Plant development and growth are extremely plastic in response to changes in ambient light conditions. Light is not only the ultimate energy source for photosynthesis; its physical parameters, such as quality, intensity, direction, and duration, also serve as key environmental and time cues (Chen et al., 2004). Therefore, it is vital for plants to closely monitor and precisely respond to changes in light properties in order to optimize growth under a wide range of ecological environments and to synchronize developmental transitions with diurnal and seasonal time. Plants have evolved to “see” the light spectrum between 280 and 750 nm, which spans UV-B, UV-A, and visible light, through five classes of photoreceptors. In the reference plant Arabidopsis (Arabidopsis thaliana), they include the newly determined UV-B receptor UV RESISTANCE LOCUS8 (UVR8; Kaiserli and Jenkins, 2007; Rizzini et al., 2011); three types of UV-A/blue light receptors, including cryptochromes (cry1 to 3; Kleine et al., 2003; Yu et al., 2010; Chaves et al., 2011), phototropins (phot1 and 2; Briggs and Christie, 2002), and the family of ZEITLUPE/FLAVIN-BINDING, KELCH, F-BOX1 (FKF1)/LOV KELCH PROTEIN2 (LKP2; Nelson et al., 2000; Somers et al., 2000; Schultz et al., 2001); and the red and far-red sensing phytochromes (phyA to E; Quail, 2010). Collectively, these photoreceptors regulate almost every facet of plant development and growth from seed germination to floral initiation (Franklin and Quail, 2010; Kami et al., 2010).
Many light responses are mediated by alterations in gene expression. Among the 15 photoreceptors discovered so far, 10 of them, UVR8 (Kaiserli and Jenkins, 2007; Favory et al., 2009), cry1 (Wu and Spalding, 2007), cry2 (Kleiner et al., 1999; Yu et al., 2009), FKF1 (Sawa et al., 2007; Fornara et al., 2009), LKP2 (Yasuhara et al., 2004), and phyA to E (Sakamoto and Nagatani, 1996; Yamaguchi et al., 1999; Kircher et al., 2002), have been shown to localize to the nucleus, where they regulate gene expression in a light-dependent manner (Jiao et al., 2007). An emerging common mechanism for such light-dependent gene expression is through regulating the stability of key transcriptional regulators (discussed below; Yi and Deng, 2005; Brown et al., 2009; Chen and Chory, 2011; Leivar and Quail, 2011).
At the cellular level, photoactivation of photoreceptors triggers the rapid localization of a number of them, including phyA to E, cry2, and possibly cry1 and UVR8, to discrete subnuclear foci or nuclear bodies (Kleiner et al., 1999; Yamaguchi et al., 1999; Wang et al., 2001; Kircher et al., 2002; Favory et al., 2009; Yu et al., 2009; Gu et al., 2011; Lian et al., 2011; Liu et al., 2011). Nuclear bodies are morphologically distinct subnuclear domains that provide microenvironments for the regulation of protein dynamics, gene expression, and DNA replication and repair in both plant and animal cells (Shaw and Brown, 2004; Spector, 2006). The general principles of nuclear body function and assembly are still largely unknown. The photoreceptor-containing nuclear bodies, or “photobodies,” found in plants are a unique type of subnuclear domain whose size, number, and potentially function are directly regulated by external light cues (Chen and Chory, 2011). Observations of these speckle-shaped photobodies have raised many questions. How is the formation of photobodies regulated? What are the functions of the photobodies? What are the factors required for photobody formation? These are some of the key questions that are being actively investigated. Further understanding of photobody function and regulation will not only be important for understanding the subcellular organization of light signaling events in plants but also could potentially uncover general principles governing subnuclear domains in higher eukaryotes.
In this review, we will summarize recent developments related to phy-containing photobodies, touch briefly on photobodies containing crys, and discuss the potential functions of photobodies in relationship to protein degradation and gene expression. For more comprehensive coverage of light signaling in plants, the reader is referred to the following recent reviews (Henriques et al., 2009; Jenkins, 2009; Chory, 2010; Franklin and Quail, 2010; Kami et al., 2010; Lau and Deng, 2010; Möglich et al., 2010; Nagatani, 2010; Rockwell and Lagarias, 2010; Yu et al., 2010; Chaves et al., 2011; Chen and Chory, 2011; Leivar and Quail, 2011; Ulijasz and Vierstra, 2011).
LIGHT-DEPENDENT DYNAMICS OF PHYTOCHROME PHOTOBODY FORMATION
Phys in higher plants are red (R) and far-red (FR) light receptors that use a linear tetrapyrrole, phytochromobilin, as their chromophore. Phys can interconvert between two relatively stable conformers: an R light-absorbing inactive Pr form (λmax = 660) and a FR light-absorbing active Pfr form (λmax = 730; Rockwell et al., 2006). Although it was initially thought that phys localized and functioned primarily in the cytoplasm, a series of studies performed over a decade ago using GUS-tagged and fluorescent protein-tagged phys convincingly demonstrated that photoactivation from the Pr to the Pfr form results in the rapid translocation of phys from the cytoplasm to the nucleus (Sakamoto and Nagatani, 1996; Kircher et al., 1999, 2002; Yamaguchi et al., 1999; Kim et al., 2000). This change in localization is one of the earliest phy responses to light; for both phyA and phyB, the most prominent phys in Arabidopsis, nuclear accumulation is required for most of their downstream responses (Huq et al., 2003; Genoud et al., 2008).
