Abstract
The molecular mechanism of how the histone deacetylase HDA6 participates in maintaining transposable element (TE) silencing in Arabidopsis (Arabidopsis thaliana) is not yet defined. In this study, we show that a subset of TEs was transcriptionally reactivated and that TE reactivation was associated with elevated histone H3 and H4 acetylation as well as increased H3K4Me3 and H3K4Me2 in hda6 mutants. Decreased DNA methylation of the TEs was also detected in hda6 mutants, suggesting that HDA6 silences the TEs by regulating histone acetylation and methylation as well as the DNA methylation status of the TEs. Similarly, transcripts of some of these TEs were also increased in the methyltransferase1 (met1) mutant, with decreased DNA methylation. Furthermore, H4 acetylation, H3K4Me3, H3K4Me2, and H3K36Me2 were enriched at the coregulated TEs in the met1 and hda6 met1 mutants. Protein-protein interaction analysis indicated that HDA6 physically interacts with MET1 in vitro and in vivo, and further deletion analysis demonstrated that the carboxyl-terminal region of HDA6 and the bromo-adjacent homology domain of MET1 were responsible for the interaction. These results suggested that HDA6 and MET1 interact directly and act together to silence TEs by modulating DNA methylation, histone acetylation, and histone methylation status.
Transposable elements (TEs) constitute a major part of the complex genomes of plants and animals. The genome sequence analysis of Arabidopsis (Arabidopsis thaliana) has revealed that TEs are enriched in the centromeric region of chromosomes, which is highly methylated and packed into heterochromatin (Arabidopsis Genome Initiative, 2000). TEs are classical models for epigenetic inheritance, and silent transposons can be activated and inherited in the active state (Lippman et al., 2003). Recent studies revealed that DNA methylation, histone deacetylation, histone methylation, and RNA interference (RNAi) are involved in the activation or silencing of TEs (Murfett et al., 2001; Johnson et al., 2002). DNA methylation in mammalian genomes occurs predominantly in the context of the CG sequence and is maintained by the DNMT1 methyltransferase. In Arabidopsis, the DNMT1 homolog, METHYLTRANSFERASE1 (MET1), plays vital roles in maintaining cytosine methylation (Murfett et al., 2001). Antisense suppression or mutation of MET1 in Arabidopsis causes a global reduction in cytosine methylation, particularly at CG sites (Finnegan et al., 1996; Ronemus et al., 1996), and induces the release of TEs and transcriptional gene silencing (Kankel et al., 2003; Saze et al., 2003). In addition to DNA methylation, TEs are also subjected to the regulation mediated by histone deacetylation and methylation and RNAi (Lippman et al., 2003).
DNA methylation and histone deacetylation are two major epigenetic marks that contribute to the stability of gene expression status (MacDonald and Roskams, 2009). In several cases, gene silencing has been reported to be relieved by treatment with either histone deacetylase (HDAC) inhibitors or an inhibitor of DNA methylation (Chen and Pikaard, 1997; Pikaart et al., 1998; Selker, 1998). Inhibiting cytosine methylation induces histone acetylation, whereas inhibiting histone deacetylation causes the loss of cytosine methylation (Lawrence et al., 2004). In mammals, HDACs and DNMTs were suggested to act in the same protein complexes. HDACs can be recruited by high DNA methylation levels, via association with methyl-DNA-binding domain-containing proteins (Nan et al., 1998) or via direct recruitment by the DNA methyltransferase DNMT1 (Fuks et al., 2000), suggesting a tight interplay between histone deacetylation and DNA methylation.
In Arabidopsis, the histone deacetylase HDA6, a class I RPD3-like HDAC, is required for TE and rRNA gene silencing and cytosine methylation maintenance (Lippman et al., 2003; Earley et al., 2006, 2010). HDA6 was first identified to be involved in transgene silencing through an auxin-responsive element mutant screening (Murfett et al., 2001). HDA6 mutant alleles axe1-1 to axe1-5 displayed increased expression of the auxin-responsive reporter genes in the absence of auxin treatment, suggesting a role of HDA6 in gene silencing (Murfett et al., 2001). Another hda6 mutant allele, sil1, was also identified in a screen for mutations releasing transgene silencing, indicating that HDA6 is required for the maintenance of transcriptional gene silencing (Probst et al., 2004). Furthermore, HDA6 was also identified as an essential component in RNA-directed DNA methylation (RdDM; Aufsatz et al., 2002, 2007). The HDA6 mutant allele rts1 exhibits a reactivation of RdDM-silenced promoters and results in reduced cytosine methylation in symmetric sequence contexts, highlighting a function for HDA6 in methylation maintenance (Aufsatz et al., 2002, 2007). More recently, To et al. (2011) reported that HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. In addition, HDA6 and MET1 cotarget to the heterochromatin sites and maintain heterochromatin silencing (To et al., 2011). However, the molecular mechanism underlying the function of HDA6 in gene silencing is still unclear.
In this study, we found that a subset of TEs was reactivated in the hda6 mutants axe1-5 and sil1. In addition, the histone acetylation, histone methylation, and DNA methylation status of these TEs were affected in hda6 mutants. Direct protein-protein interaction between HDA6 and MET1 was detected by yeast two-hybrid, bimolecular fluorescence complementation (BiFC), and glutathione S-transferase (GST) pull-down assays. Furthermore, the landscapes of histone modification and DNA methylation at the TEs were also studied in met1-3 and axe1-5met1-3 plants. Our results indicate that HDA6 and MET1 interact directly and act together to maintain TE silencing by modulating their histone acetylation, methylation, and DNA methylation status.
