Abstract
The CONSTITUTIVE EXPRESSOR OF PATHOGENESIS-RELATED GENES5 (CPR5) gene of Arabidopsis (Arabidopsis thaliana) encodes a putative membrane protein of unknown biochemical function and displays highly pleiotropic functions, particularly in pathogen responses, cell proliferation, cell expansion, and cell death. Here, we demonstrate a link between CPR5 and the GLABRA1 ENHANCER BINDING PROTEIN (GeBP) family of transcription factors. We investigated the primary role of the GeBP/GeBP-like (GPL) genes using transcriptomic analysis of the quadruple gebp gpl1,2,3 mutant and one overexpressing line that displays several cpr5-like phenotypes including dwarfism, spontaneous necrotic lesions, and increased pathogen resistance. We found that GeBP/GPLs regulate a set of genes that represents a subset of the CPR5 pathway. This subset includes genes involved in response to stress as well as cell wall metabolism. Analysis of the quintuple gebp gpl1,2,3 cpr5 mutant indicates that GeBP/GPLs are involved in the control of cell expansion in a CPR5-dependent manner but not in the control of cell proliferation. In addition, to our knowledge, we provide the first evidence that the CPR5 protein is localized in the nucleus of plant cells and that a truncated version of the protein with no transmembrane domain can trigger cpr5-like processes when fused to the VP16 constitutive transcriptional activation domain. Our results provide clues on how CPR5 and GeBP/GPLs play opposite roles in the control of cell expansion and suggest that the CPR5 protein is involved in transcription.
Pleiotropic genes participate in many seemingly unrelated traits and play a key role in fundamental aspects of life such as evolution, development, and aging. In Arabidopsis (Arabidopsis thaliana), the CONSTITUTIVE EXPRESSOR OF PATHOGENESIS-RELATED GENES5 (CPR5; CPR5/HYS1/OLD1) gene is involved in highly pleiotropic developmental processes. Although initially identified based on their constitutive pathogen response phenotype, mutants of CPR5 possess striking phenotypes including (1) enhanced constitutive expression of pathogen-related (PR) genes associated with increased pathogen resistance (Bowling et al., 1994; Boch et al., 1998); (2) defects in cell division, expansion, endoreduplication, and cell wall biogenesis associated with a reduced stature (Kirik et al., 2001; Brininstool et al., 2008); (3) spontaneous lesions mimicking cell death and accelerated leaf senescence (Jing et al., 2002, 2007; Yoshida et al., 2002); and (4) modified hormonal and metabolic responses in the salicylic acid, jasmonic acid, abscisic acid, ethylene, and sugar-sensing pathways (Clarke et al., 2000; Aki et al., 2007; Jing and Dijkwel, 2008). Thus CPR5 appears to play a broad role in plant growth and development.
Genetic analysis has revealed that CPR5 independently controls many of these developmental processes, as blocking a particular signaling pathway does not affect the alterations in other signaling pathways in cpr5 mutants (Bowling et al., 1997; Aki et al., 2007; Jing et al., 2008). This suggests that CPR5 is a key factor that regulates the activity of distinct subpathways, modules, or subsets (Brininstool et al., 2008; Jing and Dijkwel, 2008). In the well-known pathogen-response pathway, CPR5 appears to act just downstream of pathogen recognition and displays both NON EXPRESSOR OF PR GENES1 (NPR1)-dependent and NPR1-independent disease resistance (Bowling et al., 1997) while it regulates PR gene expression in the RPS2-mediated pathway (Boch et al., 1998). One specific visible phenotype of cpr5 mutants not found in the phenotype of any of the other constitutive pathogen response mutants is abnormal trichome cell development (Kirik et al., 2001; Brininstool et al., 2008). While most trichome cells on wild-type Arabidopsis leaves display multiple branches, cpr5 leaves exhibit both reduced trichome branching and reduced trichome number. This phenotype is reminiscent of loss-of-function mutant phenotypes in genes involved in trichome initiation such as GLABRA1 (GL1) or GL3. Genetic studies suggest that at least some of these trichome regulators are dependent on CPR5 function (Brininstool et al., 2008). This is consistent with the recently described role of GL1 in systemic acquired resistance in defense to both bacterial and fungal pathogens, a role that might be due to the regulation of cuticle development (Xia et al., 2010).
The CPR5 protein is well conserved throughout the plant kingdom and is predicted to be a type IIIa membrane protein (Kirik et al., 2001). A well-defined bipartite nuclear localization signal (NLS) in the nonmembrane region predicts nuclear targeting. Although its intracellular localization and mode of action are unknown, several authors have proposed that CPR5 might be anchored in a membrane, such as that of the endoplasmic reticulum or the inner nuclear envelope, and might function upon proteolytic cleavage as a transcription (co)factor (Kirik et al., 2001; Brininstool et al., 2008).
We have previously identified a family of transcription factors whose founding member, GL1 ENHANCER BINDING PROTEIN (GeBP), is a putative regulator of GL1. The four GeBP/GeBP-like (GPL) genes encode unconventional Leu-zipper transcription factors (Curaba et al., 2003; Chevalier et al., 2008) and redundant roles of GeBP/GPLs in the indirect regulation of some cytokinin response genes have been shown (Chevalier et al., 2008). However, GeBP/GPLs primary role remains unknown. We show here that the GeBP/GPL family fulfills a subset of CPR5 functions. In this study we performed a transcriptomic profiling of the quadruple gebp gpl1,2,3 mutant and one overexpressing line and found that GeBP/GPLs control genes involved in defense responses and cell wall metabolism that overlap with a subset of CPR5-regulated genes. Our genetic analysis demonstrates that GeBP/GPL genes play a repressive role in cell expansion by counteracting the positive role of CPR5 in this process. In contrast to CPR5, GeBP/GPLs do not regulate cell proliferation or endoreduplication. Finally, we show that CPR5 is a nuclear protein and that its putative nucleosolic domain alone is sufficient for transcriptional regulatory responses. These results demonstrate that GeBP/GPL genes have a role in CPR5-dependent processes, such as stress and cell expansion, and suggest that CPR5 may directly participate in transcription through a proteolytic activation mechanism.
RESULTS
GeBP/GPL Downstream Genes Represent a Subset of CPR5-Regulated Genes
We have previously shown that GeBP/GPL genes play a redundant role in the indirect regulation of the cytokinin negative feedback loop (Chevalier et al., 2008). However, their primary function was unknown. To assess the function of GeBP/GPL genes in Arabidopsis development, we constructed the quadruple gebp gpl1,2,3 mutant and performed a transcriptomic analysis, which we compared to that of wild-type Arabidopsis. A transgenic line overexpressing a version of GPL2 with constitutive transcriptional activation activity (VP16:GPL2 line; Chevalier et al., 2008) was also included in the analysis. Three-week-old rosettes were used to prepare RNAs, a developmental stage at which all GeBP/GPL genes are expressed. At this stage, the gebp gpl1,2,3 mutant showed no visible phenotype but the VP16:GPL2 line showed retarded growth, early senescence, and necrotic lesion phenotypes (see below). The CATMA v2 arrays, which represent 24,576 gene sequence tags, were used to hybridize three independent biological replicates for each genotype and data were treated as previously described (Gagnot et al., 2008). In the quadruple gebp gpl1,2,3 mutant, the transcripts of 88 genes were reproducibly misregulated (37 up- and 51 down-regulated genes), while in the VP16:GPL2 line, the transcripts of 332 genes were affected (251 up- and 81 down-regulated genes; Supplemental Tables S1 and S2). The datasets were mined with the software tools Genevestigator (Calikowski et al., 2003), Mapman (Thimm et al., 2004; Rotter et al., 2007), and EasyGO/AgriGO (Ashburner et al., 2000) to search for specific correlations. These analyses indicated an enrichment of genes involved in response to stress (10.39-fold; hypergeometric test: P value = 0.0) and cell wall metabolism (26.5-fold; P value < 5.6 10−9) pathways (Fig. 1A) and revealed, more specifically, that transcriptomic alterations in both gebp gpl1,2,3 and VP16:GPL2 plants largely overlapped with one specific set of publicly available microarray data (Fig. 1B). This data set is from a series of mutants that all share a mutation in the CPR5 gene (NASCARRAYS-355 performed by Dr. Yang). Among the 409 genes misregulated both in gebp gpl1,2,3 and VP16:GPL2, 20% were included in cpr5 transcriptomic data (P value < 1.0 10−19), indicating that a substantial fraction of GeBP/GPL-dependent genes are found in the CPR5 pathway. Transcriptomic alterations induced by multiple abiotic stresses (Zeller et al., 2009) showed no overlap with gpl1,2,3 and a limited overlap with VP16:GPL2 data sets, suggesting that the GeBP/GPL pathway is closer to CPR5 than to a more general stress-related pathway (Supplemental Fig. S1). Genes that were down-regulated in the gebp gpl1,2,3 mutant were often up-regulated in cpr5 plants (Fig. 1B; Supplemental Fig. S1). This negative correlation was also true for genes that were up-regulated in the gebp gpl1,2,3 mutant, suggesting that GeBP/GPL and CPR5 genes are involved, directly or indirectly, in opposite regulation of the same downstream genes.
