Abstract
Nitric oxide (NO) is a key redox-active, small molecule involved in various aspects of plant growth and development. Here, we report the identification of an NO accumulation mutant, nitric oxide excess1 (noe1), in rice (Oryza sativa), the isolation of the corresponding gene, and the analysis of its role in NO-mediated leaf cell death. Map-based cloning revealed that NOE1 encoded a rice catalase, OsCATC. Furthermore, noe1 resulted in an increase of hydrogen peroxide (H2O2) in the leaves, which consequently promoted NO production via the activation of nitrate reductase. The removal of excess NO reduced cell death in both leaves and suspension cultures derived from noe1 plants, implicating NO as an important endogenous mediator of H2O2-induced leaf cell death. Reduction of intracellular S-nitrosothiol (SNO) levels, generated by overexpression of rice S-nitrosoglutathione reductase gene (GSNOR1), which regulates global levels of protein S-nitrosylation, alleviated leaf cell death in noe1 plants. Thus, S-nitrosylation was also involved in light-dependent leaf cell death in noe1. Utilizing the biotin-switch assay, nanoliquid chromatography, and tandem mass spectrometry, S-nitrosylated proteins were identified in both wild-type and noe1 plants. NO targets identified only in noe1 plants included glyceraldehyde 3-phosphate dehydrogenase and thioredoxin, which have been reported to be involved in S-nitrosylation-regulated cell death in animals. Collectively, our data suggest that both NO and SNOs are important mediators in the process of H2O2-induced leaf cell death in rice.
Nitric oxide (NO) is a redox-active molecule that plays a key role in a broad spectrum of physiological and developmental functions throughout the plant life cycle. Thus, NO is thought to contribute to the control of germination, leaf expansion, lateral root development, flowering, stomatal closure, and defenses against biotic and abiotic stresses (Neill et al., 2002; He et al., 2004; Hong et al., 2008; Wilson et al., 2008; Leitner et al., 2009). NO has also been implicated as a key regulator of senescence and leaf cell death in higher plants (Guo and Crawford, 2005; Zago et al., 2006). While descriptions of NO-mediated processes are accumulating, the details of how this small molecule regulates its target signaling networks remain largely unknown (Wang et al., 2010b). To date, numerous potential sources for NO production have been described in plants (Gupta et al., 2011), including a nitric oxide synthase (NOS)-like activity (Delledonne et al., 1998; Durner et al., 1998; Besson-Bard et al., 2008), nitrate reductase (NR; Stöhr and Stremlau, 2006; Stoimenova et al., 2007; Seligman et al., 2008), xanthine oxidoreductase (Corpas et al., 2008), and polyamine/hydroxylamine-mediated NO production (Rümer et al., 2009). Archetypal NOS enzymes found in animals (Palmer et al., 1988; Knowles and Moncada, 1994) and also the green alga Ostreococcus tauri (Foresi et al., 2010) catalyze the NADPH-dependent oxidation of l-Arg, producing l-citrulline and NO. A similar activity has been reported in higher plants, which is blunted by classical mammalian NOS inhibitors (Delledonne et al., 1998; Durner et al., 1998; Jasid et al., 2006; Vandelle and Delledonne, 2008; Corpas et al., 2009). However, no archetypal NOS-encoding genes have been identified so far in higher plants. Another potential source of NO production is NR, which catalyzes the production of this gas by the successive reduction of nitrite and nitrate. In Arabidopsis (Arabidopsis thaliana), NR is encoded by two genes, NIA1 and NIA2 (Yamasaki and Sakihama, 2000).
It has been shown that NO regulates a number of physiological processes directly by affecting gene transcription (Huang et al., 2002; Wang et al., 2002; Polverari et al., 2003; Parani et al., 2004; Shoulars et al., 2008). NO can also regulate specific physiological and developmental processes through its interplay with other small biomolecules. For example, NO can scavenge hydrogen peroxide (H2O2) and protects plant cells from damage (Beligni et al., 2002; Crawford and Guo, 2005). It was also reported that NO and H2O2 function in combination to elaborate cell death associated with the hypersensitive response (HR) following pathogen recognition (Delledonne et al., 2001). Furthermore, S-nitrosylation, the addition of an NO moiety to a Cys thiol to form an S-nitrosothiol (SNO), is emerging as a key redox-based posttranslational modification in plants and a pivotal mechanism to convey NO bioactivity. S-Nitrosylation may be integral to NO function during a variety of cellular processes. For example, S-nitrosylation of peroxiredoxin IIE during the defense response regulates the antioxidant function of this key enzyme and might contribute to the HR (Romero-Puertas et al., 2007). In animals, SNO formation has been reported to be involved in the regulation of apoptosis at multiple steps (Sawa et al., 1997; Hara et al., 2005; Mannick, 2007). In this context, the S-nitrosylation of GOSPEL (Sen et al., 2009), caspase 3 (Tsang et al., 2009), cyclooxygenase 2 (Carreras and Poderoso, 2007), FLICE (for FADD-like ICE) inhibitory protein (Chanvorachote et al., 2005), and apoptosis signal-regulating kinase1 (ASK1; Park et al., 2004) are important features of cell death in animals.
The global levels of S-nitrosylation are regulated by S-nitrosoglutathione reductase1 (GSNOR1) in Arabidopsis (Feechan et al., 2005). Loss-of-function mutations in GSNOR1 increased total cellular SNO content and compromised both nonhost and resistance (R) gene-mediated protection and also disabled basal defense responses (Feechan et al., 2005). Furthermore, this line was also perturbed in thermotolerance and responses to paraquat (Lee et al., 2008; Chen et al., 2009).
To generate insights into the potential role of S(NO) in the process of rice (Oryza sativa) leaf cell death and identify associated S-nitrosylated proteins, we conducted a forward genetic screen utilizing a large T-DNA-tagged population to identify mutants with perturbed (S)NO homeostasis. This approach uncovered the nitric oxide excess1 (noe1) mutant, which accumulated more (S)NO in comparison with wild-type plants. Employing noe1, we implicate NR-dependent NO production in rice leaf cell death in response to high light. Furthermore, our findings suggest a function for (S)NO and H2O2 in rice leaf cell death. Through the biotin-switch assay and nanoliquid chromatography-tandem mass spectrometry (LC-MS/MS), we identified S-nitrosylated proteins during cell death triggered by high light. These results provide initial insights into the mechanisms underpinning this process in rice leaves.
RESULTS
Identification of noe1
In order to shed light on the role of (S)NO in leaf programmed cell death (PCD), we performed a large-scale screen to identify SNO content-altered mutants using the Saville-Griess assay from our T-DNA insertional mutant population (Ma et al., 2009). To simplify the assay with a large number of samples, we used 96-well plates and a Tecan Infinite M200 (Tecan Group) to undertake the photometrical measurement at 540 nm. Among the 380 screened mutants that show a PCD-like phenotype, 15 mutants with higher SNO content and four mutants with lower SNO content were selected for further analysis. One of them, designated noe1, was identified because its SNO content was significantly higher than that of the wild type (Fig. 1A). No signs of damage were developed in the shaded parts of the leaves of noe1 grown in the field under natural growth conditions (high light; 1,600 μmol m−2 s−1) or under low light (400 μmol m−2 s−1; Supplemental Fig. S1, A and C). However, leaf damage, characterized by white variegated areas and cell death, developed when noe1 was cultivated under high light (Fig. 1, B and C; Supplemental Fig. S1B). When the 10-d-old low-light-grown noe1 plants were transferred to high light, the obvious leaf bleaching and cell death were developed in noe1 plants on the 2nd and 5th d, respectively (Supplemental Fig. S2A). Furthermore, no cell death was observed in noe1 plants cultivated under low light with different temperatures or photoperiods (Supplemental Fig. S2B). By contrast, exposure to high light together with different temperatures (20°C, 25°C, and 30°C) or photoperiods (light/dark: 6 h/18 h, 10 h/14 h, and 14 h/10 h) promoted cell death in all noe1 plants but not in wild-type controls (Supplemental Fig. S2C). These data implied that the observed cell death in noe1 plants is dependent on high light.
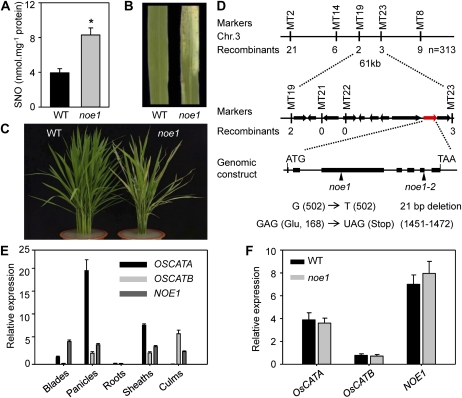
Figure 1.
Phenotypes of noe1 and map-based cloning of NOE1/OsCATC. A to C, SNO content (A) and phenotypes (B and C) of 3-month-old wild-type (WT) and noe1 plants grown under high light (1,600 μmol m−2 s−1). D, Map-based cloning of NOE1/OsCATC. The NOE1/OsCATC gene was mapped in the interval of MT2 and MT8 on the short arm of chromosome 3 and was delimited to a 61-kb region with 10 candidate genes between the MT19 and MT23 markers; the mutated sites of two distinct allelic mutants are shown in the gene LOC_Os03g03910. E, Expression patterns of OsCATA, OsCATB, and NOE1/OsCATC in different tissues analyzed by real-time quantitative PCR. F, Expression levels of OsCATA, OsCATB, and NOE1/OsCATC in wild-type and noe1 leaves. These data were obtained from three independent replicates. * P < 0.05 (t test).