RESULTS
HDA6 Maintains TE Silencing through Modulating Histone H3 and H4 Acetylation as Well as Histone H3K4 Methylation
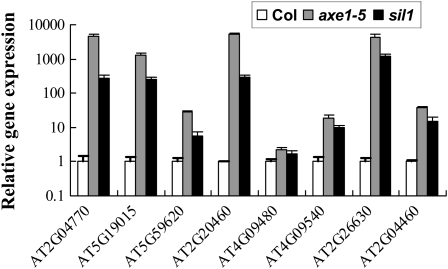
Our previous study revealed that a subset of TEs was up-regulated in axe1-5 and HDA6-RNAi plants (Yu et al., 2011). An additional hda6 mutant allele, sil1, in the Columbia (Col) background (Probst et al., 2004), was also obtained, and the expression of these transposons in sil1 and axe1-5 was compared. As shown in Figure 1, all eight transposons were remarkably activated in both axe1-5 and sil1 mutants, further supporting the notion that HDA6 is required for the silencing of TEs.
Figure 1.
Gene expression analysis of transposons in axe1-5 and sil1 mutants. Total RNA samples were extracted from 18-d-old seedlings growing under long-day conditions.
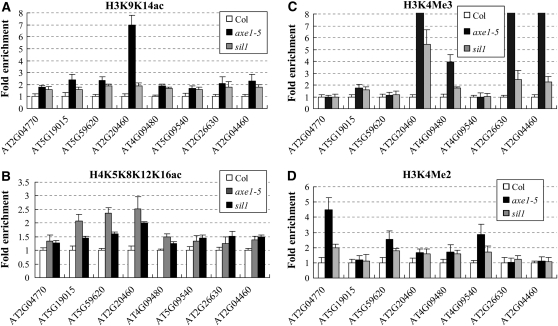
To investigate whether the reactivation of TEs was caused by histone acetylation, we measured the histone H3 and H4 acetylation levels of the TEs surrounding their transcription starting sites in axe1-5 and sil1 mutants by chromatin immunoprecipitation (ChIP) assay using antibodies specific for acetylated histone H3K9K14 and H4K5K8K12K16, respectively (Koch et al., 2008). As shown in Figure 2, A and B, the histone H3 and H4 acetylation levels of these TEs were elevated in axe1-5 and sil1 mutants compared with the wild type, suggesting that HDA6 may silence TEs by histone deacetylation.
Figure 2.
ChIP analysis of histone acetylation and methylation levels of the up-regulated TEs in axe1-5 and sil1 mutants. The immunoprecipitated DNA was quantified by real-time PCR. H3K9K14Ac (A), H4K5K8K12K16Ac (B), H3K4Me3 (C), and H3K4Me2 (D) levels of the regions surrounding the transcription starting sites were analyzed by normalization to an internal control (ACTIN2).
Since an increase in histone acetylation is often correlated with the methylation at Lys-4 of histone H3 (H3K4; Strahl and Allis, 2000), we further analyzed H3K4Me3 and H3K4Me2 levels of the up-regulated TEs in axe1-5 and sil1 plants. As shown in Figure 2C, in axe1-5 and sil1 mutants, the H3K4Me3 levels of AT5G19015, AT2G20460, AT4G09480, AT2G26630, and AT2G04460 were highly enriched, while no substantial changes were detected at AT2G04770, AT5G59620, and AT4G09540 loci. Furthermore, the H3K4Me2 levels of AT2G04770, AT5G59620, AT2G20460, AT4G09480, and AT4G09540 were elevated in axe1-5 and sil1 mutants (Fig. 2D). These data demonstrated that either H3K4Me3 or H3K4Me2 was accumulated in axe1-5 and sil1 plants. Therefore, the hyperacetylation of the TEs in hda6 mutants is correlated with an increase in H3K4 methylation. Taken together, our results suggested that HDA6 silences the TEs by modulating the histone H3 and H4 acetylation as well as H3K4 methylation levels.
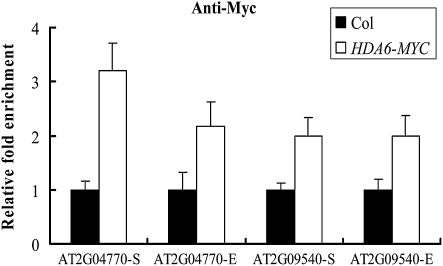
To investigate whether HDA6 directly regulates TEs in vivo, Arabidopsis plants overexpressing HDA6-MYC (35S:HDA6-TAP) were used to perform a ChIP assay with an anti-MYC antibody. Real-time PCR was used to analyze the relative abundance of HDA6 surrounding the transcription starting sites and coding regions of the representative TEs (AT2G04770 and AT2G09540). As seen in Figure 3, HDA6 was recruited to the transcription starting sites and coding regions of AT2G04770 and AT2G09540, suggesting that these TEs are direct targets of HDA6.
Figure 3.
HDA6 directly targets to TEs. Transgenic plants expressing HDA6-MYC were subjected to ChIP analysis using an anti-MYC antibody. Wild-type plants were used as negative controls. The relative fold enrichment of a target surrounding the transcription starting sites was calculated by dividing the amount of DNA immunoprecipitated from the HDA6-MYC transgenic plants by that from the negative control plants and comparing with input DNA. “S” means the transcriptional starting site, and “E” means the coding region.
HDA6 Functions in Maintaining CG, CHG, and CHH Methylation of TEs
To assess whether the reactivation of the TEs in the hda6 mutant is associated with changes in DNA methylation, a McrBC-PCR assay was used to analyze the DNA methylation status of TEs. McrBC is a DNA methylation-sensitive enzyme that preferentially cuts methylated DNA, and high levels of methylation result in increased McrBC digestion and, consequently, reduced amplification by PCR.
