Abstract
Benzoxazinones (Bxs) are major defensive secondary metabolites in wheat (Triticum aestivum), rye (Secale cereale), and maize (Zea mays). Here, we identified full sets of homeologous and paralogous genes encoding Bx glucosyltransferase (GT) and Bx-glucoside glucosidase (Glu) in hexaploid wheat (2n = 6x = 42; AABBDD). Four GT loci (TaGTa–TaGTd) were mapped on chromosomes 7A, 7B (two loci), and 7D, whereas four glu1 loci (Taglu1a–Taglu1d) were on chromosomes 2A, 2B (two loci), and 2D. Transcript levels differed greatly among the four loci; B-genome loci of both TaGT and Taglu1 genes were preferentially transcribed. Catalytic properties of the enzyme encoded by each homeolog/paralog also differed despite high levels of identity among amino acid sequences. The predominant contribution of the B genome to GT and Glu reactions was revealed, as observed previously for the five Bx biosynthetic genes, TaBx1 to TaBx5, which are separately located on homeologous groups 4 and 5 chromosomes. In rye, where the ScBx1 to ScBx5 genes are dispersed to chromosomes 7R and 5R, ScGT and Scglu were located separately on chromosomes 4R and 2R, respectively. The dispersal of Bx-pathway loci to four distinct chromosomes in hexaploid wheat and rye suggests that the clustering of Bx-pathway genes, as found in maize, is not essential for coordinated transcription. On the other hand, barley (Hordeum vulgare) was found to lack the orthologous GT and glu loci like the Bx1 to Bx5 loci despite its close phylogenetic relationship with wheat and rye. These results contribute to our understanding of the evolutionary processes that the Bx-pathway loci have undergone in grasses.
Benzoxazinones (Bxs) are one of the better studied classes of plant secondary metabolites in terms of their distribution, biological activities, as well as their biosynthesis from biochemical and molecular genetic aspects (for review, see Niemeyer, 1988, 2009; Sicker et al., 2000; Frey et al., 2009). Bxs are produced in many species of Poaceae, including the major agricultural crops wheat (Triticum aestivum), maize (Zea mays), and rye (Secale cereale). Reported functions include defense against microbial attack or herbivore predation as well as allelopathic agents. DIBOA (2,4-dihydroxy-1,4-benzoxazin-3-one) and its C7 methoxy derivative DIMBOA are the predominant forms of Bxs in plants. They are stored in the vacuole as 2-O-β-d-glucopyranosides (Bx-Glcs), designated DIBOA-Glc and DIMBOA-Glc.
DIBOA is biosynthesized in five sequential reactions starting with indole-3-glycerol phosphate derived from the Trp pathway (Fig. 1; Frey et al., 1997, 2000; Melanson et al., 1997). The genes involved have been isolated in maize (ZmBx1–ZmBx5; Frey et al., 1995, 1997), wild barley (Hordeum lechleri; HlBx1–HlBx5; Grün et al., 2005), wild diploid wheat (Triticum boeoticum; TbBx1–TbBx5; Nomura et al., 2007a), and hexaploid wheat (TaBx1A–TaBx5A, TaBx1B–TaBx5B, TaBx1D–TaBx5D; Nomura et al., 2002, 2003, 2005). Of the five biosynthetic genes, Bx2 to Bx5 encode cytochrome P450 monooxygenases of the CYP71C subfamily. In maize, two genes (ZmBx8 and ZmBx9), each encoding a UDP-Glc:Bx glucosyltransferase (GT), have been identified (von Rad et al., 2001). It has long been unclear whether the conversion of DIBOA to DIMBOA occurs on the aglycone or the glucoside, but recently it was shown that ZmBX6, a 2-oxoglutarate-dependent dioxygenase, accepts DIBOA-Glc but not DIBOA as a substrate. This indicates that the 2-O-glucosylation of DIBOA by the GTs precedes the 7-hydroxylation, which is followed by 7-O-methylation of TRIBOA-Glc by the O-methyltransferase ZmBX7 to form DIMBOA-Glc (Frey et al., 2003; Jonczyk et al., 2008; Fig. 1). Although genes orthologous to ZmBx6 to ZmBx9 have not yet been identified in other plants, the same reactions likely occur. In wheat, Bx-GT has been characterized enzymatically (Sue et al., 2000a). The Bx-Glcs, which have reduced toxicity compared with the aglycones, are stored in the vacuole, and the toxic aglycones are released by a specific β-glucosidase (Glu) existing in the plastid when cells are disrupted by wounding and/or infection (Fig. 1). The Bx-Glc Glus have been identified from maize (Cicek and Esen, 1999; Czjzek et al., 2000; Verdoucq et al., 2003), wheat (Sue et al., 2000c, 2005, 2006), and rye (Sue et al., 2000b; Nikus et al., 2003).
Figure 1.
The Bx biosynthetic pathway and deglucosylation by β-glucosidase (Glu). The enzyme catalyzing each reaction is shown in a box. Black boxes indicate glucosyltransferase (GT) and Glu examined in this study. BX6 and BX7, which catalyze sequential 7-hydroxylation and 7-O-methylation of DIBOA-Glc to DIMBOA-Glc, have been identified in maize but not in wheat or rye. The GTs we identified have been designated TaGTs and ScGT in wheat and rye, respectively. The corresponding GTs in maize have been designated BX8 and BX9.
Common wheat is hexaploid with the genome constitution AABBDD (2n = 6x = 42), which originated through successive chromosome doubling of hybrids involving three ancestral diploid species (2n = 2x = 14): the A genome came from Triticum urartu (AA), the B genome from Aegilops speltoides (SS) or another species classified in the genus Aegilops (Sitopsis section), and the D genome from Aegilops tauschii (Huang et al., 2002; Feldman and Levy, 2005; Salse et al., 2008b). Allopolyploidization leads to the generation of duplicated homeologous genes (homeologs). Consequently, the hexaploid wheat genome contains triplicated homeologs for most genes derived from the diploid progenitors, but elimination and/or amplification also occur for some homeologs (Ozkan et al., 2001). In addition, even though triplicated homeologs are retained through allopolyploidization, they do not always function equally due to biased transcription, including homeolog-specific silencing, and nonfunctionalization of a specific homeolog caused by structural alteration (Bottley et al., 2006; Shitsukawa et al., 2007; Akhunova et al., 2010). Previously, three sets of the five TaBx genes were all identified in hexaploid wheat, and their chromosomal locations were determined (Nomura et al., 2002, 2003, 2005). TaBx1 and TaBx2 homeologs were located in the same chromosomal bin on homeologous group 4 chromosomes (4A, 4B, and 4D), while TaBx3 to TaBx5 homeologs existed in the same chromosomal bin on group 5 chromosomes (5A, 5B, and 5D). Transcription of TaBx1 to TaBx5 is coordinated, but levels vary depending on the genome, where B-genome homeologs are transcribed preferentially (Nomura et al., 2005). In addition to transcript levels, enzymatic activities also vary with genome despite extremely high sequence identity. Based on the differences in transcript levels and enzymatic activities among the three homeologs of TaBx1 to TaBx5, it has been suggested that the B genome contributes most to Bx biosynthesis in hexaploid wheat (Nomura et al., 2005). Moreover, it was also proposed that differential transcription of the three homeologs in hexaploid wheat originated during diploidy and was retained through polyploidization.
In rye (2n = 2x = 14; RR), the TaBx1 to TaBx5 orthologs, ScBx1 to ScBx5, were also shown to be located on two distinct chromosomes, 7R (ScBx1 and ScBx2) and 5R (ScBx3–ScBx5; Nomura et al., 2003). In cultivated barley (Hordeum vulgare; 2n = 2x = 14; HH), however, none of the five Bx loci was present (Nomura et al., 2003) despite close relationships between species in the tribe Triticeae, which evolved from a common ancestor, share the same basic chromosome number, and have highly similar gene sequences (Devos and Gale, 1997; Huang et al., 2002). In contrast to wheat and rye, maize genes ZmBx1 to ZmBx5 form a cluster on the short arm of chromosome 4 (Frey et al., 1995, 1997). In addition, genes ZmBx6 to ZmBx8 are also included in the cluster (von Rad et al., 2001; Jonczyk et al., 2008), whereas ZmBx9, a highly identical homolog of ZmBx8, is situated on chromosome 1 (von Rad et al., 2001), and Zmglu1 and Zmglu2 are on chromosome 10 (http://www.maizesequence.org/index.html). In general, genes for most metabolic pathways are not clustered in plants, but evidence that genes for secondary metabolic pathways are clustered has recently emerged: the maize ZmBx genes, the diploid oat (Avena strigosa) avenacin biosynthetic genes (Papadopoulou et al., 1999; Qi et al., 2004), the rice (Oryza sativa) momilactone (Wilderman et al., 2004; Shimura et al., 2007) and phytocassane (Swaminathan et al., 2009) biosynthetic genes, and the Arabidopsis (Arabidopsis thaliana) thalianol pathway genes (Field and Osbourn, 2008). Gene clustering is thought to facilitate not only the inheritance of beneficial gene combinations but also the coordinated transcription of pathway genes by enabling localized changes in chromatin structure (Wegel et al., 2009). It has been demonstrated that Bx-Glc levels peak soon after germination and then decrease to a constant level in wheat (Nomura et al., 2005), maize (Ebisui et al., 1998), and rye (Sue et al., 2000b). At the same time, enzymatic activities of GT and Glu also occur concomitantly with the accumulation profiles of Bxs (Ebisui et al., 1998, 2001; Sue et al., 2000a, 2000b, 2000c). Consistent with this, transcript levels of all Bx-pathway genes increase transiently in seedlings and decrease to a lower constant level as plants grow (Frey et al., 1995; von Rad et al., 2001; Nomura et al., 2005; Sue et al., 2006; Jonczyk et al., 2008).
