Chromosomes are key building blocks of eukaryotic genomes. Studies on chromosome organization and dynamics not only address questions of how chromosomes behave and what mechanisms control this behavior but also examine how chromosome organization and dynamics affect gene expression and genome maintenance. A number of important studies on chromosome organization and dynamics have been conducted in plants in the past few years. Many of them have been made possible by recent advances in cytogenetics tools, including improvements in fluorescent in situ hybridization (FISH) protocols and development of live-imaging techniques. To the most significant discoveries belong elucidating the chromosome arrangement in interphase nuclei in Arabidopsis (Arabidopsis thaliana) and finding that interphase chromosome organization is controlled by both genetic and environmental factors. Other notable studies included elucidation of the role of the Pairing homoeologous1 (Ph1) locus in chromosome interactions in somatic and meiotic cells in wheat (Triticum aestivum), identification of the link between homologous chromosome pairing in meiosis and recombination, and discovery of rapid chromosome movements in meiotic prophase.
Investigations of chromosome organization and arrangement in the nucleus have been conducted since the invention of the light microscope. With the development of molecular cytogenetics tools, these studies matured from mostly descriptive to more mechanism driven that aim to elucidate factors controlling chromosome organization and dynamics. Until very recently, chromosomes, particularly in plants and other multicellular eukaryotes, were mostly examined in fixed cells. These observations, although static themselves, provided indications that chromosome behavior is quite dynamic. Introduction of new microscopy methods that allow observations of chromosomes in live cells has confirmed the dynamic nature of chromosomes and enabled better understanding of the complexities of chromosome behavior. In this review, we mainly focus on two aspects of chromosome organization and dynamics that have received the most attention in the past few years in plant studies: chromosome organization in interphase nuclei and organization and dynamics of chromosomes during the prophase of meiosis.
CHROMOSOME ORGANIZATION AND DYNAMICS IN THE INTERPHASE NUCLEUS
Although organization of chromatin in interphase nuclei has been a subject of speculations for several decades, the past few years have brought much better understanding of this issue. In plants, studies conducted during the last 10 years resulted in elucidating interphase chromatin organization in Arabidopsis. A driving force behind this research is the desire to understand how organization of interphase chromosomes affects gene expression, although such studies are only now beginning in plants and other multicellular eukaryotes.
Chromosome Territories
During interphase, chromosomes assume a largely decondensed state. However, chromatin is still nonrandomly arranged within the nuclear space. Each chromosome occupies a limited, exclusive nuclear subdomain, known as a chromosome territory. The concept of chromosome territories was proposed by Carl Rabl in 1885, based on his observation of salamander cell division. Existence of chromosome territories was confirmed in the 1980s in human cells using FISH with chromosome-specific DNA probes (Manuelidis and Borden, 1988). Chromosome territories in plants were first visualized in Arabidopsis using chromosome-specific bacterial artificial chromosome FISH probes (Lysak et al., 2001).
Rabl Configuration
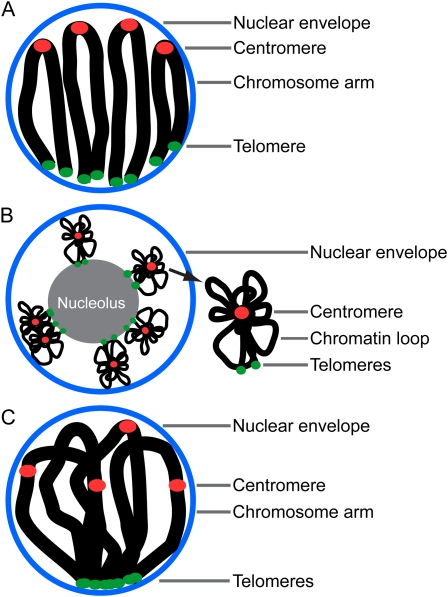
In many plant species with relatively large genomes, chromosomes during interphase adopt Rabl configuration (Cowan et al., 2001). This term describes an interphase chromosome arrangement in which centromeres and telomeres are located at opposite sides of the nucleus (Fig. 1A). This configuration is thought to be a remnant of a preceding anaphase. In some of these species, such as wheat or barley (Hordeum vulgare), Rabl configuration persists in all cells throughout the plant (Anamthawat-Jonsson and Heslop-Harrison, 1990). In rice (Oryza sativa), on the other hand, Rabl is only observed in certain tissues, such as xylem cells in the root and undifferentiated cells in the anther (Prieto et al., 2004a; Santos and Shaw, 2004). Other plant species, such as maize (Zea mays) and sorghum (Sorghum bicolor), despite having fairly large genomes, are not known to exhibit Rabl configuration at all (Dong and Jiang, 1998). In these species, chromosomes lose their polarized anaphase distribution of centromeres and telomeres after entering interphase.
Figure 1.
Patterns of chromosome arrangement in the nucleus. A, Rabl configuration found in interphase nuclei of many large-genome plant species. B, Rosette-like organization of chromosomes in interphase nuclei in Arabidopsis. C, Telomere bouquet.