It was first reported by Akira Nagatani and colleagues that photoactivated phyB-GFP was not only localized to the nucleus but also further compartmentalized to subnuclear speckle-like photobodies (Yamaguchi et al., 1999). Parallel studies by Eberhard Schäfer, Ferenc Nagy, and colleagues demonstrated that all five Arabidopsis phys, phyA to E, localize to photobodies in the light and that phy photobody localization is conserved in both dicotyledonous and monocotyledonous plants (Kircher et al., 1999, 2002; Kim et al., 2000). Although most studies on phy photobodies were conducted using transgenic lines that overexpressed fluorescent protein-tagged phys, both native pea (Pisum sativum) phyA and Arabidopsis phyB photobodies have been observed using immunocytochemistry, suggesting that the formation of photobodies is not an artifact of phy overexpression (Hisada et al., 2000; Kircher et al., 2002).
The translocation of phys to photobodies happens very quickly during the dark-to-light transition; photobodies containing both phyA and phyB can be observed after 1 to 2 min of R light exposure (Bauer et al., 2004). PhyB photobody localization is triggered by R light (Yamaguchi et al., 1999; Kircher et al., 2002). In contrast, phyA photobody localization is triggered by R, FR, and blue light (Kim et al., 2000). These “early” photobodies are transient and disappear after 1 h of light exposure (Bauer et al., 2004). Phy photobodies reappear after 2 h in R light and remain present in the light (Yamaguchi et al., 1999; Bauer et al., 2004). These “late” photobodies contain mainly phyB, because phyA is rapidly degraded in R light (Kim et al., 2000; Kircher et al., 2002).
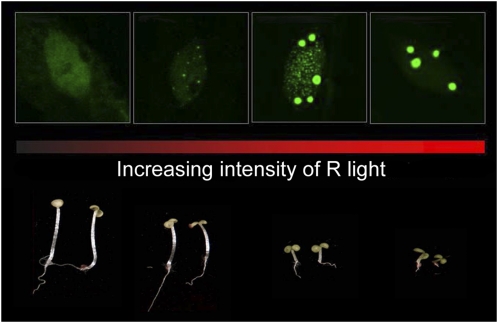
Joanne Chory and colleagues showed that the steady-state pattern (size and number) of phyB photobodies under continuous R light is determined by the percentage of phyB in the Pfr form at a given moment (Chen et al., 2003). Light conditions that shift the Pr/Pfr equilibrium to the Pfr side or stabilize the Pfr form will promote large phy photobody formation. Consistent with this notion, under high-intensity R light, which drives the equilibrium to the Pfr form, phyB appears to be localized exclusively to a few large photobodies with diameters between 1 and 2 μm (Chen et al., 2003, 2010b). By contrast, under dim R light or light with a low R-to-FR ratio, where more phyB stays in the Pr form, phyB tends to localize to many smaller photobodies or localizes diffusely in the nucleoplasm (Fig. 1; Chen et al., 2003). The formation of large phyB photobodies correlates tightly with the light-dependent hypocotyl inhibition response. The fact that the steady-state pattern of phyB-GFP is predictable and can be precisely manipulated by external light quantity and quality makes it an excellent visible readout for genetic screens (discussed below; Chen, 2008). Although phyB photobodies appear to be morphologically stable, they are actually quite dynamic subnuclear domains; fluorescence recovery after photobleaching experiments on phyB-yellow fluorescent protein (YFP) photobodies showed that photobody-associated phyB-YFP is rapidly exchanged with nucleoplasmic phyB-YFP (Rausenberger et al., 2010).
Figure 1.
The morphology of phy photobodies is directly regulated by light. Confocal images of phyB-GFP localization patterns and corresponding PBG seedlings under increasing intensities of red light are shown. PhyB-GFP is evenly distributed under dim (0.5 μmol m−2 s−1) R light. With increasing R light intensity (1 μmol m−2 s−1 and 2 μmol m−2 s−1), phyB-GFP starts to form exclusively small, or both small and large photobodies, respectively. Under strong R light (above 8 μmol m−2 s−1), phyB-GFP localizes exclusively to large photobodies. The localization of phyB-GFP correlates with the degree of hypocotyl inhibition in the light.
Some phy photobodies might also contain the blue light receptor cry2. Crys are photolyase-like photoreceptors that use FAD as the chromophore. When phyB-GFP and cry2-red fluorescent protein were coexpressed in BY-2 protoplasts, not only did they colocalize on photobodies, they could also be coimmunoprecipitated (Más et al., 2000). Because cry2 is photolabile and rapidly degraded in blue light, it could be colocalized with phyB in early photobodies during the dark-to-light transition. Consistent with this hypothesis, cry2 photobody localization is also a rapid light response, as Arabidopsis cry2 is translocated to photobodies within 15 min after blue light exposure (Yu et al., 2009). Activated cry1 has also been suggested to localize to nuclear bodies (Wang et al., 2001; Gu et al., 2011; Lian et al., 2011; Liu et al., 2011). Because cry1 and cry2 were colocalized with COP1 and SPA1 on nuclear bodies (Gu et al., 2011; Lian et al., 2011; Liu et al., 2011; Zuo et al., 2011), it is likely that cry1, similar to cry2, could also localize to phy-containing photobodies in the light.