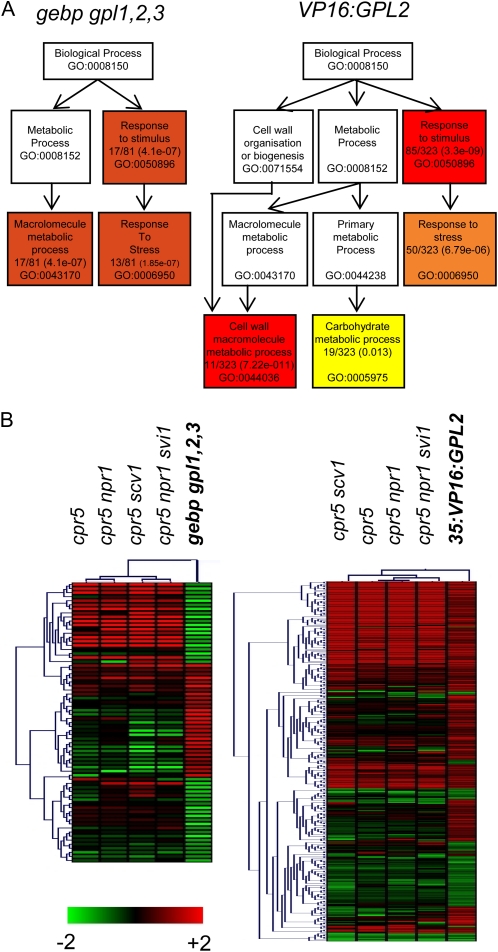
Figure 1.
Gene ontology of gebp gpl1,2,3 and VP16:GPL2 transcriptomic data and transcriptomic similarities with the cpr5 mutant series. A, Gene ontology of gebp gpl1,2,3 (left) and VP16:GPL2 (right) transcriptomic data. Distribution of gene sets among functional biological pathways using singular enrichment analysis of AgriGO are shown. Colors are as in AgriGO, and only the most significant pathways are shown. The ratio of genes involved in each pathway and P values are indicated within boxes together with the gene ontology accession number. The highest probabilities are for the stimuli/stress response (gebp gpl1,2,3 and VP16:GPL2) and cell wall process (VP16:GPL2) pathways. Several entries were not associated to GO terms. Hence 81 genes instead of 88 were used in this analysis for the gebp gpl1,2,3 data and 323 genes instead of 332 for the VP16:GPL2 data. B, Hierarchical clusterings of genes misregulated in the gebp gpl1,2,3 (left) or VP16:GPL2 (right) and cpr5 mutant series. Graphics were generated using Genevestigator and MultiExperiment viewer software.
The overlap between transcriptomic alterations in gebp gpl1,2,3 and cpr5 mutants suggests that GeBP/GPL transcription factors control a subset of CPR5-regulated genes.
GeBP/GPL Genes Are Involved in Several, But Not All, CPR5-Like Pathways
In addition to their substantial overlapping transcriptomic profiles, strong phenotypic similarities were observed between cpr5-2 and VP16:GPL2 plants. As shown in Figure 2A, both types of plant displayed a reduced stature relative to the wild type, together with spontaneous necrotic lesions on cotyledons and leaves. A similar phenotype was observed in transgenic lines that overexpress a stable form of GeBP fused to the VP16 domain (Supplemental Fig. S2). Interestingly, while cpr5 mutant plants made smaller trichomes (Kirik et al., 2001), trichomes of VP16:GPL2 plants were indistinguishable from those of the wild type (Fig. 2A). Trichomes of wild-type Arabidopsis have an average DNA content of 32C due to endoreduplication, a cell cycle bypass that permits replication of the DNA without mitosis (Sugimoto-Shirasu and Roberts, 2003; John and Qi, 2008). We measured the DNA levels in the trichomes of wild-type, cpr5-2, and VP16:GPL2 plants (Supplemental Fig. S3). Relative to the wild type, trichomes of cpr5 had a lower DNA level as previously reported (Kirik et al., 2001), while trichomes of VP16:GPL2 plants reached a DNA level similar to the wild type. This indicates that, in contrast to CPR5, GeBP/GPL genes are not involved in the control of endoreduplication, at least in trichomes.
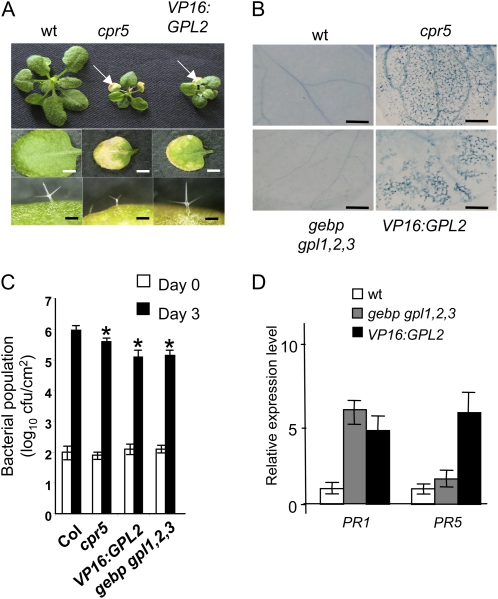
Figure 2.
Phenotypic similarities between VP16:GPL2 and cpr5-2 mutants. A, Top row, rosettes of 3-week-old wild-type, cpr5-2, and VP16:GPL2 plants grown in soil. White arrows indicate early senescing leaves. Middle row, individual third leaves of 4-week-old plants. Control lines expressing the VP16 domain alone showed no visible phenotypes as previously described (Chevalier et al., 2008). Scale bars: 1 mm. Bottom row, sizes of trichomes. Scale bars: 100 μm. B, Trypan blue staining of wild-type, cpr5-2, gebp gpl1,2,3, and VP16:GPL2 leaves. Scale bars: 500 μm. C, Bacterial populations in wild-type, cpr5-2, VP16:GPL2, and gebp gpl1,2,3 plants. Inoculations with Pst DC3000 strain were performed on leaves without lesions with a bacterial suspension at 2 × 105 cfu mL−1. Bacterial populations were measured at 0 (white bars) and 3 d (dark bars) postinoculation. Mean bacterial densities are shown (three to five replicates with corresponding sds) for one representative experiment from two or three independent experiments. Asterisks denote significantly different values from bacterial number in the wild type according to the Student’s t test (P ≤ 0.05). D, Transcript levels of PR1 and PR5 genes in gebp gpl1,2,3 and VP16:GPL2 relative to the wild type. Real-time RT-PCR was performed on 3-week-old rosettes. [See online article for color version of this figure.]