Map-Based Cloning of NOE1/OsCATC
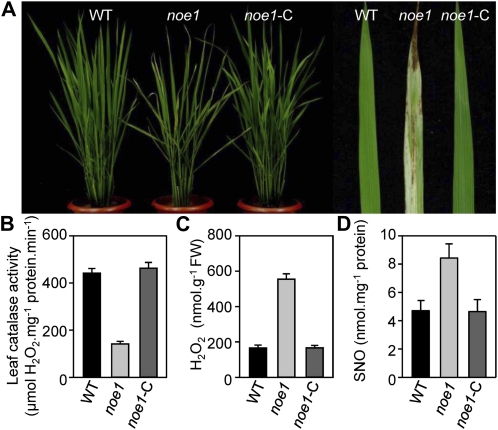
Despite being identified from a T-DNA insertion population, the SNO overaccumulation and cell death phenotypes of noe1 did not cosegregate with a T-DNA insertion. Thus, the noe1 line without a T-DNA insertion was isolated by backcrossing and used for further analysis. An F2 mapping population was established using a cross between noe1 (japonica) and Minghui 63 (indica). In the F2 population, we investigated 1,227 individual plants. Among them, 313 plants exhibited the noe1 mutant phenotype and 914 plants exhibited wild-type phenotypes, suggesting that the noe1 mutant was controlled by a single recessive gene. Rough mapping delimited the NOE1 locus at about 950 kb on chromosome 3 with genetic markers M2 and M8 (Fig. 1D). Further fine-mapping was performed using insertion-deletion molecular markers (Supplemental Table S1). The NOE1 locus was narrowed down to a 61-kb region between MT19 and MT23 (Fig. 1D). After sequencing all 10 candidate genes within this region, a single-nucleotide G-to-T transition at the 168th position, which caused a Glu (GAG)-to-stop codon (TAG) change, was found in LOC_Os03g03910 of noe1. Subsequently, we identified another allelic mutant (confirmed by allelic test), noe1-2, with a similar aberrant phenotype in leaves and the same higher SNO content compared with the wild type. After sequencing the NOE1 (LOC_Os03g03910) locus, a 21-bp deletion was found, which led to a frame-shift mutation (Fig. 1D). NOE1 encodes the rice catalase domain-containing protein and has extremely high sequence similarity with catalase isozyme A (OsCATA) and catalase isozyme B (OsCATB) in rice and three catalase isozymes in Arabidopsis (Supplemental Fig. S3). As in the rice genome, there are only three catalase-encoding genes, so NOE1 should be OsCATC. Real-time quantitative PCR analysis revealed that the expression of OsCATA, OsCATB, and NOE1/OsCATC varied in different tissues (Fig. 1E). NOE1/OsCATC was mainly expressed in leaf blades, panicles, leaf sheaths, and culms, but expression was extremely low in roots. In contrast, OsCATA was expressed at a high level in leaf blades, panicles, and leaf sheaths but weakly expressed in roots and culms. As for OsCATB, high expression was observed in the panicles, leaf sheaths, and culms. In leaves, OsCATA and NOE1/OsCATC were highly expressed but OsCATB was much less abundant (Fig. 1E). In addition, no compensatory induction of OsCATA and OsCATB transcripts was observed in noe1 plants (Fig. 1F). These results implied that NOE1/OsCATC, but not OsCATA or OsCATB, was responsible for the redox homeostasis in leaves. To further validate NOE1/OsCATC, genetic complementation was carried out by introducing the entire cDNA of NOE1/OsCATC into noe1 plants. As expected, the leaf cell death phenotype was not observed in all noe1-C transgenic lines (Fig. 2A). Furthermore, enzyme activity assays showed that the catalase activities of complemented plants were also restored to wild-type levels (Fig. 2B). Consequently, H2O2 contents in noe1-C plants were also decreased to that of the wild type (Fig. 2C) as well as the content of SNO (Fig. 2D). Consistent with the impairment of NOE1/OsCATC, total catalase activity in noe1 plants was reduced dramatically to about 30% of that of the wild type (Fig. 2B), and more H2O2 and cell death accumulated (Fig. 2C; Supplemental Fig. S4). After the addition of 200 μm CAT (an H2O2 scavenger), the leaf cell death in noe1 plants did not develop at the 2nd or 5th d under high light (Supplemental Fig. S5, A and C). Furthermore, cell death in noe1 suspension cells was reduced from 53.9% to 16.6% after treatment with 200 μm CAT (Supplemental Fig. S5E). These data suggest that leaf cell death in noe1 may be caused by H2O2 overaccumulation. Taken together, loss-of-function mutations in NOE1/OsCATC are responsible for the phenotypes displayed in the noe1 mutants.
Figure 2.
Phenotypes of noe1 were complemented by NOE1/OsCATC. A, Phenotypic comparison among 3-month-old wild-type (WT), noe1, and complemented transgenic noe1-C plants. B to D, Total extractable leaf catalase activities and H2O2 and SNO contents in 3-month-old wild type, noe1, and noe1-C plants grown under high light (1,600 μmol m−2 s−1). These data were obtained from three independent replicates. FW, Fresh weight.
H2O2 Stimulates NO Production
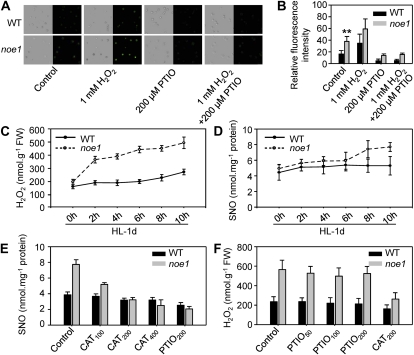
Labeling NO with the cell-permeable, NO-specific fluorescent probe 4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate (DAF-FM DA) and imaging by confocal microscopy revealed that the NO content in noe1 suspension cells was about 2.3-fold greater than that of the wild type (Fig. 3, A and B). To elucidate the relationship between H2O2 and NO, treatment of wild-type and noe1 suspension cells with 1 mm H2O2 was performed, and DAF-FM DA analysis showed that H2O2 induced NO production significantly (Fig. 3, A and B). The seedlings of wild-type and noe1 plants grown under low light for 10 d were transferred to high light, then H2O2 and SNO contents were measured at the 6th, 8th, and 10th d under low light and each day under high light. When grown under low light, there was no significant difference between wild-type and noe1 plants with regard to H2O2 and SNO contents, which increased gradually with time (Supplemental Fig. S6). After transfer to high light, however, H2O2 and SNO were produced more rapidly within the 1st d in noe1 plants as compared with wild-type plants (Supplemental Fig. S6). These findings indicated that the rapid increase of H2O2 and SNO detected in noe1 plants is dependent on high light. Further detailed analysis was made every 2 h on the 1st d under high light. The rapid production of H2O2 and SNO in noe1 occurred at the 2nd and 8th h, respectively (Fig. 3, C and D). In addition, treatment with different concentrations of CAT revealed that the generation of SNO was inhibited in a dose-dependent manner (Fig. 3E). In contrast, H2O2 content was not altered after treatment of high-light-grown noe1 plants with 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide (PTIO; an NO scavenger; Fig. 3F). These results suggested that H2O2-stimulated NO production may cause the increased SNO levels in leaves of noe1 plants under high light.
Figure 3.
Accumulated H2O2 stimulated NO production and caused the increase of SNO level. A and B, NO level (A) and relative fluorescence intensity (B; control or with treatment of 1 mm H2O2 or 200 μm PTIO for 30 min at 28°C) in wild-type (WT) and noe1 suspension cells were monitored by labeling NO using the specific NO dye DAF-FM DA. C and D, H2O2 (C) and SNO (D) contents in wild-type and noe1 seedlings at 2-h intervals on the first day following exposure to high light (HL). E, SNO content in wild-type and noe1 seedlings treated with 100, 200, and 400 μm catalase (CAT; an H2O2 scavenger) for 2 d under high light. F, H2O2 content in wild-type and noe1 seedlings treated with 50, 100, and 200 μm PTIO for 2 d under high light. Both 10-d-old wild-type and noe1 seedlings cultivated under low light (400 μmol m−2 s−1) were transferred to high light (1,600 μmol m−2 s−1). These data were obtained from three independent replicates. ** P < 0.01 (t test). FW, Fresh weight.
H2O2-Induced NR-Dependent Generation of NO
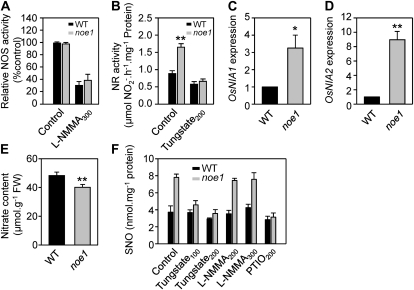
To elucidate the main source responsible for NO production in noe1 plants under high light, the activities of NOS and NR were measured. No difference in NOS activity was observed between wild-type and noe1 plants (Fig. 4A). NR activity in noe1 plants, however, was approximately twice that of the wild type (Fig. 4B). Quantitative real-time PCR analysis revealed that the expression of OsNIA1 and OsNIA2 increased 3- and 9-fold, respectively, in noe1 plants (Fig. 4, C and D), in line with the increase of NR activity. As a result, the content of the NR substrate, nitrate, was decreased in noe1 plants (Fig. 4E). To further confirm that NO was mainly produced from NR activity, tungstate (a NR inhibitor) and NG-monomethyl-l-arginine, monoacetate salt (l-NMMA; an NOS inhibitor) were used, and SNO contents were monitored. The content of SNOs was inhibited in a dose-dependent manner when noe1 seedlings were treated with tungstate, while no change in SNO content was observed in noe1 seedlings treated with l-NMMA (Fig. 4F). These results implied that the excess NO in noe1 was mainly generated from NR. Presumably, this increased NO concentration subsequently resulted in greater SNO levels.
Figure 4.
NR was mainly responsible for the S(NO) accumulation in noe1. A to E, NOS (A) and NR (B) activities, relative expression levels of OsNIA1 (C) and OsNIA2 (D) by quantitative RT-PCR (with the wild-type value [WT] set as 1.0), and nitrate content (E) in wild-type and noe1 plants. FW, Fresh weight. F, Effects of tungstate (an NR inhibitor) and l-NMMA (an NOS inhibitor) on endogenous NO production in wild-type and noe1 plants. Both 10-d-old wild-type and noe1 seedlings cultivated under low light (400 μmol m−2 s−1) were treated with tungstate or l-NMMA under high light (1,600 μmol m−2 s−1) for 2 d. These data were obtained from three independent replicates. Tungstate100 and Tungstate200, 100 and 200 μm tungstate; l-NMMA200 and l-NMMA300, 200 and 300 μm l-NMMA; PTIO200, 200 μm PTIO. * P < 0.05, ** P < 0.01 (t test).