Interphase Chromosome Organization in Arabidopsis
Interphase chromosomes in Arabidopsis do not display Rabl configuration but exhibit a strikingly different type of chromatin arrangement. In this species, telomeres cluster around the nucleolus while centromeres are located at the nuclear periphery (Armstrong et al., 2001; Fransz et al., 2002). Arabidopsis centromeric heterochromatin forms distinct, dense bodies called chromocenters. Chromocenters contain the majority of genomic repeats and exhibit epigenetic marks of inactive chromatin (Fransz et al., 2002). From the chromocenters, euchromatic loops of 0.2 to 2 Mb in length emanate, resulting in a rosette-like structure of Arabidopsis chromosome territories (Fig. 1B). Chromocenters of most Arabidopsis chromosomes do not seem to show preferential positioning relative to each other (Pecinka et al., 2004; Berr and Schubert, 2007; de Nooijer et al., 2009). Exceptions to this rule are chromosomes carrying nucleolar organizing regions (NORs), which contain tandemly arranged copies of rRNA genes (Pecinka et al., 2004). Physical association of NORs with the nucleolus is likely responsible for the nonrandom association of the NOR-bearing chromosomes. Although centromeres in Arabidopsis interphase nuclei do not cluster, telomeres show persistent clustering at the nucleolus (Armstrong et al., 2001). This phenomenon is not related to Rabl configuration but, similarly to Rabl, results in bringing certain chromosome regions into close vicinity of each other, which may have direct effects on interchromosome interactions and dynamics.
Factors Affecting Interphase Chromosome Organization
The arrangement of chromosome territories within the nucleus exhibits dynamic changes in response to various internal and external conditions. Histone modification and DNA methylation patterns are expected to affect chromosome organization, although data on this subject are still scarce. Nevertheless, it has been shown that in rice DNA demethylation causes chromatin decondensation and induces Rabl configuration in those tissues in which Rabl is not normally present (Santos et al., 2011). Changes in the ploidy level generated by endoreduplication have been shown to affect chromosome arrangement in Arabidopsis (Berr and Schubert, 2007). The shape and size of the nucleus is also related to chromosome arrangement, although it is not clear whether chromosome organization is the cause or a result of altered nuclear size and/or shape (Berr and Schubert, 2007; Dittmer et al., 2007).
Chromosome organization has been shown to change during plant development and in response to the environment. Chromocenters become smaller in leaves prior to the transition to reproductive development and recover to their former size after the elongation of the floral stem (Tessadori et al., 2007). Both processes are affected by light conditions. Furthermore, Arabidopsis genotypes acclimated to different latitudes exhibit genetically programmed levels of chromatin compaction, depending on the light intensity of their original habitats (Tessadori et al., 2009). In rice, heat stress has been shown to induce chromatin decondensation (Santos et al., 2011).
Functional Implications of Interphase Chromosome Organization
In the past few years, there has been a growing interest in understanding how chromosome and chromatin arrangement in interphase nuclei affect gene activity. Arrangement of chromosome territories that brings certain chromosome regions together has the potential to contribute to regulation of gene expression. This notion has lead to development of the concept of transcriptional factories, discrete sites in the nucleus where gene transcription is particularly active (Sutherland and Bickmore, 2009). Hundreds of such factories are proposed to be present in each nucleus and they are thought to be anchored to a nuclear substructure. On the other hand, there might be also heterochromatic neighborhoods in which gene expression is silenced. It has been proposed that physical interactions between gene copies located on different chromosomes may contribute to gene silencing (Lanctôt et al., 2007).
Effects of chromatin organization on gene expression are poorly understood in plants. In Arabidopsis, the majority of genes are located on euchromatic loops stretching out of the chromocenters (Fransz et al., 2002). However, it is unclear if there are particular nucleus regions that are occupied by highly expressed genes. We anticipate that the near future will bring a more complete picture of interphase chromosome arrangement and dynamics during growth and development as well as under various environmental conditions in plants. These data will be a starting point for understanding how interphase chromosome arrangement affects gene expression.
Interphase Chromosome Dynamics
In the past few years new tools have been developed to facilitate investigations of interphase chromosome dynamics in live cells in Arabidopsis (Fang and Spector, 2005; Matzke et al., 2005, 2008, 2010). However, so far, it appears that interphase chromosomes only display mostly limited, diffusive movements (Kato and Lam, 2003; Fang and Spector, 2005). Interstitial chromosome regions generally exhibit more movements than centromeres. Interestingly, endoreduplication-driven polyploidy has been found to reduce chromosome movement speed but increase the freedom of movement, i.e. the area within the nucleus to which movement of a chromosome segment is constrained (Kato and Lam, 2003). Overall, chromosome dynamics in interphase nuclei is still quite poorly understood. Further development of live-imaging tools should lead to substantial progress in this area, particularly in understanding the implications of interphase chromosome motility for gene activity as well as for genome maintenance processes, such as DNA replication or repair.
CHROMOSOME DYNAMICS IN MITOSIS
Even though chromosome segregation in mitosis is one of the most obvious and easily observable types of nuclear dynamics, patterns and mechanisms of mitotic chromosome segregation have, so far, been relatively poorly researched in plants. Nevertheless, live imaging of chromosomes during mitosis in root meristematic cells in Arabidopsis have yielded several interesting data (Fang and Spector, 2005). Chromosomes in mitosis show the most dynamic behavior during their congression to metaphase plate at the transition from prophase and metaphase and during their segregation in anaphase. During the prophase to metaphase transition, after breakdown of the nuclear envelope, condensed chromosomes relocate to the center of the cell and their centromeric regions gradually rotate to become oriented perpendicular to the metaphase plate (Fang and Spector, 2005). In anaphase, chromosomes move, centromere first, toward the opposite poles. This movement is not synchronous among all centromeres in the cell. Furthermore, a centromere may first start moving to one of the poles and later change direction and move to the other pole (Fang and Spector, 2005). Following anaphase, chromosomes assume the interphase configuration. However, chromosome positions and chromocenter arrangement in the nucleus in the daughter cells are not the same as in the mother cell (Fang and Spector, 2005; Berr and Schubert, 2007). On the other hand, chromosome positions in the two daughter cells often show mirror symmetry immediately after mitosis (Berr and Schubert, 2007).