STRUCTURAL BASIS OF PHYTOCHROME PHOTOBODY LOCALIZATION
The intramolecular requirements for photobody localization have been most extensively studied using Arabidopsis phyB. Two general approaches have been taken to dissect the phy subdomains required for photobody localization: one approach is to examine the localization pattern of phy truncation fragments; the other approach is to characterize the localization pattern of loss-of-function or gain-of-function alleles of phy.
The domain structure of phys has been well defined. Phys can form either homodimers or heterodimers (Clack et al., 2009); each monomer is an approximately 125-kD polypeptide. The phy protein can be divided into two domains: an N-terminal photosensory and signaling domain and a C-terminal dimerization domain, with a hinge region connecting the two (Fig. 2; Nagatani, 2010). The N-terminal domain comprises four subdomains: an N-terminal extension (NTE); a PAS (for PER, ARNT, and SIM) domain; a GAF (for cGMP phosphodiesterase, adenylate cyclase, and FhlA) domain, which contains a conserved Cys residue forming a thioether linkage with the A ring of the chromophore phytochromobilin; and a PHY (for phytochrome-specific GAF-related) domain (Rockwell et al., 2006). The crystal structure of the PAS/GAF domain of the bacteriophytochrome DrBph1 from Deinococcus radiodurans revealed the presence of a light-sensing knot, which plays an important role in signaling. For example, amino acid residues located in the knot are involved in the interaction with phytochrome-interacting factors (PIFs; Oka et al., 2008; Kikis et al., 2009; Nagatani, 2010; Ulijasz and Vierstra, 2011). The C-terminal half of phys contains two subdomains, a PAS-related domain (PRD) containing two PAS domains (PAS-A and PAS-B) and a His kinase-related domain (HKRD; Rockwell et al., 2006).
Figure 2.
Structural basis for phyB localization and signaling. Phys can be divided into the N-terminal light-sensing and signaling domain and the C-terminal dimerization and localization domain (Nagatani, 2010). The GAF and PHY domains of phyB physically interact with the PRD to mask nuclear localization signals located in the PRD (Chen et al., 2005). Additionally, although the PRD alone is sufficient for nuclear import, the PRD and HKRD together are required for normal phyB photobody localization (Matsushita et al., 2003; Chen et al., 2005). Mutations in red are those that have apparently normal photobody localization but fail to complement a null phyB mutant, possibly due to reduced interaction with PIF proteins (Oka et al., 2008; Kikis et al., 2009). The Y276H mutation, in green, causes constitutive phyB localization to large photobodies and constitutive phytochrome signaling (Su and Lagarias, 2007). The G767R mutation, in brown, results in the inability of phyB to localize to the nucleus (Matsushita et al., 2003). All other mutations, represented in blue, show abnormal photobody localization as well as impaired light signaling (Kircher et al., 2002; Chen et al., 2003; Oka et al., 2008). Specifically, mutant G118R does not incorporate the chromophore; mutants C327Y and A587T show slightly faster dark reversion; and mutant A372T shows a slightly red-shifted spectrum in addition to highly accelerated dark reversion. The chromophore is represented by four consecutive squares.
Truncation studies have revealed that the C-terminal half of phyB localizes to photobodies independently of light (Chen et al., 2003; Matsushita et al., 2003). Similarly, the photobody localization of phyA also requires its C-terminal half (Wolf et al., 2011). Within the C-terminal domain of phyB, the PRD is both required and sufficient for nuclear localization, suggesting that it either possesses a nuclear localization signal (NLS) or is able to bind to an unidentified shuttle protein containing a NLS (Matsushita et al., 2003; Chen et al., 2005). Both the PRD and HKRD are required for normal photobody localization (Chen et al., 2005). Consistent with this notion, several missense mutations that result in defective nuclear or photobody localization have been identified within the PRD (Fig. 2; Kircher et al., 2002; Chen et al., 2003; Matsushita et al., 2003). The role of the HKRD in photobody localization is still a mystery; although truncations lacking the entire HKRD do not localize to photobodies, a phyB truncation lacking a portion of the HKRD could still form smaller photobodies (Chen et al., 2005). Because nuclear and photobody localization are both light dependent, it raises the question of how the N-terminal photosensory domain regulates the C-terminal NLS/photobody localization signals. The current model is that, in the Pr form, C-terminal localization signals are masked by the N-terminal domain through an interaction between the GAF-PHY subdomains and the PRD, whereas both the putative NLS and photobody localization signals are exposed in the Pfr form as a result of light-dependent conformational changes (Chen, 2008; Fankhauser and Chen, 2008). The “open” conformation of the Pfr form could also expose domains required for interacting with other signaling components. Consistent with the notion that the conformation of Pfr is important for photobody localization, the NTE, which plays a role in stabilizing the Pfr form, is also required for phyB photobody formation (Chen et al., 2005).
Localization studies of missense phyB alleles have further demonstrated that the Pfr form of phyB is required for photobody localization. N-terminal missense loss-of-function phyB alleles that are defective in photobody localization also have less stable Pfr, and some have abnormal light absorption spectra (Fig. 2; Chen et al., 2003; Oka et al., 2008). By contrast, YHB, a constitutively active phyB mutant, localizes to photobodies regardless of light conditions (Su and Lagarias, 2007). Moreover, loss-of-function mutations that only affect signaling but not the absorption properties of phyB have normal photobody localization patterns (Oka et al., 2008; Kikis et al., 2009), which further suggests that the photobody localization depends on phyB being in the Pfr form and is not a consequence of phy signaling.