A common consequence of defense responses to infection or stimuli is the hypersensitive response (HR), a local and rapid cell death that helps halt the spread of pathogens. Necrotic lesions in VP16:GPL2 plants were similar to those observed in the cpr5 mutant, mimicking HR in the absence of pathogen. To determine whether the ectopic immune response suggested by the gene expression profiles correlated with HR-like cell death, we first stained leaves of VP16:GPL2, cpr5, and quadruple gebp gpl1,2,3 plants with Trypan blue, an indicator of dead cells. Leaves of VP16:GPL2 and cpr5 showed staining, indicating spontaneous cell death, while the quadruple gebp gpl1,2,3 mutant was indistinguishable from the wild type (Fig. 2B).
To confirm unambiguously a role for GeBP/GPL genes in immune responses, we assayed the response of the gebp gpl1,2,3 mutant and the VP16:GPL2 line to the pathogen Pseudomonas syringae pv tomato (Pst) DC3000. Plant resistance was evaluated by the measurement of in planta bacterial growth 3 d after leaf infiltration. As shown in Figure 2C, in planta pathogen growth was significantly decreased in VP16:GPL2 as well as cpr5 relative to wild-type plants. The gebp gpl1,2,3 mutant also showed a similar phenotype (Fig. 2C). This suggests that pathogen response pathways were activated in both gebp gpl1,2,3 and VP16:GPL2 plants more strongly than in the wild type. Transcript levels of the pathogen response marker genes PR1 and PR5 were measured using real-time reverse transcription (RT)-PCR and were indeed increased over the wild-type levels in both types of plant (Fig. 2D). The observation that PR transcript levels are increased in the quadruple mutant suggests that GeBP/GPLs are repressors of PR gene expression. An up-regulation of PR genes in both gebp gpl1,2,3 and in VP16:GPL2 plants is not surprising since GPL2 can act as a transcriptional activator in the presence of the VP16 domain. Therefore the VP16:GPL2 fusion should behave as a dominant negative gain-of-function allele on GPL2 negatively regulated genes such as PR genes, a situation already described for LEAFY:VP16 and its target gene TERMINAL FLOWER1 (Parcy et al., 2002).
Taken together, these data indicate that the GeBP/GPL genes are likely involved in cell death and defense pathways but not in trichome development or endoreduplication, in contrast to CPR5.
Epistatic Relationship between GeBP/GPL and CPR5 Pathways
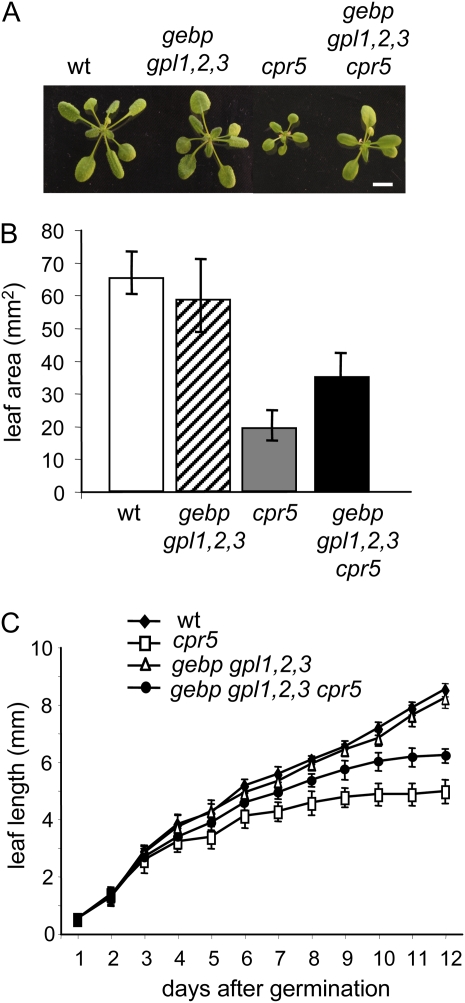
To analyze the role of GeBP/GPLs within the CPR5 pathway, we generated the gebp gpl1,2,3 cpr5 quintuple mutant and looked for epistatic relationships. As shown in Figure 3A, the growth defect of the cpr5 mutant was suppressed, at least partially, in the gebp gpl1,2,3 cpr5 mutant, indicating that gebp/gpl mutations are epistatic to the cpr5 mutation. Leaf area in the gebp gpl1,2,3 cpr5 mutant was 62% larger, on average, than the cpr5 leaf area (Fig. 3B). This leaf growth increase was mainly caused by a suppression of the cpr5 premature growth arrest phenotype (Kirik et al., 2001), as leaf elongation of the gebp gpl1,2,3 cpr5 quintuple mutant continued after the cpr5 leaf elongation stopped (Fig. 3C).
Figure 3.
Vegetative growth of wild-type, cpr5-2, and gebp gpl1,2,3 cpr5 mutants. A, Rosettes of wild type, gebp gpl1,2,3 quadruple mutant, cpr5-2, and gebp gpl1,2,3 cpr5 quintuple mutant grown in soil for 3 weeks. Scale bar: 3 mm. B, Leaf area of the wild type, gebp gpl1,2,3 quadruple mutant, cpr5-2, and gebp gpl1,2,3 cpr5 quintuple mutant. C, Leaf elongation rate in wild type, cpr5-2, gebp/gpl quadruple mutant, and gebp/gpl cpr5-2 quintuple mutant. Plants were grown in soil, and measurements of the third leaf were taken at daily intervals. Initial growth rates were similar in all types of plant. [See online article for color version of this figure.]
To determine whether the suppression of the cpr5 growth arrest phenotype by gebp/gpl mutations was due to the restoration of cell proliferation, we first analyzed the sensitivity of the wild type and mutants to the DNA replication-blocking agent aphidicolin, an inhibitor of early S phase (Fig. 4A). When grown in the presence of aphidicolin, wild-type plants showed a slight reduction of growth. While the quadruple gebp gpl1,2,3 mutant displayed a similar degree of sensitivity, the growth of cpr5 plants was completely inhibited, providing experimental indication for a cpr5 defect in the cell cycle or cell proliferation, although we cannot exclude that aphidicolin affects additional replication-independent processes. The growth of the gebp gpl1,2,3 cpr5 quintuple mutant was similar to that of the cpr5 single mutant, suggesting that gebp/gpl mutations do not restore cell proliferation in the gebp gpl1,2,3 cpr5 quintuple mutant.
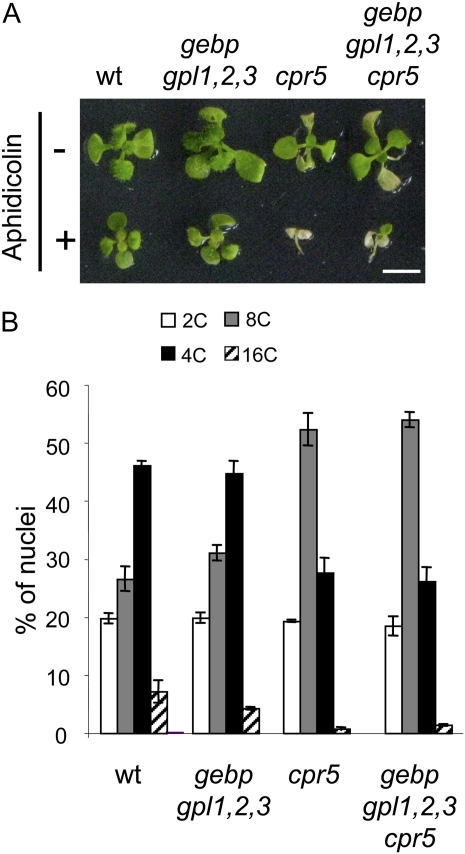
Figure 4.
Aphidicolin sensitivity assay and DNA levels in wild type, gebp gpl1,2,3 quadruple mutant, cpr5-2, and gebp gpl1,2,3 cpr5 quintuple mutant. A, Aphidicolin sensitivity assay. Plants were grown in vitro for 3 weeks in the absence (−) or presence (+) of aphidicolin (12 μg mL−1). Scale bars: 5 mm. B, Distribution of nuclei according to DNA content in cells of third rosette leaves. [See online article for color version of this figure.]
