Abstract
Plant organ development is important for adaptation to a changing environment. Genetic and physiological studies have revealed that plant hormones play key roles in lateral root formation. In this study, we show that MIZU-KUSSEI1 (MIZ1), which was identified originally as a regulator of hydrotropism, functions as a novel regulator of hormonally mediated lateral root development. Overexpression of MIZ1 (MIZ1OE) in roots resulted in a reduced number of lateral roots being formed; however, this defect could be recovered with the application of auxin. Indole-3-acetic acid quantification analyses showed that free indole-3-acetic acid levels decreased in MIZ1OE roots, which indicates that alteration of auxin level is critical for the inhibition of lateral root formation in MIZ1OE plants. In addition, MIZ1 negatively regulates cytokinin sensitivity on root development. Application of cytokinin strongly induced the localization of MIZ1-green fluorescent protein to lateral root primordia, which suggests that the inhibition of lateral root development by MIZ1 occurs downstream of cytokinin signaling. Surprisingly, miz2, a weak allele of gnom, suppressed developmental defects in MIZ1OE plants. Taken together, these results suggest that MIZ1 plays a role in lateral root development by maintaining auxin levels and that its function requires GNOM activity. These data provide a molecular framework for auxin-dependent organ development in Arabidopsis (Arabidopsis thaliana).
Terrestrial plants are sessile and therefore need to adapt to the surrounding environment. Plants modulate their root system architecture according to soil conditions, adapting to changes such as water deprivation and nutrient starvation. Morphological plasticity allows plants to make the best use of natural resources, even under nonideal conditions. Therefore, the optimization of root system architecture is important for agricultural productivity. The root system contains a primary root, lateral roots, and adventitious roots. Among these, the lateral roots are formed postembryogenically from pericycle cells in the primary root. Plant hormones and/or environmental signals coordinately regulate the development of lateral roots.
The phytohormone auxin controls many aspects of plant development, and several lines of evidence have indicated that it is involved in lateral root development (Fukaki and Tasaka, 2009; Benková and Bielach, 2010). Exogenous auxin promotes cell division in the pericycle, resulting in the production of numerous lateral roots. Genetic analyses of auxin-related mutants have also shown the importance of auxin for lateral root development, which is impaired in mutants that reduce auxin synthesis or responsiveness and increased in those that overproduce auxin (Celenza et al., 1995; Fukaki et al., 2002; Yamada et al., 2009). For lateral roots to develop properly, an auxin gradient needs to be established in lateral root primordia (LRP; Benková et al., 2003), which is dependent upon polar auxin transport via PINFORMED (PIN) auxin efflux carriers. Genetic and pharmacological analyses show that inhibition of polar auxin transport leads to defects in lateral root development (Geldner et al., 2001; Benková et al., 2003).
In contrast to the activities of auxin, the effects of cytokinin on lateral root development remain less well understood. Li et al. (2006) showed that exogenous cytokinin inhibits lateral root formation by arresting cell cycling in the pericycle. In addition, exogenous cytokinin perturbs LRP patterning by interfering with the expression of PIN genes, preventing the formation of an auxin gradient (Laplaze et al., 2007). These findings indicate that cytokinin inhibits lateral root formation via the modulation of PIN activity.
The direction that each root grows is determined by tropic responses; thus, these responses are important factors that shape the root system architecture. Plant roots show tropisms in response to environmental stimuli such as gravity, light, and touch. Since lateral root formation can be initiated by the gravitropism-dependent generation of auxin maxima (De Smet et al., 2007; Ditengou et al., 2008), it is likely that tropic responses directly modulate root system architecture. Roots also exhibit hydrotropism, which is a directional growth response toward high moisture (Takahashi et al., 2009). Although hydrotropism and developed root system architecture contribute toward effective water uptake, the relationship between these factors has not yet been described.
Previously, we identified two genes essential for hydrotropism from genetic analyses of the ahydrotropic mutant mizu-kussei (miz). Positional cloning revealed that MIZ1 encoded a protein of unknown function, whereas MIZ2 encoded GNOM, a GDP/GTP exchange factor for ADP-ribosylation factors (ARF-GEF). MIZ1 comprises 297 amino acids and contains a domain of unknown function (DUF617) that we termed the MIZ domain (Kobayashi et al., 2007). Although DUF617-containing proteins are widely conserved among terrestrial plants, including moss and fern, their molecular functions and physiological characteristics remain unidentified (Kobayashi et al., 2007; Takahashi et al., 2009). GNOM is known as a key regulator for vesicle trafficking, and GNOM-dependent vesicle trafficking is required for establishing the polar localization of PIN proteins on the plasma membrane (Steinmann et al., 1999; Geldner et al., 2003). The coordinated polar localization of PIN1 and PIN2 proteins is impaired in strong alleles of gnom mutants (Steinmann et al., 1999; Kleine-Vehn et al., 2008). As a consequence, embryogenic and postembryogenic auxin gradients are disturbed, causing defects in the creation of an apical-basal axis, tropic responses, and organogenesis, including lateral root development (Steinmann et al., 1999; Geldner et al., 2004). In contrast to the severe phenotypes observed in strong gnom alleles, the miz2 mutant does not exhibit defects in development (Miyazawa et al., 2009b), which suggests that a novel GNOM-dependent vesicle trafficking system regulates the hydrotropic response.
This study demonstrates that, in addition to the regulation of the hydrotropic response, MIZ1 regulates root system architecture via the modulation of auxin and cytokinin action. It also shows that MIZ1-dependent regulation of lateral root development requires GNOM activity.
RESULTS
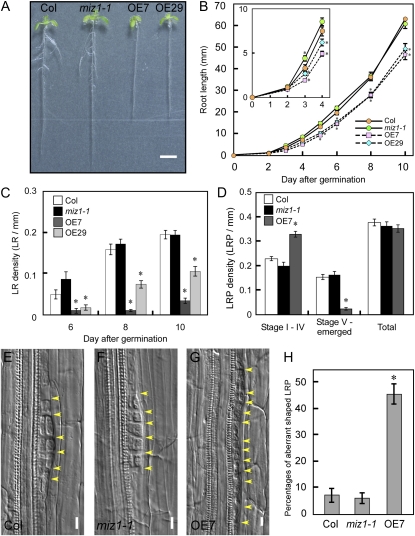
Root Development Is Defective in Plants Overexpressing MIZ1
To gain insight into the functions of MIZ1, we generated transgenic plants expressing MIZ1 cDNA driven by the cauliflower mosaic virus 35S promoter. MIZ1 mRNA accumulation was examined in these transgenic plants, and the lines OE7 and OE29 were chosen for use in this study (Y. Miyazawa, T. Moriwaki, M. Uchida, A. Kobayashi, and H. Takahashi, unpublished data). Although the MIZ1 loss-of-function mutant miz1-1 showed no defect in root system architecture, plants overexpressing MIZ1 (MIZ1OE) exhibited a short-root phenotype and drastically reduced lateral root formation (Fig. 1A). To understand this phenotype in detail, the root development of MIZ1OE plants was analyzed. Following germination, MIZ1OE plants showed slow root growth kinetics, and their lateral root densities were severely reduced (Fig. 1, B and C). To test that the phenotypes of MIZ1OE plants were dependent on MIZ1 mRNA abundance, we investigated the root phenotype of a more mildly overexpressed line, OE4. MIZ1 mRNA levels of OE7, OE29, and OE4 were 345-, 209-, and 44-fold when compared with the wild type, respectively. Root developmental phenotypes of OE4 were mild when compared with those of OE29 (Supplemental Fig. S1). These results suggested the possibility that the more severe defects in lateral root formation observed in MIZ1OE seedlings corresponded to MIZ1 mRNA accumulation.
