Abstract
Binary vectors are an indispensable component of modern Agrobacterium tumefaciens-mediated plant genetic transformation systems. A remarkable variety of binary plasmids have been developed to support the cloning and transfer of foreign genes into plant cells. The majority of these systems, however, are limited to the cloning and transfer of just a single gene of interest. Thus, plant biologists and biotechnologists face a major obstacle when planning the introduction of multigene traits into transgenic plants. Here, we describe the assembly of multitransgene binary vectors by using a combination of engineered zinc finger nucleases (ZFNs) and homing endonucleases. Our system is composed of a modified binary vector that has been engineered to carry an array of unique recognition sites for ZFNs and homing endonucleases and a family of modular satellite vectors. By combining the use of designed ZFNs and commercial restriction enzymes, multiple plant expression cassettes were sequentially cloned into the acceptor binary vector. Using this system, we produced binary vectors that carried up to nine genes. Arabidopsis (Arabidopsis thaliana) protoplasts and plants were transiently and stably transformed, respectively, by several multigene constructs, and the expression of the transformed genes was monitored across several generations. Because ZFNs can potentially be engineered to digest a wide variety of target sequences, our system allows overcoming the problem of the very limited number of commercial homing endonucleases. Thus, users of our system can enjoy a rich resource of plasmids that can be easily adapted to their various needs, and since our cloning system is based on ZFN and homing endonucleases, it may be possible to reconstruct other types of binary vectors and adapt our vectors for cloning on multigene vector systems in various binary plasmids.
To date, biotechnological improvement of plant species has largely been limited to the introduction of single novel traits into the genomes of target plants. However, many agronomic traits may depend on complex interactions between several proteins, and biotechnological improvement of a particular species may thus require the delivery and expression of whole, complex metabolic pathways (Halpin et al., 2001; Daniell and Dhingra, 2002; Lyznik and Dress, 2008; Naqvi et al., 2010). In addition, transgenic modification of commercially important plant species also calls for the development of novel tools for the removal, addition, and replacement of existing transgenes in plant cells (Hanin and Paszkowski, 2003; Porteus, 2009; Moon et al., 2010; Weinthal et al., 2010). Thus, two of the major challenges that are still to be addressed in plant biotechnology are the development of a technology to combine several transgenic traits in a single plant by stacking a number of genes in the same chromosomal locus into a single multigene array and the successive manipulation of this array by genome editing (Dafny-Yelin and Tzfira, 2007; Lyznik and Dress, 2008; Taverniers et al., 2008; Naqvi et al., 2010). The main strategies for the introduction of multiple genes into plant cells include retransformation, cotransformation, sexual crossing, and transformation of multigene constructs (for review, see Dafny-Yelin and Tzfira, 2007; Naqvi et al., 2010). While proven useful for the production of transgenic plants with novel traits, retransformation, cotransformation, and sexual crossing approaches all suffer from several flaws (Dafny-Yelin and Tzfira, 2007; Naqvi et al., 2010). Retransformation and sexual crossing, for example, are time consuming and rely on the use of different selectable marker genes for each transformation/crossing cycle. In cotransformation, it is virtually impossible to predict the number of insertions and the distribution of the inserted genes across the plant genome. In addition, cotransformation may result in complex integration patterns that may hinder the use of such plants for commercial purposes, for which single and well-characterized integration events are required. While sexual crosses may be simplified by using marker-free transgenic plants, the delivery of multiple genes as a single, well-defined, multigene array would perhaps be the simplest and most reliable method for the production of multigene transgenic plants. Furthermore, multigene arrays may also offer the advantage of simplifying successive manipulations of multigene assays in transgenic plants by genome-editing technologies (Porteus, 2009; Weinthal et al., 2010). Yet, while multigene constructs have been successfully used in several studies (Bohmert et al., 2000, 2002; Wu et al., 2005; Zhong et al., 2007; Fujisawa et al., 2009), the assembly of multigene constructs remains challenging, being nearly impossible to achieve by traditional cloning methods (Dafny-Yelin and Tzfira, 2007; Naqvi et al., 2010).
Only a handful of dedicated vector assembly systems have been developed in the past several years for the assembly of multigene transformation vectors (Cheo et al., 2004; Sasaki et al., 2004; Karimi et al., 2005; Chen et al., 2006, 2010; Wakasa et al., 2006). Most of these vector assembly systems have a rather limited capacity and hence have been utilized for the delivery of only a small number (i.e. up to five) of transgenes in a single array. Noteworthy here is the assembly system of Lin et al. (2003), in which a combination of the Cre/loxP recombination system and two homing endonucleases is used for successive cloning of a potentially infinite number of genes onto a transformation-competent artificial chromosome-based vector. Lin et al. (2003) used their system for the delivery of eight different genes from two independent T-DNA molecules that were launched from a single binary vector into the rice (Oryza sativa) genome. However, a crucial limitation of the ingenious approach of Lin et al. (2003) was that, in contradiction to classical cloning by restriction enzymes, once assembled into the binary vector, new DNA fragments can no longer be removed or replaced by others.
Another important contribution to the field is the unique multiple-round in vivo site-specific assembly system of Chen et al. (2010) that enables in vivo recombination-based assembly of multigene vectors. The system was used to construct several multitransgene binary vectors and led to the production of transgenic plants with eight transgenes (Chen et al., 2010). A number of other recombination-based cloning strategies have also been developed for the construction of multigene plant transformation vectors (Cheo et al., 2004; Sasaki et al., 2004; Karimi et al., 2005; Chen et al., 2006, 2010; Wakasa et al., 2006). However, here too, the irreversible nature of the recombination-based reactions does not enable the modification of existing binary vectors, and users of such systems may be required to completely rebuild their transformation vectors to give different gene combinations. Multigene transformation vectors have also been assembled by using both traditional cloning methods. Wu et al. (2005), for example, combined the use of type II restriction enzymes and Gateway-mediated cloning to construct several binary vectors with up to 10 different expression cassettes and used the combination to reconstitute the docosahexaenoic acid biosynthetic pathway in Indian mustard (Brassica juncea). Fujisawa et al. (2009) combined the use of type II restriction enzymes and homing endonucleases to construct a seven-transgene-long T-DNA molecule, which they applied to genetically modify the carotenoid biosynthesis pathway in Brassica napus. Other examples include the engineering of soybean (Glycine max), potato (Solanum tuberosum), rice, Arabidopsis (Arabidopsis thaliana), and several other plant species by using four- to five-transgene-long transformation vectors (for review, see Naqvi et al., 2010; Peremarti et al., 2010). While proven successful for the metabolic engineering of various plant species, such vectors were mostly custom designed to their specific tasks, and their modification for other metabolic pathways and multigene traits may prove to be difficult or even impossible to achieve.
A versatile and modular system for the assembly of multigene binary vectors has been developed by Goderis et al. (2002), who exploited a set of homing endonucleases to construct a vector system facilitating the successive cloning of independent plant expression cassettes. The principles of the assembly system and successor satellite (pSAT) vector system were previously reported by Tzfira et al. (2005). An important advantage of this method, over the approaches described above, is its modularity. Plant expression cassettes can easily be removed or replaced from existing binary vectors. Nevertheless, the capacity of the vector system and its successors is limited by the very small number of commercially available homing endonucleases.
We have recently shown that zinc finger nucleases (ZFNs), engineered restriction enzymes that can be designed to bind and cleave long stretches of DNA sequences (Mani et al., 2005), can be used for molecular cloning, and we have used such enzymes for the construction of dual-gene binary vectors (Zeevi et al., 2008). In this paper, we describe a modular binary vector assembly system that represents a fundamental improvement over the original design of Goderis et al. (2002) in that it supports the construction of multigene transformation vectors not only by homing endonucleases but also by designed ZFNs. We describe the design of our system and demonstrate its use by cloning nine different DNA fragments onto a modified binary transformation vector. We also show that such vectors can be used for the production of multigene transgenic plants and for the transient expression of multiple genes in plant protoplasts. The advantage of using ZFNs for the construction of multigene transformation systems is discussed.
