Abstract
We have investigated the location of the Psb27 protein and its role in photosystem (PS) II biogenesis in the cyanobacterium Synechocystis sp. PCC 6803. Native gel electrophoresis revealed that Psb27 was present mainly in monomeric PSII core complexes but also in smaller amounts in dimeric PSII core complexes, in large PSII supercomplexes, and in the unassembled protein fraction. We conclude from analysis of assembly mutants and isolated histidine-tagged PSII subcomplexes that Psb27 associates with the “unassembled” CP43 complex, as well as with larger complexes containing CP43, possibly in the vicinity of the large lumenal loop connecting transmembrane helices 5 and 6 of CP43. A functional role for Psb27 in the biogenesis of CP43 is supported by the decreased accumulation and enhanced fragmentation of unassembled CP43 after inactivation of the psb27 gene in a mutant lacking CP47. Unexpectedly, in strains unable to assemble PSII, a small amount of Psb27 comigrated with monomeric and trimeric PSI complexes upon native gel electrophoresis, and Psb27 could be copurified with histidine-tagged PSI isolated from the wild type. Yeast two-hybrid assays suggested an interaction of Psb27 with the PsaB protein of PSI. Pull-down experiments also supported an interaction between CP43 and PSI. Deletion of psb27 did not have drastic effects on PSII assembly and repair but did compromise short-term acclimation to high light. The tentative interaction of Psb27 and CP43 with PSI raises the possibility that PSI might play a previously unrecognized role in the biogenesis/repair of PSII.
PSII is the multisubunit chlorophyll (Chl)-binding protein complex of cyanobacteria, algae, and plants that catalyzes electron transfer from water to plastoquinone (Barber, 2006). The native functional form of the PSII complex is most probably a dimer, and in the latest, most detailed structural model of cyanobacterial PSII, each monomer contains 16 intrinsic and three extrinsic protein subunits, 35 Chl a molecules, two pheophytins, 11 β-carotenes, two haem molecules, one nonhaem iron, four calcium ions, three chloride ions, two plastoquinones, more than 20 lipid molecules, and an Mn4Ca metal cluster that oxidizes water to dioxygen (Umena et al., 2011).
At the center of the complex are two homologous reaction center (RC) subunits called D1 and D2, encoded by the psbA and psbD genes, respectively. These subunits form a heterodimer that binds most of the cofactors involved in electron transfer within PSII. Unlike other PSII proteins, D1 is synthesized as a precursor with a C-terminal extension (Marder et al., 1984; Takahashi et al., 1988). In the cyanobacterium Synechocystis sp. PCC 6803 (hereafter called Synechocystis 6803), the extension consists of 16 amino acid residues, which are cleaved by the CtpA protease (Anbudurai et al., 1994) in two steps (Komenda et al., 2007) to allow assembly of the Mn4Ca cluster (Nixon et al., 1992).
Two Chl-binding inner antenna proteins, CP47 and CP43, are located symmetrically on either side of the D1-D2 heterodimer (Zouni et al., 2001) and deliver excitation energy to the RC, driving electron transfer. CP43 also provides an amino acid ligand to the Mn4Ca cluster (Ferreira et al., 2004). On the periphery of these large subunits are 13 small, mostly single-helix, subunits. The lumenal part of PSII is shielded by three extrinsic subunits, PsbO, PsbU, and PsbV, which stabilize the Mn4Ca cluster (for review, see Roose et al., 2007).
N-terminal sequencing and mass spectrometric analyses of proteins found in a His-tagged PSII preparation from Synechocystis 6803 have revealed the presence of several proteins that are absent in the PSII crystal structures (Kashino et al., 2002). One of these, the Psb27 subunit, is enriched in a His-tagged PSII preparation from a ctpA null mutant of Synechocystis 6803 unable to mature the D1 protein (Roose and Pakrasi, 2004). Consequently, Roose and Pakrasi (2004) suggested that Psb27 might be associated with the lumenal side of the complex, where the cleavable extension of the D1 precursor is located. This was subsequently confirmed in a later study in the thermophilic cyanobacterium Thermosynechococcus elongatus, which revealed that Psb27 is a lipoprotein associated with a monomeric PSII complex lacking the PsbO, PsbV, and PsbU extrinsic proteins (Nowaczyk et al., 2006) and also lacking a functional Mn4Ca cluster (Mamedov et al., 2007). Dimeric PSII-Psb27 complexes, inactive for water splitting, can also be isolated from T. elongatus grown at low temperature (Grasse et al., 2011).
Psb27 has been implicated in the assembly of the Mn4Ca cluster (Roose and Pakrasi, 2008), possibly during PSII repair (Nowaczyk et al., 2006). Two Psb27 homologs are found in Arabidopsis (Arabidopsis thaliana): One has been proposed to function in PSII repair (Chen et al., 2006), whereas the second is required for efficient maturation of D1 precursor (Wei et al., 2010). The solution structure of the soluble domain of Psb27 from Synechocystis 6803 has been determined by NMR spectroscopy (Cormann et al., 2009; Mabbitt et al., 2009), but as yet, the binding site of Psb27 within PSII is unknown.
Here, we report the localization of Psb27 within PSII and an analysis of its putative role in biogenesis of PSII in Synechocystis 6803. Our results show that Psb27 binds to and stabilizes unassembled CP43 and also indicate that during the PSII assembly pathway, both proteins bind to the CP47-D1-D2 complex together, forming the monomeric PSII core complex. Data also suggest the possible binding of Psb27 to PSI complexes.
RESULTS
The Psb27 Protein Is Associated with the CP43 Inner Antenna Complex
To gain more information about the location of Psb27 within PSII, we analyzed the membrane proteins of several strains of Synechocystis 6803 using two-dimensional (2D) blue native (BN)/SDS-PAGE and identified Psb27 using an antibody raised to Psb27 from Synechocystis 6803. Approximately 70% of the overall Psb27 level in the wild-type strain was detected in the monomeric PSII complex [RCC(1), Fig. 1, left, band 4], approximately 20% was detected in large supercomplexes close to the edge of the gel (bands 1 and 2), and the remaining Psb27 was found in the PSII dimer [RCC(2), band 3] and in a small complex (band 5) with mobility similar to that of unassembled CP43 or CP47. Similarly sized complexes, with the exception of band 2, were also detected in a strain lacking PSI (see Supplemental Fig. S1 online).
Figure 1.
Localization of the Psb27 protein by 2D-BN/SDS-PAGE in membranes of Synechocystis wild-type (WT) cells cultivated under 40 μmol photons m−2 s−1 and then exposed to 2,000 μmol photons m−2 s−1 for 20 min in the presence of chloramphenicol (WT 20 min HL + CAP). Membrane proteins were separated by 2D electrophoresis, the gel was either stained by Sypro Orange (Stained gel) or blotted to a PVDF membrane (Blot), and CP43 and Psb27 were detected by specific antibodies. The identity of other designated proteins on the stained gel was verified by MS. Designation of complexes: RCCS1 and RCCS2, PSII supercomplexes; RCC(2) and RCC(1), dimeric and monomeric PSII core complexes, respectively; PSI(3) and PSI(1), trimeric and monomeric PSI; RC47, PSII core complex lacking CP43; u.CP43 and horizontal arrows, unassembled CP43; oblique arrows with numbers, Psb27-containing complexes. Small PSI subunits PsaD and PsaF are also designated by dots.