To complete the elucidation of the Bx biosynthetic system in hexaploid wheat, the mechanisms of (1) Bx-pathway gene coexpression in each of the three genomes and (2) differential transcription and catalytic activity among the three genomes need to be determined. Moreover, considering that the Bx biosynthetic genes identified so far are clustered in maize, but not in wheat and rye, the Bx pathway is an excellent model to investigate the biological and molecular genetic significance of gene clusters for secondary pathways in plants. In wheat and rye, however, molecular characterization of the GT and glu genes, and the genes involved in the conversion of DIBOA-Glc to DIMBOA-Glc, has not yet been completed. In this study, we focused on the GT and glu genes in hexaploid wheat and rye. Through cDNA isolation, chromosome assignment, and transcriptional and enzymatic characterizations, we demonstrate the differential contribution of the three genomes of hexaploid wheat to the GT and Glu reactions and propose an evolutionary process for the Bx-pathway loci in grasses, in particular in the tribe Triticeae including the genera Triticum, Aegilops, Secale, and Hordeum. We also discuss a molecular basis for the coexpression of Bx genes and their biased expression among the three genomes of hexaploid wheat as well as the significance of gene cluster formation in secondary metabolite biosynthesis.
RESULTS
Isolation of GT and glu cDNAs from Hexaploid Wheat and Rye
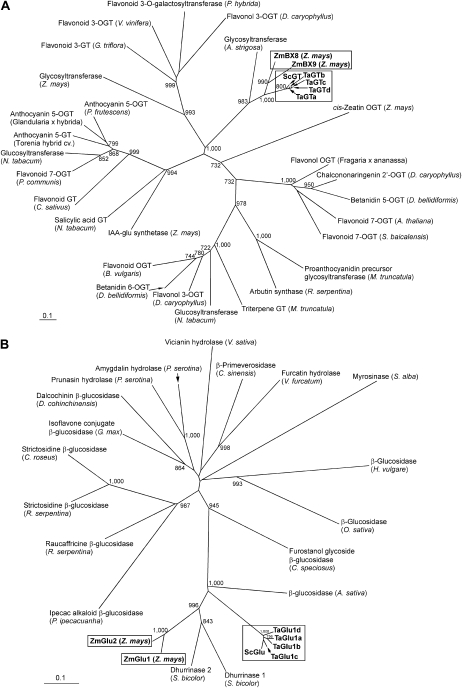
Screening of the cDNA library prepared from young seedlings of hexaploid wheat (cv Chinese Spring [CS]) using the maize ZmBx8 cDNA as a probe and additional reverse transcription (RT)-PCR resulted in the isolation of four TaGT cDNAs, TaGTa to TaGTd (GenBank accession nos. AB547237–AB547240). The cDNA library prepared from young rye seedlings was screened using TaGTa cDNA as a probe to obtain ScGT cDNA (GenBank accession no. AB548283), the TaGT ortholog of rye. TaGTa to TaGTd shared 96.3% to 98.5% identity with each other at the amino acid level (Supplemental Table S1; Supplemental Fig. S1) and had high identity (91.6%–94.3%) to ScGT. Amino acid identities of the TaGTs to the maize ortholog ZmBX8 were 68.2% to 68.8%, slightly higher than those to ZmBX9 (66.2%–67.0%; Supplemental Table S1). Phylogenetic analysis showed a close relationship among the GTs of wheat, rye, and maize (Fig. 2A).
Figure 2.
Unrooted phylogenetic trees of GTs (A) and Glus (B). Full-length amino acid sequences were aligned using ClustalW. The phylogenetic tree was built on calculations using the neighbor-joining method (Saitou and Nei, 1987) with bootstrap analysis of 1,000 replicates and was visualized with Treeview. Numbers at each node are bootstrap values (over 70%) per 1,000 trials. Scale bars indicate substitutions per site. GTs and Glus of Bx glucosylation and deglucosylation, respectively, in wheat, rye, and maize are shown within a box in each tree. Accession numbers of proteins used for constructing the trees are listed in Supplemental Materials and Methods S1.
We previously isolated three Taglu1 cDNAs (Taglu1a–Taglu1c) from hexaploid wheat (Sue et al., 2006). As described below, however, they were localized to chromosomes 2B and 2D. Since most genes are localized in the three genomes in hexaploid wheat, the presence of another homeolog in the A genome was expected. PCR of genomic DNA followed by RT-PCR resulted in the isolation of the novel Taglu1d cDNA (GenBank accession no. AB548284). Taglu1d encoded a polypeptide of 564 amino acids with a plastid-targeting transit peptide similar to those found in TaGlu1a to TaGlu1c. Amino acid sequences of the four TaGlu1s shared 91.8% to 95.1% identity with each other and approximately 92% and 60%, respectively, with their orthologs in rye (ScGlu; AAG00614) and maize (ZmGlu1 and ZmGlu2; AAA65946 and AAD09850, respectively; Supplemental Table S2; Supplemental Fig. S2). Phylogenetic analysis of plant family 1 glycoside hydrolases (Fig. 2B) showed that the TaGlu1s and ScGlu are closely related to each other and also to ZmGlu1 and ZmGlu2, as well as to dhurrinases in sorghum (Sorghum bicolor; AAC49177 and AAK49119) and β-glucosidase in oat (CAA55196).
BLAST searches against the maize sequence database (http://www.maizesequence.org/blast) using TaGT and Taglu1 sequences as queries detected ZmBx8/ZmBx9 and Zmglu1/Zmglu2, respectively, as sequences of best matches. In addition, searches against the wheat sequence database (http://www.nbrp.jp/) detected only the TaGT and Taglu1 sequences identified, including sequences having interspecific single nucleotide polymorphisms, with significant E-values and coverage rates. These results indicate that the four cDNAs for each of the TaGT and Taglu1 genes cover their functionally expressing loci in CS wheat.
Chromosomal Assignment of GT and glu Genes in Hexaploid Wheat and Rye
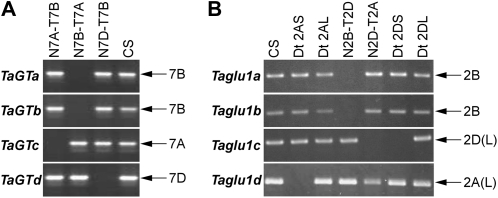
Genomic PCR of the aneuploid lines of CS wheat using primers specific to TaGTa, TaGTb, TaGTc, or TaGTd amplified no PCR products from N7B-T7A, N7B-T7A, N7A-T7B, and N7D-T7B, respectively (Fig. 3A), indicating that the TaGTa, TaGTb, TaGTc, and TaGTd loci are located on chromosomes 7B, 7B, 7A, and 7D, respectively, in hexaploid wheat. Chromosomal locations of the four Taglu1 loci were determined using the same procedure. Specific PCR products for each of the four Taglu1s were missing in N2B-T2D, N2B-T2D, N2D-T2A, and Dt2AS, respectively (Fig. 3B). A Taglu1c-specific product was also absent in Dt2DS. These results showed that the Taglu1a, Taglu1b, Taglu1c, and Taglu1d loci are located on chromosomes 2B, 2B, 2DL (where L represents long arm), and 2AL, respectively. These results indicate that four loci of each of the TaGT and Taglu1 genes are composed of three homeologs (one homeolog on each genome) and one paralog on the B genome, but it remains unclear which of the two loci on the B genome is the original locus or the paralogous locus that arose by duplication of the original locus for both TaGT and Taglu1 genes.