POSSIBLE FUNCTIONS OF PHOTOBODIES IN LIGHT SIGNALING
Ever since the initial observation of phy photobodies, there has been much speculation about their function. One hypothesis is that the photobodies are storage depots for active photoreceptors but are not required for light signaling (Fig. 3A). In this model, photobodies serve as a valve to regulate the amount of active phy in the nucleoplasm. Consistent with this model, the N terminus of phyB fused to a dimerization domain and a NLS is active in mediating light responses but does not localize to photobodies (Matsushita et al., 2003; Palágyi et al., 2010). However, accumulating evidence from localization and colocalization studies on light signaling components supports the idea that photobodies might be the sites of light signaling.
Figure 3.
Alternative models of photobody function. A, The storage depot model. In this model, the amount of photoactivated nucleoplasmic phys is regulated by sequestering them within photobodies. These photobodies serve as storage depots that later release the phys into the nucleoplasm to carry out their signaling functions, resulting in the regulation of light-responsive genes. B, The degradation model. In this model, the photobodies are sites for the ubiquitylation and degradation of key transcriptional regulators. C, The transcription model. Transcriptional regulators localize to photobodies, bringing their target DNA with them. The expression of the target genes is regulated within or in the vicinity of the photobodies. E3, E3 ubiquitin ligase; TR, transcriptional regulator.
Most phy-mediated responses require global reprogramming of the transcriptome (Tepperman et al., 2006; Jiao et al., 2007; Hu et al., 2009; Leivar et al., 2009; Shin et al., 2009). Two emerging signaling mechanisms suggest that the key signaling events regulating gene expression work by modulating the stability of either positively or negatively acting transcription factors. The positively acting transcription factors include the basic leucine zipper (bZIP) transcription factor HY5 (for elongated hypocotyl 5; Koornneef et al., 1980; Oyama et al., 1997), the MYB factor LAF1 (for long after far-red light 1; Ballesteros et al., 2001), the helix-loop-helix (HLH) factor HFR1 (for long hypocotyl in far-red 1; Fairchild et al., 2000), and some members of the B-box zinc finger family (BBX; Kumagai et al., 2008; Khanna et al., 2009), including CONSTANS (CO)/BBX1 (Laubinger et al., 2006; Liu et al., 2008), COL3 (for CONSTANS-like 3)/BBX4 (Datta et al., 2006), LZF1 (for light-regulated zinc finger protein 1)/STH3 (for salt tolerance homolog 3)/BBX22 (Chang et al., 2008, 2011; Datta et al., 2008), and BBX21/STH2 (Datta et al., 2007). These proteins are degraded in the dark by the E3 ubiquitin ligase COP1 (for constitutively photomorphogenic 1) and/or the cullin4-DDB1 (for damaged DNA-binding protein 1)-COP1-SPA (for suppressor of phytochrome A-105) E3 ubiquitin ligase complex, where COP1 and members of the SPA family of proteins form the substrate recognition complex (Osterlund et al., 2000; Seo et al., 2003; Duek et al., 2004; Jang et al., 2005; Yang et al., 2005; Datta et al., 2006, 2007; Laubinger et al., 2006; Liu et al., 2008; Chen et al., 2010a; Chang et al., 2011). COP1 is also involved in the turnover of both phyA and phyB, partly as a mechanism to attenuate phy signaling in the light (Seo et al., 2004; Jang et al., 2010). The current model is that phys promote the stability of this group of positively acting transcription regulators by repressing E3 ubiquitin ligases (Chen and Chory, 2011). Although the molecular mechanism of how phys repress COP1 and/or the COP1 E3 complex is still unclear, recently it has been shown that crys directly regulate either the formation of the substrate receptor COP1/SPA1 complex or the interaction between the substrate receptor COP1/SPA complex and its target proteins (Lian et al., 2011; Liu et al., 2011; Zuo et al., 2011). It is quite possible that phys could utilize a similar mechanism to regulate the activity of COP1.
Besides positively acting transcriptional regulators, there are also transcriptional regulators that are antagonistic to phy signaling. Some of the well-studied members of this group are the bHLH transcription factor called PIFs (Ni et al., 1998; Oh et al., 2007; Shen et al., 2008; Lorrain et al., 2009; Shin et al., 2009; Leivar and Quail, 2011). Phys bind directly to PIFs and trigger their phosphorylation and subsequent degradation in the light (Al-Sady et al., 2006; Leivar and Quail, 2011). The rapid turnover of PIFs in the light is a key mechanism to turn on phy-mediated responses (Leivar et al., 2009; Shin et al., 2009).
One widely proposed hypothesis is that photobodies are sites for protein degradation (Fig. 3B). This model is supported by the fact that many signaling components are localized to photobodies prior to their degradation (Table I). For example, during the dark-to-light transition in seedling development, both phyA and PIF3 colocalize to early phy photobodies before their degradation (Al-Sady et al., 2006). In addition, the positively acting transcriptional regulators, including HY5, LAF1, HFR1, and some BBX proteins, also colocalize with COP1 on nuclear bodies (Table I). Moreover, members of the SPA protein family have also been colocalized with COP1 on nuclear bodies (Seo et al., 2003; Zhu et al., 2008). These results suggest that these transcriptional regulators could at least be ubiquitylated on photobodies. In addition, cry2-containing photobodies may be associated with cry2 degradation (Yu et al., 2009).