CHROMOSOME DYNAMICS IN MEIOTIC PROPHASE
Prophase of the first division of meiosis is a period of some of the most dynamic chromosome behavior. During this time, chromatin undergoes major reorganization that includes: (1) chromosome condensation and establishment of meiotic chromosome structure, (2) pairing of homologous chromosomes, and (3) dynamic chromosome movements. The result of these activities is formation of stable chromosome pairs, the bivalents, which is essential for ensuring correct chromosome segregation at the end of meiosis.
Chromosome Condensation
Condensation is the most noticeable change in chromosome appearance in early meiosis and serves as the main criterion for dividing meiosis prophase into substages (Fig. 2). Chromatin condensation in leptotene, in addition to making chromosomes more compact, leads to establishment of a meiosis-specific chromosome structure. Adoption of meiosis chromosome structure is required for key processes of meiotic prophase I (Dawe et al., 1994). Studies in maize lacking ABSENCE OF FIRST DIVISION1, an α-kleisin participating in formation of the chromosome axis at the onset of meiosis, showed that proper chromosome structure is essential for meiotic recombination as well as chromosome pairing (Golubovskaya et al., 2006). Meiosis-specific patterns of chromatin remodeling have been also implicated in preconditioning specific chromosome regions to become sites of meiotic recombination events in mouse (Mus musculus) and budding yeast (Saccharomyces cerevisiae) studies (Borde et al., 2009; Baudat et al., 2010). Transcriptome analyses of Arabidopsis meiocytes showed that a staggering number of genes are expressed during meiotic prophase I (Chen et al., 2010). These gene expression patterns are also presumably results of chromatin remodeling in early meiosis. It remains to be seen whether all these genes are indeed needed for meiosis progression and their large number reflects the complexity of meiotic prophase, or whether the massive gene expression is a by-product of genome-wide chromatin opening to facilitate chromosome pairing and recombination.
Figure 2.
A diagram showing major events and processes of meiotic prophase I. Only two chromosomes, each in different color, are shown in the diagram on the left.
Pairing of Homologous Chromosomes
Chromosome pairing is a process in which two homologous chromosome copies find each other among all chromosomes in the nucleus and juxtapose. Pairing includes bringing chromosomes together into a close proximity as well as an intimate homology search to recognize the correct pairing partner. Pairing interactions at select locations are followed by alignment along the entire length of the chromosomes.
Homologous chromosome pairing in plants generally proceeds de novo at the onset of meiotic prophase and there is little evidence for persistent pairing of homologous chromosomes prior to meiosis. Some elements of interphase chromosome arrangement may, however, facilitate meiotic pairing. The Rabl-induced interphase centromere clustering in polyploid wheat affects progression of homologous pairing (Martinez-Perez et al., 2001). Similarly, the interphase telomere association with the nucleolus in Arabidopsis has been hypothesized to act in prealigning chromosomes and aiding pairing interactions in telomeric and subtelomeric regions (Armstrong et al., 2001). The basis of chromosome homology recognition in most species, including plants, is the DNA sequence along the entire chromosome. However, this rule does not exclude a potential for a role of chromatin states and modification patterns in chromosome pairing.
Although considerable progress has been made during the past decade in understanding the biological nature of chromosome pairing, it is still one of the least-explored aspects of meiosis. Several meiotic processes are known to contribute to homologous chromosome pairing, including meiotic recombination, chromosome motility in early substages of meiotic prophase, and formation of the telomere bouquet (see below).
Homologous Chromosome Pairing and Recombination
Studies in a variety of eukaryotes, including plant model species Arabidopsis and maize, suggest that successful completion of homologous chromosome pairing is tightly linked to the progression of meiotic recombination (Franklin et al., 1999; Li et al., 2004; Pawlowski et al., 2004; Ronceret et al., 2009). This intimate dependence of pairing on recombination exists also in fungi and mammals (Baudat et al., 2000; Peoples-Holst and Burgess, 2005), but, interestingly, not in Drosophila melanogaster or Caenorhabditis elegans (Dernburg et al., 1998; McKim et al., 1998).
