One of the most remarkable biological insights in the past 30 years has been that many genetic programs for complex traits, such as flower or limb development, are shared across broad groups of organisms. These conserved pathways in turn can be tuned to produce tremendous phenotypic differences, not only between, but also within species. Intraspecific variation is often quantitative, one example being the onset of flowering, although there is also qualitative variation, such as in the ability to resist pathogens.
While many tools for quantitative genetics were developed by breeders, the model plant Arabidopsis (Arabidopsis thaliana) was adopted for studying the genetic architecture of quantitative traits soon after molecular markers for mapping became available (Chang et al., 1988; Nam et al., 1989). The species belongs to a small genus with nine members. Different from most of its congeners, Arabidopsis is self-compatible, and its life cycle can be as short as 6 weeks, both properties that greatly facilitate genetic studies. Its native range is considered to be continental Eurasia and North Africa (Al-Shehbaz and O'Kane, 2002), but it has been introduced throughout much of the rest of the world, especially around the northern hemisphere.
The potential of genetic variation to inform many different areas of Arabidopsis biology was most strongly advocated by Maarten Koornneef and his students. From the mid-1990s, they published both an impressive number of original research articles on this subject and a series of influential review articles that advertised the impact that the study of natural genetic variation could have on questions of both development and physiology (Alonso-Blanco and Koornneef, 2000; Koornneef et al., 2004).
Today, the study of natural variation in Arabidopsis continues to reveal new biology. In addition, the entire genus is increasingly being used to address fundamental questions of evolution (Mitchell-Olds and Schmitt, 2006; Bergelson and Roux, 2010). Some of the problems studied are: How, and how frequently, do new variants arise? Why do some variants rise to high frequency, while others are eliminated? And why are certain combinations of new variants incompatible with each other? Here, I will first give an overview of the tools and resources available for the study of natural variation in Arabidopsis. Next, I will present a few examples of how our knowledge of important biological processes has been improved through insights obtained from varieties other than the common laboratory accessions. Where similar or contrasting findings have been made in other species of the Brassicaceae, to which Arabidopsis belongs, I will mention these. The article concludes with a discussion of recent work that aims to integrate evolutionary and ecological studies with functional tests.
A final introductory note: Natural accessions of Arabidopsis have in the past often been referred to as “ecotypes.” This term implies that a line has a unique ecology and is adapted to specific environments, as opposed to differing only in genotype from other varieties (Turesson, 1922b). Preferable is the neutral term accession, which merely means that a unique identifier in a collection has been assigned (Alonso-Blanco and Koornneef, 2000).
GENETIC TOOL KIT FOR THE STUDY OF NATURAL VARIATION
Experimental Populations for Genetic Mapping
Accessions of Arabidopsis vary in a number of traits (Fig. 1; Table I). The most general way to identify genes is by crossing two accessions, which may or may not have a different phenotype, but produce nonuniform F2 progeny. In the F2 or later generations, specific phenotypes are then associated with segregating genetic markers that distinguish the contributions from the parental genomes. When phenotypic classes are not discrete, this is done using the methods of quantitative trait locus (QTL) mapping (Falconer and Mackay, 1996).
Figure 1.
Gross morphological variation in Arabidopsis and relatives. A, Variation between Arabidopsis accessions. On top, vegetative rosettes of accessions grown for 4 weeks in long days are shown. They vary in rosette diameter and compactness, leaf shape, and tissue necrosis or onset of senescence. Similarly, variation in size and shape of individual leaves, in this case the sixth in the rosette, is apparent in the 10 examples shown on the bottom left. Finally, differences in overall architecture are illustrated with five plants. On the left is an early flowering accession with few rosette leaves. The next two flower later, but the second one from the left has reduced apical dominance. Finally, the two accessions on the right have similarly tall main inflorescences but differ in the number of secondary inflorescences. The appearance on the far right is common among wild-grown plants. B, Some characters, such as flower size and fruit shape, vary relatively little within Arabidopsis, but more dramatic variation is found in comparison with closely related taxa, such as Capsella rubella (left) and A. lyrata (right). Images courtesy of Eunyoung Chae, Sang-Tae Kim, and George Wang.
Table I. Traits studied by natural variation in Arabidopsis.
For references, see Supplemental Table S1.
| Trait | Gene(s) Cloned?a |
| Aluminum content | N |
| Autonomous endosperm development | N |
| Auxin response | N |
| Carbohydrate availability and content | N |
| Cell wall composition | N |
| Chiasma frequency | N |
| Chromatin compaction | N |
| Circadian clock | C |
| Copper tolerance | Y |
| Crowding response | C |
| Disease resistance | Y |
| Drought response | N |
| Editing and processing of mitochondrial transcripts | Y |
| Elemental composition | Y/N |
| Flowering time | Y |
| Freezing tolerance | Y |
| Fruit number | C |
| Genetic robustness | N |
| Glucosinolate content | Y |
| Inflorescence replacement (mimicking grazing) | N |
| Jasmonate response | N |
| Leaf senescence | N |
| Leaf, inflorescence, and flower morphology | Y |
| Lethality in interploidy crosses | N |
| Life history traits other than flowering and growth | N |
| Light response | Y |
| Molybdenum content | Y |
| Nitrogen availability response | N |
| Oil content | N |
| Osmotic and salt stress tolerance | N |
| Phosphate content | N |
| Phytate content | N |
| Recruitment of bacterial rhizosphere communities | N |
| Root hydraulics | N |
| Root system size | N |
| Salicylic acid response | N |
| Salinity tolerance | N |
| Seed dormancy | Y |
| Seed germination, longevity | N |
| Seed lipids | N |
| Seed mucilage composition | Y |
| Sinapoylmalate biosynthesis | Y |
| Sodium accumulation | Y |
| Stomata density | N |
| Submergence tolerance | N |
| Sulfate content | Y |
| Terpene biosynthesis | Y |
| Thermal dissipation | N |
| Trichome density | Y |
| Zinc response | Y |
Y, Yes; N, no; C, likely candidates.
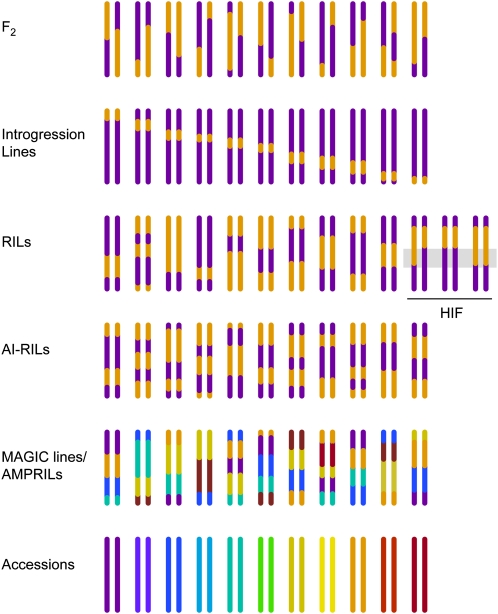
Because marker analysis used to be very tedious and expensive, substantial efforts were invested early on into producing recombinant inbred lines (RILs), which constitute immortal populations in which recombinant chromosomes have been fixed through inbreeding (Reiter et al., 1992; Lister and Dean, 1993; Fig. 2). RILs, which were first developed in mice (Bailey, 1971), have the advantage that they need to be genotyped only once but can be phenotyped repeatedly for many different traits and under different environmental conditions. An advantage of Arabidopsis is its self-compatibility, so that inbred lines can be easily generated by selfing and single-seed descent. Around 60 RIL populations are available from the stock centers as of the time this article is written (end of 2011; http://www.inra.fr/internet/Produits/vast/RILs.htm, http://www.arabidopsis.org/ and http://www.arabidopsis.info/). Importantly, the lengthy inbreeding process can now be bypassed through a revolutionary technology introduced by the laboratory of Simon Chan. This method allows the facile production of doubled haploid plants from recombinant populations (Ravi and Chan, 2010).
Figure 2.
Populations for mapping genes causing trait variation. Colors indicate contribution from different parental accessions. Only one chromosome pair is shown for each individual. HIF individuals are derived from RILs, in which a small portion of the genome is still heterozygous.
Even after five or six generations of inbreeding, which is customary for RILs, a small percent of the genome remains heterozygous. This turns out to have its own benefits. In such a heterogeneous inbred family (HIF), only a small portion of the genome segregates for the two parental alleles (Tuinstra et al., 1997). Additional recombinants that further reduce an interval of interest are easily derived from heterozygous HIF individuals, as are near isogenic lines (NILs) that are homozygous for either parental allele at this locus. A disadvantage of HIF-derived NILs is that each HIF has a unique genome composition and that one can therefore not easily place several QTL in a common genetic background.
NILs that carry only a small genomic region from one parent in a background that is otherwise composed of the genome of the other parent can also be generated directly by repeated backcrosses (Fig. 2). Such NILs, pioneered in crops where they are also called introgression lines (Seevers et al., 1971; Rhodes et al., 1989; Eshed and Zamir, 1995), are powerful for systematic analyses of interactions between genes from different genomes, although epistatic interactions among alleles from the introgressed genome are mostly lost. The properties of NIL sets are in many ways complementary to those of RILs, and they are particularly useful when introgression is performed in two directions (Törjék et al., 2008). NILs can identify QTL of smaller effect but with lower resolution than RIL populations (Falconer and Mackay, 1996; Keurentjes et al., 2007).
Although the genomes of RILs already contain more recombination events than F2 populations and therefore afford higher mapping resolution, this can be further increased with advanced intercross RILs, in which individuals from the F2 and later generations are intermated before inbred lines are derived (Darvasi and Soller, 1995; Balasubramanian et al., 2009). Other approaches involve the use of multiple parents, as in the MAGIC (for multiple advanced generation intercross) and AMPRIL (for Arabidopsis multiparent RIL) populations (Fig. 2; Kover et al., 2009; Huang et al., 2011). The MAGIC design is more elaborate and generates more recombination events per line than the AMPRIL strategy, but the founder genomes are less evenly represented in the final lines. Mapping in either population is more complex than with RILs, but with a sufficiently high density of intermediate frequency markers, one can infer the most likely local founder genotype. Even more so than simple F2 or RIL populations, AMPRILs and MAGIC lines are likely to contain genotypic combination not found in the wild.
QTL mapping accuracy increases with the MAGIC and AMPRIL populations, but not all possible QTL that can be found in pairwise crosses between some of the parents are detected. An alternative would be to combine the most informative subsets of RIL populations and to perform a joint QTL analysis. Especially when genotyped with common markers, a joint analysis can confirm common QTL (Bentsink et al., 2010; Salomé et al., 2011b).
Some of the advantages of using RIL-type populations will continue to apply in the future. Trait values, especially those with low heritability, can be estimated more precisely due to replication (Soller and Beckmann, 1990; Mackay, 2001). Perhaps most importantly, one can study correlations between different traits, which can reveal fitness trade-offs, and reaction norms, the response of a specific genotype to different environments. However, not every geographic region where Arabidopsis is found is fairly represented in the available RIL populations because geographic sampling of Arabidopsis has so far been rather uneven (Fig. 3). Thus, forward genetics in additional material, even if composed mostly of F2 populations, will likely be informative. Fortunately, with reduced representation approaches such as restriction-associated DNA sequencing (RAD-seq) or genotyping-by-sequencing (Baird et al., 2008; Elshire et al., 2011) and multiplexing of genomic DNA from many individuals (currently, at least 96), costs for interrogating thousands of markers have dropped to a few U.S. dollars.
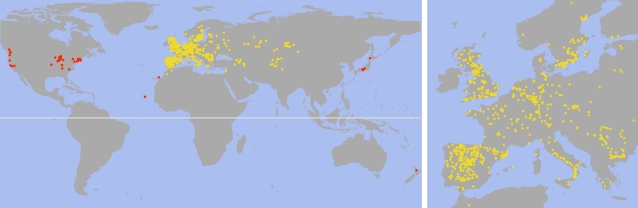
Figure 3.
Distribution of over 7,000 Arabidopsis accessions collected from the wild and available in the stock center or soon-to-be-released collections. Western and southern Europe, including Great Britain, is heavily overrepresented, although sampling is not even. Accessions from the presumed native range are in yellow and likely introductions in red. Whether the distribution across China to Japan is continuous with the native range is unclear. Arabidopsis has been reported in additional locales, such as South Korea, and several African countries (Alonso-Blanco and Koornneef, 2000). Maps courtesy of George Wang.
Finally, a general caveat when performing conventional genetic mapping is that chiasma frequencies differ between accessions (Sanchez-Moran et al., 2002). Data from F2 populations also support the conclusion that recombination rates vary depending on the cross (Salomé et al., 2011a). Thus, the ease with which loci are mapped will differ from cross to cross, even more so if structural variants interfere with recombination near the loci of interest.
Identification and Validation of Causal Genes and Polymorphisms
After a genomic interval underlying phenotypic differences has been identified, there are various options to track down the responsible gene, assuming that only a single gene is causal. Different from induced mutations, simply resequencing a region with dozens or more genes is on its own generally not informative because of the high number of polymorphisms that distinguish an arbitrary pair of accessions, about 1 in every 200 bp. Fortunately, compared to other multicellular organism in which natural variation is studied, Arabidopsis has the enormous advantage that almost all accessions are quite easily transformed by dipping flowering plants into a suspension of Agrobacterium tumefaciens containing a T-DNA vector with the transgene of interest (Clough and Bent, 1998).
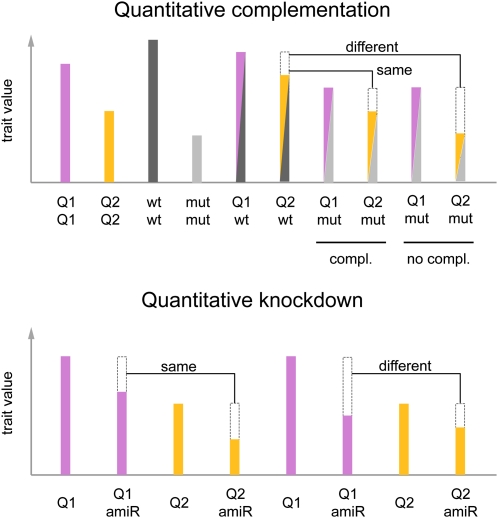
If the final mapping interval does not contain a gene previously implicated in the trait of interest, one of the first steps will often be to investigate whether null alleles affect this trait. For the vast majority of genes, T-DNA insertion lines in the reference Columbia-0 (Col-0) background are available from the stock centers (http://arabidopsis.org, http://arabidopsis.info; for review, see Alonso and Ecker, 2006). The most straightforward approach to investigate the activity of individual genes in other genetic backgrounds is gene silencing, and collections of vectors for knocking down a large fraction of genes present in the reference genome are available, both for conventional hairpin RNA interference and artificial microRNAs (amiRNAs; for review, see Ossowski et al., 2008b). Gene silencing is a convenient tool to test the relative activity of alleles, an approach that we have called quantitative knockdown (Schwartz et al., 2009). It is conceptually related to quantitative complementation, where different alleles are examined in the hemizygous state, by crossing a homozygous strain to a tester that carries a knockout allele of the gene of interest (Mackay, 2001; Fig. 4).
