Abstract
In seeds, glutamate decarboxylase (GAD) operates at the metabolic nexus between carbon and nitrogen metabolism by catalyzing the unidirectional decarboxylation of glutamate to form γ-aminobutyric acid (GABA). To elucidate the regulatory role of GAD in seed development, we generated Arabidopsis (Arabidopsis thaliana) transgenic plants expressing a truncated GAD from Petunia hybrida missing the carboxyl-terminal regulatory Ca2+-calmodulin-binding domain under the transcriptional regulation of the seed maturation-specific phaseolin promoter. Dry seeds of the transgenic plants accumulated considerable amounts of GABA, and during desiccation the content of several amino acids increased, although not glutamate or proline. Dry transgenic seeds had higher protein content than wild-type seeds but lower amounts of the intermediates of glycolysis, glycerol and malate. The total fatty acid content of the transgenic seeds was 50% lower than in the wild type, while acyl-coenzyme A accumulated in the transgenic seeds. Labeling experiments revealed altered levels of respiration in the transgenic seeds, and fractionation studies indicated reduced incorporation of label in the sugar and lipid fractions extracted from transgenic seeds. Comparative transcript profiling of the dry seeds supported the metabolic data. Cellular processes up-regulated at the transcript level included the tricarboxylic acid cycle, fatty acid elongation, the shikimate pathway, tryptophan metabolism, nitrogen-carbon remobilization, and programmed cell death. Genes involved in the regulation of germination were similarly up-regulated. Taken together, these results indicate that the GAD-mediated conversion of glutamate to GABA during seed development plays an important role in balancing carbon and nitrogen metabolism and in storage reserve accumulation.
Efficient assimilation of carbon (C) and nitrogen (N) is essential for optimal plant growth, productivity, and yield (Stitt, 1999), particularly in seeds in which the content of essential amino acids is low. Hence, a comprehensive understanding of seed development and metabolism is central to the enhancement of crop yield and quality. Seed development can be divided into three phases: cell division, maturation (accumulation of food reserves), and desiccation (Weber et al., 2005). The shift from one phase to the other is affected by sugars (Suc, hexoses, trehalose) and abscisic acid. In the early stages of seed development, maternal regulation is maintained via assimilate unloading and supply of nutrients, thereby sustaining cell division. During maturation, seed metabolism changes and storage reserves accumulate in expanding cells. At this time, photosynthetic activity is initiated in the seed, which is believed to improve oxygen supply and the energy state in the seed, thus counteracting increasingly hypoxic conditions (Borisjuk and Rolletschek, 2009). The maturation stage is followed by a phase termed “maturation drying,” in which the metabolism of Arabidopsis (Arabidopsis thaliana) seeds shifts from a general decrease in unbound metabolites to the accumulation of a set of specific metabolites, including γ-aminobutyric acid (GABA; Fait et al., 2006; Angelovici et al., 2010).
GABA is a four-C nonprotein amino acid generally abundant in plants whose levels change significantly in response to a wide array of endogenous and exogenous factors, including biotic and abiotic stresses (Bouché et al., 2003; Bouché and Fromm, 2004; Bown et al., 2006), to the diurnal cycle (Masclaux-Daubresse et al., 2002), and to developmental processes, such as seed maturation (Fait et al., 2006), flower fertilization (Palanivelu et al., 2003), and fruit development (Rolin et al., 2000; Carrari et al., 2006; Akihiro et al., 2008; Yin et al., 2010). GABA is derived from Glu via the action of glutamate decarboxylase (GAD), the enzyme responsible for the first step in the GABA shunt metabolic pathway (Fig. 1; Bown and Shelp, 1997). In plant species such as petunia (Petunia hybrida; Chen et al., 1994; Arazi et al., 1995), soybean (Glycine max; Snedden et al., 1995), tobacco (Nicotiana tabacum; Baum et al., 1996), and Arabidopsis (Turano and Fang, 1998; Zik et al., 1998), the GAD polypeptides contain C-terminal extensions, ranging between 30 and 50 amino acids, that are not present in the bacterial GAD enzyme and function as Ca2+-calmodulin-binding domains (Baum et al., 1993). Thus, GAD catalyzes the decarboxylation of Glu to GABA in a calmodulin-dependent manner. Scientists have thus hypothesized both a protective role for GABA during stress events and a signaling function (Shelp et al., 1999; Bouché et al., 2003; Bouché and Fromm, 2004; Chevrot et al., 2006; Fait et al., 2008). While the latter role is under debate, a number of investigations have suggested a primary role of the GABA shunt in central C-N metabolism. The impact of a constitutive and systemically deregulated GABA biosynthesis has been shown in tobacco (Baum et al., 1996; McLean et al., 2003): transgenic plants overexpressing a mutant form of GAD displayed a severely decreased Glu content (Baum et al., 1996), which is generally maintained steady within the cell (Stitt and Fernie, 2003; Forde and Lea, 2007).
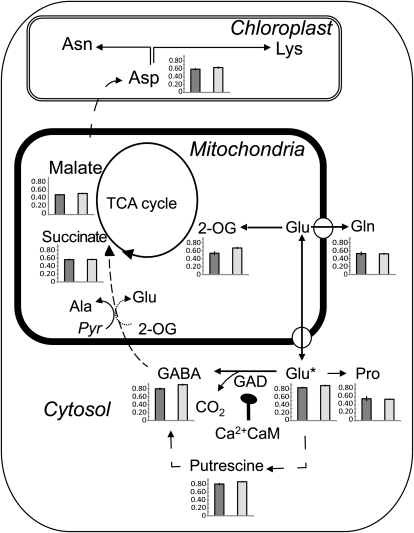
Figure 1.
Fate of 13C-labeled Glu (Glu*) supplied to stratified (dark gray bars) and 1-DAS (light gray bars) seeds of wild-type Arabidopsis (ecotype Wassilewskija). The relative isotope redistribution of Glu is described in a schematic presentation of the GABA shunt and related major metabolic pathways. Bars represent the C fractional enrichment [13C/(13C + 12C)] of metabolites labeled. C fractional enrichment was calculated by evaluating the labeled portion of the various pools as a proportion of the various pool sizes, following an incubation period of 6 h of stratified and 1-DAS seeds. Values are means ± sd from six determinations per time point.
It has been suggested that GABA constitutes a readily accessible nontoxic reserve of C and N for amino acid metabolism and tricarboxylic acid (TCA) cycle activity, which is of particular relevance during stress events (Breitkreuz et al., 1999; Solomon and Oliver, 2002; Beuvé et al., 2004; Bouché and Fromm, 2004). However, only recently has direct evidence, integrating metabolite and transcript profiling and stable-isotope labeling, emerged for the metabolic significance of the GABA shunt (Studart-Guimarães et al., 2007). In transgenic plants deficient in succinyl-CoA ligase, the enzyme that catalyzes the conversion of succinyl-CoA to succinate (Studart-Guimarães et al., 2005), succinate production was partly maintained via the GABA shunt (Studart-Guimarães et al., 2007). Earlier molecular studies and radioisotope experiments that demonstrated rapid degradation of Glu to GABA and thereafter to succinate also indicated a metabolic role for GABA in developing and germinating seeds (Vandewalle and Olsson, 1983; Tuin and Shelp, 1994). These results, taken together with the more recent metabolic profiling of developing seeds, suggest that GABA accumulation in the dry seed supports early metabolic reorganization at germination. However, a comprehensive understanding of the interregulation between the GABA shunt and central C-N metabolism is lacking. Moreover, there is a need for additional research to assess the function of GABA during seed development (i.e. at a time when metabolic processes are under tight concerted regulation; Weber et al., 2005; Holdsworth et al., 2008). To address this need, a development-inducible system was used that has previously been successfully applied to study the metabolic regulation of Lys biosynthesis (Zhu and Galili, 2003; Angelovici et al., 2009, 2010). In the system used in this study, the seed-specific phaseolin storage protein promoter drives the expression of a deregulated GAD from petunia in developing Arabidopsis seeds. The developmentally induced production of a deregulated GAD in maturing seeds provides us with a model system in which the perturbation in central metabolism can be monitored and the consequent effect on concertedly regulated metabolic processes during seed maturation can be assessed. We subjected developing seeds to metabolite profiling and genome-wide gene expression analysis with the aim to investigate the effect of an increasingly deregulated conversion of Glu to GABA on seed metabolism. Our results demonstrated that Glu-to-GABA deregulated conversion has a profound effect on the C-N balance of the seed, despite the existence of metabolite-recycling routes (i.e. GABA transaminase and the TCA cycle). The results are discussed with respect to the current understanding of seed development metabolism and its implications for seed germination.
RESULTS
Stable Isotope-Labeling Experiments Suggest That the GABA Shunt Represents a Considerable Source of C for the TCA Cycle during Early Stages of Seed Germination
Earlier radiolabeling studies and more recent metabolite profiling suggest that GABA plays a role in seed metabolism, accumulating in the dry seed and decreasing upon germination (Vandewalle and Olsson, 1983; Fait et al., 2006). Although informative, radiolabeling studies are limited in their applicability to monitoring the degree of labeling across a range of metabolites; as a result, stable isotopes coupled with mass spectrometry (MS) or NMR have been increasingly used in the last decade for pathway elucidation and flux analysis (Sauer, 2006, and refs. therein). To elucidate whether the GABA shunt operates as a nexus between C and N metabolism upon seed imbibition prior to seed germination, we evaluated the metabolic fate of Glu in Arabidopsis seeds freshly stratified and 1 d after stratification (DAS; when germination has been initiated); an adapted gas chromatography (GC)-MS-based protocol (Roessner-Tunali et al., 2004) was used in combination with a feeding 13C-substrate. The relative redistribution of 13C was measured by assessing (1) the incorporation of the isotope into each intermediate within the relevant pathways (Fig. 1) and (2) the proportional labeling of 13C and 12C (C fractional enrichment) in metabolites closely associated with Glu. In stratified seeds supplied with [13C]Glu, we detected significant and equivalent incorporation of 13C in GABA and in putrescine, followed by succinate, Asp, and 2-oxoglutarate (2-OG). In 1-DAS seeds, 13C incorporation increased significantly, particularly in GABA and 2-OG, followed by putrescine and Asp (Fig. 1). This finding indicates that the proportion of 13C derived from Glu that entered the GABA pool in early germinating seeds (prior to germination sensu strictu) exceeded that entering the pools of Pro, Gln, or 2-OG. The data also indicate the existence in seeds of a tighter relation between Glu-GABA metabolism and the polyamine putrescine than currently realized. Different pathways could potentially sustain the conversion of Glu-GABA to putrescine (1) by the action of putrescine:oxygen oxidoreductase (EC 1.4.3.10) on the aldehyde form of GABA, (2) via the Arg decarboxylase pathway, or (3) via spermidine metabolism. While it is not possible to mathematically deduce the absolute input fluxes into the succinate pool (Baxter et al., 2007), the above data expand on previous work suggesting that the GABA shunt constitutes an important metabolic route sustaining the TCA cycle and early amino acid biosynthesis in germinating seeds.