RESULTS
Design of the Binary Vector and Novel pSATs
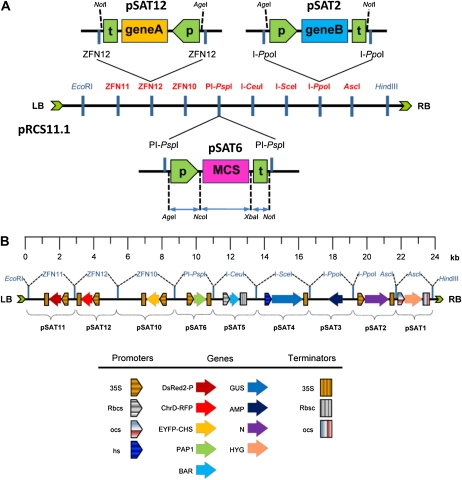
We followed the basic design of the pAUX (Goderis et al., 2002) and its successor pSAT (Chung et al., 2005; Tzfira et al., 2005; Dafny-Yelin et al., 2007; Dafny-Yelin and Tzfira, 2007) family of plasmids to facilitate the assembly of multiple genes into a single binary plasmid. In the pSAT system, functional plant expression cassettes are individually cloned into different pSAT plasmids (e.g. gene B in pSAT2; Fig. 1A), and an expression cassette from each type of pSAT plasmid can be cloned onto pPZP-RCS1- or pPZP-RCS2-based (Goderis et al., 2002) Agrobacterium tumefaciens binary plasmids by using matching homing endonucleases (Fig. 1A). As was the case for the original set of pAUX plasmids, the original set of pSAT plasmids was composed of seven different versions, in which the expression cassettes were flanked by AscI (pSAT1), AscI and I-PpoI (pSAT2), I-PpoI (pSAT3), I-SceI (pSAT4), I-CeuI (pSAT5), PI-PspI (pSAT6), and PI-TliI (pSAT7). Here, we subsequently expanded this set to include pSAT10, pSAT11, and pSAT12, in which the expression cassettes were flanked by ZFN10, ZFN11, and ZFN12 (3 × 3 finger ZFNs), respectively (e.g. gene A in pSAT12; Fig. 1A). We also constructed pRCS11.1, which is a modification of pPZP-RCS1 that was engineered to include, in addition to the original recognition sites of AscI, I-PpoI, I-SceI, I-CeuI, PI-PspI, and PI-TliI, recognition sites to the ZFN10, ZFN11, and ZFN12 (Fig. 1A). This system was subsequently used for the construction of several multigene binary plasmids, as described below.
Figure 1.
General features of the ZFN- and homing endonuclease-mediated multigene binary vector assembly system. A, The general structure of a typical pSAT vector is exemplified by pSAT6, in which the promoter is flanked by the unique AgeI and NcoI sites, the terminator is flanked by the unique XbaI and NotI sites, the gene of interest is cloned into an extended MCS, and the entire plant expression cassette is flanked by recognition sites for ZFNs (e.g. ZFN12 in pSAT12) or homing endonuclease (e.g. PI-PspI in pSAT6). The ZFN and homing endonuclease recognition sites are shown on the binary pRCS11.1. Using ZFNs and homing endonucleases, up to nine expression cassettes can be transferred from the pSAT vectors into the T-DNA region of pRCS11.1. LB, Left border; P, promoter sequence; RB, right border; T, terminator sequence. B, Structure and scale of the pRCS11.1[1.HYG][2.N][3.AMP][4.GUS][5.BAR][6.PAP1][10.EYFP-CHS][11.DsRed2-P][12.ChrD-RFP] nine-transgene-long plant transformation binary vector. The general structure and direction of each expression cassette is shown. Promoters and terminators are as follow: 35S, double CaMV 35S; Rbcs, Rubisco small subunit; ocs, octopine synthase; hs, hsp18.1 heat shock. Proteins are as follow: DsRed2-P, P protein of SYNV fused to DsRed2; ChrD-RFP, chromoplast-specific carotenoid-associated protein ChrD fused to RFP; EYFP-CHS, chalcone synthase fused to EYFP; PAP1, Arabidopsis transcription factor production of anthocyanin pigment 1; BAR, Basta resistance-encoding gene; AMP, ampicillin resistance; N, N protein of SYNV; HYG, hygromycin resistance.
Construction of Multigene Vectors by Exploiting ZFNs
We constructed a set of pSAT plasmids carrying a wide variety of genes. Table I lists the different pSAT plasmids constructed, the genes they carry, and the enzymes used for their cloning into pRCS11.1. We started by cloning the constitutive expression cassette of the endoplasmic reticulum-bound chalcone synthase (CHS) gene (Pelletier and Shirley, 1996) tagged with enhanced yellow fluorescent protein (EYFP) from pSAT10.EYFP-CHS by using ZFN10 to produce pRCS11[10.EYFP-CHS]. Next, we added the constitutive expression cassette of the phospho (P) protein of Sonchus yellow net rhabdovirus (SYNV; Goodin et al., 2001, 2002) tagged with DsRed2 from pSAT11.DsRed2-P by using ZFN11, to produce pRCS11.1[10.EYFP-CHS][11.DsRed2-P].
Table I.
Genes, vectors, and enzymes used for the construction of multigene vectors, in order of their assembly in pRCS11.1
| pSAT | Protein | Enzyme(s) | Phenotype |
| 10 | EYFP-CHS | ZFN10 | Decoration of endoplasmic reticulum by yellow fluorescence |
| 11 | DsRed2-P | ZFN11 | Decoration of the cytoplasm and nucleus by red fluorescence |
| 12 | ChrD-RFP | ZFN12 | Decoration of the chloroplasts by red fluorescence |
| 2 | N | AscI-I-PpoI | Directing DsRed2-P into subnuclear compartments |
| 1 | HYG | AscI | Hygromycin resistance |
| 3 | AMP | I-PpoI | Bacterial ampicillin resistance gene and origin of replication |
| 5 | BAR | I-CeuI | Basta resistance |
| 4 | GUS | I-SceI | Heat shock-induced GUS expression |
| 6 | PAP1 | PI-PspI | Activation of anthocyanin expression |
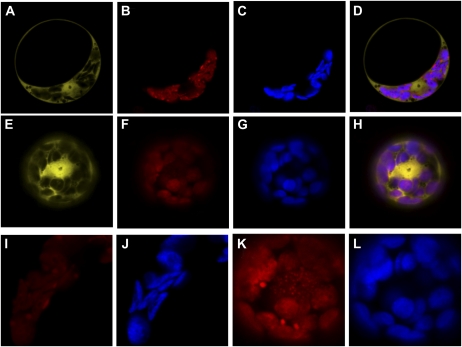
The key to the versatility of the original pSAT/pRCS2 multigene assembly system lies in the ability it offers not only to add but also to remove and replace DNA fragments by homing endonucleases during the construction of the various binary plasmids (Tzfira et al., 2005). Thereafter, we demonstrated that ZFNs too can also be used to remove and replace DNA fragments from existing binary constructs. We started by adding the plasmid backbone from pSAT12.MCS into pRCS11.1[10.EYFP-CHS][11.DsRed2-P] and produced pRCS11.1[10.EYFP-CHS][11.DsRed2-P][12.AMP]. As expected, the resultant plasmid conferred resistance to ampicillin in Escherichia coli cells. We also observed that a DNA preparation of this plasmid from an overnight E. coli culture resulted in much higher DNA yield than that obtained with its progenitor or with other pRCS2-based vectors, most probably due to the presence of the ColE1 origin of replication on the pSAT12.MCS backbone. We then used ZFN12 to remove the pSAT12.MCS backbone and replaced it with a constitutive expression cassette of the chromoplast-specific carotenoid-associated protein ChrD from Cucumis sativus (Vishnevetsky et al., 1996, 1999) tagged with red fluorescent protein (RFP) from pSAT12.ChrD-RFP to produce pRCS11.1[10.EYFP-CHS][11.DsRed2-P][12.ChrD-RFP]. As expected, this plasmid no longer conferred resistance to ampicillin in E. coli cells. We tested the expression of the three reporter genes from pRCS11.1[10.EYFP-CHS][11.DsRed2-P][12.ChrD-RFP] in Arabidopsis protoplasts. As expected, the EYFP-tagged CHS associated with the rough endoplasmic reticulum throughout the cell, around the protoplasts, and around the nucleus (Fig. 2A); DsRed2-P was observed in the cytoplasm clustered around the chloroplasts; and ChrD-RFP, which has been characterized previously as a chloroplast-associated protein (Ben Zvi et al., 2008), indeed localized in the chloroplasts (Fig. 2B). The clustering of DsRed2-P was probably due to the tendency of DsRed2 to aggregate in living cells. We next added a fourth plant expression cassette for the SYNV nucleocapsid (N) protein and produced pRCS11.1[2.N][10.EYFP-CHS][11.DsRed2-P][12.ChrD-RFP]. Interaction of the SYNV N protein with the SYNV P protein is required to translocate it into subnuclear compartments in Nicotiana benthamiana cells (Goodin et al., 2001, 2002). Indeed, nuclear localization and subnuclear compartmenting of the DsRed2-P signal was observed in pRCS11.1[2.N][10.EYFP-CHS][11.DsRed2-P][12.ChrD-RFP]-infected protoplasts (Fig. 2, F and K) but not in pRCS11.1[10.EYFP-CHS][11.DsRed2-P][12.ChrD-RFP]-infected protoplasts (Fig. 2, B and I). Our data thus show that designed ZFNs can be used for the construction of multigene vectors and that such vectors can be used to drive the simultaneous expression of several genes in plant cells.
