Abstract
To elucidate the genetic and biochemical regulation of elicitor-induced p-coumaraldehyde accumulation in plants, we undertook a multifaceted approach to characterize the metabolic flux through the phenylpropanoid pathway via the characterization and chemical analysis of the metabolites in the p-coumaryl, coniferyl, and sinapyl alcohol branches of this pathway. Here, we report the identification and characterization of four cinnamyl alcohol dehydrogenases (CADs) from cucumber (Cucumis sativus) with low activity toward p-coumaraldehyde yet exhibiting significant activity toward other phenylpropanoid hydroxycinnamaldehydes. As part of this analysis, we identified and characterized the activity of a hydroxycinnamoyl-coenzyme A:shikimate hydroxycinnamoyl transferase (HCT) capable of utilizing shikimate and p-coumaroyl-coenzyme A to generate p-coumaroyl shikimate. Following pectinase treatment of cucumber, we observed the rapid accumulation of p-coumaraldehyde, likely the result of low aldehyde reductase activity (i.e. alcohol dehydrogenase in the reverse reaction) of CsCAD enzymes on p-coumaraldehyde. In parallel, we noted a concomitant reduction in the activity of CsHCT. Taken together, our findings support the hypothesis that the up-regulation of the phenylpropanoid pathway upon abiotic stress greatly enhances the overall p-coumaryl alcohol branch of the pathway. The data presented here point to a role for CsHCT (as well as, presumably, p-coumarate 3-hydroxylase) as a control point in the regulation of the coniferyl and sinapyl alcohol branches of this pathway. This mechanism represents a potentially evolutionarily conserved process to efficiently and quickly respond to biotic and abiotic stresses in cucurbit plants, resulting in the rapid lignification of affected tissues.
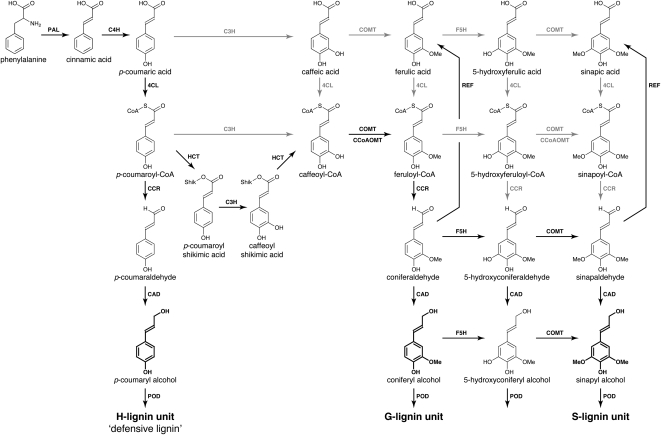
The induction of stress and defense responses in higher plants is regulated by a broad spectrum of genetic and biochemical processes. At the cellular level, activation requires the coordinated activity of key enzymes, the function of which often depends upon the availability of substrates to drive the chemical reactions in which they participate. The lignin biosynthetic pathway (Fig. 1) is one of the major biosynthetic processes in plants and, like the entire phenylpropanoid pathway, responds to stress (Dixon and Paiva, 1995; Naoumkina et al., 2010). In cucumber (Cucumis sativus), for example, both biotic and abiotic stresses elicit the synthesis of a variety of phenolic compounds that have p-rimary functions in the activation of defense signaling (Stange et al., 1999, 2001; Daayf et al., 2000; McNally et al., 2003), such as cell wall lignification (Naoumkina et al., 2010) and the production of antimicrobial compounds (Nicholson and Hammerschmidt, 1992). As part of this general defense response, lignin-related defense responses have been hypothesized to be associated with the enhancement of physical barriers, the decreased sensitivity of plant cell walls to cell wall-degrading enzymes, as well as the production of toxic intermediates produced during lignin biosynthesis (Ride, 1975). In the early 1960s, Hijwegen (1963) reported that pretreatment of cucumber seedlings with phenyl-Ser induces resistance to the scab pathogen Cladosporium cucumerinum. This resistance was associated with the deposition of phloroglucinol-positive materials, which suggested a role for lignification in defense. Further studies reported on the deposition of lignin as part of the systemic acquired resistance of cucumber plants to fungal pathogens (for review, see Hammerschmidt, 2009) as well as part of genetic resistance to C. cucumerinum (Robertsen, 1987; Robertsen and Svalheim, 1990).
Figure 1.
The lignin biosynthetic pathway in cucumber. PAL, Phe ammonia-lyase; 4CL, 4-coumarate:CoA ligase; CCoAOMT, caffeoyl-CoA O-methyltransferase; F5H, ferulate-5-hydroxylase; COMT, caffeate O-methyltransferase; POD, peroxidase. Major known in vivo pathways are shown with the enzymes in black lettering with black arrows. Pathways that may have been demonstrated in vitro or in vivo, but that are not considered to be primary pathways, are shown in gray. (Adapted from Boerjan et al., 2003.)
The chemical nature of the lignin-like material produced by cucumber and other cucurbits following biotic and abiotic stress induction has been studied by a number of approaches. For example, histochemical observations of infected or elicited tissues revealed the deposition of a wall-associated insoluble material positively stained by phloroglucinol-HCl (Hammerschmidt et al., 1984; Robertsen and Svalheim, 1990). Cupric oxide oxidation of cucurbit cell wall components from plants that have been elicited by, or are actively defending against, fungal infection yields increased amounts of simple p-phenols (e.g. p-hydroxybenzoic acid and p-hydroxybenzaldehyde; Hammerschmidt et al., 1985; Robertsen and Svalheim, 1990) as compared with methoxylated analogs (e.g. vanillic acid and vanillin). Similarly, acidolysis of cell walls from squash (Cucurbita maxima) fruit previously elicited with pectinase resulted in the release of p-coumaraldehyde (Stange et al., 2001). These observations suggested that the “lignin” polymer produced during this response was different from that found in, for example, xylem tissues. Indeed, early analysis suggested that lignin could be defined according to its chemical composition, such as defensive lignin (H-lignin) and developmental lignins (S- and G-lignin; for review, see Whetten et al., 1998). The p-hydroxyphenylpropanoid nature of defense-associated lignin in cucurbits was further supported by Stange et al. (1999), who reported the accumulation of free p-coumaraldehyde in polygalacturonase-elicted squash fruit. Taken together, these results suggest that the biosynthesis of lignin precursors in pathogen-challenged cucurbits is directed toward the synthesis of p-coumaryl derivatives rather than the more highly substituted methoxylated analogs. This suggests that pathogen infection, or treatment with elicitors, leads to a shift in phenylpropanoid biosynthesis, favoring 4-hydroxycinnamic acid derivatives. This seems reasonable in the sense that the most expedient monomers for polymerization are those made with the fewest biosynthetic steps: each addition of a methoxyl requires the action of a hydroxylase and an O-methyltransferase (Boerjan et al., 2003).
Here, we demonstrate that changes in the regulation (i.e. gene expression and enzyme activity) of key monolignol biosynthetic enzymes are responsible for the production and accumulation of p-coumaraldehyde. It has long been known that cinnamyl alcohol dehydrogenases (CADs) are responsible for the conversion of p-hydroxycinnamaldehydes to p-hydroxycinnamyl alcohols. In this study, we have identified and characterized four CAD enzymes from cucumber, characterizing their mRNA expression levels and enzymatic activities, both in vitro and in vivo, following elicitation by abiotic stress. First, using an in vitro approach coupling biochemical and genetic analyses, we demonstrate that the four CsCADs have low activity to p-coumaraldehyde. Next, we describe the isolation and functional characterization of a hydroxycinnamoyl-coenzyme A:shikimate hydroxycinnamoyl transferase (HCT) from cucumber responsible for the conversion of p-coumaric acid (via its CoA thioester) and shikimic acid to p-coumaroyl shikimate. Finally, our results of monitoring mRNA transcript and metabolite accumulation in parallel with the enzyme and substrate availability, in vivo and in vitro, suggest that even though CAD activity was induced, the CAD’s low affinity for p-coumaraldehyde, coupled with the concomitant down-regulation of HCT, leads to overall enhancement of the p-coumaryl branch of the phenylpropanoid monolignol pathway and an accumulation of p-coumaraldehyde, which may function as a phytoalexin (Stange et al., 1999). The work described here is, to our knowledge, the first extensive in vivo analysis of the regulation of the phenylpropanoid pathway following abiotic stress elicitation in cucumber.
