Abstract
The membrane protein AtTLP18.3 of Arabidopsis (Arabidopsis thaliana) contains a domain of unknown function, DUF477; it forms a polysome with photosynthetic apparatuses in the thylakoid lumen. To explore the molecular function of AtTLP18.3, we resolved its crystal structures with residues 83 to 260, the DUF477 only, and performed a series of biochemical analyses to discover its function. The gene expression of AtTLP18.3 followed a circadian rhythm. X-ray crystallography revealed the folding of AtTLP18.3 as a three-layer sandwich with three α-helices in the upper layer, four β-sheets in the middle layer, and two α-helices in the lower layer, which resembles a Rossmann fold. Structural comparison suggested that AtTLP18.3 might be a phosphatase. The enzymatic activity of AtTLP18.3 was further confirmed by phosphatase assay with various substrates (e.g. p-nitrophenyl phosphate, 6,8-difluoro-4-methylumbelliferyl phosphate, O-phospho-l-serine, and several synthetic phosphopeptides). Furthermore, we obtained the structure of AtTLP18.3 in complex with O-phospho-l-serine to identify the binding site of AtTLP18.3. Our structural and biochemical studies revealed that AtTLP18.3 has the molecular function of a novel acid phosphatase in the thylakoid lumen. DUF477 is accordingly renamed the thylakoid acid phosphatase domain.
Chloroplasts in higher plants are the center of photosynthesis. In recent years, proteomic studies of the chloroplastic envelope membrane (Ferro et al., 2003; Froehlich et al., 2003), thylakoid membrane (Friso et al., 2004; Peltier et al., 2004), and luminal fraction (Peltier et al., 2002; Schubert et al., 2002; Goulas et al., 2006) have provided clearer knowledge of the chloroplastic proteome of Arabidopsis (Arabidopsis thaliana). The most abundant protein complexes, such as PSII and PSI, cytochrome b6f, and ATP synthase, are photosynthetic apparatuses located at the thylakoid membrane for biogenesis. Intensive research into the photosynthetic protein complexes resolved the crystal structures of PSII/I, cytochrome b6f, and ATP synthase (Jordan et al., 2001; Zouni et al., 2001; Groth, 2002; Stroebel et al., 2003; Loll et al., 2005; Amunts et al., 2007; Umena et al., 2011). Combined with biochemical and functional analyses, structural studies have provided the detailed orientation of the cofactors and the molecular mechanism for photosynthesis. However, a number of auxiliary proteins, including protein translocons, kinases, phosphatases, proteases, chaperones, and many proteins with unknown function, have been isolated by proteome analyses. The proportion of proteins with unknown function is about 25% for the 384 thylakoid proteins (Peltier et al., 2004). The unknown auxiliary proteins are challenging targets because of their low abundance or transient expression at a particular stage (Klimmek et al., 2006). An alternative approach to exploring a molecular function for an unknown protein is to resolve its three-dimensional structure (Hwang et al., 1999; Lee et al., 2001).
The AtTLP18.3 protein located at the thylakoid lumen is 18.3 kD (Peltier et al., 2002; Schubert et al., 2002; Friso et al., 2004). On the basis of bioinformatics analysis, the AtTLP18.3 protein can be divided into three regions: a transit peptide for importing into the chloroplast and thylakoid lumen, residues 1 to 82; a domain of unknown function 477 (DUF477), residues 83 to 235; and a potential transmembrane helix (TMH), residues 260 to 282. The mature form of AtTLP18.3 consists of DUF477 and TMH for anchoring at the thylakoid membrane. In previous studies, the knockout mutant of AtTLP18.3 did not show an observable phenotype under normal growth conditions but showed retarded growth under fluctuating light (Sirpiö et al., 2007; Mulo et al., 2008). The protein was proposed to provide partial protection for the PSII complex to resist photoinhibition under fluctuating high-light conditions (Sirpiö et al., 2007). Furthermore, except for research involving microarrays and proteomics that has alluded to this gene, no research has focused on the function of this gene. Investigating the molecular function of AtTLP18.3 will provide valuable insights into its role in the thylakoid lumen.
DUF477 represents a superfamily of uncharacterized proteins and can be found in eukarya and eubacteria (Finn et al., 2010). We aimed to resolve the three-dimensional structure of AtTLP18.3 to explore its molecular function by structural comparison and functional characterization. We prepared and crystallized a truncated AtTLP18.3 from residues 83 to 260, without the TMH, then structurally compared the resolved structure of AtTLP18.3 and showed a folding similar to that of phosphatases. The phosphatase activity of AtTLP18.3 was further analyzed. In addition, the structure of AtTLP18.3 in complex with O-phospho-l-serine (pSer) was resolved to confirm its substrate binding site and acid phosphatase activity. We discuss the possible role of AtTLP18.3 in photosystem repair in chloroplasts.
RESULTS
Transcripts of AtTLP18.3 Followed a Diurnal Rhythm
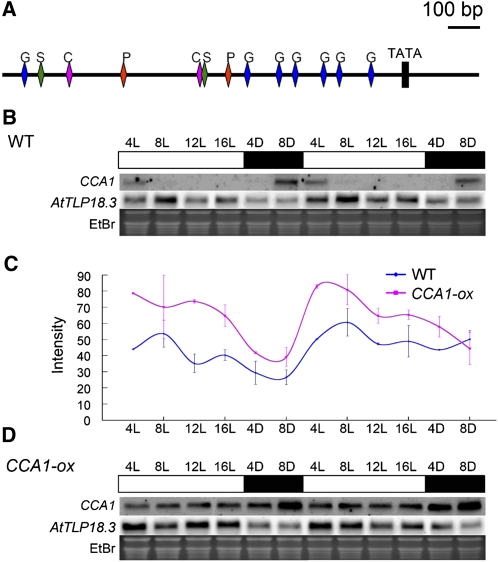
We analyzed the promoter region 1,500 bp upstream of the start codon by using both the PLACE (Higo et al., 1998, 1999) and PlantCARE (Rombauts et al., 1999; Lescot et al., 2002) software. As shown in Figure 1A, the existence of cis-acting regulatory elements of a sugar-repressive element (S), a phytochrome-regulation element (P), a CCA1-binding site (C), and a GA-responsive element (G) implied that AtTLP18.3 might be controlled by the circadian rhythm, Suc, and GA. We compared the diurnal expression patterns of AtTLP18.3 in CIRCADIAN CLOCK-ASSOCIATED1 (CCA1)-overexpressing (CCA1-ox) mutants and wild-type Arabidopsis. Northern-blot analysis revealed AtTLP18.3 with a diurnally fluctuating expression pattern in wild-type plants; the transcripts showed two high expression peaks, at 8 and 16 h after lights were turned on (Fig. 1, B and C). In the CCA1-ox mutant, the diurnal rhythmic expression pattern was still observable but slightly disrupted (Fig. 1, C and D).