Figure 2.
Multiple gene expression in protoplasts from triple- and quadruple-gene long binary vectors. EYFP-CHS expression was targeted to the rough endoplasmic reticulum (shown in yellow in A and E), while ChrD-RFP was targeted to the chloroplasts (shown in red in B, F, I, and K) and overlapped with the chloroplast autofluorescence (shown in blue in C, G, J, and L). DsRed2-P was targeted to the cytoplasm (shown in red in B and I) and was redirected into the nucleus in the presence of free N protein to form subnuclear aggregates (shown in red in F and K). D and H present merged signals of EYFP, DsRed2, RFP, and plastid autofluorescence. I and J are magnifications of B and C; K and L are magnifications of F and G. All panels are single confocal sections.
Assembly of Multigene Binary Vectors for Stable Transformation
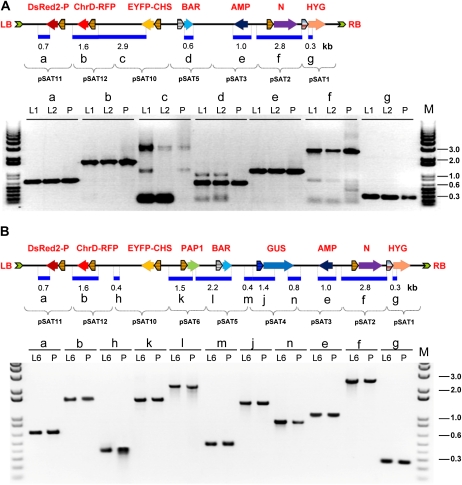
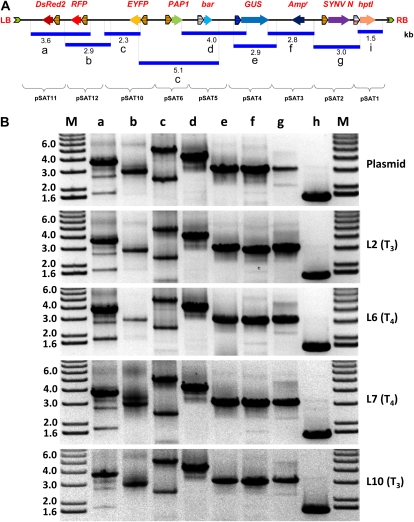
pRCS11.1[2.N][10.EYFP-CHS][11.DsRed2-P][12.ChrD-RFP] carried four pairs of repetitive elements (i.e. cauliflower mosaic virus [CaMV] dual 35S promoters and CaMV 35S terminators). We next tested whether the four transgenes and two additional genes (i.e. a hygromycin resistance gene driven by the control of the octopine synthase promoter and terminator and the Basta resistance gene driven by the control of the Rubisco small subunit promoter and terminator) can be stably transformed and expressed in transgenic plants. Many commonly used binary vectors have been designed with the selection marker cassette cloned next to the T-DNA’s left border (van Engelen et al., 1995; Hellens et al., 2000). We elected to clone the hygromycin-resistant expression cassette near the T-DNA’s right border. This design, while having the potential to produce transgenic plants with truncated T-DNAs, allowed us to examine whether repetitive elements have a negative impact on the T-DNA structure during transformation. To achieve the above design, we first produced pRCS11.1[1.HYG][2.N][10.EYFP-CHS][11.DsRed2-P][12.ChrD-RFP], in which the hygromycin-resistance gene expression cassette was cloned into the AscI site of pRCS11.1[3.N][10.EYFP-CHS][11.DsRed2-P][12.ChrD-RFP]. We then added the plasmid backbone from pSAT3.MCS into the I-PpoI site to facilitate future cloning and subsequent isolation of T-DNA-plant junction sequences by plasmid rescue and the Basta hygromycin-resistant expression cassette into the I-CeuI site of pRCS11.1[3.N][10.EYFP-CHS][11.DsRed2-P][12.ChrD-RFP]. Finally, we used the seven-transgene-long pRCS11.1[1.HYG][2.N][3.AMP][5.BAR][10.EYFP-CHS][11.DsRed2-P][12.ChrD-RFP] binary plasmid (Fig. 3A) to produce transgenic Arabidopsis plants. Confocal microscopy analysis revealed that nine of our 27 transgenic lines exhibited yellow and red fluorescence expression patterns similar to the patterns of protoplasts infected with pRCS11.1[2.N][10.EYFP-CHS][11.DsRed2-P][12.ChrD-RFP] (data not shown). Molecular analysis of two randomly selected transgenic lines revealed that PCR amplification of their T-DNA regions was similar to that of the seven-transgene-long binary vector (Fig. 3A); these findings indicate that our transgenic lines carried all the T-DNA molecule-encoded transgenes within their genome.
Figure 3.
PCR analysis of multigene binary T-DNAs in transgenic plants. A, PCR analysis of seven-transgene-long T-DNA inserts in a binary plasmid (P) and two transgenic plants (L1 and L2). B, PCR analysis of nine-transgene-long T-DNA inserts in a transgenic plant (L6) and in a binary plasmid (P). The general structure and the expected sizes (in kb) of the PCR bands are shown. Abbreviations are as in Figure 1.
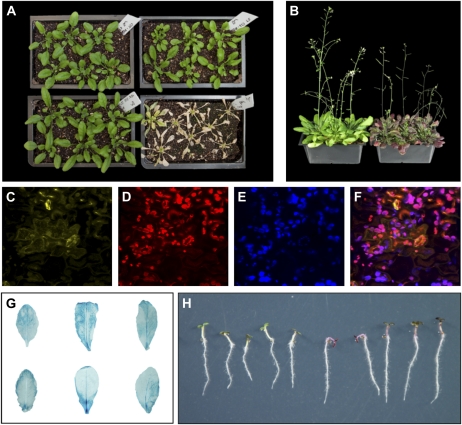
We then allowed several randomly selected transgenic lines to mature and set seed. Shown in Figure 4 are examples of hygromycin- and Basta-resistant seedlings. Collectively, our data indicate that the T-DNA-encoded seven transgenes were stably expressed and inherited.