To help localize Psb27, we exposed wild-type cells for 20 min to high irradiance (2,000 μmol photons m−2 s−1) in the presence of the protein synthesis inhibitor chloramphenicol to disassemble the large PSII complexes into smaller complexes. This treatment resulted in a partial reduction in the amount of RCC(2) and RCC(1) and increased levels of the PSII monomer lacking CP43 (designated RC47) and unassembled forms of CP47 and CP43 (Fig. 1, right). These changes to PSII were accompanied by a partial disappearance of Psb27 from RCC(1), paralleled by its appearance in the small complex (band 5) at the position of unassembled CP47 and CP43. No Psb27 signal was detected in the RC47 complex. There was also an increase in the amount of large complexes containing Psb27 and CP43.
To determine which protein formed a complex with Psb27 in the region of unassembled CP47 and CP43, we analyzed PSII complexes in the ΔCP47 strain accumulating an increased level of unassembled CP43 (Fig. 2A) and in the ΔCP43 strain containing RC47 and unassembled CP47 (Fig. 2B; Komenda et al., 2004). The data clearly supported the presence of Psb27 in the unassembled CP43 complex and its absence in free CP47, RC47, and RC complexes lacking both CP47 and CP43.
Figure 2.
Localization of the Psb27 protein in the membrane complexes of CP47-less (A) and CP43-less (B) mutants of Synechocystis. Membrane proteins from psbB deletion mutant, ΔCP47, and psbC deletion mutant, ΔCP43, were separated by 2D PAGE, blotted to a PVDF membrane, and CP43, CP47, D1, PsaD, and Psb27 proteins were detected by specific antibodies. Designation of complexes as in Fig. 1; RC47(1) and RC47(2), monomeric and dimeric form of the PSII core complex lacking CP43, respectively; RC* and RCa, RC complexes lacking both CP47 and CP43; u.CP47 and u.CP43, unassembled CP47 and CP43, u.CP43’ modified unassembled CP43; vertical arrows, Psb27-containing complexes related to PSI; iD1, D1 intermediate. [See online article for color version of this figure.]
Further evidence for a specific interaction between the CP43 complex and Psb27 was obtained by analysis of PSII complexes isolated from various strains accumulating His-tagged versions of unassembled CP43, unassembled CP47, RC47, and PSII core complexes (Boehm et al., 2011). Psb27 was detected only in the isolated CP43-His complex and in the His-tagged PSII core complex, whereas the CP47-His and RC47-His preparations were free of Psb27 (Fig. 3).
Figure 3.
Presence of Psb27 in various PSII complexes isolated using His-tagged CP43 and CP47. PSII complexes CP47-His, CP43-His, RC47-His, and complete core complexes (PSII-His) were isolated by a combination of Ni-metal affinity and size exclusion chromatography. The complexes were analyzed by SDS-PAGE in an 18% gel. Wild-type thylakoid membranes (0.5 μg of Chl a) and final samples of CP47-His, RC47-His, CP43-His, and PSII-His (1 μg of Chl a) were loaded on the gels, and proteins were either stained by Coomassie Blue (Stained gel) or blotted onto a PVDF membrane (Blot) and probed with antibody against T. elongatus Psb27. The arrow designates a CP43 fragment.
The level of Psb27 also correlated very well with the amount of unassembled CP43 complex found in various PSII mutants. In particular, the amount of Psb27 was approximately 4 times higher in ΔCP47 than in the wild type and was barely detectable in the ΔCP43 strain (Fig. 4).
Figure 4.
Accumulation of Psb27 in strains differing in the level of unassembled CP43 analyzed by Western blotting. Membranes from the wild type (WT), psbC deletion mutant ΔCP43, and psbB deletion mutant ΔCP47 were analyzed by denaturing SDS-PAGE, and D1 and Psb27 were detected using specific antibodies. Correct protein loading was shown by immunodetection with PsaD-specific antibody. One, 0.5, and 0.25 μg of Chl were loaded onto the gel for each sample.
The Psb27 Protein May Also Interact with PSI
Strikingly, 2D protein analyses of the membranes from the PSII− mutants, ΔCP47 and ΔCP43, indicated that the presence of Psb27 is not exclusively limited to PSII complexes (Fig. 2). Namely, low levels of Psb27 comigrated with trimeric [PSI(3)] and especially monomeric [PSI(1)] PSI complexes (Figs. 1, 2, and 5). The Psb27 signals comigrating with PSI in the ΔCP43 strain disappeared when PSI was additionally deleted (in the ΔCP43/ΔPSI strain lacking psaA, psaB, and psbC), suggesting a specific interrelationship between Psb27 and PSI (see Supplemental Fig. S2 online).
Figure 5.
Accumulation of CP43 in the psbB deletion mutant ΔCP47 and the double mutant ΔCP47/ΔPsb27 analyzed by 2D BN/SDS-PAGE. Membranes of ΔCP47 (left) and ΔCP47/ΔPsb27 (right) were analyzed by 2D BN/SDS-PAGE in combination with immunoblotting. Top, Sypro Orange-stained gels. Bottom, Corresponding blots obtained using antibodies specific for CP43 and Psb27. Five micrograms of Chl was loaded for each sample. Designation of complexes as in Fig. 1; oblique arrows, fragment of CP43; vertical arrows, CP43-free complexes of Psb27 with PSI complexes; asterisks, cross-reactions of the anti Psb27 antibody; dots, small PSI subunits PsaD and PsaF. [See online article for color version of this figure.]
To find further support for the unexpected interaction of Psb27 with PSI, we isolated the PSI complex using the strain expressing a His-tagged PsaF subunit of PSI protein (the F-His strain; Kubota et al., 2010). The complex isolated by single-step nickel-affinity chromatography was analyzed by 2D BN/SDS-PAGE, and Psb27 was detected in the trimeric form of PSI as well as in the region of unassembled protein, most probably because of its release from PSI during native PAGE (see Supplemental Fig. S3, right).
The third line of support for an interaction between Psb27 and PSI was obtained by yeast two-hybrid analyses using the split ubiquitin system (Pasch et al., 2005; Komenda et al., 2008). In this system, protein interaction-dependent reporter gene activation leads to His prototrophy of yeast cells. Supplemental Figure S4 online shows that on medium lacking His, yeast cells can grow when Psb27 and the PsaB subunit of PSI are coexpressed. By contrast, no growth was detected in a control in which the unrelated yeast eR-protein Alg5 was coexpressed with Psb27 and YCF48 with PsaB. These data provide evidence for an association of Psb27 with PSI via the PsaB subunit.