RESULTS
p-Coumaraldehyde and a Lignin-Like Material Accumulate in Cucumber Hypocotyls upon Pectinase Treatment
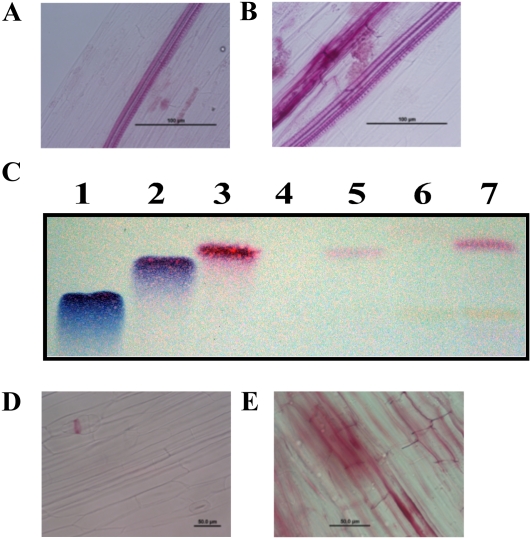
Previous research has demonstrated that the fruits of squash and cucumber hypocotyls rapidly accumulate a phloroglucinol-positive material following pectinase treatment and fungal infection (Hammerschmidt et al., 1984; Robertsen and Svalheim, 1990; Stange et al., 2002). To visualize the accumulation of lignin-like materials in planta, phloroglucinol staining of cucumber hypocotyls following pectinase treatment was performed (Fig. 2). As shown in Figure 2A, staining of cucumber hypocotyls in the absence of pectinase treatment resulted in little or no visible response. In contrast, phloroglucinol staining of hypocotyls following pectinase treatment revealed the rapid accumulation of a lignin-like material (Fig. 2B). To determine if the phloroglucinol-reactive material in pectinase-elicited hypocotyls was composed, at least in part, of p-coumaraldehyde, methanol extractions of the treated and control tissues were performed, followed by analysis using thin-layer chromatography (TLC). As shown in Figure 2C, visualization with phloroglucinol revealed a single (red) band that migrated to an RF value of 0.45, which was identical to that of the p-coumaraldehyde standard (Fig. 2C; lane 3, standard; lane 5, cucumber hypocotyls). As expected, p-coumaraldehyde was not detected (or was weakly present) in untreated hypocotyls (Fig. 2, A and C, lane 4). Next, the methanol extract from cucumber hypocotyls was subjected to mild acid hydrolysis, and using this method, we detected the release of additional amounts of p-coumaraldehyde (Fig. 2C, lane 7). This result suggests that p-coumaraldehyde is either weakly bound or present in different conjugated forms or chemical derivatives. Similar observations were previously made in the fruits of squash (Stange et al., 2001); in this study, we also observed the release of p-coumaraldehyde in cucumber fruits (data not shown), which suggests that this might be a general mechanism in cucurbits.
Figure 2.
Pectinase treatment induces the rapid and tissue-specific induction of p-coumaraldehyde in cucumber hypocotyls. Cucumber hypocotyls were treated with pectinase, and epidermal tissues were stained with phloroglucinol to visualize the production of p-coumaraldehyde. A, Untreated cucumber hypocotyl. B, Pectinase-treated cucumber hypocotyl (48 h). Bars = 100 μm. C, TLC silica plate visualized/developed with phloroglucinol staining. Total extracts were prepared from both wounded (i.e. pectinase) and infected samples and analyzed by TLC. Lane 1, Sinapaldehyde control; lane 2, coniferaldehyde control; lane 3, p-coumaraldehyde control; lane 4, methanol extraction of untreated cucumber hypocotyls; lane 5, methanol extraction of 48-h pectinase-treated cucumber hypocotyl; lane 6, methanol extract of untreated cucumber hypocotyl after mild acid hydrolysis; lane 7, methanol extract of 48-h pectinase-treated cucumber hypocotyl after mild acid hydrolysis. D, Untreated Arabidopsis hypocotyl stained with phloroglucinol. E, Arabidopsis hypocotyl 24 h post treatment stained with phloroglucinol. Bars = 50 μm.
Staining of Arabidopsis (Arabidopsis thaliana) hypocotyls yielded a result similar to that observed in cucumber, with untreated tissues accumulating little or no lignin-like material (Fig. 2D), while pectinase treatment resulted in the visible red staining of the epidermal tissue (Fig. 2E); however, as noted below, Arabidopsis did not accumulate p-coumaraldehyde. We presume in this case that positive phloroglucinol staining is due to the accumulation of lignin-like materials derived from coniferaldehyde and/or sinapaldehyde. Taken together, these data confirm that the accumulation of lignin-like materials is a rapid response in plants associated with the initiation of stress-specific responses but that the accumulation of p-coumaraldehyde is a feature of cucurbits.
p-Coumaraldehyde Accumulation in Cucumber Hypocotyls following Pectinase Treatment Represents an Overall Enhancement of the Phenylpropanoid Pathway
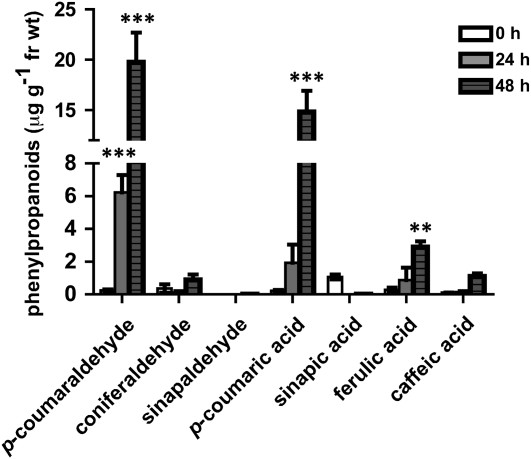
While phloroglucinol staining is a valid method for the visualization of lignin and lignin-like materials in plant tissues (Jensen, 1962), it is for the most part purely qualitative. To this end, p-coumaraldehyde was quantified in planta following pectinase treatment using ultraperformance liquid chromatography coupled to tandem mass spectrometry (UPLC MS/MS). As shown in Figure 3, p-coumaraldehyde content was increased approximately 25-fold at 24 h following treatment with pectinase, and by 48 h, this level was approximately 78-fold over untreated samples. Using these measurements, we calculated the amount of p-coumaraldehyde in planta to be approximately 20 μg g−1 fresh weight. Measurements were also made for additional phenylpropanoids, including p-coumaric acid (more than 67-fold increase), coniferaldehyde, and ferulic and caffeic acids (Fig. 1), each with an approximately 8-fold increase. While attempted, we could not detect sinapaldehyde in cucumber hypocotyls, although the compound could be detected in Arabidopsis hypocotyls (Supplemental Fig. S1). Interestingly, p-coumaraldehyde was not detected in Arabidopsis hypocotyls, suggesting possible differences in phenylpropanoid-based defense regulation between these two plant species.
Figure 3.
Quantification of p-coumaraldehyde, coniferaldehyde, sinapaldehyde, p-coumaric acid, sinapic acid, ferulic acid, and caffeic acid in pectinase-treated hypocotyls of cucumber. Cucumber hypocotyls were treated with pectinase, and the accumulation of major phenylpropanoid metabolites was monitored (μg g−1 fresh weight [fr wt]). Pectinase-treated hypocotyls were collected at 24 and 48 h post treatment. Untreated hypocotyls were used for baseline measurements. Statistical significance was determined using two-way ANOVA followed by Bonferroni post tests, where ** P < 0.01 and *** P < 0.001. Numeric values are listed in Supplemental Table S1.