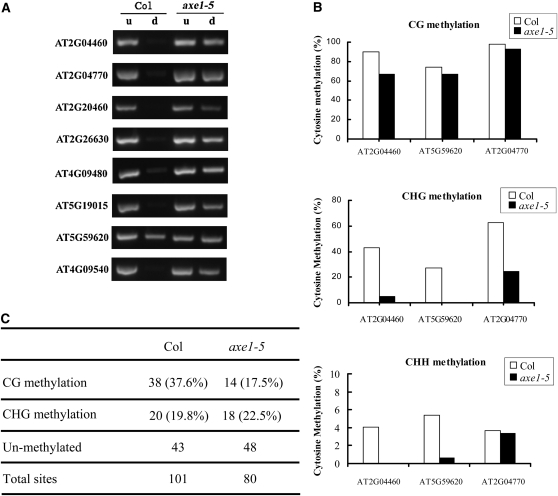
As shown in Figure 4A, in the wild type, no obvious DNA bands were detected in McrBC-digested samples of AT2G04770, AT5G19015, AT2G20460, AT4G09480, AT4G09540, AT2G26630, and AT2G04460, indicating that these HDA6-regulated TEs are highly methylated. In contrast, a strong DNA band was detected in a digested sample of AT5G59620, suggesting that this TE is relatively hypomethylated in the wild type. In the axe1-5 mutant, strong bands were scored in McrBC-digested samples of all the TEs, indicating a loss of cytosine methylation of the TEs. Taken together, these data demonstrate that HDA6 affects the DNA methylation status of the TEs.
Figure 4.
Cytosine methylation analyses of the up-regulated TEs in axe1-5. A, McrBC-PCR analysis of the DNA cytosine methylation status of the TEs in wild-type and axe1-5 plants. d, McrBC-digested DNA input; u, undigested DNA input. B, Bisulfite sequencing analysis of three representative TEs in axe1-5. C, Comparison of CG methylation and CHG methylation levels in CCGG sites in the wild type and axe1-5 by MSAP.
The DNA methylation status of three representative TEs, AT5G59620, AT2G04460, and AT2G04470, was further analyzed by bisulfite sequencing. As shown in Figure 4B and Supplemental Figure S1, the CG sites at these loci were heavily methylated in the wild type. In comparison, the methylation levels at CHG and CHH sites in these loci were much lower than those at CG sites in the wild type (Fig. 4B). An obvious decrease of CG methylation in AT2G04460 was detected in the axe1-5 mutant. Furthermore, substantial decreases of CHG and CHH were detected in AT2G04460 and AT5G59620, and the CHG methylation of AT2G04770 was also lost in the axe1-5 mutant. These data demonstrate that a loss of function of HDA6 results in a loss of cytosine methylation in CG, CHG, and CHH sites in TEs, further suggesting that HDA6 plays an important role in maintaining CG, CHG, and CHH methylation.
To explore the effect of HDA6 on genomic cytosine methylation, we employed methylation-sensitive amplification polymorphism (MSAP) to analyze the landscapes of this DNA modification in axe1-5 mutants. The MSAP assay is a modified version of amplified fragment length polymorphism analysis incorporating methylation-sensitive restriction enzymes in an efficient procedure to reveal genome-wide DNA methylation alterations in a locus-specific manner (Zhang et al., 2009). HpaII and MspI recognize the same restriction site (5′-CCGG) but have different sensitivities to methylation of the cytosines. HpaII does not cut if either of the cytosines is fully (double strand) methylated, whereas MspI does not cut if the external cytosine is fully or hemi (single) methylated. Thus, full methylation of the internal cytosine, or hemimethylation of the external cytosine at the assayed CCGG sites, can be unequivocally identified by MSAP. For clarity, we herein refer to these two types of patterns as CG and CHG methylation, respectively (Dong et al., 2006). By using eight pairs of selected EcoRI + HpaII/MspI primer combinations (Supplemental Table S1), 101 and 80 clear and reproducible bands were scored for the wild type and the axe1-5 mutant, respectively (Fig. 4C; Supplemental Fig. S2). A total of 37.6% (38) of the bands were detected to be CG methylated in the wild type. In contrast, only 22.5% (18) of CG methylation sites were found in the axe1-5 mutant, suggesting that the hda6 mutation induces a decrease in genomic cytosine methylation. These data further support the role of HDA6 in maintaining cytosine methylation.
Interactions of HDA6 with MET1 Both in Vitro and in Vivo
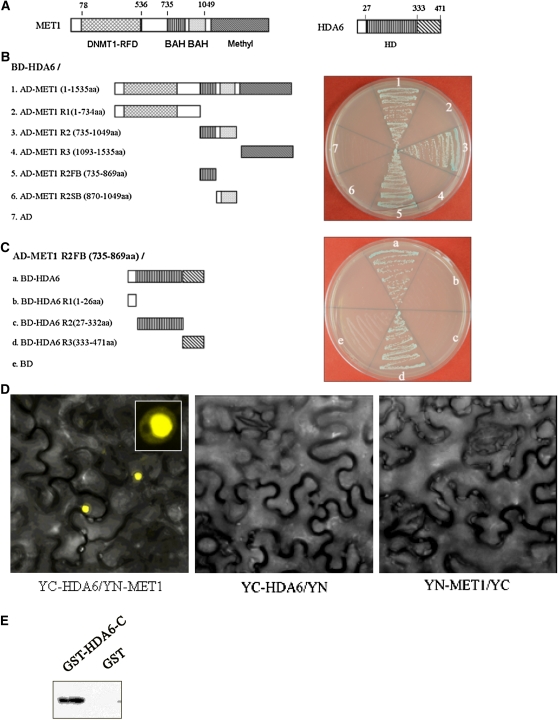
In Arabidopsis, the DNMT1 homolog MET1 plays vital roles in maintaining cytosine methylation (Finnegan et al., 1996). The finding that HDA6 is required for CG, CHG, and CHH methylation of TEs prompted us to investigate the interaction between HDA6 and MET1. Using the yeast two-hybrid assay, we found that HDA6 can interact with MET1 in yeast cells (Fig. 5B). Deletion analysis identified that the first bromo-adjacent homology (BAH) domain (amino acids 735–869) of MET1 and the C-terminal region (amino acids 333–471) of HDA6 were responsible for their interaction (Fig. 5, A–C).
Figure 5.