Meiotic recombination is universally initiated by formation of double-strand breaks (DSBs) in chromosomal DNA (Fig. 2) by a conserved topoisomerase-like protein SPO11 (Lichten, 2001). Subsequently, the DSBs are resected, leading to formation of single-stranded DNA overhangs. Single-stranded DNA ends, which are several-hundred base pair long, invade dsDNA regions on the homologous chromosomes. This process, known as single-end invasion (SEI), is thought to be the basis of homology recognition during chromosome pairing in plants, fungi, and mammals (Bozza and Pawlowski, 2008). Defects in chromosome pairing have been observed in plant mutants in genes controlling DSB formation and resection, as well as SEI (Pawlowski and Cande, 2005). In most eukaryotes with relatively large genomes, including plants, the number of SEI sites is far greater than the number of crossovers. In maize, there are, on average, about 20 crossover sites per meiocyte, but as many as 500 SEI sites. These sites can be identified by immunolocalizing proteins that facilitate the SEI process, such as RAD51 (Fig. 2). RAD51 forms distinct foci on chromosomes in zygotene and pachytene. Studies in maize suggest that most, if not all, of the SEI sites are required to facilitate correct chromosome pairing. Mutants that exhibit reduced number of RAD51 foci show chromosome pairing defects as well (Pawlowski et al., 2003; Ronceret et al., 2009).
Chromosome Pairing and Genome Complexity
Although the dependence of pairing on recombination has been recognized, the exact nature of this link is not yet fully understood. Particularly, it is not clear how ectopic pairing is prevented between repetitive genome regions. For example, about 85% of the maize genome consists of repetitive DNA elements, many of which are several kilobase pair long (Schnable et al., 2009). These data suggest that there must be mechanisms that coordinate pairing along the entire length of chromosomes so that bivalents are only formed between homologous chromosome partners. However, the nature of these mechanisms remains unknown.
Polyploidy, which is frequent in many plant families, adds another level of complication to the process of pairing. While in autopolyploids chromosome pairing is generally disturbed and may lead to formation of multivalents, allopolyploid species have evolved mechanisms that can distinguish between homologous and homeologous chromosomes (i.e. chromosomes derived from different progenitors that are similar but not identical). Studies in polyploid wheat have demonstrated that homeologous associations between chromosomes from different genomes are suppressed by the Ph1 locus (Moore and Shaw, 2009). Absence of Ph1 leads to incorrect chromosome pairing (Al-Kaff et al., 2008). Ph1 is proposed to act by controlling remodeling of chromatin structure at the onset of meiosis (Prieto et al., 2004b; Colas et al., 2008). The chromatin conformational change affects the homology search and, in particular, the specificity of interactions between wheat centromeres (Martinez-Perez et al., 2001). In the presence of Ph1, associations between centromeres of homeologous chromosomes become disrupted and only homologous centromere interactions remain.
The Ph1 locus has been defined to a single wheat chromosome region that contains a cluster of genes related to the cell cycle regulator Cyclin-dependent kinase2 (Cdk2) gene (Griffiths et al., 2006). Cdk2 is known to control meiosis progression, expression of meiotic genes, meiotic DSB formation, as well as chromatin structure (Yousafzai et al., 2010). The function of Ph1 can be mimicked by application of okadaic acid, a drug known to induce chromosome condensation and affect meiosis progression by altering phosphorylation of the H1 histone (Knight et al., 2010). These data imply that histone phosphorylation and chromosome condensation may affect the chromosome pairing dynamics.
The presence of the Cdk2 gene cluster appears to be specific to tetraploid and hexaploid wheat (Griffiths et al., 2006). This observation implies that, even though they could exploit the same aspect of chromosome dynamics, mechanisms for preventing homeologous pairing have likely evolved many times independently in different polyploid taxa. Although the mechanism of Ph1 function still remains to be fully elucidated, it suggests existence, at least in some plant species, of chromatin-level homology recognition mechanisms that operate in addition to the DSB-dependent mechanism of homology search (Moore and Shaw, 2009).
Chromosome Motility in Meiotic Prophase I
Live-imaging observations in a number of species of plants, animals, and fungi, most of them conducted during the past few years, have demonstrated that early stages of meiotic prophase are a period of extremely dynamic chromosome movements (Koszul and Kleckner, 2009; Sheehan and Pawlowski, 2009; Baudrimont et al., 2010). In plants, studies using intact live anthers of maize showed that meiotic chromosomes exhibit complex and stage-specific motility patterns in zygotene and pachytene (Sheehan and Pawlowski, 2009). During zygotene, short chromosome segments adjacent to chromosome ends exhibit robust short-range movements, while movements of interstitial chromosome segments are more restrained. At the same time, the entire chromatin in the nucleus rotates back and forth in a coordinated manner at angles ranging from 7° to 10°, but sometimes as much as 90°. In pachytene, the rapid short-range chromosome end movements are replaced by slower but long-distance movements of much larger chromosome segments. The rotational movements, in contrast, persist through pachytene. Prophase chromosome movements in maize appear more complex in comparison to other species, as they include both coordinated chromatin rotations as well as movements of individual chromosome segments. In contrast to maize, only uncoordinated movements of individual chromosomes or chromosome segments are seen in budding yeast, while only coordinated movements of the entire chromatin have been reported in fission yeast (Schizosaccharomyces pombe) and rat (Rattus norvegicus) spermatocytes (Sheehan and Pawlowski, 2009).
The significance of the prophase chromosome movements is not yet entirely understood. It has been suggested that zygotene chromosome movements may aid homologous chromosome pairing by facilitating interchromosome interactions and disrupting associations of nonhomologous chromosomes (Koszul and Kleckner, 2009; Sheehan and Pawlowski, 2009). The pachytene movements, on the other hand, may help resolve chromosome entanglements (known as interlocks) that form during chromosome pairing in zygotene. Interlocks have to be disentangled prior to further chromosome condensation and segregation or chromosome breakage may occur.