Putative Interaction of CP43 with PSI
We also detected in the ΔCP47 strain low levels of a complex apparently containing CP43 and PSI(1) [Fig. 5, PSI(1)-CP43 band]. Support for this initial assignment came from comigration of CP43 with a green band containing the small PSI proteins PsaD and PsaF plus absence of this CP43 signal in the double mutant ΔCP47/ΔPsb27. A specific interaction between CP43 and PSI was subsequently confirmed in pull-down experiments using His-tagged complexes. Monomeric PSI complexes copurified with CP43-His (but not with CP47-His; see Supplemental Fig. S5 online), and CP43, but not D1, was detected in His-tagged PSI (see Supplemental Fig. S3, right).
A close relationship between CP43 and PSI was also suggested following radioactive pulse-labeling experiments using a ΔPSI strain, which showed a much lower level of labeling of the CP43 apopolypeptide than in the wild-type cells grown under the same conditions (see Supplemental Fig. S6 online). In addition, ΔPSI also accumulated much more newly synthesized RC47 with highly labeled D2 and D1, indicating lack of CP43 to accomplish the de novo assembly of the core complexes.
Psb27 Stabilizes Unassembled CP43
Identification of Psb27 as a binding partner of unassembled CP43 suggested a possible role for Psb27 in the biogenesis of the CP43 during de novo assembly of PSII. To test this, levels of unassembled CP43 were analyzed in the ΔCP47 strain and in the ΔCP47 strain lacking Psb27 (ΔCP47/ΔPsb27). In the ΔCP47 mutant, CP43 accumulates to approximately 70% of the wild-type level but cannot be incorporated into PSII complexes and remains unassembled (Komenda et al., 2004). Deletion of the psb27 gene in this strain led to an approximate 50% decrease in the accumulation of CP43 as estimated by the densitometric scanning of the CP43 band on a 2D gel (Fig. 5). Moreover, a significant fraction of the protein was cleaved and detected as a 28-kD fragment (Fig. 5, oblique arrow), similar in size to that detected previously in isolated His-tagged CP43 complexes (Fig. 4; see Supplemental Fig. S7, arrows; Boehm et al., 2011).
Characterization of the Isolated CP43-His Complex
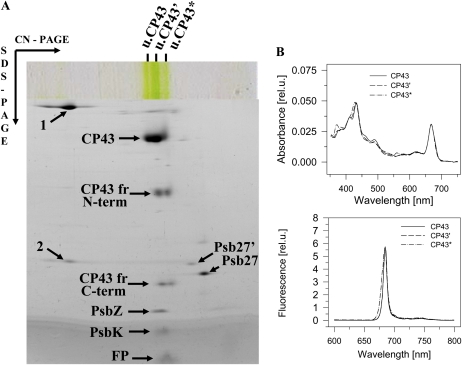
To characterize CP43 fragmentation in more detail, we analyzed the His-tagged CP43 complex isolated by single-step immobilized nickel-affinity chromatography. Using clear-native (CN)-PAGE, three Chl-binding bands were separated in the CP43-His preparation (Fig. 6A). We used this type of electrophoresis because, unlike BN-PAGE, it enables spectroscopic characterization of resolved pigment proteins. The two slower migrating complexes had similar mobilities to the CP43 and CP43’ bands detected in cells of the ΔCP47 strain (Fig. 2A). Analysis of their subunit composition using SDS-PAGE in the second direction coupled to mass spectrometry (MS) analysis of resolved protein spots confirmed that both bands contained the CP43 apopolypeptide and the low-molecular mass (LMM) subunits, PsbZ and PsbK. By contrast, the fastest migrating band, termed CP43*, contained PsbK but not PsbZ. In the CP43’ and CP43* bands, we detected the aforementioned N-terminal 28-kD fragment of the CP43 apoprotein and also a C-terminal 8-kD fragment, which did not react with our anti-CP43 antibody. By contrast, the CP43 apopolypeptide was not fragmented in the CP43 band. Detailed MS analysis of the CP43 fragments revealed that the CP43 apopolypeptide had been cleaved in the lumenal region between the fifth and sixth transmembrane helices and surprisingly that the CP43 fragments were not complementary, with approximately 84 to 87 amino acid residues missing (Table I). We unequivocally determined the N-terminal residue of the smaller C-terminal fragment as S390; by contrast, two different C-terminal residues, T303 and V306, were detected in the 28-kD N-terminal fragment. Thus, the proteolysis of the CP43 apoprotein resulted in the loss of the major part of the lumenal region between helices 5 and 6, including almost all the helical motifs.
Figure 6.
Separation of different forms of His-tagged CP43 by 2D-CN/SDS-PAGE (A) and their absorption and 77 K fluorescence spectra (B). A, The CP43-His complex isolated by Ni-metal affinity chromatography was analyzed by 2D CN/SDS-PAGE, and separated proteins were identified by MS. The numbered spots with arrows are not specific to CP43-His and represent CbbL (1) and CbbS (2). The other proteins designated by arrows are specific for CP43-His and represent PSII subunits or their fragments (Table I). FP, Free pigments. B, Room temperature absorption and 77K Chl fluorescence spectra of His-tagged CP43 forms measured directly in green bands excised from the gel. [See online article for color version of this figure.]
Table I. MS identification of proteins associated with the CP43-His protein and separated by CN/SDS-PAGE.
| Protein | Protease | No. of Peptides | Sequence Coverage | MALDI-TOF Tandem MS Fragmentation |
| % | ||||
| CP43a | Trypsin | 39 | 65 | 296ASQSQAFTFLVRD308 |
| Glu-C | 379AAEYMTHAPLGSLN SVGGVITDVNSFNYVSPR410 | |||
| CP43a N terminal | Trypsin | 11 | 53 | 296ASQSQAFT303 |
| Glu-C | 296ASQSQAFTFLV306 | |||
| CP43a C terminal | Trypsin | 4 | 63 | 390SLNSVGGVITDVNSFNYVSPR410 |
| Glu-C | ||||
| Psb27’b | Trypsin | 8 | 46 | S-(sn-1-Glyceryl)-1CDSGTGLTGNYSQDTLTVIATLR23 |
| Psb27b | Trypsin | 9 | 50 | Not applied |
| PsbZb | Trypsin | 1 | 12 | 28ASPQNWDR35 |
| PsbKb | Chymotrypsin | 5 | 51 | 1KLPEAYQIF9 29VWQAAVGFK37 |
Identified by liquid chromatography-ESI-FT MS. bIdentified by MALDI-FT MS.
We also excised the Chl-protein bands from the gel and measured their absorption and 77K Chl fluorescence spectra (Fig. 6B). The CP43 band showed a similar fluorescence spectrum to CP43’, with a maximum emission at 685 nm. The CP43* band contained a lower level of carotenoid and also exhibited a fluorescence maximum at 685 nm, but in addition, there was a shoulder corresponding to a Chl species emitting at around 679 nm (indicated by the second derivative spectrum, not shown). The CP43 apopolypeptide of CP43* also showed a smeared characteristic, indicating protein oxidation (Lupínková and Komenda, 2004). All three separated Chl-protein bands were free of Psb27, which was detected in the region of unassembled proteins. However, horizontal smearing of the Psb27 band starting from the mobility region of CP43 complexes suggested its release from CP43 complexes during electrophoresis. Indeed, when we analyzed the isolated His-tagged CP43 using BN/SDS-PAGE, a portion of Psb27 remained attached to CP43 (see Supplemental Fig. S8 online). Two spots of Psb27 were resolved by 2D CN/SDS-PAGE (Fig. 6A). The spot labeled Psb27’, of lower mobility, contained a glycerol molecule bound to the N-terminal Cys (Table I), whereas the Psb27 spot probably represented the protein with its full complement of lipids, as previously detected in T. elongatus (Nowaczyk et al., 2006). Unfortunately, we were not able to confirm this because the lipidated N-terminal peptide of the protein was always missing in the analyzed MS spectra, most probably due to its highly hydrophobic character and insufficient ionization.