HDA6 interacts with MET1 both in vitro and in vivo. A, Diagram of MET1 and HDA6 protein domains. B, HDA6 interacts with MET1 and the first BAH domain of MET1 in yeast cells. C, The BAH domain of MET1 interacts with the C-terminal region of HDA6 in yeast cells. D, HDA6 interacts with MET1 in tobacco epidermal cells. E, The HDA6 C terminus interacts with the MET1 BAH domain in vitro. The GST-HDA6 C terminus and GST were incubated with MET1 R2 (BAH)-His and GST affinity resin, and the bound proteins were then eluted from resin and probed with the anti-His antibody.
The interaction of HDA6 and MET1 was further confirmed by the BiFC assay. MET1 and HDA6 were fused to the N-terminal 174-amino acid portion of yellow fluorescent protein (YFP) in the pEarleyGate201 vector (pEarleyGate201-YN) and the C-terminal 66-amino acid portion of YFP in the pEarleyGate202 vector (pEarleyGate202-YC), respectively (Lu et al., 2010). The corresponding fused constructs were codelivered into tobacco (Nicotiana benthamiana) leaves by infiltration with Agrobacterium tumefaciens GV3101 (Kerppola, 2006), and fluorescence was observed using a confocal microscope. As shown in Figure 5D, strong signal was observed in the nuclei of tobacco epidermal cells, suggesting a tight interaction between the HDA6 and MET1 proteins. In addition, the constructs were also codelivered into Arabidopsis protoplasts. The direct interaction was observed in the nucleus of Arabidopsis protoplasts (Supplemental Fig. S3).
Furthermore, we determined the interaction between HDA6 and MET1 by using an in vitro pull-down assay. The C-terminal region of HDA6 (amino acids 333–471) and the double BAH domain of MET1 (amino acids 735–1,049) were fused with GST and His tag, respectively (Eckner et al., 1994). The purified proteins were mixed and incubated in vitro, and the elution was detected with an anti-His antibody. As illustrated in Figure 5E, GST-HDA6-C (for C terminus) was pulled down by His-MET1-BAH, suggesting a direct association of HDA6 with MET1. Taken together, we conclude that HDA6 and MET1 interact both in vitro and in vivo and that the first BAH domain of MET1 and the C terminus of HDA6 are responsible for the interaction.
HDA6 and MET1 Act Together to Manipulate DNA Methylation, Histone Acetylation, and Histone Methylation of TEs
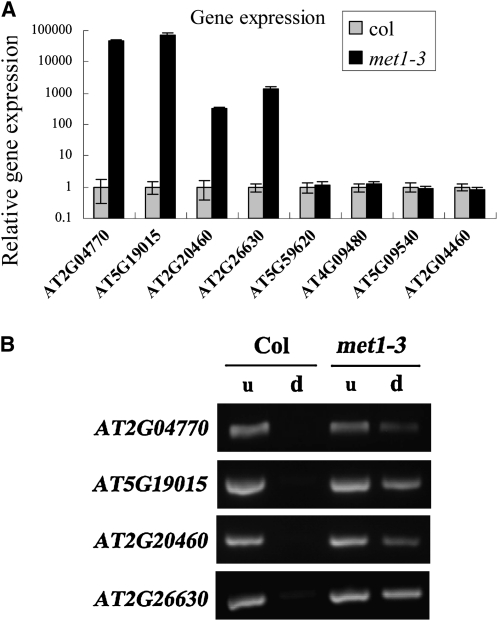
We further examined the genetic interaction between HDA6 and MET1 in regulating transposon silencing. We analyzed the expression of TEs in a MET1-null mutant, met1-3 (Saze et al., 2003). Among the eight TEs up-regulated in axe1-5 and sil1, four TEs (AT5G19015, AT2G26630, AT2G20460, and AT2G04770) were highly activated in met1-3 plants (Fig. 6A), indicating that they are regulated by both HDA6 and MET1. Three of these TEs, AT2G04770, AT5G19015, and AT2G26630, were also identified by To et al. (2011) to be coregulated by HDA6 and MET1. McrBC-PCR analysis shows that the DNA methylation levels of AT5G19015, AT2G26630, AT2G20460, and AT2G04770 were obviously decreased in met1-3 plants (Fig. 6B). These TEs were also hypomethylated in axe1-5 (Fig. 4A), suggesting that MET1 and HDA6 may act together to maintain the DNA methylation of these TEs.
Figure 6.
Gene expression (A) and cytosine methylation (B) analyses of HDA6-regulated TEs in met1-3 plants. d, McrBC-digested DNA input; u, undigested DNA input.
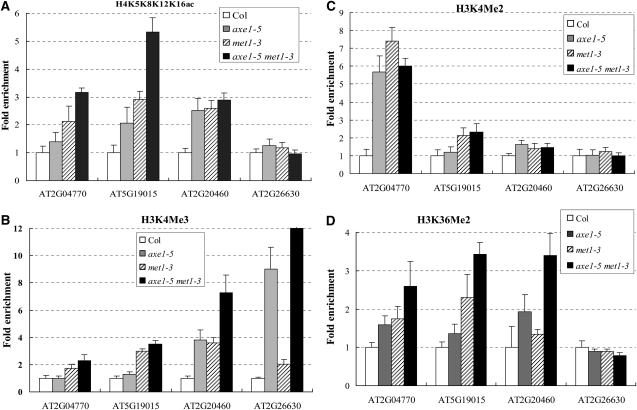
We further analyzed the axe1-5met1-3 double mutant and the histone landscapes of the four coregulated TEs by ChIP assay. Increased expression of coregulated TEs was observed in the hda6met1 double mutant (Supplemental Fig. S5). As shown in Figure 7A, histone H4 acetylation levels of AT2G04770, AT5G19015, and AT2G20460 were elevated in met1-3 mutant and axe1-5met1-3 double mutant plants. In addition, histone H4 acetylation levels of AT2G04770 and AT5G19015 were additively accumulated in axe1-5met1-3. These data indicate that HDA6 acts additively with MET1 to regulate the H4 acetylation of TEs.
Figure 7.
