GENE101 (SAG101), whose combined activities and mutually stabilizing effects are essential for coordinating responses to multiple environmental stress stimuli (Wiermer et al., 2005). Together, EDS1, PAD4, and SAG101 constitute a family of plant-specific proteins with intriguing homology to eukaryotic lipases, although no lipid-related enzymatic activity has been reported for these proteins and their biochemical mode of action remains unknown. EDS1 transiently associates in the cytoplasm and in the nucleus with its coregulator partners. While EDS1 homodimers are mostly cytoplasmic, EDS1-PAD4 heterodimers are found in both cytoplasm and nucleus and EDS1-SAG101 complexes are exclusively nuclear (Feys et al., 2005). Pathogen recognition triggers an early increase in the nuclear EDS1 pool, which directs transcriptional reprogramming of salicylic acid (SA)- and defense-related genes (García et al., 2010). Importantly, such perturbations of nuclear EDS1 levels appear to become sensed and balanced with the EDS1 cytoplasmic pool, which is also required for full resistance. How nuclear EDS1 may affect defense-associated transcription is still unclear, but EDS1 interaction with a number of TFs in yeast (Saccharomyces cerevisiae) suggests that EDS1-containing nuclear complexes may work by binding TFs and/or repressors in the nucleoplasm to guide activities and associations with the DNA (García et al., 2010).
Coordination between the EDS1 cytoplasmic and nuclear pools via the NPC trafficking machinery is needed to condition full plant resistance. For example, EDS1-dependent constitutive resistance displayed by the suppressor of npr1-1, constitutive 1 mutant, which carries a gain-of-function mutation in an R gene, is associated with changes in relative amounts of cytoplasmic and nuclear EDS1 (García et al., 2010). In addition, the EDS1 nuclear pool increases during the RPS4-triggered immune response and associated transcriptional reprogramming, even though a cytoplasmic EDS1 pool is retained during defense. Redirecting EDS1 to the cytoplasm using a nuclear export signal fusion reduced RPS4-triggered resistance and basal defense responses. Thus, EDS1 functions through balanced nuclear and cytoplasmic activities to mediate the induction and repression of particular defense-related genes, thereby allowing the plant to mount an appropriate immune response (García et al., 2010).
A recent report shows that EDS1 does not bridge between PAD4 and SAG101 in a ternary complex (Rietz et al., 2011). Thus, resistance signaling is more likely to be achieved by separate complexes of EDS1 with either SAG101 or PAD4, which are expected to have distinct functions in plant defense. A model has been proposed in which EDS1 molecular transitions between complexes determine different stages of EDS1 signal relay. After pathogen perception, the presence of EDS1 and PAD4, but not their physical interaction, is sufficient to start the resistance response by activating rapid, localized host cell death, which leads to pathogen containment at infection sites. In contrast, in cells neighboring the death foci, EDS1-PAD4 complexes serve a different function necessary for basal resistance to virulent pathogen infection, which correlates with transcriptional up-regulation of PAD4. This enables the spread of resistance to systemic tissues and mobilization of SA-mediated defenses during systemic acquired resistance (SAR), an inducible form of plant defense conferring broad-spectrum immunity to secondary infection. In this model, EDS1-SAG101 complexes would link the HR at infection sites with the reinforcement of resistance at the edges of the local HR. EDS1-SAG101 complexes would thus represent a transition state between the initial triggering of cell death (requiring separate EDS1 and PAD4) and the mobilization of resistance in surrounding tissues (requiring EDS-PAD4 complexes; Rietz et al., 2011).
TRANSCRIPTION FACTORS AND REGULATORS
Transcriptional reprogramming is a key step of plant defense as up to 25% of Arabidopsis genes respond to pathogen infection by altering their transcript levels (Maleck et al., 2000; Tao et al., 2003). This dynamic regulation of gene expression relies on an intricate network of TFs and regulators and directs adaptive plasticity of plants in highly variable environments. Members of several TF families, such as WRKY, ERF, TGA, Whirly, and MYB factors, have been shown to bind to promoter elements of individual defense-related genes and to regulate their expression (Eulgem and Somssich, 2007). As illustrated by the following examples, cytoplasmic retention of inactive TFs or transcriptional regulators, followed by their activation and signal-dependent translocation into the nucleus, allows plants to rapidly connect signal perception at the cell surface and cytoplasmic signal transduction to defense gene activation in a stimulus-dependent manner.