Figure 4.
Phenotypic analysis of seven-transgene-long transgenic Arabidopsis plants. A, Hygromycin resistance in two independent transgenic T1 Arabidopsis plants (L1 and L2). w.t., Wild-type plants. B, Basta resistance in a hygromycin-resistant transgenic Arabidopsis line (left). Wild-type plants are shown on the right.
Production of Nine-Transgene-Long Transgenic Plants
We constructed a nine-transgene-long binary vector by adding the GUS and PAP1 (for Arabidopsis transcription factor production of anthocyanin pigment; Borevitz et al., 2000; Ben Zvi et al., 2008) expression cassettes into the I-SceI and PI-PspI sites of pRCS11.1[1.HYG][2.N][3.AMP][5.BAR][10.EYFP-CHS][11.DsRed2-P][12.ChrD-RFP]. In the final binary vector, pRCS11.1[1.HYG][2.N][3.AMP][4.GUS][5.BAR][6.PAP1][10.EYFP-CHS][11.DsRed2-P][12.ChrD-RFP], GUS expression is controlled by the hsp18.1 heat shock promoter (Takahashi and Komeda, 1989), while PAP1 expression is controlled by the CaMV dual 35S promoter. We used this vector to produce over 30 hygromycin-resistant transgenic Arabidopsis plants, of which about half exhibited PAP1-related reddish hypocotyls and cotyledons during germination. We further characterized these lines and determined whether they are likely to harbor the full T-DNA molecule by monitoring DsRed2-P expression, which was located next to the left border. Twelve T0 plants of the PAP1 transgenic plants also expressed DsRed2-P, which indicated that they are likely to harbor at least one full T-DNA copy. Molecular analysis of several lines revealed that PCR amplification of several regions on their T-DNA was similar to that of the nine-transgene-long binary vector, as demonstrated, for example, in Figure 3B. These observations indicate that the plants carried the entire T-DNA molecule-encoded genes.
We allowed seven of the transgenic lines to mature and set seed and determined the inheritance and stability of the T-DNA in the next generations. T0 plants of lines L6, L7, and L10 exhibit a plausible single-insert segregation pattern when grown on a hygromycin-containing medium (Table II). T1 seedlings of lines L2, L6, L7, and L10, T2 seedlings that derived from homozygous (i.e. L6-1) and heterozygous (i.e. L6-2 and L10-2) parents, exhibited resistance to Basta, as did T2 seedlings derived from lines L2 and L3 (Table II; Fig. 5A). Confocal microscopy analysis revealed that all T0 lines and their progeny (Table II) exhibited red and yellow fluorescence, as shown, for example, for line L6-1 (Fig. 5, C–F). Analysis of several T2 lines revealed that they all expressed heat shock-induced GUS expression (Fig. 5G). GUS expression was not observed in untreated plants. In addition, all T1, T2, and T3 hygromycin-resistant plants exhibited PAP1 expression (Table I), as shown, for example, for L3-3-1 (Fig. 5B) and L6-2-1-1 (Fig. 5F) plants. The segregation analyses suggest that a single transgene locus was obtained in some of the analyzed lines (i.e. L6, L7, and L10).
Table II.
Analysis of transgenic Arabidopsis plants across several generations
| T0 |
T1 |
T2 |
T3 |
||||||||||||||||
| Line | PAP1 | DsRed2 | HYGa | BARb | Line | PAP1 | DsRed2 | EYFP | RFP | GUS | HYGa | BARb | Line | PAP1 | GUS | HYGa | Line | PAP1 | DsRed2 |
| % | % | % | |||||||||||||||||
| L1 | +c | + | 100 | NDd | |||||||||||||||
| L1-1 | + | + | + | + | + | ND | |||||||||||||
| L2 | + | + | 100 | + | |||||||||||||||
| L2-1 | + | + | + | + | + | 100 | + | ||||||||||||
| L2-1-1 | + | + | |||||||||||||||||
| L2-1-2 | + | + | |||||||||||||||||
| L2-1-3 | + | + | |||||||||||||||||
| L2-2 | + | + | + | + | + | 100 | + | L2-2-1 | + | + | |||||||||
| L3 | + | + | 100 | ND | |||||||||||||||
| L3-1 | + | + | + | + | + | 100 | |||||||||||||
| L3-3 | + | + | + | + | + | 100 | + | L3-3-1 | + | ND | |||||||||
| L6 | + | + | 75 | + | |||||||||||||||
| L6-1 | + | + | + | + | + | 100 | + | ||||||||||||
| L6-1-1 | + | + | 100 | ||||||||||||||||
| L6-1-1-1 | + | + | |||||||||||||||||
| L6-1-2 | + | + | ND | ||||||||||||||||
| L6-1-3 | + | + | ND | ||||||||||||||||
| + | L6-2 | + | + | + | + | + | 75 | + | |||||||||||
| L6-2-1 | + | + | 75 | ||||||||||||||||
| L6-2-1-1 | + | + | |||||||||||||||||
| L6-2-2 | + | + | 100 | ||||||||||||||||
| L6-2-2-1 | + | + | |||||||||||||||||
| L7 | + | + | 75 | + | |||||||||||||||
| L7-1 | + | + | + | + | + | 75 | ND | ||||||||||||
| L7-1-1 | + | ND | 75 | ||||||||||||||||
| L7-1-1-1 | + | + | |||||||||||||||||
| L7-1-2 | + | ND | 75 | ||||||||||||||||
| L7-1-3 | + | ND | 100 | ||||||||||||||||
| + | L7-2 | + | + | + | + | + | 75 | ND | |||||||||||
| L7-2-1 | + | ND | |||||||||||||||||
| L7-2-2 | + | ND | 75 | ||||||||||||||||
| L8 | + | + | 100 | ND | L8-1 | + | + | + | + | + | ND | ||||||||
| L10 | + | + | 75 | + | |||||||||||||||
| L10-1 | + | + | + | + | + | 100 | ND | ||||||||||||
| L10-1-1 | + | ND | |||||||||||||||||
| L10-1-2 | + | ND | |||||||||||||||||
| L10-2 | + | + | + | + | + | ||||||||||||||
| 75 | + | ||||||||||||||||||
| L10-2-1 | + | + | |||||||||||||||||
| L10-2-2 | + | + | |||||||||||||||||
| L10-3 | + | + | + | + | + | ND | |||||||||||||
| L10-4 | + | + | + | + | + | ND | |||||||||||||
| L10-5 | + | + | + | + | + | ND | |||||||||||||
Percentage of hygromycin-resistant plantlets growing on hygromycin-containing germination medium.
Several hygromycin-resistant plantlets were transferred to soil and sprayed with Basta.
+, Positive phenotype.
ND, Not determined.
Figure 5.
Phenotypic analysis of nine-transgene-long transgenic Arabidopsis plants. A, Basta resistance in three independent transgenic lines. Clockwise from bottom left: offspring of T0 L10 plants, offspring of T1 L6-1 plants, offspring of T1 L7 plants, and wild-type plants. B, PAP1 phenotype in offspring of T2 L3-3-1 plants (right). Wild-type plants are shown on the left. C to F, EYFP-CHS expression (in yellow), ChrD-RFP and DsRed2-P expression (in red), plastid autofluorescence (in blue), and merged signals in leaf of T1 L6-1 plants. All panels are single confocal sections. G, Heat shock-induced GUS expression in transgenic leaves. Clockwise from top left: samples from L2-1-1, L2-2-1, L6-1-1, L10-2-1, L10-1-1, and L6-2-2. H, PAP1 phenotype in seedlings of offspring of T2 L6-2-2 plants (left). Seedlings of wild-type plants are shown on the right.
We further analyzed four transgenic lines to demonstrate the stability and integrity of the T-DNA inserts in T3 (lines L2 and L10) and T4 (lines L6 and L7) hygromycin-resistant plants by PCR amplification of overlapping fragments of their T-DNA region (Fig. 6). When combined with segregation analysis, our molecular data further supported the notion that our transgenic plants carry full T-DNA inserts. No less important, our data clearly show that multigene-expressing plants can be produced using our multigene transformation vectors.