Figure 1.
cis-Acting regulatory and expression pattern of AtTLP18.3. A, cis-Acting regulatory element analyzed by PLACE and PlantCARE. Nucleic acid sequence analysis revealed the sugar-repressive element (S), the CCA1-binding site (C), the phytochrome-regulation element (P), and the GA-responsive element (G) within 1,500 bp upstream of the translation start codon of the AtTLP18.3 gene. The block indicates a TATA box. The bar represents 100 bp of nucleic acids. B, Expression pattern of AtTLP18.3 in wild-type (WT) Arabidopsis. C, Comparison of the expression pattern of AtTLP18.3 in wild-type plants and the CCA1-ox mutant. Error bars represent se. D, Expression pattern of AtTLP18.3 in the CCA1-ox mutant. The duration under light (L) and dark (D) are denoted at the top. White bars denote lighted intervals and black bars denote darkness. The expression of CCA1 was used as an internal control. The expression pattern of AtTLP18.3 is shown. Ethidium bromide (EtBr) was used as a loading control. The signal of AtTLP18.3 on northern blots was normalized with 28S rRNA.
The Protein AtTLP18.3 with DUF477 Is Conserved from Cyanobacteria and Algae to Higher Plants
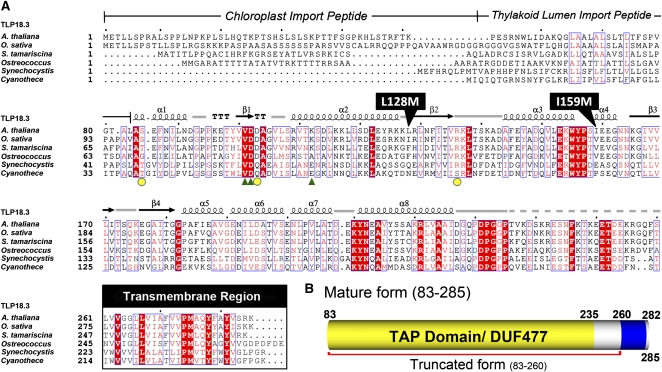
We used the amino acid sequence of AtTLP18.3 to search for homologs with the National Center for Biotechnology Information BLASTP program against the nonredundant protein sequence database (Altschul et al., 1997). No homologs could be found in animal cells. The protein AtTLP18.3 is conserved from cyanobacteria and algae to green plants and is found in photosynthetic organisms. AtTLP18.3 belongs to an orthologous group specific to algae and green plants. The N terminus of AtTLP18.3 (residues 1–82) was predicted as the transit peptide for importing into the chloroplast and thylakoid lumen (Fig. 2A). The mature form of AtTLP18.3 (residues 83–285) comprises DUF477 (residues 83–235) and the TMH (residues 260–282). Amino acid sequence alignment revealed the mature form of the AtTLP18.3 protein with 83% identity and 96% similarity to the homolog of Oryza sativa (japonica cultivar group), 66% identity and 86% similarity to that of Selaginella tamariscina, 47% identity and 66% similarity to that of Ostreococcus lucimarinus CCE9901, 32% identity and 51% similarity to that of Synechocystis sp. PCC 6803, and 31% identity and 52% similarity to that of Cyanothece sp. PCC 7424 (Fig. 2A).
Figure 2.
Sequence alignment of AtTLP18.3 and its homologs. A, Structure-based multiple sequence alignment of AtTLP18.3 among plants, algae, and cyanobacteria homologs. The homologs of AtTLP18.3 show conserved residues and similar domain architecture. Positions of identically conserved residues are shown in white on red, and regions of similarly conserved residues are boxed. The secondary structure elements are depicted above the sequence alignment: α-helices are denoted by coils and β-strands by arrows. Representation of secondary structure elements and numbering are based on the AtTLP18.3 structure. From the sequence alignment, the predicted transit peptide and transmembrane region are delimited in the corresponding boxes. Two point mutations, L128M and I159M, were introduced for the single-wavelength anomalous dispersion method by site-directed mutagenesis. The residues for Ser binding are shown by green triangles and those for calcium ion binding by yellow circles. Species for the sequences are as follows: A. thaliana, Arabidopsis thaliana; O. sativa, Oryza sativa (japonica cultivar group); S. tamariscina, Selaginella tamariscina; Ostreococcus, Ostreococcus lucimarinus CCE9901; Synechocystis, Synechocystis sp. PCC 6803; and Cyanothece, Cyanothece sp. PCC 7424. B, The mature form of AtTLP18.3 in the thylakoid lumen. From the sequence analysis, the mature AtTLP18.3 could be predicted with two major regions: a domain of unknown function (DUF477) and a transmembrane α-helix (TMH).
Structure Determination and Overall Structure of AtTLP18.3 Reveal Its α/β-Folds
Because the phenotype of AtTLP18.3 could not be observed by knockout or overexpression experiments under normal growth conditions (Sirpiö et al., 2007), investigating the molecular structure suggested an alternative approach to explore its function. Therefore, we aimed to resolve the structure of AtTLP18.3 by x-ray crystallography and propose its molecular function by comparing the structure with the known functions of other proteins (Lee et al., 2001). We constructed the truncated AtTLP18.3 from residues 83 to 260 (Fig. 2B). Because AtTLP18.3 lacks a Met residue, we introduced five Met residues into AtTLP18.3 by site-directed mutagenesis (Supplemental Table S1; i.e. L107M, L128M, I133M, I159M, and L202M) for solving the phase by x-ray crystallography. The selected point mutations were located near the loop region by predicting the secondary structure with the PSIPRED server (McGuffin et al., 2000). After screening, we could obtain only the crystals of the protein with the double mutation L128M and I159M (TLP_M45; Fig. 2A).
The crystal of TLP_M45 belongs to an orthorhombic space group, P212121, with unit-cell parameters a = 46.9 Å, b = 49.8 Å, c = 76.7 Å, α = β = γ = 90°, and diffraction to a resolution of 2.6 Å. Two seleno-Met (Se-Met) residues were found after a search by use of SOLVE/RESOLVE (Terwilliger and Berendzen, 1999) with the method of peak wavelength in single-wavelength anomalous dispersion (SAD). After model rebuilding and refinement, the final structure of TLP_M45 reached 20.1% R-factor and 26.8% Rfree. Because the crystals of native AtTLP18.3 were isomorphous to TLP_M45, the structure could be adapted from the structure of TLP_M45. The resolution of native AtTLP18.3 was further diffracted to 1.6 Å, and the structure of AtTLP18.3 was refined to 20.3% R-factor and 22.6% Rfree. All crystallography statistics are shown in Table I.