Expression of a Truncated GAD from Petunia Lacking the Calmodulin-Binding Domain Stimulates GABA Accumulation in Dry Seeds
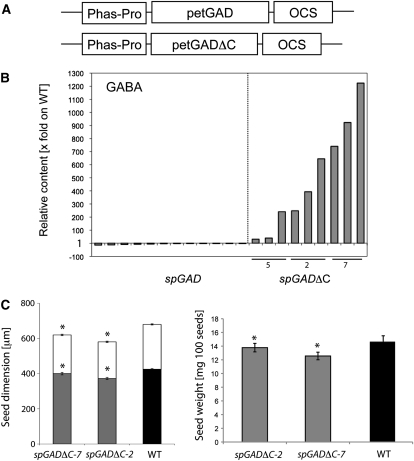
Two types of transgenic Arabidopsis plant (spGAD and spGADΔC) were generated (Fig. 2A), expressing either the full-length GAD or a truncated GAD cDNA from petunia, under the control of the bean (Phaseolus vulgaris) phaseolin promoter (see “Materials and Methods”), producing a constitutively active, although developmentally induced, GAD enzyme. Three and four independent transformants of spGADΔC and spGAD, respectively, were eventually isolated on antibiotic plates and tested for the segregation of T2 seeds. Thereafter, bulked T3 seeds were tested for segregation to confirm homozygosity and evaluated for GABA content (Fig. 2B). The GABA content in the dry seeds of all isolated spGAD lines was similar to that in the wild type. In contrast, the dry seeds of all spGADΔC lines were characterized by hyperaccumulation of GABA. Besides the metabolic phenotype of the seed, these plants showed no significant differences in vegetative traits as compared with Wassilewskija plants (data not shown). However, the dimensions of mature T3 seeds of spGADΔC-2 and spGADΔC-7, the two strongest transgenic lines with respect to GABA accumulation in the dry seed, were found to be slightly but significantly smaller than those of the wild type (Fig. 2C, left). The measured elliptic area, based on the width and length of the seeds in the above two transgenic lines, was shown to be 18.8% and 28.5%, respectively, smaller than that of the wild type. The reduction in area was partially reflected in the seed weight, with a statistically significant decrease of 7.3% and 15.5% in spGADΔC-2 and spGADΔC-7 as compared with the wild type, respectively (Fig. 2C, right). In germination tests of all genotypes under standard conditions for Arabidopsis (seeds of the control and of the transgenics were harvested at the same time and stored together; see “Materials and Methods”), stratified transgenic seeds displayed only 50% of the germination of wild-type seeds. Staining with tetrazolium confirmed that the transgenic seeds displayed reduced vigor (data not shown).
Figure 2.
A, Schematic diagram of the chimeric gene utilized in this study. Phas-Pro is the bean phaseolin storage protein promoter, spGAD and spGADΔC are the coding DNA sequences of the wild type and mutant for GAD from petunia, and OCS is the DNA sequence of the octopine synthase 3′ terminator. B, GABA relative content in mature T3 seeds of transgenic plants harboring spGAD and spGADΔC constructs, collected in three independent bulks from 30 plants each. GABA was measured by a GC-MS-based protocol (Roessner et al., 2001). Values represent fold change relative to the wild type (WT). Transgenic lines spGADΔC-5, spGADΔC-2, and spGADΔC-7 are indicated. C, Seed dimensions and weight. The seed dimension values are means ± sd of 30 individual measurements repeated for three independent bulks of 10 (T3) plants each. The seed weight values are means ± sd of 100 bulked seeds from 10 plants each and repeated 10 times. Seeds were viewed with a Leica MZ12.5 stereomicroscope (Leica DFC420 digital camera) and measured using Leica Application Suite software. Asterisks represent significant differences (P < 0.05) as compared with the wild type.
GABA Content Is Differentially Regulated during the Late Seed Maturation-to-Desiccation Stage, and Its Accumulation Is Indicative of a Shift toward N Metabolism
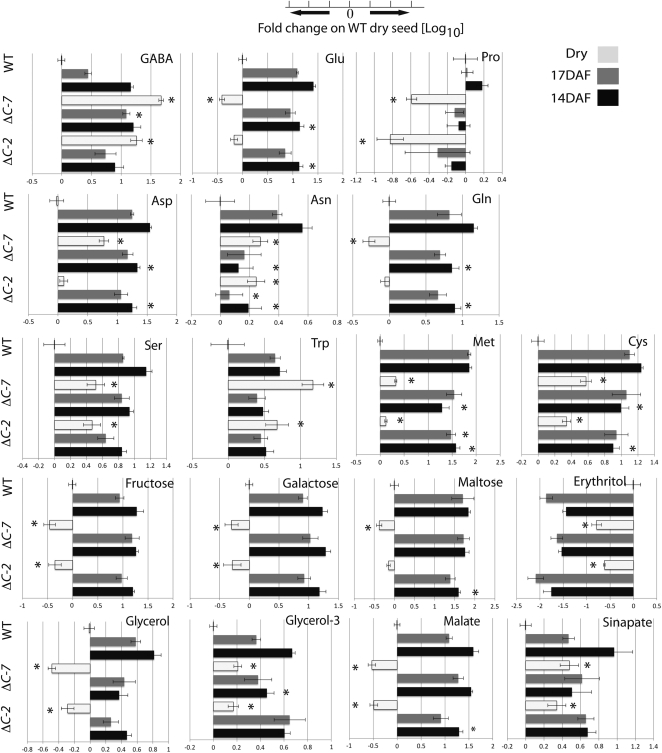
To assess the overall metabolite response to the maturation-induced gradual accumulation of GABA, we performed a developmental analysis of the metabolic changes in transgenic spGADΔC T4 seeds. Siliques of spGADΔC-2 and spGADΔC-7 and of the untransformed wild type were successively collected from tagged flowers at 14 and 17 d after flowering (DAF) and at full maturity. Seeds were extracted and analyzed using an established GC-MS-based protocol (Roessner et al., 2001; Fait et al., 2006). Figure 3 shows changes in the metabolite content of seeds of the transgenics relative to the wild type. Only metabolites showing significant (P < 0.01) and consistent changes are shown; the complete data set is given in Supplemental Table S1. While the phaseolin promoter activity is known to commence during early maturation of the seed (van der Geest and Hall, 1996), our results revealed that GABA gradually accumulated in the transgenic seeds both at late maturation and to a considerably greater extent in the dry seed (Fig. 3). Increased GABA levels were associated with a small but significant (P < 0.05) decrease in the content of Glu at 14 DAF and later in the dry seed. Similarly affected were the levels of biosynthetically related amino acids such as Gln and Pro, the latter displaying a marked decrease (one-fourth to one-sixth) in the dry seeds of spGADΔC as compared with the wild type. That said, the general effect of the constitutive activity of GAD was a decrease in the content of the vast majority of measured amino acids at 14 DAF (Fig. 3; Supplemental Table S1) and the accumulation of several of these amino acids in the dry seeds of both transgenic lines (Fig. 3; Supplemental Table S1). Increased levels were measured for Asp, Asn, Met, Cys, and Ser, and a particularly significant increase was found for Trp in the dry seeds (Fig. 3; Supplemental Table S1). In line with the results from the stable isotope experiments (Fig. 1), GABA accumulation in the dry seeds was accompanied by a decrease in the content of the polyamine putrescine (Supplemental Table S1), one of the possible GABA precursors (Fait et al., 2008).
Figure 3.
Relative content of metabolites in developing seeds of spGADΔC-2 (ΔC-2), spGADΔC-7 (ΔC-7), and the wild type (WT). Relative content of free metabolites was measured in developing seeds at 14 and 17 DAF and in dry seeds as described in “Materials and Methods.” The 14- and 17-DAF stages reflect mid and late maturation, respectively, under the condition of growth used (Fait et al., 2006; Angelovici et al., 2009). Bar graphs indicate the mean fold change of each metabolite at 14 DAF (black bars), 17 DAF (gray bars), and in the dry seeds (white bars) relative to its content in the dry seeds of the wild type. Asterisks denote statistically significant (two-tailed Student’s t test; P < 0.01) differences between spGADΔC and the wild type for the metabolite content at each given developmental stage. Only metabolites that presented significant and consistent changes in content in the transgenics as compared with the wild type are shown. The entire data set is given in Supplemental Table S1.
In contrast, when estimating the effect of an enhanced Glu-to-GABA conversion on carbohydrate and carboxylic acid metabolism, we found decreases in the concentrations of a number of sugars and organic acids (Fig. 3; Supplemental Table S1). For instance, in the dry transgenic seeds, Fru was decreased to 30% of its content in the dry seed of the wild type (Fig. 3). Gal, maltose, and erythritol followed a similar trend. The glycolysis-derived intermediates of glycerolipid metabolism, glycerate, glycerol, and the related compound myoinositol, were consistently decreased in the transgenic seeds (Fig. 3; Supplemental Table S1). Among the TCA cycle intermediates, succinate, a product of GABA catabolism, accumulated in the dry seeds of the transgenics by 1.5-fold more than in those of the wild type but citrate content was similar in the transgenics and the wild type (Supplemental Table S1). Notably, malate content was one-third in the dry seeds of the transgenics than in those of the wild type (Fig. 3).
14C-Labeling Experiments Reveal Decreased TCA Cycle Activity But Increased Glu Catabolism in the GABA-Hyperaccumulating Transgenic Lines
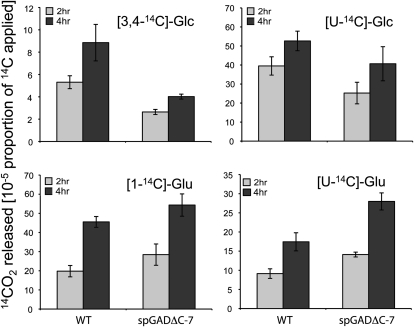
We next sought to evaluate the effect during seed development of a constitutively enhanced conversion of Glu to GABA on the activity of the TCA cycle and on glycolysis. For this purpose, we evaluated the relative rates of carbohydrate oxidation in the transformants by measuring the evolution of 14CO2 following the incubation of siliques in solutions of positionally labeled molecules. Maturing siliques (14 and 17 DAF) from the wild type and spGADΔC-7 were incubated with [1-14C]Glc, [3;4-14C]Glc, or [U-14C]Glc or with [U-14C]Glu. The 14CO2 evolved during the incubation period was collected following 2 and 4 h of incubation. Since carbon dioxide can be released from the C3:4 positions of Glc via enzymes associated with mitochondrial respiration (ap Rees and Beevers, 1960), carbon dioxide evolution from C3:4 positions of Glc provides an indication of the relative rate of the TCA cycle (Nunes-Nesi et al., 2005). In maturing siliques, the evolution of labeled CO2 from [3;4-14C]Glc was 50% lower in the representative transgenic line spGADΔC-7 than in the wild type (Fig. 4). 14CO2 can be released from [1-14C]Glu by the decarboxylation of Glu to GABA (Fig. 5): monitoring the evolution of labeled CO2 from maturing siliques fed with [1-14C]Glu confirmed the up-regulation of the GABA shunt (Fig. 4). [U-14C]Glu feeding gave a comparable increase in CO2 evolution, suggesting that Glu-derived CO2 was generated mainly via the activity of the GABA shunt rather than from incorporation into and respiration through the TCA cycle.