Table I. Photobody constituents.
A list of light signaling components that have been shown to localize to photobodies or photobody-like subnuclear domains. These include not only photoreceptors but a number of transcriptional regulators and their E3 ubiquitin ligases, suggesting that photobodies are involved in light-regulated protein degradation and/or transcription.
In mammalian cells, components of the ubiquitin-proteasome pathway have been shown to localize to subnuclear foci called clastosomes (Lafarga et al., 2002). However, it has not been demonstrated whether the proteasome colocalizes with photobodies in plants. In fact, the Arabidopsis proteasome components AtS6A and AtS9 were instead shown to localize to the nucleoplasm (Kwok et al., 1999). CUL4 and DDB1, the other two key components of the CUL4-DDB1-COP1-SPA complex, also localize to the nucleoplasm (Zhang et al., 2008). In addition, COP9, which is the key component of the COP9 signalosome that regulates the activity of cullin-based E3 ubiquitin ligases, localizes to the nucleoplasm in both light- and dark-grown Arabidopsis cotyledon and hypocotyl protoplasts (Chamovitz et al., 1996; Staub et al., 1996). Taken together, these results suggest the possibility that the protein substrates are modified on photobodies and are subsequently degraded in the nucleoplasm. Future investigation into the constituents of photobodies will further clarify this model.
Although photobodies are likely involved in protein degradation, it is quite clear that not all of their constituents are subject to protein degradation. For example, PIF7 is localized to phy photobodies but is stable in the light (Leivar et al., 2008). What other functions could photobodies have besides protein degradation? One possibility is that photobodies are involved in transcriptional regulation (Fig. 3C). This is supported by the fact that many of the photobody constituents are transcriptional regulators, which could bring their targeted genes to the vicinity of photobodies. The link between nuclear bodies and transcriptional regulation is well documented (Zhao et al., 2009). For example, Promyelocytic Leukemia (PML) protein bodies have been shown to organize the higher order chromatin structures of the Major Histocompatibility Complex (MHC) class I locus, and they regulate the expression of MHC class I genes in mammalian cell lines by direct interactions between PML and Special AT-Rich Sequence Binding Protein1 (Kumar et al., 2007). Likewise, phy photobodies could serve as organization centers involved in the regulation of light-responsive genes (Fig. 3C). This model and the degradation model in Figure 3B are not mutually exclusive, as the degradation of some transcriptional activators has been shown to be coupled with their transcriptional activity (Lipford et al., 2005; Collins and Tansey, 2006). Therefore, it is also possible that photobodies are sites for both the degradation of transcriptional regulators and the regulation of transcription.
GENETIC DISSECTION OF PHOTOBODY FUNCTION
Although nuclear bodies have been extensively studied, particularly in mammalian systems, and key constituents of a few nuclear bodies have been successfully identified by cell biology and proteomic approaches (Gall, 2000; Bernardi and Pandolfi, 2007), the precise function and regulation of nuclear bodies are still poorly understood. Arabidopsis represents an ideal organism to dissect the function of nuclear bodies by the combination of molecular genetic and cell biological approaches (Shaw and Brown, 2004; Collier et al., 2006; Fang and Spector, 2010). Because their steady-state pattern can be precisely manipulated by external light quality and quantity, phyB-containing photobodies provide an excellent model system to investigate the function and regulatory mechanisms of nuclear bodies in relation to signaling events in the nucleus (Chen, 2008).
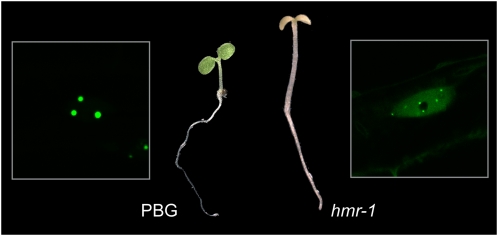
We recently reported a forward genetic screen aimed at isolating mutants defective in phyB-GFP photobody localization in the light (Chen et al., 2010b). This screen identified a novel phy signaling component, HEMERA (HMR), which itself is localized to the periphery of phyB photobodies. The hmr mutant represents a new class of light signaling mutants that are albino and tall in R and FR light. In addition, phyB-GFP localizes to smaller photobodies in hmr (Fig. 4). More interestingly, in hmr mutants, phyA, PIF1, and PIF3 accumulate in the light (Chen et al., 2010b). HMR is predicted to be structurally similar to RAD23, which is a multiubiquitin receptor that delivers multiubiquitylated proteins to the proteasome for degradation, suggesting that HMR could play a similar role in phyA, PIF1, and PIF3 degradation in the light (Chen et al., 2010b). Taken together, these results provide genetic evidence supporting the model in which photobodies are sites for protein degradation (Fig. 3B; Chen et al., 2010b). Further investigation of the biochemical functions of HMR as well as the identification of other genes from the same genetic screen will likely provide greater insight into the link between photobodies and protein degradation.
Figure 4.
The hmr mutant. Images of 4-d-old PBG (the parental type of hmr-1) and hmr-1 mutant seedlings grown under 8 μmol m−2 s−1 light. The hmr seedling has both long-hypocotyl and albino phenotypes. Confocal images show that phyB-GFP is localized to large photobodies in PBG seedlings. By contrast, phyB-GFP fails to form large photobodies and is instead localized to smaller photobodies in hmr-1.