Table I. Crystallography statistics and refinement for AtTLP18.3.
| Characteristic | TLP | TLP_M45 | TLP-pSer Complex |
| Beamlines at the National Synchrotron Radiation Research Center | 13B1 | 13B1 | 13C1 |
| Diffraction data statistics | |||
| Space group | P212121 | P212121 | P212121 |
| Cell constants (Å and °) | a = 46.98, b = 49.84, c = 76.67; α = β = γ = 90° | ||
| Wavelength (Å) | 0.97315 | 0.9788 | 0.9762 |
| Resolution range (Å) | 30.0–1.6 | 30.0–2.6 | 30.0–2.1 |
| Observed reflections | 343,890 | 57,179 | 58,760 |
| Unique reflections | 24,243 | 5,797 | 10,968 |
| Completeness (%) | 99.3 (100.0) | 97.8 (99.6) | 99.5 (99.9) |
| I/σ(I) | 58.6 (8.7) | 30.8 (38.4) | 26.4 (3.7) |
| Rmerge (%)ab | 3.5 (31.6) | 4.6 (6.2) | 6.3 (49.6) |
| Phasing statistics | |||
| No. of selenium sites | 2 | ||
| Signal-noise ratio | 1.55 | ||
| Figure of merit | 0.42 | ||
| Refinement statistics | |||
| Resolution range (Å) | 30.0–1.6 | 30.0–2.6 | 30.0–2.1 |
| Reflections (working/test) | 21,263/2,346 | 9,945/552 | 9,319/1,068 |
| R-factor/Rfree (%) | 19.5/23.0 | 19.9/27.0 | 18.9/24.8 |
| No. of atoms | |||
| Protein atoms | 1,174 | 1,174 | 1,173 |
| Solvent atoms | 180 | 67 | 150 |
| Root mean square deviations | |||
| Bond length (Å) | 0.022 | 0.015 | 0.012 |
| Bond angle (°) | 2.2 | 1.6 | 1.5 |
| Average B-factor (Å2) | |||
| B-factor (protein) | 28.9 | 31.3 | 39.7 |
| B-factor (solvent) | 51.5 | 37.1 | 57.8 |
| B-factor (ligand) | 71.24 | ||
| Ramachandran plot (%) | |||
| Most favored | 91.7 | 91.7 | 93.2 |
| Additional allowed | 7.5 | 8.3 | 6.8 |
| Generously allowed | 0.8 | 0.0 | 0.0 |
| Disallowed | 0.0 | 0.0 | 0.0 |
Rmerge = 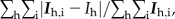 where Ih is the mean intensity of i observations for a given reflection h.
where Ih is the mean intensity of i observations for a given reflection h.
The last shell values are indicated in parentheses.
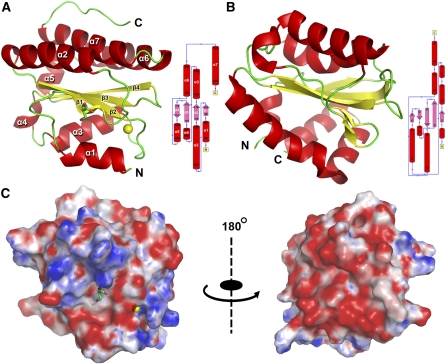
The final structure of truncated AtTLP18.3 contained only a domain with a globular shape from residues 83 to 235 and the C-terminal region from residues 236 to 260, which had no electron density for model building. The domain showed eight α-helices and four β-sheets. The domain architecture belongs to an α/β-folding similar to a “hamburger” shape (Fig. 3A). The lower layer consists of two helices: α1 from Ser-83 to Asp-92 and α3 from Asp-143 to Tyr-156. The upper layer consists of three α-helices: (1) α2 from Ser-108 to Lys-127, (2) α6 from Glu-191 to Asp-208, and (3) α7 from Lys-210 to Asp-227. The middle layer is formed by four-stranded β-sheets with (1) β1 from Tyr-99 to Asp-103, (2) β2 from Arg-129 to Val-135, (3) β3 from Asn-164 to Thr-172, and (4) β4 from Glu-176 to Gly-181. Two short α-helices, α4 from Ser-158 to Asn-163 and α5 from Gly-182 to Gly-190, are perpendicular to the plane of the middle layer and provide the connection for overall structure of the domain (Fig. 3A). We calculated the molecular surface with electrostatic potential (Fig. 3C). In the left panel of Figure 3C, this side was proposed to be the active site in the center, with the positive charge surrounding the negative charge. By rotating the molecular surface 180° (right panel of Fig. 3C), this side showed almost the entire surface with a negative charge.
Figure 3.
Overall structure and topology diagram of AtTLP18.3. A, The crystal structure of AtTLP18.3 was built from residues 83 to 235, which is the thylakoid acid phosphatase domain only. The structure contains the mixed α/β Rossmann fold with a parallel four-stranded twisted β-sheet (topology β1, β2, β3, β4) flanked by two helices on one side (α1, α3) and three helices on the other side (α2, α6, α7). B, The solution structure of mannitol-specific cryptic phosphotransferase enzyme II A CmtB (PDB identifier 2OQ3) from E. coli shares a similar structure and topology with AtTLP18.3. In comparison with CmtB, the RMSD of AtTLP18.3 is 4 Å, with a lower amino acid sequence identity of 12%. AtTLP18.3 also shows a similar structure, with a low structural similarity to other phosphatases. Secondary structure elements are distinguished by color: α-helix by red, β-sheet by yellow, and loops by green. C, Electrostatic surface potential contoured from −10 kT (red) to 10 kT (blue). Two views related by a 180° rotation are shown.
Structure Comparison Suggests That AtTLP18.3 Functions as a Phosphatase
To investigate the molecular function of AtTLP18.3 from the resolved structure, we submitted the coordinates of AtTLP18.3 to the CATH server (Kawabata, 2003; Greene et al., 2007) to reveal its structure classification. The results indicated that the folding of AtTLP18.3 belongs to a Rossmann fold (CATH code 3.40.50) with an architecture of a three-layer (αβα) sandwich. For advanced searching by structure comparison, we submitted the coordinates of AtTLP18.3 to the programs Matras (Kawabata, 2003) and DALI (Holm and Sander, 1998). AtTLP18.3 shared the highest structural similarity in protein size with a mannitol-specific cryptic phosphotransferase enzyme IIA, CmtB from Escherichia coli (Protein Data Bank [PDB] identifier 2OQ3; Fig. 3B). The Z-score between AtTLP18.3 and CmtB was 12.25, and the root mean square deviation was 4 Å. These two proteins have low sequence identity (12%). High Z-scores in the Matras list provided more evidence to suggest that AtTLP18.3 might be a phosphatase, such as the exopolyphosphatase-related protein (PDB identifier 3DMA; Z-score = 14.9, sequence identity = 13.2%) and the cytosolic exopolyphosphatase (PDB identifier 2QB7; Z-score = 11, sequence identity = 13.2%). AtTLP18.3 might function as an enzyme that transfers or removes a phosphate group from a protein. Therefore, we proposed that AtTLP18.3 might be a phosphatase that removes the phosphate group from phosphorylated proteins.