Four Enzymes from Cucumber Exhibit CAD Activity
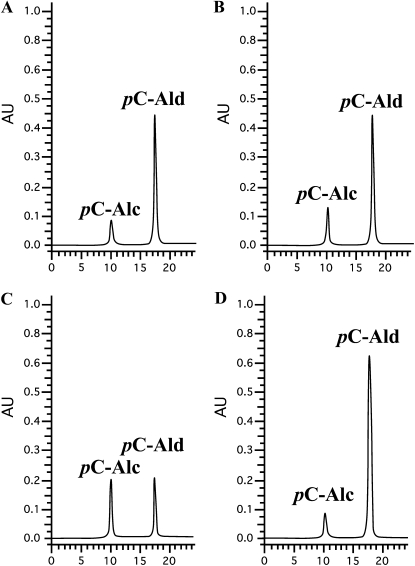
BLAST analysis of the cucumber genome (Huang et al., 2009) identified six genes as having high homology (greater than 50% amino acid sequence similarity) with known CADs from Arabidopsis (Supplemental Fig. S2; Kim et al., 2004). To evaluate the role of the cucumber CADs in the accumulation of p-coumaraldehyde following stress induction, all six CsCAD genes were heterologously expressed in Escherichia coli, and the purified proteins were tested for activity toward all three aforementioned hydroxycinnamaldehydes (i.e. p-coumaraldehyde, coniferaldehyde, and sinapaldehyde) and the related hydroxycinnamyl alcohols. Of the six CsCADs that we identified and isolated, four were experimentally confirmed as being able to convert p-coumaraldehyde to the corresponding alcohol (Fig. 4). Using clustering analysis based on sequence homology of the CsCADs with known, characterized CADs from Arabidopsis, we observed a distinct partitioning of the AtCADs and CsCADs into two distinct clades (Supplemental Fig. S2). In short, this clustering correlates with the functional diversification of the CAD family described previously (Somssich et al., 1996; Kim et al., 2004). For example, CsCAD1 and CsCAD2 cluster with AtCAD4 and AtCAD5, suggestive of a similar, conserved activity in cucumber as compared with Arabidopsis. Likewise, CsCAD3 and CsCAD4 segregated to a second cluster that includes ELI3 from Arabidopsis, a well-characterized CAD associated with plant defense signaling (Somssich et al., 1996). Based on the output of this analysis, as well as the previous observation of the enzymatic properties of ELI3 (Somssich et al., 1996), 2-methoxybenzyl alcohol and its corresponding aldehyde were included as possible CAD substrates.
Figure 4.
Purified recombinant cucumber CAD enzymes are capable of converting p-coumaraldehyde to p-coumaryl alcohol. A, CsCAD1. B, CsCAD2. C, CsCAD3. D, CsCAD4. Data shown are representative of analyses obtained from six independent experiments. Alc, p-Coumaryl alcohol; Ald, p-coumaraldehyde. The x axis shows retention time (min) and the y axis shows absorbance units (AU).
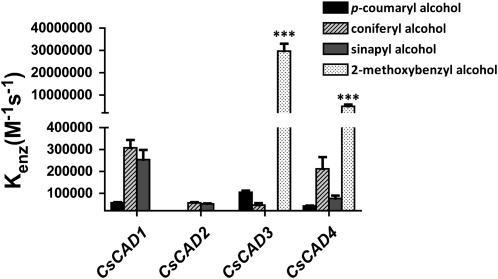
As shown in Figure 5, CsCAD3 and CsCAD4 each has a high specific activity toward 2-methoxybenzaldehyde yet much lower activity toward the hydroxycinnamaldehydes. Moreover, and as predicted based on inference from our phylogenetic analysis, CsCAD1 and CsCAD2 were found to have high specific activities to both sinapyl and coniferyl alcohol. Conversely, both enzymes were found to have significantly lower activity toward p-coumaryl alcohol and no detected activity toward 2-methoxybenzaldehyde/2-methoxybenzyl alcohol (Fig. 5). All four enzymes showed low activity to p-coumaraldehyde as well as toward p-coumaryl alcohol. CsCAD5 and CsCAD6 did not have any detectable activity, as monitored by HPLC (e.g. no reduction of p-coumaraldehyde) and by spectrophotometry (e.g. oxidation of p-coumaryl alcohol to its corresponding aldehyde), with any of the tested substrates (Fig. 5). Based on these results, we conclude that CsCAD5 and CsCAD6 activity is extremely low, below the limits of detection, as was observed with several members of the Arabidopsis CAD family (Kim et al., 2004).
Figure 5.
Four CADs from cucumber exhibit activity to hydroxycinnamyl alcohols. The relative substrate activity of each recombinant CsCAD isoform against p-coumaryl alcohol, coniferyl alcohol, sinapyl alcohol, and 2-methoxybenzyl alcohol is presented as Kenz (m−1 s−1), where Kenz is calculated by dividing Kcat by Km. Statistical significance was determined using two-way ANOVA followed by Bonferroni post tests, where *** P > 0.001 refers to differences between the CsCADs for each substrate.
While we can conclude that CsCAD1 and CsCAD2 proteins are bona fide CADs, they have very different properties in terms of substrate specificities compared with AtCAD4 and AtCAD5 (Fig. 5). For example, AtCAD4 and AtCAD5 were previously shown to have the highest activities, in vitro, toward p-coumaraldehyde, followed by lesser activities to sinapaldehyde and coniferaldehyde (Kim et al., 2004). In contrast, our data demonstrate that the activities of both CsCAD1 and CsCAD2, when compared with CsCAD3 and CsCAD4, although having high specific activities toward hydroxycinnamaldehydes, demonstrated even higher affinity toward 2-methoxybenzaldehyde/2-methoxybenzyl alcohol. This result is similar to that observed for ELI3 (i.e. AtCAD7; Somssich et al., 1996).
To further evaluate the substrate specificities of the purified CsCAD enzymes, the steady-state kinetics of each were analyzed in vitro (Table I). For the majority of the enzymes, the apparent Km values for sinapyl alcohol and coniferyl alcohol were in the range of 1 to 200 μm. CsCAD3 and CsCAD4 had the lowest Km values (2 and 4 μm, respectively) with 2-methoxybenzaldehyde as a substrate, whereas all bona fide CsCADs (i.e. CsCAD1, -2, -3, and -4) had the highest Km values (ranging from 0.5 to 1.2 mm) with p-coumaraldehyde as a substrate. For comparison, AtCAD5 had an apparent Km of 20 μm for p-coumaraldehyde.
Table I. Enzyme kinetics of recombinant CsCAD proteins.
