Abstract
Snakin-1 (SN1) is an antimicrobial cysteine-rich peptide isolated from potato (Solanum tuberosum) that was classified as a member of the Snakin/Gibberellic Acid Stimulated in Arabidopsis protein family. In this work, a transgenic approach was used to study the role of SN1 in planta. Even when overexpressing SN1, potato lines did not show remarkable morphological differences from the wild type; SN1 silencing resulted in reduced height, which was accompanied by an overall reduction in leaf size and severe alterations of leaf shape. Analysis of the adaxial epidermis of mature leaves revealed that silenced lines had 70% to 90% increases in mean cell size with respect to wild-type leaves. Consequently, the number of epidermal cells was significantly reduced in these lines. Confocal microscopy analysis after agroinfiltration of Nicotiana benthamiana leaves showed that SN1-green fluorescent protein fusion protein was localized in plasma membrane, and bimolecular fluorescence complementation assays revealed that SN1 self-interacted in vivo. We further focused our study on leaf metabolism by applying a combination of gas chromatography coupled to mass spectrometry, Fourier transform infrared spectroscopy, and spectrophotometric techniques. These targeted analyses allowed a detailed examination of the changes occurring in 46 intermediate compounds from primary metabolic pathways and in seven cell wall constituents. We demonstrated that SN1 silencing affects cell division, leaf primary metabolism, and cell wall composition in potato plants, suggesting that SN1 has additional roles in growth and development beyond its previously assigned role in plant defense.
Snakin-1 (SN1) is an antimicrobial Cys-rich peptide isolated from potato (Solanum tuberosum) tubers that has been found to be active against different plant pathogens in vitro (Segura et al., 1999). Recently, we demonstrated that overexpression of the SN1 gene in potato plants enhances resistance to Rhizoctonia solani and Erwinia carotovora, showing that it also has in vivo antifungal and antibacterial activities (Almasia et al., 2008). SN1 was classified as a member of the Snakin/Gibberellic Acid Stimulated in Arabidopsis (GASA) protein family based on its amino acid sequence homology with peptides of the Arabidopsis (Arabidopsis thaliana) GASA family (Berrocal-Lobo et al., 2002). Homologous genes have been identified in tomato (Solanum lycopersicum; GAST; Shi et al., 1992), petunia (Petunia hybrida; GIP; Ben-Nissan and Weiss, 1996), gerbera (Gerbera hybrida; GEG; Kotilainen et al., 1999), rice (Oryza sativa; OsGASR; Furukawa et al., 2006), strawberry (Fragaria spp.; FaGAST; de la Fuente et al., 2006), and maize (Zea mays; Zimmermann et al., 2010), among other species. All these genes encode small proteins with a putative signal peptide at the N terminus and a C-terminal region of approximately 60 amino acids containing 12 Cys residues in conserved positions. Despite their common features, there is no apparent consensus in the roles they play, and little is known about their mode of action. Based on their sequences, the phenotypic characterization of transgenic plants, and studies of their expression patterns, it has been suggested that these peptides are involved in diverse cellular processes. Expression analyses of GIP genes suggest that GIP1 and GIP2 act during cell elongation, while GIP4 and GIP5 act in cell division (Ben-Nissan and Weiss, 1996; Ben-Nissan et al., 2004). Interestingly, it has been suggested that GIP2 affects flowering and stem elongation by modulating the levels of reactive oxygen species (Wigoda et al., 2006). In line with these findings, Roxrud et al. (2007) suggested that GASA4 might be involved in cell division of flower bud, and Zhang et al. (2009) demonstrated that GASA5 is implicated in controlling flowering time and stem growth. Recently, Rubinovich and Weiss (2010) reported that GASA4 is involved in the promotion of GA responses, acting as a redox regulator through its Cys-rich domain. Altogether, these results suggest that GASA family genes play important roles in the development programs of higher plants. Even though the two potato members of this family isolated to date (snakin-1 and snakin-2) were reported to have antimicrobial activity (Segura et al., 1999; Berrocal-Lobo et al., 2002; Almasia et al., 2008), their involvement in other plant cellular processes is not incompatible with this defense role. Examples of such dual-function peptides have been recently reported; some defensins (i.e. MsDef1, MtDef2, RsAFP2, and DEF2), small Cys-rich antimicrobial proteins, have been shown to have additional roles in plant growth and development (Allen et al., 2008; Stotz et al., 2009).
Plant development results from the ordered expansion of cells that proliferate in meristems. The formation of a plant organ can be regarded as a result of cell multiplication through cell division and subsequent cell expansion; hence, the final morphology of a plant is achieved through a tight regulation of both processes (Meyerowitz, 1997). Inhibition of leaf cell proliferation in Arabidopsis transgenic lines resulted in alterations of organ growth and shape, suggesting the involvement of cell division in morphogenesis (Wang et al., 2000; Verkest et al., 2005). The determination of cell morphology and size during plant development is also conditioned by the highly complex structure of the cell wall. The cell wall constitutes a dynamic and responsive structure that regulates plant responses to stresses and external stimuli, including interaction with diverse pathogens (Humphrey et al., 2007), and relays information about the environment to the cell cytoplasm via signal transduction pathways (Hématy et al., 2009). The growing cell wall has a fiberglass-like structure with crystalline cellulose microfibrils that are embedded in a matrix of complex polysaccharides, which are divided into two classes, hemicelluloses and pectins (Cosgrove, 2005), as well as proteins. Hemicelluloses are branched polysaccharides containing backbones of neutral sugars, whereas pectins are defined by the presence of uronic acids as major components (Somerville et al., 2004). Even though the synthesis of cell wall polysaccharides is poorly understood, it is known that many of the nucleotide sugars needed for cell wall formation are also intermediates of essential biosynthetic pathways, such as ascorbate biosynthesis (Gilbert et al., 2009; Di Matteo et al., 2010). Ascorbic acid is synthesized and accumulated in large amounts in leaves. It has multiple roles in metabolism, electron transport, and plant responses to pathogens and abiotic stress, and it is also considered to influence plant growth and development through its effects on the cell cycle and cell elongation (Olmos et al., 2006).
In this work, we describe the characterization of transgenic potato lines with altered expression of SN1 to gain a better understanding of the role of this gene in planta. We demonstrate that SN1 silencing affects cell division, primary metabolism, and cell wall composition in leaves, suggesting that SN1 has additional roles in growth and development beyond its recognized role in defense.
RESULTS
Molecular Characterization of SN1 Transgenic Potato Lines
Our group has previously obtained SN1-overexpressing lines that showed enhanced antimicrobial resistance and were phenotypically indistinguishable from the wild-type control (Almasia et al., 2008). In this work, cv Kennebec transgenic potato lines expressing an antisense construct of this gene (pPZPK-SN1Antisense) were produced. Five out of 16 PCR-confirmed independent transformation events (A1–A5) showed an abnormal phenotype with reduced height and alterations of leaf shape (Fig. 1A). Southern blot revealed that A1 carried multiple insertions; therefore, it was left aside for subsequent analyses. A2 to A5 lines had one or two copies of the transgene (data not shown). Since the most severe phenotype lines (A2 and A3) also had the lowest levels of SN1 transcript, they were selected for further characterization. As shown in Figure 1B, overexpressing lines (S3 and S5) accumulated high levels of SN1 transcript, while A2 and A3 exhibited substantially reduced levels compared with the wild type.
Figure 1.