Figure 4.
Quantitative complementation and knockdown to determine whether QTL are allelic to a candidate gene. Both tests rely on quantitative comparisons between genotypes; the dashed boxes indicate phenotypic differences to the genotype to the left. In a quantitative complementation test, one determines whether the two QTL alleles, Q1 and Q2, are differentially affected when heterozygous with the wild-type (wt) or mutant (mut) allele of a candidate gene (Mackay, 2001). If the QTL alleles respond differently, i.e. if in this example only Q1 complements the mutant phenotype, the candidate gene and the QTL are probably allelic. Similarly, in a quantitative knockdown experiment, a differential effect of an amiRNA (amiR) against the candidate gene indicates that the Q1 allele has lower activity than Q2 and that the candidate gene is likely responsible for the QTL.
As an alternative, one can introduce genomic fragments spanning the region of interest to identify the gene(s) affecting the trait under investigation. Transgenic complementation also allows the examination of chimeric genes in different backgrounds to pinpoint the causal region, or even nucleotide, within an allele. A possible complication arises from the fact that the addition of an extra wild-type copy of an independent gene in the same pathway can quantitatively affect the phenotype and thus confound the interpretation of the observed phenotypes. An attractive feature of amiRNAs is that one can engineer transgenes that do not change the encoded protein but do not respond to silencing by a specific amiRNA anymore (Palatnik et al., 2003). One can thus use an amiRNA to knock out the endogenous gene and at the same time introduce a variant copy of the gene that is not affected by the amiRNA. This allows in essence the functional replacement of one allele with another.
A final word of caution: Spontaneous mutations are not as rare as one might think, with direct measurements indicating about one new single base pair mutation per haploid genome and generation (Ossowski et al., 2010). Thus, not every genetic variant that distinguishes accessions must be a natural variant in the sense that it was present in nature. Indeed, there are now several reports of mutations with large phenotypic effects that were segregating in an accession and may only have arisen after the accession was collected. Two of these cases affect parents of commonly used RIL populations, Landsberg erecta-0 and Bayreuth-0 (Doyle et al., 2005; Loudet et al., 2008; Laitinen et al., 2010). Thus, even if misidentification of an accession has been ruled out, which is not uncommon (Anastasio et al., 2011; Simon et al., 2011), there can be true genetic and phenotypic differences between accessions that share recent common ancestry.
WHOLE-GENOME RESOURCES FOR THE STUDY OF NATURAL VARIATION
Enabling Genome-Wide Association Studies
Genetic mapping in crosses is greatly facilitated when genome-wide polymorphisms, or better yet the entire genome sequences, of the investigated accessions are known. If a sufficient number of genome sequences is available, one can even dispense with experimental crosses and exploit shared ancestry to directly identify common alleles that are responsible for phenotypic variation in the entire population. This approach was first proposed for human, already before the first finished human genome sequence was in sight (Lander, 1996; Risch and Merikangas, 1996). Because obtaining complete genome sequences for many individuals of the same species was out of question at the time, it was proposed to rely on linkage disequilibrium (LD). LD refers to the fact that in most species there has not been enough historic recombination to produce all possible combinations of physically adjacent polymorphisms, but rather that sequence variants are normally found in haplotype blocks of various lengths. Thus, a causal polymorphism can in principle be identified indirectly through its association with any of the other sequence variants in its haplotype block (Kruglyak, 1999; Jorde, 2000). The term that is normally used today for this experimental strategy is genome-wide association study (GWAS). A shortcut that reduces the required genotyping effort has been to make use of prior information and to first focus on genes already shown to affect a trait of interest (Long et al., 1998; Caicedo et al., 2004; Olsen et al., 2004; Balasubramanian et al., 2006; Ehrenreich et al., 2009), but this has become largely obsolete today.
While the principles of GWAS are easy to understand, important limitations arise from population structure, that is, not all investigated individuals being equally distantly related to each other. Powerful methods have been developed to correct for population structure, but how to reliably detect alleles that are largely fixed between populations remains a challenge. Other issues are allelic heterogeneity, that is, alleles at a single locus with similar effects on gene function having arisen repeatedly; or complex genetic architecture, where many different genes affect the same trait. A recent article by Myles et al. (2009) provides an excellent primer of the challenges for GWAS.
As with RIL analyses, the selfing nature of Arabidopsis is a boon for GWAS, since each accession needs to be genotyped or sequenced only once but can be phenotyped many times. Magnus Nordborg almost single-handedly convinced the Arabidopsis community of the feasibility and usefulness of GWAS approaches, even before high-density genotype information was available (Aranzana et al., 2005; Zhao et al., 2007). While initial estimates of LD in Arabidopsis were too high (Nordborg et al., 2002, 2005), it finally turned out that LD in the global population extends over not more than about 5 to 10 kb, or one to two genes, which is very convenient for GWAS (Kim et al., 2007). It is thought that the relatively low LD reflects a history of frequent outcrossing together with rapid dispersal enabled by the selfing mode of reproduction.
The first enterprise with the goal of finding a large fraction of sequence variants used high-density custom arrays with almost one billion unique oligonucleotides to interrogate the genomes of 20 accessions, including the Col-0 reference accession (Clark et al., 2007). This set was chosen to be maximally diverse based on a previous analysis of 96 accessions, from which about 1,000 short fragments distributed throughout the genome had been dideoxy sequenced (Nordborg et al., 2005). The most important information to come from the array-based resequencing study was a collection of hundreds of thousands of nonsingleton single nucleotide polymorphisms (SNPs) that could be used for GWAS (Kim et al., 2007). About 216,000 SNPs, or one every 0.5 kb, have been subsequently typed in over 1,000 accessions (Horton et al., 2012), chosen from a larger panel of more than 5,000 accessions for which information from 149 intermediate frequency markers was available (Platt et al., 2010). The high density of SNPs meant that a typical haplotype block was tagged with several SNPs, which made GWAS in Arabidopsis right away more powerful than in humans. Despite similar LD characteristics, GWAS in human initially used only about 1 SNP per 6 kb (Wellcome Trust Case Control Consortium, 2007).
Prospects of GWAS in Arabidopsis
Several proof-of-concept examples have now been published, indicating that GWAS will often be successful in Arabidopsis. In the first comprehensive study, over 100 different morphological, physiological, and molecular traits were analyzed in 96 to 192 accessions (Atwell et al., 2010). In several cases, known genes were rediscovered, and in many others, plausible candidates were identified with high precision. The most impressive results, in agreement with previous pilot studies (Aranzana et al., 2005), were obtained for disease resistance, which is often controlled by single genes with very large effects. This is in contrast with humans, where effect sizes of QTL detected by GWAS are often small (McCarthy et al., 2008; Manolio et al., 2009).
The utility of GWAS can be increased by making use of prior information, such as functional data from mutant studies, gene annotation, or membership of genes in specific regulatory networks to prioritize GWAS candidates (Aranzana et al., 2005; Schadt et al., 2005; Atwell et al., 2010; Chan et al., 2011). Similarly, QTL mapping in experimental populations can greatly reduce the portion of the genome that one has to consider for the location of GWAS QTL (Brachi et al., 2010; Nemri et al., 2010). This approach becomes particularly powerful when both strategies are directly integrated using experimental populations with several parents, so that alleles pinpointed by GWAS are represented in multiple founder backgrounds. The term nested association mapping has been coined for this approach, which was pioneered in maize (Zea mays; Yu et al., 2008; McMullen et al., 2009). Arabidopsis populations, such as the MAGIC lines and AMPRILs, serve a similar purpose (Kover et al., 2009; Huang et al., 2011). An alternative will be to examine several independent RIL populations. An advantage of using RIL sets over F2 individuals in this case is that for each set of founders, the lines can be chosen to be maximally informative in terms of contribution of the founder genomes, thus greatly reducing phenotyping efforts (Xu et al., 2005; Simon et al., 2008).
Because of the plasticity of plant development and physiology, the influence of genes on the phenotype is very often dependent on the environment, often codified as gene-by-environment or GxE interaction. Similarly, the effects of individual genes are often modified by other genes in the genome because genes do not act on their own but form more or less complex functional networks. When genes have nonadditive effects, this is called GxG or more commonly an epistatic interaction. While the identification of epistatic QTL is standard fare for mapping in experimental populations (Mackay, 2001), this continues to be a major challenge for GWAS. This has been suggested to be computationally and statistically feasible several years ago (Marchini et al., 2005), and several computational strategies have been developed since (Mitchell-Olds, 1995; Cordell, 2009; Kam-Thong et al., 2011). However, I am not aware of an example where all variants were used in a GWAS to detect epistatic loci. Here again, mapping in experimental populations, perhaps in combination with network reconstruction (Rowe et al., 2008; Jiménez-Gómez et al., 2010; Kerwin et al., 2011), should help to reduce the search space for GWAS of epistatic loci.
A Proliferation of Genome Sequences
In addition to the anonymous SNPs for the first generation of GWAS in Arabidopsis, array-based resequencing revealed tens of thousands of amino acid replacements along with hundreds of more drastic mutations that are likely to eliminate the function of many genes in various accessions. In addition, a large percentage of the reference genome was found to be missing in each accession (Borevitz et al., 2007; Clark et al., 2007; Zeller et al., 2008; Plantegenet et al., 2009). This implied that, conversely, the reference accession Col-0 likely lacked a substantial portion of genes present in other accessions. The analysis of individual loci had already shown that some gene families could differ greatly between accessions. Foremost are the disease resistance genes of the nucleotide-binding site-Leu-rich repeat (NB-LRR) class, with both presence/absence polymorphisms and highly divergent alleles in different accessions (Grant et al., 1995; Caicedo et al., 1999; Noël et al., 1999; Stahl et al., 1999; Rose et al., 2004). A logical next step was therefore to scrutinize the genomes of accessions for sequences not represented in the reference genome. With the advent of new sequencing technologies, this goal became attainable at a reasonable cost. Even before these methods were exploited to the same end for human genomes, it was shown that they not only gave an accurate account of small-scale polymorphisms in Arabidopsis genomes but that they could also be used to detect copy number variants and to assemble sequences absent from the reference (Ossowski et al., 2008a).
The 1001 Genomes Project for Arabidopsis was announced in 2007 (Nordborg and Weigel, 2008; Weigel and Mott, 2009). The initial proposal was to pursue a two-pronged hierarchical strategy for defining the pangenome of Arabidopsis. The first hierarchical aspect was a sampling of accessions throughout the range of Arabidopsis such that diversity could be analyzed at global, regional, and local scales. Thus, rather than equidistant distribution of samples, it was envisioned that the project would include regional populations separated by distances measured in kilometers as well as individuals from within local stands spaced only meters apart. The second hierarchical aspect was to produce genome sequences at different levels of accuracy and completeness such that a relatively small number of highly accurate and complete genomes would inform the analysis of a much larger number of genomes that had not been completely assembled. The rationale behind this proposal was that mere lists of sequence variants that result from simple resequencing approaches, in which sequence reads are only aligned to a target genome, can be misleading. Specifically, because of false-negative problems, trying to reconstruct contiguous sequences by superimposing known isolated polymorphisms on the reference genome information can be problematic. To overcome these limitations, two groups have introduced reference-guided assembly approaches (Gan et al., 2011; Schneeberger et al., 2011), in which the Col-0 reference genome (Arabidopsis Genome Initiative, 2000) is first used to identify portions of the genome that are conserved in other accessions. Gaps are then filled in by assembling sequence reads and anchoring them to the known bits. As expected, multiple out-of-phase insertions or deletions in coding sequences can combine to restore open reading frames (Schneeberger et al., 2011). Similarly, additional mutations can make up for defects in splice acceptor or donor sites, as can be inferred from transcriptome analysis by RNA sequencing (Gan et al., 2011). The error rates of these reference-guided assemblies in single-copy regions were close to what was deemed as the lower acceptable bound in the initial reference genome sequencing project, about 1 in 10,000 bp (although final error rates in the reference genome were probably only about one-fifth; Ossowski et al., 2008a).
As expected from previous resequencing studies, up to 2% of reference positions were judged to be absent from the new assemblies. Conversely, up to 0.6% of the new assemblies represented sequences not found in the reference genome (Gan et al., 2011; Schneeberger et al., 2011). Because the new sequencing technologies generate more error-prone and shorter reads, and the insert sizes for paired-end sequencing libraries are generally smaller as well (Metzker, 2010), there are limits to closing gaps between regions that are well conserved relative to the reference genome. That bases present in the reference, but missing from a nonreference accession, outnumber the opposite class severalfold indicates the shortcomings of the reference-guided assemblies, since it should be equally likely that insertions and deletions occur on either lineage. We are thus currently faced with a paradox: >90% of the euchromatic portion of an accession’s genome can be sequenced for a few hundred dollars, but the remainder can only be recovered when investing many hundred or thousand times that amount. This is particularly relevant because some of the most interesting genes in the genome, such as many disease resistance genes, reside in highly variable gene clusters with often nearly identical tandem repeats that are even challenging for assembly from dideoxy sequenced bacterial artificial chromosomes or fosmid clones (Noël et al., 1999).
While the most common approach for the identification and annotation of variants has been comparison against the reference, a multiple alignment consensus benefits the evaluation of complex alleles (Gan et al., 2011). However, with the rapid increase in the number of genome sequences, simple all-against-all comparisons will soon not be feasible anymore because of the time required to perform them. It has therefore been proposed to represent the pangenome, that is, the collection of all possible sequence variants along each chromosome, in a single data structure as a graph, which would both facilitate the identification of polymorphisms in newly sequenced genomes and their classification as shared or unique (Schneeberger et al., 2009).
Insights from Comparing Genome Sequences
Apart from supporting forward genetic studies in Arabidopsis, genome sequences have increased our understanding of the evolutionary history of the species. Array-based comparison of 20 accessions revealed only a single large region in the genome that was shared by the majority of accessions, indicative of this region having experienced recent and strong selection in many different populations (Clark et al., 2007). Remarkably, the much more fine-grained information from short-read sequencing of 80 lines did not substantially change this picture of strong selective sweeps being rare, even though population differentiation along the genome is not uniform (Cao et al., 2011).