Figure 6.
PCR analysis of T-DNA inserts in offspring of transgenic Arabidopsis plants. A, Structure of the nine-gene-long T-DNA molecule and sizes and locations of the overlapping amplified PCR fragments. Abbreviations are as in Figure 1. B, PCR analysis of binary plasmid and T3 (lines L2 and L10) and T4 (lines L6 and L7) hygromycin-resistant plants. Note the two distinct fragments in lane c, produced using a single pair of primers, due to the presence of repetitive elements on the nine-gene-long T-DNA molecule. The molecular marker (M) and expected sizes of the PCR fragments are given in kb.
DISCUSSION
We demonstrated that by using engineered ZFNs, we can overcome the limitation imposed by the small number of commercial homing endonucleases to the construction of multigene binary vectors. We used several ZFN- and homing endonuclease-constructed binary vectors for transient and stable genetic transformation of plant cells. We then showed that transgenic plants that had been stably transformed by nine-transgene-long T-DNA stably expressed the cloned genes across several generations. To the best of our knowledge, our report is one of just a small number of studies in which a very large number of independently expressed genes were delivered using a single T-DNA molecule into plant cells and, moreover, is the only report in which a multigene vector has been assembled by a modular, step-by-step construction method (for review, see Naqvi et al., 2010). Thus, our approach represents an important technical leap in the construction of multigene plant transformation vectors.
There are three main advantages of our system over other multigene construction systems, as detailed below. First, we demonstrated that ZFNs can be used in a manner similar to the technique that relies on homing endonucleases and that ZFNs can be exploited not only for adding but also for replacing DNA fragments from existing multigene binary vectors. Thus, in contrast to many other multigene vector assembly systems (Lin et al., 2003; Karimi et al., 2005; Wakasa et al., 2006; Chen et al., 2010), our system has the dual advantage of being modular and of enabling the modification of existing plant expression cassettes at any stage during the construction of the transformation vector. In addition, the cloning capacity of existing pRCS11.1-based binary vectors can be increased by reengineering their multiple cloning sites (MCSs) for additional features (which may include, for example, recognition sites for new ZFNs and homing endonucleases or even sites for Cre/lox and Gateway recombination systems) not only prior to but also during the process of assembling multigene vectors. This can be achieved by cloning an expanded ZFN-recognition MCS into the growing pRCS11.1-based binary vectors, which can then be used for successive additions of new expression cassettes. This feature further adds to the modularity of our ZFN-based vector assembly system, as compared with the more rigid design of other binary vector assembly systems.
Second, because ZFNs can be designed to target and digest an extremely large number of target sequences (Maeder et al., 2008), novel ZFNs can be developed and used to expand our nine-gene-long system beyond its current capacity. Since most pSAT vectors share a similar basic structure (Tzfira et al., 2005), it is easy to convert existing pSAT vectors into novel vectors by creating pSAT backbones with new ZFN recognition sites. Furthermore, since we used semipalindromic ZFN recognition target sequences, it may be feasible to adapt existing ZFNs (i.e. those that have been developed for targeting experiments in various nonplant eukaryotic cells [Urnov et al., 2005; Carroll et al., 2008; Zimmerman et al., 2008; Geurts et al., 2009; Takasu et al., 2010]) for cloning purposes. Indeed, we have previously shown that ZFNs can be expressed and purified to the level of molecular biology reagents by using relatively simple expression and purification steps (Zeevi et al., 2010; Tovkach et al., 2011). While specific modifications may be required to adapt our expression and purification system for novel ZFNs, the simplicity of the process and the availability of dedicated ZFN assembly and expression vectors for bacterial expression and for plant genome editing (Tovkach et al., 2010; Zeevi et al., 2010) may further facilitate the use of our system for the assembly of multigene vector systems.
Third, our system supports the use of a very large family of pSAT- and pAUX-based plasmids. The pSAT family of plasmids is composed of plasmids that have been designed to facilitate (1) the overexpression of target genes under the control of various promoters and terminators (Chung et al., 2005; Tzfira et al., 2005); (2) the analysis of protein-protein interactions by using the bimolecular fluorescence complementation and multicolor bimolecular fluorescence complementation assays (Citovsky et al., 2006; Lee et al., 2008); (3) the fusion of target genes to various autofluorescence proteins (Tzfira et al., 2005); (4) RNA interference-mediated down-regulation (Dafny-Yelin et al., 2007); (5) the expression of ZFNs (Tovkach et al., 2009, 2010); (6) Gateway-mediated gene cloning (Tzfira et al., 2005; Chakrabarty et al., 2007); and (7) the expression of epitope-tagged proteins (T. Tzfira, unpublished data). Thus, users of our system can enjoy a rich resource of plasmids that can be easily adapted to their various needs. Furthermore, since our cloning system is based on ZFN and homing endonucleases, it may be simple to transfer, or reconstruct, the ZFN and homing endonuclease MCS from pRCS11.1 into any other type of binary vectors and adapt our pSAT set of vectors for cloning on multigene vector systems in various binary plasmids. Such binary plasmids can, for example, be based on the transformation-competent artificial chromosome (Liu et al., 1999; Lin et al., 2003) and BIBAC (Hamilton, 1997; Frary and Hamilton, 2001) vectors, which can facilitate the cloning and transfer of an extremely large number of independent plant expression cassettes.
An important feature of our multigene vector assembly system lies in the unique structure of the final array of transgenes, in which each expression cassette or gene is flanked by pairs of unique ZFNs or homing endonucleases. This structure may facilitate genomic editing of multitransgene arrays in transgenic plants by harnessing the cell’s nonhomologous end-joining (NHEJ) DNA-repair pathway. Both homing endonucleases and ZFNs have been used for gene targeting in plant cells (i.e. site-specific mutagenesis, gene addition, deletion, and/or replacement; Salomon and Puchta, 1998; Chilton and Que, 2003; Tzfira et al., 2003; Cai et al., 2009; Shukla et al., 2009; Townsend et al., 2009). Yet, while most ZFN-mediated genome-editing systems rely on homologous recombination between the donor DNA and the target genome, the presence of repetitive elements (i.e. promoter and terminator sequences) in multitransgene arrays may hinder the homologous recombination-mediated genome editing of such structures. Thus, genome editing of multitransgene arrays (i.e. removal, replacement, and addition of expression cassettes by NHEJ) can potentially be achieved by transient ZFN expression in target cells, with or without the addition of a donor DNA molecule (Weinthal et al., 2010). Indeed, both I-SceI- and I-CeuI-mediated transgene addition (Salomon and Puchta, 1998; Chilton and Que, 2003; Tzfira et al., 2003) and ZFN-mediated transgene deletion (Petolino et al., 2010) have been reported in plant cells. In addition, we have recently demonstrated that ZFN- and NHEJ-mediated gene replacement is also feasible in plant species (D. Weinthal, T. Taylor, and T. Tzfira, unpublished data). Worth noting is the fact that by flanking each expression cassette with semipalindromic sequences, the editing process may be technically simplified, since, in contrast to most homologous recombination-mediated gene-replacement methods, it will require the expression of just one ZFN monomer and not pairs of ZFNs. Also worth noting is that other types of engineered enzymes, such as engineered meganucleases (Gao et al., 2010) and TALENs (Cermak et al., 2011; Mahfouz et al., 2011), have also been used for genome editing in plant cells. Developing procedures for the expression and purification of engineered meganucleases and TALENs for cloning purposes will further extend the repertoire of enzymes suitable for the assembly of multigene vector systems and their successive manipulation in plant cells.