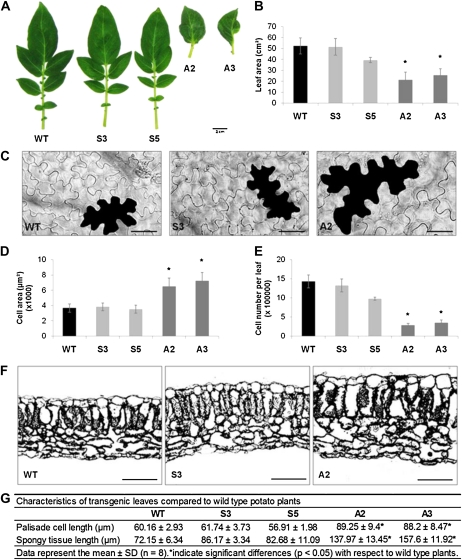
A, Phenotypes SN1-overexpressing and -silenced lines. B, Total SN1 mRNA accumulation in transgenic and wild-type plants by northern blot. The bottom panel shows an ethidium bromide (EtBr)-stained gel. WT, Wild type; S3 and S5, SN1-overexpressing lines; A2 and A3, SN1-silenced lines.
SN1 Silencing Affects Leaf Morphology
In order to further characterize the leaf phenotype, selected overexpressing and silenced lines were clonally propagated and grown under greenhouse conditions along with the wild type, and the fifth leaf (counted down from the apex) from 8-week-old plants was used for further analyses. Silenced lines showed malformed leaves that exhibited no leaflets accompanied by an overall reduction in leaf size (Fig. 2A). Measurements of fully expanded leaves revealed that SN1-silenced lines had a leaf area reduction of 50% to 60% with respect to the wild type (Fig. 2B).
Figure 2.
Leaf phenotypic characterization of wild-type and transgenic lines. A, Overall size and shape of fully expanded leaves. Bar = 2 cm. B, Average total area of 10 leaves. C, Representative images of adaxial epidermal cells from wild-type, S3, and A2 plants. Bars = 50 μm. D, Average area of 100 to 150 epidermal cells. E, Estimated total number of epidermal cells per leaf. F, Light micrographs of fully expanded leaf cross-sections. Bars = 100 μm. G, Measurement of palisade cell length and spongy tissue length of leaf cross-sections. Data represent means ± sd. Asterisks indicate significant differences (P < 0.05) with respect to the wild type. WT, Wild type; S3 and S5, SN1-overexpressing lines; A2 and A3, SN1-silenced lines.
As leaf size is determined by cell proliferation and expansion, we next assessed whether the reduced leaf area observed in silenced lines correlated with a reduction in cell size and/or in cell number. Analyses of adaxial epidermis of mature leaves revealed that SN1-silenced lines had much larger cells than wild-type plants (Fig. 2C). Cell size quantification from confocal images showed that A2 and A3 plants had 70% to 90% increases in mean cell size with respect to wild-type leaves (Fig. 2D).
Next, the average cell number per leaf was estimated by dividing total leaf area by average cell area. The number of epidermal cells on the adaxial side of the leaves was significantly reduced in silenced lines (Fig. 2E). In contrast, overexpression of SN1 did not result in significant changes in leaf area or cell size of adaxial epidermis; therefore, no differences in estimated cell number per leaf were observed in S3 and S5 lines (Fig. 2, B, D, and E).
To determine whether SN1-altered expression affected leaf histological structure, cross-sections of wild-type and transgenic line leaves were examined. The arrangement of parenchyma cell layers remained largely unaffected in all transgenic lines; however, leaves from silenced lines exhibited an increased thickness (Fig. 2F). For quantitative comparisons, the parenchyma tissue lengths were measured, showing that palisade and spongy tissues were significantly increased in silenced lines, while no significant changes were found in overexpressing lines (Fig. 2G).
SN1 Transgenic Lines Differ in Leaf Primary Metabolism
Given the above-mentioned results, we next focused on the analysis of leaf primary metabolism by applying gas chromatography coupled to mass spectrometry (GC-MS) and spectrophotometric techniques. Fully expanded leaves from 8-week-old plants grown under greenhouse conditions were harvested in the middle of the light period, and crude extracts from the soluble fraction were prepared as described by Roessner et al. (2001) and Fernie et al. (2001). These targeted analyses allowed a detailed examination of the changes occurring in 46 intermediate compounds from primary metabolic pathways (Supplemental Table S1) as a consequence of SN1 overexpression or silencing in transgenic potato lines.
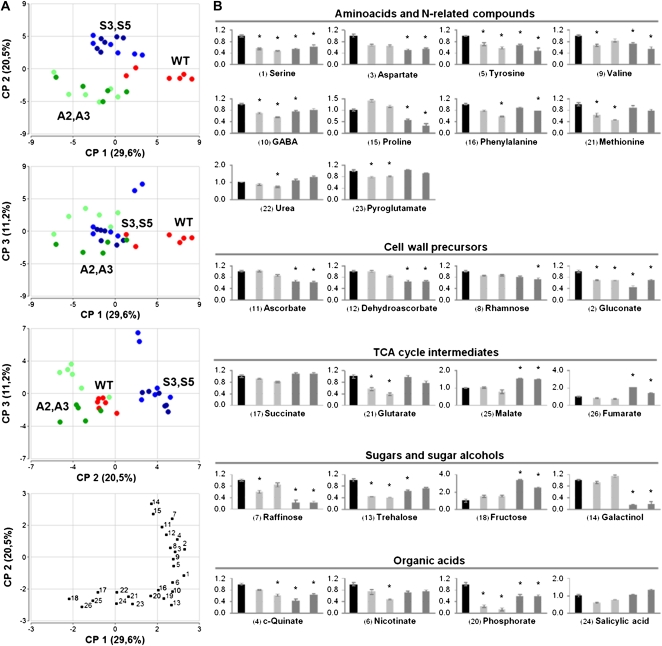
In order to interpret the metabolic differences occurring in SN1 transgenic lines, a nonparametric analysis was applied. A model of principal component analysis (PCA) explains 61% of the data variance, showing that the first and second components discriminate SN1 transgenic from wild-type samples (Fig. 3A). Investigation of the relative contribution (loadings) of individual variables in these two dimensions highlighted 26 metabolites with major impact on genotype separation by principal components (PCs) PC1 and PC2 (0.18 or greater and −0.18 or less). All of these variables corresponded to sugars and sugar alcohols (Fru, trehalose, raffinose, and galactinol), tricarboxylic acid (TCA) cycle intermediates (glutarate, malate, fumarate, and succinate), amino acids and nitrogen-related compounds (Phe, Met, pyro-Glu, γ-aminobutyrate, Ser, Asp, Tyr, Val, Pro, and urea), organic acids (quinate, nicotinate, salicylate, and phosphorate), and, interestingly, four metabolites directly involved in cell wall metabolism (Rha, gluconate, ascorbate, and dehydroascorbate; Fig. 3B).
Figure 3.
Metabolic profiles of wild-type (WT) and transgenic potato leaves. A, PCA of metabolic profiles of leaf samples from wild-type and transgenic potato plants. The percentage of variance explained by each component is shown in parentheses. Samples represent leaves from the wild type (red circles), overexpressing lines S3 and S5 (light blue and blue circles), and silenced lines A2 and A3 (light green and green circles). Each data point represents an independent sample. The bottom panel shows 26 specific metabolites with major impact on genotype separation by PC1 and PC2 (0.18 or greater and −0.18 or less). B, Relative contents of the 26 specific metabolites identified and quantified by GC-MS in the bottom panel in A. Bars represent the following lines: wild type (black), S3 and S5 (light gray), and A2 and A3 (dark gray). Significant differences from the wild type according to Student’s t test are indicated with asterisks (P ≤ 0.05).