In addition to local polymorphism patterns that are shaped by selection and demography, there are consistent chromosomal-scale differences that are probably caused by molecular and genetic factors, such as mutation, recombination, and biased gene conversion. One of these is an excess of polymorphisms in regions adjacent to the centromeres (Borevitz et al., 2007; Clark et al., 2007), which has also been reported in Medicago truncatula and rice (Oryza sativa), but not in maize (Gore et al., 2009; Huang et al., 2010c; Branca et al., 2011). The interpretation of polymorphism patterns in Arabidopsis has also benefited from the high-quality reference sequence available now for the close relative Arabidopsis lyrata (Hu et al., 2011). In agreement with lack of conservation between the two species reflecting either that sequences are dispensable or subject to species-specific positive selection, regions found only in Arabidopsis are more polymorphic than shared regions (Cao et al., 2011).
Finally, Arabidopsis accessions harbor extensive variation in mitochondrial genomes (Forner et al., 2005; Arrieta-Montiel et al., 2009), in subtelomeric regions (Kuo et al., 2006; Wang et al., 2010), and in heterochromatic repeats, including retrotransposons and rDNA (Fransz et al., 2000; Davison et al., 2007; Ito et al., 2007). Structural differences between mitochondrial genomes can be revealed relatively easily by new sequencing methods (Davila et al., 2011). Furthermore, although read lengths and insert sizes are insufficient for long-range reconstruction of highly repetitive regions of the genome, read coverage and sequence variation in individual reads can be exploited to determine differences in genome size and repeat content (James et al., 2009; Tenaillon et al., 2011).
Utility of Genome Sequences
As of the time that this article was written (end of 2011), over 100 genome sequences for Arabidopsis had been published. In addition, sequence data for over 300 additional accessions were already publicly available. In aggregate, commitments for over 700 accessions had been made, indicating that the initial goal of 1,001 genome sequences would be reached well before the end of 2012 (http://1001genomes.org).
Several of the Arabidopsis genome sequences were immediately useful. For example, the Landsberg erecta accession is commonly used for mutant screens, and its genome sequence is facilitating the mapping and analysis of induced mutations. Similarly, several of the accessions are parents of RIL populations (Ossowski et al., 2008a; Schneeberger et al., 2009, 2011; Gan et al., 2011), and their genome sequences are aiding the identification of polymorphisms responsible for QTL. Genome sequences also provide an inventory of potential knockout mutations, which is informative given that a considerable fraction of natural genetic variation is due to loss-of-function alleles. Examples are new alleles of PHYTOCHROME D (PHYD) and FRIGIDA LIKE1 (FRL1), for which before only single alleles were known (Aukerman et al., 1997; Schläppi, 2006; Cao et al., 2011).
In addition, the 1001 Genomes Project is advancing GWAS. As discussed above, the first phase of GWAS in Arabidopsis has been based on a set of 216k tag SNPs, which were estimated to predict >90% of all common variants (Kim et al., 2007; Horton et al., 2012). It is simple to call the same SNPs in any of the accessions of the 1001 Genomes Project and to include any line that has not been array genotyped into GWAS projects that makes use of the 216k tag SNP array data. Furthermore, it is possible to accurately impute common variants identified by whole-genome sequencing in array genotyped accessions and GWAS with imputed data detects additional polymorphisms linked to traits under consideration (Cao et al., 2011).
Apart from increasing the chances that sequence differences directly responsible for trait variation are found by GWAS, a major advantage of complete genome sequences is that they support the prediction of activity differences between potentially causal alleles. For example, in coding regions, mutations that disrupt the open reading frame or affect splicing are more likely to affect gene function than codon or silent changes. And among amino acid substitutions, one can estimate how probable it is that a mutation has deleterious effects based on conservation of that amino acid in other species (Ng and Henikoff, 2006).
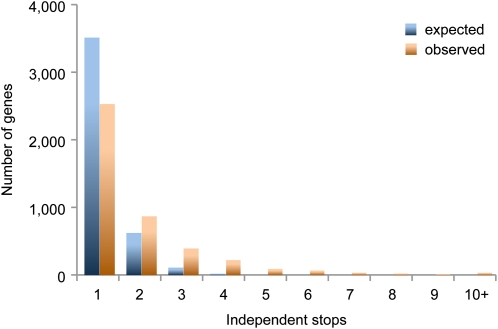
Complete genome sequences will thus help to tackle one of the major challenges of GWAS, allelic heterogeneity, where several different alleles have similar effects on the trait of question. That independent alleles at the same locus can have the same phenotypic consequences has been known for a quarter of a century, since the first genes responsible for genetic disorders or cancer in humans were cloned (Royer-Pokora et al., 1986; Clark et al., 1989; Botstein and Risch, 2003). In Arabidopsis, the flowering regulators FRIGIDA (FRI) and FLOWERING LOCUS C (FLC) are often partially or completely inactivated, with many of these alleles being found only in single accessions (Johanson et al., 2000; Le Corre et al., 2002; Gazzani et al., 2003; Michaels et al., 2003; Lempe et al., 2005; Shindo et al., 2005; Méndez-Vigo et al., 2011). Drastic mutations that prematurely terminate or partially delete the same open reading frame are found more often than expected by chance in the genomes of different accessions (Cao et al., 2011; Fig. 5). This might be the outcome of positive selection, as is the case for FRI and FLC (Toomajian et al., 2006), or purifying selection being weak or absent. In either case, the presence of multiple alleles with similar effects on a particular phenotype makes the detection of such loci in GWAS analyses difficult since each polymorphism is considered separately (Myles et al., 2009). If, instead, all alleles with similar predicted activity differences were combined or, better yet, if alleles were considered according to their relative degree of activity, this hurdle could be overcome.
Figure 5.
Comparison of expected and observed occurrences of 8,133 independent premature stops in 4,263 protein coding genes, considering all genes with >90% coverage in 75 out of 80 accessions. Data are from Cao et al. (2011).
The methods discussed in the preceding paragraph would be a considerable improvement over the strategy that is gaining popularity in humans: the search for an excess of rare variants in candidate genes. In rare-variant-burden methods, rare variants are combined for the purposes of contrasting phenotypically distinct classes of individuals, but functional effects of alleles are ignored, and these methods are not integrated into standard GWAS (Asimit and Zeggini, 2010).
Epigenomic Variation
GWAS in humans, where it is not unusual that tens of thousands of individuals are analyzed, has been successful in detecting many alleles, even with very small effects, but the fraction of the total variation explained by these variants is often only small. This also has been the case for traits such as height that are known to be highly heritable from family studies. Some possibilities are that genetic architecture may be more complex, with many interacting loci, or that rare alleles are more important than anticipated (see above). An alternative explanation, which is en vogue in many circles, is that epigenetic variation unlinked to sequence variants and, hence, not detectable by conventional GWAS is responsible for many phenotypic differences (McCarthy et al., 2008; Manolio et al., 2009).
Epigenetic differences can have obvious consequences in plants. In several species, including Arabidopsis, spontaneously occurring epialleles with overt phenotypes have been described (Jacobsen and Meyerowitz, 1997; Cubas et al., 1999; Hollick et al., 2000; Soppe et al., 2000; Stam et al., 2002; Manning et al., 2006; Martin et al., 2009). The epialleles often show increased cytosine methylation of the promoter and strongly reduced RNA expression. In several cases, the epialleles are associated with structural changes, such as the g mutation in melon, which is apparently caused by the insertion of a transposon and spread of DNA methylation into adjacent sequences.
Tiling array analyses comparing two different pairs of Arabidopsis accessions have shown that these differ in the extent of methylation at individual cytosines. That there are fewer differences in transposable element than genic methylation between natural accessions (Vaughn et al., 2007) agrees with transposable element methylation being more stable in inbred lines (Becker et al., 2011; Schmitz et al., 2011). Methylation differences seem to be largely stable in F1 hybrids (Woo and Richards, 2008; Zhang et al., 2008; Groszmann et al., 2011), but methylation patterns can change at relatively high rates, around 1% or more, in subsequent generations (Vaughn et al., 2007). The fluidity of the genomic methylation landscape after crosses is consistent with RNA-dependent DNA methylation mediated by short interfering RNAs being able to target other loci in trans, as long as these harbor sufficient levels of sequence similarity (Melquist and Bender, 2003). This is substantiated by nonadditive expression levels of short interfering RNAs and correlated effects on DNA methylation in F1 hybrids (Groszmann et al., 2011).
Importantly, although epialleles with phenotypic effects are largely stable and can be inherited over many generations, most revert occasionally to the wild-type form (Jacobsen and Meyerowitz, 1997; Cubas et al., 1999; Hollick et al., 2000; Soppe et al., 2000; Stam et al., 2002; Manning et al., 2006; Martin et al., 2009). The stability of DNA methylation in inbred Arabidopsis lines has recently been examined directly (Becker et al., 2011; Schmitz et al., 2011). While loss and gain of methylation at individual sites occurred much more often than mutations in the nucleotide sequence (Ossowski et al., 2010), changes in larger methylated regions similar to the ones that distinguish epialleles identified by forward genetics were rare. However, both types of methylation changes were distinguished from DNA mutations in that the same positions were affected in independent lines much more often than expected by chance and that there was an appreciable rate of reversions.
Crosses of wild-type lines to mutant strains with largely demethylated genomes have also revealed a wide range in the stability of epialleles after the causal mutations had been segregated away (Reinders et al., 2009; Teixeira et al., 2009). Consistent with the more labile nature of epialleles, heritability estimates in such lines are considerably lower than they are in natural accessions for the same traits (Roux et al., 2011). Thus, while the large majority of DNA methylation differences is sufficiently stable to account for inheritance within a limited number of generations, it remains unclear how often epialleles can become subject to Darwinian selection and thus make a contribution to long-term evolution. If reversion rates exceed the selective advantage conferred by an epiallele, its frequency in the population will be largely determined by the equilibrium of forward and reverse epimutation rates (Slatkin, 2009; Johannes and Colomé-Tatché, 2011).
In summary, although natural epialleles are often due to nearby structural variation, crosses between divergent accessions can induce new epialleles in trans. While the first class does not pose a problem for conventional GWAS, as such alleles should be tagged by linked sequence polymorphisms, the second class would only be revealed if GWAS would be extended to directly include information on DNA methylation profiles. A different question is whether epialleles are equally, more, or less likely than DNA alleles to reflect adaptation to the local environment.
LEARNING NEW BIOLOGY FROM THE STUDY OF NATURAL VARIATION
While knowledge about the origin and phenotypic effects of sequence polymorphisms is central to understanding how species adapt to their natural environment, most studies of genetic variation in Arabidopsis have probably been motivated by the desire to identify regulatory and other genes that are not present in the common laboratory accessions. An especially original use of natural variation has been the search for second site modifiers of ABA insensitive3 and leafy cotyledon1 mutant phenotypes. Both mutants suffer from impaired seed maturation, and seed viability declines much more rapidly than in wild-type plants. Introgression of the mutant alleles into other accessions identified natural modifiers that can partially suppress the mutant phenotypes, possibly pointing to new regulators of seed maturation (Sugliani et al., 2009). In a similar manner, the CAULIFLOWER (CAL) gene was discovered serendipitously as an enhancer of the apetala1 (ap1) mutant phenotype. CAL and AP1 turned out to be paralogs with an asymmetrical relationship: While AP1 can compensate for loss of CAL activity, the reverse is not true. Thus, in contrast with induced ap1 mutations, natural loss-of-function alleles of CAL have no overt phenotype on their own and are only noticed if AP1 is inactive as well (Bowman et al., 1993; Kempin et al., 1995).
Arabidopsis was used early on to identify genes that control seed dormancy (van Der Schaar et al., 1997). For ease of cultivation, common laboratory accessions had been selected to be early flowering (more below) and to have little dormancy, meaning that seeds would germinate relatively quickly after harvest. The DELAY OF GERMINATION1 (DOG1) locus, the first dormancy QTL cloned, encodes the prototype of a small gene family of unknown molecular function. There is extensive variation in DOG1 expression levels between accessions, suggesting the presence of many functionally distinct alleles of DOG1 (Bentsink et al., 2006). Arabidopsis accessions also remain an important resource for functional and evolutionary analyses of large-effect resistance genes (Staskawicz et al., 1995). This is a large area for which there are several recent in-depth reviews (Nishimura and Dangl, 2010).
Below, I will discuss three naturally variable traits in some more detail: trichome density, which provides a paradigm for how information from multiple genome sequences can be used to pinpoint causal polymorphisms; glucosinolate content, which has an underlying biochemical pathway with variation at almost every step; and the onset of flowering, a developmental trait with a well-understood molecular basis.
Trichome Density
Early studies by Rodney Mauricio and Mark Rausher came to the conclusion that both physical defenses in the form of leaf hairs (trichomes) and chemicals (glucosinolates) reduce herbivore damage to Arabidopsis in the field but that these are not without costs (Mauricio and Rausher, 1997; Mauricio, 1998). Several genes have been identified as affecting trichome density of natural Arabidopsis accessions. The most dramatic effects are seen in accessions that are glabrous, that is, lack trichomes completely, and at least two different loss-of-function mutations at GLABRA1 (GL1) have been found. Whether a fitness trade-off, as suggested for other defense traits, underpins the GL1 polymorphisms is unknown. Balancing selection, however, which is often taken as a sign of trade-offs, does not appear to be responsible for maintaining different GL1 alleles (Hauser et al., 2001). Glabrousness caused by inactivating mutations in GL1 also segregates in A. lyrata and Arabidopsis halleri populations (Hauser et al., 2001; Kärkkäinen and Ågren, 2002; Kivimäki et al., 2007; Kawagoe et al., 2011).
A less extreme phenotype of reduced trichome density is caused in some Arabidopsis accessions by a nonsynonymous substitution in MYC1 (Symonds et al., 2011). As another warning to population geneticists, one of the exons was found to exhibit a strong signal of divergent selection, with many amino acid substitutions. However, this signal was not correlated with trichome density.
Other accessions have increased trichome number relative to the Col-0 reference accession, and ENHANCER OF TRY AND CPC2 (ETC2) has been identified as the causal gene (Hilscher et al., 2009). ETC2, MYC1, and GL1 all encode transcription factors, with GL1 promoting and ETC2 repressing trichome formation by competing for interaction with common partners, a group of basic helix-loop-helix proteins that includes GL3 and MYC1 (Ishida et al., 2008). In contrast with MYC1, the high- and low-activity variants of ETC2 segregate at intermediate frequencies, indicating that ETC2 is a major determinant of natural variation in trichome number. ETC2 very likely corresponds to one of the first QTL that was mapped in Arabidopsis, REDUCED TRICHOME NUMBER (Larkin et al., 1996), and consistent with alleles of different activity being common, ETC2 can also be detected by GWAS (Atwell et al., 2010). Notably, it had initially been suggested that ETC2 has only a minor role in trichome formation, a conclusion that came from studies done with common accessions that have an ETC2 allele without obvious disruptions but with nevertheless low activity.