Histone modification landscapes of the TEs coregulated by HDA6 and MET1. ChIP analysis is shown for histone H4K5K8K12K16Ac (A), H3K4Me3 (B), H3K4Me2 (C), and H3K36Me2 (D) levels of the TEs in axe1-5, met1-3, and the axe1-5met1-3 double mutant.
Furthermore, H3K4Me3 and/or H3K4Me2 of these four TEs were also enriched in met1-3 mutant and axe1-5met1-3 double mutant plants (Fig. 7, B and C). H3K4Me3 of AT2G20460 and AT2G26630 in axe1-5met1-3 were higher than those of axe1-5 and met1-3 single mutants. Moreover, H3K36Me2 of AT2G04470, AT5G19015, and AT2G20460 were elevated in axe1-5 and met1-3 plants and additively increased in axe1-5met1-3 plants. These data suggest that HDA6 and MET1 coregulate the histone methylation profiles of TEs. Taken together, our results suggest that HDA6 and MET1 act together to regulate DNA methylation, histone acetylation, and histone methylation of the TEs in Arabidopsis.
DISCUSSION
HDA6 Silences TEs by Modulating Histone Acetylation, Histone Methylation, and DNA Methylation
Plant genomes contain many TEs, most of which are inactivated or “silenced” by chromatin-remodeling and DNA methylation factors (Okamoto and Hirochika, 2001). A subset of TEs is transcriptionally reactivated in hda6 mutants, suggesting that HDA6 is required for the silencing of TEs. Activation marks, such as histone H3 and H4 acetylation, H3K4 trimethylation or dimethylation, and H3K36 dimethylation, are increased, whereas cytosine methylation is decreased at the TEs in hda6 plants, suggesting that HDA6 silences the TEs by regulating these modifications. HDA6, therefore, may play important roles in the interplay among histone deacetylation, histone demethylation, and DNA methylation in transcriptional regulation.
HDA6 is a trichostain A-sensitive HDAC capable of removing acetyl groups from multiple Lys residues of histone (Earley et al., 2006, 2010). HDA6, therefore, may repress the TEs through direct deacetylating of their chromatin. The accumulation of H3K4Me3, H3K4Me2, and H3K36Me2 marks at the TEs in hda6 mutants also suggests a role for HDA6 in the repression of active histone methylation. The cross talk between histone deacetylation and demethylation has previously been implicated to modulate gene expression in mammalian cells (Shi et al., 2005; Lee et al., 2006). The histone demethylase LSD1 is an integral component of histone deacetylase corepressor complexes, in which HDACs and LSD1 may cooperate to remove activating acetyl and methyl histone modifications (Lee et al., 2006). More recently, we found that HDA6 physically associates with Flowering Locus D (FLD), a plant homolog of mammalian LSD1 (Yu et al., 2011). HDA6, therefore, may recruit a histone demethylase such as FLD to regulate the histone methylation of TEs.
Cytosine methylation levels in CHG and CHH sites were reduced in axe1-5 plants, which is associated with the loss of the H3K9Me2 mark (Supplemental Fig. S4). Previous studies revealed that methylation maintained by CHROMOMETHYLASE3 (CMT3) is dependent on H3K9Me2 (Johnson et al., 2002; Lindroth et al., 2004), indicating that HDA6 may act indirectly to maintain CHG and CHH methylation through the regulation of H3K9Me2 of TEs.
HDA6 Directly Interacts with MET1
The interplay between histone deacetylation and DNA methylation in gene silencing has been documented (Nan et al., 1998; Fuks et al., 2000; Lippman et al., 2003; To et al., 2011). In mammal cells, the DNA methyltransferase DNMT1 represses gene expression via an association with the methyl-CG-binding domain protein MBD, which in turn recruits histone deacetylase activities (Nan et al., 1998). Based on this model, MBD proteins act as a bridge in DNA methylation and histone deacetylation in gene silencing. A more direct connection between DNA methylation and deacetylation has also been demonstrated by the finding that the mammalian DNMT1 binds to HDAC1 using the N-terminal noncatalytic domain to repress gene transcription through histone deacetylase activity (Fuks et al., 2000; Rountree et al., 2000). Furthermore, DNMT1 also interacts directly with HDAC2 to form a repression complex at replication foci during late S phase (Rountree et al., 2000). Two de novo methyltransferases, DNMT3A and DNMT3B, interact with HDACs through their PHD-like motif (Bachman et al., 2001). These associations may help to establish the chromatin in late S phase and to maintain the repressive heterochromatin state throughout the cell cycle (Rountree et al., 2000; Bachman et al., 2001).
In this study, we demonstrated that the Arabidopsis DNMT1 homolog MET1 interacts with HDA6, suggesting a direct connection between DNA methylation and deacetylation in plants through the association of MET1 and HDA6. The direct interaction of HDA6 and MET1 may be necessary for the establishment and maintenance of the proper maturation of heterochromatin in Arabidopsis cells. Deletion analysis by yeast two-hybrid and GST pull-down assay showed that the BAH domain of MET1 and the C-terminal region of HDA6 were responsible for this interaction. The C-terminal region of HDA6 contains a SANT-like domain (Aufsatz et al., 2007), which is known to mediate DNA binding as well as protein-protein interactions (Aasland et al., 1996). Thus, the C terminus of HDA6 is an important region in maintaining cytosine methylation through interaction with MET1.