Comparison of the relative quantities of soluble compounds revealed a high number of changes occurring in the levels of primary nitrogen and carbon intermediate metabolites in transgenic lines with respect to their wild type (Supplemental Table S1). Amino acid levels were lower both in overexpressing and silenced lines, and TCA cycle intermediates also were altered, these changes being more pronounced in the silenced lines. Among sugars and sugar alcohols, Glc and Fru showed more than 2-fold increases in the A2 and A3 SN1-silenced lines, while raffinose, galactinol, and the precursor of cell wall Rha were reduced in these lines. In agreement with these results is the finding that the precursor of ascorbic acid biosynthesis, gluconate 1,4-lactone, decreased in A2 and A3, as did the levels of ascorbate and its end product, dehydroascorbate.
SN1-Silenced Lines Differ in Cell Wall Composition
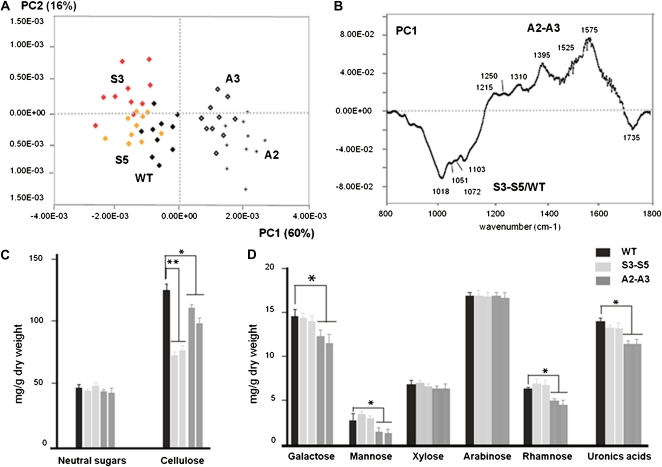
Cell walls from overexpressing and silenced lines as well as wild-type leaves were subjected to Fourier transform infrared (FTIR) spectroscopy analysis to obtain qualitative spectratypes (i.e. cell wall phenotypes). The overall comparison of averaged FTIR spectra from two biological replicates (n = 10) showed strong differences between wild-type/S3-S5 and A2-A3 plants (data not shown). Exploratory PCA showed that most of the total sample variation (60%) was explained by PC1 (Fig. 4A), where overexpressing lines segregated closer to the wild type and were clearly distant from A2 and A3. Absorbances in PC1 at 1,018, 1,051, 1,072, and 1,103 cm−1, common in the fingerprint for several cell wall polysaccharides (Kacuráková et al., 2000), were comparatively more abundant in S3-S5/wild-type samples than in silenced lines, whereas bands at 1,215, 1,250, 1,310, 1,395, 1,525, and 1,575 cm−1 seemed to increase in A2-A3 (Fig. 4, A and B). The complexity of the PC1 spectrum suggests that the cell wall differences between A2-A3 lines and wild-type plants cannot be assigned to a defect in a single cell wall polymer. This also suggests that reduced SN1 levels are directly or indirectly affecting the cell wall composition.
Figure 4.
FTIR-PCA and monosaccharide analyses of SN1-overexpressing lines (S3-S5), SN1-silenced lines (A2-A3), and wild-type (WT) samples. A, Variability of cell wall samples of potato analyzed by FTIR-PCA. Biplot PC1-PC2 shows the separation of S3-S5/wild-type samples from A2-A3 samples in PC1. Mid-IR spectra were analyzed by the covariance-matrix approach for PCA (Kemsley, 1996). B, Loading of PC1, where major IR bands are shown. C, Neutral sugars and cellulose content (mg g−1 dry weight) of wild-type (black bars), S3-S5 (light gray bars), and A2-A3 (dark gray bars) cell walls. Data represent average values ± se of three replicates. Statistical analysis of the data was performed using Student’s t test (* P ≤ 0.05, ** P ≤ 0.001). D, Neutral monosaccharide composition by alditol acetates coupled to GC-MS of cell wall samples. Significant differences were analyzed by Student’s t test (* P ≤ 0.001).
To obtain a more quantitative analysis of the cell wall changes in SN1-overexpressing and -silenced lines, we compared the noncellulosic neutral monosaccharide composition as well as the cellulose and uronic acid leaf contents (Reiter et al., 1993). Cellulose was significantly lower in both overexpressing (P ≤ 0.001) and silenced (P ≤ 0.05) lines when compared with wild-type cell walls (Fig. 4C; Supplemental Table S1). Interestingly, the amounts of Gal, Rha, Man, and uronic acids in A2 and A3 were significantly lower than those of wild-type samples, while no significant changes were observed in Xyl and Ara in these lines (Fig. 4D; Supplemental Table S1). In parallel, no significant changes were observed between overexpressing lines and wild-type plants in the amounts of any neutral monosaccharide analyzed or uronic acid contents (Fig. 4, C and D; Supplemental Table S1).
SN1 Localizes in Plasma Membranes and Self-Interacts in Vivo
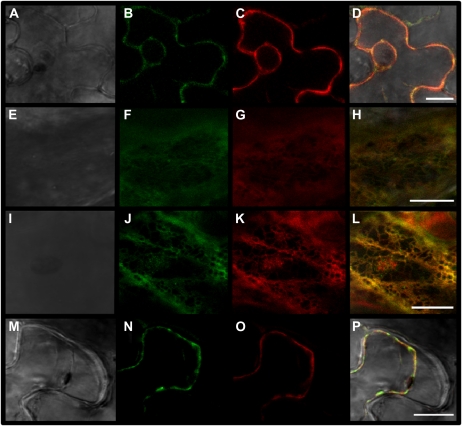
In order to experimentally determine the subcellular localization of SN1, the complete coding region was fused, at the C-terminal end, to an enhanced GFP coding sequence (Egfp) under the control of the cauliflower mosaic virus 35S promoter (SN1SP-Egfp). To test whether the N-terminal sequence of SN1 determines its localization, a truncated fusion protein lacking the putative signal peptide (Segura et al., 1999) called SN1ΔSP-Egfp was produced. Transient transformations were carried out by agroinfiltration of Nicotiana benthamiana leaves, and samples of agroinfiltrated zones were inspected using confocal microscopy. Green fluorescence was observed around the nucleus and at the cell periphery in cross-sections of leaves infiltrated with the SN1SP-Egfp construct (Fig. 5B). This pattern overlapped with that observed in coagroinfiltrated leaves with the plasma membrane marker pm-rk, a construct carrying AtPIP2A fused to mCherry (Nelson et al., 2007; Fig. 5, C and D). Cortical views of these cells revealed colocalization of SN1SP-Egfp and pm-rk (Fig. 5, E–H), with a Pearson’s coefficient of 0.82 to 0.87. In order to confirm whether SN1 follows the secretory pathway, we further evaluated the colocalization of SN1SP-Egfp and ER-rk, a construct that carries mCherry fused to the signal peptide of AtWAK2 and the HDEL retention peptide, which ensures its localization to the endoplasmic reticulum (Nelson et al., 2007; Fig. 5, I–L). As shown in Figure 5L, this study revealed colocalization of SN1SP-Egfp and ER-rk, with a Pearson’s coefficient of 0.71 to 0.8. Finally, plasmolysis treatment to coinfiltrated leaves with SN1SP-Egfp and pm-rk demonstrated that SN1 was retained in the plasma membrane and was not localized to the cell wall or apoplast (Fig. 5, M–P). Transmembrane prediction programs (DAS, TMHMM TMpred, and TopPred 0.01 from the Expasy site) identified a potential hydrophobic region at the N terminus, as expected for the signal peptide, whereas no transmembrane segment was predicted when considering the mature peptide.