Figure 4.
14CO2 release from siliques incubated in specifically labeled isotopes. Equivalent samples of maturing siliques collected between 14 and 18 DAF were incubated in 1 μCi mL−1 [1-14C]Glu, [U-14C]Glu, [3,4-14C]Glc, or [U-14C]Glc as described in “Materials and Methods.” Values are presented as cumulative 14CO2 release and are means ± se of four biological replicates. WT, Wild type.
Figure 5.
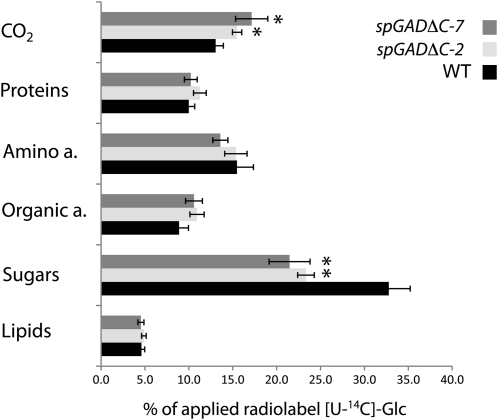
Effect of deregulated Glu decarboxylation during seed maturation on the metabolism of [U-14C]Glc by stratified seeds. Tissue was fractionated exactly as described in “Materials and Methods.” Seeds were preincubated in 200 μL of MES-KOH buffer (pH 6.4) containing 2 mm unlabeled precursor. After 1 h of acclimation, 200 μL of buffer containing specific radiolabel was added. 14CO2 was collected using CO2 traps containing 200 μL of 10 mm KOH as described by ap Rees and Beevers (1960) and Nunes-Nesi et al. (2005). Collected seeds were frozen immediately in liquid N after washing two times with 1.5 mL of MES-KOH buffer. The material was extracted using combined and modified methods as described in “Materials and Methods.” Values are expressed as percentages of the total radiolabel metabolized and are means ± se of four biological replicates. Asterisks indicate values that were judged to be significantly different from the control (P < 0.05) when tested by Student’s t test. WT, Wild type.
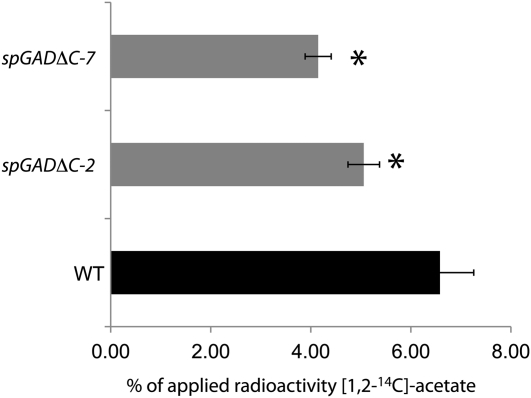
To specifically describe the changes in C allocation in the seed, we next supplemented dry seeds with [U-14C]Glc and measured the incorporation of the radiolabel in the polar neutral, acidic, and basic fractions and in the lipid fraction of the seed extracts. The metabolism of the radiolabeled compound in the transgenic lines was found to be markedly different from that in the wild type (Fig. 5): the proportion of radiolabel incorporated into sugars was significantly lower in both spGADΔC lines than in the wild type, a finding that could suggest increased use of added Glc in the transgenic lines to produce compounds other than sugars. The latter scenario is supported by increased incorporation of radiolabel in organic acids and CO2 evolution in the transgenic seeds (Fig. 5). For seeds fed with [1,2-14C]acetate, the label incorporation into lipids in the transgenic lines was up to 30% lower than its incorporation in the wild type, which suggested a decrease in C flow toward lipid biosynthesis (Fig. 6), while the amounts of radiolabel metabolized to compounds other than lipids were equivalent to those in the wild type (Supplemental Table S2).
Figure 6.
Effect of deregulated Glu decarboxylation during seed maturation on the incorporation of [1,2-14C]acetate in lipids by stratified seeds. Tissue was fractionated exactly as described in “Materials and Methods.” Values are expressed as percentages of the total radiolabel metabolized and are means ± se of four biological replicates. Asterisks indicate values that were judged to be significantly different from the wild type (WT; P < 0.05).
Up-Regulation of the GABA Shunt Leads to the Alteration of Storage Reserve Accumulation and Fatty Acid Metabolism
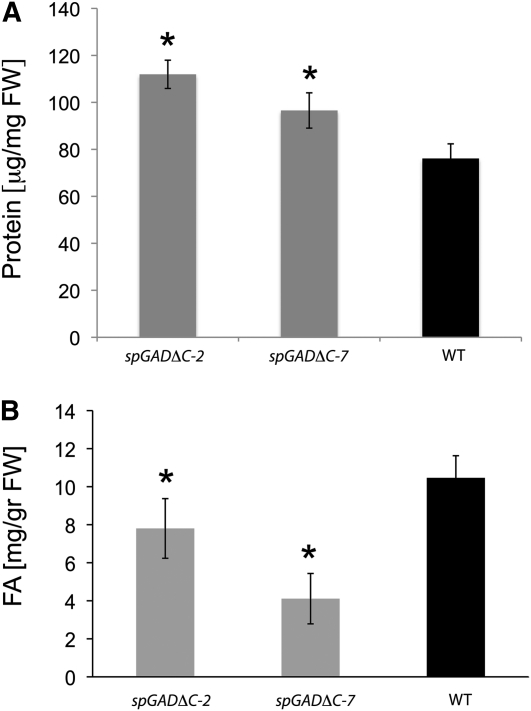
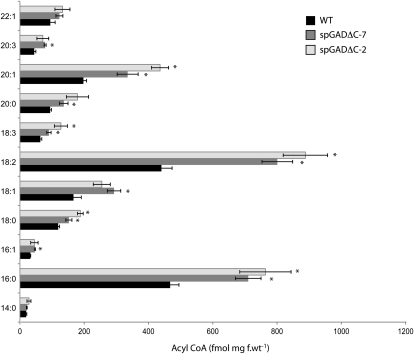
Accumulation of storage reserves in dry seeds was evaluated in terms of the contents of protein, total fatty acids (TFA), and CoA derivatives in spGADΔC-2 and spGADΔC-7 versus the wild type. In keeping with the general effect on amino acids, transgenic seeds displayed considerable accumulation of protein at maturity (Fig. 7A; i.e. 26.8% and 47% higher protein content in spGADΔC-2 and spGADΔC-7 than in the wild type, respectively). In contrast, HPLC analysis of TFA revealed markedly reduced levels of TFA in the transgenic seeds, with spGADΔC-2 and spGADΔC-7 displaying on average 30% and 70% less TFA than the wild type (Fig. 7B). However, there was a general accumulation of acyl-CoAs in the dry seeds, with a notable 2-fold increase for 16:0, 18:2, 20:1, and 20:3 fatty acids in both transgenics as compared with the wild type (Fig. 8).
Figure 7.
Protein (A) and TFA (B) content in mature seeds of wild-type (WT) and transgenic plants. The effect of up-regulated GABA metabolism on the content of seed proteins and TFAs was measured by standard Bradford protocol and a GC-based protocol as described in “Materials and Methods.” Results are presented as means ± se of at least four biological replicates of 20 mg of seeds. FW, Fresh weight.
Figure 8.
Content of acyl-etheno-CoA derivatives in mature seeds of wild-type (WT) and transgenic plants. Twenty-milligram portions of material were frozen in liquid N and extracted for subsequent quantitative analysis of fluorescent acyl-etheno-CoA derivatives by HPLC (Larson and Graham, 2001; Larson et al., 2002; Sayanova et al., 2007). Values represent means ± sd of four biological replicates. Asterisks represent significant (P < 0.05) differences with respect to the wild type calculated using Student’s t test. f.wt, Fresh weight.
Genome-Wide Expression Analysis of spGADΔC Transgenics Highlights the Occurrence of Targeted Changes in Genes Involved in Amino Acid, Mitochondrial Electron Transport, and the TCA Cycle and Storage Reserve Metabolism
To understand the role of transcriptional regulation in the changes observed at the metabolite level and to estimate the impact of enhanced GABA production on the gene transcriptional program of the seed, we performed microarray analysis on the dry stored seeds, transgenics versus the wild type, from the same material collected for metabolite profiling using the Affymetrix AtH1 chip. One-way ANOVA of the microarray data indicated that changes in the level of transcripts included 149 genes significantly (P < 0.001; false discovery rate [FDR] < 0.05) up-regulated and 55 genes down-regulated in the transgenics as compared with the wild type, showing at least 1.5-fold change. The detailed list of genes showing statistically significant changes in expression following ANOVA between the dry seeds of the transgenic line and the wild type, and the calculated fold changes, are provided in Supplemental Tables S3 and S4. The entire data set is provided in Supplemental Table S5. In summary, genes related to metabolism were abundant, and the vast majority were up-regulated. Moreover, overrepresentation analysis using the tools embedded in the PageMan software (http://mapman.mpimpgolm.mpg.de/general/ora/ora.shtml) indicated that among the up-regulated genes (Supplemental Table S3) in the transgenic seeds, there was overrepresentation of the following categories: biotic stress (six genes), redox state (three genes), calcium-associated signaling (six genes), and cell wall metabolism in the transgenic seeds (Table I [overrepresentation analysis]; for the list of up-regulated genes, see Supplemental Table S3). Among the genes associated with metabolism, the most markedly affected were Glu dehydrogenase, Gln synthetase (ATGSR2), Ala:glyoxylate aminotransferase, and S-adenosyl Met 1 synthetase as well as several genes associated with aromatic amino acid metabolism (anthranilate synthase α-subunit 1, anthranilate phosphoribosyltransferase, shikimate kinase) and polyamine catabolism (polyamine oxidase; for fold change changes, see Supplemental Table S3). In contrast, the expression of genes associated with Ser catabolism (Ser hydroxymethyltransferase 4 and Trp synthase) was down-regulated (Supplemental Table S4).