Role of Psb27 in PSII Repair
To test the physiological importance of Psb27 for photoprotection and PSII repair, we assessed the sensitivity of a deletion mutant, ΔPsb27 (see Supplemental Fig. S9 online), to photoinhibition by following the time course of light-induced inhibition of oxygen evolution in cultures of the wild type and ΔPsb27 subjected to white light of 500 μmol photons m−2 s−1, either in the absence or presence of the protein synthesis inhibitor lincomycin. In both the absence and the presence of lincomycin, the initial decrease of activity was slightly faster in the mutant than in the wild type, but subsequently, the time course of the PSII activity was very similar in both strains (Fig. 7A). In agreement with this result, growth of the wild type and mutant cells was identical under low irradiance, but upon exposure to increased irradiance, the ΔPsb27 mutant showed a short lag phase in growth (Fig. 8). Thus, the presence of the Psb27 protein seems to be advantageous for rapid acclimation to high light. Pulse-chase experiments (Fig. 7B) showed that the turnover of D1 in ΔPsb27 was somewhat faster than in the wild type. Thus, PSII activity seems to be maintained in the mutant at increased irradiance by more frequent replacement of D1.
Figure 7.
PSII repair (A) and degradation of the PSII proteins (B) in the wild type (WT) and ΔPsb27 strains under high irradiance. A, Autotrophic wild-type cells (left) and ΔPsb27 (right) were illuminated with 500 μmol photons m−2 s−1 of white light for 180 min without antibiotic (black symbols) or in the presence of 100 μg mL−1 lincomycin (white symbols). During illumination, PSII oxygen-evolving activity was assayed in whole cells. The initial values of oxygen evolution were 672 ± 10 μmol O2 mg Chl−1 h−1 and 708 ± 60 μmol O2 mg Chl−1 h−1 for the wild type and ΔPsb27, respectively. B, Autotrophic cells of both strains were pulse-labeled with [35S]Met/Cys, and then the chase of the label was followed at irradiance of 500 μmol photons m−2 s−1 of white light for 6 h. Thylakoids were isolated and run on an SDS-PAGE gel. The gel was stained (Gel stain), dried, and radioactive labeling of the proteins was visualized using a PhosphorImager (Autorad).
Figure 8.
Growth curves of the Synechocystis wild type (WT) and ΔPsb27 cells under 10 or 100 μmol photons m−2 s−1. Wild-type cells were grown autotrophically under 40 μmol photons m−2 s−1, diluted to the initial OD750nm of 0.005, and transferred to microtitration plates exposed to the given irradiances; plotted values are means of 11 measurements ± sd.
When we compared the distribution of the different types of PSII complex in the wild type and ΔPsb27 using 2D CN/SDS-PAGE, we noticed that ΔPsb27 exhibited a higher ratio of RCC(2)/RCC(1) (a value of 1–1.5 compared with 0.5–0.7 in the wild type) and that the level of large CP43-containing complexes was higher in the mutant, whereas unassembled CP43 was more abundant in the wild type (see Supplemental Fig. S10 online).
DISCUSSION
Location of Psb27 within PSII
We have obtained four lines of evidence to support a close structural relationship between the CP43 antenna and Psb27: the first comes from comigration of Psb27 and CP43 in 2D native gels of wild type and PSII mutants (Figs. 1 and 2), the second from pull-down experiments using different types of His-tagged PSII complexes with Psb27 found in the CP43-His complex (Fig. 3), the third from the interdependence of the Psb27 and CP43 cellular levels (Fig. 4), and the fourth from the destabilization of unassembled CP43 in the absence of Psb27 (Fig. 5). In addition, parallel work has shown that Psb27 can be cross-linked to CP43 in isolated His-tagged Psb27-containing PSII core complexes (Liu et al., 2011a).
It is important to mention that the binding of Psb27 to CP43 was more stabilized under conditions of BN- than CN-PAGE. In BN-PAGE, the substance giving negative charge to complexes is Coomassie Blue, which contains two distant sulfo groups. By contrast, the detergent deoxycholate used in CN-PAGE contains only a single carboxyl group. We speculate that the two strongly acidic sulfo groups of Coomassie Blue can form a bridge stabilizing mutual binding of Psb27 and CP43 because of interactions with positively charged residues in both subunits. The destabilization of Psb27 binding during CN-PAGE was apparent both in the membranes of the wild type (see Supplemental Fig. S10 online, compare with Fig. 1) and in isolated His-tagged CP43 (Fig. 6, compare with Supplemental Fig. S8 online). In contrast with BN-PAGE, the majority of Psb27 in the wild type was identified as a smeared band in the region of unassembled proteins in the CN gel.
Further information on the Psb27 binding site can be inferred from the study of the ΔCP47/ΔPsb27 strain. The 2D analysis of the membrane proteins from this strain showed extensive fragmentation of unassembled CP43 (Fig. 5). A similarly sized N-terminal fragment was also detected in the isolated His-tagged CP43 complex (Fig. 6; see Supplemental Fig. S7 online). MS analysis identified cleavage sites in the long lumenal loop of the protein between the fifth and sixth transmembrane helix, so that the region between residues CP43-Thr-303 and CP43-Ser-390 was removed. It therefore seems likely that Psb27 is bound to CP43 in the vicinity of this loop region and that binding might prevent access of a protease to the loop. Such a location is in agreement with the recent detection of a cross-link between CP43-Asp-321 and Psb27-Lys-63 (Liu et al., 2011a). Earlier in silico docking models had proposed binding sites for Psb27 close to that of the bulk of PsbO (Nowaczyk et al., 2010; Fagerlund and Eaton-Rye, 2011). However, these binding sites are not easily reconciled with the parallel binding of PsbO and Psb27 to monomeric PSII (Liu et al., 2011b).
The isolated “unassembled” His-tagged CP43 complex to which Psb27 binds also contains the PsbK, PsbZ, and Ycf12/Psb30 subunits (Boehm et al., 2011; Table I). In principle, Psb27 might also interact with these LMM subunits. According to the latest, most detailed PSII structural models (Ferreira et al., 2004; Guskov et al., 2009; Umena et al., 2011), these three LMM subunits are bound in proximity to the first two helices of CP43, relatively far from the proposed binding site of Psb27 within the lumenal loop connecting transmembrane helices 5 and 6. In line with this, Psb27 still associates with monomeric and dimeric PSII core complexes in a mutant lacking PsbK (see Supplemental Fig. S3, left). A role for PsbZ and Psb30 in binding Psb27 seems equally unlikely, given that the absence of PsbK in T. elongatus also destabilizes binding of PsbZ and Ycf12/Psb30 to CP43 (Iwai et al., 2010).