Figure 5.
SN1 localization determined by confocal microscopy analysis of SN1-Egfp fusion protein in agroinfiltrated N. benthamiana epidermal leaf cells. Expression of SN1SP-Egfp (B, F, J, and N), plasma membrane (pm-rk; C, G, and O), or endoplasmic reticulum (ER-rk; K) markers is shown in cross-sections of cells without (A–D) or with (M–P) plasmolysis. Cortical views of the coagroinfiltrated cells are shown in E to L). Bright-field (A, E, I, and M) and merged images of GFP and red channels (D, H, L, and P) are given. Bars = 10 μm.
In turn, leaves expressing the control truncated fusion protein (SN1ΔSP-Egfp) yielded a strong fluorescence uniformly distributed throughout the cytoplasm and within the nucleus, as expected due to simple diffusion of small proteins (Supplemental Fig. S1B). In addition, coexpression of SN1ΔSP-Egfp and pm-rk (Supplemental Fig. S1, A–H) or ER-rk (Supplemental Fig. S1, I–L) showed low colocalization rates (0.30–0.35 and 0.30–0.50, respectively).
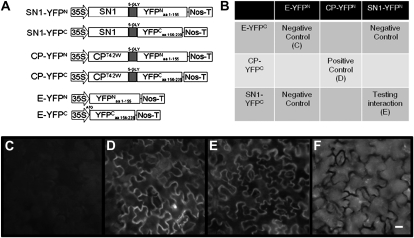
We next performed bimolecular fluorescence complementation (BiFC) assays. Sequences encoding yellow fluorescent protein (YFP) amino acids 1 to 155 and amino acids 156 to 239 (YFPN and YFPC, respectively) were fused to sequences encoding the open reading frame of SN1 to produce SN1-YFPN and SN1-YFPC. CP-YFPN/CP-YFPC (CP for Coat Protein) and E-YFPN/E-YFPC (E for Empty) were used as positive and negative controls, respectively, as described by Bazzini et al. (2007; Fig. 6A). N. benthamiana leaves were infiltrated with different combinations of Agrobacterium tumefaciens suspensions harboring the BiFC constructs (Fig. 4B) and further inspected by fluorescence microscopy. Coexpression of SN1-YFPN and SN1-YFPC completely restored the fluorescence, revealing specific interactions between SN1 proteins (Fig. 6E). As expected, coexpression of E-YFPN and E-YFPC or combinations of them with constructs encoding SN1 did not produce detectable signals (Fig. 6C), while YFP fluorescence was detected after coinfiltration of CP-YFPN and CP-YFPC (Fig. 6D).
Figure 6.
SN1 self-interacted in vivo by BIFC. A, Schematic representation of the cassette vectors used for agroinfiltration assays: SN1-YFPN, SN1-YFPC, CP-YFPN, CP-YFPC, E-YFPN, and E-YFPC. SN1, SN-1 open reading frame; 35S, cauliflower mosaic virus-35S promoter; Nos-T, nopaline synthase terminator; CP, tobacco mosaic virus coat protein (CP)T42W (a variant of CP); E, Empty; YPFN, fragment of YPF encoding amino acids 1 to 155; YPFC, fragment of YPF encoding amino acids 155 to 239. Diagrams are not to scale. B, Scheme of the combination of constructs coexpressed in BiFC assays. C to F, YFP epifluorescence microscopy images of N. benthamiana epidermal cells of agroinfiltrated leaves with a mixture of Agrobacterium strains harboring constructs encoding fusion proteins. Each image is representative of several experiments: negative controls (C), positive control (D), SN1 tested interaction (E), and epidermal cells under white light (F). Bar = 10 μm.
DISCUSSION
SN1 Is Involved in Plant Growth and Development
In order to gain further insight into the physiological function of SN1, we accomplished a detailed characterization of transgenic potato lines with altered expression of this gene. Northern-blot analysis confirmed SN1 overexpression in S3 and S5 and significant down-regulation of the endogenous gene in A2 and A3 lines. Overexpressing lines did not show visual morphological differences from wild-type plants. However, silenced lines exhibited reduced height and altered leaf morphology, suggesting that SN1 plays additional roles beyond its assigned function in plant defense. In agreement, it has been recently reported that several antimicrobial peptides are also involved in plant growth and development. For example, Stotz et al. (2009) demonstrated that overexpression of the tomato defensin DEF2 enhances foliar resistance to the fungal pathogen Botrytis cinerea and pleiotropically alters the growth of various organs. Furthermore, antisense suppression or constitutive overexpression of this gene reduces pollen viability and seed production. Interestingly, germinating Arabidopsis seeds exogenously treated with defensins showed root growth inhibition (Allen et al., 2008).
SN2 is the other antimicrobial peptide isolated from potato that represents a quite divergent (38% conserved residues) snakin/GASA subfamily (Berrocal-Lobo et al., 2002). Expression analysis of SN2 in our transgenic potato plants revealed that it was unaffected by the presence of the SN1 antisense construct in the phenotypically altered silenced plants (data not shown). Thus, it is reasonable to conclude that the abnormal phenotype is attributable to partial gene silencing of the endogenous SN1 and not to unintended altered SN2 expression.
SN1 Affects Cell Division
In this work, we found that leaf areas of SN1-silenced lines are 50% to 60% smaller than in the wild type. Since the final size of a leaf is regulated by cell division and cell expansion (Gonzalez et al., 2010), the smaller leaf size observed in the silenced lines might be the result of either a lower cell number due to the inhibition of division or smaller cells due to the inhibition of cell growth. Analyses of cell area in mature leaves revealed that SN1-silenced lines showed a significant increase in the mean cell size compared with wild-type plants, thus indicating a decrease in total cell number per leaf. These results suggest that SN1 is involved in the leaf cell division process. In accordance, it has been previously suggested that the SN1 homologous genes GIP4, GIP5, and GASA4 act in cell division, based on their expression in meristematic tissue (Aubert et al., 1998; Ben-Nissan et al., 2004; Roxrud et al., 2007). Moreover, Furukawa et al. (2006) demonstrated that the expression of OsGASR genes is synchronized with cell proliferation in suspension-cultured rice cells and that OsGASR1 and OsGASR2 are strongly expressed in meristems, which in turn suggests that both genes are involved in cell division (Furukawa et al., 2006). Interestingly, phylogenetic trees depicting the evolutionary relations of SN1 homologous genes and their amino acid sequence analyses revealed a close relationship between SN1 and OsGASR1 (Furukawa et al., 2006; Peng et al., 2010). On the other hand, the presence of fewer but larger cells in the leaves of SN1-silenced transgenic plants might be a consequence of a compensation effect. This phenomenon, which can occur when cell proliferation is inhibited in growing leaves or related lateral organs, partially compensates the decrease of total leaf size by increasing cell expansion (Tsukaya and Beemster, 2006; Tsukaya, 2008). Moreover, mutants and transgenic plants exhibiting compensation effects usually have smaller and abnormally shaped leaves (Wang et al., 2000; De Veylder et al., 2001; Horiguchi et al., 2005; Verkest et al., 2005; Ferjani et al., 2007).