PERSPECTIVE
Photoreceptor-containing photobodies in plants are unique and fascinating subnuclear domains whose assembly and function are directly regulated by light. The localization of photoreceptors, including phys and crys, to photobodies is triggered by a light-induced conformational switch to their active states. It is likely that the exposure of certain domains in the active state, such as the C-terminal domain of phyB, facilitates new protein-protein interactions and the “formation” or recruitment of photoreceptors and other signaling molecules to photobodies. The detailed mechanism of photobody assembly is still elusive. Localization studies of light signaling components and recent genetic evidence support the model that photobodies are sites for light signaling events, such as light-dependent turnover of key transcriptional regulators. However, we have only begun to understand the regulatory mechanisms and functions of photobodies. Many key questions remain to be answered. It is still not clear whether photobody-localized transcriptional regulators are degraded or only modified on photobodies. We still do not know whether photobodies are directly involved in transcriptional regulation and whether they are associated with chromatin. Accumulating evidence shows a convergence between the light signaling pathway and other signaling pathways, including those of temperature, hormones, and the circadian clock, on shared downstream signaling molecules (Kami et al., 2010; Lau and Deng, 2010; Leivar and Quail, 2011). Could photobodies serve as a hub for the interaction between these signaling pathways? We anticipate that by using a combination of molecular genetic, cell biological, proteomic, and genomic approaches, studies over the next few years promise to answer some of these questions and uncover new mechanisms of function and regulatory mechanisms for photobodies in light signaling.
References
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH. (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1: 213–222 [DOI] [PubMed] [Google Scholar]
- Ballesteros ML, Bolle C, Lois LM, Moore JM, Vielle-Calzada JP, Grossniklaus U, Chua NH. (2001) LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev 15: 2613–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D, Viczián A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adám E, Fejes E, Schäfer E, et al. (2004) Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP. (2007) Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 8: 1006–1016 [DOI] [PubMed] [Google Scholar]
- Briggs WR, Christie JM. (2002) Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci 7: 204–210 [DOI] [PubMed] [Google Scholar]
- Brown BA, Headland LR, Jenkins GI. (2009) UV-B action spectrum for UVR8-mediated HY5 transcript accumulation in Arabidopsis. Photochem Photobiol 85: 1147–1155 [DOI] [PubMed] [Google Scholar]
- Chamovitz DA, Wei N, Osterlund MT, von Arnim AG, Staub JM, Matsui M, Deng XW. (1996) The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86: 115–121 [DOI] [PubMed] [Google Scholar]
- Chang CS, Li YH, Chen LT, Chen WC, Hsieh WP, Shin J, Jane WN, Chou SJ, Choi G, Hu JM, et al. (2008) LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J 54: 205–219 [DOI] [PubMed] [Google Scholar]
- Chang CS, Maloof JN, Wu SH. (2011) COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol 156: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M. (2011) The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62: 335–364 [DOI] [PubMed] [Google Scholar]
- Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee JH, Zhu D, et al. (2010a) Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. (2008) Phytochrome nuclear body: an emerging model to study interphase nuclear dynamics and signaling. Curr Opin Plant Biol 11: 503–508 [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J. (2011) Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol 21: 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C. (2004) Light signal transduction in higher plants. Annu Rev Genet 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Chen M, Galvão RM, Li M, Burger B, Bugea J, Bolado J, Chory J. (2010b) Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141: 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Schwab R, Chory J. (2003) Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc Natl Acad Sci USA 100: 14493–14498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Tao Y, Lim J, Shaw A, Chory J. (2005) Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr Biol 15: 637–642 [DOI] [PubMed] [Google Scholar]
- Chory J. (2010) Light signal transduction: an infinite spectrum of possibilities. Plant J 61: 982–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T, Shokry A, Moffet M, Liu P, Faul M, Sharrock RA. (2009) Obligate heterodimerization of Arabidopsis phytochromes C and E and interaction with the PIF3 basic helix-loop-helix transcription factor. Plant Cell 21: 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S, Pendle A, Boudonck K, van Rij T, Dolan L, Shaw P. (2006) A distant coilin homologue is required for the formation of cajal bodies in Arabidopsis. Mol Biol Cell 17: 2942–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins GA, Tansey WP. (2006) The proteasome: a utility tool for transcription? Curr Opin Genet Dev 16: 197–202 [DOI] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi C, Johansson H, Holm M. (2007) SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19: 3242–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi GH, Deng XW, Holm M. (2006) Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18: 70–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Johansson H, Hettiarachchi C, Irigoyen ML, Desai M, Rubio V, Holm M. (2008) LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20: 2324–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek PD, Elmer MV, van Oosten VR, Fankhauser C. (2004) The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol 14: 2296–2301 [DOI] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev 14: 2377–2391 [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Spector DL. (2010) Live cell imaging of plants. Cold Spring Harb Protoc 2010: pdb top68 [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Chen M. (2008) Transposing phytochrome into the nucleus. Trends Plant Sci 13: 596–601 [DOI] [PubMed] [Google Scholar]
- Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, et al. (2009) Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G. (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17: 75–86 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. (2000) Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol 16: 273–300 [DOI] [PubMed] [Google Scholar]