The Role of Telomeres during Early Meiotic Prophase
Chromosome ends (telomeres) play a critical role in chromosome dynamics during meiotic prophase. In many species of plants, animals, and fungi, telomeres attach to the nuclear envelope and cluster within a small region, leading to formation of the telomere bouquet (Figs. 1C and 2; Harper et al., 2004). In yeast and mammals, telomeres cluster at the microtubule-organizing center. Plants do not have microtubule-organizing centers but immunocytological studies in rye (Secale cereale) showed that telomeres cluster to form the bouquet opposite a band of microtubules in early zygotene nucleus (Cowan et al., 2002). While telomeres cluster, centromeres do not, although they are generally located on the opposite side of the nucleus from the bouquet. This organization results in an overall polarization of the nucleus that is somewhat similar to Rabl configuration. However, the mechanism of the bouquet formation and the function of the bouquet are very different than those of Rabl configuration.
Based on analyses of mutants defective in the bouquet formation, it has been speculated that telomere clustering facilitates homologous pairing by bringing chromosome ends together (Harper et al., 2004). The plural abnormalities of meiosis1 (pam1) mutant of maize is one of the best-studied bouquet mutants in plants (Golubovskaya et al., 2002). Cytological analysis of pam1 showed that in early meiotic prophase telomeres in mutant meiocytes associate with the nuclear envelope but fail to cluster. The mutant also exhibits a reduction in pairing of homologous chromosomes and shows unresolved chromosome interlocks, all presumably resulting from the telomere bouquet formation defect.
Arabidopsis belongs to a small group of species (also including C. elegans and Drosophila) that do not form telomere bouquets (Harper et al., 2004). However, the clustering of Arabidopsis telomeres at the nucleolus present in interphase tends to persist into early meiosis, although the telomeres dissociate from the nucleolus during the course of leptotene and become widely dispersed within the nucleus (Armstrong et al., 2001). Subtelomeric chromosome regions begin to homologously pair prior to telomere detachment from the nucleolus. Based on this sequence of events, Armstrong et al. (2001) suggested that the premeiotic and early meiotic association of telomeres with the nucleolus in Arabidopsis may play a role similar to that of the bouquet in other species. Although Arabidopsis telomeres are not attached to the nuclear envelope during their nucleolus association, they do become transiently associated with the nuclear envelope during zygotene and occasionally exhibit loose clustering, although not classical bouquet formation.
Telomere Attachment to the Nuclear Envelope in Meiotic Prophase
Attachment of telomeres to the nuclear envelope during formation of the telomere bouquet is the basis of meiotic prophase chromosome dynamics. The telomere-nuclear envelope attachment is mediated by a multiprotein complex (Fig. 3). Proteins involved in this complex bridge the double-membrane nuclear envelope, tethering telomeres on the inner side of the nuclear envelope and linking them to the cytoskeleton on the outside. Several proteins involved in this complex have been identified in a variety of species. The best studied of them are proteins containing the conserved SUN (for Sad1p, UNC-84) domain. Homologs of these proteins have been identified in budding yeast and fission yeast mammals, C. elegans, as well as plants, maize, and Arabidopsis. SUN domain proteins bridge the inner membrane of the nuclear envelope. On their N terminus, they interact with telomere-binding proteins, while the C terminus is located in the lumen between the inner and outer nuclear membrane (Schmitt et al., 2007). The Arabidopsis genome encodes two SUN domain proteins, AtSUN1 and AtSUN2 (Graumann et al., 2010). Similarly to the SUN domain proteins from other in species, AtSUN1 and AtSUN2 localize to the inner nuclear membrane. However, this localization pattern has only been demonstrated so far in somatic cells and it remains to be shown whether the two proteins also function in meiosis.
Figure 3.
A diagram showing the telomere-nuclear envelope attachment involved in formation of the telomere bouquet.
Several telomere-binding proteins that interact with SUN domain proteins have been identified in fission yeast (Chikashige et al., 2006). However, homologs of these proteins have not been found yet in plants or other species as their sequences are fairly poorly conserved. It is also unclear what specific role the actual telomeres play in chromosome end attachment to the nuclear envelope and the bouquet formation. In the mouse, lack of the telomerase enzyme, which maintains telomeres and preserves their length, leads to defects in telomere bouquet formation and chromosome behavior (Liu et al., 2004), suggesting that presence of telomeric DNA repeats is important for bouquet function.
In yeast and mammals, it has been shown that the C-terminal part of SUN domain proteins interacts with another type of transmembrane proteins known as KASH (for Klarsicht, ANC-1, Syne Homology; Chikashige et al., 2007). On their cytoplasmic sides, KASH proteins interact with proteins that bind the cytoskeleton. Presumably, KASH proteins are also present in plants, although so far, this fact has not been directly demonstrated. The amino acid sequence of KASH proteins is much less conserved that the sequence of SUN domain proteins so identifying KASH protein homologs by sequence alone is difficult.