Phosphatase Activity Assay of AtTLP18.3
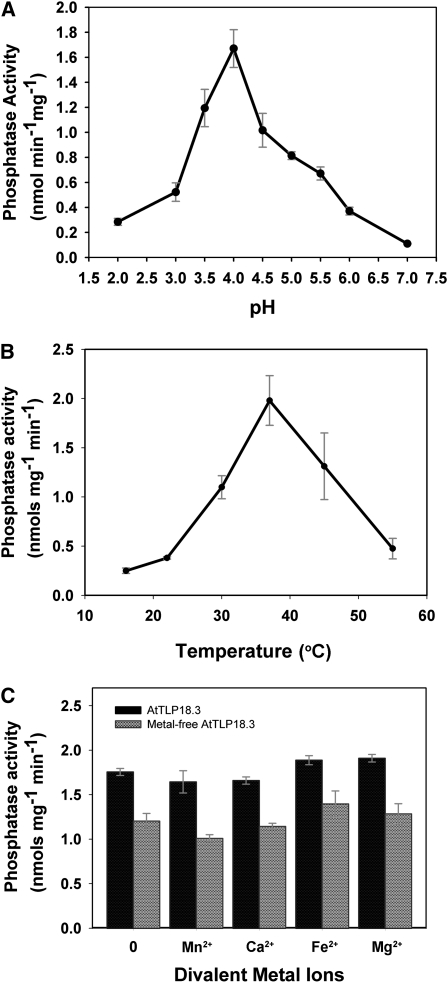
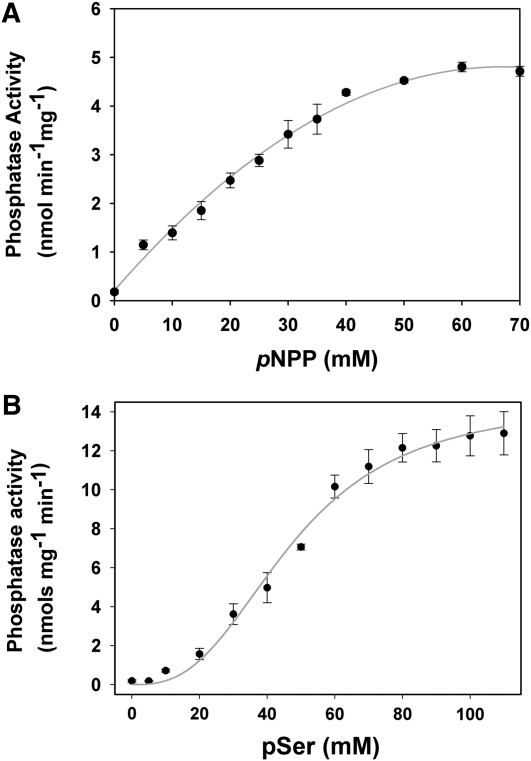
We first analyzed the phosphatase activity of AtTLP18.3 by use of the general substrate p-nitrophenyl phosphate (pNPP), a nonproteinaceous and nonspecific substrate for acid and alkaline phosphatase assays. Preliminary results indicated that AtTLP18.3 contains phosphatase activity. Consequently, we further determined the optimal conditions and enzymatic kinetics of AtTLP18.3 for pNPP. AtTLP18.3 showed the highest activity at pH 3.5 to 4.5 (Fig. 4A) and 30°C to 45°C (Fig. 4B). The pNPP substrate-binding affinity (Km) was 42.56 mm, and the catalytic velocity (Vmax) was 8.11 nmol min−1 mg−1 at the optimal condition of 37°C, pH 4.0 (Fig. 5A; Table II). Furthermore, several divalent ions (i.e. Mn2+, Ca2+, Fe2+, and Mg2+) were added to and removed from the buffer to check whether AtTLP18.3 is a divalent ion-dependent phosphatase. The metal-free AtTLP18.3 showed reduced specific activity (about 25%). Therefore, the phosphatase activity of AtTLP18.3 would be maintained by divalent ions but reduced with divalent ions removed (Fig. 4C).
Figure 4.
Biochemical characterization of AtTLP18.3 phosphatase activity. A, Characterization of AtTLP18.3 for optimal pH. Optimal pH was between 3.5 and 4.5. B, Characterization for optimal temperature. Maximal activity was found at 37°C. Phosphatase activity with pNPP was used as a substrate. Phosphatase activity was assayed by incubating samples of the protein and 7.5 mm pNPP in 0.1 m sodium acetate buffer (pH 4.0). Reactions were initiated by adding purified AtTLP18.3 and incubated for 30 min at room temperature. C, Effect of divalent metal ion on the phosphatase activity. Divalent ions were removed by adding EDTA to all buffers during purification. The metal-free AtTLP18.3 reduced the specific activity of AtTLP18.3 by about 25%. However, similar activity levels were observed in the presence or absence of divalent ions. Error bars represent se.
Figure 5.
Enzymatic kinetics of phosphatase activity with two different substrates, pNPP and pSer. A, Michaelis-Menten plot of AtTLP18.3 with pNPP used as the substrate. The Km for pNPP is 42.56 mm, and the Vmax is 8.11 nmol min−1 mg−1. B, Michaelis-Menten plot of AtTLP18.3 with pSer used as the substrate. The Km for pSer is 47.51 mm, and the Vmax is 14.55 nmol min−1 mg−1. The kinetics assay of AtTLP18.3 shows a classical hyperbolic saturation with pNPP but an allosteric sigmoid with pSer. Error bars represent se.
Table II. Kinetics of phosphatase activity of AtTLP18.3.
| Sample | Km | Vmax |
| nmol min−1 mg−1 | ||
| Substrates | ||
| pNPP | 42.56 mm | 8.10 |
| pSer | 47.51 mm | 14.55 |
| DiFMUP | 49.56 μm | – |
| Phosphopeptides | ||
| Ac-(pT)AILER | 12.49 mm | 3.15 |
| Ac-(pT)IALGK | 16.57 mm | 4.73 |
| RRA(pT)VA | 11.89 mm | 4.97 |
| KR(pT)IRR | 12.56 mm | 4.02 |
| Ac-RK(pS)AGKPKN | 13.88 mm | 7.43 |
To further determine the phosphatase activity of AtTLP18.3, we examined several different substrates, such as 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP) and pSer and five synthetic phosphorylated oligopeptides (Table II). We used the optimal condition (37°C, pH 4.0) determined by use of the substrate pNPP. Assay of AtTLP18.3 kinetics showed a classical hyperbolic saturation with pNPP but an allosteric sigmoid with pSer. The Km and Vmax from the substrate pSer were 47.51 mm and 14.55 nmol min−1 mg−1, respectively (Fig. 5B; Table II). In addition, kinetics assay with the substrate DiFMUP revealed the classical hyperbolic saturation (Supplemental Fig. S2).
We tested the phosphatase activity of AtTLP18.3 against five synthetic phosphorylated oligopeptides: Ac-(pT)AILER and Ac-(pT)IALGK are the phosphorylation sites of the major phosphopeptides of D1 and D2 proteins from Arabidopsis thylakoids (Vener et al., 2001); RRA(pT)VA and KR(pT)IRR are good substrates of the Ser/Thr phosphatases (Donella-Deana et al., 1991); and Ac-RK(pS)AGKPKN is the phosphorylation site of the phosphopeptides from spinach (Spinacia oleracea) thylakoids (Michel et al., 1991). The highest Km of phosphopeptide RRA(pT)VA was 11.89 mm. The highest Vmax of phosphopeptide Ac-RK(pS)AGKPKN was 7.43 nmol min−1 mg−1. The kinetics results for AtTLP18.3 are given in Table II. Thus, AtTLP18.3 functions as an acid phosphatase by removing the phosphate group from pSer or phospho-Thr (pThr).