Table I. Functional categories overrepresentation analysis.
Genes that were significantly up-regulated in the mutant compared with the wild type (Supplemental Table S3) were subjected to a functional categories overrepresentation analysis. Overrepresentation analysis was performed with the PageMan tool (see “Materials and Methods”). Only functional categories with two or more genes are shown.
| Main Category | Subcategory | No. of Genes | Pa | Ratiob |
| Miscellaneous | 19 | 0.0001 | 2.6 | |
| Metal handling | 5 | 0.0002 | 9.7 | |
| Binding, chelation, and storage | 4 | 0.0006 | 10.9 | |
| Amino acid metabolism | Trp anthranilate synthase | 2 | 0.0003 | 66.9 |
| Trp | 3 | 0.0004 | 20.1 | |
| Not assigned | 34 | 0.0010 | 0.6 | |
| Protein degradation | Cys protease | 4 | 0.0015 | 8.0 |
| Lipid metabolism | Fatty acid synthesis and fatty acid elongation: long-chain fatty acid CoA ligase | 2 | 0.0020 | 28.7 |
| Signaling | Calcium | 6 | 0.0021 | 4.6 |
| Redox | Glutathione S-transferases | 3 | 0.0030 | 10.4 |
| Stress | Biotic | 6 | 0.0030 | 4.2 |
| Secondary metabolism | N miscellaneous alkaloid like | 3 | 0.0032 | 10.0 |
Wilcoxon’s P value, the probability of whether the response of the genes in a given group is significantly different from that of other genes in the whole genome set (Usadel et al., 2006; http://mapman.mpimp-golm.mpg.de/pageman).
Ratio between the number of genes from a certain functional category that were found significantly up-regulated in spGADΔC compared with the wild type and the expected number of genes when randomly chosen (overrepresentation analysis; http://mapman.mpimpgolm.mpg.de/general/ora/ora.shtml).
The parallel metabolite and transcript analysis indicated that the decrease in the concentrations of a number of TCA cycle intermediates in the seeds of spGADΔC lines was also associated with changes in TCA gene expression patterns. As shown in Supplemental Table S3, changes in the level of TCA cycle-associated transcripts included the up-regulation of succinate dehydrogenase and 2-oxoglutarate dehydrogenase. The transcripts of malic enzyme, which is involved in the conversion of malate to pyruvate, the direct precursor of acetyl-CoA, were up-regulated in the transgenic line. CoA metabolism in relation to fatty acid metabolism was indeed altered at the transcript level, as shown in the up-regulation of long-chain fatty acid CoA ligase and long-chain acyl-CoA synthetase and condensing enzyme 3-ketoacyl-CoA synthase 1, which is involved in fatty acid elongation, and by a down-regulation of fatty acid transport (ATP-binding cassette transporter). In seeds of the transgenic plants, we also found a significant up-regulation of the genes (Supplemental Table S3) involved in the degradation of protein and cellular components (Cys protease and RESPONSIVE TO DESICCATION21 [RD21]) and in N remobilization (ATGSR2), which could explain the increased amino acid content in these seeds. Among the genes up-regulated in the transgenic seeds, a small number are associated with the mobilization of glycosyl groups (e.g. UDP-glycosyltransferase/transferase, glycosyl hydrolase family 3 protein). Among other functions, these enzymes are involved in the metabolism of certain disaccharides and polysaccharides. Other up-regulated genes included those involved in the mitochondrial electron transport chain (i.e. alternative oxidase and FAD-binding domain-containing protein [AT1G26380]). With respect to energy, the expression of transcripts of GTP- and ATP-binding proteins was down-regulated (AT5G56500 for the GTP-binding protein and AT5G57960 and AT1G30960 for the ATP-binding proteins; Supplemental Table S4). Gene Ontology analysis for enrichment in biological processes (http://david.abcc.ncifcrf.gov/) indicated a significant enrichment of genes associated with the electron transport process, which included 13 up-regulated genes and six down-regulated genes (Supplemental Tables S3 and S4). Among these, there was a 3.6-fold down-regulation of cytochrome p450 (CYP71A20 and CYP71A19) and a 2-fold down-regulation at the MATERNAL EFFECT EMBRYO ARREST23 electron carrier involved in embryo development and seed dormancy. Arabidopsis NAC domain-containing protein 92 (ANAC092/ATNAC2/ATNAC6) and DELAY-OF-GERMINATION1 (DOG1), associated with the regulation of germination, were up-regulated in the transgenic seeds.
DISCUSSION
In plants, Glu metabolism is pivotal for efficient N incorporation; therefore, its levels are maintained under tight regulation (Stitt and Fernie, 2003; Forde and Lea, 2007). In tobacco leaves, for example, a mutated GAD from petunia was shown to significantly reduce Glu levels (Baum et al., 1996). GAD is the entry enzyme of the GABA shunt and catalyzes the decarboxylation of Glu to GABA (Bown and Shelp, 1997). Recently, GABA was shown to accumulate to high levels in dry seeds of Arabidopsis, a finding implying a developmental role in seeds for this nonproteogenic amino acid (Fait et al., 2006). Labeling experiments conducted in our study directly confirm that GABA is strongly associated with early seed germination metabolic processes, replenishing the TCA cycle and contributing to amino acid biosynthesis (Fig. 1). These early metabolic events can be of considerable relevance to the imbibing seed, whose level of energy is low and whose storage reserves have not yet been degraded (Lunn and Madsen, 1981; Al-Ani et al., 1985; Borisjuk and Rolletschek, 2009). Regulation of GABA levels probably represents a tuning of the balance between C metabolism and N metabolism. In this study, we investigated the perturbation in seed metabolism following the seed-targeted up-regulation of Glu conversion to GABA during seed maturation. We employed the up-regulation strategy to challenge the seed metabolic network in a development-induced manner with the aim of revealing regulatory aspects of seed metabolism (Angelovici et al., 2009). Our results showed developmental differences in the response of seed metabolism to a deregulated GAD, probably caused by the changing cellular environment between seed maturation and desiccation (Fig. 9). We further showed that enhancing Glu-to-GABA decarboxylation causes a major alteration in the C-N balance and in reserve accumulation, unexpectedly in favor of N. Nonetheless, hyperaccumulation of GABA in the dry seed was not followed by a genome-wide transcriptional response, and the changes in transcript levels were generally in accordance with the metabolic data and included N mobilization, aromatic amino acid metabolism and fatty acid CoA metabolism, electron transport, and energy balance. Integration of our metabolite and transcript data suggests a tighter link between amino acid metabolism and genes involved in the regulation of germination than was previously perceived.
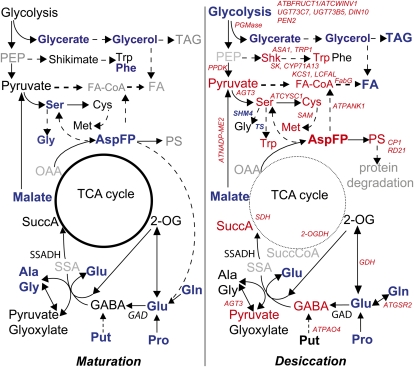
Figure 9.
Schematic summary of major metabolic processes altered in maturing (left) and desiccating (right) spGADΔC seeds. Deregulated Glu-to-GABA metabolism affects differently the metabolic network at seed maturation and seed desiccation. GABA does not accumulate during seed maturation, but amino acid content is generally reduced. In the dry seed, GABA accumulation is associated with an increased content of amino acids, an increased protein content, and a reduction of TFA. Font colors red and blue indicate increase and decrease, respectively, as compared with the control wild type, and gray indicates nonmeasured compounds. Dashed arrows indicate multiple-step pathways. The thickness of the TCA cycle circular scheme refers to the reduced TCA cycle activity during seed desiccation. Clark et al. (2009) suggest that 2-OG-dependent GABA-T activity is not present in plants. In italics are given the genes associated with metabolic processes whose expression was significantly changed in the transgenics (Supplemental Tables S3 and S4). Abbreviations not used in the text are as follows: AGT3, Ala:glyoxylate aminotransferase 3; ASA1, anthranilate synthase α-subunit 1; AspFP, Asp family pathway; ATBFRUCT1/ATCWINV1, Arabidopsis cell wall invertase 1; ATCYSC1, Cys synthase C1; ATGSR2, Gln synthetase 2; ATNADP-ME2, NADP-malic enzyme 2; ATPANK1, pantothenate kinase 1; ATPAO4, polyamine oxidase 4; CP1, Cys proteinase 1; CYP71A13, indoleacetaldoxime dehydratase; DIN10, dark-inducible 10 hydrolase; FA, fatty acids; FabG, fatty acid biosynthesis, 3-oxoacyl-(acyl-carrier protein) reductase; GDH, Glu dehydrogenase; LCFAL, long-chain fatty acid-CoA ligase/long-chain acyl-CoA synthetase; OAA, oxaloacetate; 2-OGDH, 2-oxoglutarate dehydrogenase E2 component (dihydrolipoamide succinyltransferase); PEN2, penetration 2 hydrolase; PGMase, phosphoglycerate mutase; PPDK, pyruvate orthophosphate dikinase; PS, storage protein; SAM1, S-adenosyl-Met synthetase 1; Shk, shikimate; SHM4, Ser hydroxymethyltransferase 4; SK, shikimate kinase; SSA, succinic semialdehyde; SuccA, succinate; SuccCoA, succinyl-CoA; TAG, triacylglycerol; TRP1, anthranilate phosphoribosyltransferase; TS, Trp synthase; UGT73C7 and UGT73B5, UDP-glycosyltransferase.
Up-regulation of N metabolism in the transgenic seeds was associated with decreased levels of malate and of some sugars and sugar alcohols in the dry seed as well as significantly lower TFAs. Taken together, these findings indicate that a finely regulated GABA shunt in maturing seeds is the key to maintaining C-N equilibrium. Moreover, while the link between the TCA cycle and the GABA shunt is well known (Busch and Fromm, 1999; Studart-Guimarães et al., 2007; Bunik and Fernie, 2009), we show here that Glu is metabolized preferentially via GABA (as compared with the alternative Glu-to-2-OG conversion). Our data hence suggest that the Glu-to-GABA conversion represents the main route for Glu incorporation into the TCA cycle. That said, we could not rule out the possibility that a high conversion of 2-OG and its biosynthesis from isocitrate would lead to an underestimation of Glu incorporation into the TCA cycle via 2-OG.