The work on ETC2 is noteworthy because of how the causal polymorphism was first pinpointed using a strategy that should be broadly applicable. To triangulate the causal region in the final mapping interval, accessions with either very high or very low trichome densities were selected, and the extent of haplotype sharing in each group was compared, which identified a small region with only two candidate polymorphisms (Hilscher et al., 2009). Transformation with chimeric transgenes provided conclusive support that one of the variants, a nonsynonymous mutation, was reducing the activity of ETC2. With the resources of the 1001 Genomes Project, these types of local association studies should become a common strategy for the endgame in identifying QTL after conventional mapping in F2 or similar populations.
Glucosinolate Content
In addition to the gene-for-gene resistance loci that are effective against individual pathogen strains (for review, see Nishimura and Dangl, 2010), Arabidopsis accessions also show quantitative variation in resistance, in particular against herbivorous insects. As with trichomes, chemical defenses in the form of a Brassicaceae-specific class of secondary metabolites, the glucosinolates, can reduce herbivore damage (Blau et al., 1978). There are considerable inter- and intraspecific differences in the repertoire of glucosinolates, which are hydrolyzed by the enzyme myrosinase into the active defense compounds (Kliebenstein et al., 2005). In Arabidopsis, METHYLTHIOALKYLMALATE SYNTHASE (MAM) and AOP are the two major loci responsible for variation in glucosinolate biosynthesis, with additional contributions from the GSL-OH locus (Kliebenstein et al., 2001; Kroymann et al., 2001, 2003). Hydrolysis of the glucosinolates is further affected by the polymorphic EPITHIOSPECIFIER PROTEIN and EPITHIOSPECIFIER MODIFIER1 loci (Lambrix et al., 2001; Zhang et al., 2006). In other Brassicaceae, several of the same genes are responsible for intraspecific variation in glucosinolate content, including A. lyrata (Li and Quiros, 2003; Heidel et al., 2006).
Notably, both the MAM and AOP loci are complex, with several tandem arrayed genes that vary in presence, enzyme activity, or expression level between accessions, giving rise to more than two alternative allelic states, processes that are apparently driven by positive selection (Kliebenstein et al., 2001; Kroymann et al., 2001, 2003). At least MAM shows a similar pattern of diversity created by gene duplication and neofunctionalization between other members of the Arabidopsis genus as well as closely related genera (Benderoth et al., 2006).
The detailed understanding of the control of glucosinolate accumulation in turn supports research into broader questions of genetic variation, such as the importance of stochastic variation, which was found to be genetically encoded as well (Jimenez-Gomez et al., 2011).
Flowering Time
Seed production is one of the most important components of fitness, and to optimize seed set, plants need to flower at the right time of year. In agreement with Arabidopsis is found in places with very different growing seasons, natural accessions differ greatly in their flowering behavior. Beginning with Laibach (1943, 1951), several investigators reported flowering variation not only in inbred accessions, but also in individuals collected from the wild (Napp-Zinn, 1957; Cetl et al., 1968; Jones, 1971; Westerman, 1971). That this trait is under selection has also been inferred from population genomics analyses (Flowers et al., 2009) and from the finding of latitudinal and altitudinal clines, likely due to covariation of flowering time with climatic factors (Caicedo et al., 2004; Stinchcombe et al., 2004; Lempe et al., 2005).
The first natural allele to be mapped with molecular markers in Arabidopsis was at the FRI locus, which segregates in a Mendelian manner in crosses between late- and early-flowering accessions (Lee et al., 1993; Clarke and Dean, 1994). The first QTL mapped in Arabidopsis were also ones controlling flowering (Kowalski et al., 1994; Clarke et al., 1995), followed by many additional QTL studies (for review, see Koornneef et al., 2004; Shindo et al., 2007). Mapping in crosses and GWAS have shown that flowering time variation can be explained by relatively few large-effect QTL (Atwell et al., 2010; Brachi et al., 2010; Li et al., 2010; Salomé et al., 2011b; Strange et al., 2011), which is very different from maize (Buckler et al., 2009).
FRI and the epistatically acting FLC gene are responsible for a large fraction of flowering time variation in Arabidopsis accessions when these are not exposed to a winter-like vernalization treatment. FRI promotes expression of the FLC transcription factor, which directly represses genes with positive roles in flowering (Li et al., 2008; Deng et al., 2011). Allelic variation at FLC likely accounts for flowering time differences in other Brassicaceae as well, including Capsella bursa-pastoris and some, but not all, Brassica species (Long et al., 2007; Razi et al., 2008; Slotte et al., 2009; Zhao et al., 2010). A role for FRI in flowering time variation in A. lyrata and Brassica napus has been inferred from association studies (Kuittinen et al., 2008; Wang et al., 2011).
Strikingly, there are many alleles at both FRI and FLC (Michaels and Amasino, 1999; Johanson et al., 2000; Le Corre et al., 2002; Gazzani et al., 2003; Lempe et al., 2005; Shindo et al., 2005; Méndez-Vigo et al., 2011). Because of the convenience of early flowering, commonly used laboratory accessions have a loss-of-function allele at one or both loci. However, while low-activity FRI alleles typically have disrupted open reading frames, FLC alleles are predominantly characterized by noncoding structural variation. During vernalization, FLC becomes epigenetically silenced, and natural alleles differ in the duration of vernalization needed for stably switching off FLC expression (Shindo et al., 2006). In addition to its repressive effects on flowering, high-activity alleles of FLC promote germination in the cold, which in turn allows plants to experience the longer cold period required for flowering when FLC is active (Chiang et al., 2009). The FRI homologs FRL1 and FRL2 along with the FLC homologs FLM/MAF1 and MAF2 provide additional routes to flowering time variation (Werner et al., 2005; Schläppi, 2006; Caicedo et al., 2009; Rosloski et al., 2010).
Flowering time control is one of the most intensively investigated developmental processes in Arabidopsis, and well over 100 genes are known to affect flowering, with many having substantial pleiotropic effects on plant growth (Srikanth and Schmid, 2011). Remarkably, only one gene with very few nonflowering phenotypes, the central flowering activator FT, has been shown to contribute extensively to flowering time variation between Arabidopsis accessions (Schwartz et al., 2009; Li et al., 2010; Huang et al., 2011; Salomé et al., 2011b; Strange et al., 2011). QTL studies have implicated FT as being the cause of flowering time differences also in B. napus (Long et al., 2007).
Several other genes responsible for flowering time variation in Arabidopsis have multiple functions during plant development, including the photoreceptor encoding genes CRYPTOCHROME2, PHYC, and PHYD (Aukerman et al., 1997; El-Din El-Assal et al., 2001; Balasubramanian et al., 2006; Méndez-Vigo et al., 2011). In addition, there is functional allelic variation at PHYA and PHYB. Both regulate flowering (Srikanth and Schmid, 2011), although the effects of the natural alleles on flowering have not been studied (Maloof et al., 2001; Filiault et al., 2008). Two other pleiotropically acting, naturally variable flowering regulators are FY (Adams et al., 2009) and HUA2. In addition to affecting flowering time, a natural HUA2 change-of-function allele has a dramatic effect on plant architecture that had not been anticipated from mutant studies (Alonso-Blanco et al., 1998a; Wang et al., 2007; Huang et al., 2011; Strange et al., 2011). Finally, additional loci responsible for flowering time regulation have been identified by growing plants under variable conditions (Weinig et al., 2002; Brachi et al., 2010; Li et al., 2010).
TOWARD AN UNDERSTANDING OF THE FORCES SHAPING GENETIC VARIATION
Apart from extending our knowledge of biological mechanisms and pathways in Arabidopsis, a major motivation for studying genetic variation is to understand how a species adapts to different local environments, which traces adaptation leaves in the genome, and how this leads to the formation of new species. In this section, I describe how genome analyses have provided insights into the history of the species, what is being learned about epistatic interactions between alleles from different genomes, and how evidence for local adaptation is emerging.
Geographic Distribution of Population Diversity
Until a decade ago, the vast majority of the few hundred Arabidopsis accessions available from the stock centers came from western Europe. In the past years, collections have been substantially expanded, with more than 2,000 genotypically distinct accessions having been described (Schmuths et al., 2006; Beck et al., 2008; Picó et al., 2008; Montesinos et al., 2009; Bomblies et al., 2010; Lewandowska-Sabat et al., 2010; Platt et al., 2010; Cao et al., 2011; Méndez-Vigo et al., 2011). With whole-genome data, the pattern of isolation-by-distance that had been deduced from more sparse data before came into even sharper focus. In addition, it was found that geographic regions differ greatly both with respect to the total number of polymorphisms distinguishing accessions within a region from each other and from other regions and the relative frequency of variants that are shared with other regions.
There is an overall gradient from west to east: The greatest diversity is found at the western end of the native range, in the Iberian Peninsula, including North Africa, while the most uniform regions are in Central Asia. This is consistent with the view that Arabidopsis populations in the west are the oldest, with later expansion into the eastern end of its native distribution, along with recently colonized regions, such as the Alps, in the center of the range (Sharbel et al., 2000; Nordborg et al., 2005; Schmid et al., 2005; Ostrowski et al., 2006; Beck et al., 2008; Picó et al., 2008; Platt et al., 2010; Cao et al., 2011). In addition, there is also altitudinal stratification within regions, with populations from high altitude being overall less diverse than those from lower altitude (Montesinos et al., 2009; Lewandowska-Sabat et al., 2010; Gomaa et al., 2011). It has also been suggested that there is evidence for migration from east to west, accompanying the spread of agriculture (François et al., 2008); however, knowing that the Iberian Peninsula is the most diverse region, it is unclear what to make from this. The regional differences have certainly important implications for the design of GWAS, since LD extends further in less diverse regions (Cao et al., 2011).
In continental Eurasia, identical multilocus genotypes are almost exclusively found only in the same local patches of Arabidopsis individuals (Picó et al., 2008; Bomblies et al., 2010; Lewandowska-Sabat et al., 2010; Platt et al., 2010). Exceptions are the British Isles and North America. In both regions, one specific genotype is found in many different places. For North America, recent and widespread, but uneven, introduction by European settlers has been suggested as the most likely cause; this scenario is compatible with the absence of genetic isolation by distance in North America (Platt et al., 2010).
Epistatic Interactions between Genomes
Despite its selfing nature, and contrary to what early analyses had suggested, stands of Arabidopsis plants can include several different multilocus genotypes. Moreover, outcrossing rates of Arabidopsis in nature can be several percent, and heterozygous individuals are thus not that rare (Stenøien et al., 2005; Bakker et al., 2006; Jorgensen and Emerson, 2008; Bomblies et al., 2010; Platt et al., 2010).
Superior performance in heterozygous F1 hybrids is known as heterosis or hybrid vigor. Heterosis in Arabidopsis is generally not as dramatic as in other species, but heterotic QTL for biomass and metabolites have been identified by backcrossing RILs derived from two inbred accessions to the founders (Syed and Chen, 2005; Kusterer et al., 2007; Lisec et al., 2009; Meyer et al., 2010). There is also extensive evidence for nonadditive, or epistatic, effects on gene expression in intra- and interspecific hybrids (Wang et al., 2006; Zhang and Borevitz, 2009; Zhang et al., 2011). In both stable allotetraploids and F1 hybrids of Arabidopsis × arenosa, circadian gene expression programs are altered, and a similar trend is apparent in F1 hybrids between two Arabidopsis accessions that exhibit hybrid vigor. The heterotic effects are mediated by central regulators of the circadian clock (Ni et al., 2009), although the proximate causes that alter activity of these regulators, and their relationship to the heterosis QTL identified in the same cross before, remain unknown.
Inferior performance of F1 hybrids is known as hybrid weakness or incompatibility, with extreme cases presenting as hybrid sterility or lethality. In addition, a decline in fitness of later generations is called hybrid breakdown or inbreeding depression (Hochholdinger and Hoecker, 2007; Charlesworth and Willis, 2009; Bomblies, 2010) A commonly observed incompatibility phenomenon is cytoplasmic male sterility (CMS), due to a mismatch between nuclear genes that encode proteins active in mitochondria and the mitochondrial genome (Fujii and Toriyama, 2008). Despite well over 1,000 different interaccession crosses having been examined (Bomblies et al., 2007), CMS has not yet been reported in Arabidopsis, even though weak CMS has been observed in A. lyrata (Leppälä and Savolainen, 2011). The most common obvious defect in F1 hybrids of Arabidopsis appears to be an autoimmune syndrome, hybrid necrosis, that is also known from many other plants.
Hybrid necrosis can often be explained by one or two epistatically interacting loci (Bomblies et al., 2007; Bomblies and Weigel, 2007). At least one of the genes causal for hybrid necrosis in Arabidopsis encodes an immune receptor of the NB-LRR class (Bomblies et al., 2007), consistent with the identification of immune genes underlying hybrid necrosis in other species (Krüger et al., 2002; Jeuken et al., 2009; Yamamoto et al., 2010). The NB-LRR family is the most variable gene family in plants, with genes often being found in clusters that have a complex history of gene duplication, deletion, and gene conversion. NB-LRR genes are engaged in recognition of diverse proteins (Nishimura and Dangl, 2010), providing an intuitive explanation for why hybrid necrosis is so common. In a broader context, hybrid necrosis is a manifestation of the costs of disease resistance (Tian et al., 2003).
In some instances, hybrid necrosis becomes only expressed in the F2 generation (Alcázar et al., 2009). In one such case, one of the causal genes encodes a receptor kinase homolog, with evidence of positive selection for disease resistance having increased the frequency of this allele in Central Asia (Alcázar et al., 2010). A receptor-kinase-like gene of a different class is responsible for an incompatibility that primarily causes growth defects. This specific case involves an interaction between alleles at a single locus with similar properties as many NB-LRR loci, namely being composed of a highly variable tandem array of genes (Smith et al., 2011). Notably, not every highly variable gene family appears to cause problems in hybrids. Cytochrome P450s, which are important for plant insect defense and are produced by one of the most highly variable gene families (Clark et al., 2007; Cao et al., 2011), have so far not been tied to hybrid weakness, perhaps because they are not designed to interact with a diverse set of other proteins.
Most F2 incompatibilities were not discovered because of overt phenotypic effects but were deduced from segregation distortion, that is, the absence of certain genotypic combinations, in F2 or RIL populations (Lister and Dean, 1993; Mitchell-Olds, 1995; Alonso-Blanco et al., 1998b; Loudet et al., 2002; Werner et al., 2005; Törjék et al., 2006; Simon et al., 2008; Balasubramanian et al., 2009; Salomé et al., 2011a). For RILs, this can be due to inadvertent selection, e.g. because late-germinating lines are eliminated, but several cases are associated with lethality of specific segregants. One example involves a pair of paralogs that arose from a very recent ectopic duplication event and that independently sustained inactivating mutations in different lineages (Bikard et al., 2009). About three-quarters of accessions carry inactive copies of one or the other paralog, suggesting that increased dosage is disfavored. A similar situation of reciprocally mutated paralogs explains an epistatic interaction affecting shoot growth (Vlad et al., 2010). Both cases differ from other examples of complex duplication and mutation events, where the paralogs have become neofunctionalized and have now distinct activities (Kliebenstein et al., 2001; Kroymann et al., 2003; Huang et al., 2010a).