Figure 1.
MIZ1 overexpression disturbs the development of root system architecture. A, Overview showing 8-d-old seedlings from wild-type (Columbia [Col]), miz1, OE7, and OE29 plants grown on half-strength MS medium under continuous light conditions. Bar = 5 mm. B to D, Phenotypic characterization of wild-type, miz1-1, and MIZ1OE plants. B, Time course of primary root elongation (n = 18–58). The inset shows a magnified image of root growth at 0 to 4 d after germination. C, Lateral root (LR) density (n = 10–20). D, Distribution of lateral root stages of 8-d-old seedlings. The number of primordia at each developmental stage was counted in 10 roots per line. E to G, Stage II LRP division patterns in wild-type (E), miz1 (F), and OE7 (G) plants. Yellow arrowheads indicate cell division. Bars = 10 μm. H, Percentages of aberrantly shaped primordia in stage I and II LRP. Primordia were observed from 10 roots. Asterisks indicate statistically significant differences determined using Student’s t test (P < 0.05). Error bars represent se.
To determine the stages where lateral root development were disrupted by MIZ1 overexpression, the number and the distribution of LRP at stages I to IV and stages V to emerged were determined. Although there were no significant differences in the total number of LRP among wild-type, miz1-1 mutant, and OE7 seedlings, the density of LRP at stages V to emerged was significantly reduced in OE7 (Fig. 1D). In contrast, the density of LRP at stages I to IV was significantly increased in OE7 roots (Fig. 1D). Therefore, our results suggested that MIZ1 overexpression delayed or suppressed LRP development at stages I to IV but that it did not affect lateral root initiation. Detailed morphological analysis revealed abnormal cell divisions in the LRPs of MIZ1OE plants (Fig. 1G). In contrast to wild-type and miz1-1 mutant plants, MIZ1OE plants exhibited ectopic anticlinal divisions in stage II primordia (Fig. 1, E–G) and also stage I primordia (data not shown), and disorganized LRP were observed frequently (Fig. 1H). However, these ectopic divisions were not observed at late developmental stages, including the stage III and stage IV primordia (Supplemental Fig. S2). This result suggested that lateral root formation was stopped at stages I and II in abnormally shaped primordia observed in MIZ1OE. Thus, MIZ1 might regulate an early developmental stage of lateral root formation, since its overexpression affects this phase specifically.
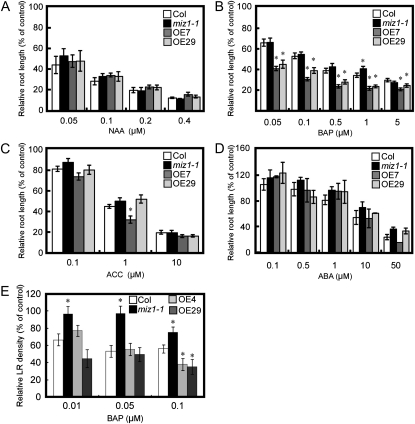
MIZ1 Regulates the Cytokinin Sensitivity of Root Development
Lateral root formation is promoted by auxin but negatively regulated by cytokinin and abscisic acid (ABA). Ethylene affects lateral root formation in a dose-dependent manner. Genetic analyses have shown that many lateral root development mutants exhibit altered hormonal responses and vice versa (Fukaki and Tasaka, 2009). To investigate whether the hormonal response was altered in MIZ1OE plants, the effects of various plant hormones on root growth were measured (Fig. 2). The synthetic cytokinin benzylaminopurine (BAP) caused greater inhibition of root growth in MIZ1OE than in the wild type, irrespective of the concentration used (Fig. 2B). The inhibitory effects of cytokinin were similar in OE7 and OE29. In contrast, miz1-1 showed a slightly resistant phenotype for high concentration of BAP (Fig. 2B). Sensitivity to 1-aminocyclopropane-1-carboxylic acid (ACC), a precursor of ethylene, was enhanced in OE7 but not in OE29 (Fig. 2C). In contrast, responsiveness to the synthetic auxin naphthaleneacetic acid (NAA) and ABA on root growth of MIZ1OE plants was unaltered (Fig. 2, A and D). Therefore, it was suggested that the overexpression of MIZ1 altered the sensitivity to cytokinin on root growth but not that to auxin, ethylene, and ABA.
Figure 2.
Inhibition of wild-type, miz1-1, and MIZ1OE root development by plant growth hormones. A to D, Four-day-old seedlings were transferred to medium containing several concentrations of NAA (A), BAP (B), ACC (C), or ABA (D). Root growth was expressed relative to growth without hormone (n = 15–30). Asterisks indicate statistically significant differences from the wild type (Col) determined using Student’s t test (P < 0.01). E, Lateral root (LR) densities of wild-type, miz1-1, and MIZ1OE seedlings grown on BAP-containing medium. Lateral root densities were counted 6 d after transplantation (n = 23–25). Asterisks indicate statistically significant differences between Col and miz1-1 as determined by Student’s t test (P < 0.05). Error bars represent se.
To analyze the relationship among MIZ1, lateral root formation, and cytokinin, we monitored lateral root densities of wild-type, miz1-1, and MIZ1OE seedlings with or without BAP. Lateral root formation was inhibited by cytokinin in wild-type and MIZ1OE seedlings, and miz1-1 was more insensitive to cytokinin on lateral root formation than the wild type (Fig. 2E). In contrast, lateral root formation of two MIZ1OE lines was significantly inhibited when compared with that of the wild type in the presence of 0.1 μm BAP (Fig. 2E). These results indicated that MIZ1 regulates cytokinin-dependent lateral root formation.
Since MIZ1 was originally identified as a regulator of root hydrotropism, we investigated whether MIZ1 also regulates lateral root formation under osmotic stress conditions. In the presence of 60 mm mannitol, lateral root densities of the wild type and OE29 were reduced (Supplemental Fig. S3). In contrast, miz1 showed a slight osmotic stress-tolerant phenotype on lateral root formation (Supplemental Fig. S3). These results suggested that MIZ1 also regulates the root system under the stressed conditions.