Transcriptional activation of VLCFA-related genes by the Arabidopsis TF AtMYB30 is required to mount an efficient defense response during bacterial infection (Raffaele et al., 2008). Several data indicate that tight control of AtMYB30 transcriptional activity is exerted during plant defense. In addition to the inhibition of AtMYB30 activity by the bacterial effector XopD that leads to impaired resistance, AtMYB30 transcriptional activation is also regulated by the plant via the secreted phospholipase AtsPLA2-α. Indeed, AtsPLA2-α was identified in a yeast two-hybrid screen as an AtMYB30-interacting protein (Froidure et al., 2010). As expected for a secreted protein, AtsPLA2-α was localized intracellularly in Golgi-associated vesicles and later secreted to the extracellular space. However, when AtsPLA2-α and AtMYB30 are transiently coexpressed in N. benthamiana, targeting of AtsPLA2-α is partially modified from cytoplasmic vesicles to the plant cell nucleus, where the physical interaction between both proteins has been demonstrated. This protein interaction leads to repression of AtMYB30-mediated transcriptional activity and negative regulation of plant HR and defense responses (Froidure et al., 2010; Fig. 2A). These data identified AtsPLA2-α as a negative regulator of AtMYB30-mediated defense signaling and highlight the importance of dynamic protein trafficking to the nucleus for the regulation of defense-related transcription.
As previously mentioned, the R. solanacearum effector PopP2 interacts with the Arabidopsis R protein RRS1-R in the nucleus, although the effect of this protein interaction on host transcription is still unknown. Interestingly, PopP2 induces nuclear targeting of the Arabidopsis Cys protease RESPONSIVE TO DEHYDRATATION19 (RD19), otherwise localized to mobile vacuole-associated vesicles and destined to the lytic vacuole (Bernoux et al., 2008). RD19, whose expression is induced by Ralstonia infection, interacts with PopP2, but not RRS1-R, in the plant cell nucleus. Since RD19 is required for Arabidopsis resistance to Ralstonia, it was proposed that RD19 associates with PopP2 to form a nuclear complex that is required for activation of the plant resistance response (Fig. 1D). Similarly to RD19, the tomato (Solanum lycopersicum) LeCp vacuolar protease is relocalized to the nucleus in response to a fungal elicitor (Matarasso et al., 2005). In the nucleus, LeCp acts as a TF to activate the expression of the ACC synthase gene, although the mechanism involved is still unknown. Therefore, it is possible that nuclear RD19 functions as a transcriptional activator and/or competes with RRS1-R for similar or overlapping cis-elements in the promoters of defense-related genes (Bernoux et al., 2008).
RD19, LeCp, or AtsPLA2-α do not contain a consensus NLS and the molecular mechanism that allows their nuclear recruitment is still unknown. However, different hypotheses have been proposed to explain the nuclear targeting of these proteins. In the case of LeCp and RD19, it was proposed that an elicitor-/effector-induced membrane permeabilization process would trigger a collapse of the vacuolar membrane. This would lead to the respective release of LeCp and RD19 from vacuole-associated compartments into the cytoplasm, where they may become available for SUMOylation (Matarasso et al., 2005; Bernoux et al., 2008). Similarly, AtMYB30 might release AtsPLA2-α from vesicle-associated compartments to the nucleus through an unknown mechanism that may involve AtsPLA2-α SUMOylation (Froidure et al., 2010). Indeed, Lys residues with high probability of being SUMOylated are present in LeCp, RD19, and AtsPLA2-α, and LeCp interacts with SUMO in yeast. Therefore, it has been suggested that SUMOylation of these proteins may generate the signal required for their nuclear translocation. Alternatively, AtMYB30 may intercept AtsPLA2-α on its way to the extracellular compartment through retrograde transport from the endomembrane system, which has some continuity with the nuclear envelope. A similar mechanism was previously proposed for PopP2-induced nuclear translocation of RD19. Finally, binding of AtMYB30 to AtsPLA2-α may mask AtsPLA2-α vesicle targeting signal or AtMYB30 may indirectly disrupt global vesicle sorting affecting the translocation of AtsPLA2-α (Froidure et al., 2010).