Kenz was calculated by dividing Kcat by Km.
| Enzyme | Substrate (Alcohol) | Km | Kcat | Kenz |
| μm | s−1 | m−1 s−1 | ||
| CsCAD1 | p-Coumaryl alcohol | 889 ± 2.3 | 0.84 | 59,100 |
| Coniferyl alcohol | 16 ± 0.2 | 4.25 | 267,295 | |
| Sinapyl alcohol | 10.7 ± 1 | 2.17 | 202,402 | |
| 2-Methoxybenzyl alcohol | No activity | No activity | No activity | |
| CsCAD2 | p-Coumaryl alcohol | 1,200 ± 8 | 0.00133 | 1.11 |
| Coniferyl alcohol | 56 ± 2.3 | 2.96 | 52,884 | |
| Sinapyl alcohol | 46 ± 0.8 | 2.44 | 53,242 | |
| 2-Methoxybenzyl alcohol | No activity | No activity | No activity | |
| CsCAD3 | p-Coumaryl alcohol | 70.2 ± 5 | 1.38 | 98,809 |
| Coniferyl alcohol | 200 ± 5 | 3.58 | 50,469 | |
| Sinapyl alcohol | 70 ± 0.2 | 7.71 | 9,635 | |
| 2-Methoxybenzyl alcohol | 2.2 ± 0.1 | 61 | 26,707,407 | |
| CsCAD4 | p-Coumaryl alcohol | 489 ± 7 | 44 | 43,500 |
| Coniferyl alcohol | 66 ± 1 | 10 | 151,332 | |
| Sinapyl alcohol | 70 ± 0.5 | 1.28 | 68,996 | |
| 2-Methoxybenzyl alcohol | 3.8 ± 0.1 | 19.2 | 5,030,621 |
Cucumber CAD mRNA Transcripts Are Induced upon Pectinase Treatment
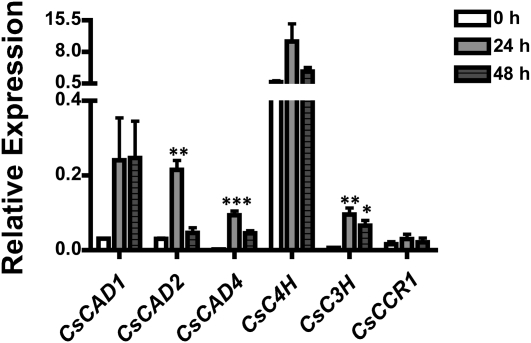
To evaluate changes in gene expression of the key enzymes from the phenylpropanoid pathway involved in lignin formation, real-time PCR (RT-PCR) was performed. In total, we monitored the expression of three cucumber CAD mRNAs (i.e. CsCAD1, -2, and -4), cinnamate 4-hydroxylase (CsC4H), p-coumarate 3-hydroxylase (CsC3H), and cinnamoyl-CoA reductase-1 (CsCCR1) following treatment of cucumber hypocotyls with pectinase. As predicted, the pathway is highly transcriptionally regulated, and many of the genes encoding the key enzymes involved in lignin biosynthesis were induced upon pectinase treatment (Fig. 6). For example, all three CsCAD mRNAs were induced at 24 h post treatment, with CsCAD1 mRNA expression remaining induced above resting levels at 48 h. Similarly, CsC4H and CsC3H were also induced 24 h post treatment, with CsC4H expression at 48 h remaining increased above resting levels. CsCCR1 mRNA was not significantly induced, nor was CsCAD3 (data not shown). Coupled with our biochemical and metabolic analyses shown in Figure 3, our data support the hypothesis that the overall regulation of the phenylpropanoid pathway in cucumber following abiotic stress perception occurs at both the transcriptional and enzymatic levels.
Figure 6.
Pectinase treatment results in the rapid induction of mRNA expression levels of the primary biosynthetic genes in the phenylpropanoid pathway. Cucumber hypocotyls were treated with pectinase, and samples were taken at 24 and 48 h post treatment. RT-PCR was used to measure the mRNA accumulation of CsCAD1, CsCAD2, CsCAD4, CsC4H, CsC3H, and CsCCR1 following treatment. Actin was used as an endogenous control for PCR. Values represent means ± se from three technical replicates of three biological replications. Statistical significance was determined using one-way ANOVA followed by Tukey post tests, where * P < 0.05, ** P < 0.005, and *** P < 0.001.
C4H is considered a key point for entering the monolignol pathway (Fig. 1), and in agreement with the induction of mRNA accumulation in pectinase-treated plants, we also detected increased levels of p-coumaric acid in pectinase-treated hypocotyls of cucumber (Fig. 3). Based on this observation, we hypothesized that CsCAD1, CsCAD2, and, to a lesser extent, CsCAD4 are the primary enzymes responsible for the generation of hydroxycinnamyl alcohol formation in cucumber hypocotyls. In agreement with this hypothesis, CsCAD3 showed higher activity toward p-coumaraldehyde in comparison with the other CsCADs; however, CsCAD3 mRNA was not detected in cucumber hypocotyls, suggesting that this enzyme is likely not expressed in hypocotyls and therefore does not contribute to the accumulation of the hydroxycinnamaldehydes in the epidermis of the hypocotyl. Although an increase in the expression levels of mRNAs of all genes described above was observed, the possibility still exists that this level does not compensate for, nor reflect, the full enzyme capacity required to convert all of the accumulated p-coumaraldehyde into its alcohol.
HCT Enzyme Activity Is Down-Regulated following Pectinase Treatment
BLAST analysis of the cucumber genome did not identify a gene with homology to HCT; however, based on our metabolomics analyses above, we hypothesized that this gene may have been missed in the original assembly of the genome. To this end, degenerate primers were synthesized for the amplification of a cucumber HCT gene based on BLAST searches of the melon EST database (http://www.icugi.org/). In short, an approximately 500-bp fragment from the 5′ end of a candidate CsHCT cDNA was successfully amplified and isolated. To complete the sequence, 3′ RACE was performed, and the full-length clone was confirmed by DNA sequencing (Supplemental Fig. S3; BankIt1454952 CsHCT JN005932).
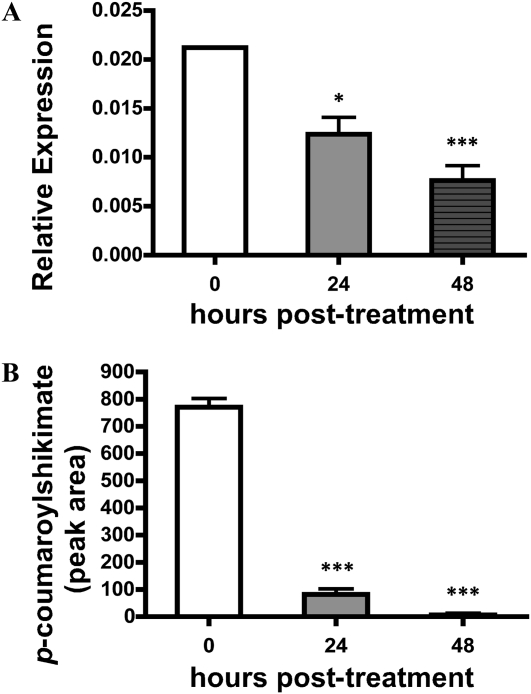
To complement the mRNA expression analyses shown in Figure 6, we next monitored the expression of CsHCT mRNA. As shown in Figure 7A, CsHCT mRNA expression was found to be reduced approximately 2-fold at 24 h post treatment, and a nearly 3-fold reduction was observed at 48 h, compared with the expression level in the untreated control. Similarly, the mRNA expression of the Arabidopsis HCT gene was analyzed (Supplemental Fig. S4), and the overall level of HCT mRNA, compared with that in cucumber, was significantly lower, yet the trend of mRNA reduction in treated versus untreated tissues was the same. Conversion of p-coumaric acid and shikimic acid to p-coumaroyl shikimate by CsHCT was analyzed from total protein extracts of cucumber hypocotyls by UPLC MS/MS (Supplemental Fig. S5). Consistent with our mRNA expression data, a down-regulation (approximately 6-fold) in CsHCT activity 24 h after treatment was observed; by 48 h, CsHCT activity was largely undetectable (Fig. 7B; Supplemental Fig. S5). No p-coumaroyl shikimate was detected in negative control reactions (Supplemental Fig. S6).
Figure 7.
CsHCT activity is reduced following pectinase treatment. A, RT-PCR analysis of the mRNA expression of CsHCT in cucumber hypocotyls following pectinase treatment. Amplification of actin was used as an endogenous control. B, Quantification of p-coumaroylshikimate obtained after a 30-min incubation of total plant protein extract with p-coumaroyl-CoA following pectinase treatment. Statistical significance was determined using one-way ANOVA followed by Tukey post tests, where * P < 0.05 and *** P < 0.001.
Pectinase treatment resulted in a reduction of HCT activity in cucumber hypocotyl extracts. As shown in Figure 7B and Supplemental Figure S5, a down-regulation (approximately 6-fold) of CsHCT was observed 24 h after treatment; by 48 h, the level of CsHCT activity was largely undetectable. Based on this, we hypothesize that CsHCT is one of the key enzymes that regulates changes in the phenylpropanoid pathway in cucumber following abiotic stress induction.