Cellular Conditions Characterizing the Desiccating Seed Lead to the Accumulation of GABA in GAD Transgenic Seeds
A recent review (Angelovici et al., 2010) presents robust evidence that metabolites, transcripts, and the enzymatic machinery accumulate in the dry seed, possibly to ensure the initiation of cellular processes upon water imbibition. There is thus a general shift in seed metabolism during seed desiccation. In this context, our results indicate differential regulation of GABA metabolism at seed maturation (14–17 DAF) and at desiccation (17 DAF to dry seed), which most probably depends on the modulation of GABA catabolism under the changing cellular conditions between maturation and desiccation (Angelovici et al., 2010). The regulation of the GABA shunt is driven by Ca2+, the redox state, and energy constraints (Baum et al., 1996; Busch and Fromm, 1999). Succinic semialdehyde dehydrogenase (SSADH) is the final enzyme of the shunt and as such represents the entry enzyme into the TCA cycle. Indeed, as simplified in the model in Figure 9, in the maturing transgenic seed, the deregulated GAD under the regulation of the phaseolin promoter is active as early as 12 DAF. At this stage, photosynthesis is still taking place in Arabidopsis seeds (Fait et al., 2006; Angelovici et al., 2009) and in the seeds of many other species (Vigeolas et al., 2003; Ruuska et al., 2004; Rolletschek et al., 2005; Tschiersch et al., 2011), with oxygen being evolved at the surface and translocated to the inner part of the seed (Borisjuk and Rolletschek, 2009). This process enables the concomitant production of ATP to sustain metabolic activity (Borisjuk and Rolletschek, 2009). As such, it is likely that the catabolism of GABA via the action of SSADH, supplying succinate to the TCA cycle (Fig. 9, left scheme) during maturation of the seed, prevents the accumulation of GABA in the transgenic seeds. Nonetheless, the effect of a deregulated Glu-to-GABA conversion was reflected in the decreased levels of the vast majority of the free amino acids measured. We thus conclude that the increased use of Glu to produce GABA prevents the maintenance of amino acid levels.
At late maturation and desiccation, the conditions within the seed become increasingly restrictive, reducing conductance and hence limiting gas exchange; the inner part of the endosperm and embryo becomes more and more hypoxic and lacking in energy (Rolletschek et al., 2005). Such conditions inhibit portions of some metabolic pathways, including sections of the TCA cycle (Tretter and Adam-Vizi, 2000; Rocha et al., 2010), thereby inducing a metabolic shift in the developing seeds (Fait et al., 2006). The changed cellular environment is probably the cause of the accumulation of GABA between late maturation and desiccation (Fig. 9, right scheme). Indeed, regulation of the GABA shunt enzymes was shown previously to be highly dependent on the redox state of the cell: Shelp et al. (1995) demonstrated that GABA shunt activity is associated with a hypoxic environment, and the activity of SSADH was proved to be dependent on favorable redox and energy state (Busch and Fromm, 1999). A similar effect was observed in coffee (Coffea arabica) seeds, in which a differential regulation of GABA metabolism was documented across the drying process (Kramer et al., 2010) and GABA accumulation was found to be associated with maximum expression of the dehydrin gene. Although a protective role of GABA during seed desiccation cannot be ruled out, our results suggest that enhanced Glu-to-GABA conversion is a consequence of a metabolic bottleneck at the entrance of the TCA cycle. GABA-deregulated production occurs in the desiccating seed at the expense of another stress-related amino acid, Pro, which directly competes for the same Glu pools. However, surprisingly, hyperaccumulation of GABA in the dry seeds eventually results in the accumulation of numerous amino acids, including a substantial but unexpected up-regulation of Trp metabolism via the shikimate pathway (Fig. 9), coupled with a significant accumulation of Asp, Asn, Ser, Cys, Met, Trp, and total protein. Generally higher levels of the various proteogenic amino acids were also shown in GABA-enriched rice (Oryza sativa) grains expressing a truncated OsGAD2 (Akama et al., 2009). It is possible that GABA-induced amino acid biosynthesis occurs via the TCA cycle, as suggested by our labeling experiments. Alternatively, accumulating GABA could be transaminated by GABA-T to produce Ala from pyruvate, or Gly from glyoxylate to Gly (Clark et al., 2009), thus contributing to amino group incorporation into the C skeleton and partly explaining the depletion of intermediates of C metabolism and fatty acid biosynthesis. This interpretation receives support from the up-regulation of Ala-glyoxylate transaminase. Nonetheless, we found pyruvate content to be higher in the transgenic seeds and pyruvate biosynthesis to be up-regulated (Fig. 9). A third and more plausible hypothesis is that increased amino acid content could be the result of protein degradation, as suggested by our transcript data (see below and Fig. 9), concomitant with increased protein content in the seed. Indirect evidence supporting this hypothesis may be drawn from the induction of protein degradation in response to accumulating GABA, which was shown in our study (Fig. 9; Supplemental Table S3) and that of Roberts (2007) to activate stress-associated protein degradation processes.
Interaction between Storage Reserve Accumulation and GABA Shunt Regulation
Our results show that in dry transgenic Arabidopsis seeds, the increase in amino acids and the depletion of TCA cycle intermediates are accompanied by an increased protein content, a decrease in TFA, and an accumulation of short- to medium-chain fatty acid CoAs. Impaired fatty acid metabolism in the transgenic seeds could be associated with increased GABA production in three different ways. First, a reduction of TCA cycle activity would probably result in impaired production of CoA moieties. Second, the low levels of malate associated with increased GABA accumulation could have a detrimental effect on fatty acid elongation and fatty acid buildup. Among the TCA cycle intermediates measured in our analysis, malate showed the most marked drop in concentration. It has previously been suggested that malate is the preferred substrate for fatty acid elongation not requiring ATP or a reductant (Smith et al., 1992; Pleite et al., 2005), which are limited in desiccating seeds. Confirming the findings of Smith et al. (1992), Kendrick and Ratledge (1992) showed that a structural analog of malate, tartronic acid, which is an inhibitor of malic enzyme, could inhibit the malate-induced stimulation of fatty acyl group desaturation and elongation in the microsomal membranes of Mucor circinelloides. In our study, an impaired TCA cycle was also reflected in the alteration of genes associated with mitochondrial electron transport and particularly in the up-regulation of alternative oxidase genes. In further support of our metabolite and transcript data, extensive analysis of transgenic tomato (Solanum lycopersicum) plants deficient in the expression of various enzymes of the TCA cycle showed impaired electron transport and photosynthesis (Nunes-Nesi et al., 2011). A third and more speculative possibility is supported by evidence from radiolabeling experiments in germinating lupines. A direct interaction between fatty acid metabolism and amino acids was shown in yellow lupine (Lupinus luteus) seedlings (Borek et al., 2003). It was demonstrated that, in the seedling axis, C atoms from acetate were incorporated mainly into amino acids upon germination. This direct metabolic cross talk might have had a feedback inhibition on the accumulation of fatty acids during seed development. It is intriguing that the same acyl-CoA derivatives found to significantly accumulate in this study were shown to change in a similar direction when a Brassica napus aminoalcoholphosphotransferase (AAPT1) was overexpressed in Arabidopsis (Qi et al., 2003). AAPT1 governs a reaction downstream of Ser catabolism, converting CDP-ethanolamine + 1,2-diacyl-sn-glycerol into CMP + α-phosphatidylethanolamine. The accumulation of Ser in our transgenic lines might have led to the availability of increased substrate and thus mimicked the effect of the overexpression reported by Qi et al. (2003). Last, Asp can be converted to pantothenate via β-Ala and support CoA biosynthesis. Our parallel metabolite and transcript analyses indicated an increase in Asp and pantothenate kinase 1, the latter encoding the enzyme downstream of pantothenate biosynthesis toward CoA production. As such, the increase in acyl-CoAs can be directly linked to the up-regulation in amino acid metabolism induced by the accumulation of GABA. The increased total protein content in the transgenic seeds can be explained by the higher availability of amino acids. Branched amino acids have been shown to induce the initiation of translation and to stimulate protein synthesis in the skeletal muscles of animals (Yoshizawa, 2004). Unfortunately, we are aware of only a few studies comprehensively attempting to link changes in single amino acids with global metabolism and protein content, particularly in seeds. In rice, transgenic seeds accumulating high levels of Trp did not exhibit substantial changes in the content of other amino acids (Dubouzet et al., 2007). That said, the seminal study of Petrie and Wood (1938) revealed positive associations between levels of amino acids and proteins in illuminated leaves. It is clear, however, that additional studies are required to elucidate the link between amino acids and the metabolism of storage reserves and to assess their relevance during seed germination. While speculative, the relation between these lines of evidence is supported by the similar regulation shared by key chloroplastic enzymes of starch, lipid, and amino acid synthesis (Geigenberger et al., 2005, and refs. therein).
Finally, in our study, the metabolic changes driven by the hyperaccumulation of GABA during seed maturation were found to be associated with a reduced germination phenotype. The increased expression of DOG1, which is one of the major regulators of seed dormancy (Bentsink et al., 2010), and ANAC092/ATNAC2/ATNAC6 in the spGADΔC transgenic seeds are probably involved in the altered germination phenotype (Balazadeh et al., 2010) and are indicative of a link between GABA accumulation and the regulation of germination. In addition, genes associated with remobilization processes (ATGSR2, RD21) and senescence (SENESCENCE-ASSOCIATED GENE21 [SAG21]) were up-regulated. For example, RD21 is an Arabidopsis Cys protease that was identified as dehydration responsive in Arabidopsis. Its pattern of expression, initiated upon imbibition in radish (Raphanus sativus; Kikuchi et al., 2008), hints at a role in the degradation of cellular materials feeding the growing embryo (Rojo et al., 2003). RD21 has been found to be associated with senescence and with SAG21, a gene associated with senescence initiation and responsive to Glc (Weaver et al., 1998); SAG21 was also increased in the transgenic seeds. Last, the up-regulation of polyamine catabolism (Fig. 9) and Gln synthetase (the plastidial form GS2, ATGSR2) was found to be linked to the remobilization of N, probably triggered by a decreased content of Glu and Gln (Glass et al., 2002; Moschou et al., 2008).
CONCLUSION
Deregulated Glu-to-GABA catabolism can lead to dramatic alterations in fatty acid metabolism and in the balance between N and C moieties during seed maturation. Unexpectedly, we found that the balance was directed toward a developmentally induced increased N-to-C ratio. Although, according to the scientific literature, up to 95% of seed protein is derived from amino acids that are exported to the seed after degradation of existing protein in the leaves, we showed that altered activity of this pathway during seed maturation could cause metabolic processes specific to the embryo (driven by the phaseolin promoter) to change the C-N balance in storage reserves. The parallel metabolite and transcript profiles indicate an intriguing link between the induction of nonprotein amino acid metabolism and fatty acid assembly, N remobilization, programmed cell death, and the major regulatory processes of germination. The above lines of evidence open new directions for research on the molecular mechanisms involved and their concerted regulation in seed maturation, which will contribute to our current understanding of seed biology.