Biogenesis of the Cyanobacterial CP43 Antenna
Our data provide strong evidence for the participation of Psb27 in the biogenesis of CP43. The CP43-Psb27 complex was identified not only in the strain lacking CP47 (Fig. 2) but also in wild-type cells (Fig. 1), which indicates that association of Psb27 with CP43 is not an artifact caused by lack of CP47. A functional role for Psb27 in stabilizing the unassembled CP43 complex comes from the observation that levels of CP43 are reduced significantly in the ΔCP47 strain when Psb27 is absent and that CP43 is prone to cleavage (Fig. 5; see Supplemental Fig. S7 online). This stabilizing role of Psb27 would help explain why levels of CP43 in mutants lacking either D1 or D2 are significantly higher than those of unassembled CP47 (Komenda et al., 2004, 2006). For the wild type, levels of unassembled CP43 are also reduced in the absence of Psb27 (see Supplemental Fig. S10 online).
Role of Psb27 in PSII Repair
The main evidence to support a role for Psb27 in PSII repair has come from [15N]-pulse labeling experiments in the cyanobacterium T. elongatus. Nowaczyk et al. (2006) found that the D1 protein in the isolated monomeric RCC(1)-Psb27 complex was the most intensively labeled protein in the complex followed by D2, CP43, and CP47, consistent with a population of PSII complexes that had undergone repair. However, preferential labeling of D1 is not a totally unambiguous marker for PSII repair. For instance, variations in rates of translation and the pool sizes of unassembled PSII subunits in the membrane mean that PSII complexes assembled de novo might still show preferential D1 labeling. Thus, Psb27-containing PSII core complexes might be a product of both PSII repair and de novo assembly. It also remains unclear whether the RCC(1)-Psb27 complex lies on the main assembly/repair pathway or can exist as a distinct subpopulation of PSII.
Our measurement of the rate of PSII inactivation in the ΔPsb27 mutant in the absence and presence of lincomycin did not provide evidence for a dramatic role for Psb27 in PSII repair under the experimental conditions used. What was observed was a subtle effect on maintaining PSII activity on exposing cells to high-light irradiances. Under these conditions, the ΔPsb27 mutant showed a faster loss of PSII activity than the wild type in the first 30 min, which appeared to be compensated for at later times by more frequent D1 replacement (Fig. 7A). Growth of the ΔPsb27 strain was also slightly impaired upon onset of high-light treatment (Fig. 8). Therefore, one plausible physiological role for PSII complexes containing Psb27, not previously discussed, might be to act as a pool or reservoir of preassembled complexes lacking a functional Mn4Ca cluster that can be rapidly photoactivated, forming a fully functional oxygen-evolving complex, to replace damaged PSII complexes when the rate of PSII damage suddenly exceeds its rate of repair, either through increased rates of damage (e.g. exposure to high-light intensities) or impaired efficiency of repair (e.g. exposure to low temperature). Such a mechanism would be fast-acting and would allow PSII homeostasis to be maintained in response to a fluctuating environment.
The Tentative Interaction of Psb27 and CP43 with PSI
We have shown here that Psb27 is associated not only with monomeric RCC(1) complexes but also with dimeric RCC(2) complexes and even larger complexes, possibly including PSII-PSI supercomplexes (Fig. 1). We have also obtained experimental support for the binding of both CP43 and Psb27 to PSI. The physiological significance of these interactions with PSI is presently unclear. Binding of CP43 to PSI might to some extent resemble the binding of the CP43 homolog, IsiA, to PSI under iron stress (Bibby et al., 2001; Boekema et al., 2001), where it functions as an extremely effective light-harvesting antenna (Melkozernov et al., 2003). By analogy, docking of CP43 to PSI would permit efficient excitation energy transfer (or spillover) from PSII to PSI, thereby helping to protect PSII from photodamage. Indeed, unlike the unassembled CP43 complex, which strongly fluoresces, CP43 in complex with the monomeric PSI complex does not exhibit significant fluorescence at room temperature (see Supplemental Fig. S5A online), consistent with efficient energy transfer from CP43 to PSI. As indicated in Figure 5, Psb27 seems to facilitate binding of CP43 to PSI and in this way may help protect CP43 from light-induced damage. Spillover could also protect PSII core complexes during the light-driven assembly of the Mn4Ca cluster during de novo assembly or repair. Therefore, the correct regulation of the interaction between PSII and PSI components would be important during PSII biogenesis and repair, and in this respect Psb27, if it is able to bind to both PSI and PSII, could play an important role.
MATERIALS AND METHODS
Construction and Cultivation of Cyanobacterial Strains
The strains used in the study were derived from the Glc-tolerant strain of Synechocystis sp. PCC 6803 (Williams, 1988) referred to as the wild type. The following, previously described strains were used in the study: (1) the CP43-less strain, ΔCP43 (Vermaas et al., 1988); (2) the CP47 deletion strain, ΔCP47, in which the psbB gene is replaced by a spectinomycin-resistance cassette (Eaton-Rye and Vermaas, 1991); (3) the PSI-less mutant ΔPSI, with both psaA and psaB replaced by chloramphenicol-resistance cassette (Shen et al., 1993) and its derivative lacking also the psbC gene (ΔCP43/ΔPSI); (4) the psbK deletion strain, ΔPsbK, in which the psbK gene is replaced by a kanamycin-resistance cassette (Zhang et al., 1993); (5) His-tagged strains accumulating unassembled CP47-His, unassembled CP47-His, and intact His-tagged PSII core complexes (PSII-His; Boehm et al., 2011); and (6) strain F-His expressing a His-tagged derivative of the PsaF subunit of PSI (Kubota et al., 2010).
The ΔPsb27 strain was prepared using overlap extension PCR. Approximately 500-bp DNA fragments upstream and downstream of psb27 (slr1645) were amplified as the flanking sequences for homologs recombination. The flanking sequences were then fused together using overlap extension PCR with primer 1-F and 4-R, and an EcoRV site was designed to replace the open reading frame of Psb27 (see Supplemental Fig. S8 online). The resulting PCR fragment was then cloned into the pGEM-T Easy vector backbone to create vector pPsb27. A chloramphenicol resistance cassette was used as selectable marker and was inserted to the EcoRV site to generate pPsb27 CamA vector. The orientation of the resistance cassette is the same as the orientation of the psb27 gene. The pPsb27CamA plasmid was then used to transform the wild-type cells. Mutants were selected for chloramphenicol resistance and were examined for segregation by PCR. The ΔCP47/ΔPsb27 double mutant was obtained by transforming the ΔPsb27 mutant with chromosomal DNA isolated from ΔCP47 (Vermaas et al., 1988). Its selection and segregation were based on additional resistance to spectinomycin. The His-tagged RC47 strain was constructed by transforming the CP47-His strain with a plasmid construct containing the psbC gene interrupted by an erythromycin-resistance cassette (M. Boehm, unpublished data).