DISCUSSION
In this study, we investigated the biochemical mechanism(s) underlying stress-induced accumulation of p-coumaraldehyde in cucumber. Based on previous observations (Stange et al., 2002), a role for p-coumaraldehyde as a defense compound in cucurbits was proposed, likely responsible for the rapid lignification response observed in wounded tissues. However, that work did not fully investigate the mechanism(s) or activities of the enzymes involved in p-coumaraldehyde accumulation. To this end, we chose to investigate this mechanism using seedlings of cucumber cv Straight 8, which is well characterized for its ectopic accumulation of p-coumaraldehyde at the site of wounding and infection (Robertsen and Svalheim, 1990). As previously demonstrated, p-coumaraldehyde is an important substrate required for the formation of p-coumaryl alcohol, a lignin precursor (Kim et al., 2003). To date, lignin analyses in dicot species have shown that the levels of H-units are always dramatically lower than those of S- and G-subunits and that H-lignin subunits are derived from p-coumaryl alcohol, which itself is derived from p-coumaraldehyde (Boerjan et al., 2003; Ralph and Landucci, 2010). In Arabidopsis, H-type lignin subunits represent less than 2% of the total lignin aromatic nuclei, compared with the G- and S-subunits, which represent the remainder in ratios of approximately 2:1 and 4:1, respectively (Abdulrazzak et al., 2006; Vanholme et al., 2010a, 2010b; Weng et al., 2011). Based on this observation, it is reasonable to hypothesize that the regulation and accumulation of the defense-specific phenylpropanoid pathway metabolites in cucurbits is highly dependent upon the primary substrate, p-coumaraldehyde, and is thus governed by a rate-limiting reaction controlling the accumulation of the alcohol precursor for lignin (Fig. 1).
To study the conversion of p-coumaraldehyde into p-coumaryl alcohol, we chose cucumber as an in vivo model for stress-induced aldehyde accumulation. To simulate pathogen infection and the subsequent induction of stress, we used pectinase treatment as an elicitor of p-coumaraldehyde (Stange et al., 2002) to analyze the biochemical and genetic regulation of monolignol synthesis in plants. As the first step in this analysis, and to monitor the de novo synthesis of free aldehydes, we used phloroglucinol staining to assess the local accumulation of stress-induced p-coumaraldehyde (Fig. 2, A and B). Using this approach, we were able to detect the presence of p-coumaraldehyde and, moreover, quantify these levels in planta using UPLC MS/MS (Fig. 3; Supplemental Table S1). Based on these initial observations, we eliminated substrate availability as the rate-limiting step controlling the formation of H-lignin following pectinase treatment. To this end, we hypothesized that the rate-limiting step in the formation of H-lignin might be the reduction of cinnamaldehydes to alcohols, a reaction catalyzed by CAD.
In Arabidopsis, several functional CAD enzymes have been characterized (Somssich et al., 1996; Kim et al., 2004), and their activities and functions have been demonstrated to drive numerous developmental and stress-associated responses. To determine the roles and subsequent activities of this family of enzymes from cucumber, a candidate gene approach was taken to identify CsCADs. In total, six genes were isolated, all of which showed between 45% and 76% protein sequence identity to CAD enzymes from other plant species (Supplemental Fig. S2; Kim et al., 2004; Sibout et al., 2005). Enzymatically, however, only four of the six cucumber CADs showed activity to hydroxycinnamaldehydes, all of them with low activity, in vitro, to the substrate p-coumaraldehyde (Fig. 4; Table I). These results are in contrast to published in vitro Arabidopsis CAD activities (Kim et al., 2004), which showed that all of the Arabidopsis enzymes have very high specificity for p-coumaraldehyde as well as for sinapaldehyde and coniferaldehyde. Based on our observations here, we conclude that the CAD enzymes in cucumber hypocotyls are restricted in their ability to convert p-coumaraldehyde to p-coumaryl alcohol. This is in agreement with our observations of increased accumulation of p-coumaraldehyde in cucumber hypocotyls upon pectinase treatment.
In this study, we observed that CsCAD1 is more active than CsCAD2, and based on this, we hypothesize that CsCAD1 is largely responsible for the reduction of hydroxycinnamaldehydes to hydroxycinnamyl alcohols, ultimately leading to lignin biosynthesis. Indeed, the two most active cucumber enzymes, CsCAD1 and CsCAD2, are clustered together with AtCAD4 and AtCAD5, the primary CAD enzymes responsible for lignin stem formation in Arabidopsis (Supplemental Fig. S2; Kim et al., 2004; Sibout et al., 2005). CsCAD3 and CsCAD4 have high protein sequence similarity to ELI3 (i.e. AtCAD7), which has been shown to play a role in plant defense (Somssich et al., 1996). Enzymatically, ELI3 exhibits a high specific activity to a number of substrates in vitro, including 2-methoxybenzaldehyde, benzaldehyde, and salicylaldehyde, as well as to monolignols. In this study, and consistent with published ELI3 activity, we observed a high activity for CsCAD3 and CsCAD4 toward 2-methoxybenzaldehyde and less activity to hydroxycinnamaldehydes. Based on this observation, we hypothesize that CsCAD3 and CsCAD4 likely play roles in host defense, similar to that described for ELI3. Taken together, our data strongly support the hypothesis that CsCAD1 and CsCAD2 are the primary biosynthetic enzymes responsible for the conversion of hydroxycinnamaldehydes to hydroxycinnamyl alcohols, which ultimately leads to lignin formation in cucumber hypocotyls. However, we cannot exclude some measure of functional redundancy among all members of the CAD family of proteins in cucumber. For example, redundancy among all members of Arabidopsis CAD proteins has been described, most recently in the study of the cadC/cadD double loss-of-function mutant (Sibout et al., 2005). Interestingly, in the cadC/cadD double mutant, a loss in function of two enzymes appeared to be partially compensated through an increase in the enzymatic activity of the remaining CADs (Tronchet et al., 2010), thus illustrating the complex regulatory and enzymatic activities governing this pathway.
Our work presented here demonstrates that all four characterized cucumber CADs show significantly lower activity to p-coumaraldehyde than that demonstrated for AtCAD4 and AtCAD5 (Fig. 5). Similarly, RT-PCR analysis of most of the phenylpropanoid biosynthetic genes from the p-coumaryl branch reveals that, upon pectinase treatment, mRNA transcripts are strongly up-regulated (Fig. 6). This would suggest de novo biosynthesis of p-coumaryl-type phenylpropanoids, which is in agreement with our metabolic analyses (Fig. 3).
In Arabidopsis, when elicited with pectinase, we could not detect p-coumaraldehyde, suggestive of a divergent phenylpropanoid response to that in cucurbits. However, we could clearly identify high levels of sinapic acid (Supplemental Fig. S1), a substrate for sinapoylmalate, a major defensive compound for many members of the Brassicaceae family (Chapple et al., 1992; Lehfeldt et al., 2000; Franke et al., 2002). Based on this, we hypothesized that an enzyme involved in late monolignol biosynthesis is differentially expressed in cucumber as compared with Arabidopsis. Here, we present the identification and characterization of the cucumber HCT gene, defining both the enzymatic activity and mRNA expression pattern in vivo. In total, our data suggest that CsHCT activity, as well as its mRNA expression level, are significantly reduced (approximately 2-fold for mRNA and approximately 6-fold for enzyme activity; Fig. 7, A and B, respectively) in pectinase-treated tissue. In contrast, in Arabidopsis, HCT activity was not significantly different at 0 and 24 h post pectinase treatment (Supplemental Figs. S4 and S5). Based on these observations, we can conclude that these treatments elicit differing effects on the enzyme activity in cucumber, as compared with other plant species, such as Arabidopsis. Our observations support the hypothesis that HCT is a control point for the synthesis of H-lignin in plants as well as a key point of interaction between otherwise independent monolignol branches (Hoffmann et al., 2004).