To reinforce this observation, we analyzed endoreduplication levels in leaf cells of wild-type and mutant plants using flow cytometry. The ploidy profiles of wild-type and quadruple gebp gpl1,2,3 mutant plants were similar, with a majority of nuclei having an 8C DNA content. In the cpr5 mutant, the ploidy profile was shifted toward lower values, with a main peak at 4C and a decrease of 8C and 16C populations. Similarly, in the gebp gpl1,2,3 cpr5 quintuple mutant, the population of 4C nuclei was increased at the expense of 8C and 16C populations, confirming that the role of CPR5 in endoreduplication is independent of GeBP/GPL genes. Finally, we introgressed the VP16:GPL2 construct into lines expressing the cell cycle markers PcycB1:GUS and PcdkA:GUS and measured transcript levels of cycD3 in wild type and VP16:GPL2 (Supplemental Fig. S4). No changes in GUS expression patterns or in transcript levels were observed in the VP16:GPL2 background, in agreement with our transcriptomic analysis, which detected no cell cycle genes that are known to be transcriptionally regulated.
Altogether, these results indicate that GeBP/GPL genes display a role in CPR5-dependent leaf expansion and plant growth but are not involved in control of the cell cycle or endoreduplication.
Epidermal Cell Size Is Controlled by GeBP/GPL Genes in the Absence of CPR5
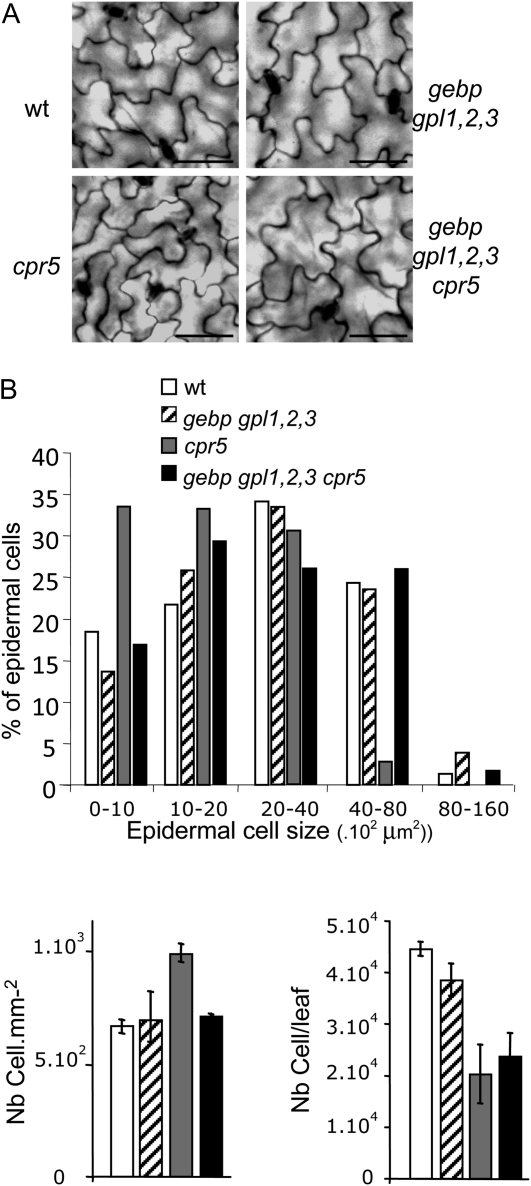
To determine whether the suppression of cpr5 growth defects in the quintuple mutant was due to an increase in cell expansion rather than in cell number, we looked at cell size in leaves of wild-type and mutant lines. As previously reported for cpr5 mutants (Kirik et al., 2001), pavement cells of cpr5 were smaller compared to the wild type (Fig. 5A). Pavement cell size in the gebp gpl1,2,3 quadruple mutant as well as the gebp gpl1,2,3 cpr5 quintuple mutant was not distinguishable from the wild type. To quantify this observation, we looked at cell size distribution in wild type and mutants. Class distribution of pavement cell size indicated that cpr5 had an excess of smaller cells and was deficient in larger cells (Fig. 5B) while the gebp gpl1,2,3 mutant was similar to wild type. On the other hand, the gebp gpl1,2,3 cpr5 quintuple mutant exhibited pavement cell sizes whose class distribution was similar to the wild type, indicating that the suppression of the cpr5 growth defect in the quintuple mutant was mainly due to an increase in cell size. This increase did not occur in all cell types as palisade cells of the quintuple mutant had a size similar to palisade cells of cpr5 (Supplemental Fig. S5). The estimated total number of epidermal cells in cpr5 and quintuple mutant leaves were within the same range (Fig. 5B), reinforcing the conclusion that GeBP/GPL genes do not play a major role in cell proliferation.
Figure 5.
Size and number of adaxial pavement cells in leaves of wild-type and mutant plants. Epidermal cell size and number in wild type, gebp gpl1,2,3 quadruple mutant, cpr5-2, and gebp gpl1,2,3 cpr5 quintuple mutant. A, Adaxial pavement cell size of third leaves of 3-week-old plants grown in soil. Scale bars: 200 μm. B, Class distribution of epidermal cell size (top histogram), epidermal cell density (bottom left), and estimated epidermal cell number per leaf (bottom right). Class distribution of cell size was performed by measuring cell area from third leaves of 3-week-old plants grown in soil (Columbia, n = 147; quadruple mutant, n = 149; cpr5, n = 294; quintuple mutant, n = 191) using ImageJ software. The total number of epidermal cells per leaf was estimated by dividing the leaf area by the average cell area.
CPR5 Is a Nuclear Protein Involved in Transcription
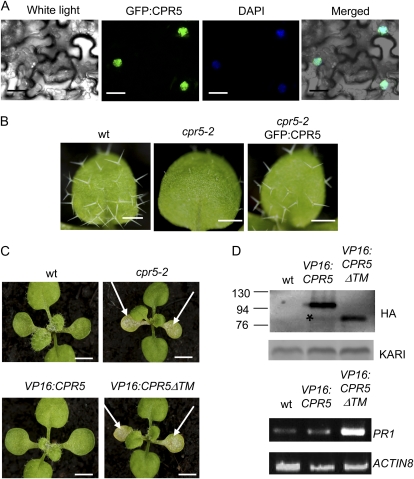
One of the key unsolved issues in understanding the function of CPR5 is its intracellular localization. To address this point, the GFP coding sequence was fused upstream of the full-length CPR5 cDNA under the control of the 35S promoter and transient transformation of tobacco (Nicotiana tabacum) cells was performed. As shown in Figure 6A, the GFP signal was present in the nuclei of plant cells. These results indicate that CPR5 is a nuclear protein. To determine whether the fusion was functional, this construct was introduced into a cpr5 mutant background. As illustrated in Figure 6B, the full-length fusion complemented many aspects of the cpr5 phenotype (dwarfism, spontaneous lesions, trichome development) except the early senescence of cotyledons, which was delayed but not completely abolished.
Figure 6.
Intracellular localization and functional analysis of CPR5 and CPR5ΔTM proteins. A, Subcellular localization of GFP:CPR5 protein in tobacco cells under confocal microscopy. Scale bars: 10 μm. B, Trichome development in plants transformed with 35S:GFP:CPR5 on a cpr5-2 mutant background. Young rosette leaves are shown. Scale bars: 1 mm. C, Phenotype of plants transformed with 35S:HA:VP16:CPR5 or 35S:HA:VP16:CPR5ΔTM on a wild-type background. Young rosettes are shown. White arrows indicate early senescing cotyledons. Scale bars: 3 mm. D, Detection of VP16 fusion proteins in western blots (two top sections) and transcript levels of PR1 in wild-type and VP16 plants using semiquantitative RT-PCR (two bottom sections) in wild-type and VP16 transgenic lines. The star indicates a weak band in the VP16:CPR5 lane corresponding to a protein with a size similar to that of the VP16:CPR5ΔTM protein. Contrast has been increased to better visualize this band. VP16 fusions were detected with a monoclonal anti-HA antibody (Roche). The KARI protein used as a loading control was detected with a polyclonal anti-KARI antibody.