Structure Determination of the AtTLP18.3-pSer Complex
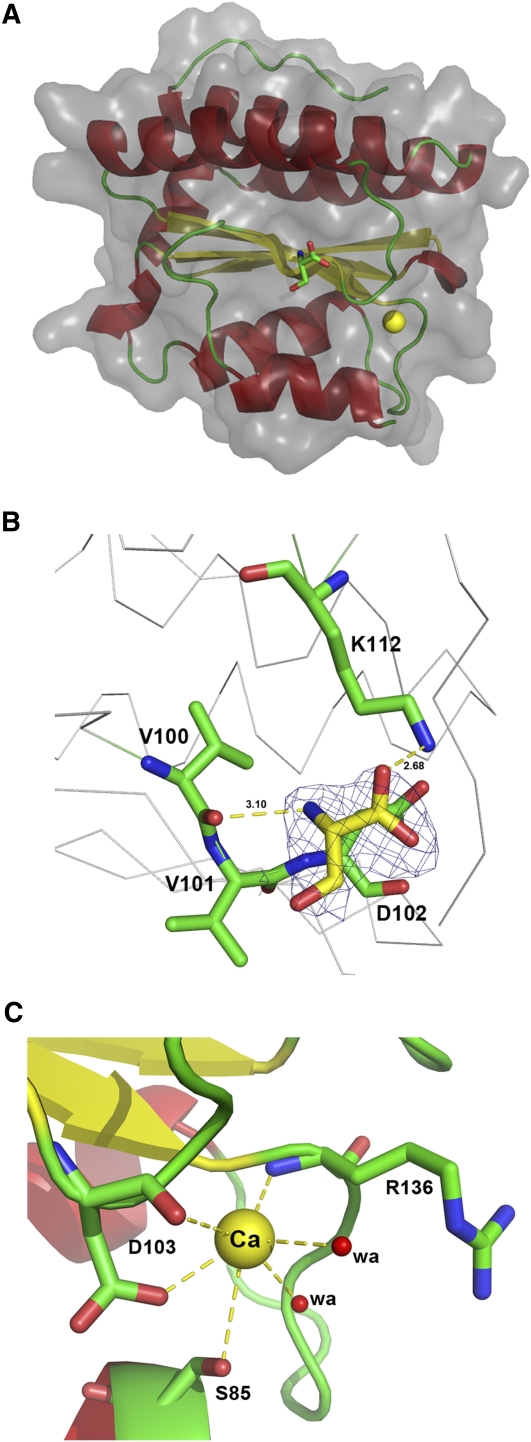
The enzymatic activity of AtTLP18.3 could remove the phosphate group from pSer or pThr; therefore, we analyzed the structure of the AtTLP18.3-pSer complex to examine the active site of AtTLP18.3. Crystals of AtTLP18.3 were grown and transferred to the same buffer containing an additional 20 mm pSer for 7 d. After soaking with pSer, the crystal of the AtTLP18.3-pSer complex was diffracted to 2.1 Å resolution (Table I). The phosphate group of pSer was removed from the structure, and the residue Ser remained in the binding site (Fig. 6A). Electron density analysis of the residue Ser showed the formation of a hydrogen bond with the residues Val-101, Asp-102, and Lys-112 (Fig. 6B). The residues Val-101 and Asp-102 are conserved, and the residue Lys-112 is conserved only in land plants (Fig. 2A). We identified a binding site of calcium ion with six hydrogen bonds formed by the oxygen atom of Ser-85, the carbonyl group and the carboxylic group of Asp-103, the carbonyl group of Arg-136, and two water molecules (Fig. 6C).
Figure 6.
The structure of the AtTLP18.3-pSer complex. A, The binding site of AtTLP18.3 can be identified with a dephosphorylated pSer and a calcium ion in the structure of AtTLP18.3. A bound calcium ion is represented by the yellow sphere, and a Ser is represented by a stick. B, The substrate-binding site of AtTLP18.3. The residue Ser is contoured by a 1.0 σ 2Fo-Fc map and is surrounded by the residues V101, D102, and K112. The yellow dashes represent hydrogen bonds. C, Hydrogen bonding for calcium ion in AtTLP18.3. Six coordinates for calcium ion (Ca) were formed by an oxygen atom of S85, a carbonyl group and a carboxylic group of D103, a carbonyl group of R136, and two water molecules (denoted by wa). The yellow dashes represent hydrogen bonds.
DISCUSSION
AtTLP18.3 Is an Auxiliary Protein Present on the Lumen Side of the Thylakoid Membrane, and Its Gene Expression Follows a Circadian Rhythm
In the past several years, different proteomic approaches have revealed hundreds of chloroplast proteins, including those with unknown function, located at the thylakoid membrane and lumen (Schubert et al., 2002; Friso et al., 2004; Goulas et al., 2006; Sirpiö et al., 2007). These proteomic studies provide a detailed overview of photosynthetic apparatuses and the auxiliary proteins for maintaining the photosystem in the chloroplast. These studies revealed the functions of most proteins isolated from the polysome in the thylakoid membrane as involved in photosynthetic apparatuses or translational complexes. Some potential auxiliary proteins with unknown function were also identified. AtTLP18.3 is one of the proteins associated with the polysome, and only a low amount can be found in the thylakoid preparation. Furthermore, the amount of AtTLP18.3 protein remains constant and is not regulated by light (Sirpiö et al., 2007).
AtTLP18.3 showed a diurnal rhythm of gene expression under a 16-h-light/8-h-dark growth photoperiod in Arabidopsis. The transcripts of AtTLP18.3 showed high expression in the daylight and low expression in the dark (Fig. 1). In contrast to the gene expression, the protein level remained constant (Sirpiö et al., 2007). The reasons for the differential expression might involve posttranscriptional control for the gene expression of AtTLP18.3 and the stability of the protein globular architectures maintaining the amount of AtTLP18.3 protein. Further experiments are needed to confirm this possibility.
AtTLP18.3 Contains a Novel Acid Phosphatase Domain
Phosphatase activity in chloroplasts has been characterized by previous studies (Sun et al., 1989; Vener et al., 1999; Trotta et al., 2011). The catalytic domain of some kinds of phosphatases, such as homologs of protein phosphatase 2A, may be exposed to the stroma face (Vener et al., 1999). However, the phosphatase located at the thylakoid lumen has not been reported. From our structural comparison with several known proteins, AtTLP18.3 seemed to be a novel acid phosphatase in the thylakoid lumen. We further analyzed the phosphatase activity of AtTLP18.3 with different substrates, such as pNPP, DiFMUP, pSer, and five phosphorylated oligopeptides. AtTLP18.3 showed phosphatase activity with two artificial substrates, pNPP and DiFMUP. To explore the possible cellular substrate activity, we confirmed the acid phosphatase activities with pSer and several designed phosphorylated oligopeptides from PSII (Vener et al., 2001). Theoretically, because the phosphorylated oligopeptides are located at the N terminus of PSII in the chloroplast stroma, their dephosphorylation by AtTLP18.3 should not occur in the thylakoid lumen. Further effort is needed to search for substrates on the lumen side for AtTLP18.3. Because AtTLP18.3 exhibited phosphatase activity on pSer and pThr (Table II), DUF477 should be renamed the thylakoid acid phosphatase domain (Fig. 2B). As compared with one undefined acid phosphatase with 10.4 μmol min−1 mg−1 specific activity of pNPP in thylakoid membranes (Rengasamy et al., 1981), AtTLP18.3 showed much lower phosphatase activity (Table II), which indicates another kind of acid phosphatase in the thylakoid lumen.