In Arabidopsis, Medicago sativa, Nicotiana benthamiana, and Pinus radiata (a softwood), silencing of HCT resulted in an enhancement of the p-coumaryl branch of the phenylpropanoid pathway under normal developmental conditions (Hoffmann et al., 2004; Wagner et al., 2007; Gallego-Giraldo et al., 2011). In cucumber, our data support the possibility that HCT regulation has evolved as a mechanism for the rapid adaptation to biotic and abiotic stress. Through quantifying the enzyme activity in planta, as well as monitoring the genetic regulation (i.e. mRNA accumulation) of the main biosynthetic enzymes in the pathway, our analyses suggest that HCT is likely the primary control point leading to the accumulation of the defensive compound p-coumaraldehyde. Taken together, we posit that p-coumaraldehyde accumulation in elicited cucumber is the result of substrate inhibition and/or low CAD activity. While the fate of p-coumaraldehyde is not known, the simplest hypothesis is that excess p-coumaraldehyde is oxidatively coupled to an insoluble material that is visible after phloroglucinol staining; coniferaldehyde and sinapaldehyde couple reasonably efficiently into syringyl lignins (Kim et al., 2003), and homopolymers of coniferaldehyde have been examined (Higuchi et al., 1994). In summary, our results provide a partial biochemical and molecular explanation for the accumulation of p-coumaraldehyde in cucurbits that are responding to an elicitor. Although how this process is regulated by the plant or induced by pectinase is not known, we have established a framework to further understand pathogenesis-related monolignol biosynthesis in cucurbits and how this stress-associated metabolism differs from developmentally regulated phenolic compound and lignin synthesis.
MATERIALS AND METHODS
Plant Growth and Treatment
Seeds of cucumber (Cucumis sativus ‘Straight 8’) were surface sterilized in 50% bleach, washed thoroughly with sterile water, and grown on wet germination paper for 7 d in the dark at room temperature. Following germination, hypocotyls were separated from the cotyledons and roots, wounded with a razor blade, and sprayed with 3.6 units mL−1 pectinase aqueous solution and 15 mg mL−1 CaCl2 as described by Stange et al. (1999).
Arabidopsis (Arabidopsis thaliana ecotype Columbia) seedlings were grown for 10 d in the dark on nylon mesh on Murashige and Skoog medium containing 10% dextrose following surface sterilization with 50% bleach and 0.05% Tween 20. Pectinase treatment of Arabidopsis hypocotyls was performed as described above.
Chemical Preparation and Aldehyde/Alcohol Synthesis
p-Coumaryl alcohol was prepared from ethyl p-coumarate according to published methods (Quideau and Ralph, 1992), and p-coumaraldehyde was prepared from p-coumaryl alcohol by 2,3-dichloro-5,6-dicyanobenzoquinone oxidation as described previously (Becker et al., 1980).
p-Coumaric acid, sinapic acid, caffeic acid, and the corresponding alcohols and aldehydes (except p-coumaraldehyde and p-coumaryl alcohol) and 2-methoxybenzaldehyde/2-methoxybenzyl alcohol were purchased from Sigma-Aldrich.
Quantification of p-Coumaraldehyde and Other Phenylpropanoids by MS
Approximately 300 mg of hypocotyls from 7-d-old cucumber seedlings was frozen in liquid nitrogen, ground, and extracted with 2 mL of methanol:water (80:20, v/v) containing 0.1% formic acid. Following grinding, the extraction was moved to 4°C for 24 h. After 24 h, homogenates were mixed by inverting and centrifuged at 12,000g for 10 min at 4°C. The resultant supernatants were removed and filtered through a 0.2-μm polytetrafluoroethylene membrane (Millipore) and transferred to autosampler vials. Five microliters was separated on an UPLC BEH C18 column (2.1 × 50 mm, 1.7 μm) attached to an Acquity UPLC system interfaced to a Quattro Premier XE mass spectrometer (Waters). For acid hydrolysis, we followed the procedure described by Stange et al. (2001) with minor modifications. In brief, extractions were performed in 2 mL of 2 n HCl and incubated at 90°C for 3 h. For further extraction, ethyl acetate was used (phenyl acetate was added as an internal standard for the quantification of p-coumaraldehyde). Homogenates were mixed and centrifuged at 12,000g for 10 min at 4°C. The extracts were evaporated at room temperature under N2 gas. Remaining residues were dissolved in 0.3 mL of 70% methanol-water (v/v) and filtered through a 0.2-μm filter. Again, 5 μL was separated on an UPLC BEH C18 column (1.7 μm, 2.1 × 50 mm) attached to an Acquity UPLC system and Quattro Premier XE mass spectrometer (Waters). A gradient of 0.15% aqueous formic acid (solvent A) and methanol (solvent B) was applied over 9 min with a mobile phase flow rate of 0.4 mL min−1. Transitions from [M-H]− to product ions were monitored using negative mode electrospray ionization for p-coumaraldehyde mass-to-charge ratio (m/z) 147 > 119 (precursor ion > product), cone (V) 34, collision (V) 16 for most of the investigated metabolites, dwell time of 0.09 s; transcinnamic acid m/z 147 > 103, cone (V) 28; p-coumaric acid m/z 163 > 119, cone (V) 22; coniferaldehyde m/z 177 > 162, cone (V) 28; caffeic acid m/z 179 > 135, cone (V) 28; ferulic acid m/z 193 > 134, cone (V) 28; sinapaldehyde m/z 207 > 177, cone (V) 28, collision (V) 22; sinapic acid m/z 223 > 164, cone (V) 28; p-coumaroyl shikimate m/z 319 > 119, cone (V) 39, collision (V) 25. Peak areas were integrated, and the analytes were quantified based on calibration curves using 10 different concentrations of the analyte. Data acquisition and processing were performed using Masslynx 4.1 (Waters).
TLC
Samples for TLC were prepared as described above. Extracted samples were loaded onto a silica gel TLC plate (silica gel 60; EMD Chemicals) and developed in hexane:ethyl acetate (1:1) as described by Stange et al. (2001). Aldehydes were visualized by spraying with phloroglucinol (1% phloroglucinol solubilized in ethanol), air dried, and then sprayed with concentrated HCl (Stange et al., 2001).
Heterologous Expression of CAD and HCT Enzymes
For heterologous expression of CADs and HCT, the open reading frame of each CsCAD and the single CsHCT was cloned into Champion pET101/D-TOPO (Invitrogen), yielding a C-terminal His tag fusion construct. His fusions constructs were confirmed by sequencing and transformed into Escherichia coli strain BL21 (DE3) cells. For protein expression, E. coli strains expressing the His fusions were grown at 37°C to an optical density at 600 nm of 0.6. Isopropylthiogalactopyranoside (Sigma) was added to a final concentration of 1 mm, and the cultures were incubated for 16 h at 22°C with shaking at 250 rpm. Induced cells were collected by centrifugation, resuspended in extraction buffer (100 mm Tris, pH 7.0, 0.5 mm phenylmethylsulfonyl fluoride, and 10% [v/v] glycerol), and disrupted by a 4 × 30 s treatment with a sonicator. Cell debris was removed by centrifugation at 14,000g, and the resultant supernatant was collected. The His-tagged CsCAD enzymes were further purified on a nickel agarose column (Qiagen) according to the manufacturer’s instructions.