It has been proposed that CPR5 might function similarly to membrane-bound transcription factors that are kept in a dormant state when anchored in membranes and are released by proteolytic cleavage, enabling them to directly regulate downstream genes (Kirik et al., 2001; Seo et al., 2008). To determine whether CPR5 or its soluble portion might be directly involved in transcriptional mechanisms, the VP16 transcriptional activation domain was fused in frame to the coding sequence of wild-type CPR5 or a CPR5 deletion mutant (CPR5ΔTM) lacking the transmembrane domain, and transgenic lines were generated. While all (23/23) VP16:CPR5 lines displayed a wild-type phenotype, nearly all (18/21) of the VP16:CPR5ΔTM lines displayed a cpr5 mutant phenotype with early senescing cotyledons, reduced trichome number, reduced growth (Fig. 6C), and up-regulation of PR1 gene expression (Fig. 6D). Western-blot analysis indicated that both fusion proteins were expressed at similar levels (Fig. 6D). In VP16:CPR5 lines, we also detected very low levels of a protein with a molecular weight similar to the VP16:CPR5ΔTM protein, suggesting that the full-length fusion can give rise to a transmembrane domain truncated form in vivo.
These results suggest that the CPR5 protein is located in the nucleus of plant cells, supporting the hypothesis that CPR5 can be involved in transcriptional processes.
DISCUSSION
GeBP/GPL Genes Are Involved in Stress Responses
Stress in plants can trigger a wide variety of physiological and molecular responses, leading to HR-like responses, reactive oxygen species production, hormonal disorders, and, ultimately, programmed cell death. We show here that GeBP/GPLs play a role in pathogen resistance and can induce HR-like responses when overexpressed as a constitutively active form. Our transcriptomic analysis indicates that GeBP/GPL genes control a relatively small number of genes, part of which are involved in stress-related pathways. This suggests that GeBP/GPL genes have a specific function among mechanisms triggered by stress. We have shown previously that GeBP/GPL genes are involved in the indirect regulation of cytokinin response genes, such as ARR6 (Chevalier et al., 2008), which was also highlighted in our microarray data. Several links between cytokinins and biotic and abiotic stresses have been described: Cytokinin treatment increases transcript levels of PR genes (Memelink et al., 1987) and can directly trigger cell death, and a genetic link between plant defense response and cytokinins has been shown by mutations in defense genes (Igari et al., 2008). Therefore, it is possible that GeBP/GPL genes have a specific role in the link between stress and the cytokinin pathway. In addition, the cpr5 mutant shows reduced cytokinin sensitivity under conditions of stress, such as the presence of a high concentration of Glc (Aki et al., 2007).
Transcriptomic, phenotypic, and genetic relationships between GeBP/GPLs and CPR5 demonstrate that they are involved in similar processes, although GeBP/GPL functions are a subset of CPR5 functions. These relationships suggest that GeBP/GPL genes act downstream of CPR5 or independently of CPR5. Transcript levels of GeBP/GPLs are similar on wild-type and cpr5 backgrounds (data not shown), suggesting that CPR5 does not regulate GeBP/GPL transcript level.
A Role for GeBP/GPL Genes in Cell Expansion
The control of final organ size implies a complex network of both promoters and inhibitors of cell proliferation and cell expansion, some of which have been identified (Mizukami, 2001; Autran et al., 2002; Hu et al., 2003; Churchman et al., 2006; Disch et al., 2006; Busov et al., 2008). Our genetic analysis shows that GeBP/GPL genes play a repressive role in final organ size determination by counteracting the positive role of CPR5 in this process. GeBP/GPLs do not seem to regulate cell proliferation, as the estimated epidermal cell number in leaves of gebp gpl1,2,3 and gebp gpl1,2,3 cpr5 plants was similar to that of wild-type plants and cpr5 plants, respectively. This observation is reinforced by the absence of modifications in the transcript levels of cell cycle marker genes in gebp gpl1,2,3 and VP16:GPL2 plants. In addition, DNA content measurements and aphidicolin sensitivity assays suggest that endoreduplication and cell cycle mechanisms are not controlled by GeBP/GPLs. Our phenotypic analysis revealed a role for GeBP/GPLs in cell size regulation, at least in epidermal cells. This role of GeBP/GPLs was observed on a cpr5 mutant background, where cell number was reduced compared to the wild type. Compensatory effects between cell number and cell expansion in leaves have been reported in many studies using growth mutants or ecotypes (Tsukaya and Beemster, 2006; Ferjani et al., 2007; Tisné et al., 2008), ectopic expression of cell-cycle-related genes (Hemerly et al., 1995; Cockcroft et al., 2000; Wang et al., 2000; De Veylder et al., 2001), or environmental stresses (Granier et al., 2000; West et al., 2004; Aguirrezabal et al., 2006; Cookson et al., 2006, 2007). This compensatory system allows, for example, a decrease in cell number to be balanced totally or partially by an increase in cell size, so that the final size of the organ is maintained. Although mechanisms involved in this process are not well understood, a general consensus exists that cell-autonomous-like programming (e.g. the classical cellular theory; Francis, 1992, 2007; Granier et al., 2000) is not sufficient to systematically account for organ development. Several works support this view and suggest that an unknown, nonautonomous signal from the epidermal layer triggers plant growth in Arabidopsis (Savaldi-Goldstein et al., 2007; Savaldi-Goldstein and Chory, 2008; Kawade et al., 2010). It is possible that GeBP/GPLs play a role in the restriction of epidermal cell size under conditions of stress.
In addition, the opposite regulation of cell expansion by GeBP/GPL and CPR5 genes could be due to more complex mechanisms. Previous work (Vanacker et al., 2001) has shown that salicylic acid whose level is increased in cpr5 mutants can influence cell expansion in opposite manner depending on the genetic background. Therefore we cannot exclude that the GeBP/GPL role in cell expansion might be different in other pathogen-resistant mutant backgrounds.
Based on the abnormal composition of cell walls in cpr5 mutants, Brininstool et al. (2008) suggested that CPR5 is involved in some cell-wall-related mechanism, as cell wall metabolism plays roles in cell expansion, pathogen response signaling, and cell death (Ellis et al., 2002; Somerville et al., 2004; Ahn et al., 2006). Several genes downstream of GeBP/GPLs and CPR5 are known to play a role in cell elongation or cell wall metabolism, such as the expansin gene family, whose activation is sufficient to induce the entire process of leaf development. This supports the concept of cell-division-independent mechanisms controlling organ development (Pien et al., 2001). Therefore, the cell-wall-related pathways could be controlled by CPR5 and GeBP/GPL, while other aspects, such as cell proliferation, could be regulated by CPR5 only.
It is interesting to notice that the partial suppression of cpr5 phenotype by gebp/gpl mutations concerns the growth defect phenotype while both gebp/gpl and cpr5 mutants display a similar defense response phenotype. Thus, GeBP/GPLs and CPR5 genes regulate plant and organ size in an opposite manner while they both act as repressors of defense response.