Experimental Ecology and Ecological Genomics
The worldwide distribution of Arabidopsis can be well described by climatic range boundaries; these indicate that laboratory conditions commonly used for growth of Arabidopsis are at the extreme end of its normal habitats, which are normally much cooler and drier (Hoffmann, 2002). This has important implications for interpreting phenotypic differences observed in the greenhouse. For example, strains with differential activity of the key flowering regulators FRI and FLC, known to vary in many accessions, only differ strongly in their flowering behavior outdoors when germinated at specific times of the year, with a critical period in early fall having a disproportionately large effect on flowering time, namely, whether plants overwinter (Wilczek et al., 2009). Such knowledge is essential if one wants to predict responses to a changing climate (Wilczek et al., 2010). Furthermore, by culturing plants in seminatural settings, in which either variable light and temperature conditions are reproduced in climate chambers or plants are germinated in the greenhouse, then transplanted outdoors, one can detect QTL that are not found when plants are grown in a uniform environment. Whether either type of QTL is more relevant is unclear and can only be addressed by phenotyping truly naturally growing individuals. Nevertheless, analysis in seminatural conditions provides insights into the genetic basis of traits considered to be indicative of fitness, such as germination, survival, fruit and seed number, or competitiveness (Weinig et al., 2002, 2003a, 2003b; Stinchcombe et al., 2004; Donohue et al., 2005; Li et al., 2006; Brachi et al., 2010; Huang et al., 2010b; Li et al., 2010; Fournier-Level et al., 2011).
Different experimental approaches are beginning to reveal local adaptation in Arabidopsis. When 74 accessions were monitored in the greenhouse under different temperatures, it was found that accessions from cold regions respond in their growth more strongly to elevated temperatures than accessions from warm regions, which are only moderately inhibited by colder temperatures (Hoffmann et al., 2005). Systematic correlation of phenotypes with environmental gradients can indicate adaptation (Endler, 1977), and there are also latitudinal clines in light sensitivity and altitudinal clines in flowering-related traits (Maloof et al., 2001; Méndez-Vigo et al., 2011). It has been similarly proposed that populations of Arabidopsis near oceans or saline soils are more likely to carry an allele at the HKT1 locus that increases sodium accumulation in leaves (Baxter et al., 2010). However, the accessions investigated were unevenly sampled, information about soil salinity at the places of origin was not available, and the relationship between compromised activity of HKT1 and salt tolerance is complex (Mäser et al., 2002; Berthomieu et al., 2003). Thus, the conclusions about adaptation to salinity should be taken with the proverbial grain of salt.
Reciprocal transplantation experiments have produced evidence for local adaptation in A. lyrata (Leinonen et al., 2009, 2011). Somewhat surprisingly, this approach, a gold standard in ecology (Turesson, 1922a), has so far only been sparingly applied in Arabidopsis. This has recently been remedied, with an impressive study in which hundreds of accessions were grown at several different places in the native range of the species (Fournier-Level et al., 2011). Alleles associated with superior fitness at each site were most likely to be found in accessions originating near that site. GWAS identified several candidates for survival and fruit number, although only one, the photoreceptor gene PHYB, which affects light response, can be easily connected to local adaptation based on prior knowledge. Additional evidence for local adaptation comes from GWAS for climate variables at the place of origin combined with fitness tests at a single site (Hancock et al., 2011). Both of these studies were carried out predominantly with accessions from the western European and Scandinavian part of the native range, and it will be interesting to repeat these experiments with a broader spectrum of accessions and test locales.
OUTLOOK
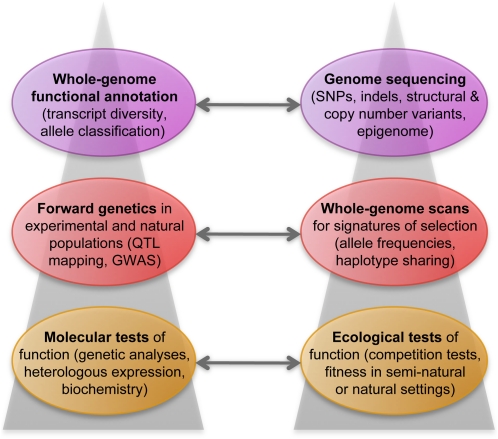
Our knowledge of natural variation in Arabidopsis has advanced tremendously in the past decade, with an impressive set of genetic and genomic approaches and resources that are now available (Fig. 6). In the near future, the simultaneous application of different strategies will lead to genetic variation increasingly informing basic plant biology. Combined analyses of global transcript and metabolite levels and biomass across accessions and RIL populations is supporting the reconstruction of functional networks (Wentzell et al., 2007; Lisec et al., 2008; Rowe et al., 2008; Sulpice et al., 2009, 2010). Integration of QTL data with such information has shown that in addition to biosynthetic and metabolic enzymes, upstream transcription factors of the MYB class contribute to diversity in glucosinolate content (Sønderby et al., 2007) and that the clock gene ELF3 has a role in shade avoidance (Jiménez-Gómez et al., 2010). Another instructive example of how natural variation can help to discover a new regulatory pathway comes from the study of xylem expansion (Sibout et al., 2008). The authors noted that the xylem expansion loci colocalized with flowering time QTL, which led them to hypothesize that the onset of flowering causes xylem expansion in both the shoot and the root. They subsequently confirmed such a model by transiently inducing the activity of a central floral regulator. There is similarly great promise in GWAS with the same material to identify cases of pleiotropic action of natural sequence variants.
Figure 6.
Relationship between approaches to the study of genetic variation.
I have also highlighted the many opportunities Arabidopsis offers for the study of interactions between divergent genomes, which may both promote or reduce outcrossing, and thereby affect the partitioning of genetic diversity into different lineages (and ultimately into different species). So far, the parents for the investigated crosses have largely been chosen randomly. With increasing information about the genome-wide and population-specific distribution of sequence polymorphisms, more judicious and systematic choices of genotype combinations should accelerate the pace with which we can obtain insights into the fascinating questions of hybrid performance.
Another important direction will be to phenotype naturally growing plants in situ over several years (Montesinos et al., 2009). Genotyping of very large numbers of wild plants has become very affordable with next-generation sequencing methods, which will facilitate linking genotype and phenotype even on an individual basis (Baird et al., 2008; Elshire et al., 2011). An example for such strategies is a study that monitored over 4 years the load of five different viruses that had been known before to infect wild Brassicaceae (Pagán et al., 2010). Such experiments are required to test claims about fitness trade-offs between disease resistance and growth (Tian et al., 2003; Todesco et al., 2010). Finally, selection experiments are a tool that should not be underestimated for their potential to provide insights into favorable allele combinations (Ungerer et al., 2003; Ungerer and Rieseberg, 2003; Scarcelli and Kover, 2009; Fakheran et al., 2010).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Full references for Table 1.
Acknowledgments
I thank Eunyoung Chae, Sang-Tae Kim, and George Wang for plant images; Joy Bergelson, Carlos Alonso-Blanco, Jun Cao, Karl Schmid, and George Wang for help in producing the map of Arabidopsis accessions; and Annie Schmitt and Joy Bergelson for preprints. I am especially grateful to three anonymous reviewers, who provided insightful comments and helped to correct several oversights in the original manuscript.
References
- Adams S, Allen T, Whitelam GC. (2009) Interaction between the light quality and flowering time pathways in Arabidopsis. Plant J 60: 257–267 [DOI] [PubMed] [Google Scholar]
- Alcázar R, García AV, Kronholm I, de Meaux J, Koornneef M, Parker JE, Reymond M. (2010) Natural variation at Strubbelig Receptor Kinase 3 drives immune-triggered incompatibilities between Arabidopsis thaliana accessions. Nat Genet 42: 1135–1139 [DOI] [PubMed] [Google Scholar]
- Alcázar R, García AV, Parker JE, Reymond M. (2009) Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc Natl Acad Sci USA 106: 334–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Ecker JR. (2006) Moving forward in reverse: genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat Rev Genet 7: 524–536 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, El-Assal SE, Coupland G, Koornneef M. (1998a) Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics 149: 749–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Koornneef M. (2000) Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci 5: 22–29 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Peeters AJ, Koornneef M, Lister C, Dean C, van den Bosch N, Pot J, Kuiper MT. (1998. b) Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J 14: 259–271 [DOI] [PubMed] [Google Scholar]
- Al-Shehbaz I, O'Kane S., Jr (2002) Taxonomy and phylogeny of Arabidopsis (Brassicaceae). The Arabidopsis Book 1: 1–22, doi/10.1199/tab.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasio AE, Platt A, Horton M, Grotewold E, Scholl R, Borevitz JO, Nordborg M, Bergelson J. (2011) Source verification of mis-identified Arabidopsis thaliana accessions. Plant J 67: 554–566 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Aranzana MJ, Kim S, Zhao K, Bakker E, Horton M, Jakob K, Lister C, Molitor J, Shindo C, Tang C, et al. (2005) Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet 1: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta-Montiel MP, Shedge V, Davila J, Christensen AC, Mackenzie SA. (2009) Diversity of the Arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics 183: 1261–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asimit J, Zeggini E. (2010) Rare variant association analysis methods for complex traits. Annu Rev Genet 44: 293–308 [DOI] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA. (1997) A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell 9: 1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey DW. (1971) Recombinant-inbred strains. An aid to finding identity, linkage, and function of histocompatibility and other genes. Transplantation 11: 325–327 [DOI] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3: e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker EG, Stahl EA, Toomajian C, Nordborg M, Kreitman M, Bergelson J. (2006) Distribution of genetic variation within and among local populations of Arabidopsis thaliana over its species range. Mol Ecol 15: 1405–1418 [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Schwartz C, Singh A, Warthmann N, Kim MC, Maloof JN, Loudet O, Trainer GT, Dabi T, Borevitz JO, et al. (2009) QTL mapping in new Arabidopsis thaliana advanced intercross-recombinant inbred lines. PLoS ONE 4: e4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Agrawal M, Michael TP, Wessinger C, Maloof JN, Clark R, Warthmann N, Chory J, Weigel D. (2006) The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat Genet 38: 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Brazelton JN, Yu D, Huang YS, Lahner B, Yakubova E, Li Y, Bergelson J, Borevitz JO, Nordborg M, et al. (2010) A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet 6: e1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JB, Schmuths H, Schaal BA. (2008) Native range genetic variation in Arabidopsis thaliana is strongly geographically structured and reflects Pleistocene glacial dynamics. Mol Ecol 17: 902–915 [DOI] [PubMed] [Google Scholar]
- Becker C, Hagmann J, Müller J, Koenig D, Stegle O, Borgwardt K, Weigel D. (September 20, 2011) Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480: 245–249 [DOI] [PubMed] [Google Scholar]
- Benderoth M, Textor S, Windsor AJ, Mitchell-Olds T, Gershenzon J, Kroymann J. (2006) Positive selection driving diversification in plant secondary metabolism. Proc Natl Acad Sci USA 103: 9118–9123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Hanson J, Hanhart CJ, Blankestijn-de Vries H, Coltrane C, Keizer P, El-Lithy M, Alonso-Blanco C, de Andrés MT, Reymond M, et al. (2010) Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proc Natl Acad Sci USA 107: 4264–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M. (2006) Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA 103: 17042–17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J, Roux F. (2010) Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat Rev Genet 11: 867–879 [DOI] [PubMed] [Google Scholar]
- Berthomieu P, Conéjéro G, Nublat A, Brackenbury WJ, Lambert C, Savio C, Uozumi N, Oiki S, Yamada K, Cellier F, et al. (2003) Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J 22: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, Patel D, Le Metté C, Giorgi V, Camilleri C, Bennett MJ, Loudet O. (2009) Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science 323: 623–626 [DOI] [PubMed] [Google Scholar]
- Blau PA, Feeny P, Contardo L, Robson DS. (1978) Allylglucosinolate and herbivorous caterpillars: a contrast in toxicity and tolerance. Science 200: 1296–1298 [DOI] [PubMed] [Google Scholar]
- Bomblies K. (2010) Doomed lovers: mechanisms of isolation and incompatibility in plants. Annu Rev Plant Biol 61: 109–124 [DOI] [PubMed] [Google Scholar]
- Bomblies K, Lempe J, Epple P, Warthmann N, Lanz C, Dangl JL, Weigel D. (2007) Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol 5: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Weigel D. (2007) Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat Rev Genet 8: 382–393 [DOI] [PubMed] [Google Scholar]
- Bomblies K, Yant L, Laitinen RA, Kim ST, Hollister JD, Warthmann N, Fitz J, Weigel D. (2010) Local-scale patterns of genetic variability, outcrossing, and spatial structure in natural stands of Arabidopsis thaliana. PLoS Genet 6: e1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Hazen SP, Michael TP, Morris GP, Baxter IR, Hu TT, Chen H, Werner JD, Nordborg M, Salt DE, et al. (2007) Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 12057–12062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Risch N. (2003) Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet (Suppl) 33: 228–237 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR. (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743 [Google Scholar]
- Brachi B, Faure N, Horton M, Flahauw E, Vazquez A, Nordborg M, Bergelson J, Cuguen J, Roux F. (2010) Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet 6: e1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branca A, Paape TD, Zhou P, Briskine R, Farmer AD, Mudge J, Bharti AK, Woodward JE, May GD, Gentzbittel L, et al. (2011) Whole-genome nucleotide diversity, recombination, and linkage disequilibrium in the model legume Medicago truncatula. Proc Natl Acad Sci USA 108: E864–E870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, et al. (2009) The genetic architecture of maize flowering time. Science 325: 714–718 [DOI] [PubMed] [Google Scholar]
- Caicedo AL, Richards C, Ehrenreich IM, Purugganan MD. (2009) Complex rearrangements lead to novel chimeric gene fusion polymorphisms at the Arabidopsis thaliana MAF2-5 flowering time gene cluster. Mol Biol Evol 26: 699–711 [DOI] [PubMed] [Google Scholar]