Protein Extraction and Assay
Plant material (i.e. hypocotyl; 1 g for cucumber, 200 mg for Arabidopsis) was ground in liquid nitrogen and then suspended in 3 mL of extraction buffer containing 100 mm phosphate buffer, pH 7.0, 10% glycerol (v/v) at 4°C, and one tablet of protease inhibitor cocktail (P-9599; Sigma). The extract was centrifuged at 10,000g for 20 min, and the supernatant was desalted on a Sephadex 50 column (GE Healthcare) into fresh extraction buffer (4°C). The sample was concentrated to a final volume of 200 μL and assayed for HCT activity (50 mm phosphate buffer, pH 7.0, 3 mm shikimic acid, 1 mm p-coumaroyl-CoA, and 25 mm ascorbic acid). For a control reaction, water was added instead of shikimic acid (thiolesterase activity) to monitor the release of free p-coumaric acid from p-coumaroyl-CoA. Fifty microliters of the enzyme extract or purified protein from E. coli was added to initiate the reaction. The assay was performed for 30 min and 1 h at 30°C. The enzymatic products were loaded on a 1-mL ENVI-18 solid-phase extraction column equilibrated with 3 × 1 mL of methanol and 3 × 1 mL of 0.1% acetic acid in water, pH adjusted to 2.75 with HCl, as described by Sullivan (2009). After drying, the extract was suspended in 50% methanol-water, and 5 μL was loaded for analysis on an UPLC BEH C18 column (1.7 μm, 2.1 × 50 mm) coupled to an Acquity UPLC system and Quattro Premier XE mass spectrometer (Waters).
Enzyme Characterization
Reduction of Aldehydes
Standard assays contained 100 mm Bis-Tris-propane buffer (pH 6.25; 1 μg of purified CAD in Tris-HCl [20 mm, pH 7.5], 0.4 mm aldehyde as a substrate, and 0.4 mm NADPH) in a total volume of 250 μL. Enzymatic reactions were conducted at 30°C for 4 min. Reactions were stopped by the addition of 10 μL of glacial acetic acid. An aliquot (80 μL) of each assay mixture was loaded on a reverse-phase C18 column (150 × 3.9 mm i.d.; Novapak; Waters) for HPLC analysis (Alliance 2690 HPLC; Waters) with a flow rate of 1 mL min−1. Detection of the substrate and the product was performed at 280 nm using an isocratic solvent system A:B (CH3CN-3% HOAc in water; 8:92) for the first 2 min, followed by a linear A:B gradient from 8:92 to 25:75 between 2 and 20 min. Assays with individual substrates were performed in triplicate or tetraplicate, with controls performed in the absence of NADPH.
Oxidation Reaction of Alcohols
A standard reaction monitored spectroscopically was used for the determination of enzyme kinetics according to the method described by Wyrambik and Grisebach (1975). In brief, the assay contained 100 mm Tris (pH 8.5), 1 μg of protein, 1 mm NADP+, and 15 different concentrations (ranging from 1 to 200 μm up to 1.5 mm, depending on the activity of the purified proteins) of the alcohol substrates The reaction was monitored on a Beckman Coulter spectrometer at 30°C for 4 min at 400 nm. The pH optimum was determined for each CsCAD candidate against each of the four alcohol substrates using spectrometric standard assay conditions (Wyrambik and Grisebach, 1975). For pH optima, incubations were carried out at 30°C with 100 mm MES buffer (pH 5.1–6.8), 100 mm Bis-Tris-propane (pH 6.2–8.0), or 100 mm Tris-HCl (pH 8.5–10) and monitored by UV spectrometry at 400/320 nm. The molar extinction coefficients used for enzyme calculations were as follows: p-coumaraldehyde, 4.8 × 103 m−1 cm−1; coniferaldehyde, 20 × 103 m−1 cm−1; sinapaldehyde, 15.8 × 103 m−1 cm−1 at 400 nm and 6.3 × 103 m−1 cm−1 at pH 6.5, NADPH at 340 nm.
p-Coumaroyl-CoA and Caffeoyl-CoA Biosynthesis
Caffeoyl- and p-coumaroyl-CoA were prepared using recombinant tobacco (Nicotiana tabacum) 4CL1 protein (Beuerle and Pichersky, 2002) and were expressed and purified in E. coli BL21 (DE3). Purification of thioesters was performed as described by Beuerle and Pichersky (2002). The concentration of thioesters was determined spectrophotometrically using extinction coefficients of 21 and 18 mm−1 cm−1 for p-coumaroyl-CoA (λmax = 333 nm) and caffeoyl-CoA (λmax = 346 nm), respectively, as described previously (Stoeckigt and Zenk, 1975).
RNA Isolation and RT-PCR Analysis
Total RNA from hypocotyl epidermal cells was extracted using a modified cetyl-trimethyl-ammonium bromide extraction protocol (Doyle and Doyle, 1987; Kim et al., 2005). RNA was further purified using the RNeasy Mini Kit (Qiagen) and treated with DNA-free (Ambion) to remove contaminating DNA. First-strand cDNA was synthesized from 500 ng of total RNA using SuperScript III reverse transcriptase (Invitrogen). Quantitative RT-PCR was performed on a Mastercycler ep Realplex RT-PCR system using SYBR Green PCR Master Mix (Applied Biosystems). Relative expression was determined using the following equation: relative expression = 2(−ΔCt), where ΔCt (change in critical threshold cycle) = Ctgene of interest – Cthouse keeping gene. The housekeeping genes actin and UBI10 (Tian et al., 2009) were used as endogenous controls for cucumber and Arabidopsis, respectively. All DNA primers used in this study are listed in Supplemental Table S2.
Statistical Analysis
Data were analyzed using GRAPHPAD PRISM software. Values are represented as means ± se. Statistical analysis was performed using one-way ANOVA followed by the Tukey post test or two-way ANOVA followed by the Bonferroni post test. Values of P ≤ 0.05 are considered significant.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number JN005932 (CsHCT).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Measurement of metabolite changes in untreated and pectinase-treated samples.
Supplemental Figure S2. Identification of cucumber CAD enzymes.
Supplemental Figure S3. Sequence and phylogenetic analysis of the HCT amino acid sequences from Arabidopsis, cucumber, and tobacco.
Supplemental Figure S4. The expression of AtHCT mRNA is reduced following treatment with pectinase.
Supplemental Figure S5. CsHCT activity is reduced, over time, following treatment with pectinase.
Supplemental Figure S6. Product identification of CsHCT recombinant protein.
Supplemental Table S1. Quantification of p-coumaraldehyde, coniferaldehyde, sinapaldehyde, p-coumaric acid, sinapic acid, ferulic acid, and caffeic acid in pectinase-treated hypocotyls of cucumber.
Supplemental Table S2. Primers used for quantitative RT-PCR.
Acknowledgments
We acknowledge technical support provided by the Michigan State University Mass Spectrometry Facility. We thank Joseph Leykam, Gregg Howe, Marco Herde, and Elizabeth Savory for instrumentation support and helpful comments and Eran Pichersky for the tobacco 4CL construct. In addition, we thank Saunia Withers and Curtis G. Wilkerson for providing assistance in the synthesis of p-coumaroyl-CoA.