Figure 3.
Genomic PCR of nullisomic-tetrasomic (N-T) and ditelosomic (Dt) lines of CS for chromosomal assignment of TaGT (A) and Taglu1 (B) loci in hexaploid wheat. PCR was performed with primer sets that specifically amplify each of the four loci of TaGT and Taglu1 genes. Results of CS aneuploid lines of homeologous group 7 and group 2 chromosomes are represented for TaGT (A) and Taglu1 (B). The chromosome on which each locus is located is indicated on the right. For Taglu1c and Taglu1d, chromosomal arm location was determined to be the long arm (L) using Dt lines.
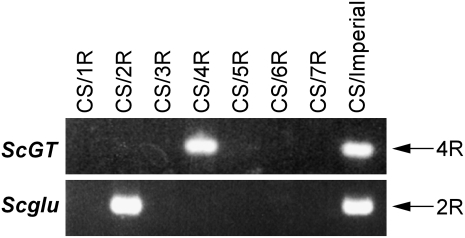
Chromosomal locations of the rye orthologs, ScGT and Scglu, were assigned by specific PCR of the wheat (CS)-rye (cv Imperial) chromosome addition lines. ScGT-specific amplification gave a PCR band in the CS/Imperial amphidiploid, which possesses whole rye chromosomes in the CS wheat genetic background, and in the 4R chromosome addition line (Fig. 4). This showed that the ScGT gene is located on chromosome 4R in rye. Scglu-specific amplification was detected in the 2R chromosome addition line as well as in the CS/Imperial amphidiploid (Fig. 4), indicating that the Scglu gene is located on chromosome 2R.
Figure 4.
Genomic PCR of wheat (CS)-rye (Imperial) chromosome addition lines for chromosome assignment of ScGT and Scglu genes in rye. PCR was performed with primer sets that specifically amplify ScGT and Scglu but not their orthologs in CS wheat. CS/Imperial has whole sets of rye chromosomes, and CS/1R-CS/7R each has a pair of respective rye chromosomes in the wheat genetic background. Chromosomes corresponding to PCR products derived from the rye genome are indicated on the right.
Catalytic Activities of TaGT and TaGlu1 Enzymes
Kinetic parameters of each of the TaGTa to TaGTd enzymes were determined using purified recombinant enzymes (Table I). All four TaGT enzymes showed higher reaction efficiencies (kcat/Km) for DIMBOA than for DIBOA, from 1.9-fold for TaGTd to 3.3-fold for TaGTa. Km and kcat values differed among the four enzymes within a 3-fold range for both DIBOA and DIMBOA. TaGTa exhibited the highest reaction efficiencies, where the kcat/Km values of TaGTa were approximately two and three times higher than those of TaGTb to TaGTd for DIBOA and DIMBOA, respectively.
Table I. Kinetic parameters of TaGT enzymes.
| Enzyme | DIBOA |
DIMBOA |
||||
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | |
| μm | s−1 | s−1 mm−1 | μm | s−1 | s−1 mm−1 | |
| TaGTa | 14.4 | 11.2 | 778 | 11.3 | 29.4 | 2,600 |
| TaGTb | 14.4 | 5.1 | 354 | 23.8 | 19.6 | 824 |
| TaGTc | 27.8 | 8.8 | 317 | 13.7 | 11.1 | 810 |
| TaGTd | 21.2 | 9.0 | 425 | 16.6 | 13.3 | 801 |
Newly isolated TaGlu1d was expressed in Escherichia coli after truncation of its plastid-targeting transit peptide, as described previously for TaGlu1a to TaGlu1c (Sue et al., 2006). Kinetic parameters of TaGlu1d for DIBOA-Glc and DIMBOA-Glc were determined and compared with those of TaGlu1a to TaGlu1c (Sue et al., 2006). As shown in Table II, Km and kcat values differed among the four enzymes. For DIBOA-Glc, TaGlu1d showed the highest reaction efficiency (313 s−1 mm−1), which was 9-fold higher than that of TaGlu1a (34.6 s−1 mm−1). For DIMBOA-Glc, TaGlu1b exhibited the highest reaction efficiency (4,141 s−1 mm−1), which was 10-fold higher than that of TaGlu1d. Notably, TaGlu1a to TaGlu1c preferentially accepted DIMBOA-Glc, where the reaction efficiencies of TaGlu1a, TaGlu1b, and TaGlu1c for DIMBOA-Glc were 27-, 28-, and 15-fold, respectively, higher than those for DIBOA-Glc. In contrast, the reaction efficiency of TaGlu1d for DIMBOA-Glc was only 1.3-fold higher than that for DIBOA-Glc, showing higher reactivity with DIBOA-Glc than the other TaGlu1s. A similar catalytic property was observed in the rye glucosidase (ScGlu; Sue et al., 2006). This is attributable to the mature enzyme amino acid residues Gly-464 and Ser-465 shared by TaGlu1d and ScGlu, which are involved in distinguishing DIBOA-Glc from DIMBOA-Glc (Sue et al., 2006). The counterpart residues in the TaGlu1a to TaGlu1c enzymes that preferentially hydrolyzed DIMBOA-Glc were Ser-464 and Leu-465 (Supplemental Fig. S2).
Table II. Kinetic parameters of TaGlu1 enzymes.
| Enzyme | DIBOA-Glc |
DIMBOA-Glc |
||||
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | |
| mm | s−1 | s−1 mm−1 | mm | s−1 | s−1 mm−1 | |
| TaGlu1aa | 1.40 | 48.8 | 34.6 | 0.36 | 338 | 939 |
| TaGlu1ba | 1.44 | 214 | 149 | 0.29 | 1,201 | 4,141 |
| TaGlu1ca | 1.05 | 137 | 131 | 0.39 | 773 | 1,982 |
| TaGlu1db | 1.83 | 572 | 313 | 0.79 | 330 | 418 |
Data taken from Sue et al. (2006).
This study.
Transcript Profiles of TaGT and Taglu1 Genes in Hexaploid and Tetraploid Wheat
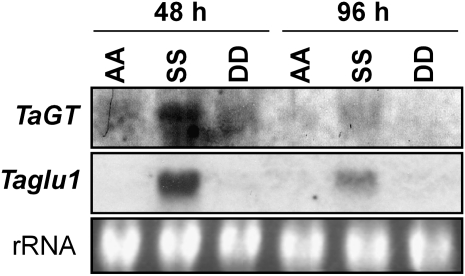
We first examined changes in the transcript levels of the TaGT and Taglu1 genes in young shoots of hexaploid wheat by northern-blot analysis. The levels of TaGT and Taglu1 transcripts peaked 48 h after seeding and then decreased to lower levels (Fig. 5A). This pattern correlated well with those of TaBx1 to TaBx5 genes as well as those for Bx content (Nomura et al., 2005). Since northern analysis cannot distinguish transcripts from each of the four homeologous and paralogous loci due to their high identities, we performed locus-specific quantitative (q)RT-PCR analysis to compare transcript levels of four loci of the TaGT and the Taglu1 genes in hexaploid wheat shoots 48 h (high Bx content) and 96 h (low Bx content) after seeding; Bx contents (total of DIBOA-Glc and DIMBOA-Glc) in 48- and 96-h-old shoots are 15.7 and 5.7 μmol g−1 fresh weight, respectively (Nomura et al., 2005). TaGTa located on the B genome was transcribed at the highest ratio in both 48-h-old (63.5%) and 96-h-old (75.5%) shoots (Fig. 5B). Transcripts of the D-genome homeolog TaGTd were detected at a ratio comparable to that of the TaGTb on the B genome in 48-h-old shoots but decreased to the lower ratio (3.0%) in 96-h-old shoots. The A-genome homeolog TaGTc was transcribed at the lowest ratio in both 48-h-old (1.7%) and 96-h-old (0.8%) shoots.
Figure 5.
Transcript levels of TaGT and Taglu1 genes in CS and Tetra-CS. A, Northern-blot analysis of 24- to 120-h-old CS shoots. Ethidium bromide-stained rRNA was used as a loading control. B to E, Transcript ratios of four loci in 48-h-old (black bars) and 96-h-old (white bars) shoots analyzed by qRT-PCR. B, TaGTs in CS. C, Taglu1s in CS. D, TaGTs in Tetra-CS. E, Taglu1s in Tetra-CS. The chromosomal location of each locus is shown in parentheses. Note that data for each growth stage (B–E) are presented as the transcript ratio of each locus to the sum of four loci. The absence of transcripts of TaGTd and Taglu1c in Tetra-CS (D and E, respectively) is due to lack of the D genome in Tetra-CS. Data are expressed as means of triplicate experiments with sd. n.d., Not detected.