Is AtTLP18.3 Involved in the Repair Cycle of the Photosystem?
In the chloroplasts, a transthylakoid proton motive force can be established across the thylakoid membrane by photosynthetic proton transfer, and the lumen becomes acidic during photosynthesis (Pottosin and Schönknecht, 1996; Kieselbach et al., 1998). A previous study indicated that the pH of the thylakoid lumen is between 5.8 and 6.5 under normal conditions and decreases to become strongly acidic, less than 5, under stress conditions (Kramer et al., 1999). From our results, the gene expression of AtTLP18.3 in the lumen follows a diurnal rhythm, with the protein level constant. In addition, the optimal pH for the acid phosphatase activity of AtTLP18.3 was between 3.5 and 4.5. Thus, the phosphatase activity of AtTLP18.3 in the lumen might be activated in the daylight and under acidic stress. The activity would be deactivated in the dark when the lumen was in a neutral condition. In the stress condition, the more acidic condition might present an activated environment for AtTLP18.3 removing the phosphate group from damaged proteins.
AtTLP18.3 may be involved in regulating the repair cycle of the photosystem (Sirpiö et al., 2007). Phosphorylation and dephosphorylation are important regulation mechanisms in a wide range of cellular processes. The thylakoid membrane contains many photosynthetic apparatuses. Many proteins in the PSII reaction center undergo posttranslational phosphorylation under high-light radiation. One of the core proteins in the PSII reaction center, D1, is regulated by reversible phosphorylation (Koivuniemi et al., 1995). Phosphorylation of D1 is a signal for migration of the photodamaged PSII core complex from grana membranes to stroma lamellae, and then the photodamaged D1 protein is digested by protease and replaced by new D1 protein. Without dephosphorylation, phosphorylated D1 is a poor substrate for degradation (Ebbert and Godde, 1996). However, the phosphorylation of D1 protein occurs in the N terminus of D1, toward the stroma side of the thylakoid membrane. The phosphorylated sites of D1 in the stroma and AtTLP18.3 in the thylakoid lumen will not physically interact because of different localization. This situation raises the question of how the AtTLP18.3 protein can repair the photosystem after photodamage. AtTLP18.3 might not be directly involved in the repair cycle of the photosystem. The thylakoid lumen may contain some unknown phosphoproteins that undergo dephosphorylation by the phosphatase activity of AtTLP18.3.
In conclusion, we used a structure approach to explore the molecular function of AtTLP18.3. The truncated AtTLP18.3 with only the domain of unknown function, DUF477, was crystallized and diffracted at resolution 1.6 Å. Because the native AtTLP18.3 contained no Met residues, we introduced two Met residues into AtTLP18.3 by site-directed mutagenesis. From the double mutation of AtTLP18.3, the crystal structure of the AtTLP18.3 mutant was resolved by the SAD method. The structure of truncated AtTLP18.3 contains a domain with an α/β-fold that belongs to a Rossmann fold. Structure comparison indicated that AtTLP18.3 may function as a phosphatase in the thylakoid lumen. We used the standard reagents for phosphatase, such as pNPP, DiFMUP, pSer, and phosphopeptides of the chloroplast, to confirm the phosphatase activity. Enzymatic kinetics and biochemical analyses revealed an optimal condition for expression at 37°C and pH 4 and identified the acid phosphatase activity. Therefore, the DUF477 of AtTLP18.3 is renamed the thylakoid acid phosphatase domain. In addition, the substrate-binding site in the structure of AtTLP18.3 was determined by use of an AtTLP18.3-pSer complex. The acid phosphatase activity of AtTLP18.3 in the thylakoid lumen may be associated with dephosphorylation under daylight and stress.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Columbia ecotype of Arabidopsis (Arabidopsis thaliana) was used in this study. Seeds of the CCA1-ox mutant were a gift from Dr. E.M. Tobin (University of California, Los Angeles). Seeds were sown in soil, acclimated at 4°C for 2 to 4 d, and then transferred to a thermo-controlled growth chamber. Seedlings were imbibed and grown under a 16-h-light/8-h-dark photoperiod at 23°C and a light intensity of 100 to 120 μmol m−2 s−1. For 1-week-old seedlings, nutrients were further supplied twice a week.
cDNA Cloning of AtTLP18.3
The cDNA (At1g54780) of Arabidopsis was prepared from total RNA with the use of SuperScript III (Invitrogen). The DNA fragment of AtTLP18.3 excluding the N-terminal transit peptide and the C-terminal transmembrane domain was amplified from full-length cDNA with the forward TLP-BamHI primer (5′-CGACGCGAATTCGCCTCTGAGTTCAATATC-3′) and the reverse TLP-XhoI primer (5′-CGATGCCTCGAGTTAACTGAACTGTCCTCGCTT-3′), in which the restriction sites of EcoRI and XhoI sites were introduced at the 5′ and 3′ ends. The obtained DNA fragment was cloned into a pGEM-T Easy vector (Promega) and amplified in the host Escherichia coli strain DH5. After confirmation by DNA sequencing, the fragment was subcloned into the pGEX6P1 expression vector (GE Healthcare).
Secondary Structure Prediction and Site-Directed Mutagenesis
Secondary structure prediction was performed by use of the PSIPRED protein structure prediction server (http://bioinf.cs.ucl.ac.uk/psipred/; Jones, 1999; McGuffin et al., 2000) for assigning the topology of AtTLP18.3. After prediction, several residues were chosen for Met substitution based on previous reports (Gassner and Matthews, 1999; Ohmura et al., 2001). Point mutations involved use of the site-directed mutagenesis protocol of whole-plasmid synthesis PCR (Weiner et al., 1994) with the designated primers (Supplemental Table S1) and the plasmid of pGEX6P1-AtTLP18.3 as a template. The above complementary primers were optimized by use of the Web-based primer design program PrimerX (http://bioinformatics.org/primerx/) and synthesized by Mission Biotech. The conditions of PCR for site-directed mutagenesis consisted of a 5-min denaturation at 95°C followed by 16 cycles of 30 s at 95°C, 60 s at 55°C, and 5.5 min at 72°C. After amplification, the reactions were digested with DpnI for 1 h at 37°C to remove template plasmid DNA. The mutants containing a single point mutation were first made individually and expressed as for overexpression of the wild-type protein for checking protein solubility. The mutant with a double point mutation, L128M and I159M, was finally chosen and generated from the soluble proteins. All plasmids containing point mutations were confirmed for substitution of the target residues by DNA sequencing.