Liquid cultures were grown in 100 to 200 mL of BG11 containing 5 mm Glc using 500-mL Erlenmeyer flasks, aerated using an orbital shaker, irradiated with 40 μmol photons m−2 s−1 white light at 29°C, and were used when they reached a Chl concentration of 2 to 3 μg mL−1. Solid medium also contained 10 mm TES/NaOH, pH 8.2, 1.5% agar, and 0.3% sodium thiosulfate (Pakrasi et al., 1988). For the large-scale cultivation used for isolation of PSII complexes, cultures were grown in 10-L flasks (culture volume 6–8 L) or 20-L carboys, stirred by a magnetic stirrer, and bubbled with air.
Measurement of growth curves was performed in a 96-well microtitration plate illuminated at 10 or 100 μmol photons m−2 s−1 and intensively shaken at 29°C. The cultures were initially diluted to an optical density of 750 nm (OD750 nm) of 0.005 (Chl concentration 0.015 μg mL−1), and OD750 nm was measured every 6 h using microplate reader (Tecan Sunrise). Values plotted against time were used for calculation of the doubling time.
Polarographic Methods
The light-saturated steady-state rate of oxygen evolution in cell suspensions was measured polarographically in BG-11 medium containing 10 mm HEPES/NaoH, pH 7.0, using 0.5 mm p-benzoquinone and 1 mm potassium ferricyanide as artificial electron acceptors.
Preparation of Membranes and Their Protein Analyses
Cyanobacterial membranes were prepared by breaking the cells using glass beads (Komenda and Barber, 1995) with the modifications described by Dobáková et al. (2009). The large-scale isolation of PSII complexes for chromatographic purification was performed using a procedure described by Boehm et al. (2011).
The protein complexes isolated from the thylakoid membranes were solubilized in 1% n-dodecyl-β-d-maltoside and analyzed by BN electrophoresis at 4°C in a 4% to 14% gradient polyacrylamide gel according to Schägger and von Jagow (1991). Alternatively, a CN variant of the method was used in which Coomassie Blue was omitted from all solutions, and upper electrophoretic buffer contained 0.05% sodium deoxycholate and 0.02% n-dodecyl-β-d-maltoside. Samples with the same Chl content (usually 5 μg for gel staining and 1 μg for Western blot) were loaded onto the gel.
The protein composition of the complexes was analyzed by electrophoresis in a denaturing 12% to 20% linear gradient polyacrylamide gel containing 7 m urea (Komenda et al., 2002). Complete lanes from the native gel were excised, incubated for 30 min in 25 mm Tris/HCl, pH 7.5, containing 1% SDS (w/v), and placed on top of the denaturing gel; two lanes were analyzed in a single denaturing gel. One-dimensional SDS-PAGE for analysis of pulse-chase labeled proteins was carried out in the same 12% to 20% polyacrylamide gel containing 7 m urea. Proteins separated in the gel were stained either by Coomassie Blue or by Sypro Orange and then subsequently transferred onto a polyvinylidene difluoride (PVDF) membrane. Membranes were incubated with specific primary antibodies and then with secondary antibody-horseradish peroxidase conjugate (Sigma-Aldrich). The primary antibodies used in the study were raised in rabbits against (1) residues 58 to 86 of the spinach D1 polypeptide, (2) the last 12 residues of plant D2 (Komenda et al., 2004), (3) residues 380 to 394 of barley CP47, (4) whole isolated CP43 from Synechocystis 6803, (5) the whole isolated PsaD from Synechocystis spp., and (6) Psb27 from either Synechocystis 6803 (antiserum kindly provided by Julian Eaton-Rye) or Thermosynechococcus elongatus, both expressed in Escherichia coli. For autoradiography, the gel or the membrane with labeled proteins was exposed to x-ray film at laboratory temperature for 2 to 3 d or to a PhosphorImager plate (GE Healthcare) overnight.
Samples with the same Chl content were used for the direct comparison and quantification of stained or labeled proteins and were run on a single gel. Quantification of bands was performed using ImageQuant 5.2 software (GE Healthcare).
Radioactive Labeling of Cells
Radioactive pulse (for 2D analysis) and pulse-chase (for one-dimensional analysis) labeling of the cells was performed using a mixture of [35S]Met and [35S]Cys (Translabel; MP Biochemicals) as described previously (Dobáková et al., 2009).
Isolation of PSII and PSI Complexes
PSII complexes containing His-tagged CP43 or CP47 and PSI complexes containing His-tagged PsaF were isolated from thylakoids of appropriate strains either using just affinity chromatography on immobilized Ni2+ ions or in combination with gel filtration as described in Boehm et al. (2011).
Yeast Two-Hybrid Analyses
Interaction studies in yeast including appropriate controls were essentially performed as previously described by using the split ubiquitin system (Pasch et al., 2005; Komenda et al., 2008). The open reading frame slr1645 was PCR amplified from genomic Synechocystis DNA by using oligonucleotides psb27fordt (ATTGGATCCTGCGACAGCGGCACAGGA9) and psb27rev (ATTGAATTCT TACGTCACACGCCCCGTTCAATG) and cloned into the BamHI and EcoRI sites of the vector pADSL (Dualsystems Biotech AG). This construct lacks the N-terminal transit sequence of Psb27 comprising 24 amino acids to avoid localization problems of the NubG-Psb27 fusion protein in yeast. The pTFB1-psaB construct has been described previously (Komenda et al., 2008).
Enzymatic In-Gel Digestion and Protein Identification by MS
Silver-stained protein spots were excised from the gel, cut into small pieces, and washed with freshly prepared solution of 30 mm potassium ferricyanide and 100 mm sodium thiosulfate (mixed in 1:1 ratio). After complete destaining, the gel was washed with water, shrunk by dehydration in acetonitrile (MeCN), and reswelled again in water. The supernatant was removed, and the gel was partly dried in a SpeedVac concentrator. The gel pieces were then rehydrated in a cleavage buffer containing 25 mm 4-ethylmorpholine acetate, 5% MeCN, and sequencing-grade protease (20 ng of Glu-C or chymotrypsin, Roche; or 100 ng of trypsin, Promega) and incubated overnight at 37°C. The resulting peptides were desalted on a peptide microtrap (Michrom BioResources) according to the manufacturer instructions. Mass spectra were measured on a APEX-Qe 9.4T FT-MS instrument equipped with a Combi electrospray ionization (ESI)/matrix-assisted laser-desorption ionization (MALDI) ion source (Bruker Daltonics). In the liquid chromatography-MS experiment, the peptides were separated on capillary Magic C18 column (Michrom BioResources) interfaced to the ESI source of the Fourier transform (FT)-MS instrument. For the matrix-assisted laser-desorption ionization (MALDI) experiment, 0.5 μL of the sample was deposited on the MALDI target and allowed to air-dry at room temperature. After complete evaporation, 0.5 μL of the matrix solution (α-cyano-4-hydroxycinnamic acid in 50% MeCN/0.1% trifluoroacetic acid; 5 mg/mL) was added. Tandem MS spectra were acquired on an Ultraflex III MALDI-TOF/TOF instrument (Bruker Daltonics). The mass spectra were searched against SwissProt 2011_04 database subset of Synechocystis proteins using the in-house MASCOT search engine.