CPR5 Is a Nuclear Protein Involved in Transcription
CPR5 is predicted to be a type IIIa membrane protein with five transmembrane domains and a potential cytoplasmic N-terminal domain that contains a bipartite NLS. While the potential biochemical function of the CPR5 protein remains speculative, two hypotheses have been proposed to account for the prediction that it represents a membrane protein with a NLS: (1) CPR5 may be localized in membranes outside the nucleus, and its cytoplasmic domain may be proteolytically cleaved and transported into the nucleus (Kirik et al., 2001), a signaling process for which there is substantial precedent in plants (Seo et al., 2008); or (2) CPR5 may be targeted to the inner nuclear membrane through an NLS-dependent mechanism (Brininstool et al., 2008), as demonstrated for some proteins in yeast (Saccharomyces cerevisiae; Lusk et al., 2007), a hypothesis that does not exclude the possibility that its nucleosolic domain is proteolytically cleaved and participates in transcriptional processes. Our work supports the latter hypothesis, as we show that the full-length CPR5 protein is localized in the nucleus and the nonmembrane portion of the protein can induce a cpr5 mutant phenotype when fused to the VP16 activation domain. Although we cannot exclude that CPR5 is also present outside the nucleus, this suggests that CPR5 or its cleavage product might be directly required in the nucleus for the proper functioning of transcriptional processes. This does not imply that CPR5 is a transcription factor as it could participate to transcriptional complexes without being involved in transcription regulation per se.
Although the CPR5 gene has been isolated more than 10 years ago, we have provided here, to our knowledge, the first functional evidence that CPR5 is a nuclear protein that might play a role in transcriptional processes. In addition, we have tackled the CPR5-dependent cell expansion pathway and provide a link between the CPR5 and GeBP/GPL pathways.
In several organisms, highly pleiotropic genes such as CPR5 participate in many biological processes through distribution of the protein in more cellular components and involvement in more protein-protein interactions compared to nonpleiotropic genes (He and Zhang, 2006; Zou et al., 2008). It will be of interest to determine whether CPR5 behaves as a shuttling membrane-bound (co)transcription factor interacting with multiple partners and to characterize in detail the molecular link between CPR5 and GeBP/GPL genes.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds were sown on soil or surface sterilized and grown in petri dishes on Murashige and Skoog basal salt mixture medium (Sigma). Plants were grown at 22°C in long-day photoperiod (16 h of 100 μE light). The Arabidopsis (Arabidopsis thaliana) Columbia-0 ecotype was the wild type used. Agrobacterium tumefaciens C58 pGV3121 was used for stable and transient transformation of Arabidopsis using the floral-dip technique (Clough and Bent, 1998) or transient expression in Nicotiana benthamiana leaves (Lavy et al., 2002). The PcycB1:GUS line was provided by Dr. Murray (Cambridge, UK) and the PcdkA:GUS line was a gift of Dr. Faure (Versailles, France). For GUS staining, plants were vaccum infiltrated and incubated 2 to 12 h with GUS substrate at 37°C and destained as described (Gallagher, 1992).
Microarray Analysis
Plants used for microarray analysis were grown in vitro under long-day conditions for 21 d. Whole rosettes and roots were used. RNA samples were processed in triplicate from pooled samples (6–8 rosettes). Total RNA was prepared using the total RNA isolation kit (Qiagen). Hybridization on chips and data analysis were performed by Partnerchip. Enrichment analyses were done with the singular enrichment analysis tool at AgriGO (http://bioinfo.cau.edu.cn/agriGO/analysis.php). Comparative analysis with publicly available microarray data were performed with Genevestigator software (https://www.genevestigator.com/gv/index.jsp). Graphic outputs were made with MultiExperiment viewer software (http://www.tm4.org/).
The enrichment in Gene Ontology terms was calculated as the ratio of the proportion of genes involved in a pathway at the genome level over the proportion of genes involved in the same pathway in our microarray data. For instance the response-to-stress pathway (GO: 0006950) comprises 456 genes among the 31,819 genes of Arabidopsis. Of these 456, 13 genes are found among the 88 misregulated genes in the gebp/gpl quadruple mutant. Therefore the enrichment is calculated as the ratio of 13/88 over 456/31,819 = 10.30. A similar enrichment (10.47-fold) is found for the VP16:GPL2 line giving an average of 10.38 enrichment for both type of plants.
Data of cpr5 microarrays were obtained at the NASCArray Web site (http://affymetrix.arabidopsis.info/) under the NASCArrays experiment reference number NASCARRAYS-355.
Real-time RT-PCR was performed as described (Curaba et al., 2003) using the following oligonucleotides: 5′-TCTTGTAGGTGCTCTTGTTCTTCC-3′ and 5′-CAACCCTCTCGTCCCACTGC-3′ for PR1, 5′-TTGAATTGACTCCAGGTGCTTCC-3′ and 5′-GCCAGAGTGACGGGAGGAAC-3′ for PR5, and 5′-AGCCTTACAACGCTACTCTGTCTGTC-3′ and 5′-CACCAGACATAGTAG-CAGAAATCAAG-3′ for TUBULIN.
Semiquantitative RT-PCR for PR1 was performed as previously described (Chevalier et al., 2008) with 26, 28, and 30 PCR cycles to ensure that the amplification was linear. Cycles 30 and 28 are shown for PR1 and ACTIN8, respectively. The following primers were used: 5′-ATGAATTTTACTGGCTATTCTCGATTTTTAATCG-3′ and 5′-TTAGTATGGCTTCTCGTTCACATAATTC-3′ for PR1 and 5′-AATCAGATGTGGATCTCTAAGGCA-3′ and 5′-TCCGAGTTTGAAGAGGCTAC-AAAC-3′ for ACTIN8.
Molecular Cloning
The CPR5 cDNA, kindly provided by Viktor Kirik, was sequenced and cloned as an Acc65I-SalI fragment between the Acc65I and XhoI sites of pENTR4 vector (Invitrogen) in frame with attB sites to give p672 vector. The CPR5ΔTM version was made by cloning the Acc65I-BsrBI fragment of CPR5 between the Acc65I and EcoRV sites of the pENTR4 vector to give p673. Translational fusions with GFP were made using the pK7WGF2.0 destination vector (Plant Systems Biology, Vlaams Interuniversitair Instituut voor Biotechnologie-Ghent University) using the attL-attR (LR) clonase (Invitrogen) with entry vectors p672 and p673 to give p680 and p674, respectively. Translational fusions with the VP16 activation domain were made using Alligator1 (Bensmihen et al., 2004), kindly provided by François Parcy (Grenoble, France) using the LR clonase with entry vectors p672 and p673 to give p677 and p681, respectively. Expression of VP16 fusion proteins in transgenic lines was assessed by western blot with a monoclonal anti-HA antibody (Roche) while the loading control ketol-acid reducto-isomerase (KARI) protein was detected with a polyclonal antibody (Dumas et al., 2001).
The N-terminal deletion in GeBP was done by deleting the EcoRI-BamHI region in the gateway entry vector p497 and ligating the adapter made by annealing the two following oligonucleotides: AATTCAAGAAGAAATTGG and GATCCAATTTCTTCTTG. This deletion removes the first 55 amino acids and was subsequently cloned in the VP16 destination vector as previously described (Chevalier et al., 2008).
Tissue Preparation and Microscopy
For confocal microscopy analysis, p680 and p674 expression vectors were introduced separately into Agrobacterium and independently infiltrated into tobacco (Nicotiana tabacum) leaves as previously described (Lavy et al., 2002) except that 4,6-diamidinophenylindole was added to the cell suspension at 1 μg mL−1 before infiltration. Observations were made with a Leica confocal microscope and data were analyzed with the Leica LCS 2.61 software. Laser excitation was done in the sequential mode in between frames first with an argon laser (515 nm) and then with a UV laser (351–364 nm). Spectra were analyzed to confirm the specificity of GFP emission.
Trichome nuclear DNA contents were measured as described previously by Brininstool et al. (2008). Images were captured with a SensiCam QE 12-bit, cooled CCD camera, and analyzed with Slidebook software from 3I. Care was taken when setting image capture parameters that the nuclei with the highest DNA content in a group of samples did not saturate the dynamic range of the images. Samples were normalized to a wild-type control, assuming a mean value of 32C for wild-type trichome nuclei. Nonparametric statistics (Kruskal-Wallis one-way ANOVA and Dunn’s all pairwise multiple comparison) were performed using SigmaStat.