MATERIALS AND METHODS
Chemicals
All chemicals were purchased from Sigma-Aldrich Israel, with the exception of N-methyl-N-[trimethylsilyl] trifluoroacetamide, which was obtained from Macherey-Nagel.
Production of Transgenic Plants
A strategy identical to the one described by Karchi et al. (1994) was used to transform wild-type Arabidopsis (Arabidopsis thaliana) with a chimeric gene encoding a GAD isolated from petunia (Petunia hybrida; Baum et al., 1993) fused to a promoter derived from the bean (Phaseolus vulgaris) phaseolin storage protein (Phas). cDNA coding either the 58-kD petunia calmodulin-binding GAD (Baum et al., 1993) or a mutant GAD lacking 27 amino acids at the C terminus (Arazi et al., 1995) was introduced downstream of the Phas promoter and upstream of a 3′ untranslated region of the octopine synthase gene from Agrobaterium tumefaciens, essentially as described by Karchi et al. (1994). Transgenic Arabidopsis plants harboring the chimeric DNA constructs were prepared by inflorescence-dipping transformation (Clough and Bent, 1998). Seeds obtained from the transformed plants were considered as T1 seeds. These seeds were collected and surface sterilized for 10 min in diluted bleach solution containing a few drops of Tween 20, followed by three rinses with sterile distilled water. The seeds were then plated on a selective medium containing kanamycin and carbenicillin. The resistant T1 seeds were grown in soil to raise T1 plants, which were also grown to maturity. The T2 seeds were collected. Using the same procedure, we obtained the T3 and T4 generations.
Plant Growth and Seed Collection
Collection of seeds and siliques was performed essentially as described previously (Fait et al., 2006; Angelovici et al., 2008). Arabidopsis Ws seeds of the wild type and spGAD and spGADΔC transgenic lines were germinated on soil and grown for two consecutive rounds in a greenhouse (21°C) under the same light regime with a minimum of 250 μmol photons m–2 s–1. Flowers were tagged on the day of anthesis and at 14 ± 1 and 17 ± 1 DAF. Mature siliques were collected and cryolyophilized to dryness, and seeds were then dissected from the dried siliques and immediately frozen in liquid N. Mature dry seeds were collected at the end of the desiccation period, allowed to dry in paper bags for 4 weeks at room temperature, and stored at 4°C until further analysis. Seed viability was assayed by germinating seeds on agar plates, following 72 h of stratification. The tetrazolium assay was performed exactly as described by Wharton (1955).
GC-MS Analysis of Seed Metabolites
Siliques representing each developmental stage were collected as described above from at least 30 individual plants divided into at least four groups unless otherwise stated. Approximately 10 mg of seeds was utilized for each extraction. Metabolites were extracted and analyzed with a GC-MS-based protocol optimized for Arabidopsis (Lisec et al., 2006) by adjusting the extraction protocol to seed material as described by Fait et al. (2006). Relative metabolite content was calculated as described by Roessner et al. (2001) following peak identification using TagFinder (Luedemann et al., 2008). Substances were identified by comparison with mass spectral tags represented in query database (Kopka et al., 2005; Erban et al., 2007). The protein concentration of ethanolic seed extracts was determined by the assay of Bradford (1976).
Measurements of Total Protein and TFAs, and HPLC Analysis of Acyl-Etheno-CoA Derivatives
Twenty-milligram portions of material were frozen in liquid N and extracted for subsequent quantitative analysis of fluorescent acyl-etheno-CoA derivatives by HPLC. Analysis of acyl-CoA was performed using an Agilent 1100 LC system [Phenomenex LUNA 150 3 2 mm C18(2) column]; the methodology and gradient conditions have been described previously (Larson and Graham, 2001; Larson et al., 2002, Sayanova et al., 2007). The concentration of protein extracted in boiling ethanol was quantified using the assay of Bradford (1976). Fatty acid methyl esters were quantified by GC with pentadecanoic acid (15:0) as the internal standard (Browse et al., 1986).
Labeling of Siliques and Seed Material
For radiolabeling of siliques and seeds, 1 μCi mL−1 solutions of the following radiolabeled precursors were used: [U-14C]Glc (specific activity, 11.1 GBq mmol−1), [U-14C]acetate (specific activity, 2.07 GBq mmol−1), [U-14C]Glu (specific activity, 9.36 GBq mmol−1), and [C1-14C]Glu (specific activity, 2.22 GBq mmol−1). To measure the release of 14CO2 during the maturation period, flowers were tagged at anthesis, and siliques were collected between 14 and 18 DAF from at least 10 plants per genotype. Intact siliques were submerged in a solution of 200 μL of MES-KOH buffer (pH 6.4) containing 2 mm nonlabeled precursor. After 1 h of acclimation, 200 μL of buffer containing the specific radiolabel was added. To measure the release of 14CO2 from imbibed seeds, 10 mg of dried seeds was stratified (1 mL of double-distilled water, 4°C, darkness) for 72 h. Four hours prior to harvesting, the water was replaced with 200 μL of MES-KOH buffer (pH 6.4) containing 2 mm nonlabeled precursor. After 1 h of acclimation, 200 μL of buffer containing the specific radiolabel was added. CO2 was collected 2 and 4 h following incubation by using CO2 traps containing 200 μL of 10 mm KOH, as described by ap Rees and Beevers (1960) and Nunes-Nesi et al. (2005).
Fractionation of 14C-Labeled Seed Extracts
Collected seeds were frozen immediately in liquid N after washing twice with 1.5 mL of MES-KOH buffer. The material was extracted using a modification of the method described by Roessner et al. (2001), Lisec et al. (2006), and Bligh and Dyer (1959). To inhibit enzyme activity, the seeds were ground with 950 μL of a methanol-chloroform mixture in a ratio of 2:1. After shaking the homogenate for 15 min at room temperature, the extract was centrifuged for 10 min at 14,000 rpm. The procedure was repeated for a second time, and each supernatant was transferred to a 4-mL glass vial. The supernatant was mixed with 625 μL of chloroform and 625 μL of water. For phase separation of the apolar and polar phases, the mixture was centrifuged for 30 min at 4,000 rpm. To analyze the lipid fraction, 800 μL of the lower apolar phase was dried on a filter paper with a N sample concentrator to prevent the quenching effects of the chloroform. For the fractionation of the upper polar phase, a 1,140-μL aliquot was taken and dried under vacuum overnight. To prevent contamination by the interphase, the remainder of the polar phase was discarded. After drying, the almost invisible pellet was suspended in 2 mL of water. Afterward, it was fractionated into a neutral fraction containing the sugars, an acidic fraction containing the organic acids, and a basic fraction containing the amino acids, as described by Quick et al. (1989) and Geigenberger et al. (1997). To this end, 1 mL of the aqueous solution was filtered over in-house-made columns containing specific anion- and cation-exchange resins. The columns were washed three times, each time with 1 mL of double-distilled water, and the solutions that passed through the columns were combined to give the neutral fraction. To extract the acidic fraction, the anion-exchange column was washed three times, each time with 1 mL of 4 m formic acid. To obtain the basic fraction, the cation-exchange column was washed three times, each time with 1 mL of 1 m NH4OH. For sugar determination, an aliquot of 200 μL of the neutral fraction was treated with 4 units mL−1 hexokinase (25°C, 4 h). In parallel, another aliquot of the same fraction was treated with 1 unit mL−1 Glc oxidase and 32 units mL−1 peroxidase (25°C, 6 h). After heat inactivation of the enzymes (95°C, 5 min), the solution was also fractionated into neutral, anionic, and cationic fractions.
The pellet remaining after the extraction of the methanol-chloroform-soluble fraction was dried and resuspended in 0.5 mL of double-distilled water. An aliquot of 200 μL was treated with 10 units of amyloglucosidase (37°C, overnight) followed by 10 units of pronase (37°C, overnight). The enzymes were inactivated by heat treatment (95°C, 5 min), and the mixture was filtered and fractionated into a neutral fraction containing starch, an acidic fraction containing the cell wall fragments, and a basic fraction containing the proteins. Fractionation on ion-exchange columns was performed as described before (Runquist and Kruger, 1999).
13C Feeding Experiments
For 13C feeding experiments, seeds (10 mg for each of six replicates) were stratified in 10 mm MES-KOH for 72 h at 4°C in the dark and then transferred to continuous light at 21°C for 24 h. Six hours prior to the end of the stratification period and at the end of the 24-h germination period (i.e. 1 DAS), imbibed seeds were fed with a substrate according to a modified method of Morcuende et al. (1998) in a solution containing 10 mm MES-KOH (pH 6.5) and 15 mm [U-13C]Glu, and the incubation was left to proceed for 6 h. At the end of this time, seeds were removed, washed carefully with 10 mm MES-KOH (pH 6.5), allowed to stand briefly on filter paper to remove the solution, weighed, frozen in liquid N, and stored at −80°C until further analysis. Samples were then extracted and evaluated exactly as described by Roessner-Tunali et al. (2004). Label redistribution was determined as described by Giegé et al. (2003) and Tieman et al. (2006).
RNA Extraction and Microarray Analysis
All experiments analyzing RNA expression levels were carried out using two replicates of seed materials, obtained from two independent seed lots, from plants grown under controlled-environment conditions. Total RNA was extracted from stored dry seeds as described previously (Angelovici et al., 2009). Total RNA was treated with DNase RQ-1 (Promega); thereafter, RNA was amplified by using two-cycle Affymetrix labeling according to the standard Affymetrix protocol. Hybridization, labeling, scanning, and data extraction were performed according to the standard Affymetrix protocols. Transcriptome analysis was carried out with Partek Genome Suite software (www.partek.com). Preprocessing was carried out using the Robust Microarray Averaging algorithm (Irizarry et al., 2003). One-way ANOVA was performed. Overrepresentation/underrepresentation analysis was performed by PageMan (http://mapman.mpimp-golm.mpg.de/general/ora/ora.shtml; Usadel et al., 2006). Enriched categories were identified using the Database for Annotation, Visualization, and Integrated Discovery version 6.7 Web-accessible program (http://david.abcc.ncifcrf.gov/). Interpretation of the gene expression data set with respect to metabolic pathways and other functional categories was aided by visualizing changes in gene expression via the MapMan software (Usadel et al., 2005).