SN1 Affects Leaf Primary Metabolism and Cell Wall Composition
Given the abnormal phenotype of the SN1-silenced lines, we next analyzed shifts in the levels of a wide range of metabolites using the combination of an established GC-MS, FTIR spectroscopy, and spectrophotometric techniques. It has been reported that transgenic plants with altered leaf metabolism show changes in leaf and shoot morphology, suggesting that the plant developmental program can be modulated by the metabolic status of the leaf. However, the mechanism involved in mediating these changes has not yet been defined (Raines and Paul, 2006). We demonstrated here that SN1-silenced lines exhibited major changes in their primary metabolism. Levels of certain TCA cycle intermediates as well as components of the ascorbic acid pathway were significantly altered. These metabolites, in turn, are those that better explain the applied PCA model separating the transgenic lines from wild-type plants. Two relevant studies support a strong functional link between respiration and ascorbate metabolism. Millar et al., (2003) reported that l-galatono-1,4 lactone-dehydrogenase (l-GalDH), the enzyme that catalyzes the last steps in ascorbate biosynthesis, is associated with complex I of the mitochondrial electron transport chain. In turn, Alhagdow et al. (2007) described that tomato plants with reduced levels of l-GalDH have significant changes in mitochondrial function, show altered ascorbate redox state, and display severe defects in leaf growth as a consequence of reduced cell expansion. Ascorbate content has also been associated with cell division. It has been reported that cell proliferation is restrained in ascorbic acid-deficient transgenic BY-2 cells by antisense expression of the GalDH gene (Tabata et al., 2001). More recently, Gilbert et al. (2009) showed that transgenic tomato lines with reduced ascorbate levels exhibited growth defects affecting both cell division and cell expansion. The fact that SN1-silenced lines show affected cell division and reduced ascorbate levels supports the proposed role of this molecule in regulating that process and suggests that SN1 might play an important role in cell division by regulating ascorbate accumulation.
In addition, Gilbert et al. (2009) described an intimate linkage between ascorbate and noncellulosic cell wall polysaccharide biosynthetic pathways. On the other hand, it has also been reported that breakdown of cell wall polymers increases the availability of precursors for ascorbate biosynthesis in the l-galactonic acid pathway (Di Matteo et al., 2010). Cell walls provide more than just mechanical support, because, together with the process of cell division, they are largely responsible for controlling plant and tissue morphology (Cosgrove, 2005). Here, we showed that SN1 silencing not only results in significant alterations of cell division but also in leaf cell wall composition. The content of noncellulosic neutral sugars, in particular those of the pectin fraction (comprising Gal and Rha), as well as the uronic acid content were significantly lower in A2 and A3 cell walls, suggesting a down-regulation of rhamnogalacturonan-I structures in silenced lines.
The cell wall is not only associated with a variety of developmental events but is also important in relaying information from external stimuli (Hématy et al., 2009). Certain changes in cell wall composition can alter the defense responses to different types of pathogens (Ellis and Turner, 2001; Vogel et al., 2002, 2004; Hernández-Blanco et al., 2007; Sánchez-Rodríguez et al., 2009). For example, Hernández-Blanco et al. (2007) demonstrated that impairment of CESA proteins, required for the synthesis of secondary cell wall cellulose in irx mutants, leads to specific activation of novel defense pathways. Here, we found that cellulose levels are significantly decreased in the cell wall of SN1 transgenic plants, these differences being more pronounced in S3 and S5. These changes in overexpressing lines, previously shown to be more resistant against fungal and bacterial infections (Almasia et al., 2008), could also be linked to some extent to the irx-mediated resistance. Along the same line, galactinol, which was recently pointed out as a signaling component of the induced systemic resistance mechanism in plants (Kim et al., 2008), showed a massive decrease in SN1-silenced plants. When assaying the effect of crude leaf protein extracts on the growth of Sclerotinia sclerotiorum, overexpressing SN1 lines were able to reduce fungal growth, whereas extracts of the silenced lines did not exhibit any inhibitory activity. Moreover, extracts from A2 and A3 lines allowed higher fungal growth than extracts from the wild-type ones, further supporting the recognized role in the defense of SN1 (Supplemental Fig. S2).
In addition, it has been suggested that galactinol as well as raffinose, which also was significantly reduced in A2 and A3, act as scavengers of reactive oxygen species to protect plant cells from oxidative damage caused by methyl viologen treatment, salinity, or chilling (Nishizawa et al., 2008). It has been reported that inhibition of 1-d-myoinositol 3-phosphate synthase (MIPS) resulted in strongly reduced levels of inositol, galactinol, and raffinose and altered phenotype in transgenic potato plants (Keller et al., 1998). Although MIPS antisense plants share some phenotypic alterations with SN1-silenced lines (reduced height, increased leaf thickness), they still exhibit compound leaves and do not show reduced leaf size, as was described for SN1 antisense plants. Even though the down-regulation of raffinose family sugars (galactinol and raffinose) in SN1 antisense lines probably contributes to the observed phenotype, it is fair to assume that morphological alterations are the result of the whole metabolite changes caused by SN1 silencing.
SN1 Localizes in Plasma Membrane and Self-Interacts in Vivo
Since protein functions rely on appropriate subcellular compartmentalization, determining their distribution within the cell is an important step toward elucidating their biological role. According to our results, SN1 is located in the plasma membrane. SN1 homologous localization was determined experimentally in the endoplasmic reticulum membrane (Ben Nissan et al., 2004) or in the apoplasm/cell wall (Furukawa et al., 2006; Zhang et al., 2009). Potential hydrophobic segments were not predicted in the mature peptide, suggesting that membrane localization could be due to electrostatic interactions, covalent bonds to membrane lipids (lipidation), or attachment/interaction with other proteins. It has been reported that a snakin-like peptide containing a GASA domain from French bean (Phaseolus vulgaris) is associated with a Pro-rich protein (PRP), resulting in a two-component protein complex involved in plant-pathogen interactions (Bindschedler et al., 2006). PRPs are a large group of plant cell wall proteins also implicated in plant development and environmental stress (Peng et al., 2010). Whether SN1 could act as a plasmalemma-wall interface-associated protein through PRP interactions (or with other proteins) remains to be studied. Recently, Rubinovich and Weiss (2010) showed that GASA4 is involved in the promotion of GA responses and acts as a redox regulator. The authors suggested that GAST-like proteins function in regulating the redox status and, thus, the activity of specific components, for example cell wall proteins, to translate the GA signal into physiological and developmental responses. In this work, we determined that at least two SN1 molecules are able to self-interact by BiFC assays. It is still unclear whether this interaction is necessary for SN1 to play its roles in development and/or defense, presumably by regulating signal transduction from the cell periphery to the cytoplasm.
CONCLUSION
Even when overexpressing SN1, potato lines did not show remarkable morphological differences from the wild type; silenced lines presented reduced height and alterations in leaf shape and area. The smaller leaf size in silenced lines is directly related to a decrease in the number of cells per leaf, a reduction that is partially compensated by an increase in the final cell size. Subcellular analyses of a SN1-GFP fusion protein showed targeting to the plasma membrane, and BiFC assays suggested that SN1 self-interacts in vivo. Moreover, profiles of soluble and insoluble metabolites in potato leaves highlighted the impact of SN1 function on cellular metabolism. We demonstrate here that SN1 silencing affects cell division, leaf metabolism, and cell wall composition, suggesting that SN1 has additional roles in plant growth and development beyond its assigned role in plant defense.