For flow cytometric analysis, the third pair of rosette leaves from 3-week-old plants grown on soil were used. Tissues were chopped with a razor blade in 900 μL of 45 mm MgCl2, 30 mm sodium citrate, 20 mm MOPS, pH 7, and 0.1% Triton X-100 supplemented with 1% polyvinylpyrrolidone 10,000, 5 mm metabisulfite, and 5 μg mL−1 RNase from a stock solution at 50 units/mg. Propidium iodide was added to the filtered supernatants at 50 μg mL−1. The nuclei were analyzed with the CyFlow SL cytometer using FloMax (Partec) software, or with an EPICS/ELITE cytometer and its software from Beckman-Coulter.
Trypan blue staining of cells was done as previously described (Lu et al., 2009). For aphidicolin sensitivity assay, plants were grown in vitro for 3 weeks in the absence or presence of aphidicolin at 12 μg mL−1 as described previously (De Schutter et al., 2007).
For leaf area measurements and cell number determination, leaves were harvested at 21 d after sowing, cleared overnight in ethanol, stored in lactic acid for microscopy, and observed with a microscope fitted with differential interference contrast optics (Leica). The total leaf blade area was determined from images taken with a digital camera (Zeiss) mounted on a binocular (Leica). Total number of pavement and guard cells, from which the average cell area was calculated, were obtained from at least 40 pictures of the adaxial epidermis located 25% and 75% from the distance between the tip and the base of the leaf, halfway between the midrib and the leaf margin. The total number of cells per leaf was estimated by dividing the leaf area by the average cell area. Epidermal cell size measurement was done using ImageJ software. Palisade cells were treated as previously described (Horiguchi et al., 2006) and cell size measurement was done using ImageJ software.
Pathogen Assay
The Pst DC3000 was grown at 29°C on King’s B medium supplemented with rifampicin 50 μg mL−1. Four- or 5-week-old plants were used for bacterial inoculation. For this objective, they were kept at high humidity 12 h before experiments for syringe infiltration facilities and then grown under daylight under the following conditions: 9-h light/15-h dark and 40%/70% humidity. Leaves without lesions from plant developing lesions were infiltrated with a bacterial suspension of 2 × 105 colony forming units mL−1. Determination of in planta bacterial growth was performed as previously described (Lorrain et al., 2004).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Overlaps between cpr5, gebp/gpl1,2,3, VP16:GPL2, and stress-induced transcriptomic alterations.
Supplemental Figure S2. A stable form of the founding member GeBP fused to the VP16 domain mimics the VP16:GPL2 phenotype.
Supplemental Figure S3. DNA level in trichomes of wild-type, cpr5, and VP16:GPL2 plants.
Supplemental Figure S4. Expression of cell cycle markers in the VP16:GPL2 background.
Supplemental Figure S5. Size of palisade cells in leaves of wild-type and mutant plants.
Supplemental Table S1. List of misregulated genes in gebp gpl1,2,3 quadruple mutant.
Supplemental Table S2. List of misregulated genes in VP16:GPL2 plants.
Acknowledgments
We thank Viktor Kirik (Standford) for providing the CPR5 cDNA. We thank Renaud Dumas (Grenoble, France) for the gift of the anti-KARI antibody. We thank The Arabidopsis Information Resource (http://arabidopsis.org) for providing Arabidopsis data. We thank Dr. Philip Zimmermann (Zurich, Switzerland) for his help in microrarray data analysis. We thank Ginger Brininstool (Baton Rouge, LA) for assistance with the DNA content measurements of trichome nuclei.
References
- Aguirrezabal L, Bouchier-Combaud S, Radziejwoski A, Dauzat M, Cookson SJ, Granier C. (2006) Plasticity to soil water deficit in Arabidopsis thaliana: dissection of leaf development into underlying growth dynamic and cellular variables reveals invisible phenotypes. Plant Cell Environ 29: 2216–2227 [DOI] [PubMed] [Google Scholar]
- Ahn JW, Verma R, Kim M, Lee JY, Kim YK, Bang JW, Reiter WD, Pai HS. (2006) Depletion of UDP-D-apiose/UDP-D-xylose synthases results in rhamnogalacturonan-II deficiency, cell wall thickening, and cell death in higher plants. J Biol Chem 281: 13708–13716 [DOI] [PubMed] [Google Scholar]
- Aki T, Konishi M, Kikuchi T, Fujimori T, Yoneyama T, Yanagisawa S. (2007) Distinct modulations of the hexokinase1-mediated glucose response and hexokinase1-independent processes by HYS1/CPR5 in Arabidopsis. J Exp Bot 58: 3239–3248 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000) Gene ontology: tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autran D, Jonak C, Belcram K, Beemster GT, Kronenberger J, Grandjean O, Inzé D, Traas J. (2002) Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J 21: 6036–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensmihen S, To A, Lambert G, Kroj T, Giraudat J, Parcy F. (2004) Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett 561: 127–131 [DOI] [PubMed] [Google Scholar]
- Boch J, Verbsky L, Robertson L, Larkin J, Kunkel B. (1998) Analysis of resistance gene-mediated defense responses in Arabidopsis thaliana plants carrying a mutation in CPR5. Mol Plant Microbe Interact 11: 1196–1206 [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X. (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6: 1845–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brininstool G, Kasili R, Simmons LA, Kirik V, Hülskamp M, Larkin JC. (2008) Constitutive Expressor Of Pathogenesis-Related Genes5 affects cell wall biogenesis and trichome development. BMC Plant Biol 8: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busov VB, Brunner AM, Strauss SH. (2008) Genes for control of plant stature and form. New Phytol 177: 589–607 [DOI] [PubMed] [Google Scholar]
- Calikowski TT, Meulia T, Meier I. (2003) A proteomic study of the Arabidopsis nuclear matrix. J Cell Biochem 90: 361–378 [DOI] [PubMed] [Google Scholar]
- Chevalier F, Perazza D, Laporte F, Le Hénanff G, Hornitschek P, Bonneville JM, Herzog M, Vachon G. (2008) GeBP and GeBP-like proteins are noncanonical leucine-zipper transcription factors that regulate cytokinin response in Arabidopsis. Plant Physiol 146: 1142–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman ML, Brown ML, Kato N, Kirik V, Hülskamp M, Inzé D, De Veylder L, Walker JD, Zheng Z, Oppenheimer DG, et al. (2006) SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell 18: 3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X. (2000) Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cockcroft CE, den Boer BG, Healy JM, Murray JA. (2000) Cyclin D control of growth rate in plants. Nature 405: 575–579 [DOI] [PubMed] [Google Scholar]
- Cookson SJ, Chenu K, Granier C. (2007) Day length affects the dynamics of leaf expansion and cellular development in Arabidopsis thaliana partially through floral transition timing. Ann Bot (Lond) 99: 703–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson SJ, Radziejwoski A, Granier C. (2006) Cell and leaf size plasticity in Arabidopsis: what is the role of endoreduplication? Plant Cell Environ 29: 1273–1283 [DOI] [PubMed] [Google Scholar]
- Curaba J, Herzog M, Vachon G. (2003) GeBP, the first member of a new gene family in Arabidopsis, encodes a nuclear protein with DNA-binding activity and is regulated by KNAT1. Plant J 33: 305–317 [DOI] [PubMed] [Google Scholar]
- De Schutter K, Joubès J, Cools T, Verkest A, Corellou F, Babiychuk E, Van Der Schueren E, Beeckman T, Kushnir S, Inzé D, et al. (2007) Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19: 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inzé D. (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disch S, Anastasiou E, Sharma VK, Laux T, Fletcher JC, Lenhard M. (2006) The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr Biol 16: 272–279 [DOI] [PubMed] [Google Scholar]