Statistical Analysis
Statistical analysis was performed on the data sets obtained from metabolite profiling with the software package TMEV (Saeed et al., 2003). Prior to the analysis, data were log transformed. In addition, to test statistical significance between specific time points, t tests were performed using the algorithm incorporated into Microsoft Excel with Bonferroni correction of the critical P value for multiple comparisons.
Supplemental Data
Supplemental Table S1. Log10-transformed mean fold change in the metabolite content of developing seeds of spGADΔC transgenics and the wild type relative to that in wild-type dry seeds.
Supplemental Table S2. The incorporation of [1,2-14C]acetate in the polar neutral, acidic, and basic fractions and in the lipid fraction of the seed extracts.
Supplemental Table S3. Calculated ratios of average expression levels of genes exhibiting significantly up-regulated expression in spGADΔC-2 as compared with the wild type in dry seeds (P < 0.05 after FDR).
Supplemental Table S4. Calculated ratios of average expression levels of genes exhibiting significantly down-regulated expression in spGADΔC-2 as compared with the wild type in dry seeds (P < 0.05 after FDR).
Supplemental Table S5. Full data set of genome-wide gene expression profiles of dry seed of spGADΔC-2 and the wild type.
Acknowledgments
We thank Hillel Fromm for critical reading of the manuscript.
References
- Akama K, Kanetou J, Shimosaki S, Kawakami K, Tsuchikura S, Takaiwa F. (2009) Seed-specific expression of truncated OsGAD2 produces GABA-enriched rice grains that influence a decrease in blood pressure in spontaneously hypertensive rats. Transgenic Res 18: 865–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akihiro T, Koike S, Tani R, Tominaga T, Watanabe S, Iijima Y, Aoki K, Shibata D, Ashihara H, Matsukura C, et al. (2008) Biochemical mechanism on GABA accumulation during fruit development in tomato. Plant Cell Physiol 49: 1378–1389 [DOI] [PubMed] [Google Scholar]
- Al-Ani A, Bruzau F, Raymond P, Saint-Ges V, Leblanc JM, Pradet A. (1985) Germination, respiration, and adenylate energy charge of seeds at various oxygen partial pressures. Plant Physiol 79: 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Fait A, Zhu XH, Szymanski J, Feldmesser E, Fernie AR, Galili G. (2009) Deciphering transcriptional and metabolic networks associated with lysine metabolism during Arabidopsis seed development. Plant Physiol 151: 2058–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Galili G, Fernie AR, Fait A. (2010) Seed desiccation: a bridge between maturation and germination. Trends Plant Sci 15: 211–218 [DOI] [PubMed] [Google Scholar]
- Arazi T, Baum G, Snedden WA, Shelp BJ, Fromm H. (1995) Molecular and biochemical analysis of calmodulin interactions with the calmodulin-binding domain of plant glutamate decarboxylase. Plant Physiol 108: 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Köhler B, Mueller-Roeber B. (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62: 250–264 [DOI] [PubMed] [Google Scholar]
- Baum G, Chen Y, Arazi T, Takatsuji H, Fromm H. (1993) A plant glutamate decarboxylase containing a calmodulin binding domain: cloning, sequence, and functional analysis. J Biol Chem 268: 19610–19617 [PubMed] [Google Scholar]
- Baum G, Lev-Yadun S, Fridmann Y, Arazi T, Katsnelson H, Zik M, Fromm H. (1996) Calmodulin binding to glutamate decarboxylase is required for regulation of glutamate and GABA metabolism and normal development in plants. EMBO J 15: 2988–2996 [PMC free article] [PubMed] [Google Scholar]
- Baxter CJ, Liu JL, Fernie AR, Sweetlove LJ. (2007) Determination of metabolic fluxes in a non-steady-state system. Phytochemistry 68: 2313–2319 [DOI] [PubMed] [Google Scholar]
- Bentsink L, Hanson J, Hanhart CJ, Blankestijn-de Vries H, Coltrane C, Keizer P, El-Lithy M, Alonso-Blanco C, de Andrés MT, Reymond M, et al. (2010) Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proc Natl Acad Sci USA 107: 4264–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuvé N, Rispail N, Laine P, Cliquet J-b, Ourry A, Le Deunff E. (2004) Putative role of γ-aminobutyric acid as a long-distance signal in up-regulation of nitrate uptake in Brassica napus L. Plant Cell Environ 27: 1035–1046 [Google Scholar]
- Bligh EG, Dyer WJ. (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Borek S, Ratajczak W, Ratajczak L. (2003) A transfer of carbon atoms from fatty acids to sugars and amino acids in yellow lupine (Lupinus luteus L.) seedlings. J Plant Physiol 160: 539–545 [DOI] [PubMed] [Google Scholar]
- Borisjuk L, Rolletschek H. (2009) The oxygen status of the developing seed. New Phytol 182: 17–30 [DOI] [PubMed] [Google Scholar]
- Bouché N, Fromm H. (2004) GABA in plants: just a metabolite? Trends Plant Sci 9: 110–115 [DOI] [PubMed] [Google Scholar]
- Bouché N, Lacombe B, Fromm H. (2003) GABA signaling: a conserved and ubiquitous mechanism. Trends Cell Biol 13: 607–610 [DOI] [PubMed] [Google Scholar]
- Bown AW, Macgregor KB, Shelp BJ. (2006) Gamma-aminobutyrate: defense against invertebrate pests? Trends Plant Sci 11: 424–427 [DOI] [PubMed] [Google Scholar]
- Bown AW, Shelp BJ. (1997) The metabolism and functions of γ-aminobutyric acid. Plant Physiol 115: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Breitkreuz KE, Shelp BJ, Fischer WN, Schwacke R, Rentsch D. (1999) Identification and characterization of GABA, proline and quaternary ammonium compound transporters from Arabidopsis thaliana. FEBS Lett 450: 280–284 [DOI] [PubMed] [Google Scholar]
- Browse J, McCourt PJ, Somerville CR. (1986) Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem 152: 141–145 [DOI] [PubMed] [Google Scholar]
- Bunik VI, Fernie AR. (2009) Metabolic control exerted by the 2-oxoglutarate dehydrogenase reaction: a cross-kingdom comparison of the crossroad between energy production and nitrogen assimilation. Biochem J 422: 405–421 [DOI] [PubMed] [Google Scholar]
- Busch KB, Fromm H. (1999) Plant succinic semialdehyde dehydrogenase: cloning, purification, localization in mitochondria, and regulation by adenine nucleotides. Plant Physiol 121: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Baxter C, Usadel B, Urbanczyk-Wochniak E, Zanor MI, Nunes-Nesi A, Nikiforova V, Centero D, Ratzka A, Pauly M, et al. (2006) Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol 142: 1380–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Baum G, Fromm H. (1994) The 58-kilodalton calmodulin-binding glutamate decarboxylase is a ubiquitous protein in petunia organs and its expression is developmentally regulated. Plant Physiol 106: 1381–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrot R, Rosen R, Haudecoeur E, Cirou A, Shelp BJ, Ron E, Faure D. (2006) GABA controls the level of quorum-sensing signal in Agrobacterium tumefaciens. Proc Natl Acad Sci USA 103: 7460–7464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SM, Di Leo R, Dhanoa PK, Van Cauwenberghe OR, Mullen RT, Shelp BJ. (2009) Biochemical characterization, mitochondrial localization, expression, and potential functions for an Arabidopsis gamma-aminobutyrate transaminase that utilizes both pyruvate and glyoxylate. J Exp Bot 60: 1743–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dubouzet JG, Ishihara A, Matsuda F, Miyagawa H, Iwata H, Wakasa K. (2007) Integrated metabolomic and transcriptomic analyses of high-tryptophan rice expressing a mutant anthranilate synthase alpha subunit. J Exp Bot 58: 3309–3321 [DOI] [PubMed] [Google Scholar]
- Erban A, Schauer N, Fernie AR, Kopka J. (2007) Nonsupervised construction and application of mass spectral and retention time index libraries from time-of-flight gas chromatography-mass spectrometry metabolite profiles. Methods Mol Biol 358: 19–38 [DOI] [PubMed] [Google Scholar]
- Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochniak E, Fernie AR, Galili G. (2006) Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol 142: 839–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A, Fromm H, Walter D, Galili G, Fernie AR. (2008) Highway or byway: the metabolic role of the GABA shunt in plants. Trends Plant Sci 13: 14–19 [DOI] [PubMed] [Google Scholar]
- Forde BG, Lea PJ. (2007) Glutamate in plants: metabolism, regulation, and signalling. J Exp Bot 58: 2339–2358 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Kolbe A, Tiessen A. (2005) Redox regulation of carbon storage and partitioning in response to light and sugars. J Exp Bot 56: 1469–1479 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Reimholz R, Geiger M, Merlo L, Canale V, Stitt M. (1997) Regulation of sucrose and starch metabolism in potato tubers in response to short-term water deficit. Planta 201: 502–518 [Google Scholar]
- Giegé P, Heazlewood JL, Roessner-Tunali U, Millar AH, Fernie AR, Leaver CJ, Sweetlove LJ. (2003) Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15: 2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass ADM, Britto DT, Kaiser BN, Kinghorn JR, Kronzucker HJ, Kumar A, Okamoto M, Rawat S, Siddiqi MY, Unkles SE, et al. (2002) The regulation of nitrate and ammonium transport systems in plants. J Exp Bot 53: 855–864 [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJJ. (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Karchi H, Shaul O, Galili G. (1994) Lysine synthesis and catabolism are coordinately regulated during tobacco seed development. Proc Natl Acad Sci USA 91: 2577–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick A, Ratledge C. (1992) Desaturation of polyunsaturated fatty acids in Mucor circinelloides and the involvement of a novel membrane-bound malic enzyme. Eur J Biochem 209: 667–673 [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Saika H, Yuasa K, Nagahama M, Tsuji A. (2008) Isolation and biochemical characterization of two forms of RD21 from cotyledons of daikon radish (Raphanus sativus). J Biochem 144: 789–798 [DOI] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. (2005) GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Kramer D, Breitenstein B, Kleinwächter M, Selmar D. (2010) Stress metabolism in green coffee beans (Coffea arabica L.): expression of dehydrins and accumulation of GABA during drying. Plant Cell Physiol 51: 546–553 [DOI] [PubMed] [Google Scholar]
- Larson TR, Edgell T, Byrne J, Dehesh K, Graham IA. (2002) Acyl CoA profiles of transgenic plants that accumulate medium-chain fatty acids indicate inefficient storage lipid synthesis in developing oilseeds. Plant J 32: 519–527 [DOI] [PubMed] [Google Scholar]