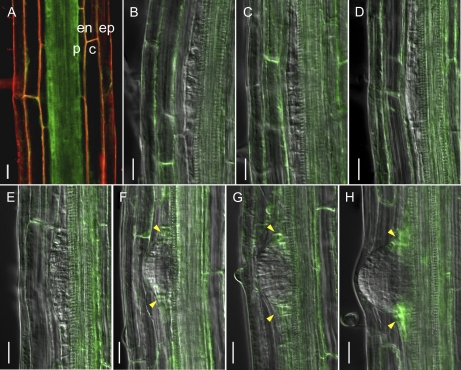
MIZ1-GFP Localizes in Lateral Root Primordia in a Cytokinin-Dependent Manner
To analyze the mechanisms underlying MIZ1-dependent lateral root development, MIZ1 localization patterns were analyzed using GFP-tagged MIZ1. Transgenic miz1-1 mutants expressing wild-type MIZ1-GFP protein under the control of the native MIZ1 promoter exhibited a normal hydrotropic response (Supplemental Fig. S4). This finding suggests that the MIZ1-GFP fusion protein is a functional equivalent for MIZ1. In the mature region of primary roots, MIZ1-GFP fluorescence was observed in all cell layers (i.e. the epidermis, cortex, endodermis, and stele including pericycle; Fig. 3A). Kobayashi et al. (2007) demonstrated that the promoter activity of MIZ1 was detected in root tips and in the root mature zone. At this point, the reasons for these differences are not clear. However, our generated MIZ1-GFP line could complement miz1, and MIZ1-GFP localization patterns were consistent with the MIZ1 expression patterns predicted from root digital in situ analysis based on microarray studies (Birnbaum et al., 2003; Brady et al., 2007; Dinneny et al., 2008). Furthermore, we observed four individual lines expressing MIZ1-GFP, and all of them showed equivalent GFP localization patterns (data not shown). For these reasons, we concluded that the generated MIZ1-GFP protein could be used to visualize the localization patterns of native MIZ1 in root tissues. MIZ1-GFP was not expressed in founder cells or LRP of stages II to IV (Fig. 3, B–E). Since MIZ1-GFP was not detected in founder cells, changes in the MIZ1-GFP localization pattern occurred at lateral root initiation (Fig. 3B). MIZ1-GFP was observed in primordia from stage V onward (Fig. 3, F–H), where it was expressed in the boundary region between the LRP and pericycle cells. Since MIZ1 overexpression inhibited lateral root development, LRP development might be activated by a reduction in MIZ1 at the pericycle.
Figure 3.
Localization patterns of MIZ1-GFP during lateral root development. A, MIZ1-GFP expression in the mature zone of primary roots prior to lateral root initiation. Red signal indicates counterstaining with propidium iodide. c, Cortex; en, endodermis; ep, epidermis; p, pericycle. B to H, Changes in MIZ1-GFP expression during LRP development. B, Founder cell. C, Stage II. D, Stage III. E, Stage IV. F, Stage V. G, Stage VI. H, Stage VII. Yellow arrowheads indicate MIZ1-GFP expression in the peripheral region of LRP. Bars = 20 μm.
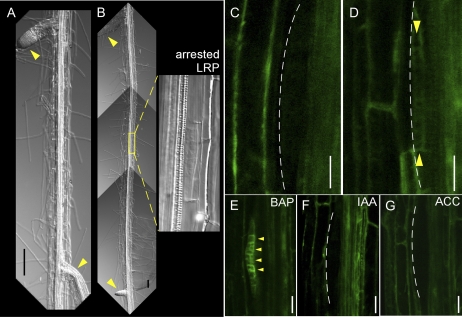
Generally, lateral root spacing is tightly regulated; thus, lateral roots are positioned in a right-to-left pattern (De Smet et al., 2007). However, the occasional arrest of LRP development leads to mispositioning of lateral roots (Fig. 4, A and B). To investigate MIZ1-dependent inhibition of LRP development and arrest, we analyzed MIZ1-GFP localization in LRP with retarded development of lateral roots. Although MIZ1-GFP localization was not observed during stages I to IV of normal LRP (Figs. 3, B–E, and 4C), MIZ1-GFP was detected in LRP arrested at an early stage (Fig. 4D). Since MIZ1 overexpression modulated the cytokinin response (Fig. 2), the effects of plant hormones on MIZ1-GFP localization patterns in LRP were examined. As shown in Figure 4E, cytokinin treatment dramatically induced the MIZ1-GFP signal at stage II of LRP development. This observation was consistent with the prediction that excess MIZ1 could inhibit lateral root development via cytokinin activity. In contrast, MIZ1-GFP localization was not affected by treatment with auxin or ACC (Fig. 4, F and G).
Figure 4.
MIZ1-GFP expression patterns correlate with the arrest of LRP development. A, Representative image of the right-to-left pattern of lateral root formation. B, Representative image of the occasional appearance of arrested LRP in the root mature zone. The right panel shows a magnified image of the area enclosed by a yellow square. Arrowheads indicate normally emerged lateral roots. C and D, MIZ1-GFP expression in stage II LRP. C, Normally developed LRP. D, Arrested LRP. E to G, Hormonal response effects on MIZ1-GFP expression in LRP. Seedlings grown on medium containing 1 μm BAP (E), 1 μm IAA (F), or 10 μm ACC (G) for 24 h are shown. Dotted lines indicate the outline of LRP. Yellow arrowheads indicate MIZ1-GFP expression in LRP. Bars = 100 μm in A and B and 20 μm in C to G.
MIZ1 Overexpression Modulates Auxin Accumulation in Plants
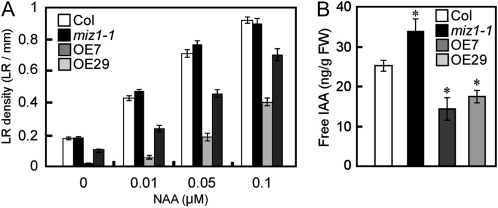
Because auxin plays an essential role in lateral root development, and cytokinin acts antagonistically to auxin in this process, we tested the effect of auxin on the lateral root formation of MIZ1OE roots. In the presence of various concentrations of NAA, lateral root formation of MIZ1OE was clearly recovered by NAA in a dose-dependent manner (Fig. 5A). It is well known that elevated ethylene levels enhance auxin biosynthesis in roots (Stepanova et al., 2005; Swarup et al., 2007). To determine whether ethylene-stimulated auxin biosynthesis could rescue lateral root formation in MIZ1OE seedlings, the plants were grown in the presence of ACC; lateral root formation was not recovered by ACC treatment (Supplemental Fig. S5). Since MIZ1OE and wild-type plants showed similar responses to ACC on root growth, it is unlikely that MIZ1-dependent lateral root development is related to ethylene-mediated alterations in auxin biosynthesis.
Figure 5.
Endogenous IAA levels in roots were modulated by MIZ1. A, Lateral root (LR) densities of wild-type (Col), miz1-1, and MIZ1OE seedlings grown on NAA-containing medium. Lateral root densities were counted 6 d after transplantation (n = 18–44). B, Changes in free IAA content in the roots of wild-type, miz1-1, and MIZ1OE plants. FW, Fresh weight (n = 5). Asterisks indicate statistically significant differences determined using Student’s t test (P < 0.05). Error bars represent se.