References
- Abdulrazzak N, Pollet B, Ehlting J, Larsen K, Asnaghi C, Ronseau S, Proux C, Erhardt M, Seltzer V, Renou JP, et al. (2006) A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroxylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiol 140: 30–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HD, Bjork A, Adler E. (1980) Quinone dehydrogenation: oxidation of benzylic alcohols with 2,3-dichloro-5,6-dicyanobenzoquinone. J Org Chem 45: 1596–1600 [Google Scholar]
- Beuerle T, Pichersky E. (2002) Enzymatic synthesis and purification of aromatic coenzyme A esters. Anal Biochem 302: 305–312 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Chapple CC, Vogt T, Ellis BE, Somerville CR. (1992) An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4: 1413–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daayf F, Ongena M, Boulanger R, Hadrami IE, Belanger RR. (2000) Induction of phenolic compounds in two cultivars of cucumber by treatment of healthy and powdery mildew-infected plants with extracts of Reynoutria sachalinensis. J Chem Ecol 26: 1579–1593 [Google Scholar]
- Dixon RA, Paiva NL. (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7: 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. (1987) A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull 19: 11–15 [Google Scholar]
- Franke R, Hemm MR, Denault JW, Ruegger MO, Humphreys JM, Chapple C. (2002) Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J 30: 47–59 [DOI] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Jikumaru Y, Kamiya Y, Tang Y, Dixon RA. (2011) Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol 190: 627–639 [DOI] [PubMed] [Google Scholar]
- Hammerschmidt R. (2009) Systemic acquired resistance. Adv Bot Res 51: 173–222 [Google Scholar]
- Hammerschmidt R, Bonnen AM, Bergstrom GC, Baker KK. (1985) Association of epidermal lignification with nonhost resistance of cucurbits to fungi. Can J Bot 63: 2393–2398 [Google Scholar]
- Hammerschmidt R, Lamport DTA, Muldoon EP. (1984) Cell wall hydroxyproline enhancement and lignin deposition as an early event in the resistance of cucumber to Cladosporium cucumerinum. Physiol Plant Pathol 24: 43–47 [Google Scholar]
- Higuchi T, Ito T, Umezawa T, Hibino T, Shibata D. (1994) Red-brown color of lignified tissues of transgenic plants with antisense CAD gene: wine-red lignin from coniferyl aldehyde. J Biotechnol 37: 151–158 [Google Scholar]
- Hijwegen T. (1963) Lignification, a possible mechanism of active resistance against pathogens. Neth J Plant Pathol 69: 314–317 [Google Scholar]
- Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C, Pollet B, Legrand M. (2004) Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16: 1446–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SW, Li RQ, Zhang ZH, Li L, Gu XF, Fan W, Lucas WJ, Wang XW, Xie BY, Ni PX, et al. (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41: 1275–1281 [DOI] [PubMed] [Google Scholar]
- Jensen WA. (1962) Botanical Histochemistry: Principles and Practice. Freeman, San Francisco [Google Scholar]
- Kim H, Ralph J, Lu FC, Ralph SA, Boudet AM, MacKay JJ, Sederoff RR, Ito T, Kawai S, Ohashi H, et al. (2003) NMR analysis of lignins in CAD-deficient plants. Part 1. Incorporation of hydroxycinnamaldehydes and hydroxybenzaldehydes into lignins. Org Biomol Chem 1: 268–281 [DOI] [PubMed] [Google Scholar]
- Kim S, Koh J, Yoo MJ, Kong H, Hu Y, Ma H, Soltis PS, Soltis DE. (2005) Expression of floral MADS-box genes in basal angiosperms: implications for the evolution of floral regulators. Plant J 43: 724–744 [DOI] [PubMed] [Google Scholar]
- Kim SJ, Kim MR, Bedgar DL, Moinuddin SG, Cardenas CL, Davin LB, Kang C, Lewis NG. (2004) Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc Natl Acad Sci USA 101: 1455–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehfeldt C, Shirley AM, Meyer K, Ruegger MO, Cusumano JC, Viitanen PV, Strack D, Chapple C. (2000) Cloning of the SNG1 gene of Arabidopsis reveals a role for a serine carboxypeptidase-like protein as an acyltransferase in secondary metabolism. Plant Cell 12: 1295–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally DJ, Wurms KV, Labbé C, Quideau S, Bélanger RR. (2003) Complex C-glycosyl flavonoid phytoalexins from Cucumis sativus. J Nat Prod 66: 1280–1283 [DOI] [PubMed] [Google Scholar]
- Naoumkina MA, Zhao Q, Gallego-Giraldo L, Dai X, Zhao PX, Dixon RA. (2010) Genome-wide analysis of phenylpropanoid defence pathways. Mol Plant Pathol 11: 829–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson RL, Hammerschmidt R. (1992) Phenolic compounds and their role in disease resistance. Annu Rev Phytopathol 30: 369–389 [Google Scholar]
- Quideau S, Ralph J. (1992) Facile large-scale synthesis of coniferyl, sinapyl, and p-coumaryl alcohol. J Agric Food Chem 40: 1108–1110 [Google Scholar]
- Ralph J, Landucci LL. (2010) NMR of lignins. Heitner C, Dimmel DR, Schmidt JA, , Lignin and Lignans: Advances in Chemistry. CRC Press, Boca Raton, FL, pp 137–234 [Google Scholar]
- Ride JP. (1975) Lignification in wounded wheat leaves in response to fungi and its possible role in resistance. Physiol Plant Pathol 5: 125–134 [Google Scholar]
- Robertsen B. (1987) Endo-polygalacturonase from Cladosporium cucumerinum elicits lignification in cucumber hypocotyls. Physiol Mol Plant Pathol 31: 361–374 [Google Scholar]
- Robertsen B, Svalheim O. (1990) The nature of lignin-like compounds in cucumber hypocotyls induced by alpha-1,4-linked oligogalacturonides. Physiol Plant 79: 512–518 [Google Scholar]
- Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Séguin A. (2005) CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17: 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich IE, Wernert P, Kiedrowski S, Hahlbrock K. (1996) Arabidopsis thaliana defense-related protein ELI3 is an aromatic alcohol:NADP+ oxidoreductase. Proc Natl Acad Sci USA 93: 14199–14203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stange RR, Alessandro R, McCollum TG, Mayer RT. (2002) Studies on the phloroglucinol-HC1 reactive material produced by squash fruit elicited with pectinase: isolation using hydrolytic enzymes and release of p-coumaryl aldehyde by water reflux. Physiol Mol Plant Pathol 60: 283–291 [Google Scholar]
- Stange RR, Jr, Ralph J, Peng JP, Sims JJ, Midland SL, McDonald RE. (2001) Acidolysis and hot water extraction provide new insights into the composition of the induced “lignin-like” material from squash fruit. Phytochemistry 57: 1005–1011 [DOI] [PubMed] [Google Scholar]
- Stange RR, Sims JJ, Midland SL, McDonald RE. (1999) Isolation of a phytoalexin, trans-p-coumaryl aldehyde, from Cucurbita maxima, Cucurbitaceae. Phytochemistry 52: 41–43 [Google Scholar]
- Stoeckigt J, Zenk MH. (1975) Chemical synthesis and properties of hydroxycinnamoyl-coenzyme A derivatives. Z Naturforsch C 30C: 352–358 [DOI] [PubMed] [Google Scholar]
- Sullivan ML. (2009) A novel red clover hydroxycinnamoyl transferase has enzymatic activities consistent with a role in phaselic acid biosynthesis. Plant Physiol 150: 1866–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Chaudhry F, Ruzicka DR, Meagher RB, Staiger CJ, Day B. (2009) Arabidopsis actin-depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB. Plant Physiol 150: 815–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronchet M, Balagué C, Kroj T, Jouanin L, Roby D. (2010) Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol Plant Pathol 11: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. (2010a) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Ralph J, Akiyama T, Lu F, Pazo JR, Kim H, Christensen JH, Van Reusel B, Storme V, De Rycke R, et al. (2010b) Engineering traditional monolignols out of lignin by concomitant up-regulation of F5H1 and down-regulation of COMT in Arabidopsis. Plant J 64: 885–897 [DOI] [PubMed] [Google Scholar]
- Wagner A, Ralph J, Akiyama T, Flint H, Phillips L, Torr KM, Nanayakkara B, Te Kiri L. (2007) Exploring lignification in conifers by silencing hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase in Pinus radiata. Proc Natl Acad Sci USA 104: 11856–11861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J-K, Akiyama T, Ralph J, Chapple C. (2011) Independent recruitment of an O-methyltransferase for syringyl lignin biosynthesis in Selaginella moellendorffii. Plant Cell 23: 2708–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetten RW, MacKay JJ, Sederoff RR. (1998) Recent advances in understanding lignin biosynthesis. Annu Rev Plant Physiol Plant Mol Biol 49: 585–609 [DOI] [PubMed] [Google Scholar]
- Wyrambik D, Grisebach H. (1975) Purification and properties of isoenzymes of cinnamyl-alcohol dehydrogenase from soybean-cell-suspension cultures. Eur J Biochem 59: 9–15 [DOI] [PubMed] [Google Scholar]