NO Is Required for H2O2-Induced Leaf Cell Death
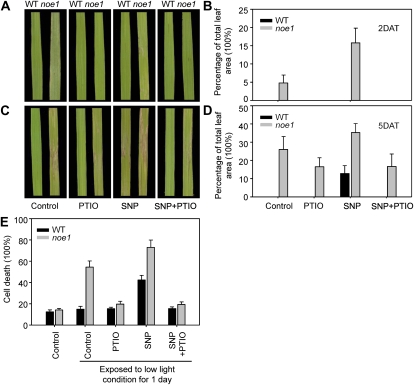
To identify the role of NO in H2O2-induced leaf cell death modulated by light (Supplemental Figs. S1 and S2), treatments of wild-type and noe1 seedlings with PTIO and sodium nitroprusside (SNP; an NO donor), both alone and in combination, were carried out. After transferring the 10-d-old low-light-grown noe1 plants to high light, obvious leaf bleaching and cell death formation occurred on the 2nd and 5th d, respectively (Fig. 5, A and C). The addition of 200 μm PTIO, which depletes NO, resulted in significantly reduced leaf cell death (Fig. 5). By contrast, leaf cell death in noe1 plants was promoted following treatment with 2.5 mm SNP (Fig. 5). Cell death could also be triggered in wild-type plants following the addition of 2.5 mm SNP for 5 d (Fig. 5, C and D), demonstrating a requirement for NO in the process of cell death. Similar treatments were performed with dark-grown rice suspension cells, in which cell death can be accurately measured. The results showed that the relative cell death ratio in noe1 suspension cells increased from 14.2% to 54.5% when exposed to low light (400 μmol m−2 s−1) for 1 d (Fig. 5E). After the addition of 200 μm PTIO, the ratio of cell death was reduced to 19.7% when exposed to low light for 1 d, and this level was similar to that found in wild-type cells (15.4%). When cultured with 2.5 mm SNP under low light for 1 d, the relative cell death ratio in wild-type and noe1 suspension cells increased to 42.4% and 73.1%, respectively, significantly greater than control values (wild type and noe1: 15.0% and 54.5%, respectively; Fig. 5E). These results suggest that NO is a pivotal mediator for H2O2-induced leaf cell death in rice.
Figure 5.
NO was required for leaf cell death. A to D, Both 10-d-old wild type (WT) and noe1 seedlings cultivated under low light (400 μmol m−2 s−1) were transferred to high light (1,600 μmol m−2 s−1). Leaf symptoms and relative cell death (percentage of total leaf area) are given for wild-type and noe1 seedlings induced by the addition of 200 μm PTIO (an NO scavenger) and 2.5 mm SNP (an NO donor) for 2 d after treatment (DAT; A and B) and 5 DAT (C and D) under high light. E, Effects of 200 μm PTIO and 2.5 mm SNP on relative cell death of wild-type and noe1 suspension cell cultures. These data were obtained from three independent replicates.
Protein S-Nitrosylation Is Involved in H2O2-Induced Leaf Cell Death
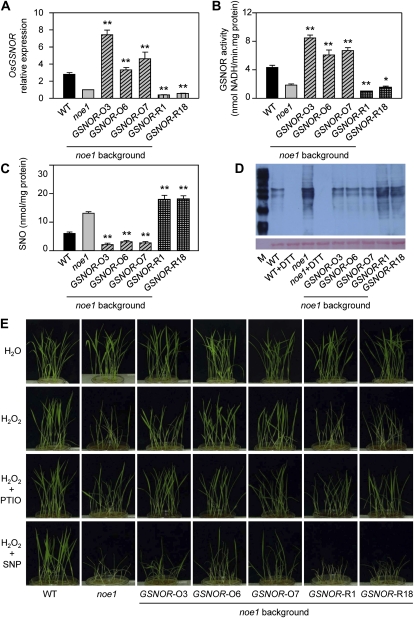
It is known that protein S-nitrosylation is a fundamental mechanism to transduce NO bioactivity (Besson-Bard et al., 2008) and that GSNOR regulates global levels of S-nitrosylation (Feechan et al., 2005). OsGSNOR is encoded by a single gene in rice that has very high sequence similarity with AtGSNOR (Supplemental Fig. S7). To investigate the potential role of OsGSNOR in leaf cell death, OsGSNOR overexpression (GSNOR-O noe1) or knockdown (GSNOR-R noe1) transgenic plants in the noe1 background were generated (Fig. 6A). GSNOR-O noe1 plants with increased GSNOR activity and GSNOR-R noe1 plants with reduced GSNOR activity were selected for further analysis (Fig. 6B). Employing the Saville-Griess assay, lower SNO content was found in GSNOR-O noe1 plants relative to noe1 lines. In contrast, GSNOR-R noe1 plants exhibited an increased level of these redox molecules (Fig. 6C). These data are consistent with those from Arabidopsis, which suggested that GSNOR controls the total level of SNOs in cells (Feechan et al., 2005). The biotin-switch assay, labeling of S-nitrosylated proteins with a biotin moiety specifically on S-nitrosylated Cys residues (Jaffrey and Snyder, 2001), further confirmed these results (Fig. 6D). When cultivated under high light, there was no difference in phenotype between either the GSNOR-O noe1 or the GSNOR-R noe1 transgenic line relative to that of noe1 plants (data not shown). However, when these lines were grown under low light for 10 d and then subsequently exposed to sustained H2O2 synthesis (generated by the Glc/Glc oxidase system, which could moderately release H2O2) for 3 d, leaf cell death in GSNOR-O noe1 plants was less than that observed in noe1 plants (Fig. 6E). Collectively, these findings imply that protein S-nitrosylation is involved in H2O2-induced leaf cell death.
Figure 6.
Protein S-nitrosylation is involved in H2O2-induced leaf cell death. A to D, OsGSNOR relative expression level (with the noe1 value set as 1.0; A), GSNOR activity (B), SNO content (C), and S-nitrosylated proteins from biotin-switch assay immunoblotted with anti-biotin antibody (D) using 4-month-old wild-type (WT), noe1, and OsGSNOR noe1 transgenic plants grown under high light (1,600 μmol m−2 s−1). E, Symptoms of 10-d-old wild-type, noe1, and OsGSNOR noe1 transgenic plants after treatment with 200 μm PTIO, 2.5 mm SNP, and H2O2 (generated by 2.5 mm Glc plus 2.5 units mL−1 Glc oxidase) for 3 d under low light (400 μmol m−2 s−1). These data were obtained from three independent replicates. * P < 0.05, ** P < 0.01 (t test). DTT, Dithiothreitol.
Proteomic Identification of S-Nitrosylated Proteins
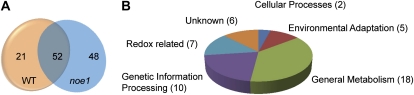
To identify possible candidates for S-nitrosylation integral to H2O2-induced leaf cell death, the biotin-switch assay and LC-MS/MS were performed. Total protein, extracted from wild type and noe1 leaves grown under low light for 10 d and subsequently transferred to high light for 2 d, were blocked with methyl methanethiosulfonate and labeled with N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (biotin-HPDP) (Wang et al., 2009); after precipitation with acetone, the resuspended mixture was first digested with trypsin before purification with affinity chromatography to harvest the biotin-labeled peptides, then S-nitrosylated peptides were judged by LC-MS/MS. Seventy-three and 100 proteins were identified from wild-type and noe1 plants, respectively, among them 52 proteins common to both wild-type and noe1 plants (Fig. 7A). Proteins identified here are involved in different processes, including general metabolism, environmental adaptation, genetic information processing, and redox regulation (Fig. 7B; Supplemental Fig. S9). Almost half the proteins identified were reported previously as being subject to S-nitrosylation (Supplemental Tables S7–S9), indicating the reliability of these results. Among the proteins identified in noe1 plants, approximately 37.5% functioned in general metabolism and 21% in genetic information processing (Fig. 7B). Proteins related to environmental adaptation and redox homeostasis accounted for 10% and 14%, respectively (Fig. 7B). Among the seven redox-related S-nitrosylated proteins identified in noe1, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and thioredoxin (TRX) were previously reported to be involved in cell death in animals, and their functions were linked with their S-nitrosylation (Sumbayev, 2003; Hara et al., 2005; Table I). These results further implied that protein S-nitrosylation is involved in H2O2-induced leaf cell death.
Figure 7.
Identification of S-nitrosylated proteins from wild-type (WT) plants and the noe1 mutant using LC-MS/MS. A, Number of S-nitrosylated proteins in wild-type and noe1 plants. B, Functional categorization of S-nitrosylated proteins identified only from noe1 plants. Both 10-d-old wild-type and noe1 seedlings cultivated under low light (400 μmol m−2 s−1) were transferred to high light (1,600 μmol m−2 s−1) for 2 d before sample collection.
Table I.