Previous work reported that exogenous auxin rescues lateral root development in auxin synthesis mutants (Celenza et al., 1995). Therefore, it is possible that endogenous auxin levels are reduced in the roots of MIZ1OE plants; to test this hypothesis, free indole-3-acetic acid (IAA) levels were measured. The free IAA levels in MIZ1OE roots were significantly lower than in the wild type (Fig. 5B). To confirm these results, activity of the auxin response reporter DR5-GFP was observed. The DR5-GFP signal was significantly weaker in both root tips and LRP of OE7 than the wild type (Supplemental Fig. S6). In contrast, maxima of DR5-GFP were correctly established in OE7 (Supplemental Fig. S6). These data support the observation that MIZ1 overexpression reduces IAA accumulation in Arabidopsis (Arabidopsis thaliana). Furthermore, these results also show that the roots of miz1 plants exhibit a slight but significant elevation in the level of free IAA in comparison with the wild type (Fig. 5B). Thus, it appears that MIZ1 functions as a negative regulator of auxin accumulation in plants.
MIZ1-Dependent Root System Formation Is Independent of PIN-Mediated Polar Auxin Transport Regulation
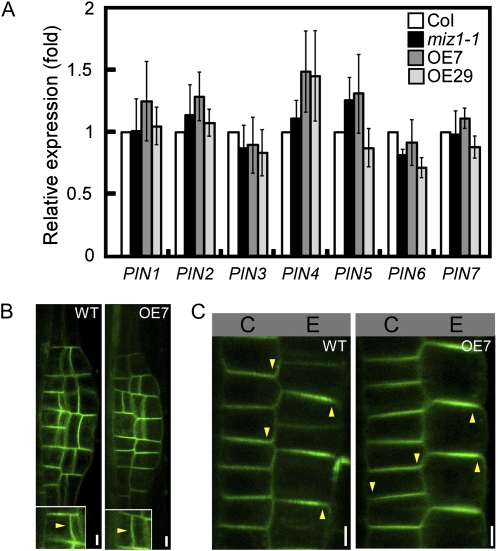
As MIZ1 overexpression impairs auxin accumulation, it is possible that polar auxin transport is also defective in MIZ1OE plants. Polar auxin transport is regulated by the activities of PIN auxin efflux carriers, and defects in the regulation or structure of PIN proteins can lead to the disruption of lateral root formation (Geldner et al., 2001, 2003; Benková et al., 2003). Recent studies show that PIN genes are regulated at both transcriptional and posttranscriptional levels (Vieten et al., 2005; Laplaze et al., 2007). To determine whether MIZ1 overexpression influences the regulation of PIN expression, quantitative analyses of PIN gene transcript levels were performed. MIZ1 overexpression or loss of function did not appear to alter the abundance of any of the PIN mRNAs analyzed (Fig. 6A). In wild-type roots, PIN1 localized to the outer periclinal cell side of the LRP and PIN2 localized to the apical sides of epidermal cells and the basal sides of cortical cells in the meristematic region (Fig. 6, B and C; Kleine-Vehn et al., 2008). Similar polar localization patterns of PIN1 and PIN2 were observed in the roots of MIZ1OE plants (Fig. 6, B and C). These data suggested that MIZ1 overexpression does not affect PIN-mediated auxin transport activity.
Figure 6.
Overexpression of MIZ1 does not affect PIN mRNA abundance or polar localization of PIN proteins. A, Relative mRNA abundance of PIN auxin transporters in whole plants of wild-type (Col), miz1, OE7, and OE29 seedlings as measured by quantitative RT-PCR analysis (n = 3–5). Error bars represent se. Note that no significant differences, determined by Student’s t test, were observed between wild-type and tested lines. B, Intercellular localization of the PIN1-GFP auxin transporter in LRP of wild-type (WT) and OE7 roots. C, Localization of PIN2-GFP in epidermal and cortical cells of wild-type and OE7 roots. Arrowheads indicate polarities of PIN accumulation. C, Cortex; E, epidermis. Bars = 5 μm.
GNOM Activity Is Required for MIZ1-Dependent Root System Formation
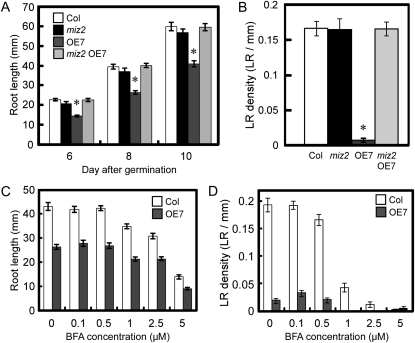
Vesicle trafficking is also important for lateral root development, and GNOM, an ARF-GEF, plays an important role in the formation of these structures (Geldner et al., 2001, 2003, 2004; Sieburth et al., 2006). GNOM regulates lateral root formation by modulating PIN polar targeting (Geldner et al., 2004), and recently, we isolated a weak gnom allele, miz2, which lacks a hydrotropic response but shows unaltered root formation and PIN1 localization (Miyazawa et al., 2009a, 2009b). Thus, miz2 represents an interesting tool for investigating GNOM functions other than the regulation of PIN trafficking. In the miz2 mutant, no significant differences in root length or lateral root number were observed (Fig. 7, A and B). Surprisingly, normal root development was also observed in a miz2 mutant overexpressing MIZ1 (miz2 OE7; Fig. 7, A and B). Reverse transcription (RT)-PCR analysis showed that the miz2 mutation did not affect MIZ1 expression (Supplemental Fig. S7). Observation of MIZ1-GFP in miz2 indicated that localization patterns of MIZ1-GFP in LRP and its cytokinin-dependent induction were not altered by miz2 mutation (Supplemental Fig. S8, A and B). Because GNOM regulates intercellular vesicle trafficking, we compared the intercellular localization of MIZ1-GFP in the wild type with that in miz2. As a result, no significant difference in MIZ1-GFP localization was detected in the miz2 mutant (Supplemental Fig. S8C). Then, we determined endogenous auxin levels in miz2 OE7 plants, since decreased auxin levels lead to the inhibition of lateral root development in MIZ1OE plants. As a result, significant differences in free IAA levels were not observed among the wild type, miz2, and miz2 OE7 (Supplemental Fig. S9). This result clearly demonstrated that GNOM function is required for MIZ1-dependent auxin control.
Figure 7.
gnommiz2 acts as a suppressor for the MIZ1-dependent formation of root system architecture. A and B, The miz2 mutation rescues the root development of MIZ1OE seedlings. A, Primary root length of wild-type (Col), miz2, OE7, and miz2 OE7 seedlings. B, Lateral root (LR) density in 8-d-old-seedlings (n = 31–37). C and D, BFA dose-dependent inhibition of primary root length (C) and lateral root density (D) in 10-d-old wild-type and OE7 seedlings (n = 26–31). Asterisks indicate statistically significant differences from the wild type determined using Student’s t test (P < 0.01). Error bars represent se.