- Caicedo AL, Schaal BA, Kunkel BN. (1999) Diversity and molecular evolution of the RPS2 resistance gene in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 302–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo AL, Stinchcombe JR, Olsen KM, Schmitt J, Purugganan MD. (2004) Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc Natl Acad Sci USA 101: 15670–15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Cetl I, Dobrovolná J, Effmertova E. (1968) The developmental character of natural populations of Arabidopsis thaliana (L.) Heynh in relation to the geographical-climatic conditions of localities. Folia Fac Sci Nat Univ Purk Brun (Biol.) 18: 37–49 [Google Scholar]
- Chan EK, Rowe HC, Corwin JA, Joseph B, Kliebenstein DJ. (2011) Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol 9: e1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Bowman JL, DeJohn AW, Lander ES, Meyerowitz EM. (1988) Restriction fragment length polymorphism linkage map for Arabidopsis thaliana. Proc Natl Acad Sci USA 85: 6856–6860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Willis JH. (2009) The genetics of inbreeding depression. Nat Rev Genet 10: 783–796 [DOI] [PubMed] [Google Scholar]
- Chiang GC, Barua D, Kramer EM, Amasino RM, Donohue K. (2009) Major flowering time gene, flowering locus C, regulates seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA 106: 11661–11666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al. (2007) Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317: 338–342 [DOI] [PubMed] [Google Scholar]
- Clark SS, Crist WM, Witte ON. (1989) Molecular pathogenesis of Ph-positive leukemias. Annu Rev Med 40: 113–122 [DOI] [PubMed] [Google Scholar]
- Clarke JH, Dean C. (1994) Mapping FRI, a locus controlling flowering time and vernalization response in Arabidopsis thaliana. Mol Gen Genet 242: 81–89 [DOI] [PubMed] [Google Scholar]
- Clarke JH, Mithen R, Brown JKM, Dean C. (1995) QTL analysis of flowering time in Arabidopsis thaliana. Mol Gen Genet 248: 278–286 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cordell HJ. (2009) Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet 10: 392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, Vincent C, Coen E. (1999) An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401: 157–161 [DOI] [PubMed] [Google Scholar]
- Darvasi A, Soller M. (1995) Advanced intercross lines, an experimental population for fine genetic mapping. Genetics 141: 1199–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila JI, Arrieta-Montiel MP, Wamboldt Y, Cao J, Hagmann J, Shedge V, Xu YZ, Weigel D, Mackenzie SA. (2011) Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol 9: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison J, Tyagi A, Comai L. (2007) Large-scale polymorphism of heterochromatic repeats in the DNA of Arabidopsis thaliana. BMC Plant Biol 7: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Ying H, Helliwell CA, Taylor JM, Peacock WJ, Dennis ES. (2011) FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc Natl Acad Sci USA 108: 6680–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue K, Dorn L, Griffith C, Kim E, Aguilera A, Polisetty CR, Schmitt J. (2005) Environmental and genetic influences on the germination of Arabidopsis thaliana in the field. Evolution 59: 740–757 [PubMed] [Google Scholar]
- Doyle MR, Bizzell CM, Keller MR, Michaels SD, Song J, Noh YS, Amasino RM. (2005) HUA2 is required for the expression of floral repressors in Arabidopsis thaliana. Plant J 41: 376–385 [DOI] [PubMed] [Google Scholar]
- Ehrenreich IM, Hanzawa Y, Chou L, Roe JL, Kover PX, Purugganan MD. (2009) Candidate gene association mapping of Arabidopsis flowering time. Genetics 183: 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Din El-Assal S, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M. (2001) A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet 29: 435–440 [DOI] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE. (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6: e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler JA. (1977) Geographic Variation, Speciation, and the Clines. Princeton University Press, Princeton, NJ: [PubMed] [Google Scholar]
- Eshed Y, Zamir D. (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakheran S, Paul-Victor C, Heichinger C, Schmid B, Grossniklaus U, Turnbull LA. (2010) Adaptation and extinction in experimentally fragmented landscapes. Proc Natl Acad Sci USA 107: 19120–19125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. (1996) Introduction to Quantitative Genetics, Ed 4. Addison Wesley Longman, Harlow, Essex, UK [Google Scholar]
- Filiault DL, Wessinger CA, Dinneny JR, Lutes J, Borevitz JO, Weigel D, Chory J, Maloof JN. (2008) Amino acid polymorphisms in Arabidopsis phytochrome B cause differential responses to light. Proc Natl Acad Sci USA 105: 3157–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers JM, Hanzawa Y, Hall MC, Moore RC, Purugganan MD. (2009) Population genomics of the Arabidopsis thaliana flowering time gene network. Mol Biol Evol 26: 2475–2486 [DOI] [PubMed] [Google Scholar]
- Forner J, Weber B, Wiethölter C, Meyer RC, Binder S. (2005) Distant sequences determine 5′ end formation of cox3 transcripts in Arabidopsis thaliana ecotype C24. Nucleic Acids Res 33: 4673–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier-Level A, Korte A, Cooper MD, Nordborg M, Schmitt J, Wilczek AM. (2011) A map of local adaptation in Arabidopsis thaliana. Science 334: 86–89 [DOI] [PubMed] [Google Scholar]
- François O, Blum MG, Jakobsson M, Rosenberg NA. (2008) Demographic history of european populations of Arabidopsis thaliana. PLoS Genet 4: e1000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz PF, Armstrong S, de Jong JH, Parnell LD, van Drunen C, Dean C, Zabel P, Bisseling T, Jones GH. (2000) Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere region. Cell 100: 367–376 [DOI] [PubMed] [Google Scholar]
- Fujii S, Toriyama K. (2008) Genome barriers between nuclei and mitochondria exemplified by cytoplasmic male sterility. Plant Cell Physiol 49: 1484–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. (2011) Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzani S, Gendall AR, Lister C, Dean C. (2003) Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol 132: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomaa NH, Montesinos-Navarro A, Alonso-Blanco C, Picó FX. (2011) Temporal variation in genetic diversity and effective population size of Mediterranean and subalpine Arabidopsis thaliana populations. Mol Ecol 20: 3540–3554 [DOI] [PubMed] [Google Scholar]
- Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J, et al. (2009) A first-generation haplotype map of maize. Science 326: 1115–1117 [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846 [DOI] [PubMed] [Google Scholar]
- Groszmann M, Greaves IK, Albertyn ZI, Scofield GN, Peacock WJ, Dennis ES. (2011) Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc Natl Acad Sci USA 108: 2617–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock AM, Brachi B, Faure N, Horton MW, Jarymowycz LB, Sperone FG, Toomajian C, Roux F, Bergelson J. (2011) Adaptation to climate across the Arabidopsis thaliana genome. Science 334: 83–86 [DOI] [PubMed] [Google Scholar]
- Hauser MT, Harr B, Schlötterer C. (2001) Trichome distribution in Arabidopsis thaliana and its close relative Arabidopsis lyrata: molecular analysis of the candidate gene GLABROUS1. Mol Biol Evol 18: 1754–1763 [DOI] [PubMed] [Google Scholar]
- Heidel AJ, Clauss MJ, Kroymann J, Savolainen O, Mitchell-Olds T. (2006) Natural variation in MAM within and between populations of Arabidopsis lyrata determines glucosinolate phenotype. Genetics 173: 1629–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilscher J, Schlötterer C, Hauser MT. (2009) A single amino acid replacement in ETC2 shapes trichome patterning in natural Arabidopsis populations. Curr Biol 19: 1747–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F, Hoecker N. (2007) Towards the molecular basis of heterosis. Trends Plant Sci 12: 427–432 [DOI] [PubMed] [Google Scholar]
- Hoffmann MH. (2002) Biogeography of Arabidopsis thaliana (L.) Heynh. (Brassicaceae). J Biogeogr 29: 125–134 [Google Scholar]
- Hoffmann MH, Tomiuk J, Schmuths H, Koch C, Bachmann K. (2005) Phenological and morphological responses to different temperature treatments differ among a world-wide sample of accessions of Arabidopsis thaliana. Acta Oecol 28: 181–187 [Google Scholar]
- Hollick JB, Patterson GI, Asmundsson IM, Chandler VL. (2000) Paramutation alters regulatory control of the maize pl locus. Genetics 154: 1827–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton M, Hancock AM, Huang YS, Toomajian C, Atwell S, Auton A, Muliyati W, Platt A, Sperone FG, Vilhkálmsson BJ, et al. (2012) Genome-wide pattern of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Abel C, Sohrabi R, Petri J, Haupt I, Cosimano J, Gershenzon J, Tholl D. (2010a) Variation of herbivore-induced volatile terpenes among Arabidopsis ecotypes depends on allelic differences and subcellular targeting of two terpene synthases, TPS02 and TPS03. Plant Physiol 153: 1293–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Paulo MJ, Boer M, Effgen S, Keizer P, Koornneef M, van Eeuwijk FA. (2011) Analysis of natural allelic variation in Arabidopsis using a multiparent recombinant inbred line population. Proc Natl Acad Sci USA 108: 4488–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Schmitt J, Dorn L, Griffith C, Effgen S, Takao S, Koornneef M, Donohue K. (2010b) The earliest stages of adaptation in an experimental plant population: strong selection on QTLs for seed dormancy. Mol Ecol 19: 1335–1351 [DOI] [PubMed] [Google Scholar]
- Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, et al. (2010c) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42: 961–967 [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T. (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Ito H, Miura A, Takashima K, Kakutani T. (2007) Ecotype-specific and chromosome-specific expansion of variant centromeric satellites in Arabidopsis thaliana. Mol Genet Genomics 277: 23–30 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Meyerowitz EM. (1997) Hypermethylated SUPERMAN epigenetic alleles in Arabidopsis. Science 277: 1100–1103 [DOI] [PubMed] [Google Scholar]
- James SA, O’Kelly MJ, Carter DM, Davey RP, van Oudenaarden A, Roberts IN. (2009) Repetitive sequence variation and dynamics in the ribosomal DNA array of Saccharomyces cerevisiae as revealed by whole-genome resequencing. Genome Res 19: 626–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeuken MJ, Zhang NW, McHale LK, Pelgrom K, den Boer E, Lindhout P, Michelmore RW, Visser RG, Niks RE. (2009) Rin4 causes hybrid necrosis and race-specific resistance in an interspecific lettuce hybrid. Plant Cell 21: 3368–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Gomez JM, Corwin JA, Joseph B, Maloof JN, Kliebenstein DJ. (2011) Genomic analysis of QTLs and genes altering natural variation in stochastic noise. PLoS Genet 7: e1002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Gómez JM, Wallace AD, Maloof JN. (2010) Network analysis identifies ELF3 as a QTL for the shade avoidance response in Arabidopsis. PLoS Genet 6: e1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes F, Colomé-Tatché M. (2011) Quantitative epigenetics through epigenomic perturbation of isogenic lines. Genetics 188: 215–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Jones ME. (1971) The population genetics of Arabidopsis thaliana. III. The effect of vernalisation. Heredity 27: 59–72 [Google Scholar]
- Jorde LB. (2000) Linkage disequilibrium and the search for complex disease genes. Genome Res 10: 1435–1444 [DOI] [PubMed] [Google Scholar]
- Jorgensen TH, Emerson BC. (2008) Functional variation in a disease resistance gene in populations of Arabidopsis thaliana. Mol Ecol 17: 4912–4923 [DOI] [PubMed] [Google Scholar]
- Kam-Thong T, Pütz B, Karbalai N, Müller-Myhsok B, Borgwardt K. (2011) Epistasis detection on quantitative phenotypes by exhaustive enumeration using GPUs. Bioinformatics 27: i214–i221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärkkäinen K, Ågren J. (2002) Genetic basis of trichome production in Arabidopsis lyrata. Hereditas 136: 219–226 [DOI] [PubMed] [Google Scholar]
- Kawagoe T, Shimizu KK, Kakutani T, Kudoh H. (2011) Coexistence of trichome variation in a natural plant population: a combined study using ecological and candidate gene approaches. PLoS ONE 6: e22184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempin SA, Savidge B, Yanofsky MF. (1995) Molecular basis of the cauliflower phenotype in Arabidopsis. Science 267: 522–525 [DOI] [PubMed] [Google Scholar]
- Kerwin RE, Jimenez-Gomez JM, Fulop D, Harmer SL, Maloof JN, Kliebenstein DJ. (2011) Network quantitative trait loci mapping of circadian clock outputs identifies metabolic pathway-to-clock linkages in Arabidopsis. Plant Cell 23: 471–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJ, Bentsink L, Alonso-Blanco C, Hanhart CJ, Blankestijn-De Vries H, Effgen S, Vreugdenhil D, Koornneef M. (2007) Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics 175: 891–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M. (2007) Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet 39: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Kivimäki M, Kärkkäinen K, Gaudeul M, Løe G, Ågren J. (2007) Gene, phenotype and function: GLABROUS1 and resistance to herbivory in natural populations of Arabidopsis lyrata. Mol Ecol 16: 453–462 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Mitchell-Olds T. (2005) The glucosinolate-myrosinase system in an ecological and evolutionary context. Curr Opin Plant Biol 8: 264–271 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T. (2001) Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D. (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55: 141–172 [DOI] [PubMed] [Google Scholar]
- Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD, Durrant C, Mott R. (2009) A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet 5: e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski SP, Lan TH, Feldmann KA, Paterson AH. (1994) QTL mapping of naturally-occurring variation in flowering time of Arabidopsis thaliana. Mol Gen Genet 245: 548–555 [DOI] [PubMed] [Google Scholar]
- Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T. (2003) Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc Natl Acad Sci USA (Suppl 2) 100: 14587–14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann J, Textor S, Tokuhisa JG, Falk KL, Bartram S, Gershenzon J, Mitchell-Olds T. (2001) A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol 127: 1077–1088 [PMC free article] [PubMed] [Google Scholar]
- Krüger J, Thomas CM, Golstein C, Dixon MS, Smoker M, Tang S, Mulder L, Jones JD. (2002) A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296: 744–747 [DOI] [PubMed] [Google Scholar]
- Kruglyak L. (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet 22: 139–144 [DOI] [PubMed] [Google Scholar]
- Kuittinen H, Niittyvuopio A, Rinne P, Savolainen O. (2008) Natural variation in Arabidopsis lyrata vernalization requirement conferred by a FRIGIDA indel polymorphism. Mol Biol Evol 25: 319–329 [DOI] [PubMed] [Google Scholar]