- Dumas R, Biou V, Halgand F, Douce R, Duggleby RG. (2001) Enzymology, structure, and dynamics of acetohydroxy acid isomeroreductase. Acc Chem Res 34: 399–408 [DOI] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG. (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferjani A, Horiguchi G, Yano S, Tsukaya H. (2007) Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol 144: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis D. (1992) The cell cycle in plant development. New Phytol 122: 1–20 [DOI] [PubMed] [Google Scholar]
- Francis D. (2007) The plant cell cycle—15 years on. New Phytol 174: 261–278 [DOI] [PubMed] [Google Scholar]
- Gagnot S, Tamby JP, Martin-Magniette ML, Bitton F, Taconnat L, Balzergue S, Aubourg S, Renou JP, Lecharny A, Brunaud V. (2008) CATdb: a public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res (Database issue) 36: D986–D990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SR. (1992) Quantitation of GUS activity by fluorometry. Gallagher SR, , GUS Protocols Using the GUS Gene as a Reporter of Gene Expression. Academic Press, Inc., San Diego, pp 47–59 [Google Scholar]
- Granier C, Turc O, Tardieu F. (2000) Co-ordination of cell division and tissue expansion in sunflower, tobacco, and pea leaves: dependence or independence of both processes? J Plant Growth Regul 19: 45–54 [DOI] [PubMed] [Google Scholar]
- He X, Zhang J. (2006) Toward a molecular understanding of pleiotropy. Genetics 173: 1885–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly A, Engler JdeA, Bergounioux C, Van Montagu M, Engler G, Inzé D, Ferreira P. (1995) Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J 14: 3925–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Fujikura U, Ferjani A, Ishikawa N, Tsukaya H. (2006) Large-scale histological analysis of leaf mutants using two simple leaf observation methods: identification of novel genetic pathways governing the size and shape of leaves. Plant J 48: 638–644 [DOI] [PubMed] [Google Scholar]
- Hu Y, Xie Q, Chua NH. (2003) The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15: 1951–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igari K, Endo S, Hibara KI, Aida M, Sakakibara H, Kawasaki T, Tasaka M. (2008) Constitutive activation of a CC-NB-LRR protein alters morphogenesis through the cytokinin pathway in Arabidopsis. Plant J 55: 14–27 [DOI] [PubMed] [Google Scholar]
- Jing HC, Anderson L, Sturre MJ, Hille J, Dijkwel PP. (2007) Arabidopsis CPR5 is a senescence-regulatory gene with pleiotropic functions as predicted by the evolutionary theory of senescence. J Exp Bot 58: 3885–3894 [DOI] [PubMed] [Google Scholar]
- Jing HC, Dijkwel PP. (2008) CPR5: a jack of all trades in plants. Plant Signal Behav 3: 562–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing HC, Hebeler R, Oeljeklaus S, Sitek B, Stühler K, Meyer HE, Sturre MJ, Hille J, Warscheid B, Dijkwel PP. (2008) Early leaf senescence is associated with an altered cellular redox balance in Arabidopsis cpr5/old1 mutants. Plant Biol (Stuttg) (Suppl 1) 10: 85–98 [DOI] [PubMed] [Google Scholar]
- Jing HC, Sturre MJ, Hille J, Dijkwel PP. (2002) Arabidopsis onset of leaf death mutants identify a regulatory pathway controlling leaf senescence. Plant J 32: 51–63 [DOI] [PubMed] [Google Scholar]
- John PC, Qi R. (2008) Cell division and endoreduplication: doubtful engines of vegetative growth. Trends Plant Sci 13: 121–127 [DOI] [PubMed] [Google Scholar]
- Kawade K, Horiguchi G, Tsukaya H. (2010) Non-cell-autonomously coordinated organ size regulation in leaf development. Development 137: 4221–4227 [DOI] [PubMed] [Google Scholar]
- Kirik V, Bouyer D, Schöbinger U, Bechtold N, Herzog M, Bonneville JM, Hülskamp M. (2001) CPR5 is involved in cell proliferation and cell death control and encodes a novel transmembrane protein. Curr Biol 11: 1891–1895 [DOI] [PubMed] [Google Scholar]
- Lavy M, Bracha-Drori K, Sternberg H, Yalovsky S. (2002) A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14: 2431–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Lin B, Auriac MC, Kroj T, Saindrenan P, Nicole M, Balagué C, Roby D. (2004) Vascular associated death1, a novel GRAM domain-containing protein, is a regulator of cell death and defense responses in vascular tissues. Plant Cell 16: 2217–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Salimian S, Gamelin E, Wang G, Fedorowski J, LaCourse W, Greenberg JT. (2009) Genetic analysis of acd6-1 reveals complex defense networks and leads to identification of novel defense genes in Arabidopsis. Plant J 58: 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk CP, Blobel G, King MC. (2007) Highway to the inner nuclear membrane: rules for the road. Nat Rev Mol Cell Biol 8: 414–420 [DOI] [PubMed] [Google Scholar]
- Memelink J, Hoge JH, Schilperoort RA. (1987) Cytokinin stress changes the developmental regulation of several defence-related genes in tobacco. EMBO J 6: 3579–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y. (2001) A matter of size: developmental control of organ size in plants. Curr Opin Plant Biol 4: 533–539 [DOI] [PubMed] [Google Scholar]
- Parcy F, Bomblies K, Weigel D. (2002) Interaction of LEAFY, AGAMOUS and TERMINAL FLOWER1 in maintaining floral meristem identity in Arabidopsis. Development 129: 2519–2527 [DOI] [PubMed] [Google Scholar]
- Pien S, Wyrzykowska J, McQueen-Mason S, Smart C, Fleming A. (2001) Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc Natl Acad Sci USA 98: 11812–11817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotter A, Usadel B, Baebler S, Stitt M, Gruden K. (2007) Adaptation of the MapMan ontology to biotic stress responses: application in solanaceous species. Plant Methods 3: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Chory J. (2008) Growth coordination and the shoot epidermis. Curr Opin Plant Biol 11: 42–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Peto C, Chory J. (2007) The epidermis both drives and restricts plant shoot growth. Nature 446: 199–202 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Kim SG, Park CM. (2008) Membrane-bound transcription factors in plants. Trends Plant Sci 13: 550–556 [DOI] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, et al. (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts K. (2003) “Big it up”: endoreduplication and cell-size control in plants. Curr Opin Plant Biol 6: 544–553 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tisné S, Reymond M, Vile D, Fabre J, Dauzat M, Koornneef M, Granier C. (2008) Combined genetic and modeling approaches reveal that epidermal cell area and number in leaves are controlled by leaf and plant developmental processes in Arabidopsis. Plant Physiol 148: 1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H, Beemster GT. (2006) Genetics, cell cycle and cell expansion in organogenesis in plants. J Plant Res 119: 1–4 [DOI] [PubMed] [Google Scholar]
- Vanacker H, Lu H, Rate DN, Greenberg JT. (2001) A role for salicylic acid and NPR1 in regulating cell growth in Arabidopsis. Plant J 28: 209–216 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhou Y, Gilmer S, Whitwill S, Fowke LC. (2000) Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J 24: 613–623 [DOI] [PubMed] [Google Scholar]
- West G, Inzé D, Beemster GT. (2004) Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiol 135: 1050–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Yu K, Navarre D, Seebold K, Kachroo A, Kachroo P. (2010) The glabra1 mutation affects cuticle formation and plant responses to microbes. Plant Physiol 154: 833–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Ito M, Nishida I, Watanabe A. (2002) Identification of a novel gene HYS1/CPR5 that has a repressive role in the induction of leaf senescence and pathogen-defence responses in Arabidopsis thaliana. Plant J 29: 427–437 [DOI] [PubMed] [Google Scholar]
- Zeller G, Henz SR, Widmer CK, Sachsenberg T, Rätsch G, Weigel D, Laubinger S. (2009) Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J 58: 1068–1082 [DOI] [PubMed] [Google Scholar]
- Zou L, Sriswasdi S, Ross B, Missiuro PV, Liu J, Ge H. (2008) Systematic analysis of pleiotropy in C. elegans early embryogenesis. PLOS Comput Biol 4: e1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]