Redox-related proteins were S-nitrosylated in noe1 plants
| Protein | Gene Accession No. in the Rice Annotation Project Database | Hints to Redox Related | Hints to Cell Death through S-Nitrosylation (*) and through Other Means (**) |
| Glyceraldehyde-3-phosphate dehydrogenase | Os04g0459500 | Rodriguez-Pascual et al. (2008) | Hara et al. (2005)* |
| Thioredoxin | Os12g0188700 | Vieira Dos Santos et al. (2006) | Sumbayev (2003)* |
| Cytosolic ascorbate peroxidase | Os07g0694700 | Andersen et al. (2009) | Vacca et al. (2004)** |
| Glutaredoxin subgroup II | Os12g0175500 | Reynaert et al. (2006) | Saeed et al. (2010)** |
| Ferredoxin-dependent Glu synthase | Os07g0658400 | García-Suárez et al. (2010) | |
| Glutathione reductase | Os02g0813500 | Landino et al. (2004) | |
| Monodehydroascorbate reductase | Os09g0567300 | Schweinzer and Goldenberg (1993) |
DISCUSSION
NO and reactive oxygen species (ROS) synthesis are routine requirements for plant cells to undergo PCD; however, in some contexts, evidence has been presented that either can induce HR independently of the other (Clarke et al., 2000). The emerging data also suggest that significant cross talk occurs between NO and ROS (Delledonne et al., 2001), although the molecular details of this interchange remain undefined. Here, we identified a rice NO accumulation mutant, noe1, following a map-based cloning approach. NOE1 was found to encode the rice catalase OsCATC. Further analysis revealed that NOE1/OsCATC is the ortholog of AtCAT2 (Iwamoto et al., 2000; Zimmermann et al., 2006). A previous report showed that cell death in cat2 plants grown in long days was linked to photoperiod, not light intensity (Queval et al., 2007). However, these data contrasted with our findings with noe1 plants. When exposed to high light, the lack of leaf catalase activity in noe1 plants resulted in an accumulation of H2O2, which further induced NO production. Conversely, the reduction of intracellular NO levels using the scavenger PTIO, or the levels of SNO by overexpression of OsGSNOR, alleviated leaf cell death triggered by H2O2 in noe1 plants. In addition, treatment of noe1 plants and OsGSNOR RNA interference (RNAi) transgenic lines with the NO donor SNP promoted cell death. Taken together, these results imply that NO and SNO are important mediators of light-dependent leaf cell death in rice. Previous work has also shown that NO acts as a pivotal positive mediator in Cd2+-induced PCD in suspension cell cultures (De Michele et al., 2009), leaf senescence (Carimi et al., 2005), and the HR (Zaninotto et al., 2006). Additionally, cell death in dicotyledonous soybean (Glycine max) suspension cultures has been reported to be regulated by the intracellular NO-ROS ratio (Delledonne et al., 2001). In contrast, it was suggested that NO acts as an antagonist to delay PCD in barley (Hordeum vulgare) aleurone layers and leaf senescence in Arabidopsis (Beligni et al., 2002; Guo and Crawford, 2005; Mishina et al., 2007). These discrepancies in the role of NO in cell death might be due to the differences in plant species, redox state, and growth conditions.
The molecular mechanism(s) of NO generation in response to various stimuli still remain controversial. However, the addition of tungstate (an NR inhibitor) dramatically blunted NO accumulation in rice following exposure to high light, suggesting that increased NO accumulation in noe1 plants in response to this stimulus was predominantly generated from NR. Moreover, the increase of NR activity in noe1 plants under high light correlated with the up-regulation of two NR-encoding genes, OsNIA1 and OsNIA2. In Arabidopsis, increased NR activity in response to light, nitrate, and Suc was also congruent with increased expression of NIA1 and NIA2 (Cheng et al., 1992; Campbell, 1999). Thus, these observations provide further circumstantial evidence of a role for NR in NO synthesis in response to high light. Interestingly, promoter analysis also revealed a 28-bp motif in the promoter region of OsNIA1 and OsNIA2, which is similar to the redox-responsive motif identified in OsSODCC1, OsTRXH, and OsGRX (Tsukamoto et al., 2005). Thus, increasing H2O2 levels following exposure to high light might increase NR expression through this redox-responsive element. Furthermore, several reports have suggested that NR activity can be modulated at the posttranscriptional level. For example, H2O2 is required for the activation of mitogen-activated protein kinase (MAPK) 6, which can subsequently increase NR activity by phosphorylation of NIA2 at Ser-627 (Wang et al., 2010a). Also, NIA2 can be phosphorylated by a calcium-dependent protein kinase at Ser-534 in the hinge 1 region in response to light and other environmental signals (Su et al., 1996). Therefore, increased cellular H2O2 concentrations following exposure to high light might also increase NR activity through the phosphorylation of either Ser-627 or Ser-534.
As S-nitrosylation is a major route for the transduction of NO bioactivity, we identified 73 and 100 S-nitrosylated proteins from wild type and noe1 plants, respectively, by employing the biotin-switch assay and LC-MS/MS. Among these proteins, there were 52 S-nitrosylated proteins from both wild-type and noe1 plants. These proteins are involved in a wide array of cellular activities, such as genetic information processing, general metabolism, environmental adaptation, and redox homeostasis. Both GAPDH and TRX, which were reported to be involved in animal cell death (Sumbayev, 2003; Hara et al., 2005), were only S-nitrosylated in noe1 plants (Table I). In mammals, GADPH is S-nitrosylated at its active site, Cys-150, which promotes its association with the E3 ligase, Siah1, resulting in nuclear translocation of the SNO-GAPDH-Siah1 complex (Mannick, 2007). Within the nucleus, SNO-GAPDH stabilizes Siah1 and facilitates the ubiquitination and degradation of nuclear proteins (Hara et al., 2005), which promotes cell death (Foster et al., 2009; Tristan et al., 2011). GAPDHs also play a central role in the carbon economy of plant cells, and higher plants possess four distinct isoforms of this protein (Hajirezaei et al., 2006; Muñoz-Bertomeu et al., 2009, 2010). A knockout line of cytosolic GAPDH shows reduced levels of oxygen uptake and ATP but increased ROS accumulation and also a higher density of trichomes (Rius et al., 2006). TRX is a key modulator of redox status (Vieira Dos Santos and Rey, 2006). During nitrosative stress, S-nitrosylation of TRX, presumed to be S-nitrosylated at Cys-32 or Cys-35 within the active site, promotes apoptosis, presumably by inhibiting the oxidoreductase function of this enzyme and by facilitating the release of sequestered ASK1 (Sumbayev, 2003; Hess et al., 2005). ASK1 is a MAPK that specifically activates the Jun N-terminal kinase and p38 MAPK signaling networks and is integral to tissue necrosis factor-α-induced apoptosis (Hess et al., 2005). Thus, S-nitrosylation of both GAPDH and TRX in noe1 but not in wild-type plants parallels the development of cell death in animal systems.
Ascorbate peroxidase (Vacca et al., 2004), glutaredoxin (Saeed et al., 2010), acyl carrier protein (Feng et al., 2009), phosphomannomutase (Hoeberichts et al., 2008), Fru-bisP aldolase isozyme (Yao et al., 2004), triose phosphate isomerase (Gnerer et al., 2006), pyruvate kinase (Steták et al., 2007), acidic 27-kD endochitinase (Shin et al., 2009), and lactate/malate dehydrogenase (Matthews et al., 2004) have all previously been reported to be involved in programmed cellular execution (Table I; Supplemental Table S4). However, the molecular mechanisms associated with the functions of these proteins in cell death remain to be established. Our findings imply that all of these proteins may also be S-nitrosylated in the rice noe1 mutant. Interestingly, although noe1 plants were not subjected to any pathogen challenge, a pathogenesis-related Bet v I family protein and PR1b were both S-nitrosylated (Supplemental Table S5), suggesting that this modification might regulate the activity of these gene products. Ribosome-related proteins (Supplemental Table S6) were also S-nitrosylated, indicating that this modification might also be involved in the regulation of protein translation. Furthermore, an elongation factor was also found to be a target for S-nitrosylation, suggesting that NO may control protein synthesis in response to H2O2 (Shenton and Grant, 2003; Lindermayr et al., 2005).
Despite a central role for GSNOR in SNO homeostasis, information on the regulation of this enzyme is limited (Besson-Bard et al., 2008). Constitutively increased GSNOR activity in Arabidopsis atgsnor1-1 and atgsnor1-2 mutants correlated with elevated AtGSNOR1 mRNA levels (Feechan et al., 2005). However, GSNOR protein abundance, rather than the magnitude of transcripts, was promoted by paraquat (Chen et al., 2009). In contrast, neither AtGSNOR1 protein levels nor transcript levels were modulated during heat acclimation, suggesting that this stimulus may control GSNOR function at the posttranscriptional level (Larkindale and Vierling, 2008; Lee et al., 2008). Our data suggest that OsGSNOR transcript accumulation in noe1 plants grown under high light levels was only one-third that of the wild type (Fig. 6A). Simultaneously, the activity of this enzyme in noe1 plants was reduced to 43% of the value determined in wild-type plants. Collectively, these data imply that OsGSNOR function during high light intensities might be controlled at the transcriptional level. However, other mechanisms, including posttranslational modification, might also be significant. In this context, S-nitrosylation would be a strong candidate, and GSNOR has indeed been found to be S-nitrosylated in the acclimation of citrus plants to salinity (Tanou et al., 2009). GSNOR loss-of-function mutations in mice, Arabidopsis, and yeast result in increased SNO levels (Liu et al., 2001, 2004; Feechan et al., 2005; Rustérucci et al., 2007). Here, over-expression of OsGSNOR in noe1 plants reduced SNO levels, consistent with a key role for this enzyme in SNO homeostasis. Moreover, our results show that no change in H2O2 content occurred in either GSNOR-O noe1 or GSNOR-R noe1 transgenic lines (Supplemental Fig. S8), suggesting that NO might function downstream of H2O2 in light-driven leaf cell death in rice.
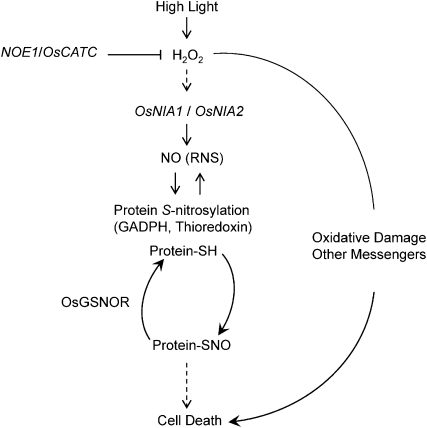
Collectively, we have identified an NO accumulation mutant, noe1, in rice. Map-based cloning revealed that NOE1 encoded OsCATC. noe1 resulted in an increase of H2O2 in the leaves, which consequently promoted NO production via the activation of NR. Our data imply that NO and SNO are important mediators of light-dependent leaf cell death in rice. Furthermore, we have uncovered a series of S-nitrosylated proteins in both noe1 and wild-type plants, which may help to reveal the role of this posttranslational modification in the control of light-mediated leaf cell death in rice (Fig. 8).