The production of transgenic plants often results in a wide range of transgene-related phenotypes, a phenomenon that is typically attributed to a transgene positional affect (Matzke and Matzke, 1998). Thus, obtaining plants with superior transgenic performance (e.g. high and stable expression levels during the plant life span and across several generations) often calls for the production and screening of a large number of transgenic plants. Increasing the number of transgenes in plants is likely to increase the number of transgenic plants that need to be produced, screened, and selected for the desired performances. This procedure may be time consuming and labor intensive, especially if the different transgenes are scattered across the genome. Linking several transgenes to the same genomic location may assist in the screening process and in maintaining superior clones across several generations. The tendency of multiple DNA molecules, derived from the cotransformation of several different Agrobacterium strains or from cobombardment of several plasmids, to integrate into the same genomic locus (De Neve et al., 1997) has been utilized as a viable method for the production of single-locus multigene transgenic plants (for review, see Naqvi et al., 2010; Peremarti et al., 2010). While this approach has been successfully used to produce transgenic plants in which up to 11 transgenes are cointegrated into the same locus (Chen et al., 1998; Zhu et al., 2008; Naqvi et al., 2009), it is impossible to predict the physical organization and the arrangement of cotransformed DNA molecules in the transgenic plants. Indeed, it has been suggested that future progress in multigene transformation may depend not only on driving the transgenes under the specific combinations of promoter and terminator sequences but also on organizing the transgenes in a predetermined pattern (Peremarti et al., 2010). Our modular system can thus be efficiently adapted for the analysis of various regulatory elements and gene organization patterns by shuffling between different pSAT vectors.
We observed a variety of phenotypes among our different transgenic lines, as was evident, for example, by differences in hygromycin resistance (Fig. 3A) and GUS expression (Fig. 5G). We also observed that transgenic lines in which all the transgenes were expressed across several generations could be obtained (Table II). Yet, we could not identify a clear correlation between the phenotypes of different transgenes on a given T-DNA or between different lines. Thus, for example, lines that exhibited strong resistance to hygromycin did not necessarily exhibit high levels of PAP1 or GUS expression, while plants that exhibited high levels of PAP1 expression did not yield high intensity of their fluorescence genes. Similarly, Fujisawa et al. (2009) reported that they could not associate the expression level of individual transgenes with the performance of three transgenic lines; similarly, they could not determine the efficiency of a specific promoter in a given multigene array due to differences in the gene expression levels, which did not correlate with their promoter types. Similarly, Chen et al. (2010) reported phenotypic variation not only between the same trait in different transgenic lines but also between individual traits, driven by the same promoter in a given multigene array, in a selected transgenic line. It thus seems that not only may the expression levels of individual genes vary among different lines but also that similar promoters, driving different genes, may behave differently in a particular multigene cluster. We are currently applying our multigene vector assembly system to construct a set of multigene binary vectors in which identical expression cassettes will be cloned to different organizations. We will use our vectors to produce collections of independent transgenic plants that will be subjected to gene expression analysis in an attempt to produce the necessary data that will assist in defining the putative rules for the organization of regulatory elements within a given construct.
MATERIALS AND METHODS
Construction of pSAT Vectors
To construct pSAT12.MCS, we PCR amplified the pSAT6.MCS (Tzfira et al., 2005) backbone by using 5′-ATAAGAATGCGGCCGCGTAAGTGTTGGTGCTGTAAGTATGGATGCAGTAATCATGGTCATAGCTGTTTCC-3′ and 5′-GACGCACCGGTAGCACCAACACTTACGTTGGTGCTGGCACTGGCCGTCGTTTTACAACG-3′ and ligated the AgeI-NotI-digested PCR product to the AgeI-NotI 1.2-kb fragment from pSAT6.MCS (Tzfira et al., 2005). The construction of pSAT10.MCS and pSAT11.MCS was described previously (Zeevi et al., 2008). pRCS11.1 was constructed by cloning the SmaI fragment of self-annealed pair of primers 5′-TCCCCCGGGTTCCCACAAACTTACTTGTGGGAAAGCACCAACACTTACGTTGGTGCTCCCGGGGGA-3′ and 5′-TCCCCCGGGAGCACCAACGTAAGTGTTGGTGCTTTCCCACAAGTAAGTTTGTGGGAACCCGGGGGA-3′, which encode the ZFN11 and ZFN12 sites, and the KpnI fragment of self-annealed pair of primers 5′-GGGGTACCTGCATCCATGTAAGTATGGATGCAGGTACCCC-3′ and 5′-GGGGTACCTGCATCCATACTTACATGGATGCAGGTACCCC-3′, which encode ZFN10 into the same sites of pRCS2. The ChrD-RFP expression cassette was transferred from pSAT6A.ChrD-RFP (Citovsky et al., 2006) as an AgeI-NotI fragment into pSAT12.MCS to produce pSAT12.ChrD-RFP. The construction of pSAT10.EYFP-CHS and pSAT11.DsRed2-P has been described previously (Zeevi et al., 2008). To produce pSAT2.N, the nucleocapsid protein (N) of the SYNV expression cassette was transferred as an AgeI-NotI fragment from pSAT3.N (Tzfira et al., 2005) into pSAT2.MCS (Tzfira et al., 2005). pSAT5A.RbcsP.BAR was constructed by transferring the BAR-encoding sequence as an XmaI-XhoI fragment from pSAT1A.ocsP.BAR (Chung et al., 2005) into the same sites of pSAT5A.RbcsP.MCS (Chung et al., 2005). pSAT4.hsP.GUS was constructed by transferring the NcoI-BamHI GUS-coding sequence from pRTL2-GUS into pSAT4.hsP.MCS (Tovkach et al., 2009). pSAT6A.PAP1 was constructed by transferring the KpnI-BamHI PAP1-coding sequence from pCHS3-PAP1 (Borevitz et al., 2000) into pSAT6A.MCS (Chung et al., 2005).
Assembly, Expression, and Purification of ZFNs
The coding sequences of Arabidopsis (Arabidopsis thaliana) ZFN10, ZFN11, and ZFN12 were assembled from overlapping oligonucleotides and cloned into the bacterial expression vector pET28-XH to produce pET28-ZFN10, pET28-ZFN11, and pET28-ZFN12. The protocol for the molecular assembly of ZFNs from overlapping oligonucleotides, and their expression and purification, have been described previously in detail (Zeevi et al., 2010). Briefly, the ZFN DNA-binding coding sequences were assembled by a single PCR from a mixture of ZFN backbone primers and ZFN finger-specific primers. Each PCR product was then cloned into pET28-XH, where it was fused with the FokI endonuclease domain and a 6xHis tag. For expression in Escherichia coli cells, the ZFN expression vectors were transferred into BL21 GOLD (DE3) PlysS cells (Stratagene). The cells were cultured, harvested, and lysed, and the extracted proteins were purified on nickel-nitrilotriacetic acid agarose beads (Qiagen) as described previously (Zeevi et al., 2010). Eluted ZFNs were stored at −20°C in 50% glycerol. Alternatively, we used the Expressway in vitro protein synthesis system (Invitrogen) for in vitro expression of ZFNs.
Construction of Binary Vectors
For ZFN-mediated digestion of pSAT expression cassettes and binary plasmids, about 200 ng each of acceptor and donor plasmids was digested with 0.05 to 1 μL of purified enzyme in 10 mm Tris (pH 8.8), 50 mm NaCl, 1 mm dithiothreitol, 100 μm ZnCl2, 50 μg mL−1 bovine serum albumin, and 100 μg mL−1 tRNA in a total reaction volume of 20 to 30 μL. The reaction was preincubated for 30 min at room temperature, followed by the addition of MgCl2 to a final concentration of 5 mm. The digestion reaction mixture was further incubated for 2 to 40 min at room temperature. For homing endonuclease-mediated digestion of pSAT expression cassettes and binary plasmids, we followed the recommended reaction conditions for each enzyme. Cleaved fragments were separated by gel electrophoresis, purified with a GFX Gel Band Purification Kit (Amersham), dephosphorylated with shrimp alkaline phosphatase (Fermentas), ligated with T4 ligase (New England Biolabs), and transferred to chemically competent DH5α E. coli cells by using standard molecular biology protocols. The order in which the different pSAT expression cassettes were assembled into pRCS11.1 is described in “Results.” Binary vectors were transformed into chemically competent EHA105 Agrobacterium tumefaciens cells as described previously (Tzfira et al., 1997). pET28 ZFN expression plasmids and the pSAT and binary plasmids described in this study are available upon request.