MATERIALS AND METHODS
Plant Transformation and Regeneration
The complete open reading frame of SN1 (EF206290) was amplified by PCR using the upSN1 and lowSN1 primers (Supplemental Table S2), cloned into the pGEM-T Easy vector (Promega; http://www.promega.com/), restricted with EcoRI, and inserted into the pBPFΩ7 vector (Coego et al., 1996). These cassettes were then subcloned into the HindIII site of the pPZPK binary vector pPZP200 (Hajdukiewicz et al., 1994), giving rise to pPZPK-SN1Sense or pPZPK-SN1Antisense. Transformation of potato (Solanum tuberosum subspecies tuberosum ‘Kennebec’) leaf discs was performed as described (Rocha-Sosa et al., 1989) via Agrobacterium tumefaciens LBA4404. Transgenic plants were maintained in vitro by periodic micropropagation in growing chambers (CMP 3244; Conviron) at 18°C to 22°C under an 8/16-h dark/light cycle. For molecular, phenotypic, and metabolic analyses, plants were transferred to soil and grown in the greenhouse at 18°C to 25°C under a 10/14-h dark/light cycle, and leaf samples of 8-week-old transgenic and wild-type plants were collected.
RNA Extraction and Northern-Blot Analysis
RNA from leaf tissue was isolated using the Trizol commercial extraction system (Invitrogen; http://www.invitrogen.com/), 10 μg was electrophoretically resolved on 1.5% agarose gels using MOPS buffer (Sigma-Aldrich; http://www.sigmaaldrich.com/) and transferred to a Hybond N+ nylon membrane (Amersham) for 16 h, and RNA was cross-linked under a UV lamp. SN1 probe was synthesized and labeled with [α-32P]dCTP using the Prime-a-Gene labeling system (Promega). Hybridizations were performed for 14 h at 42°C using ULTRAhybª hybridization solution (Ambion; http://www.ambion.com/). Membranes were washed twice in 2× SSC/0.1% SDS for 10 min at 42°C, once in 1× SSC/0.1% SDS for 20 min at room temperature, and once in 0.1× SSC/0.1% SDS for 10 min at room temperature. Membranes were exposed overnight and scanned using a Typhoon scanner (Amersham Biosciences).
Leaf and Epidermis Cell Area Measurement
The fifth leaf from 10 independent 8-week-old plants was collected and digitally photographed. Leaf area was determined with ImageJ software (http://rsb.info.nih.gov/ij/). To measure the total cell area, the fifth leaf from five independent plants was cleared and fixed overnight in ethanol:acetic acid (1:1), washed in ethanol, and placed in chloral hydrate in an extra clearing step. Cells were observed on a Leica TCS-SP5 confocal microscope (http://www.leica-microsystems.com/). The area of adaxial epidermal cells (n = 100–150) was calculated using ImageJ software. Average leaf and cell areas were used to calculate cell numbers. Statistical analyses were performed by the nonparametric Kruskal-Wallis test (InfoStat; http://www.infostat.com.ar).
Histological Analyses
Cross-sections were cut from the center of fully expanded leaf laminas of 8-week-old plants. Samples were fixed in glutaraldehyde overnight at 4°C and then washed two times in sodium phosphate buffer (pH 7.2) at 4°C. After dehydration with a graded ethanol and xylene series, samples were embedded in paraffin. Ten-nanometer-thick sections were cut with a microtome equipped with a gold blade, placed on microscope slides precoated with poly-l-Lys, and deparaffinized with xylene for 15 min. Histological sections from three replicates of the wild-type and transgenic plants (two independent experiments) were observed on a Leica TCS-SP5 confocal microscope.
Extraction, Derivatization, and Analysis of Potato Leaf Soluble Metabolites Using GC-MS
A polar metabolite fraction was obtained from 100 mg of potato leaf tissue, homogenized in 1,400 μL of 100% methanol with 60 μL of internal standard (ribitol; 0.2 mg mL−1), and extracted for 15 min at 70°C. The supernatant was mixed with 750 μL of chloroform and 1,500 μL of water and centrifuged at 3,600 rpm. Aliquots of the supernatant (150 μL) were dried in a vacuum for 4 h, redissolved in 40 μL of methoxyamine hydrochloride (20 mg mL−1 in pyridine), and derivatized for 2 h at 37°C. N-Methy-N-(trimethylsilyl) trifluoroacetamide (70 μL) and fatty acid methylesters mix (Sigma-Aldrich;7 μL) were added and left for 30 min at 37°C. GC-MS analysis was performed using a HP 5980 gas chromatograph (Agilent; http://www.agilent.com/) coupled to a Pegasus II time-of-flight mass spectrometer (Leco; http://www.leco.com/). Analyses were carried out using the Chroma Tof (Leco) and TagFinder software (Luedemann et al., 2008). t tests were performed using Microsoft Excel 7.0. Data are presented as means ± se of determinations on six individual plants per line. PCA was carried out using the software InfoStat (http://www.infostat.com.ar).
Determination of Soluble Sugars
Ten to 20 mg of leaf tissue was homogenized in 250 μL of 80% ethanol and extracted during 20 min at 80°C. After centrifugation, 150 μL of 80% ethanol was added to the pellet, mixed, and incubated for 20 min at 80°C. The second pellet was resuspended in 250 μL of 50% ethanol and extracted for 20 min at 80°C. Suc, Glc, and Fru were determined spectrophotometrically in the supernatant (Stitt, 1989).
Cell Wall Analyses
For FTIR, precleaned (by solvent extraction) and dried leaves from 8-week-old plants were pooled and homogenized. Two to three independent biological and technical replicates (n = 10) were analyzed. Fourteen FTIR spectra for each line were collected on a Thermo Nicolet Nexus 470 spectrometer (ThermoElectric) over the range 4,000 to 400 cm−1. For each spectrum, 64 scans were coadded at a resolution of 4 cm−1 for Fourier transform processing and absorbance spectrum calculation using OMNIC software (Thermo Nicolet). Using win-das software (Wiley), spectra were baseline corrected, normalized, and analyzed by PCA and the covariance matrix method (Kemsley, 1996). Cell wall monosaccharides were assayed as alditol acetate derivatives (n = 3) in two or three biological replicates (Stevenson and Furneaux, 1991) by GC performed on an SP-2330 apparatus (Bellefonte), and the results were validated as described previously (Gibeaut and Carpita, 1991). Myoinositol (Sigma-Aldrich) was added as an internal standard. Cellulose was determined on the fraction resistant to extraction with 2 m trifluoracetic acid (n = 3; Morrison, 1988) by phenol-sulfuric assay using Glc equivalents as standard (Dubois et al., 1956). Uronic acids were quantified using the soluble 2 m trifluoracetic acid fraction (n = 5; Filisetti-Cozzi and Carpita, 1991).