Figure 8.
A hypothetical model for the role of NO and SNOs in H2O2-induced leaf cell death in rice. RNS, Reactive nitrogen species; ‐SH, free thiol.
MATERIALS AND METHODS
Plant Growth and Sampling
Rice (Oryza sativa) plants were cultivated in the experimental field of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, in Beijing.
Treatments were performed using 10-d-old rice seedlings. Seeds of the wild type and noe1 were submerged in water for 2 d at 37°C, then the uniformly germinated seeds were cultured on 96-well plates supplied with half-strength Murashige and Skoog basal medium (pH 5.8) for 10 d in the growth chamber at 30°C for 14 h (low light, 400 μmol m−2 s−1) and 28°C for 10 h (dark). After the seedlings were transferred to high light (1,600 μmol m−2 s−1), leaf samples were taken on the 2nd and 5th d to measure the SNO and H2O2 contents. The chemicals used to treat the 10-d-old seedlings, SNP (an NO donor), PTIO (an NO scavenger), CAT (an H2O2 scavenger), tungstate (an NR inhibitor), and l-NMMA (an NOS inhibitor) were added with 200 mL of half-strength Murashige and Skoog medium. All chemicals were purchased from Sigma-Aldrich. GSNOR-O noe1 and GSNOR-R noe1 transgenic lines and controls were cultivated under low light for 10 d and treated with 200 μm PTIO, 2.5 mm SNP, and H2O2 (about 400 μmol L−1, generated by 2.5 mm Glc plus 2.5 units mL−1 Glc oxidase; Kim et al., 2010) for 3 d. All experiments were repeated three times.
Map-Based Cloning of NOE1/OsCATC
To map the NOE1 gene, an F2 population derived from a cross between noe1 (japonica) and Minghui 63 (indica) was constructed. The genomic DNA from 313 F2 progeny with the leaf cell death phenotype was extracted with a modified cetyl-trimethyl-ammonium bromide method described previously (Mou et al., 2000). To fine-map NOE1, some polymorphic insertion-deletion markers were generated based on the different sequences between japonica and indica. The locus was finally defined to a 61-kb region, and all 10 candidate genes in this region were sequenced and analyzed using SeqMan within the Lasergene version 5.0 software (DNASTAR). Primer sequences are listed in Supplemental Table S1.
Real-Time PCR Analysis
Total RNA was extracted using the TRIzol kit according to the user manual (GenStar; Tong et al., 2009). Two micrograms of total RNA treated with DNase I was used for cDNA synthesis with an RT kit (Promega). Real-time PCR experiments were performed using gene-specific primers in a total volume of 20 μL containing 0.3 μL of deoxyribonucleoside triphosphate, 2 μL of 10× Taq buffer, 0.2 μL of Taq enzyme, 0.5 μL of 40× SYBR Green Master Mix (Applied Biosystems), 1.5 μL of 1 mm gene-specific primers, and 1 μL of cDNA on a C1000 Thermal Cycler (CFX96 real-time system; Bio-Rad) as described before (Yang et al., 2009). The rice 18S gene was used as the internal control. Quantification consisted of at least three independent replicates. The primer sequences used are listed in Supplemental Table S2.
Complementation of noe1 and OsGSNOR Vector Construction
The binary vector pXQAct, a derivative of pCambia 2300, carrying the rice Actin1 promoter and the OCS terminator, was used for plant transformation (Sun et al., 2011). For complementation of the noe1 mutant, the 1,640-bp full-length cDNA of NOE1/OsCATC with the XbaI site amplified from Nipponbare cDNA was ligated to pXQAct, resulting in pnoe1-C vector. The 1,266-bp full-length cDNA of amplified OsGSNOR was digested with SmaI and SalI and subsequently ligated into pXQAct, resulting in pGSNOR-O vector. To generate the RNAi construct pGSNOR-R, a 318-bp fragment was amplified from OsGSNOR cDNA and sequentially cloned into XhoI/BglII and BamHI/SalI sites of pUCC-RNAi vector to target the gene in both the sense and antisense orientations (Luo et al., 2005). The whole stem-loop fragment was further cloned into pXQAct, yielding the binary GSNOR RNAi vector. These plasmids were introduced into Agrobacterium tumefaciens AGL-1 by freeze-thaw transformation. Consequently, mutant noe1 callus was transformed by an Agrobacterium-mediated method as described previously (Liu et al., 2007). The primer sequences used for vector construction are listed in Supplemental Table S3.
Cell Death Analysis
Relative cell death of suspension cells was assayed as described previously with some modifications (Delledonne et al., 2001). Briefly, after 24-h treatments with CAT, PTIO, SNP, and SNP+PTIO under low light (400 μmol m−2 s−1), the rice suspension cells were stained with 0.05% Evans blue (Sigma-Aldrich) for 15 min. The excess dye was removed by extensive washing, and dye bound to dead cells was solubilized in 50% (v/v) methanol/1% SDS in a volume of 20 mL for 30 min at 50°C and quantified by optical density at 595nm (OD595). The OD595 value of rice cell suspensions saturated with methanol for 15 min was taken as the total killed cell death. The relative cell death is expressed as an OD595 ratio of different treatments to total killed cell death. The relative cell death of wild-type and noe1 seedling leaves was analyzed using Adobe Photoshop CS and expressed as a percentage of cell death region to total leaf region.
Histochemical Detection
H2O2 was detected by 3,3′-diaminobenzidine staining as described previously with some modifications (Thordal Christensen et al., 1997). The fully expanded leaves of 2-month-old wild-type and noe1 plants were detached and infiltrated with the 0.1% 3,3′-diaminobenzidine solution. The sampled leaves were placed in a growth chamber for 5 h at 28°C and cleared in boiling ethanol (95%) for 10 min. The chlorophyll was removed by incubating in 95% (v/v) ethanol overnight before photographing.
Superoxide accumulation in rice leaves was visualized by 0.1% nitroblue tetrazolium staining as described (Fang et al., 2008). The fully expanded leaves of 2-month-old wild-type and noe1 plants were stained with nitroblue tetrazolium solution. After staining overnight, the chlorophyll was removed by incubating in 95% (v/v) ethanol overnight.
Dead cells were stained using detached leaves by a method modified previously (Wäspi et al., 2001). Leaves of 2-month-old wild-type and noe1 plants were submerged in trypan blue solution at 70°C for 10 min and then heated in boiling water for 2 min and left staining overnight. After destaining in chloral hydrate solution (25 g of chloral hydrate in 10 mL of water) for 3 d, samples were equilibrated with 70% glycerol for microphotography.
Malondialdehyde Content Assay
Malondialdehyde (MDA) was regarded as an indicator of lipid peroxidation and cell death. The content of MDA was determined with a MDA assay kit (Beyotime) with some modifications (Qian et al., 2010). Briefly, leaves (0.02 g fresh weight) were homogenized in 2 mL of phosphate buffer (pH 7.8) containing 1% polyvinylpyrrolidone and centrifuged at 2,500g for 10 min. MDA in the supernatant was determined according to the reaction with thiobarbituric acid and measured the absorbance at 450, 532, and 600 nm. According to the equation 6.45 × (OD532 − OD600) − 0.56 × OD450, the MDA content was expressed as nm MDA g−1 fresh weight.
Quantitative Measurement of H2O2 Content and Catalase Activity
The measurement of H2O2 production was performed by extracting H2O2 from leaves according to a previously described method (Huang et al., 2009) and quantifying the molecule with the Amplex Red Hydrogen Peroxidase Assay Kit (Molecular Probes, Invitrogen) according to the manufacturer’s instructions. Catalase activity was carried out with a Catalase Assay Kit (Beyotime) according to the manufacturer’s instructions.
Measurement of Ion Leakage
The measurement of ion leakage was performed according to a previously described method (Woo et al., 2001). Five different leaf discs from wild-type and noe1 plants were placed into a 100-mL beaker containing 40 mL of distilled deionized water. After shaking at 120 rpm for 3 h, the conductivity (C1) was measured. Then, the discs were boiled for 10 min and shaken for 1 h, and the conductivity (C2) was measured again. The ion leakage was calculated according to the equation (C1/C2) × 100%.
Saville-Griess Assay
SNO content was measured using the Saville-Griess assay as described with minor modifications (Feechan et al., 2005). In the Saville-Griess assay, S-NO bonds were broken by mercuric chloride, and the released NO reacted with sulfanilamide. The resulting diazonium salt is coupled with the aromatic amine N-(1-naphthyl)-ethylenediamine to form an intensely colored azo dye that can be measured at 540 nm (Feechan et al., 2005; King et al., 2005). Briefly, fine powder of plant tissues from liquid nitrogen was lysed in 600 μL of extraction buffer (50 mm Tris-HCl, pH 8.0, and 150 mm NaCl) containing 1 mm protease inhibitor phenylmethanesulfonyl fluoride (PMSF) and incubated on ice for 20 min. After centrifugation at 10,000 rpm for 15 min at 4°C, 160 μL of supernatant clear cell lysate was incubated with the same volume of 1% sulfanilamide and 0.1% N-(1-naphthyl)-ethylenediamine with or without the addition of 3.75 mm HgCl2 for 20 min in the dark. Then, SNO content was measured photometrically at 540 nm using 96-well plates with Tecan Infinite M200 (Tecan Group). The SNO content was calculated according to the absorption at 540 nm and a GSNO concentration standard curve (Feechan et al., 2005).
Determination of Endogenous NO Content of Suspension Cells with DAF-FM DA
The endogenous NO level of suspension cells was detected by imaging the cell-permeable, NO-specific fluorescent probe DAF-FM DA (Invitrogen) by confocal microscopy with some modifications (Zhao et al., 2007). Wild-type and noe1 suspension cells (untreated or treated with 1 mm H2O2 for 30 min at 28°C) were incubated with 100 μm Tris-HCl (pH 7.6) containing 5 μm DAF-FM DA at 28°C for 40 min. The cells were then washed three times with fresh Tris-HCl (100 μm, pH 7.6) to remove excess probe and incubated for an additional 20 min to allow complete deesterification of the intracellular diacetates. The incubated cells were visualized using a laser confocal scanning microscope (Leica TCS SP5). Excitation/emission wavelengths were 488/515 nm. Data are presented as mean pixel intensities. Fifty to 100 suspension cells were observed per treatment for three independent replicates.