SA is the predominant plant hormone produced during SAR. PATHOGENESIS-RELATED1 (PR1) is a marker of SAR and activation of its expression is dependent on SA production and induced by members of the TGA family of basic Leu-zipper-type TFs, which bind to two TGACG motifs in the PR1 promoter. NONEXPRESSOR OF PR GENES1 (NPR1) acts as a transcriptional coactivator of PR1 expression (Dong, 2004). In uninfected cells, NPR1 forms oligomeric complexes, masking its functional bipartite NLS and retaining the protein in the cytoplasm (Mou et al., 2003). Upon pathogen challenge, increased SA levels induce a change in the redox potential of the cell so that NPR1 oligomers are partially reduced to a monomeric state and exposure of NPR1 NLS allows its nuclear translocation. Once in the nucleus, NPR1 interacts with TGA TFs and enhances their DNA-binding activity to positive and negative regulatory elements in promoters of defense-related genes, such as PR1 and WRKY TFs (Després et al., 2000; Fig. 2B). Therefore, the cytoplasmic/nuclear ratio of NRP1 mediates the fine tuning of SA-induced responses. Interestingly, proteasomal-mediated turnover of nuclear NPR1 prevents needless stimulation of transcription in unelicited cells and allows full transcriptional activation during SAR induction (Spoel et al., 2009; Fig. 2B). Finally, a negative regulatory mechanism of defense signaling is provided by the repressor SUPPRESSOR OF NPR1, INDUCIBLE1 that inhibits PR1 expression probably by histone modifications (Mosher et al., 2006).
CONCLUSION AND PERSPECTIVES
The plant cell nucleus plays a key role in deciphering stress signals and coordinating appropriate changes in gene expression to counteract pathogen ingress while minimizing the associated fitness costs to the cell. Essential defense regulators such as TFs of the MYB or TGA families, and immunity components such as SAG101, are strictly nuclear in their localization. However, during the last decade, stimulating studies from our field have revealed an increasing number of defense-related proteins that are dynamically translocated across the nuclear envelope. For example, immune regulators such as NPR1, EDS1, and PAD4, as well as several R proteins and their cognate effectors, show dynamic, quantitative, and temporal changes in subcellular localization that are key to the modulation of their activity during defense signaling. R proteins like N, RPS4, and MLA10 require a nuclear localization for functioning, whereas Rx is activated in the cytoplasm.
Here, I have summarized recent progress in uncovering nuclear dynamics associated to plant immunity. However, despite recent stimulating discoveries, the intricate signaling mechanisms that direct changes in protein subcellular localization and activity are still to be unraveled. In this context, multidisciplinary strategies tlsb=.1pt?>using a combination of molecular, biochemical, cell biology, and genetic techniques, coupled to noninvasive, high-resolution, real-time microscopic analyses are clearly necessary. Finally, to obtain a comprehensive picture about the mechanisms by which plants elaborate appropriate defense outputs in response to changing environmental stimuli, our knowledge about nuclear trafficking of immunity components needs to be placed in the context of whole cellular dynamics, by integrating defense-related functions played by additional organelles, including chloroplasts, mitochondria, or the endoplasmic reticulum.
Acknowledgments
I apologize to all colleagues whose work could not be discussed because of space limitations.
References
- Bendahmane A, Kanyuka K, Baulcombe DC. (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11: 781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M, Timmers T, Jauneau A, Brière C, de Wit PJ, Marco Y, Deslandes L. (2008) RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell 20: 2252–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birker D, Heidrich K, Takahara H, Narusaka M, Deslandes L, Narusaka Y, Reymond M, Parker JE, O’Connell R. (2009) A locus conferring resistance to Colletotrichum higginsianum is shared by four geographically distinct Arabidopsis accessions. Plant J 60: 602–613 [DOI] [PubMed] [Google Scholar]
- Canonne J, Marino D, Jauneau A, Pouzet C, Brière C, Roby D, Rivas S. (2011) The Xanthomonas type III effector XopD targets the Arabidopsis transcription factor AtMYB30 to suppress plant defense. Plant Cell 23: 3498–3511 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Canonne J, Marino D, Noël LD, Arechaga I, Pichereaux C, Rossignol M, Roby D, Rivas S. (2010) Detection and functional characterization of a 215 amino acid N-terminal extension in the Xanthomonas type III effector XopD. PLoS ONE 5: e15773. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Caplan JL, Mamillapalli P, Burch-Smith TM, Czymmek K, Dinesh-Kumar SP. (2008) Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 132: 449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YT, Germain H, Wiermer M, Bi D, Xu F, García AV, Wirthmueller L, Després C, Parker JE, Zhang Y, et al. (2009) Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 21: 2503–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA 100: 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Rivas S. (2011) The plant cell nucleus: a true arena for the fight between plants and pathogens. Plant Signal Behav 6: 42–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C, DeLong C, Glaze S, Liu E, Fobert PR. (2000) The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12: 279–290 [PMC free article] [PubMed] [Google Scholar]