- Genoud T, Schweizer F, Tscheuschler A, Debrieux D, Casal JJ, Schäfer E, Hiltbrunner A, Fankhauser C. (2008) FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet 4: e1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu NN, Zhang YC, Yang HQ. (2011) Substitution of a conserved glycine in the PHR domain of Arabidopsis CRYPTOCHROME 1 confers a constitutive light response. Mol Plant (in press) [DOI] [PubMed] [Google Scholar]
- Henriques R, Jang IC, Chua NH. (2009) Regulated proteolysis in light-related signaling pathways. Curr Opin Plant Biol 12: 49–56 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A, Tscheuschler A, Viczián A, Kunkel T, Kircher S, Schäfer E. (2006) FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol 47: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A, Viczián A, Bury E, Tscheuschler A, Kircher S, Tóth R, Honsberger A, Nagy F, Fankhauser C, Schäfer E. (2005) Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr Biol 15: 2125–2130 [DOI] [PubMed] [Google Scholar]
- Hisada A, Hanzawa H, Weller JL, Nagatani A, Reid JB, Furuya M. (2000) Light-induced nuclear translocation of endogenous pea phytochrome A visualized by immunocytochemical procedures. Plant Cell 12: 1063–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW. (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Su YS, Lagarias JC. (2009) A light-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcriptional networks. Mol Plant 2: 166–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Quail PH. (2003) Nuclear translocation of the photoreceptor phytochrome B is necessary for its biological function in seedling photomorphogenesis. Plant J 35: 660–664 [DOI] [PubMed] [Google Scholar]
- Indorf M, Cordero J, Neuhaus G, Rodríguez-Franco M. (2007) Salt tolerance (STO), a stress-related protein, has a major role in light signalling. Plant J 51: 563–574 [DOI] [PubMed] [Google Scholar]
- Jang IC, Henriques R, Seo HS, Nagatani A, Chua NH. (2010) Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Yang JY, Seo HS, Chua NH. (2005) HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 19: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Yang SW, Yang JY, Chua NH. (2007) Independent and interdependent functions of LAF1 and HFR1 in phytochrome A signaling. Genes Dev 21: 2100–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G. (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI. (2009) Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol 60: 407–431 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW. (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Kaiserli E, Jenkins GI. (2007) UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 19: 2662–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C. (2010) Light-regulated plant growth and development. Curr Top Dev Biol 91: 29–66 [DOI] [PubMed] [Google Scholar]
- Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu SH. (2009) The Arabidopsis B-box zinc finger family. Plant Cell 21: 3416–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro S, Maruyama K, Nakashima K, Imura Y, Narusaka Y, Shinwari ZK, Osakabe Y, Fujita Y, Mizoi J, Shinozaki K, et al. (2009) The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol 151: 2046–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis EA, Oka Y, Hudson ME, Nagatani A, Quail PH. (2009) Residues clustered in the light-sensing knot of phytochrome B are necessary for conformer-specific binding to signaling partner PIF3. PLoS Genet 5: e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L, Kircher S, Toth R, Adam E, Schäfer E, Nagy F. (2000) Light-induced nuclear import of phytochrome-A:GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis. Plant J 22: 125–133 [DOI] [PubMed] [Google Scholar]
- Kircher S, Gil P, Kozma-Bognár L, Fejes E, Speth V, Husselstein-Muller T, Bauer D, Adám E, Schäfer E, Nagy F. (2002) Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schafer E, Nagy F. (1999) Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Lockhart P, Batschauer A. (2003) An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant J 35: 93–103 [DOI] [PubMed] [Google Scholar]
- Kleiner O, Kircher S, Harter K, Batschauer A. (1999) Nuclear localization of the Arabidopsis blue light receptor cryptochrome 2. Plant J 19: 289–296 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP. (1980) Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol 100: 147–160 [Google Scholar]
- Kumagai T, Ito S, Nakamichi N, Niwa Y, Murakami M, Yamashino T, Mizuno T. (2008) The common function of a novel subfamily of B-box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci Biotechnol Biochem 72: 1539–1549 [DOI] [PubMed] [Google Scholar]
- Kumar PP, Bischof O, Purbey PK, Notani D, Urlaub H, Dejean A, Galande S. (2007) Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat Cell Biol 9: 45–56 [DOI] [PubMed] [Google Scholar]
- Kwok SF, Staub JM, Deng XW. (1999) Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J Mol Biol 285: 85–95 [DOI] [PubMed] [Google Scholar]
- Lafarga M, Berciano MT, Pena E, Mayo I, Castaño JG, Bohmann D, Rodrigues JP, Tavanez JP, Carmo-Fonseca M. (2002) Clastosome: a subtype of nuclear body enriched in 19S and 20S proteasomes, ubiquitin, and protein substrates of proteasome. Mol Biol Cell 13: 2771–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW. (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Marchal V, Le Gourrierec J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U. (2006) Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133: 3213–3222 [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH. (2008) The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH. (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH. (2009) Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ. (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev 25: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford JR, Smith GT, Chi Y, Deshaies RJ. (2005) A putative stimulatory role for activator turnover in gene expression. Nature 438: 113–116 [DOI] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C. (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ. (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Trevisan M, Pradervand S, Fankhauser C. (2009) Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J 60: 449–461 [DOI] [PubMed] [Google Scholar]
- Más P, Devlin PF, Panda S, Kay SA. (2000) Functional interaction of phytochrome B and cryptochrome 2. Nature 408: 207–211 [DOI] [PubMed] [Google Scholar]