Determination of NOS and NR Activities and Nitrate Content
The activities of NOS and NR were determined with the NOS Assay Kit and the Nitrate/Nitrite Assay Kit (Beyotime) with some modifications (Xiong et al., 2009). Total protein was extracted using buffer containing 100 mm HEPES-KOH (pH 7.5), 1 mm EDTA, 10% glycerol, 5 mm dithiothreitol, 0.1% Triton X-100, 0.5 mm PMSF, 20 μm FAD, 25 μm leupeptin, 5 μm Na2MoO4, and 1% polyvinylpyrrolidone. Then, the NOS and NR activities were measured according to the manufacturer’s instructions. Nitrate content were measured using the Siever Nitric Oxide Analyzer 280i (Siever; GE Analytical Instruments) according to the manual instructions.
Glutathione-Dependent Formaldehyde Dehydrogenase/GSNOR Enzyme Activity Assay
GSNOR enzyme activity was measured spectrophotometrically at 340 nm at 25°C by monitoring the decomposition of NADH as described before (Durner et al., 1998). Glutathione‐dependent formaldehyde dehydrogenase activity was determined by incubating 100 μg of protein in 300 μL of assay mixture that contained 20 mm Tris-HCl (pH 8.0), 0.2 mm NADH, and 0.5 mm EDTA. The reaction was started by adding GSNO (Alexis Biochemicals) at a final concentration of 400 μm.
Protein Extractions
Protein extractions were performed according to a previously described method (Jaffrey and Snyder, 2001). In brief, leaf tissues (200 mg) were ground in liquid nitrogen using a mortar and pestle. Soluble proteins were extracted from the resulting powder at 4°C in 1 mL of HEN buffer (250 mm HEPES-NaOH, pH 7.7, 1 mm EDTA, 0.1 mm neocuproine, and 10 mm PMSF) in the dark. The total extracts were incubated on ice for 20 min and centrifuged at 10,000 rpm for 15 min at 4°C.
Biotin-Switch Assay and Isolation of Biotinylated Proteins
The biotin-switch assay was performed as described previously (Wang et al., 2009). Three milligrams of soluble total protein was used for the assay. After the biotin-HPDP labeling step, around 30 μg of labeled soluble protein was used for western blot according to the standard protocol. Two volumes of −20°C acetone was added to the rest of the samples, incubated for 20 min at −20°C, and then centrifuged at a minimum of 2,000g for 10 min at 4°C. The supernatant was discarded to remove biotin-HPDP, then the wall of the tube and the surface of the pellets were gently rinsed with −20°C acetone to remove traces of biotin-HPDP. The pellets were resuspend in 0.1 mL of 25 mm NH4HCO3 buffer per mg of protein in the initial protein sample and digested with trypsin (sequencing-grade modified trypsin; Promega) with a protease:protein ratio of 1:20 (w/w) at 4°C overnight. Two volumes of neutralization buffer (20 mm HEPES-NaOH, pH 7.7, 100 mm NaCl, 1 mm EDTA, and 0.5% Triton X-100) was added, then 15 μL of packed streptavidin-agarose resin per mg of protein was used in the initial protein sample to purify biotinylated proteins. The proteins were incubated with resin for 1 h at room temperature with continuous shaking, and the resin was washed five times with 10 volumes of neutralization buffer plus 600 mm NaCl and centrifuged at 200g for 5 s at room temperature between each wash. The resin was incubated with elution buffer (20 mm HEPES-NaOH, pH 7.7, 100 mm NaCl, 1 mm EDTA, and 100 mm mercaptoethanol) to recover the bound proteins. Purified protein extracts were identified by LC-MS/MS analysis to find S-nitrosylated proteins.
According to a previous report (Wang et al., 2009), LC-MS/MS experiments were carried out on a Thermo LTQ Orbitrap XL instrument (Thermo Electron) equipped with a house-made source and an Eksigent two-dimensional nanoLC system (Eksigent Technologies). Sample analyses were performed using an in-house-made reverse-phase C18 capillary column (15 cm × 100 μm i.d.). The peptides were sequentially eluted with a gradient of 0% to 80% buffer B (acetonitrile with 0.5% formic acid) in buffer A (water with 0.5% formic acid) at a flow rate of 400 nL min−1. The electrospray source parameters were 2.0 kV of electrospray voltage, the range of mass-to-charge ratio was from 300 to 1,800, and the normalized collision energy of MS/MS was 35%. The data were analyzed with Bioworks 3.31 and Sequest 2.8, and the results were filtered by Xcorr (the cross-correlation value from the search) +1 > 1.9, +2 > 2.5, +3 > 3.75, sp > 500, Δcn (the Δ correlation value) > 0.1, Rsp ≤ 5. The pathways of proteins that exert their roles were performed by searching in Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/dbget-bin/www_bfind?O.sativa).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Leaf cell death in noe1 is dependent on high light.
Supplemental Figure S2. Leaf cell death in noe1 is dependent on light intensity.
Supplemental Figure S3. NOE1/OsCATC sequence alignment with other catalase proteins from rice and Arabidopsis.
Supplemental Figure S4. More H2O2 accumulated and cell death occurred in noe1.
Supplemental Figure S5. H2O2-induced leaf cell death in noe1.
Supplemental Figure S6. H2O2 and SNO levels in wild-type and noe1 seedlings at different time points.
Supplemental Figure S7. Sequence alignment of GSNOR from rice and Arabidopsis.
Supplemental Figure S8. H2O2 contents in 4-month-old wild-type, noe1, and OsGSNOR transgenic lines grown under high light (1,600 μmol m−2 s−1).
Supplemental Figure S9. Functional categorization of S-nitrosylated proteins identified from wild-type and noe1 plants.
Supplemental Table S1. Primers used for cloning.
Supplemental Table S2. Primers used for quantitative RT-PCR.
Supplemental Table S3. Primers used for vector construction.
Supplemental Table S4. S-Nitrosylated proteins reported to trigger cell death identified from noe1 plants.
Supplemental Table S5. S-Nitrosylated proteins subjected to environmental adaptation identified from noe1 plants.
Supplemental Table S6. S-Nitrosylated proteins related to ribosome identified from noe1 plants.
Supplemental Table S7. S-Nitrosylated proteins identified from both wild-type and noe1 plants.
Supplemental Table S8. S-Nitrosylated proteins identified only from noe1 plants.
Supplemental Table S9. S-Nitrosylated proteins identified only from wild-type plants.
Acknowledgments
We thank Dr. Jeum Kyu Hong from Jinju National University for valuable advice and critical reading of the manuscript.
References
- Andersen S, Iversen F, Terpling S, Pedersen KM, Gustenhoff P, Laurberg P. (2009) More hypothyroidism and less hyperthyroidism with sufficient iodine nutrition compared to mild iodine deficiency: a comparative population-based study of older people. Maturitas 64: 126–131 [DOI] [PubMed] [Google Scholar]
- Beligni MV, Fath A, Bethke PC, Lamattina L, Jones RL. (2002) Nitric oxide acts as an antioxidant and delays programmed cell death in barley aleurone layers. Plant Physiol 129: 1642–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson-Bard A, Pugin A, Wendehenne D. (2008) New insights into nitric oxide signaling in plants. Annu Rev Plant Biol 59: 21–39 [DOI] [PubMed] [Google Scholar]
- Campbell WH. (1999) Nitrate reductase structure, function and regulation: bridging the gap between biochemistry and physiology. Annu Rev Plant Physiol Plant Mol Biol 50: 277–303 [DOI] [PubMed] [Google Scholar]
- Carimi F, Zottini M, Costa A, Cattelan I, De Michele R, Terzi M, Lo Schiavo F. (2005) NO signalling in cytokinin-induced programmed cell death. Plant Cell Environ 28: 1171–1178 [Google Scholar]
- Carreras MC, Poderoso JJ. (2007) Mitochondrial nitric oxide in the signaling of cell integrated responses. Am J Physiol Cell Physiol 292: C1569–C1580 [DOI] [PubMed] [Google Scholar]
- Chanvorachote P, Nimmannit U, Wang LY, Stehlik C, Lu B, Azad N, Rojanasakul Y. (2005) Nitric oxide negatively regulates Fas CD95-induced apoptosis through inhibition of ubiquitin-proteasome-mediated degradation of FLICE inhibitory protein. J Biol Chem 280: 42044–42050 [DOI] [PubMed] [Google Scholar]
- Chen R, Sun S, Wang C, Li Y, Liang Y, An F, Li C, Dong H, Yang X, Zhang J, et al. (2009) The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res 19: 1377–1387 [DOI] [PubMed] [Google Scholar]
- Cheng CL, Acedo GN, Cristinsin M, Conkling MA. (1992) Sucrose mimics the light induction of Arabidopsis nitrate reductase gene transcription. Proc Natl Acad Sci USA 89: 1861–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A, Desikan R, Hurst RD, Hancock JT, Neill SJ. (2000) NO way back: nitric oxide and programmed cell death in Arabidopsis thaliana suspension cultures. Plant J 24: 667–677 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Palma JM, del Río LA, Barroso JB. (2009) Evidence supporting the existence of L-arginine-dependent nitric oxide synthase activity in plants. New Phytol 184: 9–14 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Palma JM, Sandalio LM, Valderrama R, Barroso JB, Del Río LA. (2008) Peroxisomal xanthine oxidoreductase: characterization of the enzyme from pea (Pisum sativum L.) leaves. J Plant Physiol 165: 1319–1330 [DOI] [PubMed] [Google Scholar]