Phenotypic suppression of the defect in lateral root development in MIZ1OE plants may have been caused by a reduction in GEF activity, which is required for the trafficking of PIN proteins. To examine this hypothesis, MIZ1OE plants were treated with brefeldin A (BFA), an inhibitor of GEF activity of ARF-GEF. However, this defect was not rescued by BFA treatment (Fig. 7, C and D), which suggests that the miz2 mutation does not affect GEF activity. GNOM function is required for basal polarization of PIN proteins, and when seedlings are grown on BFA-containing medium, PIN2 localizes at the apical side of the plasma membrane in cortical cells (Kleine-Vehn et al., 2008; Supplemental Fig. S10A). In contrast, PIN2 localization was unaltered in the miz2 mutant (Supplemental Fig. S10B). Previously, we demonstrated the similar polarity of PIN1 localization in the wild type and the miz2 mutant (Miyazawa et al., 2009a). Thus, these results support the idea that miz2 is not defective in GEF activity.
Next, the effects of MIZ1 overexpression on vesicle trafficking were examined in OE7 root cells; however, no significant differences were observed in the internalization of the endocytotic tracer FM4-64 compared with the wild type (Supplemental Fig. S11A). Moreover, BFA treatment induced an aggregation of FM 4-64-labeled endosomes in both the wild type and OE7 (Supplemental Fig. S11B), and no differences were detected in the localization of GNOM-GFP or ARA7-GFP, which are markers of recycling and late endosomes, respectively (Supplemental Fig. S11, C and D). Thus, overexpression of MIZ1 does not interfere directly with endocytotic or endosomal vesicle trafficking activity. Taken together, these observations demonstrated that, in addition to trafficking PIN proteins, GNOM performs novel functions in root development mediated by MIZ1-dependent auxin accumulation.
DISCUSSION
MIZ1 Regulates Local Auxin Accumulation for Lateral Root Development
Numerous studies have shown that auxin plays an intrinsic role in many aspects of plant development. Here, we demonstrate that MIZ1 may be involved in maintaining the auxin abundance required for root branching and elongation by negatively regulating auxin levels. Cellular and tissue auxin levels are adjusted by several factors, including transport and synthesis. However, no alterations in the expression of PIN genes or the localization of PIN proteins were observed in MIZ1 overexpression or loss-of-function plants. Therefore, MIZ1 function is unlikely to be related to PIN-dependent auxin transport.
Auxin is also transported by certain ATP-binding cassette transporters and members of the AUX1/LAX families. Mutants of these auxin transporters show changes to the auxin content of roots and defective lateral root development (Timpte et al., 1995; Santelia et al., 2005). These auxin transporters contain transmembrane domains and localize to the plasma membrane; however, computational predictions and intercellular localization analyses of MIZ1 indicated that MIZ1 does not localize to the plasma membrane (Bennett et al., 1996; Kobayashi et al., 2007; T. Yamazaki, Y. Miyazawa, T. Moriwaki, A. Kobayashi, N. Fujii, and H. Takahashi, unpublished data). Thus, MIZ1 is more likely to act as a regulator of auxin transport than to function directly in transport activities. For example, MIZ1 might regulate the transport of auxin carriers, auxin synthesis, or auxin metabolism. Genetic work has shown that auxin synthesis is an important factor in root development (Stepanova et al., 2005; Yamada et al., 2009). Some auxin synthesis genes are expressed in LRP, and their expression patterns appear to overlap with MIZ1 localization (Ljung et al., 2005). IAA can be inactivated in several ways, and GH3 family proteins are involved in one of the important metabolic pathways. GH3 proteins synthesize amino acid-conjugated IAA, and although they may perform redundant functions, overexpression will cause alterations to IAA-conjugated levels and lateral root formation (Nakazawa et al., 2001; Staswick et al., 2005). These data indicate that both auxin metabolism and auxin synthesis are important for lateral root formation. Therefore, it is possible that MIZ1 regulates auxin accumulation via the modulation of auxin synthesis or metabolism activities. Although the analyses performed in this study did not determine whether MIZ1 functions in the transport or synthesis of auxin, they do suggest that MIZ1 plays a previously unknown regulatory role in the auxin homeostasis required for organ development.
In contrast to the severe phenotype of MIZ1OE plants, a single mutation in MIZ1 did not cause a defect in lateral root development under normal conditions. The Arabidopsis genome contains 11 homologs of MIZ1, and at least three of these genes, including MIZ1, are expressed in the pericycle or LRP (Birnbaum et al., 2003; Brady et al., 2007; Kobayashi et al., 2007). Therefore, MIZ1 and MIZ1-like proteins might function redundantly to regulate auxin and lateral root development. Unfortunately, we could not obtain a series of mutants that lacked MIZ1-like genes. Therefore, genetic analyses such as using RNA interference will provide insight into the functional redundancy of the MIZ1 family in the regulation of auxin levels and lateral root development.
Relationship between MIZ1 Function and Cytokinin Activity in Lateral Root Development
In our study, application of cytokinin induced MIZ1-GFP production in LRP, suggesting that cytokinin signaling regulates MIZ1 expression. Cytokinin functions antagonistically to auxin in plant development, and recent studies have revealed the negative regulation of lateral root development by both exogenous and endogenous cytokinins (Fukaki and Tasaka, 2009). Laplaze et al. (2007) showed that exogenous application of cytokinin leads to abnormal division patterns in the early and late developmental stages of LRP, and multiple mutations of cytokinin receptors increase the number of lateral roots formed (Riefler et al., 2006). Furthermore, we also demonstrated that MIZ1 positively regulates sensitivity to exogenous cytokinin on root development. Although MIZ1OE plants showed cytokinin hypersensitivity, we found that responses to auxin and ABA were unaltered in the roots. A similar phenotype was obtained by overexpression of AHP2, one of the positive regulators for cytokinin signaling (Suzuki et al., 2002). Taken together, these observations suggest that MIZ1 is involved with a cytokinin signal component that is induced by cytokinin and functions physiologically as a regulator for lateral root development.
Exogenous cytokinin has been shown to disrupt cell division in both the early and late phases of LRP development (Laplaze et al., 2007); however, such disorganized cell division was not observed in the stage III to emerged LRP of MIZ1OE plants. In xylem cells, enhancement of cytokinin production leads to disorganized cell division and disrupts LRP development, and these defects cannot be rescued by exogenous auxin (Li et al., 2006; Laplaze et al., 2007). Laplaze et al. (2007) also showed that exogenous application of cytokinin perturbs the expression of PIN genes, which are required for the development of the auxin gradient in LRP. These findings suggest that cytokinin disrupts the PIN-dependent auxin transport required for root development. On the other hand, application of auxin rescues the lateral root formation defect in MIZ1OE plants. Moreover, overexpression of MIZ1 leads to a reduction of auxin accumulation, rather than a modulation of PIN activity. Since elevation of cytokinin decreases the auxin level in roots (Nordström et al., 2004; Růžička et al., 2009), it is possible that MIZ1-mediated cytokinin signaling optimizes the auxin level required for lateral root development. Taken together, it is possible that cytokinins affect lateral root development via two pathways: one involving MIZ1 or MIZ1-like proteins that modulate the auxin levels required for LRP development, and the other by PIN-dependent auxin transport during lateral root formation (Supplemental Fig. S12).