The Role of Cytoskeleton in Chromosome Dynamics
The SUN-KASH protein complex provides a physical link between chromosomes and the cytoskeleton. Analyses of meiotic prophase chromosome dynamics in a number of species, including maize, indicate that the physical forces responsible for chromosome movements are generated in the cellular cytoskeleton (Koszul and Kleckner, 2009; Sheehan and Pawlowski, 2009). From there, they are conveyed onto the nuclear envelope and then, by the virtue of telomere attachment to the nuclear envelope, further onto chromosome ends. Sheehan and Pawlowski showed that treating maize anthers with cytoskeleton-disrupting drugs, colchicine, which prevents tubulin polymerization, and latrunculin B, an inhibitor of actin polymerization, leads to complete cessation of prophase chromosome movements, as well as movements of the nuclear envelope, which accompany chromosome motility (Sheehan and Pawlowski, 2009). These data suggest that both actin and tubulin cytoskeletons play critical roles in meiotic prophase chromosome dynamics. Interestingly, the link between cytoskeleton and prophase chromosome dynamics in plants had been identified indirectly even before the development of methods for live imaging of meiotic chromosomes. In the 1970s, Driscoll and Darvey showed that colchicine disrupts homologous chromosome pairing (Driscoll and Darvey, 1970). Furthermore, Cowan and Cande demonstrated that colchicine forestalls the bouquet formation (Cowan and Cande, 2002). Studies using cytoskeleton-disrupting drugs have also been conducted in species outside of plants. Interestingly, these studies uncovered that different cytoskeletal components are involved in chromosome motility in different species. In fission yeast and mammals, chromosome movements require the microtubule cytoskeleton (Salonen et al., 1982; Ding et al., 1998) whereas in budding yeast, the actin cytoskeleton is used for this purpose (Scherthan et al., 2007; Koszul et al., 2008).
How exactly the cytoskeleton generates the various types of nuclear and chromosomal movements during meiotic prophase is not yet clear. Further studies to elucidate the organization of the meiocyte cytoskeleton are needed to elucidate these dynamics. However, observations of the effects of cytoskeleton-disrupting drugs on chromosome movements, bouquet formation, and meiotic prophase progression already shed new light on the function of the telomere bouquet. These studies suggest that, rather than brining chromosome ends together, the main role of the bouquet is facilitating chromosome motility by conveying movement-generating forces from the cytoskeleton to chromosomes. Future studies combining genetic dissection of bouquet components with live-imaging observations will help elucidate this issue.
CONCLUSION AND PERSPECTIVES
Several important advances have been made in the past few years in studies on chromosome organization and dynamics in plants. These studies have addressed various aspects of chromosome organization and behavior in interphase cells, as well as mitosis and meiosis. They also examined effects that chromosome organization and dynamics have on the key nuclear functions of maintenance, transcription, and transmission to progeny of genetic material. Results of some of the studies have shown that plants exhibit patterns of chromosome organization and behavior that are similar to those found in animals and fungi, such as existence of chromosome territories or the telomere bouquet formation. Other studies, however, have revealed plant-specific modifications of universal mechanisms, or existence of mechanisms that are entirely plant specific, specific to a certain group of plants, or even to individual species. An excellent example of the latter is the proposed mechanism of the Ph1 locus function, which employs a unique way of regulating activity of the widely conserved CDK2 protein to accomplish a function specifically needed in a polyploid species with highly similar ancestral genomes.
More important than the individual discoveries, however, has been the fact that chromosome research in plants has moved from mostly descriptive studies to hypothesis-driven research addressing the mechanisms of chromosome behavior. We expect further increase in the number of such studies in the future, as more cytological and genetic tools become available. We also anticipate that future studies will address the consequences of chromosome dynamics for gene expression and genome maintenance. Finally, we hope to see more studies in areas that have so far received limited attention in plants, for example chromosome dynamics during the mitotic cell division.
References
- Al-Kaff N, Knight E, Bertin I, Foote T, Hart N, Griffiths S, Moore G. (2008) Detailed dissection of the chromosomal region containing the Ph1 locus in wheat Triticum aestivum with deletion mutants and expression profiling. Ann Bot (Lond) 101: 863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anamthawat-Jonsson K, Heslop-Harrison JS. (1990) Centromeres, telomeres and chromatin in the interphase nucleus of cereals. Caryologia 43: 205–213 [Google Scholar]
- Armstrong SJ, Franklin FC, Jones GH. (2001) Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J Cell Sci 114: 4207–4217 [DOI] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. (2010) PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. (2000) Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 6: 989–998 [DOI] [PubMed] [Google Scholar]