Content of Chl and Its Intermediates
For the measurement of Chl concentration, sedimented cells or membranes were extracted with 100% methanol, and Chl content in the extract was calculated from the absorbance at 666 and 720 nm (Wellburn and Lichtenthaler, 1984). Quantitative determination of Chl precursors in the cells was performed as described in Dobáková et al. (2009).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Localization of the Psb27 protein by 2D-BN/SDS-PAGE in membranes of the Synechocystis PSI-less strain cultivated under 5 μmol photons m−2 s−1.
Supplemental Figure S2. Localization of the Psb27 protein by 2D-BN/SDS-PAGE in membranes of the Synechocystis strains lacking CP43 and PSI.
Supplemental Figure S3. Localization of the Psb27 protein by 2D-BN/SDS-PAGE in membranes of the Synechocystis psbK deletion mutant ΔPsbK (left) and PSI-His complex isolated by a single-step nickel-affinity chromatography (PSI-His, right).
Supplemental Figure S4. Yeast two-hybrid analyses of the interaction between Psb27 and PsaB.
Supplemental Figure S5. Separation of pigment-protein complexes from isolated preparations of His-tagged CP43 and His-tagged CP47 by 2D-CN/SDS-PAGE.
Supplemental Figure S6. 2D analysis of radioactively labeled membrane protein complexes of the wild type and the PSI-less strain ΔPSI.
Supplemental Figure S7. Comparison of CP43 fragmentation in membranes of the ΔCP47 and ΔCP47/ΔPsb27 strains and in the preparation of isolated CP43-His.
Supplemental Figure S8. Analysis of His-tagged CP43 by 2D-BN/SDS-PAGE.
Supplemental Figure S9. Construction of the pPsb27 CamA vector for transformation of the Synechocystis 6803 wild-type strain (A) and confirmation of the complete segregation of the deletion mutant ΔPsb27 by PCR (B).
Supplemental Figure S10. Analysis of CP43, D1, and Psb27 distribution in complexes of the wild-type and ΔPsb27 strains by 2D-CN/SDS-PAGE.
Acknowledgments
We thank Lutz Eichacker, Roberto Barbato, John Goldbeck, and Julian Eaton-Rye for donation of specific antisera, Hajime Wada and Eva-Mari Aro for providing the F-His strain, and James Barber for providing the ΔPsbK strain of Synechocystis.
References
- Anbudurai PR, Mor TS, Ohad I, Shestakov SV, Pakrasi HB. (1994) The ctpA gene encodes the C-terminal processing protease for the D1 protein of the photosystem II reaction center complex. Proc Natl Acad Sci USA 91: 8082–8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber J. (2006) Photosystem II: an enzyme of global significance. Biochem Soc Trans 34: 619–631 [DOI] [PubMed] [Google Scholar]
- Bibby TS, Nield J, Barber J. (2001) Iron deficiency induces the formation of an antenna ring around trimeric photosystem I in cyanobacteria. Nature 412: 743–745 [DOI] [PubMed] [Google Scholar]
- Boehm M, Romero E, Reisinger V, Yu J, Komenda J, Eichacker LA, Dekker JP, Nixon PJ. (2011) Investigating the early stages of photosystem II assembly in Synechocystis sp. PCC 6803: isolation of CP47 and CP43 complexes. J Biol Chem 286: 14812–14819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekema EJ, Hifney A, Yakushevska AE, Piotrowski M, Keegstra W, Berry S, Michel KP, Pistorius EK, Kruip J. (2001) A giant chlorophyll-protein complex induced by iron deficiency in cyanobacteria. Nature 412: 745–748 [DOI] [PubMed] [Google Scholar]
- Chen H, Zhang D, Guo J, Wu H, Jin M, Lu Q, Lu C, Zhang L. (2006) A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II. Plant Mol Biol 61: 567–575 [DOI] [PubMed] [Google Scholar]
- Cormann KU, Bangert JA, Ikeuchi M, Rögner M, Stoll R, Nowaczyk MM. (2009) Structure of Psb27 in solution: implications for transient binding to photosystem II during biogenesis and repair. Biochemistry 48: 8768–8770 [DOI] [PubMed] [Google Scholar]
- Dobáková M, Sobotka R, Tichý M, Komenda J. (2009) Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 149: 1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton-Rye JJ, Vermaas WF. (1991) Oligonucleotide-directed mutagenesis of psbB, the gene encoding CP47, employing a deletion mutant strain of the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol 17: 1165–1177 [DOI] [PubMed] [Google Scholar]
- Fagerlund RD, Eaton-Rye JJ. (2011) The lipoproteins of cyanobacterial photosystem II. J Photochem Photobiol B 104: 191–203 [DOI] [PubMed] [Google Scholar]
- Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. (2004) Architecture of the photosynthetic oxygen-evolving center. Science 303: 1831–1838 [DOI] [PubMed] [Google Scholar]
- Grasse N, Mamedov F, Becker K, Styring S, Rögner M, Nowaczyk MM. (2011) Role of novel dimeric photosystem II (PSII)-Psb27 protein complex in PSII repair. J Biol Chem 286: 29548–29555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guskov A, Kern J, Gabdulkhakov A, Broser M, Zouni A, Saenger W. (2009) Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat Struct Mol Biol 16: 334–342 [DOI] [PubMed] [Google Scholar]
- Iwai M, Suzuki T, Kamiyama A, Sakurai I, Dohmae N, Inoue Y, Ikeuchi M. (2010) The PsbK subunit is required for the stable assembly and stability of other small subunits in the PSII complex in the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. Plant Cell Physiol 51: 554–560 [DOI] [PubMed] [Google Scholar]
- Kashino Y, Lauber WM, Carroll JA, Wang Q, Whitmarsh J, Satoh K, Pakrasi HB. (2002) Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41: 8004–8012 [DOI] [PubMed] [Google Scholar]
- Komenda J, Barber J. (1995) Comparison of psbO and psbH deletion mutants of Synechocystis PCC 6803 indicates that degradation of D1 protein is regulated by the QB site and dependent on protein synthesis. Biochemistry 34: 9625–9631 [DOI] [PubMed] [Google Scholar]
- Komenda J, Barker M, Kuviková S, de Vries R, Mullineaux CW, Tichý M, Nixon PJ. (2006) The FtsH protease slr0228 is important for quality control of photosystem II in the thylakoid membrane of Synechocystis sp. PCC 6803. J Biol Chem 281: 1145–1151 [DOI] [PubMed] [Google Scholar]
- Komenda J, Kuviková S, Granvogl B, Eichacker LA, Diner BA, Nixon PJ. (2007) Cleavage after residue Ala352 in the C-terminal extension is an early step in the maturation of the D1 subunit of photosystem II in Synechocystis PCC 6803. Biochim Biophys Acta 1767: 829–837 [DOI] [PubMed] [Google Scholar]
- Komenda J, Lupínková L, Kopecký J. (2002) Absence of the psbH gene product destabilizes photosystem II complex and bicarbonate binding on its acceptor side in Synechocystis PCC 6803. Eur J Biochem 269: 610–619 [DOI] [PubMed] [Google Scholar]