- Crawford NM, Guo FQ. (2005) New insights into nitric oxide metabolism and regulatory functions. Trends Plant Sci 10: 195–200 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C. (1998) Nitric oxide functions as a signal in plant disease resistance. Nature 394: 585–588 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Zeier J, Marocco A, Lamb C. (2001) Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc Natl Acad Sci USA 98: 13454–13459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Michele R, Vurro E, Rigo C, Costa A, Elviri L, Di Valentin M, Careri M, Zottini M, Sanità di Toppi L, Lo Schiavo F. (2009) Nitric oxide is involved in cadmium-induced programmed cell death in Arabidopsis suspension cultures. Plant Physiol 150: 217–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner J, Wendehenne D, Klessig DF. (1998) Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Natl Acad Sci USA 95: 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Chai C, Qian Q, Li C, Tang J, Sun L, Huang Z, Guo X, Sun C, Liu M, et al. (2008) Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J 54: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feechan A, Kwon E, Yun BW, Wang Y, Pallas JA, Loake GJ. (2005) A central role for S-nitrosothiols in plant disease resistance. Proc Natl Acad Sci USA 102: 8054–8059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Witkowski A, Smith S. (2009) Down-regulation of mitochondrial acyl carrier protein in mammalian cells compromises protein lipoylation and respiratory complex I and results in cell death. J Biol Chem 284: 11436–11445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresi N, Correa-Aragunde N, Parisi G, Caló G, Salerno G, Lamattina L. (2010) Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. Plant Cell 22: 3816–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster MW, Hess DT, Stamler JS. (2009) Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med 15: 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Suárez J, Röder M, Díaz de León J. (2010) Identification of QTLs and associated molecular markers of agronomic traits in wheat (Triticum aestivum L.) under two conditions of nitrogen fertilization. Cereal Res Commun 38: 459–470 [Google Scholar]
- Gnerer JP, Kreber RA, Ganetzky B. (2006) wasted away, a Drosophila mutation in triosephosphate isomerase, causes paralysis, neurodegeneration, and early death. Proc Natl Acad Sci USA 103: 14987–14993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FQ, Crawford NM. (2005) Arabidopsis nitric oxide synthase1 is targeted to mitochondria and protects against oxidative damage and dark-induced senescence. Plant Cell 17: 3436–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta KJ, Fernie AR, Kaiser WM, van Dongen JT. (2011) On the origins of nitric oxide. Trends Plant Sci 16: 160–168 [DOI] [PubMed] [Google Scholar]
- Hajirezaei MR, Biemelt S, Peisker M, Lytovchenko A, Fernie AR, Sonnewald U. (2006) The influence of cytosolic phosphorylating glyceraldehyde 3-phosphate dehydrogenase (GAPC) on potato tuber metabolism. J Exp Bot 57: 2363–2377 [DOI] [PubMed] [Google Scholar]
- Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, et al. (2005) S-Nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol 7: 665–674 [DOI] [PubMed] [Google Scholar]
- He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, Jing L, Yang Z, Chen L, Guo F, et al. (2004) Nitric oxide represses the Arabidopsis floral transition. Science 305: 1968–1971 [DOI] [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. (2005) Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol 6: 150–166 [DOI] [PubMed] [Google Scholar]
- Hoeberichts FA, Vaeck E, Kiddle G, Coppens E, van de Cotte B, Adamantidis A, Ormenese S, Foyer CH, Zabeau M, Inzé D, et al. (2008) A temperature-sensitive mutation in the Arabidopsis thaliana phosphomannomutase gene disrupts protein glycosylation and triggers cell death. J Biol Chem 283: 5708–5718 [DOI] [PubMed] [Google Scholar]
- Hong JK, Yun BW, Kang JG, Raja MU, Kwon E, Sorhagen K, Chu C, Wang Y, Loake GJ. (2008) Nitric oxide function and signalling in plant disease resistance. J Exp Bot 59: 147–154 [DOI] [PubMed] [Google Scholar]
- Huang X, Kiefer E, von Rad U, Ernst D, Foissner I, Durner J. (2002) Nitric oxide burst and nitric oxide-dependent gene induction in plants. Plant Physiol Biochem 40: 625–631 [Google Scholar]
- Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX. (2009) A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev 23: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Higo H, Higo K. (2000) Differential diurnal expression of rice catalase genes: the 5′-flanking region of CatA is not sufficient for circadian control. Plant Sci 151: 39–46 [Google Scholar]
- Jaffrey SR, Snyder SH. (2001) The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001: pl1. [DOI] [PubMed] [Google Scholar]
- Jasid S, Simontacchi M, Bartoli CG, Puntarulo S. (2006) Chloroplasts as a nitric oxide cellular source: effect of reactive nitrogen species on chloroplastic lipids and proteins. Plant Physiol 142: 1246–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SG, Kim ST, Wang Y, Kim SK, Lee CH, Kim KK, Kim JK, Lee SY, Kang KY. (2010) Overexpression of rice isoflavone reductase-like gene (OsIRL) confers tolerance to reactive oxygen species. Physiol Plant 138: 1–9 [DOI] [PubMed] [Google Scholar]
- King M, Gildemeister O, Gaston B, Mannick JB. (2005) Assessment of S-nitrosothiols on diaminofluorescein gels. Anal Biochem 346: 69–76 [DOI] [PubMed] [Google Scholar]
- Knowles RG, Moncada S. (1994) Nitric oxide synthases in mammals. Biochem J 298: 249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landino LM, Moynihan KL, Todd JV, Kennett KL. (2004) Modulation of the redox state of tubulin by the glutathione/glutaredoxin reductase system. Biochem Biophy Res Commun 314: 555–560 [DOI] [PubMed] [Google Scholar]
- Larkindale J, Vierling E. (2008) Core genome responses involved in acclimation to high temperature. Plant Physiol 146: 748–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee U, Wie C, Fernandez BO, Feelisch M, Vierling E. (2008) Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell 20: 786–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner M, Vandelle E, Gaupels F, Bellin D, Delledonne M. (2009) NO signals in the haze: nitric oxide signalling in plant defence. Curr Opin Plant Biol 12: 451–458 [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J. (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol 137: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS. (2001) A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410: 490–494 [DOI] [PubMed] [Google Scholar]
- Liu L, Yan Y, Zeng M, Zhang J, Hanes MA, Ahearn G, McMahon TJ, Dickfeld T, Marshall HE, Que LG, et al. (2004) Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell 116: 617–628 [DOI] [PubMed] [Google Scholar]
- Liu X, Bai X, Wang X, Chu C. (2007) OsWRKY71, a rice transcription factor, is involved in rice defense response. J Plant Physiol 164: 969–979 [DOI] [PubMed] [Google Scholar]
- Luo A, Liu L, Tang Z, Bai X, Cao S, Chu C. (2005) Down-regulation of OsGRF1 gene in rice rhd1 mutant results in reduced heading date. J Integr Plant Biol 47: 745–752 [Google Scholar]
- Ma Y, Liu L, Zhu C, Sun C, Xu B, Fang J, Tang J, Luo A, Cao S, Li G, et al. (2009) Molecular analysis of rice plants harboring a multi-functional T-DNA tagging system. J Genet Genomics 36: 267–276 [DOI] [PubMed] [Google Scholar]
- Mannick JB. (2007) Regulation of apoptosis by protein S-nitrosylation. Amino Acids 32: 523–526 [DOI] [PubMed] [Google Scholar]
- Matthews CZ, Subramanian R, Woolf EJ, Foster N, Matuszewski BK. (2004) Isolation and structural characterization of the photolysis products of etoricoxib. Pharmazie 59: 913–919 [PubMed] [Google Scholar]
- Mishina TE, Lamb C, Zeier J. (2007) Expression of a nitric oxide degrading enzyme induces a senescence programme in Arabidopsis. Plant Cell Environ 30: 39–52 [DOI] [PubMed] [Google Scholar]
- Mou Z, He Y, Dai Y, Liu X, Li J. (2000) Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell 12: 405–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J, Cascales-Miñana B, Alaiz M, Segura J, Ros R. (2010) A critical role of plastidial glycolytic glyceraldehyde-3-phosphate dehydrogenase in the control of plant metabolism and development. Plant Signal Behav 5: 67–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J, Cascales-Miñana B, Mulet JM, Baroja-Fernández E, Pozueta-Romero J, Kuhn JM, Segura J, Ros R. (2009) Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol 151: 541–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill SJ, Desikan R, Clarke A, Hancock JT. (2002) Nitric oxide is a novel component of abscisic acid signaling in stomatal guard cells. Plant Physiol 128: 13–16 [PMC free article] [PubMed] [Google Scholar]
- Palmer RMJ, Ashton DS, Moncada S. (1988) Vascular endothelial cells synthesize nitric oxide from L-arginine. Nature 333: 664–666 [DOI] [PubMed] [Google Scholar]
- Parani M, Rudrabhatla S, Myers R, Weirich H, Smith B, Leaman DW, Goldman SL. (2004) Microarray analysis of nitric oxide responsive transcripts in Arabidopsis. Plant Biotechnol J 2: 359–366 [DOI] [PubMed] [Google Scholar]
- Park HS, Yu JW, Cho JH, Kim MS, Huh SH, Ryoo K, Choi EJ. (2004) Inhibition of apoptosis signal-regulating kinase 1 by nitric oxide through a thiol redox mechanism. J Biol Chem 279: 7584–7590 [DOI] [PubMed] [Google Scholar]
- Polverari A, Molesini B, Pezzotti M, Buonaurio R, Marte M, Delledonne M. (2003) Nitric oxide-mediated transcriptional changes in Arabidopsis thaliana. Mol Plant Microbe Interact 16: 1094–1105 [DOI] [PubMed] [Google Scholar]
- Qian J, Jiang F, Wang B, Yu Y, Zhang X, Yin Z, Liu C. (2010) Ophiopogonin D prevents H2O2-induced injury in primary human umbilical vein endothelial cells. J Ethnopharmacol 128: 438–445 [DOI] [PubMed] [Google Scholar]
- Queval G, Issakidis-Bourguet E, Hoeberichts FA, Vandorpe M, Gakière B, Vanacker H, Miginiac-Maslow M, Van Breusegem F, Noctor G. (2007) Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J 52: 640–657 [DOI] [PubMed] [Google Scholar]