MIZ1 Contributes to Lateral Root Formation under Osmotic Stress Conditions
Our results also indicated that MIZ1 represses lateral root formation under osmotic stress conditions (Supplemental Fig. S3). Public microarray databases show that the expression of MIZ1 is strongly induced by osmotic stress in roots (http://www.arabidopsis.org/portals/expression/microarray/ATGenExpress.jsp). Because the overexpression of MIZ1 negatively regulates lateral root formation, it is thought that MIZ1-dependent inhibition does not work in the miz1 mutant even under drought conditions, and miz1 showed an osmotic stress-tolerant phenotype on lateral root formation. Furthermore, drought stress inhibits auxin accumulation in roots and lateral root formation in rice roots (Chhun et al., 2007). Therefore, it is possible that the osmotic stress-tolerant phenotype of miz1 on lateral root formation is somewhat dependent on its elevated auxin content.
Novel GNOM Function Is Required for MIZ1 Activity
This study demonstrates that the MIZ1 overexpression phenotype can be rescued genetically by the miz2 mutation and that the effects of MIZ1 overexpression require GNOM activity. In vivo and in silico gene expression analyses reveal that both MIZ1 and GNOM are expressed in pericycle cells (Geldner et al., 2004; Miyazawa et al., 2009a; this study). During LRP development, GNOM expresses at the whole region of LRP and MIZ1 localizes at the boundary between LRP and pericycle cells (Geldner et al., 2004; Fig. 3). Because localizations of MIZ1 and GNOM overlap at certain regions of pericycle cells and LRP, it is possible that MIZ1 and GNOM function coordinately in nature. GNOM regulates polar targeting and constitutive endosomal recycling of PIN auxin efflux carriers (Steinmann et al., 1999; Geldner et al., 2003). Polar PIN localization and auxin gradients could not be established in either partial or complete loss-of-function gnom mutants and, thus, failed to complete normal embryogenesis or development of root architectures (Steinmann et al., 1999; Geldner et al., 2004; Fischer et al., 2006; Kleine-Vehn et al., 2008). However, polar localization of PIN proteins was not affected by overexpression of MIZ1 or the miz2 mutation (Fig. 6, A and B; Supplemental Fig. S10). These results indicate that GNOM function is required for MIZ1-dependent lateral root development and that this activity is independent of its regulation of PIN polar targeting. Furthermore, IAA quantification analyses show that the miz2 mutation suppresses the reduction in auxin level caused by MIZ1 overexpression. Thus, it is possible that GNOM regulates auxin accumulation downstream of MIZ1 (Supplemental Fig. S12).
ARF-GEF proteins are responsible for exchanging ARF GDP to GTP, and this GEF activity is required for starting ARF-dependent vesicle formation. In this study, we demonstrated that the lateral root development phenotype of MIZ1OE plants was suppressed by the miz2 mutation but not by BFA treatment. This finding suggests that the miz2 mutation is not related to changes in GEF activity, at least in the case of lateral root development. Moreover, overexpression of MIZ1 did not affect GNOM localization or endosomal vesicle trafficking activity. GNOM interacts with other GNOM proteins via the SEC7-DCB domain, forming GNOM dimers (Anders et al., 2008). Since disruption of dimerization activity abolishes the membrane association of GNOM, severe developmental defects are observed in gnom mutants lacking heterotypic interactions (Anders et al., 2008). In contrast, the miz2 mutants were not defective in organ development, and the site of the miz2 mutation was not located in the GNOM dimerization region (Miyazawa et al., 2009b). Thus, it appears that MIZ1 requires a novel function of GNOM that differs from its dimerization or GEF catalytic activities.
Several lines of evidence show that auxin and cytokinin act as essential factors for the formation of root system architecture. Here, we show that MIZ1 might regulate root systems by modulating the activities of cytokinin and auxin and that MIZ1 was dependent upon GNOM function. These findings provide a novel molecular framework for the formation of the root system.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia, mutants, and transgenic plants were used. The ahydrotropic mutants miz1-1 and miz2 were isolated previously (Kobayashi et al., 2007; Miyazawa et al., 2009b). To generate an overexpression vector, the MIZ1 open reading frame was cloned into pBI121, which placed expression under the control of the 35S promoter. To generate the MIZ1-GFP construct, a fragment of synthetic GFP (sGFP) was fused to the MIZ1 coding sequence. For the GNOM-GFP construct, an sGFP fragment was introduced into the GNOM genomic sequence immediately following the predicted coding sequence (Miyazawa et al., 2009a). These GFP-tagged genes were driven by native promoters.
Plants were transformed using the floral dip method, and T4 or T5 generation transgenic seedlings were used. The ProPIN2:PIN2-GFP and ProARA7:ARA7-GFP lines were kindly provided by Dr. C. Luschnig and Dr. T. Ueda, respectively (Abas et al., 2006; Goh et al., 2007). The ProPIN1:PIN1-GFP and ProDR5:GFP lines were obtained from the Nottingham Arabidopsis Stock Centre. To introduce MIZ1 overexpression into the miz2 mutant, a transgenic line was crossed with the miz2 mutant. Seeds were surface sterilized with a solution containing 5% (v/v) sodium hypochlorite and 0.05% (v/v) Tween 20 and then germinated on half-strength Murashige and Skoog (MS) medium (Sigma Chemicals) containing 0.4% Gellan Gum (Sigma). Upon germination, plates were placed vertically under 23°C and continuous light conditions.
Phenotypic Analyses
To measure root length and the emerged lateral root number, seedlings were scanned on a flatbed scanner. Root length was measured using ImageJ 1.40 software (http://rsb.info.nih.gov/ij/), and the number of emerged lateral roots was determined by counting visible lateral roots on the digital image. The number of LRP was counted using a Nomarski microscope (IX-81; Olympus). Tissues were cleared by direct mounting with choral hydrate. Classification of LRP developmental stages was performed according to Malamy and Benfey (1997).
To determine the sensitivity of root growth or lateral root formation to hormones, 4-d-old seedlings were transferred to fresh half-strength MS medium containing various concentrations of hormones or chemicals. Following the transplantation of seedlings, root tip positions were marked and subsequent root growth was measured after 2 d. To determine the sensitivity to BFA of root growth and lateral root formation, seedlings were germinated and grown on BFA-containing medium.