Protein Expression and Purification
Both the wild type and mutant with double mutations (L128M and I159M) of AtTLP18.3 were overexpressed in E. coli BL21 (DE3). The transformant cells of wild-type AtTLP18.3 were grown at 37°C in Luria-Bertani medium containing 100 μg mL−1 ampicillin to an optical density at 600 nm of 0.5 to 0.7 before induction with 0.1 mm isopropyl β-d-1-thiogalactopyranoside for 4 h. The transformant cells containing the double mutation of AtTLP18.3 were grown in M9 medium. Se-Met at 100 μg mL−1 was supplied when A600 reached 0.6 to 0.8 and incubated at 28°C for 1 h. Finally, the recombinant protein in bacterial cells was induced with 1 mm isopropyl β-d-1-thiogalactopyranoside at 28°C for 12 to 16 h. All bacterial cells were harvested and lysed by use of a sonicator (Misonix model XL 3000) in the binding buffer (50 mm Tris-HCl, pH 7.0, and 150 mm NaCl). The supernatant containing the recombinant protein was then collected by centrifugation at 12,000g for 30 min at 4°C. The glutathione-S-transferase (GST) fusion protein was purified by use of a GSTrap FF column with an Äkta Prime FPLC system (GE Healthcare). After washing the GST affinity column with 10 column volumes of the binding buffer, the recombinant protein was further eluted with elution buffer (50 mm Tris-HCl, pH 8.8, and 10 mm reduced glutathione). The eluted protein solutions were dialyzed with the cleavage buffer (50 mm Tris-HCl, pH 7.0, 150 mm NaCl, and 1 mm dithiothreitol) for equilibration and then cleaved by use of Precission protease (GE Healthcare) with 10 units mg−1 fusion protein at 4°C for 16 h. After cleavage, the Precission protease and GST protein were removed by passage through a GSTrap FF column. The purified recombinant AtTLP18.3 protein was concentrated by ultrafiltration and dialyzed with the dialysis tube (Amicon Ultra-4; Millipore) and a buffer containing 20 mm Tris-HCl, pH 7.0, and 150 mm NaCl. The expression and purification of AtTLP18.3 by 12.5% SDS-PAGE are shown in Supplemental Figure S1. The Se-Met protein was purified with the same protocol as for the wild-type protein, except for the addition of 1 mm Tris(2-carboxyethyl)phosphine.
Northern-Blot Analysis
Total RNA was extracted with the REzol C&T reagent and analyzed on a 1.2% formaldehyde agarose gel. After electrophoresis, the RNA was transferred from the agarose gel to a nylon membrane, and then the membrane was hybridized with a digoxigenin-11-dUTP-labeled probe. Finally, the signal was captured by use of the LAX-3000 Image System (Fuji). For the CCA1 northern-blot probe, CCA1-F (5′-GAGCAAGGACCTCAGACTTATCCG-3′) and CCA1-R (5′-GGAAGGCAATTCGACCCTCG-3′) were designed for the digoxigenin-11-dUTP-labeled probe. The primers TLP-BamHI and TLP-XhoI were used for the northern-blot probe of AtTLP18.3.
Crystallization, Data Collection, and Structure Determination
The wild-type and Se-Met-labeled AtTLP18.3 proteins were crystallized by the hanging-drop vapor diffusion method (McPherson, 1990) at 23°C. Most crystals were grown in 0.2 m sodium acetate trihydrate, 0.1 m sodium cacodylate (pH 6.5), and 25% to 30% (w/v) polyethylene glycol 4000 within 2 weeks. Crystals of Se-Met-containing protein were grown under similar conditions with slightly lower pH (pH 6.0) and the addition of 2 mm Tris(2-carboxyethyl)phosphine to all solutions. For the AtTLP18.3-pSer complex, a soaking method was used. The crystals of wild-type AtTLP18.3 were first grown as described above and then transferred to the above buffer containing 20 mm pSer for 7 d.
Crystals were rapidly swept through mother liquor containing 25% (v/v) glycerol as a cryoprotectant and flash cooled in liquid nitrogen at 100 K. The diffraction data were collected by synchrotron radiation on the SPXF beamlines BL13B1 and BL13C1 equipped with CCD detectors (Q315 and Q210; Area Detector Systems Corporation) at the National Synchrotron Radiation Research Center in Taiwan. All diffraction intensities were integrated and scaled by use of the HKL2000 software package (Otwinowski and Minor, 1997). The crystal structure of AtTLP18.3 was determined by SAD, and two selenium sites were located with use of the SOLVE software (Terwilliger and Berendzen, 1999). The preliminary coordination was determined by the RESOLVE program. Manual model rebuilding involved use of the program Coot (Emsley and Cowtan, 2004), alternating with the structure refinement with the software suite Crystallography & NMR System (Brunger et al., 1998) with 5% of the observed reflections randomly selected and the Rfree value calculated. The crystallography statistics are given in Table I. All ribbon diagrams were prepared by use of PyMOL (DeLano, 2004; DeLano and Lam, 2005).
Phosphatase Activity Assay
The activity of AtTLP18.3 to hydrolyze organophosphate substrates was analyzed with use of the substrates pNPP (New England Biolabs), DiFMUP (Molecular Probes), pSer (Sigma-Aldrich), and five synthetic phosphorylated oligopeptides (Kelowna International Scientific) with the following composition, where (pX) indicates the phosphorylated residue and Ac- indicates the N-terminal acetylation of the peptides: Ac-(pT)AILER, Ac-(pT)IALGK, RRA(pT)VA, KR(pT)IRR, and Ac-RK(pS)AGKPKN.
Enzymatic kinetics of AtTLP18.3 were calculated with the substrates at a series of concentrations at 37°C. The incubated solution contained 15 μg of purified AtTLP18.3, 50 mm sodium acetate buffer (pH 4.0), and the substrates to a total volume of 0.1 mL. All reactions were initiated by adding purified AtTLP18.3 and incubating for 60 min at 37°C, with a blank reaction with AtTLP18.3 omitted. For the substrate pNPP, the reactions were terminated by the addition of 0.25 mL of 500 mm NaOH, and absorbance was measured at 405 nm by use of the spectrophotometer GeneQuant 1300 (GE Healthcare). For the substrate DiFMUP, the fluorescence of the product DiFMU was detected at 358-nm excitation and 455-nm emission. The fluorescence was recorded by use of the spectrophotometer Beckman DU640B. For the substrates pSer and phosphorylated oligopeptides, inorganic phosphate was measured colorimetrically by the modified malachite green method (Fisher and Higgins, 1994). The malachite green-ammonium molybdate reagent containing 0.045% (w/v) malachite green in 6 n H2SO4, 7.5% (w/v) ammonium molybdate in 6 n H2SO4, and 11% Tween 20 in the ratio 1:0.68:0.02 was added to the reaction buffer. Absorbance was measured at 650 nm to determine the levels of inorganic phosphate generation as compared with the standard curve of KH2PO4. Finally, the optimal activity conditions of AtTLP18.3 were determined according to the results of pNPP used as a substrate. The optimal pH for pNPP hydrolysis was measured with 50 mm concentrations of the buffers with sodium citrate (pH 2–3), sodium acetate (pH 3.5–5), MES-NaOH (pH 5.5–6), and Tris-HCl (pH 7.0). The effect of temperature was determined in the range of 10°C to 55°C.