- Kuo HF, Olsen KM, Richards EJ. (2006) Natural variation in a subtelomeric region of Arabidopsis: implications for the genomic dynamics of a chromosome end. Genetics 173: 401–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusterer B, Piepho HP, Utz HF, Schön CC, Muminovic J, Meyer RC, Altmann T, Melchinger AE. (2007) Heterosis for biomass-related traits in Arabidopsis investigated by quantitative trait loci analysis of the triple testcross design with recombinant inbred lines. Genetics 177: 1839–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laibach F. (1943) Arabidopsis thaliana (L.)Heynh. als Objekt für genetische und entwicklungsphysiologische Untersuchungen. Bot Arch 4: 439–445 [Google Scholar]
- Laibach F. (1951) Über sommer- und winterannuelle Rassen von Arabidopsis thaliana (L.) Heynh. Ein Beitrag zur Ätiologie der Blütenbildung. Beitr Biol Pflanzen 28: 173–210 [Google Scholar]
- Laitinen RA, Schneeberger K, Jelly NS, Ossowski S, Weigel D. (2010) Identification of a spontaneous frame shift mutation in a nonreference Arabidopsis accession using whole genome sequencing. Plant Physiol 153: 652–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrix V, Reichelt M, Mitchell-Olds T, Kliebenstein DJ, Gershenzon J. (2001) The Arabidopsis epithiospecifier protein promotes the hydrolysis of glucosinolates to nitriles and influences Trichoplusia ni herbivory. Plant Cell 13: 2793–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES. (1996) The new genomics: global views of biology. Science 274: 536–539 [DOI] [PubMed] [Google Scholar]
- Larkin JC, Young N, Prigge M, Marks MD. (1996) The control of trichome spacing and number in Arabidopsis. Development 122: 997–1005 [DOI] [PubMed] [Google Scholar]
- Le Corre V, Roux F, Reboud X. (2002) DNA polymorphism at the FRIGIDA gene in Arabidopsis thaliana: Extensive nonsynonymous variation is consistent with local selection for flowering time. Mol Biol Evol 19: 1261–1271 [DOI] [PubMed] [Google Scholar]
- Lee I, Bleecker A, Amasino R. (1993) Analysis of naturally occurring late flowering in Arabidopsis thaliana. Mol Gen Genet 237: 171–176 [DOI] [PubMed] [Google Scholar]
- Leinonen PH, Remington DL, Savolainen O. (2011) Local adaptation, phenotypic differentiation, and hybrid fitness in diverged natural populations of Arabidopsis lyrata. Evolution 65: 90–107 [DOI] [PubMed] [Google Scholar]
- Leinonen PH, Sandring S, Quilot B, Clauss MJ, Mitchell-Olds T, Ågren J, Savolainen O. (2009) Local adaptation in European populations of Arabidopsis lyrata (Brassicaceae). Am J Bot 96: 1129–1137 [DOI] [PubMed] [Google Scholar]
- Lempe J, Balasubramanian S, Sureshkumar S, Singh A, Schmid M, Weigel D. (2005) Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet 1: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppälä J, Savolainen O. (2011) Nuclear-cytoplasmic interactions reduce male fertility in hybrids of Arabidopsis lyrata subspecies. Evolution 65: 2959–2972 [DOI] [PubMed] [Google Scholar]
- Lewandowska-Sabat AM, Fjellheim S, Rognli OA. (2010) Extremely low genetic variability and highly structured local populations of Arabidopsis thaliana at higher latitudes. Mol Ecol 19: 4753–4764 [DOI] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H. (2008) A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Li G, Quiros CF. (2003) In planta side-chain glucosinolate modification in Arabidopsis by introduction of dioxygenase Brassica homolog BoGSL-ALK. Theor Appl Genet 106: 1116–1121 [DOI] [PubMed] [Google Scholar]
- Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO. (2010) Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 21199–21204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Roycewicz P, Smith E, Borevitz JO. (2006) Genetics of local adaptation in the laboratory: flowering time quantitative trait loci under geographic and seasonal conditions in Arabidopsis. PLoS ONE 1: e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Meyer RC, Steinfath M, Redestig H, Becher M, Witucka-Wall H, Fiehn O, Törjék O, Selbig J, Altmann T, et al. (2008) Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J 53: 960–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Steinfath M, Meyer RC, Selbig J, Melchinger AE, Willmitzer L, Altmann T. (2009) Identification of heterotic metabolite QTL in Arabidopsis thaliana RIL and IL populations. Plant J 59: 777–788 [DOI] [PubMed] [Google Scholar]
- Lister C, Dean C. (1993) Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J 4: 745–750 [DOI] [PubMed] [Google Scholar]
- Long AD, Lyman RF, Langley CH, Mackay TF. (1998) Two sites in the Delta gene region contribute to naturally occurring variation in bristle number in Drosophila melanogaster. Genetics 149: 999–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, Shi J, Qiu D, Li R, Zhang C, Wang J, Hou J, Zhao J, Shi L, Park BS, et al. (2007) Flowering time quantitative trait loci analysis of oilseed brassica in multiple environments and genomewide alignment with Arabidopsis. Genetics 177: 2433–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. (2002) Bay-0 x Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet 104: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Loudet O, Michael TP, Burger BT, Le Metté C, Mockler TC, Weigel D, Chory J. (2008) A zinc knuckle protein that negatively controls morning-specific growth in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 17193–17198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF. (2001) The genetic architecture of quantitative traits. Annu Rev Genet 35: 303–339 [DOI] [PubMed] [Google Scholar]
- Maloof JN, Borevitz JO, Dabi T, Lutes J, Nehring RB, Redfern JL, Trainer GT, Wilson JM, Asami T, Berry CC, et al. (2001) Natural variation in light sensitivity of Arabidopsis. Nat Genet 29: 441–446 [DOI] [PubMed] [Google Scholar]
- Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. (2009) Finding the missing heritability of complex diseases. Nature 461: 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Donnelly P, Cardon LR. (2005) Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet 37: 413–417 [DOI] [PubMed] [Google Scholar]
- Martin A, Troadec C, Boualem A, Rajab M, Fernandez R, Morin H, Pitrat M, Dogimont C, Bendahmane A. (2009) A transposon-induced epigenetic change leads to sex determination in melon. Nature 461: 1135–1138 [DOI] [PubMed] [Google Scholar]
- Mäser P, Eckelman B, Vaidyanathan R, Horie T, Fairbairn DJ, Kubo M, Yamagami M, Yamaguchi K, Nishimura M, Uozumi N, et al. (2002) Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett 531: 157–161 [DOI] [PubMed] [Google Scholar]
- Mauricio R. (1998) Costs of resistance to natural enemies in field populations of the annual plant Arabidopsis thaliana. Am Nat 151: 20–28 [DOI] [PubMed] [Google Scholar]
- Mauricio R, Rausher MD. (1997) Experimental manipulation of putative selective agents provides evidence for the role of natural enemies in the evolution of plant defense. Evolution 51: 1435–1444 [DOI] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. (2008) Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9: 356–369 [DOI] [PubMed] [Google Scholar]
- McMullen MD, Kresovich S, Villeda HS, Bradbury P, Li H, Sun Q, Flint-Garcia S, Thornsberry J, Acharya C, Bottoms C, et al. (2009) Genetic properties of the maize nested association mapping population. Science 325: 737–740 [DOI] [PubMed] [Google Scholar]
- Melquist S, Bender J. (2003) Transcription from an upstream promoter controls methylation signaling from an inverted repeat of endogenous genes in Arabidopsis. Genes Dev 17: 2036–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Vigo B, Picó FX, Ramiro M, Martínez-Zapater JM, Alonso-Blanco C. (2011) Altitudinal and climatic adaptation is mediated by flowering traits and FRI, FLC, and PHYC genes in Arabidopsis. Plant Physiol 157: 1942–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML. (2010) Sequencing technologies: the next generation. Nat Rev Genet 11: 31–46 [DOI] [PubMed] [Google Scholar]
- Meyer RC, Kusterer B, Lisec J, Steinfath M, Becher M, Scharr H, Melchinger AE, Selbig J, Schurr U, Willmitzer L, et al. (2010) QTL analysis of early stage heterosis for biomass in Arabidopsis. Theor Appl Genet 120: 227–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, He Y, Scortecci KC, Amasino RM. (2003) Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci USA 100: 10102–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T. (1995) Interval mapping of viability loci causing heterosis in Arabidopsis. Genetics 140: 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T, Schmitt J. (2006) Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441: 947–952 [DOI] [PubMed] [Google Scholar]
- Montesinos A, Tonsor SJ, Alonso-Blanco C, Picó FX. (2009) Demographic and genetic patterns of variation among populations of Arabidopsis thaliana from contrasting native environments. PLoS ONE 4: e7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costich DE, Buckler ES. (2009) Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell 21: 2194–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam HG, Giraudat J, Den Boer B, Moonan F, Loos WD, Hauge BM, Goodman HM. (1989) Restriction fragment length polymorphism linkage map of Arabidopsis thaliana. Plant Cell 1: 699–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napp-Zinn K. (1957) Untersuchungen über das Vernalisationsverhalten einer winterannuellen Rasse von Arabidopsis thaliana (L.) Heynh. Planta 50: 177–210 [Google Scholar]
- Nemri A, Atwell S, Tarone AM, Huang YS, Zhao K, Studholme DJ, Nordborg M, Jones JD. (2010) Genome-wide survey of Arabidopsis natural variation in downy mildew resistance using combined association and linkage mapping. Proc Natl Acad Sci USA 107: 10302–10307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. (2006) Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet 7: 61–80 [DOI] [PubMed] [Google Scholar]
- Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ. (2009) Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura MT, Dangl JL. (2010) Arabidopsis and the plant immune system. Plant J 61: 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël L, Moores TL, van Der Biezen EA, Parniske M, Daniels MJ, Parker JE, Jones JD. (1999) Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11: 2099–2112 [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Borevitz JO, Bergelson J, Berry CC, Chory J, Hagenblad J, Kreitman M, Maloof JN, Noyes T, Oefner PJ, et al. (2002) The extent of linkage disequilibrium in Arabidopsis thaliana. Nat Genet 30: 190–193 [DOI] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al. (2005) The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Weigel D. (2008) Next-generation genetics in plants. Nature 456: 720–723 [DOI] [PubMed] [Google Scholar]
- Olsen KM, Halldorsdottir SS, Stinchcombe JR, Weinig C, Schmitt J, Purugganan MD. (2004) Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles. Genetics 167: 1361–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Clark RM, Lanz C, Warthmann N, Weigel D. (2008a) Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res 18: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Lucas-Lledó JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M. (2010) The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D. (2008b) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Ostrowski MF, David J, Santoni S, McKhann H, Reboud X, Le Corre V, Camilleri C, Brunel D, Bouchez D, Faure B, et al. (2006) Evidence for a large-scale population structure among accessions of Arabidopsis thaliana: possible causes and consequences for the distribution of linkage disequilibrium. Mol Ecol 15: 1507–1517 [DOI] [PubMed] [Google Scholar]
- Pagán I, Fraile A, Fernandez-Fueyo E, Montes N, Alonso-Blanco C, García-Arenal F. (2010) Arabidopsis thaliana as a model for the study of plant-virus co-evolution. Philos Trans R Soc Lond B Biol Sci 365: 1983–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Picó FX, Méndez-Vigo B, Martínez-Zapater JM, Alonso-Blanco C. (2008) Natural genetic variation of Arabidopsis thaliana is geographically structured in the Iberian peninsula. Genetics 180: 1009–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plantegenet S, Weber J, Goldstein DR, Zeller G, Nussbaumer C, Thomas J, Weigel D, Harshman K, Hardtke CS. (2009) Comprehensive analysis of Arabidopsis expression level polymorphisms with simple inheritance. Mol Syst Biol 5: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A, Horton M, Huang YS, Li Y, Anastasio AE, Mulyati NW, Agren J, Bossdorf O, Byers D, Donohue K, et al. (2010) The scale of population structure in Arabidopsis thaliana. PLoS Genet 6: e1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi M, Chan SW. (2010) Haploid plants produced by centromere-mediated genome elimination. Nature 464: 615–618 [DOI] [PubMed] [Google Scholar]
- Razi H, Howell EC, Newbury HJ, Kearsey MJ. (2008) Does sequence polymorphism of FLC paralogues underlie flowering time QTL in Brassica oleracea? Theor Appl Genet 116: 179–192 [DOI] [PubMed] [Google Scholar]
- Reinders J, Wulff BB, Mirouze M, Marí-Ordóñez A, Dapp M, Rozhon W, Bucher E, Theiler G, Paszkowski J. (2009) Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev 23: 939–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter RS, Williams JG, Feldmann KA, Rafalski JA, Tingey SV, Scolnik PA. (1992) Global and local genome mapping in Arabidopsis thaliana by using recombinant inbred lines and random amplified polymorphic DNAs. Proc Natl Acad Sci USA 89: 1477–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D, Rich PJ, Brunk DG, Ju GC, Rhodes JC, Pauly MH, Hansen LA. (1989) Development of two isogenic sweet corn hybrids differing for glycinebetaine content. Plant Physiol 91: 1112–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K. (1996) The future of genetic studies of complex human diseases. Science 273: 1516–1517 [DOI] [PubMed] [Google Scholar]
- Rose LE, Bittner-Eddy PD, Langley CH, Holub EB, Michelmore RW, Beynon JL. (2004) The maintenance of extreme amino acid diversity at the disease resistance gene, RPP13, in Arabidopsis thaliana. Genetics 166: 1517–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosloski SM, Jali SS, Balasubramanian S, Weigel D, Grbic V. (2010) Natural diversity in flowering responses of Arabidopsis thaliana caused by variation in a tandem gene array. Genetics 186: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux F, Colomé-Tatché M, Edelist C, Wardenaar R, Guerche P, Hospital F, Colot V, Jansen RC, Johannes F. (2011) Genome-wide epigenetic perturbation jump-starts patterns of heritable variation found in nature. Genetics 188: 1015–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HC, Hansen BG, Halkier BA, Kliebenstein DJ. (2008) Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell 20: 1199–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer-Pokora B, Kunkel LM, Monaco AP, Goff SC, Newburger PE, Baehner RL, Cole FS, Curnutte JT, Orkin SH. (1986) Cloning the gene for an inherited human disorder—chronic granulomatous disease—on the basis of its chromosomal location. Nature 322: 32–38 [DOI] [PubMed] [Google Scholar]