Similarly, among the four Taglu1 loci, Taglu1a and Taglu1b on the B genome were preferentially transcribed. Transcript ratios of Taglu1a and Taglu1b in 48-h-old shoots were 55.0% and 41.2%, respectively, and those in 96-h-old shoots were 84.3% and 15.4% (Fig. 5C). The sum of ratios of the two B-genome locus transcripts reached 96.2% and 99.7% in 48- and 96-h-old shoots, respectively. In contrast, the D-genome homeolog Taglu1c and the A-genome homeolog Taglu1d were transcribed at low ratios (Fig. 5C).
To see if the transcript levels of A- and B-genome loci are affected by the D genome, we investigated the profiles in Tetra-CS, which is a tetraploid wheat generated from hexaploid CS; thus, its A and B genomes are identical to those of CS. Changes in the TaGT and Taglu1 transcripts between 48- and 96-h-old shoots, where the Bx contents are 18.4 and 10.9 μmol g−1 fresh weight, respectively (Nomura et al., 2005), were substantially the same as observed in hexaploid CS wheat (Fig. 5A). Locus-specific qRT-PCR revealed, as observed in CS, that the majority of TaGT transcripts were contributed by the B-genome loci, TaGTa and TaGTb (i.e. 78% and 20%, respectively, in both 48- and 96-h-old shoots; Fig. 5D). In contrast, the transcript ratio of the TaGTc homeolog located on the A genome was only 1.8% and 1.2% of the total in 48- and 96-h-old shoots, respectively. For the three Taglu1 loci, transcript profiles were similar to those in CS (Fig. 5E). The sum of ratios of the transcripts from the two B-genome loci (Taglu1a and Taglu1b) reached 93.3% and 99.0% in 48- and 96-h-old shoots, respectively.
Transcript Profiles of TaGT and Taglu1 Orthologs in Diploid Progenitors of Hexaploid Wheat
To determine whether biased transcription among the three genomes of hexaploid wheat (Fig. 5) is a function of polyploidization, transcript levels of TaGT and Taglu1 orthologs in the three diploid progenitors of hexaploid wheat were examined by northern analysis (Fig. 6). Blots of RNA isolated from 48- and 96-h-old shoots of each diploid progenitor were probed with orthologous sequences from hexaploid wheat. For both the TaGT and Taglu1 probes, strongest hybridization signals were detected in A. speltoides (SS), the B-genome donor to hexaploid wheat. In contrast, only faint signals were detected in T. urartu (AA) and A. tauschii (DD). The patterns correlated well with the Bx contents: 1.2 (48 h) and 0.3 (96 h) μmol g−1 fresh weight in T. urartu, 20.8 (48 h) and 9.3 (96 h) μmol g−1 fresh weight in A. speltoides, and 6.9 (48 h) and 0.7 (96 h) μmol g−1 fresh weight in A. tauschii (Nomura et al., 2005). These results suggested that the preferential transcription of B-genome TaGT and Taglu1 loci in hexaploid wheat originated in a diploid progenitor and was not caused by polyploidization.
Figure 6.
Transcript levels of TaGT and Taglu1 orthologs in diploid progenitors of hexaploid wheat. RNA isolated from 48- and 96-h-old shoots were analyzed by northern hybridization. AA, SS, and DD represent T. urartu, A. speltoides, and A. tauschii, respectively. Ethidium bromide-stained rRNA was used as a loading control.
Southern Analysis to Search for TaGT and Taglu1 Orthologs in Barley
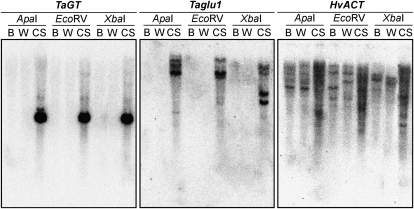
Since wheat and barley are closely related, their orthologous genes usually share about 95% identity at the nucleotide level. Therefore, wheat orthologs can be detected in barley DNA by Southern hybridization under stringent hybridization/washing conditions (Nomura et al., 2003). Southern analysis of DNA from two barley cultivars, however, revealed no hybridization when probed with wheat TaGTa or Taglu1a (Fig. 7). When the same DNA blot was probed with barley HvACT cDNA, orthologs of which are present in hexaploid wheat (Nomura et al., 2007b), clear hybridization bands were detected in both barley cultivars and in CS wheat, thus validating the negative result using wheat TaGTa and Taglu1a probes. In addition, we performed BLAST searches against the barley full-length cDNAs and ESTs (Barley DB; http://www.shigen.nig.ac.jp/barley). No barley sequences exhibiting significant identity with the TaGT and Taglu1 coding regions were found, an outcome consistent with the Southern-blot result. We conclude that neither TaGT nor Taglu1 orthologs exist in cultivated barley.
Figure 7.
Southern-blot analysis of barley cv Betzes (B) and Wasedori-nijo (W) probed with TaGTa and Taglu1a. Genomic DNA digests of CS wheat were included as a positive hybridization control. The barley HvACT gene whose orthologs are present in wheat (Nomura et al., 2007b) was used as a positive control for hybridization with barley DNA and for cross-hybridization between barley and wheat sequences.
DISCUSSION
Dispersed Chromosomal Locations of Bx-Pathway Genes in Wheat and Rye
Of the two GT genes in the Bx pathway in maize (ZmBx8 and ZmBx9), ZmBx8 is only 44 kb apart from ZmBx1 (Frey et al., 2009), which is 2.5 kb from ZmBx2. Therefore, we expected that the TaGT loci would map to the homeologous group 4 chromosomes on which the TaBx1 and TaBx2 genes are situated (Nomura et al., 2003). However, we found the TaGT loci on homeologous group 7 chromosomes 7A, 7B, and 7D (Fig. 8; Supplemental Table S3). It has been reported that segmental translocations occurred between groups 4, 5, and 7 chromosomes during the evolution of wheat, but the events involved only 4A, 5A, and 7B (Liu et al., 1992). Therefore, these translocations cannot explain the split location of the TaGT locus from TaBx1 and TaBx2 in all three genomes. It is more likely that the split location of the TaGT locus occurred prior to the divergence of the three diploid progenitors of hexaploid wheat. This scenario is supported by the fact that the ScGT gene was mapped to chromosome 4R, which is different from the ScBx1 and ScBx2 genes on chromosome 7R (Nomura et al., 2003) in rye (Fig. 8; Supplemental Table S3). The locations of TaGT loci on homeologous group 7 chromosomes and ScGT on chromosome 4R are consistent with the chromosomal synteny between wheat and rye (Devos et al., 1993). Likewise, the chromosomal locations of Taglu1 loci in wheat (homeologous group 2 chromosomes) and Scglu in rye (chromosome 2R; Fig. 8; Supplemental Table S3) follow the synteny between wheat and rye.
Figure 8.
Overview of chromosomal locations of Bx-pathway loci in Triticeae species and maize. Locations of Bx-pathway loci determined so far are shown on each chromosome arm. S and L represent short and long arms, respectively. TaGT and ScGT genes are also designated as TaBx8 and ScBx8, respectively, in parentheses. Note that orthologs of ZmBx6 and ZmBx7 have not yet been identified in wheat and rye. Arm locations of TaBx1 to TaBx5 were reported by Nomura et al. (2003, 2005) and those of Taglu1c and Taglu1d in this study. For the other loci in wheat and rye, chromosomal assignments were performed by Nomura et al. (2003; ScBx1–ScBx5) and in this study (Taglu1a, Taglu1b, TaGTs, Scglu, and ScGT), and their arm locations were estimated based on the chromosomal synteny between wheat and rye (Devos et al., 1993). For the locations of ZmBx1 to ZmBx9 in maize, see Frey et al. (1997), Jonczyk et al. (2008), and von Rad et al. (2001). Locations of Zmglu1 and Zmglu2 were found by searching the maize sequence database.