Confocal Laser Scanning Microscopy
GFP fluorescence was examined using confocal laser scanning microscopy (FV-1000; Olympus) of 5- to 7-d-old seedlings prepared without fixation. To visualize cell wall outlines, seedlings were stained with 10 μm propidium iodide (Molecular Probes). For observation of FM4-64 (Molecular Probes) uptake and the effect of BFA, seedlings were incubated in liquid half-strength MS medium containing 4 μm FM4-64 for the indicated times with or without 50 μm BFA. For hormone and drug treatments, 6-d-old seedlings were transferred to fresh medium containing the appropriate chemicals and grown for 24 h prior to observation. GFP was excited by a 488-nm laser and detected with a band-pass 500- to 550-nm filter. Propidium iodide and FM4-64 were excited by a 543-nm laser and detected with a band-pass 570- to 670-nm filter. GFP fluorescence was quantified as brightness values using the ImageJ software. GFP signals within columella cells including quiescent center cells or the whole region of stage II LRP were enclosed as the region of interest in ImageJ, and averages of brightness values within the region of interest were calculated. Values for background signals were detected from the area of no root in the same image and were subtracted from signal values of GFP. In each experiment, images used for quantification were acquired under the same setting controlled by Fluoview software (Olympus). All GFP images were confirmed so that GFP fluorescence was not saturated by Fluoview software (Olympus). Acquired images were finally assembled in Canvas 11 (ACD Systems of America) and Paint.NET (http://www.paint.net/).
Gene Expression Analysis
RNAs were extracted from 4-d-old seedlings using an RNeasy kit (Qiagen). cDNA was synthesized from 150 to 500 ng of total RNA with the Revertra Ace q-RT-PCR kit (Toyobo). Quantitative PCR was performed using MyiQ (Bio-Rad), and the results were analyzed with iQ5 software (Bio-Rad). Transcript levels were normalized against ACT2 and are expressed as relative values where the transcript level of the wild type was defined as 1.
All PCR experiments were carried out using three to five biological and two to three technical replicates. Semiquantitative RT-PCR was performed as described previously (Kobayashi et al., 2007). MIZ1 and ACT2 transcripts were detected by conventional PCR using 25 and 30 cycles of amplification, respectively. The annealing temperature was 55°C. The following primers were used to quantify gene expression levels: ACT2, 5′-TCAATCTCATCTTCTTCCGC-3′ and 5′-CAATCGTGATGACTTGCCCA-3′; PIN1, 5′-TACTCCGAGACCTTCCAACTACG-3′ and 5′-TCCACCGCCACCACTTCC-3′; PIN2, 5′-GGCGAAGAAAGCAGGAAGA-3′ and 5′-GGTGGGTACGACGGAACA-3′; PIN3, 5′-GAGGGAGAAGGAAGAAAGGGAAAC-3′ and 5′-CTTGGCTTGTAATGTTGGCATCAG-3′; PIN4, 5′-TCATTGCTTGTGGGAACTCT-3′ and 5′-CTGAACGATGGCTATACGGAG-3′; PIN5, 5′-GGAACAAGCTTGACTGTGATGG-3′ and 5′-CGAGCACAGGTAGAGAGACCAA-3′; PIN6, 5′-AGTAAGCATCGGAGGAAGCATAAC-3′ and 5′-CCACGGCGGAGGAGGAAG-3′; PIN7, 5′-ACTCCTCGTCCGTCTAATCTCAC-3′ and 5′-GAAGCCATAGCACAACTCTCCTC-3′; and MIZ1, 5′-TGACTTCTCCGGCGCGTAGC-3′ and 5′-CCAAGCTCCGTTCCACTACC-3′.
Extraction and Determination of Free IAA
IAA purification and measurement were performed as described previously by Nishimura et al. (2006) with the following minor modifications. To extract free IAA from tissues, 6-d-old Arabidopsis roots (10–35 mg fresh weight) were placed in 2.0-mL tubes with 180 μL of cold 80% methanol containing 0.1 mg mL−1 2,6-di-tert-butyl-4-methylphenol and 2.0 to 4.0 ng of [13C6]IAA (Cambridge Isotope Laboratories) as well as a small amount of glass beads. The tubes were sealed and shaken vigorously for 4 min using a Tissue Lyser (Qiagen). After centrifugation at 15,000g for 10 min at 4°C, IAA was purified directly from the supernatant using a HPLC apparatus (LC-10AD, C-R8A; Shimadzu) connected to a fluorescence detector (RF-10A; Shimadzu). Ion-charged HPLC was carried out using a Nucleosil N (CH3)2 column (5 mm i.d. × 100 mm; GL Sciences) at a flow rate of 0.8 mL min−1 with 0.3% acetic acid in methanol. The elution of IAA occurred at approximately 14 min and was monitored at 355 nm following excitation with 280-nm light. From the crude extracts, IAA was quantified with a gas chromatograph-selected ion monitoring-mass spectrometer (QP2010; Shimadzu) coupled to a gas chromatograph (GC-2010; Shimadzu) that was equipped with a capillary column (0.25 mm i.d. × 30 m, film thickness of 0.32 μm; HR-1701; Shinwa Chemical Industries). The injector temperature was set at 250°C, and the flow rate of the helium carrier gas was 1 mL min−1. The column temperature was maintained at 80°C for 1 min and then increased to 280°C at a rate of 30°C min−1. Dried samples were trimethylsilylated with N-methyl-N-trimethylsilyl-trifluoroacetamide at 60°C for 15 min. Ions with mass-to-charge ratios of 202 and 208 (target ions from native IAA and 13C6-labeled IAA internal standard, respectively) and 319 and 325 (qualifier ions and m+ +6 respectively), were monitored.
Sequence data from this article can be found in The Arabidopsis Information Resource (http://www.arabidopsis.org/) under the following accession numbers: MIZ1 (AT2G41660), GNOM (AT1G13980), ACT2 (AT3G18780), PIN1 (AT1G73590), PIN2 (AT5G57090), PIN3 (AT1G70940), PIN4 (AT2G01420), PIN5 (AT5G16530), PIN6 (AT1G77110), PIN7 (AT1G23080), and ARA7 (AT4G19460).
Supplemental Data
The following materials are available in the online version of the article.
Supplemental Figure S1. Root developmental phenotype of weak MIZ1OE seedlings.
Supplemental Figure S2. Morphological analyses of stage III to emerged LRP in the wild type, miz1-1, and OE7.
Supplemental Figure S3. Effect of osmotic stress on lateral root formation in miz1 and MIZ1OE roots.
Supplemental Figure S4. GFP-fused MIZ1 protein complements the root ahydrotropic phenotype of miz1-1.
Supplemental Figure S5. Ethylene treatment did not rescue lateral root formation in MIZ1OE seedlings.
Supplemental Figure S6. DR5-GFP expression in MIZ1OE plants.
Supplemental Figure S7. Semiquantitative RT-PCR analysis of MIZ1 gene expression in the wild type, miz2, OE7, and miz2 OE7.
Supplemental Figure S8. Relationships between MIZ1 localization and GNOM function.
Supplemental Figure S9. Free IAA contents in roots of the wild type, miz2, and miz2 OE7.
Supplemental Figure S10. PIN2-GFP localization in BFA-treated wild-type seedlings and mock-treated miz2 seedlings.