The interaction between HDA6 and MET1 in Arabidopsis was previously inferred based on genetic evidence (Lippman et al., 2003; To et al., 2011). By characterizing the chromatin modifications of a subset of transposons in mutants deficient for DNA methylation, histone deacetylation, and histone methylation, it was proposed that MET1, DDM1 (for deficient in DNA methylation 1), and HDA6 proteins may act together in a protein complex (Lippman et al., 2003). Gain of histone H3K4 methylation in met1 mutants and loss of DNA methylation in ddm1 and hda6 were found in representative transposons (Lippman et al., 2003). In this study, we also demonstrated increases of histone H4 acetylation and H3K4Me in met1-3 and a decrease of DNA methylation in axe1-5 in the TEs that are coregulated by MET1 and HDA6. More recently, To et al. (2011) identified a number of loci silenced by MET1 and/or HDA6. It was found that 81 and 733 Arabidopsis Genome Initiative loci were identified to be released in axe1-5 and met1-3, respectively, and 47 loci were common targets of HDA6 and MET1 (To et al., 2011). Far more loci show reactivation in the met1 mutant than in the hda6 mutant, suggesting that HDA6 is only required to silence some MET1 targets. The direct interaction of HDA6 and MET1 suggests that they can form a repression complex to regulate these common targets. Besides HDA6, a plant-specific histone deacetylase, HDT1, is also reported to be required for rRNA gene silencing. Blocking histone deacetylation by HDT1-RNAi induces a loss of cytosine methylation (Lawrence et al., 2004). Further research is required to determine whether MET1 may also associate other HDACs such as HDT1 to maintain the gene silencing of the other targets.
HDA6 and MET1 Act Together to Repress TEs
DNA methylation is associated with gene repression and plays a key role in protecting the genome from “selfish” DNA elements such as TEs (Chan et al., 2005). In Arabidopsis, DNA methylation occurs at both symmetric (CG and CHG) and asymmetric (CHH) sites. MET1 is responsible for the maintenance of symmetric CG cytosine methylation, while the plant-specific CMT3 is responsible for the maintenance of the CHG and asymmetric sites (Bartee et al., 2001; Cao et al., 2003). The loss of cytosine methylation in CG, CHG, and CHH sites of TEs in axe1-5 suggests that HDA6 may functionally associate with a DNA methyltransferase to maintain DNA methylation.
Treatment with the DNA methylation inhibitor aza-dC induces H3K9 acetylation, and the loss-of-function mutant of MET1 accumulates H4 acetylation of the Ta2 retrotransposon (Tariq et al., 2003; Lawrence et al., 2004). Furthermore, combined treatment with 5-AzaC and the histone deacetylation inhibitor TSA induces a more accessible chromatin structure of the METALLOTHIONEIN-I gene (Majumder et al., 2006), revealing a tight interplay between HDACs and DNMTs. In this work, we demonstrate that HDA6 mutations induce a reduction of DNA methylation and that a loss-of-function mutation of MET1 causes an increase in histone H4 acetylation of TEs, suggesting that the enzymatic activities of HDACs and DNMTs are closely linked in Arabidopsis. In addition, the axe1-5met1-3 double mutant displayed increases in histone H4 acetylation, H3K4Me2, H3K4Me3, and H3K36Me2 compared with the wild type, supporting a scenario in which HDA6 and MET1 act cooperatively to silence TEs. The additive increase of these histone modifications of some loci in the axe1-5met1-3 double mutant suggests that HDA6 and MET1 may collaborate with other chromatin-remodeling factors to modulate the chromatin landscapes. In this study, we demonstrated that HDA6 directly associates with MET1 to silence TEs by modulating DNA methylation, histone acetylation, and histone methylation status. Our study provides a new perspective to understand the interplay between histone deacetylation and DNA methylation in gene silencing.
In addition to interacting with MET1 to maintain gene silencing, HDA6 was characterized as an essential component in the RdDM pathway (Aufsatz et al., 2002; Probst et al., 2004). Among the eight up-regulated TEs in axe1-5 and sil1, AT2G04460, AT4G09540, AT4G09480, and AT5G59620 were not activated in the met1 mutant (Fig. 6A), implying that these TEs may not be regulated by MET1. A recent study revealed that the mutant RNA-directed DNA methylation4, a component in the RdDM pathway, releases the transcripts of AT2G04460 (He et al., 2009), suggesting that this TE may be silenced by the RdDM pathway.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) wild-type and hda6 mutant (axe1-5 and sil1) plants in the Col background were grown under long-day conditions (16 h of light, 8 h of darkness). axe1-5 was originally isolated based on deregulated expression of auxin-responsive transgenes and was outcrossed to the Col wild type three times (Yu et al., 2011). The sil1 mutant Col background was isolated by genetic screening for mutants showing ectopic expression of seed storage proteins in leaves, which carry a point mutation in the 46 bp after the ATG initiation codon, leading to the replacement of Gly-16 by Arg (Probst et al., 2004).
Real-Time Reverse Transcription-PCR Analysis
A total weight of 0.1 to 0.2 g of Arabidopsis leaves was ground using liquid nitrogen with a mortar and pestle and mixed with 1 mL of TRIZOL Reagent (Invitrogen) to isolate total RNA. One microgram of total RNA was used for the first-strand cDNA synthesis after DNase treatment. cDNA was synthesized in a volume of 20 μL that contained the Moloney murine leukemia virus reverse transcriptase buffer (Promega), 10 mm dithiothreitol, 1.5 μm poly(dT) primer, 0.5 mm deoxyribonucleotide triphosphates, 25 units of RNasin RNase inhibitor, and 200 units of Moloney murine leukemia virus reverse transcriptase at 37°C for 1 h.
cDNAs obtained from reverse transcription were used as templates to run real-time PCR. The following components were added to a reaction tube: 9 μL of iQ SYBR Green Supermix solution (catalogue no. 170-8882; Bio-Rad), 1 μL of 5 μm specific primers, and 8 μL of the diluted template. Thermocycling conditions were 95°C for 3 min followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 20 s, with a melting curve detected at 95°C for 1 min and 55°C for 1 min and denaturing time detected from 55°C to 95°C. Each sample was quantified at least in triplicate and normalized using UBIQUITIN10 as an internal control. The primer pairs for quantitative reverse transcription-PCR are listed in Supplemental Table S1.