In maize, genes encoding most of the Bx-pathway enzymes (ZmBx1–ZmBx8) are clustered on the short arm of chromosome 4 (Frey et al., 2009), and three (ZmBx9, Zmglu1, and Zmglu2) are located outside the cluster (Fig. 8; Supplemental Table S3). ZmBx9 is located on chromosome 1 (von Rad et al., 2001), and Zmglu1 and Zmglu2 are located on chromosome 10 (3.5 Mb apart from each other). Recent high-resolution comparative mapping in grass species revealed microcolinearity among grass chromosomes and demonstrated that wheat group 7 chromosomes (where TaGT loci are located) are partially orthologous to maize chromosomes 1 (where ZmBx9 is located) and 4 (where ZmBx8 is located; Devos, 2005; Salse et al., 2008a, 2009). Therefore, we cannot judge whether the TaGT gene corresponds to ZmBx8 or ZmBx9 only based on the chromosomal synteny. Sequence identities of the TaGTs to ZmBx8 were slightly higher than those to ZmBx9 (Supplemental Table S1). In addition, TaGT enzymes showed similar catalytic properties to ZmBX8 rather than to ZmBX9; TaGTs and ZmBX8 showed moderately higher catalytic efficiency to DIMBOA than to DIBOA, while the efficiency of ZmBX9 to DIBOA is extremely lower than that to DIMBOA (von Rad et al., 2001). Therefore, TaGT appears to correspond to ZmBx8. ZmBx9 may have originated by duplication of ZmBx8, or it may be the trace of paleotetraploidy of the maize genome (Swigonova et al., 2004; Schnable et al., 2011). Localization of the Taglu1 loci on group 2 chromosomes in hexaploid wheat coincides with the partial synteny between wheat group 2 and maize chromosome 10 (Devos, 2005; Salse et al., 2008a, 2009), on which Zmglu1 and Zmglu2 genes are located (Fig. 8). We cannot judge whether Taglu1 corresponds to Zmglu1 or Zmglu2 based on the sequence comparisons (Fig. 2B; Supplemental Table S2). However, considering that Taglu1 and Zmglu1 genes are highly expressed in young seedlings while Zmglu2 starts to express at a later stage (Cicek and Esen, 1999), Taglu1s isolated in this study appear to be the counterparts of Zmglu1. It should be noted that a BLAST search with the Taglu1 query against the wheat sequence database did not retrieve sequences other than the Taglu1s identified, suggesting that Zmglu2 also arose only in maize, as mentioned above for ZmBx9.
Salse et al. (2008a, 2009) proposed that grass genomes have evolved from a common ancestor with five protochromosomes. According to the model, parts of wheat groups 4 (where TaBx1 and TaBx2 are located) and 5 chromosomes (where TaBx3–TaBx5 are located) are derived from the same protochromosome (designated A11 in the literature), which also is an origin of a part of maize chromosome 4 (where ZmBx1–ZmBx8 are located), supporting our previous hypothesis that the Bx1 to Bx5 genes arose as a cluster and were split into two chromosomes during the evolutionary processes that rearranged the ancient grass genome into seven chromosomes of the tribe Triticeae (Nomura et al., 2003; Fig. 8). However, a part of the wheat group 7 chromosomes (where TaGT loci are located) that shows synteny with parts of maize chromosomes 1 (where ZmBx9 is located) and 4 (where ZmBx8 is located) is shown to have originated from the other protochromosome, A8, and a part of the wheat group 2 chromosomes (where Taglu1 loci are located) that shows synteny with a part of maize chromosome 10 (where Zmglu1 and Zmglu2 genes are located) is from the protochromosome A4. This implies that the GT and glu loci had not been included in the ancestral Bx-pathway gene cluster (Fig. 8). However, the fact that one of the maize GT genes, ZmBx8, is situated only 44 kb apart from ZmBx1 (Frey et al., 2009) does not allow us to exclude the possibility of the ancestral Bx-pathway gene cluster. Including the Bx6 and Bx7 loci in wheat, which remain to be elucidated, in such synteny analysis would help us to know an original form of the Bx-pathway loci.
How Did Barley Lose the Bx-Pathway Loci?
It has been reported that cultivated barley produces no Bxs due to loss of the Bx1 to Bx5 loci (Gierl and Frey, 2001; Nomura et al., 2003). Grün et al. (2005) demonstrated that one wild barley species (Hordeum spontaneum) also lacks those loci, but other wild species (e.g. H. lechleri) accumulate Bxs. It has been reported that GT activity is also not detectable in cultivated barley (Leighton et al., 1994). Our study here revealed that this is attributable to loss of the GT locus.
Even in maize, where eight Bx genes are clustered, Zmglu1 and Zmglu2 loci are situated on a chromosome different from that of the Bx gene cluster (Fig. 8). Thus, we expected to find that the glu locus still exists in cultivated barley even after the loss of all other Bx loci. However, Southern analysis revealed that the glu locus is also missing in cultivated barley. Nomura et al. (2007a) proposed that degeneration of coding sequence, silencing, or loss of one Bx locus triggers the loss of other Bx loci, which finally leads to the elimination of all Bx-pathway loci. We previously predicted that Bx-producing wild barley species would have a Bx gene cluster on a single chromosome, because it seemed unlikely that Bx loci on separate chromosomes would have been eliminated totally in cultivated barley (Nomura et al., 2003). As mentioned above, however, it now seems reasonable to suggest that the ancient Bx loci in barley were already dispersed into distinct chromosomes, as in wheat and rye, as a result of evolutionary processes that rearranged the ancient grass genome into seven chromosomes of the tribe Triticeae (Fig. 8). Presumably, elimination of the GT and glu loci, as well as the Bx1 to Bx5 loci, in barley occurred sequentially according to the same scenario found in the wild A-genome diploid wheat (Nomura et al., 2007a).
Differential Contributions of the Three Hexaploid Wheat Genomes to Reactions Catalyzed by TaGT and TaGlu1 Enzymes
Transcript levels of the four loci differed greatly for both TaGT and Taglu1 genes; B-genome loci were predominantly transcribed, a feature in common with the TaBx1 to TaBx5 genes (Nomura et al., 2005). Evidence of differential transcription does not necessarily indicate the actual contribution of each homeolog to its corresponding reaction. Differences in catalytic properties of enzymes encoded by each homeolog must also be considered (Nomura et al., 2005).
TaGT enzymes catalyze the 2-O-glucosylation of DIBOA and DIMBOA in vitro, where the reaction efficiencies for DIMBOA are approximately 2- to 3-fold higher than those for DIBOA. In the maize biosynthetic pathway (Jonczyk et al., 2008), however, 2-O-glucosylation occurs for DIBOA to form DIBOA-Glc, followed by 7-hydroxylation and 7-O-methylation to form DIMBOA-Glc. Therefore, the contribution of individual TaGT enzymes to the reaction in vivo should be estimated based on their reaction efficiencies for DIBOA. Obviously, TaGTa encoded by the B-genome locus plays the major role in the reaction, because its transcript levels are 3- to 37-fold higher in 48-h-old shoots and 4- to 92-fold higher in 96-h-old shoots than are those of the other TaGTs, and the reaction efficiency of TaGTa for DIBOA is approximately twice as high as those of the other TaGTs. Even though the reaction efficiencies of TaGTc and TaGTd are comparable to those of TaGTb, their transcript levels are notably lower than those of the B-genome loci TaGTa and TaGTb, especially in 96-h-old shoots. These results indicated that the B genome contributes most to the TaGT reaction in hexaploid wheat.
TaGlu1 functions as homohexamers and heterohexamers (Sue et al., 2000c, 2006). Judging from transcript levels of individual Taglu1 loci, the natural hexameric TaGlu1 enzymes are presumed to be composed mainly of TaGlu1a and TaGlu1b, both of which are encoded by B-genome loci. Although the reaction efficiency of the homohexamer of TaGlu1a for DIBOA-Glc is 4- and 9-fold lower than those of TaGlu1d and TaGlu1c, respectively, its transcript level is approximately 31- and 27-fold higher than those of Taglu1c and Taglu1d, respectively, in 48-h-old shoots and 320- and 8,400-fold higher than those in 96-h-old shoots. These results show that the lower reaction efficiency of TaGlu1a can readily be overcome by its substantially higher transcript level. Similarly, the 2-fold higher reaction efficiency of TaGlu1c over TaGlu1a for DIMBOA-Glc is likely canceled by the low transcript level of Taglu1c. Although the reaction efficiency of the TaGlu1b enzyme for DIBOA-Glc was comparable to that of TaGlu1c and 2-fold lower than that of TaGlu1d, its transcript level is remarkably higher than those of Taglu1c and Taglu1d. Moreover, the reaction efficiency of TaGlu1b for DIMBOA-Glc is obviously higher than those of TaGlu1c and TaGlu1d. Accordingly, we conclude that the main contribution to the deglucosylation reaction is made by the B genome in hexaploid wheat.
In tetraploid wheat, the ratio of transcripts from the A- and B-genome loci of the TaGT and Taglu1 genes was about the same as that observed in hexaploid wheat, in which the B-genome loci were predominantly transcribed. These results suggest that hexaploidization does not influence the transcript profiles of the A- and B-genome loci. In addition, among the three diploid progenitors of hexaploid wheat, transcript levels of the TaGT and Taglu1 orthologs were highest in A. speltoides (SS), the B-genome donor to hexaploid wheat. These facts suggest that the transcriptional bias of the TaGT and Taglu1 genes in hexaploid wheat originated at the diploid level and was retained through polyploidization.