- Baudrimont A, Penkner A, Woglar A, Machacek T, Wegrostek C, Gloggnitzer J, Fridkin A, Klein F, Gruenbaum Y, Pasierbek P, et al. (2010) Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans. PLoS Genet 6: e1001219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A, Schubert I. (2007) Interphase chromosome arrangement in Arabidopsis thaliana is similar in differentiated and meristematic tissues and shows a transient mirror symmetry after nuclear division. Genetics 176: 853–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V, Robine N, Lin W, Bonfils S, Géli V, Nicolas A. (2009) Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J 28: 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza CG, Pawlowski WP. (2008) The cytogenetics of homologous chromosome pairing in meiosis in plants. Cytogenet Genome Res 120: 313–319 [DOI] [PubMed] [Google Scholar]
- Chen C, Farmer AD, Langley RJ, Mudge J, Crow JA, May GD, Huntley J, Smith AG, Retzel EF. (2010) Meiosis-specific gene discovery in plants: RNA-Seq applied to isolated Arabidopsis male meiocytes. BMC Plant Biol 10: 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Haraguchi T, Hiraoka Y. (2007) Another way to move chromosomes. Chromosoma 116: 497–505 [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. (2006) Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 125: 59–69 [DOI] [PubMed] [Google Scholar]
- Colas I, Shaw P, Prieto P, Wanous M, Spielmeyer W, Mago R, Moore G. (2008) Effective chromosome pairing requires chromatin remodeling at the onset of meiosis. Proc Natl Acad Sci USA 105: 6075–6080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CR, Cande WZ. (2002) Meiotic telomere clustering is inhibited by colchicine but does not require cytoplasmic microtubules. J Cell Sci 115: 3747–3756 [DOI] [PubMed] [Google Scholar]
- Cowan CR, Carlton PM, Cande WZ. (2001) The polar arrangement of telomeres in interphase and meiosis: Rabl organization and the bouquet. Plant Physiol 125: 532–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CR, Carlton PM, Cande WZ. (2002) Reorganization and polarization of the meiotic bouquet-stage cell can be uncoupled from telomere clustering. J Cell Sci 115: 3757–3766 [DOI] [PubMed] [Google Scholar]
- Dawe RK, Sedat JW, Agard DA, Cande WZ. (1994) Meiotic chromosome pairing in maize is associated with a novel chromatin organization. Cell 76: 901–912 [DOI] [PubMed] [Google Scholar]
- de Nooijer S, Wellink J, Mulder B, Bisseling T. (2009) Non-specific interactions are sufficient to explain the position of heterochromatic chromocenters and nucleoli in interphase nuclei. Nucleic Acids Res 37: 3558–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. (1998) Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398 [DOI] [PubMed] [Google Scholar]
- Ding D-Q, Chikashige Y, Haraguchi T, Hiraoka Y. (1998) Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci 111: 701–712 [DOI] [PubMed] [Google Scholar]
- Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ. (2007) LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell 19: 2793–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Jiang J. (1998) Non-Rabl patterns of centromere and telomere distribution in the interphase nuclei of plant cells. Chromosome Res 6: 551–558 [DOI] [PubMed] [Google Scholar]
- Driscoll CJ, Darvey NL. (1970) Chromosome pairing: effect of colchicine on an isochromosome. Science 169: 290–291 [DOI] [PubMed] [Google Scholar]
- Fang Y, Spector DL. (2005) Centromere positioning and dynamics in living Arabidopsis plants. Mol Biol Cell 16: 5710–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin AE, McElver J, Sunjevaric I, Rothstein R, Bowen B, Cande WZ. (1999) Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell 11: 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I. (2002) Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc Natl Acad Sci USA 99: 14584–14589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya IN, Hamant O, Timofejeva L, Wang CJ, Braun D, Meeley R, Cande WZ. (2006) Alleles of afd1 dissect REC8 functions during meiotic prophase I. J Cell Sci 119: 3306–3315 [DOI] [PubMed] [Google Scholar]
- Golubovskaya IN, Harper LC, Pawlowski WP, Schichnes D, Cande WZ. (2002) The pam1 gene is required for meiotic bouquet formation and efficient homologous synapsis in maize (Zea mays L.). Genetics 162: 1979–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann K, Runions J, Evans DE. (2010) Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J 61: 134–144 [DOI] [PubMed] [Google Scholar]
- Griffiths S, Sharp R, Foote TN, Bertin I, Wanous M, Reader S, Colas I, Moore G. (2006) Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439: 749–752 [DOI] [PubMed] [Google Scholar]
- Harper L, Golubovskaya I, Cande WZ. (2004) A bouquet of chromosomes. J Cell Sci 117: 4025–4032 [DOI] [PubMed] [Google Scholar]
- Kato N, Lam E. (2003) Chromatin of endoreduplicated pavement cells has greater range of movement than that of diploid guard cells in Arabidopsis thaliana. J Cell Sci 116: 2195–2201 [DOI] [PubMed] [Google Scholar]
- Knight E, Greer E, Draeger T, Thole V, Reader S, Shaw P, Moore G. (2010) Inducing chromosome pairing through premature condensation: analysis of wheat interspecific hybrids. Funct Integr Genomics 10: 603–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S. (2008) Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell 133: 1188–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R, Kleckner N. (2009) Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections? Trends Cell Biol 19: 716–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. (2007) Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet 8: 104–115 [DOI] [PubMed] [Google Scholar]
- Li W, Chen C, Markmann-Mulisch U, Timofejeva L, Schmelzer E, Ma H, Reiss B. (2004) The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci USA 101: 10596–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M. (2001) Meiotic recombination: breaking the genome to save it. Curr Biol 11: R253–R256 [DOI] [PubMed] [Google Scholar]