Supplemental Figure S11. Localization of endocytosis and endosome markers in MIZ1OE seedlings.
Supplemental Figure S12. Schematic model of the MIZ1-GNOM-dependent pathway for lateral root development.
Acknowledgments
We thank Dr. Christian Luschnig (University of Natural Resources and Applied Life Sciences, Vienna) and Dr. Takashi Ueda (University of Tokyo) for sharing seeds of the PIN2-GFP and ARA7-GFP lines, and Dr. Yasuo Niwa (University of Shizuoka, Japan) for providing sGFP. We also thank Dr. Tomokazu Koshiba (Tokyo Metropolitan University) for advice on IAA quantification.
References
- Abas L, Benjamins R, Malenica N, Paciorek T, Wiśniewska J, Moulinier-Anzola JC, Sieberer T, Friml J, Luschnig C. (2006) Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat Cell Biol 8: 249–256 [DOI] [PubMed] [Google Scholar]
- Anders N, Nielsen M, Keicher J, Stierhof YD, Furutani M, Tasaka M, Skriver K, Jürgens G. (2008) Membrane association of the Arabidopsis ARF exchange factor GNOM involves interaction of conserved domains. Plant Cell 20: 142–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Bielach A. (2010) Lateral root organogenesis: from cell to organ. Curr Opin Plant Biol 13: 677–683 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Celenza JL, Jr, Grisafi PL, Fink GR. (1995) A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev 9: 2131–2142 [DOI] [PubMed] [Google Scholar]
- Chhun T, Uno Y, Taketa S, Azuma T, Ichii M, Okamoto T, Tsurumi S. (2007) Saturated humidity accelerates lateral root development in rice (Oryza sativa L.) seedlings by increasing phloem-based auxin transport. J Exp Bot 58: 1695–1704 [DOI] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frey NF, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. (2007) Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690 [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Ditengou FA, Teale WD, Kochersperger P, Flittner KA, Kneuper I, van der Graaff E, Nziengui H, Pinosa F, Li X, Nitschke R, et al. (2008) Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 18818–18823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Ikeda Y, Ljung K, Serralbo O, Singh M, Heidstra R, Palme K, Scheres B, Grebe M. (2006) Vectorial information for Arabidopsis planar polarity is mediated by combined AUX1, EIN2, and GNOM activity. Curr Biol 16: 2143–2149 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tasaka M. (2009) Hormone interactions during lateral root formation. Plant Mol Biol 69: 437–449 [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G. (2003) The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jürgens G, Palme K. (2001) Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Geldner N, Richter S, Vieten A, Marquardt S, Torres-Ruiz RA, Mayer U, Jürgens G. (2004) Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 131: 389–400 [DOI] [PubMed] [Google Scholar]
- Goh T, Uchida W, Arakawa S, Ito E, Dainobu T, Ebine K, Takeuchi M, Sato K, Ueda T, Nakano A. (2007) VPS9a, the common activator for two distinct types of Rab5 GTPases, is essential for the development of Arabidopsis thaliana. Plant Cell 19: 3504–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Dhonukshe P, Sauer M, Brewer PB, Wiśniewska J, Paciorek T, Benková E, Friml J. (2008) ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr Biol 18: 526–531 [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Takahashi A, Kakimoto Y, Miyazawa Y, Fujii N, Higashitani A, Takahashi H. (2007) A gene essential for hydrotropism in roots. Proc Natl Acad Sci USA 104: 4724–4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, Maes L, Vanneste S, Swarup R, Weijers D, Calvo V, Parizot B, Herrera-Rodriguez MB, et al. (2007) Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19: 3889–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mo X, Shou H, Wu P. (2006) Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiol 47: 1112–1123 [DOI] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Celenza J, Yamada M, Estelle M, Normanly J, Sandberg G. (2005) Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17: 1090–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Miyazawa Y, Ito Y, Moriwaki T, Kobayashi A, Fujii N, Takahashi T. (2009a) A molecular mechanism unique to hydrotropism in roots. Plant Sci 177: 297–301 [Google Scholar]
- Miyazawa Y, Takahashi A, Kobayashi A, Kaneyasu T, Fujii N, Takahashi H. (2009b) GNOM-mediated vesicular trafficking plays an essential role in hydrotropism of Arabidopsis roots. Plant Physiol 149: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M. (2001) DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J 25: 213–221 [DOI] [PubMed] [Google Scholar]
- Nishimura T, Mori Y, Furukawa T, Kadota A, Koshiba T. (2006) Red light causes a reduction in IAA levels at the apical tip by inhibiting de novo biosynthesis from tryptophan in maize coleoptiles. Planta 224: 1427–1435 [DOI] [PubMed] [Google Scholar]
- Nordström A, Tarkowski P, Tarkowska D, Norbaek R, Åstot C, Dolezal K, Sandberg G. (2004) Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin-cytokinin-regulated development. Proc Natl Acad Sci USA 101: 8039–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T. (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K, Simásková M, Duclercq J, Petrásek J, Zazímalová E, Simon S, Friml J, Van Montagu MC, Benková E. (2009) Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc Natl Acad Sci USA 106: 4284–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santelia D, Vincenzetti V, Azzarello E, Bovet L, Fukao Y, Düchtig P, Mancuso S, Martinoia E, Geisler M. (2005) MDR-like ABC transporter AtPGP4 is involved in auxin-mediated lateral root and root hair development. FEBS Lett 579: 5399–5406 [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Muday GK, King EJ, Benton G, Kim S, Metcalf KE, Meyers L, Seamen E, Van Norman JM. (2006) SCARFACE encodes an ARF-GAP that is required for normal auxin efflux and vein patterning in Arabidopsis. Plant Cell 18: 1396–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, Gälweiler L, Palme K, Jürgens G. (1999) Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286: 316–318 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM. (2005) A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17: 2230–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Ishikawa K, Yamashino T, Mizuno T. (2002) An Arabidopsis histidine-containing phosphotransfer (HPt) factor implicated in phosphorelay signal transduction: overexpression of AHP2 in plants results in hypersensitiveness to cytokinin. Plant Cell Physiol 43: 123–129 [DOI] [PubMed] [Google Scholar]
- Swarup R, Perry P, Hagenbeek D, Van Der Straeten D, Beemster GT, Sandberg G, Bhalerao R, Ljung K, Bennett MJ. (2007) Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19: 2186–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Miyazawa Y, Fujii N. (2009) Hormonal interactions during root tropic growth: hydrotropism versus gravitropism. Plant Mol Biol 69: 489–502 [DOI] [PubMed] [Google Scholar]
- Timpte C, Lincoln C, Pickett FB, Turner J, Estelle M. (1995) The AXR1 and AUX1 genes of Arabidopsis function in separate auxin-response pathways. Plant J 8: 561–569 [DOI] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wiśniewska J, Benková E, Benjamins R, Beeckman T, Luschnig C, Friml J. (2005) Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531 [DOI] [PubMed] [Google Scholar]
- Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M. (2009) The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol 151: 168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]