- Dong X. (2004) NPR1, all things considered. Curr Opin Plant Biol 7: 547–552 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE. (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, Cabral A, Parker JE. (2005) Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froidure S, Canonne J, Daniel X, Jauneau A, Brière C, Roby D, Rivas S. (2010) AtsPLA2-alpha nuclear relocalization by the Arabidopsis transcription factor AtMYB30 leads to repression of the plant defense response. Proc Natl Acad Sci USA 107: 15281–15286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García AV, Blanvillain-Baufumé S, Huibers RP, Wiermer M, Li G, Gobbato E, Rietz S, Parker JE. (2010) Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog 6: e1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García AV, Parker JE. (2009) Heaven’s Gate: nuclear accessibility and activities of plant immune regulators. Trends Plant Sci 14: 479–487 [DOI] [PubMed] [Google Scholar]
- Gassmann W, Hinsch ME, Staskawicz BJ. (1999) The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J 20: 265–277 [DOI] [PubMed] [Google Scholar]
- Hotson A, Chosed R, Shu H, Orth K, Mudgett MB. (2003) Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol Microbiol 50: 377–389 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kay S, Bonas U. (2009) How Xanthomonas type III effectors manipulate the host plant. Curr Opin Microbiol 12: 37–43 [DOI] [PubMed] [Google Scholar]
- Kay S, Hahn S, Marois E, Hause G, Bonas U. (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318: 648–651 [DOI] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA. (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet 26: 403–410 [DOI] [PubMed] [Google Scholar]
- Matarasso N, Schuster S, Avni A. (2005) A novel plant cysteine protease has a dual function as a regulator of 1-aminocyclopropane-1-carboxylic acid synthase gene expression. Plant Cell 17: 1205–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I. (2007) Composition of the plant nuclear envelope: theme and variations 58: 27–34 [DOI] [PubMed] [Google Scholar]
- Meier I, Brkljacic J. (2009) The nuclear pore and plant development. Curr Opin Plant Biol 12: 87–95 [DOI] [PubMed] [Google Scholar]
- Mosher RA, Durrant WE, Wang D, Song J, Dong X. (2006) A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell 18: 1750–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X. (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944 [DOI] [PubMed] [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y. (2009) RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J 60: 218–226 [DOI] [PubMed] [Google Scholar]
- Nissan G, Manulis-Sasson S, Weinthal D, Mor H, Sessa G, Barash I. (2006) The type III effectors HsvG and HsvB of gall-forming Pantoea agglomerans determine host specificity and function as transcriptional activators. Mol Microbiol 61: 1118–1131 [DOI] [PubMed] [Google Scholar]
- Palma K, Zhang Y, Li X. (2005) An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr Biol 15: 1129–1135 [DOI] [PubMed] [Google Scholar]
- Raffaele S, Vailleau F, Léger A, Joubès J, Miersch O, Huard C, Blée E, Mongrand S, Domergue F, Roby D. (2008) A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20: 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietz S, Stamm A, Malonek S, Wagner S, Becker D, Medina-Escobar N, Vlot AC, Feys BJ, Niefind K, Parker JE. (2011) Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol 191: 107–119 [DOI] [PubMed] [Google Scholar]
- Römer P, Hahn S, Jordan T, Strauss T, Bonas U, Lahaye T. (2007) Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318: 645–648 [DOI] [PubMed] [Google Scholar]
- Sacco MA, Mansoor S, Moffett P. (2007) A RanGAP protein physically interacts with the NB-LRR protein Rx, and is required for Rx-mediated viral resistance. Plant J 52: 82–93 [DOI] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Slootweg E, Roosien J, Spiridon LN, Petrescu AJ, Tameling W, Joosten M, Pomp R, van Schaik C, Dees R, Borst JW, et al. (2010) Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 22: 4195–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X. (2009) Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WI, Baulcombe DC. (2007) Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to Potato virus X. Plant Cell 19: 1682–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WI, Nooijen C, Ludwig N, Boter M, Slootweg E, Goverse A, Shirasu K, Joosten MH. (2010) RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell 22: 4176–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, Katagiri F. (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasset C, Bernoux M, Jauneau A, Pouzet C, Brière C, Kieffer-Jacquinod S, Rivas S, Marco Y, Deslandes L. (2010) Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog 6: e1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinthal DM, Barash I, Tzfira T, Gaba V, Teper D, Sessa G, Manulis-Sasson S. (2011) Characterization of nuclear localization signals in the type III effectors HsvG and HsvB of the gall-forming bacterium Pantoea agglomerans. Microbiology 157: 1500–1508 [DOI] [PubMed] [Google Scholar]
- Wiermer M, Feys BJ, Parker JE. (2005) Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 8: 383–389 [DOI] [PubMed] [Google Scholar]
- Wirthmueller L, Zhang Y, Jones JD, Parker JE. (2007) Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol 17: 2023–2029 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li X. (2005) A putative nucleoporin 96 is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell 17: 1306–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]