The same conclusion has been reported for the TaBx1 to TaBx5 genes (Nomura et al., 2005). In allopolyploids, there is no global genomic bias in gene transcription (i.e. genomes in which preferentially transcribed homoeoalleles are present vary from gene to gene; Adams et al., 2003). Our results here, combined with results for the TaBx1 to TaBx5 genes, imply a common mechanism allowing preferential transcription of B-genome loci of the Bx-pathway genes. Nomura et al. (2008) analyzed promoter activities of the three homeologs of TaBx3 and TaBx4 genes by transient expression of a reporter protein in wheat protoplasts, but no significant differences were detected among the three homeologs. The authors speculated that this might be due to epigenetic gene regulation related to chromatin structure, such as DNA methylation and/or histone modification. In fact, several studies of transcriptional bias in hexaploid wheat showed that epigenetic chromatin modifications were involved (Bottley et al., 2006; Shitsukawa et al., 2007). The preferential transcription of B-genome loci of all Bx-pathway genes may be controlled by such epigenetic alterations.
Is Dispersal of Bx-Pathway Genes Disadvantageous for Wheat and Rye?
Bx-pathway genes are dispersed to homeologous groups 4 (TaBx1 and TaBx2), 5 (TaBx3–TaBx5), 7 (TaGT), and 2 (Taglu1) chromosomes in hexaploid wheat and to chromosomes 7R (ScBx1 and ScBx2), 5R (ScBx3–ScBx5), 4R (ScGT), and 2R (Scglu) in rye. In contrast, all maize Bx biosynthetic genes (ZmBx1–ZmBx8) are clustered on the short arm of chromosome 4, except for Zmglu1 and Zmglu2 on chromosome 10 and ZmBx9, a ZmBx8 homolog, on chromosome 1 (Fig. 8). It is the common feature in prokaryotic actinomycetes that the biosynthetic genes of secondary metabolites are clustered (Dairi, 2005). Also, secondary metabolites are commonly synthesized by groups of genes that form metabolic gene clusters in eukaryotic filamentous fungi, where the clustered genes are not transcribed as a single mRNA, unlike bacterial operons (Osbourn and Field, 2009). In contrast, most plant genes for secondary metabolite pathways characterized so far are not clustered. But now, five examples of such clustering in plants are known: the Bx-pathway genes in maize (Frey et al., 2009), the triterpenoid avenacin pathway genes in oat (Papadopoulou et al., 1999; Qi et al., 2004), the diterpenoids momilactone (Wilderman et al., 2004; Shimura et al., 2007) and phytocassane (Swaminathan et al., 2009) pathway genes in rice, and triterpenoid thalianol pathway genes in Arabidopsis (Field and Osbourn, 2008). Clustering is thought to facilitate the inheritance of beneficial gene combinations and to promote the coordinated transcription of pathway genes by enabling localized changes in chromatin structure (Wegel et al., 2009). In fact, Zhan et al. (2006) demonstrated that neighboring genes are more frequently coexpressed than would be expected by chance. This model would fit the case of TaBx1 and TaBx2, which are 2.2 kb apart (T. Nomura, unpublished data for the A-genome homeologs) and of TaBx3 and TaBx4, which are 7.3 to 11.3 kb apart in the three genomes of hexaploid wheat (Nomura et al., 2008). Nevertheless, all Bx-pathway genes characterized so far in hexaploid wheat, which are dispersed onto four chromosomes, are coordinately transcribed in each genome despite their genome-dependent differential transcript levels, which vary according to juvenile growth stage. The coordinated transcription of Bx-pathway genes has also been observed in maize (Frey et al., 1995; von Rad et al., 2001; Jonczyk et al., 2008) and may be the case in rye, where Bx production peaks in young seedlings (Sue et al., 2000b) as in wheat and maize; transcript levels have not yet been determined for all of the Bx genes. Apparently, coordinated transcription of Bx-pathway genes during early growth does not depend on gene clustering in wheat and rye. Perhaps there are cis-elements and transcription factors common to all Bx-pathway genes. A computational survey of promoter sequences of all TaBx3 and TaBx4 homeologs in hexaploid wheat and their orthologs in diploid progenitors predicted several cis-elements in common (Nomura et al., 2008). Transcription of some or all genes in a secondary metabolite pathway can be regulated by a small number of transcription factors, such as the OsTGAP1 transcription factor involved in diterpenoid phytoalexin biosynthesis (Okada et al., 2009) and the ORCA3 transcription factor involved in terpenoid-indole alkaloid biosynthesis (van der Fits and Memelink, 2000). The identification of transcription factor(s) and chromatin-related regulatory machinery for Bx-pathway genes should give important clues regarding the metabolic significance of gene clusters in plants.
MATERIALS AND METHODS
Plant Materials
A cultivar of hexaploid wheat (Triticum aestivum; 2n = 6x = 42; genomes AABBDD), Chinese Spring, was used for cDNA cloning, genomic PCR, northern hybridization, qRT-PCR, and Southern hybridization. A tetraploid wheat derived from CS (Tetra-CS; 2n = 4x = 28; AABB; Yang et al., 1999) was used for northern hybridization and qRT-PCR. For Southern analysis, two cultivars of barley (Hordeum vulgare; 2n = 2x = 14; HH), Betzes and Wasedori-nijo, and three diploid progenitors (2n = 2x = 14) of hexaploid wheat, Triticum urartu (accession KU199-6; AA), Aegilops speltoides (KU5727, SS), and Aegilops tauschii (KU20-9, DD) were used. A cultivar of rye (Secale cereale; 2n = 2x = 14; RR), Haru-ichiban, was used for cloning ScGT cDNA. To assign chromosomal locations of GT and glu loci in hexaploid wheat and rye, we used aneuploid lines of CS wheat. Details of each line used are described in Supplemental Materials and Methods S1. Seeds of Wasedori-nijo and Haru-ichiban were purchased from Yukijirushi Shubyo. Other seed stocks were obtained from the National BioResource Project-Wheat in Japan. Seeds were germinated and grown as described by Nomura et al. (2002).
Cloning of GT and Glu cDNAs
The cDNAs for TaGTa to TaGTd and Taglu1d were isolated from 48-h-old shoots of hexaploid wheat (cv CS) by screening a cDNA library and RT-PCR using primers listed in Supplemental Table S4. ScGT cDNA was isolated from a cDNA library of 48-h-old shoots of rye (cv Haru-ichiban). Details are described in Supplemental Materials and Methods S1.
Chromosomal Assignment of GT and Glu Loci in Hexaploid Wheat and Rye
Chromosomal locations of the TaGTa to TaGTd loci and the Taglu1a to Taglu1d loci in hexaploid wheat were assigned using the procedure described by Nomura et al. (2005). Chromosomal locations of the ScGT and Scglu loci in rye were determined by genomic PCR from wheat-rye chromosome addition lines. Primers used for chromosomal assignment are listed in Supplemental Table S6. Details are described in Supplemental Materials and Methods S1.
Expression and Purification of Recombinant TaGT and TaGlu1 Enzymes
For heterologous expression of N-terminal His-tagged TaGT enzymes, the entire coding region flanked by NdeI and HindIII sites was amplified by PCR using the primers listed in Supplemental Table S5 and was ligated into the NdeI and HindIII sites of a pET28a vector (Novagen). The resulting plasmid was transferred into BL21(DE3)pLysS for protein expression. See Supplemental Materials and Methods S1 for expression and purification of the recombinant TaGT enzymes.
For expression of the N-terminal His-tagged TaGlu1d enzyme, the coding region of mature TaGlu1d enzyme flanked by NcoI and XhoI sites was prepared by PCR using the primers shown in Supplemental Table S5. Protein expression in Escherichia coli and purification were performed as described for TaGlu1a to TaGlu1c (Sue et al., 2006).