- Liu L, Franco S, Spyropoulos B, Moens PB, Blasco MA, Keefe DL. (2004) Irregular telomeres impair meiotic synapsis and recombination in mice. Proc Natl Acad Sci USA 101: 6496–6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Fransz PF, Ali HB, Schubert I. (2001) Chromosome painting in Arabidopsis thaliana. Plant J 28: 689–697 [DOI] [PubMed] [Google Scholar]
- Manuelidis L, Borden J. (1988) Reproducible compartmentalization of individual chromosome domains in human CNS cells revealed by in situ hybridization and three-dimensional reconstruction. Chromosoma 96: 397–410 [DOI] [PubMed] [Google Scholar]
- Martinez-Perez E, Shaw P, Moore G. (2001) The Ph1 locus is needed to ensure specific somatic and meiotic centromere association. Nature 411: 204–207 [DOI] [PubMed] [Google Scholar]
- Matzke AJ, Huettel B, van der Winden J, Matzke M. (2005) Use of two-color fluorescence-tagged transgenes to study interphase chromosomes in living plants. Plant Physiol 139: 1586–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke AJ, Huettel B, van der Winden J, Matzke M. (2008) Fluorescent transgenes to study interphase chromosomes in living plants. Methods Mol Biol 463: 241–265 [DOI] [PubMed] [Google Scholar]
- Matzke AJ, Watanabe K, van der Winden J, Naumann U, Matzke M. (2010) High frequency, cell type-specific visualization of fluorescent-tagged genomic sites in interphase and mitotic cells of living Arabidopsis plants. Plant Methods 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Green-Marroquin BL, Sekelsky JJ, Chin G, Steinberg C, Khodosh R, Hawley RS. (1998) Meiotic synapsis in the absence of recombination. Science 279: 876–878 [DOI] [PubMed] [Google Scholar]
- Moore G, Shaw P. (2009) Improving the chances of finding the right partner. Curr Opin Genet Dev 19: 99–104 [DOI] [PubMed] [Google Scholar]
- Pawlowski WP, Cande WZ. (2005) Coordinating the events of the meiotic prophase. Trends Cell Biol 15: 674–681 [DOI] [PubMed] [Google Scholar]
- Pawlowski WP, Golubovskaya IN, Cande WZ. (2003) Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests the involvement of RAD51 in meiotic homology recognition. Plant Cell 15: 1807–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski WP, Golubovskaya IN, Timofejeva L, Meeley RB, Sheridan WF, Cande WZ. (2004) Coordination of meiotic recombination, pairing, and synapsis by PHS1. Science 303: 89–92 [DOI] [PubMed] [Google Scholar]
- Pecinka A, Schubert V, Meister A, Kreth G, Klatte M, Lysak MA, Fuchs J, Schubert I. (2004) Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 113: 258–269 [DOI] [PubMed] [Google Scholar]
- Peoples-Holst TL, Burgess SM. (2005) Multiple branches of the meiotic recombination pathway contribute independently to homolog pairing and stable juxtaposition during meiosis in budding yeast. Genes Dev 19: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto P, Santos AP, Moore G, Shaw P. (2004a) Chromosomes associate premeiotically and in xylem vessel cells via their telomeres and centromeres in diploid rice (Oryza sativa). Chromosoma 112: 300–307 [DOI] [PubMed] [Google Scholar]
- Prieto P, Shaw P, Moore G. (2004b) Homologue recognition during meiosis is associated with a change in chromatin conformation. Nat Cell Biol 6: 906–908 [DOI] [PubMed] [Google Scholar]
- Ronceret A, Doutriaux MP, Golubovskaya IN, Pawlowski WP. (2009) PHS1 regulates meiotic recombination and homologous chromosome pairing by controlling the transport of RAD50 to the nucleus. Proc Natl Acad Sci USA 106: 20121–20126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen K, Paranko J, Parvinen M. (1982) A colcemid-sensitive mechanism involved in regulation of chromosome movements during meiotic pairing. Chromosoma 85: 611–618 [DOI] [PubMed] [Google Scholar]
- Santos AP, Ferreira L, Maroco J, Oliveira MM. (2011) Abiotic stress and induced DNA hypomethylation cause interphase chromatin structural changes in rice rDNA loci. Cytogenet Genome Res 132: 297–303 [DOI] [PubMed] [Google Scholar]
- Santos AP, Shaw P. (2004) Interphase chromosomes and the Rabl configuration: does genome size matter? J Microsc 214: 201–206 [DOI] [PubMed] [Google Scholar]
- Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, Kaback DB. (2007) Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 104: 16934–16939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt J, Benavente R, Hodzic D, Höög C, Stewart CL, Alsheimer M. (2007) Transmembrane protein Sun2 is involved in tethering mammalian meiotic telomeres to the nuclear envelope. Proc Natl Acad Sci USA 104: 7426–7431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Sheehan MJ, Pawlowski WP. (2009) Live imaging of rapid chromosome movements in meiotic prophase I in maize. Proc Natl Acad Sci USA 106: 20989–20994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland H, Bickmore WA. (2009) Transcription factories: gene expression in unions? Nat Rev Genet 10: 457–466 [DOI] [PubMed] [Google Scholar]
- Tessadori F, Schulkes RK, van Driel R, Fransz P. (2007) Light-regulated large-scale reorganization of chromatin during the floral transition in Arabidopsis. Plant J 50: 848–857 [DOI] [PubMed] [Google Scholar]
- Tessadori F, van Zanten M, Pavlova P, Clifton R, Pontvianne F, Snoek LB, Millenaar FF, Schulkes RK, van Driel R, Voesenek LA, et al. (2009) Phytochrome B and histone deacetylase 6 control light-induced chromatin compaction in Arabidopsis thaliana. PLoS Genet 5: e1000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousafzai FK, Al-Kaff N, Moore G. (2010) Structural and functional relationship between the Ph1 locus protein 5B2 in wheat and CDK2 in mammals. Funct Integr Genomics 10: 157–166 [DOI] [PubMed] [Google Scholar]