- Larson TR, Graham IA. (2001) A novel technique for the sensitive quantification of acyl CoA esters from plant tissues. Plant J 25: 115–125 [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Luedemann A, Strassburg K, Erban A, Kopka J. (2008) TagFinder for the quantitative analysis of gas chromatography-mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 24: 732–737 [DOI] [PubMed] [Google Scholar]
- Lunn G, Madsen E. (1981) ATP-levels of germinating seeds in relation to vigor. Physiol Plant 53: 164–169 [Google Scholar]
- Masclaux-Daubresse C, Valadier MH, Carrayol E, Reisdorf-Cren M, Hirel B. (2002) Diurnal changes in the expression of glutamate dehydrogenase and nitrate reductase are involved in the C/N balance of tobacco source leaves. Plant Cell Environ 25: 1451–1462 [Google Scholar]
- McLean MD, Yevtushenko DP, Deschene A, Van Cauwenberghe OR, Makhmoudova A, Potter JW, Bown AW, Shelp BJ. (2003) Overexpression of glutamate decarboxylase in transgenic tobacco plants confers resistance to the northern root-knot nematode. Mol Breed 11: 277–285 [Google Scholar]
- Morcuende R, Krapp A, Stitt M. (1998) Sucrose-feeding leads to increased rates of nitrate assimilation, increased rates of 2-oxoglutarate synthesis and increased synthesis of a wide spectrum of amino acids in tobacco leaves. Planta 206: 394–409 [Google Scholar]
- Moschou PN, Sanmartin M, Andriopoulou AH, Rojo E, Sanchez-Serrano JJ, Roubelakis-Angelakis KA. (2008) Bridging the gap between plant and mammalian polyamine catabolism: a novel peroxisomal polyamine oxidase responsible for a full back-conversion pathway in Arabidopsis. Plant Physiol 147: 1845–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Araújo WL, Fernie AR. (2011) Targeting mitochondrial metabolism and machinery as a means to enhance photosynthesis. Plant Physiol 155: 101–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, Fernie AR. (2005) Enhancing crop yield in solanaceous species through the genetic manipulation of energy metabolism. Biochem Soc Trans 33: 1430–1434 [DOI] [PubMed] [Google Scholar]
- Palanivelu R, Brass L, Edlund AF, Preuss D. (2003) Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 114: 47–59 [DOI] [PubMed] [Google Scholar]
- Petrie AHK, Wood JG. (1938) Studies on the nitrogen metabolism of plants. III. On the effect of water content on the relationship between proteins and amino-acids. Ann Bot ii: 33 [Google Scholar]
- Pleite R, Pike MJ, Garcés R, Martínez-Force E, Rawsthorne S. (2005) The sources of carbon and reducing power for fatty acid synthesis in the heterotrophic plastids of developing sunflower (Helianthus annuus L.) embryos. J Exp Bot 56: 1297–1303 [DOI] [PubMed] [Google Scholar]
- Qi Q, Huang YF, Cutler AJ, Abrams SR, Taylor DC. (2003) Molecular and biochemical characterization of an aminoalcoholphosphotransferase (AAPT1) from Brassica napus: effects of low temperature and abscisic acid treatments on AAPT expression in Arabidopsis plants and effects of over-expression of BnAAPT1 in transgenic Arabidopsis. Planta 217: 547–558 [DOI] [PubMed] [Google Scholar]
- Quick P, Siegl G, Neuhaus E, Feil R, Stitt M. (1989) Short-term water stress leads to a stimulation of sucrose synthesis by activating sucrose-phosphate synthase. Planta 177: 535–546 [DOI] [PubMed] [Google Scholar]
- Rees TA, Beevers H. (1960) Pathways of glucose dissimilation in carrot slices. Plant Physiol 35: 830–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MR. (2007) Does GABA act as a signal in plants? Hints from molecular studies. Plant Signal Behav 2: 408–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha M, Licausi F, Araújo WL, Nunes-Nesi A, Sodek L, Fernie AR, van Dongen JT. (2010) Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol 152: 1501–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR. (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U, Liu J, Leisse A, Balbo I, Perez-Melis A, Willmitzer L, Fernie AR. (2004) Kinetics of labelling of organic and amino acids in potato tubers by gas-chromatography mass-spectrometry following incubation in 13C labelled isotopes. Plant J 39: 669–679 [DOI] [PubMed] [Google Scholar]
- Rojo E, Zouhar J, Carter C, Kovaleva V, Raikhel NV. (2003) A unique mechanism for protein processing and degradation in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 7389–7394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolin D, Baldet P, Just D, Chevalier C, Biran M, Raymond P. (2000) NMR study of low subcellular pH during the development of cherry tomato fruit. Aust J Plant Physiol 27: 61–69 [Google Scholar]
- Rolletschek H, Radchuk R, Klukas C, Schreiber F, Wobus U, Borisjuk L. (2005) Evidence of a key role for photosynthetic oxygen release in oil storage in developing soybean seeds. New Phytol 167: 777–786 [DOI] [PubMed] [Google Scholar]
- Runquist M, Kruger NJ. (1999) Control of gluconeogenesis by isocitrate lyase in endosperm of germinating castor bean seedlings. Plant J 19: 423–431 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Schwender J, Ohlrogge JB. (2004) The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiol 136: 2700–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Sauer U. (2006) Metabolic networks in motion: 13C-based flux analysis. Mol Syst Biol 2: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayanova O, Haslam R, Venegas Caleron M, Napier JA. (2007) Cloning and characterization of unusual fatty acid desaturases from Anemone leveillei: identification of an acyl-coenzyme A C20 Delta5-desaturase responsible for the synthesis of sciadonic acid. Plant Physiol 144: 455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelp BJ, Bown AW, McLean MD. (1999) Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci 4: 446–452 [DOI] [PubMed] [Google Scholar]
- Shelp BJ, Walton CS, Snedden WA, Tuin LG, Oresnik IJ, Layzell DB. (1995) Gaba shunt in developing soybean seeds is associated with hypoxia. Physiol Plant 94: 219–228 [Google Scholar]
- Smith RG, Gauthier DA, Dennis DT, Turpin DH. (1992) Malate- and pyruvate-dependent fatty acid synthesis in leucoplasts from developing castor endosperm. Plant Physiol 98: 1233–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedden WA, Arazi T, Fromm H, Shelp BJ. (1995) Calcium/calmodulin activation of soybean glutamate decarboxylase. Plant Physiol 108: 543–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon PS, Oliver RP. (2002) Evidence that gamma-aminobutyric acid is a major nitrogen source during Cladosporium fulvum infection of tomato. Planta 214: 414–420 [DOI] [PubMed] [Google Scholar]
- Stitt M. (1999) Nitrate regulation of metabolism and growth. Curr Opin Plant Biol 2: 178–186 [DOI] [PubMed] [Google Scholar]
- Stitt M, Fernie AR. (2003) From measurements of metabolites to metabolomics: an ‘on the fly’ perspective illustrated by recent studies of carbon-nitrogen interactions. Curr Opin Biotechnol 14: 136–144 [DOI] [PubMed] [Google Scholar]
- Studart-Guimarães C, Fait A, Nunes-Nesi A, Carrari F, Usadel B, Fernie AR. (2007) Reduced expression of succinyl-coenzyme A ligase can be compensated for by up-regulation of the gamma-aminobutyrate shunt in illuminated tomato leaves. Plant Physiol 145: 626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studart-Guimarães C, Gibon Y, Frankel N, Wood CC, Zanor MI, Fernie AR, Carrari F. (2005) Identification and characterisation of the alpha and beta subunits of succinyl CoA ligase of tomato. Plant Mol Biol 59: 781–791 [DOI] [PubMed] [Google Scholar]
- Tieman D, Taylor M, Schauer N, Fernie AR, Hanson A, Klee H. (2006) Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc Natl Acad Sci USA 103: 8287–8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter L, Adam-Vizi V. (2000) Inhibition of Krebs cycle enzymes by hydrogen peroxide: a key role of [alpha]-ketoglutarate dehydrogenase in limiting NADH production under oxidative stress. J Neurosci 20: 8972–8979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschiersch H, Borisjuk L, Rutten T, Rolletschek H. (2011) Gradients of seed photosynthesis and its role for oxygen balancing. Biosystems 103: 302–308 [DOI] [PubMed] [Google Scholar]
- Tuin LG, Shelp AW. (1994) In situ [14C]Glu metabolism by developing soybean cotyledons. I. Metabolic routes. J Plant Physiol 143: 1–7 [Google Scholar]
- Turano FJ, Fang TK. (1998) Characterization of two glutamate decarboxylase cDNA clones from Arabidopsis. Plant Physiol 117: 1411–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Blasing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al. (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Geest AH, Hall TC. (1996) A 68 bp element of the β-phaseolin promoter functions as a seed-specific enhancer. Plant Mol Biol 32: 579–588 [DOI] [PubMed] [Google Scholar]
- Vandewalle I, Olsson R. (1983) The γ-aminobutyric acid shunt in germinating Sinapis alba seeds. Plant Sci Lett 31: 269–273 [Google Scholar]
- Vigeolas H, van Dongen JT, Waldeck P, Huhn D, Geigenberger P. (2003) Lipid storage metabolism is limited by the prevailing low oxygen concentrations within developing seeds of oilseed rape. Plant Physiol 133: 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM. (1998) A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol 37: 455–469 [DOI] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U. (2005) Molecular physiology of legume seed development. Annu Rev Plant Biol 56: 253–279 [DOI] [PubMed] [Google Scholar]
- Wharton MJ. (1955) The use of tetrazolium test for determining the viability of seeds of the genus Brassica. Proc Int Seed Test Assoc 20: 81–88 [Google Scholar]
- Yin YG, Tominaga T, Iijima Y, Aoki K, Shibata D, Ashihara H, Nishimura S, Ezura H, Matsukura C. (2010) Metabolic alterations in organic acids and γ-aminobutyric acid in developing tomato (Solanum lycopersicum L.) fruits. Plant Cell Physiol 51: 1300–1314 [DOI] [PubMed] [Google Scholar]
- Yoshizawa F. (2004) Regulation of protein synthesis by branched-chain amino acids in vivo. Biochem Biophys Res Commun 313: 417–422 [DOI] [PubMed] [Google Scholar]
- Zhu X, Galili G. (2003) Increased lysine synthesis coupled with a knockout of its catabolism synergistically boosts lysine content and also transregulates the metabolism of other amino acids in Arabidopsis seeds. Plant Cell 15: 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zik M, Arazi T, Snedden WA, Fromm H. (1998) Two isoforms of glutamate decarboxylase in Arabidopsis are regulated by calcium/calmodulin and differ in organ distribution. Plant Mol Biol 37: 967–975 [DOI] [PubMed] [Google Scholar]