ChIP Assay
The ChIP assay was carried out as described (Gendrel et al., 2005). Chromatin was extracted from 18-d-old plants growing in long-day conditions. After being fixed with formaldehyde, the chromatin was sheared to an average length of 500 bp by sonication and then immunoprecipitated with specific antibodies, including anti-acetyl-histone H3K9K14 (catalogue no. 06-599; Millipore), anti-acetyl-histone H4K5K8K12K16 (06-866; Millipore), anti-trimethyl-histone H3K4 (04-745; Millipore), anti-dimethyl-histone H3K4 (17-677; Millipore), anti-dimethyl-histone H3K9 (05-1250; Millipore), and anti-dimethyl-histone H3K36 (07-369; Millipore). The DNA cross-linked to immunoprecipitated proteins was analyzed by real-time PCR. Relative enrichments of transcriptional starting sites of transposons in axe1-5, met1-t3, and axe1-5met1-3 over Col were calculated after normalization to ACTIN2. Each of the immunoprecipitations was replicated three times with different sets of plants, and each sample was quantified at least in triplicate during real-time PCR analysis. The primers used for real-time PCR analysis in ChIP assays are listed in Supplemental Table S2.
McrBC-PCR and Bisulfite Sequencing
McrBC-PCR analysis was performed as described previously (Ding et al., 2007). A total of 100 ng of genomic DNA from 18-d-old Col, axe1-5, and met1-3 plants, growing in long-day conditions, was digested with 20 units of McrBC endonuclease (catalogue no. M0272S; New England Biolabs) for 8 h at 37°C. Following the McrBC treatment, subsequent PCR was used to analyze the methylation status of the transposons.
Bisulfite sequencing analysis was carried out with the EZ DNA Methylation-Gold Kit (catalogue no. D5005; Zymo Research). Briefly, 500-ng genomic DNA samples were treated with conversion reagents as described in the manufacturer’s instructions. The treated DNA was cleaned up and served as a template for subsequent PCR. The PCR products were cloned into pGEM-T Easy vector and then transformed into Escherichia coli. More than 15 individual clones for each genotype were sequenced and analyzed with the Web-based tool as described (Gruntman et al., 2008). The primers used for McrBC-PCR and bisulfite sequencing assays are listed in Supplemental Table S3.
MSAP
The MSAP assay was carried out as described (Zhang et al., 2009). Purified DNA was digested with enzyme combinations EcoRI/HpaII and EcoRI/MspII. Appropriate adapters (Supplemental Table S3) were ligated to the restricted ends. Each sample was subjected to primary and secondary amplification using the primers as listed in Supplemental Table S3. After denaturing, the PCR products were electrophoresed on 12% denaturing polyacrylamide gels and visualized by silver staining. Only clear and completely reproducible bands were scored.
Yeast Two-Hybrid Assay
Yeast two-hybrid assays were performed according to the instructions for the Matchmaker GAL4-based Two-Hybrid System 3 (Clontech). Constructs were generated by cloning full-length or different regions of MET1 and HDA6 cDNA fragments into pGADT7 and pGBKT7 vectors. All constructs were transformed into yeast strain AH109 by the lithium acetate method, and yeast cells were grown on minimal medium/-Leu-Trp according to the manufacturer’s instructions (Clontech). Transformed colonies were plated onto minimal medium/-Leu/Trp-His/X-α-gal containing 3-amino-1,2,4-triazole to test for possible interactions between MET1 and HDA6.
BiFC Assay
Full-length cDNA fragments of MET1 and HDA6 were subcloned into the pCR8/GW/TOPO vectors and then recombined into the pEarleyGate201-YN and pEarleyGate202-YC vectors (Lu et al., 2010). Constructed vectors were transiently transformed into Arabidopsis protoplasts (Yoo et al., 2007). Transfected cells were imaged using the Leica TCS SP5 confocal spectral microscopy imaging system.
To detect the interaction in tobacco (Nicotiana benthamiana), leaves of 2- to 4-week-old tobacco plants were infiltrated with Agrobacterium tumefaciens strain GV3101 containing HDA6 and MET1 BiFC construct pairs. Epidermal cell layers were examined 3 to 4 d after infiltration using the YFP filter.
In Vitro GST Pull-Down Assay
The GST pull-down assay was performed as described previously (Yang et al., 2008) with some modifications. GST and GST-HDA6 C-terminal recombinant proteins were incubated with 30 μL of GST resin in a binding buffer (50 mm Tris-Cl, pH 7.5, 100 mm NaCl, 0.25% Triton X-100, and 35 mm β-mercaptoethanol) for 2 h at 4°C. After the washing step with the binding buffer, the MET1 BAH domain-His recombinant protein was added and incubated for an additional 2 h at 4°C. Following extensive washing, the pulled down proteins were eluted by boiling, separated by 10% SDS-PAGE, and detected by western blotting using an anti-His antibody.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Detailed cytosine methylation profiles of three representative transposons.
Supplemental Figure S2. A representative SDS-PAGE gel image of MSAP analysis of the cytosine methylation profiles in 5′-CCGG sites in the axe1-5 mutant.
Supplemental Figure S3. HDA6 interaction with MET1 in Arabidopsis protoplasts, as detected by BiFC assay.
Supplemental Figure S4. ChIP analysis of H3K9Me2 levels of the TEs in axe1-5.
Supplemental Figure S5. Gene expression analysis of TEs coregulated by HDA6 and MET1 in axe1-5 met1-3 plants.
Supplemental Table S1. Primer pairs for quantitative reverse transcription-PCR.
Supplemental Table S2. Primers used for real-time PCR analysis in ChIP assays.
Supplemental Table S3. Primer used for DNA methylation analysis.
Acknowledgments
We are grateful to Technology Commons, College of Life Science, National Taiwan University, for the convenient use of the Bio-Rad real-time PCR system and the confocal spectral microscopy imaging system.