- Matsushita T, Mochizuki N, Nagatani A. (2003) Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424: 571–574 [DOI] [PubMed] [Google Scholar]
- Möglich A, Yang X, Ayers RA, Moffat K. (2010) Structure and function of plant photoreceptors. Annu Rev Plant Biol 61: 21–47 [DOI] [PubMed] [Google Scholar]
- Nagatani A. (2010) Phytochrome: structural basis for its functions. Curr Opin Plant Biol 13: 565–570 [DOI] [PubMed] [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B. (2000) FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101: 331–340 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G. (2007) PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, Matsushita T, Mochizuki N, Quail PH, Nagatani A. (2008) Mutant screen distinguishes between residues necessary for light-signal perception and signal transfer by phytochrome B. PLoS Genet 4: e1000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palágyi A, Terecskei K, Adám E, Kevei E, Kircher S, Mérai Z, Schäfer E, Nagy F, Kozma-Bognár L. (2010) Functional analysis of amino-terminal domains of the photoreceptor phytochrome B. Plant Physiol 153: 1834–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH. (2010) Phytochromes. Curr Biol 20: R504–R507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausenberger J, Hussong A, Kircher S, Kirchenbauer D, Timmer J, Nagy F, Schäfer E, Fleck C. (2010) An integrative model for phytochrome B mediated photomorphogenesis: from protein dynamics to physiology. PLoS ONE 5: e10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, et al. (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC. (2010) A brief history of phytochromes. ChemPhysChem 11: 1172–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC. (2006) Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57: 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JS, Kim JI, Kunkel T, Kim BC, Cho DS, Hong SH, Kim SH, Fernández AP, Kim Y, Alonso JM, et al. (2005) Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer. Cell 120: 395–406 [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Nagatani A. (1996) Nuclear localization activity of phytochrome B. Plant J 10: 859–868 [DOI] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T. (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA. (2001) A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13: 2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH. (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18: 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH. (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Brown JW. (2004) Plant nuclear bodies. Curr Opin Plant Biol 7: 614–620 [DOI] [PubMed] [Google Scholar]
- Shen H, Zhu L, Castillon A, Majee M, Downie B, Huq E. (2008) Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G. (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA. (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Spector DL. (2006) SnapShot: cellular bodies. Cell 127: 1071. [DOI] [PubMed] [Google Scholar]
- Stacey MG, von Arnim AG. (1999) A novel motif mediates the targeting of the Arabidopsis COP1 protein to subnuclear foci. J Biol Chem 274: 27231–27236 [DOI] [PubMed] [Google Scholar]
- Staub JM, Wei N, Deng XW. (1996) Evidence for FUS6 as a component of the nuclear-localized COP9 complex in Arabidopsis. Plant Cell 8: 2047–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YS, Lagarias JC. (2007) Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell 19: 2124–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian C, Kim BH, Lyssenko NN, Xu X, Johnson CH, von Arnim AG. (2004) The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: mutational analysis by bioluminescence resonance energy transfer. Proc Natl Acad Sci USA 101: 6798–6802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Hwang YS, Quail PH. (2006) phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J 48: 728–742 [DOI] [PubMed] [Google Scholar]
- Ulijasz AT, Vierstra RD. (2011) Phytochrome structure and photochemistry: recent advances toward a complete molecular picture. Curr Opin Plant Biol 14: 498–506 [DOI] [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW. (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Wolf I, Kircher S, Fejes E, Kozma-Bognár L, Schäfer E, Nagy F, Adám E. (2011) Light-regulated nuclear import and degradation of Arabidopsis phytochrome-A N-terminal fragments. Plant Cell Physiol 52: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Spalding EP. (2007) Separate functions for nuclear and cytoplasmic cryptochrome 1 during photomorphogenesis of Arabidopsis seedlings. Proc Natl Acad Sci USA 104: 18813–18818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. (1999) Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol 145: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Marquardt K, Indorf M, Jutt D, Kircher S, Neuhaus G, Rodriguez-Franco M. (2011) Nuclear localization and interaction with COP1 are required for STO/BBX24 function during photomorphogenesis. Plant Physiol 156: 1772–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lin R, Sullivan J, Hoecker U, Liu B, Xu L, Deng XW, Wang H. (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara M, Mitsui S, Hirano H, Takanabe R, Tokioka Y, Ihara N, Komatsu A, Seki M, Shinozaki K, Kiyosue T. (2004) Identification of ASK and clock-associated proteins as molecular partners of LKP2 (LOV Kelch protein 2) in Arabidopsis. J Exp Bot 55: 2015–2027 [DOI] [PubMed] [Google Scholar]
- Yi C, Deng XW. (2005) COP1: from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15: 618–625 [DOI] [PubMed] [Google Scholar]
- Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, Liu L, Zhang Y, Irigoyen ML, Sullivan JA, et al. (2008) COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell 32: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Liu H, Klejnot J, Lin C. (2010) The cryptochrome blue light receptors. The Arabidopsis Book; 8: e0135, 10.1199/tab.0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Sayegh R, Maymon M, Warpeha K, Klejnot J, Yang H, Huang J, Lee J, Kaufman L, Lin C. (2009) Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell 21: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng S, Chen F, Chen H, Wang J, McCall C, Xiong Y, Deng XW. (2008) Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell 20: 1437–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Bodnar MS, Spector DL. (2009) Nuclear neighborhoods and gene expression. Curr Opin Genet Dev 19: 172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Maier A, Lee JH, Laubinger S, Saijo Y, Wang H, Qu LJ, Hoecker U, Deng XW. (2008) Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C. (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]