Construction of the GFP Fusion Vector for Transient Expression
SN1 was amplified by PCR in two versions: including the signal peptide (SN1SP; upSN1 and low1SN1) or without it (SN1ΔSP; upΔSN1 and low1SN1; Supplemental Table S2), cloned into the pCR8/GW/TOPO TA (Invitrogen), and recombined into the pK7FWG2 vector (Karimi et al., 2002) through Gateway technology (Invitrogen), giving rise to SN1SP-Egfp and SN1ΔSP-Egfp. Nicotiana benthamiana leaves were infiltrated with A. tumefaciens GV3101 carrying SN1SP-Egfp, SN1ΔSP-Egfp, plasma membrane (pm-rk), or endoplasmic reticulum (ER-rk) markers (Nelson et al., 2007). Overnight cultures were resuspended in MES and adjusted to optical density at 600 nm = 0.8. Acetosyringone was added to bacterial suspensions (20 μm final), and they were kept in darkness for 4 h prior to agroinfiltration. Images were acquired 2 d after infiltration with a confocal TCS SP5 using a 63× water-immersion objective. Plasmolysis treatment was performed for 15 min in 0.8 m mannitol before observation.
BiFC Analyses
pMONA-YFPN, pMONB-YFPC, CP-YFPN, CP-YFPC, E-YFPN, and EYFPC were provided by Dr. S. Asurmendi. SN1 sequence was amplified using upΔSN1 and low2SN1 (including a BglII site; Supplemental Table S2). PCR products were cloned into pCR2.1-TOPO (Invitrogen), restricted with EcoRI and BglII, and cloned into pMONA-YFPN and pMONB-YFPC vectors. The resulting constructs were subsequently cloned into the NotI site of the pART binary vector, giving rise to SN1-YFPN and SN1-YFPC. Constructs were transferred to A. tumefaciens strain GV3101 and infiltrated into 4-week-old N. benthamiana plants. The YFP signal was photographed at 3 d after infection using a fluorescence microscope (Nikon ECLIPSE TS100) and a 40× objective.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number EF206290.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Confocal microscopy analysis of truncated SN1ΔSP-Egfp fusion protein expression in agroinfiltrated N. benthamiana epidermal leaf cells.
Supplemental Figure S2. Fungal growth inhibition assays.
Supplemental Table S1. Soluble and insoluble metabolite levels in leaves of 8-week-old potato plants expressing the SN-1 gene both in sense (S3 and S5) and antisense (A2 and A3) orientations relative to the wild type.
Supplemental Table S2. List of the primers used in this study.
Acknowledgments
We thank Vanesa Mongelli, Gonzalo Roqueiro, and Sara Maldonado for technical assistance and helpful discussions. We thank Teresa Cabrera, Valeria Beracochea, and Valeria Peralta for their excellent technical assistance in the production and maintenance of transgenic potato plants. We also thank Mariana del Vas and Alisdair Fernie for critically reading the manuscript and Eleonora Campos for her invaluable English assistance.
References
- Alhagdow M, Mounet F, Gilbert L, Nunes-Nesi A, Garcia V, Just D, Petit J, Beauvoit B, Fernie AR, Rothan C, et al. (2007) Silencing of the mitochondrial ascorbate synthesizing enzyme L-galactono-1,4-lactone dehydrogenase affects plant and fruit development in tomato. Plant Physiol 145: 1408–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen A, Snyder AK, Preuss M, Nielsen EE, Shah DM, Smith TJ. (2008) Plant defensins and virally encoded fungal toxin KP4 inhibit plant root growth. Planta 227: 331–339 [DOI] [PubMed] [Google Scholar]
- Almasia NI, Bazzini AA, Hopp HE, Vazquez-Rovere C. (2008) Overexpression of snakin-1 gene enhances resistance to Rhizoctonia solani and Erwinia carotovora in transgenic potato plants. Mol Plant Pathol 9: 329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert D, Chevillard M, Dorne AM, Arlaud G, Herzog M. (1998) Expression patterns of GASA genes in Arabidopsis thaliana: the GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol Biol 36: 871–883 [DOI] [PubMed] [Google Scholar]
- Bazzini AA, Hopp HE, Beachy RN, Asurmendi S. (2007) Infection and coaccumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proc Natl Acad Sci USA 104: 12157–12162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Nissan G, Lee JY, Borohov A, Weiss D. (2004) GIP, a Petunia hybrida GA-induced cysteine-rich protein: a possible role in shoot elongation and transition to flowering. Plant J 37: 229–238 [DOI] [PubMed] [Google Scholar]
- Ben-Nissan G, Weiss D. (1996) The petunia homologue of tomato gast1: transcript accumulation coincides with gibberellin-induced corolla cell elongation. Plant Mol Biol 32: 1067–1074 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Segura A, Moreno M, López G, García-Olmedo F, Molina A. (2002) Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol 128: 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindschedler LV, Whitelegge JP, Millar DJ, Bolwell GP. (2006) A two component chitin-binding protein from French bean: association of a proline-rich protein with a cysteine-rich polypeptide. FEBS Lett 580: 1541–1546 [DOI] [PubMed] [Google Scholar]
- Coego A, Vazquez R, Yamilet Coll JA, Pujol M, Menendez E, Lopez MA, Molina P, Hernandez L, Bencomo B, de la Riva G, et al. (1996) Effect of promoter-stimulatory element combination on transient reporter gene expression in tobacco protoplast using PEG treatment. Biotechnologia Aplicada 13: 147–153 [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- de la Fuente JI, Amaya I, Castillejo C, Sánchez-Sevilla JF, Quesada MA, Botella MA, Valpuesta V. (2006) The strawberry gene FaGAST affects plant growth through inhibition of cell elongation. J Exp Bot 57: 2401–2411 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inzé D. (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Matteo A, Sacco A, Anacleria M, Pezzotti M, Delledonne M, Ferrarini A, Frusciante L, Barone A. (2010) The ascorbic acid content of tomato fruits is associated with the expression of genes involved in pectin degradation. BMC Plant Biol 10: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Gilles DA, Hamilton JK, Rebers PA, Smith F. (1956) Colorimetric method of determination of sugars and related substances. Anal Chem 28: 350–356 [Google Scholar]
- Ellis C, Turner JG. (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13: 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferjani A, Horiguchi G, Yano S, Tsukaya H. (2007) Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol 144: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Roessner U, Trethewey RN, Willmitzer L. (2001) The contribution of plastidial phosphoglucomutase to the control of starch synthesis within the potato tuber. Planta 213: 418–426 [DOI] [PubMed] [Google Scholar]
- Filisetti-Cozzi TM, Carpita NC. (1991) Measurement of uronic acids without interference from neutral sugars. Anal Biochem 197: 157–162 [DOI] [PubMed] [Google Scholar]
- Furukawa T, Sakaguchi N, Shimada H. (2006) Two OsGASR genes, rice GAST homologue genes that are abundant in proliferating tissues, show different expression patterns in developing panicles. Genes Genet Syst 81: 171–180 [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Carpita NC. (1991) Tracing cell wall biogenesis in intact cells and plants: selective turnover and alteration of soluble and cell wall polysaccharides in grasses. Plant Physiol 97: 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L, Alhagdow M, Nunes-Nesi A, Quemener B, Guillon F, Bouchet B, Faurobert M, Gouble B, Page D, Garcia V, et al. (2009) GDP-D-mannose 3,5-epimerase (GME) plays a key role at the intersection of ascorbate and non-cellulosic cell-wall biosynthesis in tomato. Plant J 60: 499–508 [DOI] [PubMed] [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, Jikumaru Y, Chae E, Dhondt S, Van Daele T, De Milde L, Weigel D, Kamiya Y, et al. (2010) Increased leaf size: different means to an end. Plant Physiol 153: 1261–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hématy K, Cherk C, Somerville S. (2009) Host-pathogen warfare at the plant cell wall. Curr Opin Plant Biol 12: 406–413 [DOI] [PubMed] [Google Scholar]