Enzyme Assays
Activities of the TaGT enzymes were measured in 50 mm Tris-HCl buffer (pH 7.5) in a total volume of 500 μL. After incubation at 35°C, reactions were terminated by adding 50 μL of 1 n HCl, and reaction products were analyzed by HPLC (eluent, 24% [v/v] methanol containing 0.1% [v/v] acetic acid; column, Wakosil-II 5C18 HG [4.6 × 150 mm]; detection, 280 nm; flow rate, 0.9 mL min−1; temperature, 40°C). To determine the kinetic parameters, UDP-Glc was fixed at 0.5 mm and concentrations of DIBOA or DIMBOA were varied from 3 to 50 μm. Kinetic parameters for TaGlu1d were determined according to the method described previously (Sue et al., 2006). Km and Vmax values were calculated by fitting the data from several experiments to the Michaelis-Menten equation using SigmaPlot 11 (Systat Software).
qRT-PCR Analysis
Total RNA was isolated from 48- and 96-h-old CS and Tetra-CS using an RNeasy Plant Mini Kit (Qiagen), and the first-strand cDNA was synthesized from 2 μg of total RNA with SuperScript III reverse transcriptase (Invitrogen) and an oligo(dT) primer. Appropriately diluted RT sample (1 μL per 20 μL of PCR mixture) was subjected to real-time qPCR analysis on a MiniOpticon (Bio-Rad) with SYBR GreenER qPCR Supermix Universal (Invitrogen). Primers specific to each of the four cDNAs of TaGT and Taglu1 genes were used (Supplemental Table S7). All PCR conditions followed the manufacturer’s instructions, except the annealing temperature was 60°C. Each sample was quantified with respect to DNA standards (ranging from 102 to 106 copies per reaction tube). Specificity of the amplification was confirmed by melt-curve analysis and agarose gel electrophoresis.
Northern Analysis
Total RNA was isolated from shoots of CS, Tetra-CS, and diploid progenitors of hexaploid wheat, T. urartu, A. speltoides, and A. tauschii. See Supplemental Materials and Methods S1 for labeling of the probe, hybridization, and signal detection.
Southern Analysis
Total DNA was isolated from 5-d-old shoots of two barley cultivars (Betzes and Wasedori-nijo) and CS wheat using a DNeasy Plant Mini Kit (Qiagen). Aliquots (20 μg) of total DNA were digested individually with ApaI, EcoRV, or XbaI. See Supplemental Materials and Methods S1 for labeling of the probe, hybridization, and signal detection.
The nucleotide sequences reported in this paper have been submitted to the GenBank/EMBL/DDBJ databases with accession numbers AB547237 to AB547240, AB548283, and AB548284.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of amino acid sequences of TaGTs with their orthologs in rye (ScGT) and maize (ZmBX8 and ZmBX9).
Supplemental Figure S2. Alignment of amino acid sequences of TaGlu1s with their orthologs in rye (ScGlu) and maize (ZmGlu1 and ZmGlu2).
Supplemental Table S1. Nucleotide and amino acid identities among GT genes in hexaploid wheat, rye, and maize.
Supplemental Table S2. Nucleotide and amino acid identities among glu genes in hexaploid wheat, rye, and maize.
Supplemental Table S3. GT and glu cDNAs in hexaploid wheat, rye, and maize.
Supplemental Table S4. Primer sequences used for cloning.
Supplemental Table S5. Primer sequences used for the construction of E. coli expression plasmids.
Supplemental Table S6. Primer sequences used for chromosomal assignment.
Supplemental Table S7. Primer sequences used for qRT-PCR.
Supplemental Materials and Methods S1. Detailed experimental procedures.
Acknowledgments
We are grateful to Dr. Takashi Endo (Kyoto University) for seeds of the CS-4H (4B) substitution line.
References
- Adams KL, Cronn R, Percifield R, Wendel JF. (2003) Genes duplicated by polyploidy show unequal contribution to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA 100: 4649–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhunova AR, Matniyazov RT, Liang H, Akhunov ED. (2010) Homoeolog-specific transcriptional bias in allopolyploid wheat. BMC Genomics 11: 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottley A, Xia GM, Koebner RMD. (2006) Homoeologous gene silencing in hexaploid wheat. Plant J 47: 897–906 [DOI] [PubMed] [Google Scholar]
- Cicek M, Esen A. (1999) Expression of soluble and catalytically active plant (monocot) β-glucosidases in E. coli. Biotechnol Bioeng 63: 392–400 [DOI] [PubMed] [Google Scholar]
- Czjzek M, Cicek M, Zamboni V, Bevan DR, Henrissat B, Esen A. (2000) The mechanism of substrate (aglycone) specificity in β-glucosidases is revealed by crystal structures of mutant maize β-glucosidase-DIMBOA, -DIMBOA-Glc, and -dhurrin complexes. Proc Natl Acad Sci USA 97: 13555–13560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dairi T. (2005) Studies on biosynthetic genes and enzymes of isoprenoids produced by actinomycetes. J Antibiot 58: 227–243 [DOI] [PubMed] [Google Scholar]
- Devos KM. (2005) Updating the ‘crop circle’. Curr Opin Plant Biol 8: 155–162 [DOI] [PubMed] [Google Scholar]
- Devos KM, Atkinson MD, Chinoy CN, Francis HA, Harcourt RL, Koebner RMD, Liu CJ, Masojć P, Xie DX, Gale MD. (1993) Chromosomal rearrangements in the rye genome relative to that of wheat. Theor Appl Genet 85: 673–680 [DOI] [PubMed] [Google Scholar]
- Devos KM, Gale MD. (1997) Comparative genetics in the grasses. Plant Mol Biol 35: 3–15 [PubMed] [Google Scholar]
- Ebisui K, Ishihara A, Hirai N, Iwamura H. (1998) Occurrence of 2,4-dihydroxy-7-methoxy-1,4-benzpxazin-3-one (DIMBOA) and a β-glucosidase specific for its glucoside in maize seedlings. Z Naturforsch C 53: 793–798 [Google Scholar]
- Ebisui K, Ishihara A, Iwamura H. (2001) Purification and characterization of UDP-glucose:cyclic hydroxamic acid β-glucosyltransferases from maize seedlings. Plant Physiol Biochem 39: 27–35 [Google Scholar]
- Feldman M, Levy AA. (2005) Allopolyploidy: a shaping force in the evolution of wheat genomes. Cytogenet Genome Res 109: 250–258 [DOI] [PubMed] [Google Scholar]
- Field B, Osbourn AE. (2008) Metabolic diversification-independent assembly of operon-like gene clusters in different plants. Science 320: 543–547 [DOI] [PubMed] [Google Scholar]
- Frey M, Chomet P, Glawischnig E, Stettner C, Grün S, Winklmair A, Eisenreich W, Bacher A, Meeley RB, Briggs SP, et al. (1997) Analysis of a chemical plant defense mechanism in grasses. Science 277: 696–699 [DOI] [PubMed] [Google Scholar]
- Frey M, Huber K, Park WJ, Sicker D, Lindberg P, Meeley RB, Simmons CR, Yalpani N, Gierl A. (2003) A 2-oxoglutarate-dependent dioxygenase is integrated in DIMBOA-biosynthesis. Phytochemistry 62: 371–376 [DOI] [PubMed] [Google Scholar]
- Frey M, Kliem R, Saedler H, Gierl A. (1995) Expression of a cytochrome P450 gene family in maize. Mol Gen Genet 246: 100–109 [DOI] [PubMed] [Google Scholar]
- Frey M, Schullehner K, Dick R, Fiesselmann A, Gierl A. (2009) Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry 70: 1645–1651 [DOI] [PubMed] [Google Scholar]
- Frey M, Stettner C, Paré PW, Schmelz EA, Tumlinson JH, Gierl A. (2000) An herbivore elicitor activates the gene for indole emission in maize. Proc Natl Acad Sci USA 97: 14801–14806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierl A, Frey M. (2001) Evolution of benzoxazinone biosynthesis and indole production in maize. Planta 213: 493–498 [DOI] [PubMed] [Google Scholar]
- Grün S, Frey M, Gierl A. (2005) Evolution of indole alkaloid biosynthesis in the genus Hordeum: distribution of gramine and DIBOA and isolation of the benzoxazinoid biosynthesis genes from Hordeum lechleri. Phytochemistry 66: 1264–1272 [DOI] [PubMed] [Google Scholar]
- Huang S, Sirikhachornkit A, Su X, Faris J, Gill B, Haselcorn R, Gornicki P. (2002) Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc Natl Acad Sci USA 99: 8133–8138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonczyk R, Schmidt H, Osterrieder A, Fiesselmann A, Schullehner K, Haslbeck M, Sicker D, Hofmann D, Yalpani N, Simmons C, et al. (2008) Elucidation of the final reactions of DIMBOA-glucoside biosynthesis in maize: characterization of Bx6 and Bx7. Plant Physiol 146: 1053–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton V, Niemeyer HM, Jonsson LMV. (1994) Substrate specificity of a glucosyltransferase and an N-hydroxylase involved in the biosynthesis of cyclic hydroxamic acids in Gramineae. Phytochemistry 36: 887–892 [Google Scholar]