- Reynaert NL, van der Vliet A, Guala AS, McGovern T, Hristova M, Pantano C, Heintz NH, Heim J, Ho YS, Matthews DE, et al. (2006) Dynamic redox control of NF-kappaB through glutaredoxin-regulated S-glutathionylation of inhibitory kappaB kinase beta. Proc Natl Acad Sci USA 103: 13086–13091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rius SP, Casati P, Iglesias AA, Gomez-Casati DF. (2006) Characterization of an Arabidopsis thaliana mutant lacking a cytosolic non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase. Plant Mol Biol 61: 945–957 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Pascual F, Redondo-Horcajo M, Magan-Marchal N, Lagares D, Martinez-Ruiz A, Kleinert H, Lamas S. (2008) Glyceraldehyde-3-phosphate dehydrogenase regulates endothelin-1 expression by a novel, redox-sensitive mechanism involving mRNA stability. Mol Cell Biol 28: 7139–7155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Puertas MC, Laxa M, Matte A, Zaninotto F, Finkemeier I, Jones AM, Perazzolli M, Vandelle E, Dietz KJ, Delledonne M. (2007) S-nitrosylation of peroxiredoxin II E promotes peroxynitrite-mediated tyrosine nitration. Plant Cell 19: 4120–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rümer S, Kapuganti JG, Kaiser WM. (2009) Oxidation of hydroxylamines to NO by plant cells. Plant Signal Behav 4: 853–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustérucci C, Espunya MC, Díaz M, Chabannes M, Martínez MC. (2007) S-Nitrosoglutathione reductase affords protection against pathogens in Arabidopsis, both locally and systemically. Plant Physiol 143: 1282–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed U, Ray A, Valli RK, Kumar AM, Ravindranath V. (2010) DJ-1 loss by glutaredoxin but not glutathione depletion triggers Daxx translocation and cell death. Antioxid Redox Signal 13: 127–144 [DOI] [PubMed] [Google Scholar]
- Sawa A, Khan AA, Hester LD, Snyder SH. (1997) Glyceraldehyde-3-phosphate dehydrogenase: nuclear translocation participates in neuronal and nonneuronal cell death. Proc Natl Acad Sci USA 94: 11669–11674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligman K, Saviani EE, Oliveira HC, Pinto-Maglio CA, Salgado I. (2008) Floral transition and nitric oxide emission during flower development in Arabidopsis thaliana is affected in nitrate reductase-deficient plants. Plant Cell Physiol 49: 1112–1121 [DOI] [PubMed] [Google Scholar]
- Schweinzer E, Goldenberg H. (1993) Monodehydroascorbate reductase activity in the surface membrane of leukemic cells: characterization by a ferricyanide-driven redox cycle. Eur J Biochem 218: 1057–1062 [DOI] [PubMed] [Google Scholar]
- Sen N, Hara MR, Ahmad AS, Cascio MB, Kamiya A, Ehmsen JT, Agrawal N, Hester L, Doré S, Snyder SH, et al. (2009) GOSPEL: a neuroprotective protein that binds to GAPDH upon S-nitrosylation. Neuron 63: 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenton D, Grant CM. (2003) Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem J 374: 513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin KS, Kwon NJ, Kim YH, Park HS, Kwon GS, Yu JH. (2009) Differential roles of the ChiB chitinase in autolysis and cell death of Aspergillus nidulans. Eukaryot Cell 8: 738–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoulars K, Rodriguez MA, Thompson T, Turk J, Crowley J, Markaverich BM. (2008) Regulation of the nitric oxide pathway genes by tetrahydrofurandiols: microarray analysis of MCF-7 human breast cancer cells. Cancer Lett 264: 265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steták A, Veress R, Ovádi J, Csermely P, Kéri G, Ullrich A. (2007) Nuclear translocation of the tumor marker pyruvate kinase M2 induces programmed cell death. Cancer Res 67: 1602–1608 [DOI] [PubMed] [Google Scholar]
- Stöhr C, Stremlau S. (2006) Formation and possible roles of nitric oxide in plant roots. J Exp Bot 57: 463–470 [DOI] [PubMed] [Google Scholar]
- Stoimenova M, Igamberdiev AU, Gupta KJ, Hill RD. (2007) Nitrite-driven anaerobic ATP synthesis in barley and rice root mitochondria. Planta 226: 465–474 [DOI] [PubMed] [Google Scholar]
- Su W, Huber SC, Crawford NM. (1996) Identification in vitro of a post-translational regulatory site in the hinge 1 region of Arabidopsis nitrate reductase. Plant Cell 8: 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumbayev VV. (2003) S-Nitrosylation of thioredoxin mediates activation of apoptosis signal-regulating kinase 1. Arch Biochem Biophys 415: 133–136 [DOI] [PubMed] [Google Scholar]
- Sun C, Liu L, Tang J, Lin A, Zhang F, Fang J, Zhang G, Chu C. (2011) RLIN1, encoding a putative coproporphyrinogen III oxidase, is involved in lesion initiation in rice. J Genet Genomics 38: 29–37 [DOI] [PubMed] [Google Scholar]
- Tanou G, Job C, Rajjou L, Arc E, Belghazi M, Diamantidis G, Molassiotis A, Job D. (2009) Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J 60: 795–804 [DOI] [PubMed] [Google Scholar]
- Thordal Christensen H, Zhang ZG, Wei YD, Collinge DB. (1997) Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Tong H, Jin Y, Liu W, Li F, Fang J, Yin Y, Qian Q, Zhu L, Chu C. (2009) DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J 58: 803–816 [DOI] [PubMed] [Google Scholar]
- Tristan C, Shahani N, Sedlak TW, Sawa A. (2011) The diverse functions of GAPDH: views from different subcellular compartments. Cell Signal 23: 317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang AH, Lee YI, Ko HS, Savitt JM, Pletnikova O, Troncoso JC, Dawson VL, Dawson TM, Chung KK. (2009) S-Nitrosylation of XIAP compromises neuronal survival in Parkinson’s disease. Proc Natl Acad Sci USA 106: 4900–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto S, Morita S, Hirano E, Yokoi H, Masumura T, Tanaka K. (2005) A novel cis-element that is responsive to oxidative stress regulates three antioxidant defense genes in rice. Plant Physiol 137: 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca RA, de Pinto MC, Valenti D, Passarella S, Marra E, De Gara L. (2004) Production of reactive oxygen species, alteration of cytosolic ascorbate peroxidase, and impairment of mitochondrial metabolism are early events in heat shock-induced programmed cell death in tobacco Bright-Yellow 2 cells. Plant Physiol 134: 1100–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandelle E, Delledonne M. (2008) Methods for nitric oxide detection during plant-pathogen interactions. Methods Enzymol 437: 575–594 [DOI] [PubMed] [Google Scholar]
- Vieira Dos Santos C, Rey P. (2006) Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci 11: 329–334 [DOI] [PubMed] [Google Scholar]
- Wang EQ, Lee WI, Brazeau D, Fung HL. (2002) cDNA microarray analysis of vascular gene expression after nitric oxide donor infusions in rats: implications for nitrate tolerance mechanisms. AAPS PharmSci 4: E10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Du Y, Li Y, Ren D, Song CP. (2010a) Hydrogen peroxide-mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis. Plant Cell 22: 2981–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen C, Loake GJ, Chu C. (2010b) Nitric oxide: promoter or suppressor of programmed cell death? Protein Cell 1: 133–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YQ, Feechan A, Yun BW, Shafiei R, Hofmann A, Taylor P, Xue P, Yang FQ, Xie ZS, Pallas JA, et al. (2009) S-Nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J Biol Chem 284: 2131–2137 [DOI] [PubMed] [Google Scholar]
- Wäspi U, Schweizer P, Dudler R. (2001) Syringolin reprograms wheat to undergo hypersensitive cell death in a compatible interaction with powdery mildew. Plant Cell 13: 153–161 [PMC free article] [PubMed] [Google Scholar]
- Wilson ID, Neill SJ, Hancock JT. (2008) Nitric oxide synthesis and signalling in plants. Plant Cell Environ 31: 622–631 [DOI] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Lu H, Lu KX, Duan YX, An LY, Zhu C. (2009) Cadmium decreases crown root number by decreasing endogenous nitric oxide, which is indispensable for crown root primordia initiation in rice seedlings. Planta 230: 599–610 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Sakihama Y. (2000) Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: in vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett 468: 89–92 [DOI] [PubMed] [Google Scholar]
- Yang Z, Wu Y, Li Y, Ling HQ, Chu C. (2009) OsMT1a, a type 1 metallothionein, plays the pivotal role in zinc homeostasis and drought tolerance in rice. Plant Mol Biol 70: 219–229 [DOI] [PubMed] [Google Scholar]
- Yao DC, Tolan DR, Murray MF, Harris DJ, Darras BT, Geva A, Neufeld EJ. (2004) Hemolytic anemia and severe rhabdomyolysis caused by compound heterozygous mutations of the gene for erythrocyte/muscle isozyme of aldolase, ALDOA(Arg303X/Cys338Tyr). Blood 103: 2401–2403 [DOI] [PubMed] [Google Scholar]
- Zago E, Morsa S, Dat JF, Alard P, Ferrarini A, Inzé D, Delledonne M, Van Breusegem F. (2006) Nitric oxide- and hydrogen peroxide-responsive gene regulation during cell death induction in tobacco. Plant Physiol 141: 404–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaninotto F, La Camera S, Polverari A, Delledonne M. (2006) Cross talk between reactive nitrogen and oxygen species during the hypersensitive disease resistance response. Plant Physiol 141: 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MG, Tian QY, Zhang WH. (2007) Nitric oxide synthase-dependent nitric oxide production is associated with salt tolerance in Arabidopsis. Plant Physiol 144: 206–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Heinlein C, Orendi G, Zentgraf U. (2006) Senescence-specific regulation of catalases in Arabidopsis thaliana (L.) Heynh. Plant Cell Environ 29: 1049–1060 [DOI] [PubMed] [Google Scholar]