- Hernández-Blanco C, Feng DX, Hu J, Sánchez-Vallet A, Deslandes L, Llorente F, Berrocal-Lobo M, Keller H, Barlet X, Sánchez-Rodríguez C, et al. (2007) Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19: 890–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H. (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43: 68–78 [DOI] [PubMed] [Google Scholar]
- Humphrey TV, Bonetta DT, Goring DR. (2007) Sentinels at the wall: cell wall receptors and sensors. New Phytol 176: 7–21 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kacuráková M, Capeka P, Sasinkova V, Wellnerb N, Ebringerova A. (2000) FT-IR study of plant cell wall model compounds: pectic polysaccharides and hemicelluloses. Carbohydr Polym 23: 195–203 [Google Scholar]
- Keller R, Brearley CA, Trethewey RN, Müller-Rober B. (1998) Reduced inositol content and altered morphology in transgenic potato plants inhibited for 1D-myo-inositol 3-phosphate synthase. Plant J 16: 403–410 [Google Scholar]
- Kemsley EK. (1996) Discriminant analysis of high-dimensional data: a comparison of principal components analysis and partial least squares data reduction methods. Chemom Intell Lab Syst 33: 47–61 [Google Scholar]
- Kim MS, Cho SM, Kang EY, Im YJ, Hwangbo H, Kim YC, Ryu CM, Yang KY, Chung GC, Cho BH. (2008) Galactinol is a signaling component of the induced systemic resistance caused by Pseudomonas chlororaphis O6 root colonization. Mol Plant Microbe Interact 21: 1643–1653 [DOI] [PubMed] [Google Scholar]
- Kotilainen M, Helariutta Y, Mehto M, Pollanen E, Albert VA, Elomaa P, Teeri TH. (1999) GEG participates in the regulation of cell and organ shape during corolla and carpel development in Gerbera hybrida. Plant Cell 11: 1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedemann A, Strassburg K, Erban A, Kopka J. (2008) TagFinder for the quantitative analysis of gas chromatography-mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 24: 732–737 [DOI] [PubMed] [Google Scholar]
- Meyerowitz EM. (1997) Genetic control of cell division patterns in developing plants. Cell 88: 299–308 [DOI] [PubMed] [Google Scholar]
- Millar AH, Mittova V, Kiddle G, Heazlewood JL, Bartoli CG, Theodoulou FL, Foyer CH. (2003) Control of ascorbate synthesis by respiration and its implications for stress responses. Plant Physiol 133: 443–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison IM. (1988) Hydrolysis of plant cell walls with trifluoroacetic acid. Phytochemistry 27: 1097–1100 [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Shigeoka S. (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos E, Kiddle G, Pellny T, Kumar S, Foyer C. (2006) Modulation of plant morphology, root architecture, and cell structure by low vitamin C in Arabidopsis thaliana. J Exp Bot 57: 1645–1655 [DOI] [PubMed] [Google Scholar]
- Peng J, Lai L, Wang X. (2010) Temporal and spatial expression analysis of PRGL in Gerbera hybrida. Mol Biol Rep 37: 3311–3317 [DOI] [PubMed] [Google Scholar]
- Raines CA, Paul MJ. (2006) Products of leaf primary carbon metabolism modulate the developmental programme determining plant morphology. J Exp Bot 57: 1857–1862 [DOI] [PubMed] [Google Scholar]
- Reiter WD, Chapple CC, Somerville CR. (1993) Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science 261: 1032–1035 [DOI] [PubMed] [Google Scholar]
- Rocha-Sosa M, Sonnewald U, Frommer W, Stratmann M, Schell J, Willmitzer L. (1989) Both developmental and metabolic signals activate the promoter of a class I patatin gene. EMBO J 8: 23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie A. (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxrud I, Lid SE, Fletcher JC, Schmidt ED, Opsahl-Sorteberg HG. (2007) GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol 48: 471–483 [DOI] [PubMed] [Google Scholar]
- Rubinovich L, Weiss D. (2010) The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J 64: 1018–1027 [DOI] [PubMed] [Google Scholar]
- Sánchez-Rodríguez C, Estévez JM, Llorente F, Hernández-Blanco C, Jordá L, Pagán I, Berrocal M, Marco Y, Somerville S, Molina A. (2009) The ERECTA receptor-like kinase regulates cell wall-mediated resistance to pathogens in Arabidopsis thaliana. Mol Plant Microbe Interact 22: 953–963 [DOI] [PubMed] [Google Scholar]
- Segura A, Moreno M, Madueño F, Molina A, García-Olmedo F. (1999) Snakin-1, a peptide from potato that is active against plant pathogens. Mol Plant Microbe Interact 12: 16–23 [DOI] [PubMed] [Google Scholar]
- Shi L, Gast RT, Gopalraj M, Olszewski NE. (1992) Characterization of a shoot-specific, GA3- and ABA-regulated gene from tomato. Plant J 2: 153–159 [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, et al. (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Stevenson TT, Furneaux RH. (1991) Chemical methods for the analysis of sulphated galactans from red algae. Carbohydr Res 210: 277–298 [DOI] [PubMed] [Google Scholar]
- Stitt M. (1989) Product inhibition of potato tuber pyrophosphate:fructose-6-phosphate phosphotransferase by phosphate and pyrophosphate. Plant Physiol 89: 628–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz HU, Spence B, Wang Y. (2009) A defensin from tomato with dual function in defense and development. Plant Mol Biol 71: 131–143 [DOI] [PubMed] [Google Scholar]
- Tabata K, Oba K, Suzuki K, Esaka M. (2001) Generation and properties of ascorbic acid-deficient transgenic tobacco cells expressing antisense RNA for L-galactono-1,4-lactone dehydrogenase. Plant J 27: 139–148 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (2008) Controlling size in multicellular organs: focus on the leaf. PLoS Biol 6: 1373–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H, Beemster GT. (2006) Genetics, cell cycle and cell expansion in organogenesis in plants. J Plant Res 119: 1–4 [DOI] [PubMed] [Google Scholar]
- Verkest A, Manes CL, Vercruysse S, Maes S, Van Der Schueren E, Beeckman T, Genschik P, Kuiper M, Inzé D, De Veylder L. (2005) The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17: 1723–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Schiff C, Somerville SC. (2002) PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14: 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Somerville CR, Somerville SC. (2004) Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J 40: 968–978 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhou Y, Gilmer S, Whitwill S, Fowke LC. (2000) Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J 24: 613–623 [DOI] [PubMed] [Google Scholar]
- Wigoda N, Ben-Nissan G, Granot D, Schwartz A, Weiss D. (2006) The gibberellin-induced, cysteine-rich protein GIP2 from Petunia hybrida exhibits in planta antioxidant activity. Plant J 48: 796–805 [DOI] [PubMed] [Google Scholar]
- Zhang S, Yang C, Peng J, Sun S, Wang X. (2009) GASA5, a regulator of flowering time and stem growth in Arabidopsis thaliana. Plant Mol Biol 69: 745–759 [DOI] [PubMed] [Google Scholar]
- Zimmermann R, Sakai H, Hochholdinger F. (2010) The Gibberellic Acid Stimulated-Like gene family in maize and its role in lateral root development. Plant Physiol 152: 356–365 [DOI] [PMC free article] [PubMed] [Google Scholar]