Protoplast Transfection and Production of Transgenic Plants
The Tape-Arabidopsis Sandwich (Wu et al., 2009) method was used for protoplast isolation and transfection by multigene binary vectors. Transfected protoplasts were cultured in W5 solution on 1% bovine serum albumin-coated six-well plates for 16 to 24 h at 24°C to allow expression of the transfected DNA. Transgenic Arabidopsis plants were produced by using the standard flower-dip transformation method (Clough and Bent, 1998). Transgenic plants were selected on hygromycin selection medium. For analysis of resistance to Basta, hygromycin-resistant seedlings were transferred to soil, allowed to grow, and then sprayed with commercial Basta.
Confocal Microscopy
Protoplasts and plant tissues were viewed directly with a confocal laser-scanning microscope (TCS SP5; Leica). EYFP was excited with an argon laser at 514 nm, and fluorescence was monitored between 525 and 540 nm. DsRed2 and RFP were excited with a helium-neon laser at 561 nm, and fluorescence was monitored between 570 and 630 nm. Chlorophyll fluorescence was monitored above 660 nm.
References
- Ben Zvi MM, Negre-Zakharov F, Masci T, Ovadis M, Shklarman E, Ben-Meir H, Tzfira T, Dudareva N, Vainstein A. (2008) Interlinking showy traits: co-engineering of scent and colour biosynthesis in flowers. Plant Biotechnol J 6: 403–415 [DOI] [PubMed] [Google Scholar]
- Bohmert K, Balbo I, Kopka J, Mittendorf V, Nawrath C, Poirier Y, Tischendorf G, Trethewey RN, Willmitzer L. (2000) Transgenic Arabidopsis plants can accumulate polyhydroxybutyrate to up to 4% of their fresh weight. Planta 211: 841–845 [DOI] [PubMed] [Google Scholar]
- Bohmert K, Balbo I, Steinbüchel A, Tischendorf G, Willmitzer L. (2002) Constitutive expression of the β-ketothiolase gene in transgenic plants: a major obstacle for obtaining polyhydroxybutyrate-producing plants. Plant Physiol 128: 1282–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CQ, Doyon Y, Ainley WM, Miller JC, Dekelver RC, Moehle EA, Rock JM, Lee YL, Garrison R, Schulenberg L, et al. (2009) Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Mol Biol 69: 699–709 [DOI] [PubMed] [Google Scholar]
- Carroll D, Beumer KJ, Morton JJ, Bozas A, Trautman JK. (2008) Gene targeting in Drosophila and Caenorhabditis elegans with zinc-finger nucleases. Methods Mol Biol 435: 63–77 [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty R, Banerjee R, Chung S-M, Farman M, Citovsky V, Hogenhout SA, Tzfira T, Goodin M. (2007) PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol Plant Microbe Interact 20: 740–750 [DOI] [PubMed] [Google Scholar]
- Chen L, Marmey P, Taylor NJ, Brizard JP, Espinoza C, D’Cruz P, Huet H, Zhang S, de Kochko A, Beachy RN, et al. (1998) Expression and inheritance of multiple transgenes in rice plants. Nat Biotechnol 16: 1060–1064 [DOI] [PubMed] [Google Scholar]
- Chen QJ, Xie M, Ma XX, Dong L, Chen J, Wang XC. (2010) MISSA is a highly efficient in vivo DNA assembly method for plant multiple-gene transformation. Plant Physiol 153: 41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QJ, Zhou HM, Chen J, Wang XC. (2006) A Gateway-based platform for multigene plant transformation. Plant Mol Biol 62: 927–936 [DOI] [PubMed] [Google Scholar]
- Cheo DL, Titus SA, Byrd DR, Hartley JL, Temple GF, Brasch MA. (2004) Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: functional analysis of multi-segment expression clones. Genome Res 14: 2111–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilton M-D, Que Q. (2003) Targeted integration of T-DNA into the tobacco genome at double-stranded breaks: new insights on the mechanism of T-DNA integration. Plant Physiol 133: 956–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SM, Frankman EL, Tzfira T. (2005) A versatile vector system for multiple gene expression in plants. Trends Plant Sci 10: 357–361 [DOI] [PubMed] [Google Scholar]
- Citovsky V, Lee LY, Vyas S, Glick E, Chen MH, Vainstein A, Gafni Y, Gelvin SB, Tzfira T. (2006) Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol 362: 1120–1131 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dafny-Yelin M, Chung S-M, Frankman EL, Tzfira T. (2007) pSAT RNA interference vectors: a modular series for multiple gene down-regulation in plants. Plant Physiol 145: 1272–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafny-Yelin M, Tzfira T. (2007) Delivery of multiple transgenes to plant cells. Plant Physiol 145: 1118–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Dhingra A. (2002) Multigene engineering: dawn of an exciting new era in biotechnology. Curr Opin Biotechnol 13: 136–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Neve M, De Buck S, Jacobs A, Van Montagu M, Depicker A. (1997) T-DNA integration patterns in co-transformed plant cells suggest that T-DNA repeats originate from co-integration of separate T-DNAs. Plant J 11: 15–29 [DOI] [PubMed] [Google Scholar]
- Frary A, Hamilton CM. (2001) Efficiency and stability of high molecular weight DNA transformation: an analysis in tomato. Transgenic Res 10: 121–132 [DOI] [PubMed] [Google Scholar]
- Fujisawa M, Takita E, Harada H, Sakurai N, Suzuki H, Ohyama K, Shibata D, Misawa N. (2009) Pathway engineering of Brassica napus seeds using multiple key enzyme genes involved in ketocarotenoid formation. J Exp Bot 60: 1319–1332 [DOI] [PubMed] [Google Scholar]
- Gao H, Smith J, Yang M, Jones S, Djukanovic V, Nicholson MG, West A, Bidney D, Falco SC, Jantz D, et al. (2010) Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J 61: 176–187 [DOI] [PubMed] [Google Scholar]
- Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, et al. (2009) Knockout rats via embryo microinjection of zinc-finger nucleases. Science 325: 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goderis IJ, De Bolle MF, François IE, Wouters PF, Broekaert WF, Cammue BP. (2002) A set of modular plant transformation vectors allowing flexible insertion of up to six expression units. Plant Mol Biol 50: 17–27 [DOI] [PubMed] [Google Scholar]
- Goodin MM, Austin J, Tobias R, Fujita M, Morales C, Jackson AO. (2001) Interactions and nuclear import of the N and P proteins of Sonchus yellow net virus, a plant nucleorhabdovirus. J Virol 75: 9393–9406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. (2002) pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J 31: 375–383 [DOI] [PubMed] [Google Scholar]
- Halpin C, Barakate A, Askari BM, Abbott JC, Ryan MD. (2001) Enabling technologies for manipulating multiple genes on complex pathways. Plant Mol Biol 47: 295–310 [PubMed] [Google Scholar]
- Hamilton CM. (1997) A binary-BAC system for plant transformation with high-molecular-weight DNA. Gene 200: 107–116 [DOI] [PubMed] [Google Scholar]
- Hanin M, Paszkowski J. (2003) Plant genome modification by homologous recombination. Curr Opin Plant Biol 6: 157–162 [DOI] [PubMed] [Google Scholar]