References
- Aasland R, Stewart AF, Gibson T. (1996) The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci 21: 87–88 [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJ. (2002) HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J 21: 6832–6841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufsatz W, Stoiber T, Rakic B, Naumann K. (2007) Arabidopsis histone deacetylase 6: a green link to RNA silencing. Oncogene 26: 5477–5488 [DOI] [PubMed] [Google Scholar]
- Bachman KE, Rountree MR, Baylin SB. (2001) Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J Biol Chem 276: 32282–32287 [DOI] [PubMed] [Google Scholar]
- Bartee L, Malagnac F, Bender J. (2001) Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev 15: 1753–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao XF, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE. (2003) Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 13: 2212–2217 [DOI] [PubMed] [Google Scholar]
- Chan SWL, Henderson IR, Jacobsen SE. (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6: 351–360 [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Pikaard CS. (1997) Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev 11: 2124–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Wang X, Su L, Zhai J, Cao S, Zhang D, Liu C, Bi Y, Qian Q, Cheng Z, et al. (2007) SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice. Plant Cell 19: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong ZY, Wang YM, Zhang ZJ, Shen Y, Lin XY, Ou XF, Han FP, Liu B. (2006) Extent and pattern of DNA methylation alteration in rice lines derived from introgressive hybridization of rice and Zizania latifolia Griseb. Theor Appl Genet 113: 196–205 [DOI] [PubMed] [Google Scholar]
- Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, Neves N, Gross M, Viegas W, Pikaard CS. (2006) Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev 20: 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Pontvianne F, Wierzbicki AT, Blevins T, Tucker S, Costa-Nunes P, Pontes O, Pikaard CS. (2010) Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev 24: 1119–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner R, Arany Z, Ewen M, Sellers W, Livingston DM. (1994) The adenovirus E1A-associated 300-kD protein exhibits properties of a transcriptional coactivator and belongs to an evolutionarily conserved family. Cold Spring Harb Symp Quant Biol 59: 85–95 [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES. (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA 93: 8449–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. (2000) DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet 24: 88–91 [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Martienssen R, Colot V. (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2: 213–218 [DOI] [PubMed] [Google Scholar]
- Gruntman E, Qi Y, Slotkin RK, Roeder T, Martienssen RA, Sachidanandam R. (2008) Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Zhu S, Liu HL, Pontes O, Zhu J, Cui X, Wang CS, Zhu JK. (2009) A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes Dev 23: 2717–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Cao XF, Jacobsen SE. (2002) Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr Biol 12: 1360–1367 [DOI] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. (2003) Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163: 1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK. (2006) Visualization of molecular interactions by fluorescence complementation. Nat Rev Mol Cell Biol 7: 449–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch G, Fernandez Del Olmo M, Cheeran B, Schippling S, Caltagirone C, Driver J, Rothwell JC. (2008) Functional interplay between posterior parietal and ipsilateral motor cortex revealed by twin-coil transcranial magnetic stimulation during reach planning toward contralateral space. J Neurosci 28: 5944–5953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, Viegas W, Pikaard CS. (2004) A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell 13: 599–609 [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Bochar DA, Hakimi MA, Cooch N, Shiekhattar R. (2006) Functional interplay between histone demethylase and deacetylase enzymes. Mol Cell Biol 26: 6395–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, Schubert I, et al. (2004) Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J 23: 4286–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, May B, Yordan C, Singer T, Martienssen R. (2003) Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol 1: E67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Tang X, Tian G, Wang F, Liu K, Nguyen V, Kohalmi SE, Keller WA, Tsang EW, Harada JJ, et al. (2010) Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J 61: 259–270 [DOI] [PubMed] [Google Scholar]
- MacDonald JL, Roskams AJ. (2009) Epigenetic regulation of nervous system development by DNA methylation and histone deacetylation. Prog Neurobiol 88: 170–183 [DOI] [PubMed] [Google Scholar]
- Majumder S, Kutay H, Datta J, Summers D, Jacob ST, Ghoshal K. (2006) Epigenetic regulation of metallothionein-i gene expression: differential regulation of methylated and unmethylated promoters by DNA methyltransferases and methyl CpG binding proteins. J Cell Biochem 97: 1300–1316 [DOI] [PubMed] [Google Scholar]
- Murfett J, Wang XJ, Hagen G, Guilfoyle TJ. (2001) Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell 13: 1047–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393: 386–389 [DOI] [PubMed] [Google Scholar]
- Okamoto H, Hirochika H. (2001) Silencing of transposable elements in plants. Trends Plant Sci 6: 527–534 [DOI] [PubMed] [Google Scholar]
- Pikaart MJ, Recillas-Targa F, Felsenfeld G. (1998) Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev 12: 2852–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Fagard M, Proux F, Mourrain P, Boutet S, Earley K, Lawrence RJ, Pikaard CS, Murfett J, Furner I, et al. (2004) Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16: 1021–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus MJ, Galbiati M, Ticknor C, Chen J, Dellaporta SL. (1996) Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273: 654–657 [DOI] [PubMed] [Google Scholar]
- Rountree MR, Bachman KE, Baylin SB. (2000) DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet 25: 269–277 [DOI] [PubMed] [Google Scholar]
- Saze H, Mittelsten Scheid O, Paszkowski J. (2003) Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet 34: 65–69 [DOI] [PubMed] [Google Scholar]
- Selker EU. (1998) Trichostatin A causes selective loss of DNA methylation in Neurospora. Proc Natl Acad Sci USA 95: 9430–9435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. (2005) Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 19: 857–864 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Tariq M, Saze H, Probst AV, Lichota J, Habu Y, Paszkowski J. (2003) Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc Natl Acad Sci USA 100: 8823–8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To TK, Kim JM, Matsui A, Kurihara Y, Morosawa T, Ishida J, Tanaka M, Endo T, Kakutani T, Toyoda T, et al. (2011) Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet 7: e1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Iwasaki M, Machida C, Machida Y, Zhou X, Chua NH. (2008) BetaC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev 22: 2564–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu CW, Liu X, Luo M, Chen C, Lin X, Tian G, Lu Q, Cui Y, Wu K. (2011) HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol 156: 173–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Xu C, Yan H, Zhao N, von Wettstein D, Liu B. (2009) Limited tissue culture-induced mutations and linked epigenetic modifications in F hybrids of sorghum pure lines are accompanied by increased transcription of DNA methyltransferases and 5-methylcytosine glycosylases. Plant J 57: 666–679 [DOI] [PubMed] [Google Scholar]