- Salomé PA, Bomblies K, Fitz J, Laitinen RAE, Warthmann N, Yant L, Weigel D. (November 9, 2011a) The recombination landscape in Arabidopsis thaliana F2 populations. Heredity http://dx.doi.org/10.1038.hdy.2011.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, Bomblies K, Laitinen RA, Yant L, Mott R, Weigel D. (2011b) Genetic architecture of flowering-time variation in Arabidopsis thaliana. Genetics 188: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Moran E, Armstrong SJ, Santos JL, Franklin FC, Jones GH. (2002) Variation in chiasma frequency among eight accessions of Arabidopsis thaliana. Genetics 162: 1415–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli N, Kover PX. (2009) Standing genetic variation in FRIGIDA mediates experimental evolution of flowering time in Arabidopsis. Mol Ecol 18: 2039–2049 [DOI] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, et al. (2005) An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet 37: 710–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schläppi MR. (2006) FRIGIDA LIKE 2 is a functional allele in Landsberg erecta and compensates for a nonsense allele of FRIGIDA LIKE 1. Plant Physiol 142: 1728–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid KJ, Ramos-Onsins S, Ringys-Beckstein H, Weisshaar B, Mitchell-Olds T. (2005) A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics 169: 1601–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Lewsey MG, O'Malley RC, Urich MA, Libiger O, Schork NJ, Ecker JR. (2011) Transgenerational epigenetic instability is a source of novel methylation variants. Science 334: 369–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmuths H, Bachmann K, Weber WE, Horres R, Hoffmann MH. (2006) Effects of preconditioning and temperature during germination of 73 natural accessions of Arabidopsis thaliana. Ann Bot (Lond) 97: 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Hagmann J, Ossowski S, Warthmann N, Gesing S, Kohlbacher O, Weigel D. (2009) Simultaneous alignment of short reads against multiple genomes. Genome Biol 10: R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Ott F, Klein JD, Wang X, Lanz C, Smith LM, Cao J, Fitz J, Warthmann N, et al. (2011) Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc Natl Acad Sci USA 108: 10249–10254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C, Balasubramanian S, Warthmann N, Michael TP, Lempe J, Sureshkumar S, Kobayashi Y, Maloof JN, Borevitz JO, Chory J, et al. (2009) Cis-regulatory changes at FLOWERING LOCUS T mediate natural variation in flowering responses of Arabidopsis thaliana. Genetics 183: 723–732, 721SI–727SI [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seevers PM, Daly JM, Catedral FF. (1971) The role of peroxidase isozymes in resistance to wheat stem rust disease. Plant Physiol 48: 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharbel TF, Haubold B, Mitchell-Olds T. (2000) Genetic isolation by distance in Arabidopsis thaliana: biogeography and postglacial colonization of Europe. Mol Ecol 9: 2109–2118 [DOI] [PubMed] [Google Scholar]
- Shindo C, Aranzana MJ, Lister C, Baxter C, Nicholls C, Nordborg M, Dean C. (2005) Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol 138: 1163–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C, Bernasconi G, Hardtke CS. (2007) Natural genetic variation in Arabidopsis: tools, traits and prospects for evolutionary ecology. Ann Bot (Lond) 99: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C, Lister C, Crevillen P, Nordborg M, Dean C. (2006) Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev 20: 3079–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Plantegenet S, Hardtke CS. (2008) Flowering as a condition for xylem expansion in Arabidopsis hypocotyl and root. Curr Biol 18: 458–463 [DOI] [PubMed] [Google Scholar]
- Simon M, Loudet O, Durand S, Bérard A, Brunel D, Sennesal FX, Durand-Tardif M, Pelletier G, Camilleri C. (2008) Quantitative trait loci mapping in five new large recombinant inbred line populations of Arabidopsis thaliana genotyped with consensus single-nucleotide polymorphism markers. Genetics 178: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Simon A, Martins F, Botran L, Tisné S, Granier F, Loudet O, Camilleri C. (November 11, 2011) DNA fingerprinting and new tools for fine-scale discrimination of Arabidopsis thaliana accessions. Plant J http://dx.doi.org/10.1111/j.1365-1313X.2011.04852.x [DOI] [PubMed] [Google Scholar]
- Slatkin M. (2009) Epigenetic inheritance and the missing heritability problem. Genetics 182: 845–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T, Huang HR, Holm K, Ceplitis A, Onge KS, Chen J, Lagercrantz U, Lascoux M. (2009) Splicing variation at a FLOWERING LOCUS C homeolog is associated with flowering time variation in the tetraploid Capsella bursa-pastoris. Genetics 183: 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Bomblies K, Weigel D. (2011) Complex evolutionary events at a tandem cluster of Arabidopsis thaliana genes resulting in a single-locus genetic incompatibility. PLoS Genet 7: e1002164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller M, Beckmann JS. (1990) Marker-based mapping of quantitative trait loci using replicated progenies. Theor Appl Genet 80: 205–208 [DOI] [PubMed] [Google Scholar]
- Sønderby IE, Hansen BG, Bjarnholt N, Ticconi C, Halkier BA, Kliebenstein DJ. (2007) A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS ONE 2: e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe WJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJ. (2000) The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell 6: 791–802 [DOI] [PubMed] [Google Scholar]
- Srikanth A, Schmid M. (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68: 2013–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. (1999) Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400: 667–671 [DOI] [PubMed] [Google Scholar]
- Stam M, Belele C, Dorweiler JE, Chandler VL. (2002) Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev 16: 1906–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz BJ, Ausubel FM, Baker BJ, Ellis JG, Jones JD. (1995) Molecular genetics of plant disease resistance. Science 268: 661–667 [DOI] [PubMed] [Google Scholar]
- Stenøien HK, Fenster CB, Tonteri A, Savolainen O. (2005) Genetic variability in natural populations of Arabidopsis thaliana in northern Europe. Mol Ecol 14: 137–148 [DOI] [PubMed] [Google Scholar]
- Stinchcombe JR, Weinig C, Ungerer M, Olsen KM, Mays C, Halldorsdottir SS, Purugganan MD, Schmitt J. (2004) A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc Natl Acad Sci USA 101: 4712–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange A, Li P, Lister C, Anderson J, Warthmann N, Shindo C, Irwin J, Nordborg M, Dean C. (2011) Major-effect alleles at relatively few loci underlie distinct vernalization and flowering variation in Arabidopsis accessions. PLoS ONE 6: e19949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugliani M, Rajjou L, Clerkx EJ, Koornneef M, Soppe WJ. (2009) Natural modifiers of seed longevity in the Arabidopsis mutants abscisic acid insensitive3-5 (abi3-5) and leafy cotyledon1-3 (lec1-3). New Phytol 184: 898–908 [DOI] [PubMed] [Google Scholar]
- Sulpice R, Pyl ET, Ishihara H, Trenkamp S, Steinfath M, Witucka-Wall H, Gibon Y, Usadel B, Poree F, Piques MC, et al. (2009) Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci USA 106: 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R, Trenkamp S, Steinfath M, Usadel B, Gibon Y, Witucka-Wall H, Pyl ET, Tschoep H, Steinhauser MC, Guenther M, et al. (2010) Network analysis of enzyme activities and metabolite levels and their relationship to biomass in a large panel of Arabidopsis accessions. Plant Cell 22: 2872–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed NH, Chen ZJ. (2005) Molecular marker genotypes, heterozygosity and genetic interactions explain heterosis in Arabidopsis thaliana. Heredity 94: 295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds VV, Hatlestad G, Lloyd AM. (2011) Natural allelic variation defines a role for ATMYC1: trichome cell fate determination. PLoS Genet 7: e1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira FK, Heredia F, Sarazin A, Roudier F, Boccara M, Ciaudo C, Cruaud C, Poulain J, Berdasco M, Fraga MF, et al. (2009) A role for RNAi in the selective correction of DNA methylation defects. Science 323: 1600–1604 [DOI] [PubMed] [Google Scholar]
- Tenaillon MI, Hufford MB, Gaut BS, Ross-Ibarra J. (2011) Genome size and transposable element content as determined by high-throughput sequencing in maize and Zea luxurians. Genome Biol Evol 3: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J. (2003) Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423: 74–77 [DOI] [PubMed] [Google Scholar]
- Todesco M, Balasubramanian S, Hu TT, Traw MB, Horton M, Epple P, Kuhns C, Sureshkumar S, Schwartz C, Lanz C, et al. (2010) Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465: 632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomajian C, Hu TT, Aranzana MJ, Lister C, Tang C, Zheng H, Zhao K, Calabrese P, Dean C, Nordborg M. (2006) A nonparametric test reveals selection for rapid flowering in the Arabidopsis genome. PLoS Biol 4: e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Törjék O, Meyer RC, Zehnsdorf M, Teltow M, Strompen G, Witucka-Wall H, Blacha A, Altmann T. (2008) Construction and analysis of 2 reciprocal Arabidopsis introgression line populations. J Hered 99: 396–406 [DOI] [PubMed] [Google Scholar]
- Törjék O, Witucka-Wall H, Meyer RC, von Korff M, Kusterer B, Rautengarten C, Altmann T. (2006) Segregation distortion in Arabidopsis C24/Col-0 and Col-0/C24 recombinant inbred line populations is due to reduced fertility caused by epistatic interaction of two loci. Theor Appl Genet 113: 1551–1561 [DOI] [PubMed] [Google Scholar]
- Tuinstra MR, Ejeta G, Goldsbrough PB. (1997) Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet 95: 1005–1011 [Google Scholar]
- Turesson G. (1922a) The genotypical response of the plant species to the habitat. Hereditas 3: 211–350 [Google Scholar]
- Turesson G. (1922b) The species and the variety as ecological units. Hereditas 3: 100–113 [Google Scholar]
- Ungerer MC, Linder CR, Rieseberg LH. (2003) Effects of genetic background on response to selection in experimental populations of Arabidopsis thaliana. Genetics 163: 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer MC, Rieseberg LH. (2003) Genetic architecture of a selection response in Arabidopsis thaliana. Evolution 57: 2531–2539 [DOI] [PubMed] [Google Scholar]
- van Der Schaar W, Alonso-Blanco C, Léon-Kloosterziel KM, Jansen RC, van Ooijen JW, Koornneef M. (1997) QTL analysis of seed dormancy in Arabidopsis using recombinant inbred lines and MQM mapping. Heredity 79: 190–200 [DOI] [PubMed] [Google Scholar]
- Vaughn MW, Tanurdzić M, Lippman Z, Jiang H, Carrasquillo R, Rabinowicz PD, Dedhia N, McCombie WR, Agier N, Bulski A, et al. (2007) Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol 5: e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad D, Rappaport F, Simon M, Loudet O. (2010) Gene transposition causing natural variation for growth in Arabidopsis thaliana. PLoS Genet 6: e1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CT, Ho CH, Hseu MJ, Chen CM. (2010) The subtelomeric region of the Arabidopsis thaliana chromosome IIIR contains potential genes and duplicated fragments from other chromosomes. Plant Mol Biol 74: 155–166 [DOI] [PubMed] [Google Scholar]
- Wang J, Tian L, Lee HS, Wei NE, Jiang H, Watson B, Madlung A, Osborn TC, Doerge RW, Comai L, et al. (2006) Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Qian W, Suppanz I, Wei L, Mao B, Long Y, Meng J, Muller AE, Jung C. (2011) Flowering time variation in oilseed rape (Brassica napus L.) is associated with allelic variation in the FRIGIDA homologue BnaA.FRI.a. J Exp Bot 62: 5641–5658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sajja U, Rosloski S, Humphrey T, Kim MC, Bomblies K, Weigel D, Grbic V. (2007) HUA2 caused natural variation in shoot morphology of A. thaliana. Curr Biol 17: 1513–1519 [DOI] [PubMed] [Google Scholar]
- Weigel D, Mott R. (2009) The 1001 genomes project for Arabidopsis thaliana. Genome Biol 10: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinig C, Dorn LA, Kane NC, German ZM, Halldorsdottir SS, Ungerer MC, Toyonaga Y, Mackay TF, Purugganan MD, Schmitt J. (2003a) Heterogeneous selection at specific loci in natural environments in Arabidopsis thaliana. Genetics 165: 321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinig C, Stinchcombe JR, Schmitt J. (2003b) QTL architecture of resistance and tolerance traits in Arabidopsis thaliana in natural environments. Mol Ecol 12: 1153–1163 [DOI] [PubMed] [Google Scholar]
- Weinig C, Ungerer MC, Dorn LA, Kane NC, Toyonaga Y, Halldorsdottir SS, Mackay TF, Purugganan MD, Schmitt J. (2002) Novel loci control variation in reproductive timing in Arabidopsis thaliana in natural environments. Genetics 162: 1875–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzell AM, Rowe HC, Hansen BG, Ticconi C, Halkier BA, Kliebenstein DJ. (2007) Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet 3: 1687–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JD, Borevitz JO, Warthmann N, Trainer GT, Ecker JR, Chory J, Weigel D. (2005) Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc Natl Acad Sci USA 102: 2460–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerman JM. (1971) Genotype-environment interaction and developmental regulation in Arabidopsis thaliana. IV. Wild material; analysis. Heredity 26: 383–395 [Google Scholar]
- Wilczek AM, Burghardt LT, Cobb AR, Cooper MD, Welch SM, Schmitt J. (2010) Genetic and physiological bases for phenological responses to current and predicted climates. Philos Trans R Soc Lond B Biol Sci 365: 3129–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczek AM, Roe JL, Knapp MC, Cooper MD, Lopez-Gallego C, Martin LJ, Muir CD, Sim S, Walker A, Anderson J, et al. (2009) Effects of genetic perturbation on seasonal life history plasticity. Science 323: 930–934 [DOI] [PubMed] [Google Scholar]
- Woo HR, Richards EJ. (2008) Natural variation in DNA methylation in ribosomal RNA genes of Arabidopsis thaliana. BMC Plant Biol 8: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Zou F, Vision TJ. (2005) Improving quantitative trait loci mapping resolution in experimental crosses by the use of genotypically selected samples. Genetics 170: 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto E, Takashi T, Morinaka Y, Lin S, Wu J, Matsumoto T, Kitano H, Matsuoka M, Ashikari M. (2010) Gain of deleterious function causes an autoimmune response and Bateson-Dobzhansky-Muller incompatibility in rice. Mol Genet Genomics 283: 305–315 [DOI] [PubMed] [Google Scholar]
- Yu J, Holland JB, McMullen MD, Buckler ES. (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178: 539–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller G, Clark RM, Schneeberger K, Bohlen A, Weigel D, Rätsch G. (2008) Detecting polymorphic regions in Arabidopsis thaliana with resequencing microarrays. Genome Res 18: 918–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Borevitz JO. (2009) Global analysis of allele-specific expression in Arabidopsis thaliana. Genetics 182: 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Cal AJ, Borevitz JO. (2011) Genetic architecture of regulatory variation in Arabidopsis thaliana. Genome Res 21: 725–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shiu SH, Cal A, Borevitz JO. (2008) Global analysis of genetic, epigenetic and transcriptional polymorphisms in Arabidopsis thaliana using whole genome tiling arrays. PLoS Genet 4: e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ober JA, Kliebenstein DJ. (2006) The gene controlling the quantitative trait locus EPITHIOSPECIFIER MODIFIER1 alters glucosinolate hydrolysis and insect resistance in Arabidopsis. Plant Cell 18: 1524–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kulkarni V, Liu N, Del Carpio DP, Bucher J, Bonnema G. (2010) BrFLC2 (FLOWERING LOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa. J Exp Bot 61: 1817–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Aranzana MJ, Kim S, Lister C, Shindo C, Tang C, Toomajian C, Zheng H, Dean C, Marjoram P, Nordborg M. (2007) An Arabidopsis example of association mapping in structured samples. PLoS Genet 3: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]