- Hellens R, Mullineaux P, Klee H. (2000) Technical Focus. A guide to Agrobacterium binary Ti vectors. Trends Plant Sci 5: 446–451 [DOI] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P. (2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Lee LY, Fang MJ, Kuang LY, Gelvin SB. (2008) Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Liu YG, Xu X, Li B. (2003) Efficient linking and transfer of multiple genes by a multigene assembly and transformation vector system. Proc Natl Acad Sci USA 100: 5962–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Shirano Y, Fukaki H, Yanai Y, Tasaka M, Tabata S, Shibata D. (1999) Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning. Proc Natl Acad Sci USA 96: 6535–6540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyznik LA, Dress V. (2008) Gene targeting for chromosome engineering applications in eukaryotic cells. Recent Pat Biotechnol 2: 94–106 [DOI] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. (2008) Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell 31: 294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz MM, Li L, Shamimuzzaman M, Wibowo A, Fang X, Zhu JK. (2011) De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci USA 108: 2623–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani M, Kandavelou K, Dy FJ, Durai S, Chandrasegaran S. (2005) Design, engineering, and characterization of zinc finger nucleases. Biochem Biophys Res Commun 335: 447–457 [DOI] [PubMed] [Google Scholar]
- Matzke AJ, Matzke MA. (1998) Position effects and epigenetic silencing of plant transgenes. Curr Opin Plant Biol 1: 142–148 [DOI] [PubMed] [Google Scholar]
- Moon HS, Li Y, Stewart CN., Jr (2010) Keeping the genie in the bottle: transgene biocontainment by excision in pollen. Trends Biotechnol 28: 3–8 [DOI] [PubMed] [Google Scholar]
- Naqvi S, Farré G, Sanahuja G, Capell T, Zhu C, Christou P. (2010) When more is better: multigene engineering in plants. Trends Plant Sci 15: 48–56 [DOI] [PubMed] [Google Scholar]
- Naqvi S, Zhu C, Farre G, Ramessar K, Bassie L, Breitenbach J, Perez Conesa D, Ros G, Sandmann G, Capell T, et al. (2009) Transgenic multivitamin corn through biofortification of endosperm with three vitamins representing three distinct metabolic pathways. Proc Natl Acad Sci USA 106: 7762–7767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier MK, Shirley BW. (1996) Analysis of flavanone 3-hydroxylase in Arabidopsis seedlings: coordinate regulation with chalcone synthase and chalcone isomerase. Plant Physiol 111: 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremarti A, Twyman RM, Gómez-Galera S, Naqvi S, Farré G, Sabalza M, Miralpeix B, Dashevskaya S, Yuan D, Ramessar K, et al. (2010) Promoter diversity in multigene transformation. Plant Mol Biol 73: 363–378 [DOI] [PubMed] [Google Scholar]
- Petolino JF, Worden A, Curlee K, Connell J, Strange Moynahan TL, Larsen C, Russell S. (2010) Zinc finger nuclease-mediated transgene deletion. Plant Mol Biol 73: 617–628 [DOI] [PubMed] [Google Scholar]
- Porteus MH. (2009) Plant biotechnology: zinc fingers on target. Nature 459: 337–338 [DOI] [PubMed] [Google Scholar]
- Salomon S, Puchta H. (1998) Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J 17: 6086–6095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Sone T, Yoshida S, Yahata K, Hotta J, Chesnut JD, Honda T, Imamoto F. (2004) Evidence for high specificity and efficiency of multiple recombination signals in mixed DNA cloning by the multisite Gateway system. J Biotechnol 107: 233–243 [DOI] [PubMed] [Google Scholar]
- Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, et al. (2009) Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459: 437–441 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Komeda Y. (1989) Characterization of two genes encoding small heat-shock proteins in Arabidopsis thaliana. Mol Gen Genet 219: 365–372 [DOI] [PubMed] [Google Scholar]
- Takasu Y, Kobayashi I, Beumer K, Uchino K, Sezutsu H, Sajwan S, Carroll D, Tamura T, Zurovec M. (2010) Targeted mutagenesis in the silkworm Bombyx mori using zinc finger nuclease mRNA injection. Insect Biochem Mol Biol 40: 759–765 [DOI] [PubMed] [Google Scholar]
- Taverniers I, Papazova N, Bertheau Y, De Loose M, Holst-Jensen A. (2008) Gene stacking in transgenic plants: towards compliance between definitions, terminology, and detection within the EU regulatory framework. Environ Biosafety Res 7: 197–218 [DOI] [PubMed] [Google Scholar]
- Tovkach A, Zeevi V, Tzfira T. (2009) A toolbox and procedural notes for characterizing novel zinc finger nucleases for genome editing in plant cells. Plant J 57: 747–757 [DOI] [PubMed] [Google Scholar]
- Tovkach A, Zeevi V, Tzfira T. (2010) Validation and expression of ZFNs and in plant cells. Methods Mol Cell Biol 649: 315–336 [DOI] [PubMed] [Google Scholar]
- Tovkach A, Zeevi V, Tzfira T. (2011) Expression, purification and characterization of cloning-grade zinc finger nuclease. J Biotechnol 151: 1–8 [DOI] [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. (2009) High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 459: 442–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Frankman LR, Vaidya M, Citovsky V. (2003) Site-specific integration of Agrobacterium tumefaciens T-DNA via double-stranded intermediates. Plant Physiol 133: 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Jensen CS, Wangxia W, Zuker A, Altman A, Vainstein A. (1997) Transgenic Populus: a step-by-step protocol for its Agrobacterium-mediated transformation. Plant Mol Biol Rep 15: 219–235 [Google Scholar]
- Tzfira T, Tian G-W, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V. (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. (2005) Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435: 646–651 [DOI] [PubMed] [Google Scholar]
- van Engelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ. (1995) pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res 4: 288–290 [DOI] [PubMed] [Google Scholar]
- Vishnevetsky M, Ovadis M, Itzhaki H, Levy M, Libal-Weksler Y, Adam Z, Vainstein A. (1996) Molecular cloning of a carotenoid-associated protein from Cucumis sativus corollas: homologous genes involved in carotenoid sequestration in chromoplasts. Plant J 10: 1111–1118 [DOI] [PubMed] [Google Scholar]
- Vishnevetsky M, Ovadis M, Vainstein A. (1999) Carotenoid sequestration in plants: the role of carotenoid-associated proteins. Trends Plant Sci 4: 232–235 [DOI] [PubMed] [Google Scholar]
- Wakasa Y, Yasuda H, Takaiwa F. (2006) High accumulation of bioactive peptide in transgenic rice seeds by expression of introduced multiple genes. Plant Biotechnol J 4: 499–510 [DOI] [PubMed] [Google Scholar]
- Weinthal D, Tovkach A, Zeevi V, Tzfira T. (2010) Genome editing in plant cells by zinc finger nucleases. Trends Plant Sci 15: 308–321 [DOI] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS. (2009) Tape-Arabidopsis Sandwich: a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Truksa M, Datla N, Vrinten P, Bauer J, Zank T, Cirpus P, Heinz E, Qiu X. (2005) Stepwise engineering to produce high yields of very long-chain polyunsaturated fatty acids in plants. Nat Biotechnol 23: 1013–1017 [DOI] [PubMed] [Google Scholar]
- Zeevi V, Tovkach A, Tzfira T. (2008) Increasing cloning possibilities using artificial zinc finger nucleases. Proc Natl Acad Sci USA 105: 12785–12790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi V, Tovkach A, Tzfira T. (2010) Artificial zinc finger nucleases for DNA cloning. Methods Mol Biol 649: 209–225 [DOI] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH. (2007) Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225: 1603–1611 [DOI] [PubMed] [Google Scholar]
- Zhu C, Naqvi S, Breitenbach J, Sandmann G, Christou P, Capell T. (2008) Combinatorial genetic transformation generates a library of metabolic phenotypes for the carotenoid pathway in maize. Proc Natl Acad Sci USA 105: 18232–18237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman KA, Fischer KP, Joyce MA, Tyrrell DL. (2008) Zinc finger proteins designed to specifically target duck hepatitis B virus covalently closed circular DNA inhibit viral transcription in tissue culture. J Virol 82: 8013. [DOI] [PMC free article] [PubMed] [Google Scholar]