- Liu MD, Atkinson MD, Chinoy CN, Devos KM, Gale MD. (1992) Nonhomoeologous translocations between group 4, 5 and 7 chromosomes within wheat and rye. Theor Appl Genet 83: 305–312 [DOI] [PubMed] [Google Scholar]
- Melanson D, Chilton M-D, Masters-Moore D, Chilton WS. (1997) A deletion in an indole synthase gene is responsible for the DIMBOA-deficient phenotype of bxbx maize. Proc Natl Acad Sci USA 94: 13345–13350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer HM. (1988) Hydroxamic acids (4-hydroxy-1,4-benzoxazin-3-ones): defence chemicals in the Gramineae. Phytochemistry 27: 3349–3358 [Google Scholar]
- Niemeyer HM. (2009) Hydroxamic acids derived from 2-hydroxy-2H-1,4-bezoxazin-3 (4H)-one: key defense chemicals of cereals. J Agric Food Chem 57: 1677–1696 [DOI] [PubMed] [Google Scholar]
- Nikus J, Esen A, Jonsson LMV. (2003) Cloning of a plastidic rye (Secale cereale) β-glucosidase cDNA and its expression in Escherichia coli. Physiol Plant 118: 337–345 [Google Scholar]
- Nomura T, Ishihara A, Imaishi H, Endo TR, Ohkawa H, Iwamura H. (2002) Molecular characterization and chromosomal localization of cytochrome P450 genes involved in the biosynthesis of cyclic hydroxamic acids in hexaploid wheat. Mol Genet Genomics 267: 210–217 [DOI] [PubMed] [Google Scholar]
- Nomura T, Ishihara A, Imaishi H, Ohkawa H, Endo TR, Iwamura H. (2003) Rearrangement of the genes for the biosynthesis of benzoxazinones in the evolution of Triticeae species. Planta 217: 776–782 [DOI] [PubMed] [Google Scholar]
- Nomura T, Ishihara A, Iwamura H, Endo TR. (2007a) Molecular characterization of benzoxazinone-deficient mutation in diploid wheat. Phytochemistry 68: 1008–1016 [DOI] [PubMed] [Google Scholar]
- Nomura T, Ishihara A, Yanagita RC, Endo TR, Iwamura H. (2005) Three genomes differentially contribute to the biosynthesis of benzoxazinones in hexaploid wheat. Proc Natl Acad Sci USA 102: 16490–16495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Ishizuka A, Kishida K, Islam AKMR, Endo TR, Iwamura H, Ishihara A. (2007b) Chromosome arm location of the genes for the biosynthesis of hordatines in barley. Genes Genet Syst 82: 455–464 [DOI] [PubMed] [Google Scholar]
- Nomura T, Nasuda S, Kawaura K, Ogihara Y, Kato N, Sato F, Kojima T, Toyoda A, Iwamura H, Endo TR. (2008) Structure of the three homoeologous loci of wheat benzoxazinone biosynthetic genes TaBx3 and TaBx4 and characterization of their promoter sequences. Theor Appl Genet 116: 373–381 [DOI] [PubMed] [Google Scholar]
- Okada A, Okada K, Miyamoto K, Koga J, Shibuya N, Nojiri H, Yamane H. (2009) OsTGAP1, a bZIP transcription factor, coordinately regulates the inductive production of diterpenoid phytoalexins in rice. J Biol Chem 284: 26510–26518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn AE, Field B. (2009) Operons. Cell Mol Life Sci 66: 3755–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan H, Levy AA, Feldman M. (2001) Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13: 1735–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou K, Melton RE, Leggett M, Daniels MJ, Osbourn AE. (1999) Compromised disease resistance in saponin-deficient plants. Proc Natl Acad Sci USA 96: 12923–12928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Bakht S, Leggett M, Maxwell C, Melton R, Osbourn A. (2004) A gene cluster for secondary metabolism in oat: implications for the evolution of metabolic diversity in plants. Proc Natl Acad Sci USA 101: 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Salse J, Abrouk M, Bolot S, Guihot N, Courcelle E, Faraut T, Waugh R, Close TJ, Messing J, Feuillet C. (2009) Reconstruction of monocotyledonous proto-chromosomes reveals faster evolution in plants than in animals. Proc Natl Acad Sci USA 106: 14908–14913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salse J, Bolot S, Throude M, Jouffe V, Piegu B, Quraishi UM, Calcagno T, Cooke R, Delseny M, Feuillet C. (2008a) Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 20: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salse J, Chagué V, Bolot S, Magdelenat G, Huneau C, Pont C, Belcram H, Couloux A, Gardais S, Evrard A, et al. (2008b) New insights into the origin of the B genome of hexaploid wheat: evolutionary relationships at the SPA genomic region with the S genome of the diploid relative Aegilops speltoides. BMC Genomics 9: 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable JC, Springer NM, Freeling M. (2011) Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci USA 108: 4069–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura K, Okada A, Okada K, Jikumaru Y, Ko K-W, Toyomasu T, Sassa T, Hasegawa M, Kodama O, Shibuya N, et al. (2007) Identification of a biosynthetic gene cluster in rice for momilactones. J Biol Chem 282: 34013–34018 [DOI] [PubMed] [Google Scholar]
- Shitsukawa N, Tahira C, Kassai K, Hirabayashi C, Shimizu T, Takumi S, Mochida K, Kawaura K, Ogihara Y, Murai K. (2007) Genetic and epigenetic alteration among three homoeologous genes of a class E MADS box gene in hexaploid wheat. Plant Cell 19: 1723–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicker D, Frey M, Schulz M, Gierl A. (2000) Role of natural benzoxazinones in the survival strategy of plants. Int Rev Cytol 198: 319–346 [DOI] [PubMed] [Google Scholar]
- Sue M, Ishihara A, Iwamura H. (2000a) Occurrence and characterization of a UDP-glucose:hydroxamic acid glucosyltransferase isolated from wheat (Triticum aestivum) seedlings. Z Naturforsch C 55: 701–707 [DOI] [PubMed] [Google Scholar]
- Sue M, Ishihara A, Iwamura H. (2000b) Purification and characterization of a β-glucosidase from rye (Secale cereale L.) seedlings. Plant Sci 155: 67–74 [DOI] [PubMed] [Google Scholar]
- Sue M, Ishihara A, Iwamura H. (2000c) Purification and characterization of a hydroxamic acid glucoside β-glucosidase from wheat (Triticum aestivum L.) seedlings. Planta 210: 432–438 [DOI] [PubMed] [Google Scholar]
- Sue M, Yamazaki K, Kouyama J, Sasaki Y, Ohsawa K, Miyamoto T, Iwamura H, Yajima S. (2005) Purification, crystallization and preliminary x-ray analysis of a hexameric β-glucosidase from wheat. Acta Crystallogr Sect F Struct Biol Cryst Commun 61: 864–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sue M, Yamazaki K, Yajima S, Nomura T, Matsukawa T, Iwamura H, Miyamoto T. (2006) Molecular and structural characterization of hexameric β-d-glucosidases in wheat and rye. Plant Physiol 141: 1237–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ. (2009) CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21: 3315–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonova Z, Lai J, Ma J, Ramakrishna W, Llaca V, Bennetzen JL, Messing J. (2004) On the tetraploid origin of the maize genome. Comp Funct Genomics 5: 281–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. (2000) ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289: 295–297 [DOI] [PubMed] [Google Scholar]
- Verdoucq L, Czjzek M, Moriniére J, Bevan DR, Esen A. (2003) Mutational and structural analysis of aglycone specificity in maize and sorghum β-glucosidases. J Biol Chem 278: 25055–25062 [DOI] [PubMed] [Google Scholar]
- von Rad U, Hüttl R, Lottspeich F, Gierl A, Frey M. (2001) Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize. Plant J 28: 633–642 [DOI] [PubMed] [Google Scholar]
- Wegel E, Koumproglou R, Shaw P, Osbourn A. (2009) Cell type-specific chromatin decondensation of a metabolic gene cluster in oats. Plant Cell 21: 3926–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman PR, Xu M, Jin Y, Coates RM, Peters RJ. (2004) Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol 135: 2098–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YF, Furuta Y, Nagata S, Watanabe N. (1999) Tetra Chinese Spring with AABB genomes extracted from the hexaploid common wheat, Chinese Spring. Genes Genet Syst 74: 67–70 [Google Scholar]
- Zhan S, Horrocks J, Lukens LN. (2006) Islands of co-expressed neighbouring genes in Arabidopsis thaliana suggest higher-order chromosome domains. Plant J 45: 347–357 [DOI] [PubMed] [Google Scholar]