- Komenda J, Nickelsen J, Tichý M, Prásil O, Eichacker LA, Nixon PJ. (2008) The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J Biol Chem 283: 22390–22399 [DOI] [PubMed] [Google Scholar]
- Komenda J, Reisinger V, Müller BC, Dobáková M, Granvogl B, Eichacker LA. (2004) Accumulation of the D2 protein is a key regulatory step for assembly of the photosystem II reaction center complex in Synechocystis PCC 6803. J Biol Chem 279: 48620–48629 [DOI] [PubMed] [Google Scholar]
- Kubota H, Sakurai I, Katayama K, Mizusawa N, Ohashi S, Kobayashi M, Zhang P, Aro EM, Wada H. (2010) Purification and characterization of photosystem I complex from Synechocystis sp. PCC 6803 by expressing histidine-tagged subunits. Biochim Biophys Acta 1797: 98–105 [DOI] [PubMed] [Google Scholar]
- Liu H, Huang RYC, Chen J, Gross ML, Pakrasi HB. (2011a) Psb27, a transiently associated protein, binds to the chlorophyll-binding protein in photosystem II assembly intermediates. Proc Natl Acad Sci USA http://dx.doi.org/10.1073/pnas.1111597108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Roose JL, Cameron JC, Pakrasi HB. (2011b) A genetically tagged Psb27 protein allows purification of two consecutive photosystem II (PSII) assembly intermediates in Synechocystis 6803, a cyanobacterium. J Biol Chem 286: 24865–24871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupínková L, Komenda J. (2004) Oxidative modifications of the photosystem II D1 protein by reactive oxygen species: from isolated protein to cyanobacterial cells. Photochem Photobiol 79: 152–162 [DOI] [PubMed] [Google Scholar]
- Mabbitt PD, Rautureau GJP, Day CL, Wilbanks SM, Eaton-Rye JJ, Hinds MG. (2009) Solution structure of Psb27 from cyanobacterial photosystem II. Biochemistry 48: 8771–8773 [DOI] [PubMed] [Google Scholar]
- Mamedov F, Nowaczyk MM, Thapper A, Rögner M, Styring S. (2007) Functional characterization of monomeric photosystem II core preparations from Thermosynechococcus elongatus with or without the Psb27 protein. Biochemistry 46: 5542–5551 [DOI] [PubMed] [Google Scholar]
- Marder JB, Goloubinoff P, Edelman M. (1984) Molecular architecture of the rapidly metabolized 32-kilodalton protein of photosystem II. Indications for COOH-terminal processing of a chloroplast membrane polypeptide. J Biol Chem 259: 3900–3908 [PubMed] [Google Scholar]
- Melkozernov AN, Bibby TS, Lin S, Barber J, Blankenship RE. (2003) Time-resolved absorption and emission show that the CP43′ antenna ring of iron-stressed synechocystis sp. PCC6803 is efficiently coupled to the photosystem I reaction center core. Biochemistry 42: 3893–3903 [DOI] [PubMed] [Google Scholar]
- Nixon PJ, Trost JT, Diner BA. (1992) Role of the carboxy terminus of polypeptide D1 in the assembly of a functional water-oxidizing manganese cluster in photosystem II of the cyanobacterium Synechocystis sp. PCC 6803: Assembly requires a free carboxyl group at C-terminal position 344. Biochemistry 31: 10859–10871 [DOI] [PubMed] [Google Scholar]
- Nowaczyk MM, Hebeler R, Schlodder E, Meyer HE, Warscheid B, Rögner M. (2006) Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II. Plant Cell 18: 3121–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowaczyk MM, Sander J, Grasse N, Cormann KU, Rexroth D, Bernát G, Rögner M. (2010) Dynamics of the cyanobacterial photosynthetic network: communication and modification of membrane protein complexes. Eur J Cell Biol 89: 974–982 [DOI] [PubMed] [Google Scholar]
- Pakrasi HB, Williams JGK, Arntzen CJ. (1988) Targeted mutagenesis of the psbE and psbF genes blocks photosynthetic electron transport: evidence for a functional role of cytochrome b559 in photosystem II. EMBO J 7: 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasch JC, Nickelsen J, Schünemann D. (2005) The yeast split-ubiquitin system to study chloroplast membrane protein interactions. Appl Microbiol Biotechnol 69: 440–447 [DOI] [PubMed] [Google Scholar]
- Roose JL, Pakrasi HB. (2004) Evidence that D1 processing is required for manganese binding and extrinsic protein assembly into photosystem II. J Biol Chem 279: 45417–45422 [DOI] [PubMed] [Google Scholar]
- Roose JL, Pakrasi HB. (2008) The Psb27 protein facilitates manganese cluster assembly in photosystem II. J Biol Chem 283: 4044–4050 [DOI] [PubMed] [Google Scholar]
- Roose JL, Wegener KM, Pakrasi HB. (2007) The extrinsic proteins of photosystem II. Photosynth Res 92: 369–387 [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 199: 223–231 [DOI] [PubMed] [Google Scholar]
- Shen G, Boussiba S, Vermaas WFJ. (1993) Synechocystis sp PCC 6803 strains lacking photosystem I and phycobilisome function. Plant Cell 5: 1853–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Shiraishi T, Asada K. (1988) COOH-terminal residues of D1 and the 44 kDa CPa-2 at spinach photosystem II core complex. FEBS Lett 240: 6–8 [DOI] [PubMed] [Google Scholar]
- Umena Y, Kawakami K, Shen J-R, Kamiya N. (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473: 55–60 [DOI] [PubMed] [Google Scholar]
- Vermaas WFJ, Ikeuchi M, Inoue Y. (1988) Protein composition of the photosystem II core complex in genetically engineered mutants of the cyanobacterium Synechocystis sp. PCC 6803. Photosynth Res 17: 97–113 [DOI] [PubMed] [Google Scholar]
- Wei L, Guo J, Ouyang M, Sun X, Ma J, Chi W, Lu C, Zhang L. (2010) LPA19, a Psb27 homolog in Arabidopsis thaliana, facilitates D1 protein precursor processing during PSII biogenesis. J Biol Chem 285: 21391–21398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellburn AR, Lichtenthaler K. (1984) Formulae and programme to determine total carotenoids and chlorophyll a and b of leaf extracts in different solvents. Sybesma C, , Advances in Photosynthesis Research, Martinus Nijjhoff, Dordrecht, The Netherlands, pp 10–12 [Google Scholar]
- Williams JGK. (1988) Construction of specific mutations in PSII photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol 167: 766–778 [Google Scholar]
- Zhang ZH, Mayes SR, Vass I, Nagy L, Barber J. (1993) Characterization of the psbK locus of Synechocystis PCC 6803 in terms of photosystem II function. Photosynth Res 38: 369–377 [DOI] [PubMed] [Google Scholar]
- Zouni A, Witt HT, Kern J, Fromme P, Krauss N, Saenger W, Orth P. (2001) Crystal structure of photosystem II from Synechococcus elongatus at 3.8 A resolution. Nature 409: 739–743 [DOI] [PubMed] [Google Scholar]