Amino acid sequences were aligned by the use of ClustalW (http://www.ch.embnet.org/software/ClustalW.html). Figure 2 was prepared by use of the Web-based ESPript 2.2 (http://espript.ibcp.fr/ESPript/ESPript/).
The gene sequence of AtTLP18.3 is in the Arabidopsis Information Resource database (accession no. AT1G54780). The accession numbers of amino acid sequences in the National Center for Biotechnology Information are NP_564667 for AtTLP18.3, NP_001055486 for Oryza sativa (japonica cultivar group), ABF21087 for Selaginella tamariscina, XP_001420826 for Ostreococcus lucimarinus CCE9901, NP_441552 for Synechocystis sp. PCC 6803, and YP_002378642 for Cyanothece sp. PCC 7424. All structural factors and coordinates have been deposited in the PDB with PDB code 3PTJ for the Se-Met AtTLP18.3, 3PVH for wild-type AtTLP18.3, and 3PW9 for the AtTLP18.3-pSer complex.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression and purification of AtTLP18.3 protein.
Supplemental Figure S2. Enzymatic kinetics for phosphatase activity assay with the substrate DiFMUP.
Supplemental Table S1. Primers for site-directed mutagenesis.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A, Drory O, Nelson N. (2007) The structure of a plant photosystem I supercomplex at 3.4 Å resolution. Nature 447: 58–63 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- DeLano WL. (2004) Use of PYMOL as a communications tool for molecular science. Abstracts of Papers of the American Chemical Society 228: U313–U314 [Google Scholar]
- DeLano WL, Lam JW. (2005) PyMOL: a communications tool for computational models. Abstracts of Papers of the American Chemical Society 230: U1371–U1372 [Google Scholar]
- Donella-Deana A, Meyer HE, Pinna LA. (1991) The use of phosphopeptides to distinguish between protein phosphatase and acid/alkaline phosphatase activities: opposite specificity toward phosphoseryl/phosphothreonyl substrates. Biochim Biophys Acta 1094: 130–133 [DOI] [PubMed] [Google Scholar]
- Ebbert V, Godde D. (1996) Phosphorylation of PS II polypeptides inhibits D1 protein-degradation and increases PS II stability. Photosynth Res 50: 257–269 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Ferro M, Salvi D, Brugière S, Miras S, Kowalski S, Louwagie M, Garin J, Joyard J, Rolland N. (2003) Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol Cell Proteomics 2: 325–345 [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. (2010) The Pfam protein families database. Nucleic Acids Res 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DK, Higgins TJ. (1994) A sensitive, high-volume, colorimetric assay for protein phosphatases. Pharm Res 11: 759–763 [DOI] [PubMed] [Google Scholar]
- Friso G, Giacomelli L, Ytterberg AJ, Peltier JB, Rudella A, Sun Q, Wijk KJ. (2004) In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16: 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich JE, Wilkerson CG, Ray WK, McAndrew RS, Osteryoung KW, Gage DA, Phinney BS. (2003) Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. J Proteome Res 2: 413–425 [DOI] [PubMed] [Google Scholar]
- Gassner NC, Matthews BW. (1999) Use of differentially substituted selenomethionine proteins in x-ray structure determination. Acta Crystallogr D Biol Crystallogr 55: 1967–1970 [DOI] [PubMed] [Google Scholar]
- Goulas E, Schubert M, Kieselbach T, Kleczkowski LA, Gardeström P, Schröder W, Hurry V. (2006) The chloroplast lumen and stromal proteomes of Arabidopsis thaliana show differential sensitivity to short- and long-term exposure to low temperature. Plant J 47: 720–734 [DOI] [PubMed] [Google Scholar]
- Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, Reid A, et al. (2007) The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res 35: D291–D297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth G. (2002) Structure of spinach chloroplast F1-ATPase complexed with the phytopathogenic inhibitor tentoxin. Proc Natl Acad Sci USA 99: 3464–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Higo H. (1998) PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res 26: 358–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. (1998) Touring protein fold space with Dali/FSSP. Nucleic Acids Res 26: 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang KY, Chung JH, Kim SH, Han YS, Cho Y. (1999) Structure-based identification of a novel NTPase from Methanococcus jannaschii. Nat Struct Biol 6: 691–696 [DOI] [PubMed] [Google Scholar]
- Jones DT. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292: 195–202 [DOI] [PubMed] [Google Scholar]
- Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, Krauss N. (2001) Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411: 909–917 [DOI] [PubMed] [Google Scholar]
- Kawabata T. (2003) MATRAS: a program for protein 3D structure comparison. Nucleic Acids Res 31: 3367–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieselbach T, Hagman, Andersson B, Schröder WP. (1998) The thylakoid lumen of chloroplasts: isolation and characterization. J Biol Chem 273: 6710–6716 [DOI] [PubMed] [Google Scholar]
- Klimmek F, Sjödin A, Noutsos C, Leister D, Jansson S. (2006) Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiol 140: 793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivuniemi A, Aro EM, Andersson B. (1995) Degradation of the D1- and D2-proteins of photosystem II in higher plants is regulated by reversible phosphorylation. Biochemistry 34: 16022–16029 [DOI] [PubMed] [Google Scholar]
- Kramer DM, Sacksteder CA, Cruz JA. (1999) How acidic is the lumen? Photosynth Res 60: 151–163 [Google Scholar]
- Lee JY, Kwak JE, Moon J, Eom SH, Liong EC, Pedelacq JD, Berendzen J, Suh SW. (2001) Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family. Nat Struct Biol 8: 789–794 [DOI] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. (2005) Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438: 1040–1044 [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. (2000) The PSIPRED protein structure prediction server. Bioinformatics 16: 404–405 [DOI] [PubMed] [Google Scholar]
- McPherson A. (1990) Current approaches to macromolecular crystallization. Eur J Biochem 189: 1–23 [DOI] [PubMed] [Google Scholar]
- Michel H, Griffin PR, Shabanowitz J, Hunt DF, Bennett J. (1991) Tandem mass spectrometry identifies sites of three post-translational modifications of spinach light-harvesting chlorophyll protein II: proteolytic cleavage, acetylation, and phosphorylation. J Biol Chem 266: 17584–17591 [PubMed] [Google Scholar]
- Mulo P, Sirpiö S, Suorsa M, Aro EM. (2008) Auxiliary proteins involved in the assembly and sustenance of photosystem II. Photosynth Res 98: 489–501 [DOI] [PubMed] [Google Scholar]
- Ohmura T, Ueda T, Hashimoto Y, Imoto T. (2001) Tolerance of point substitution of methionine for isoleucine in hen egg white lysozyme. Protein Eng 14: 421–425 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Peltier JB, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Söderberg L, Roepstorff P, von Heijne G, et al. (2002) Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14: 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, Ytterberg AJ, Sun Q, van Wijk KJ. (2004) New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J Biol Chem 279: 49367–49383 [DOI] [PubMed] [Google Scholar]
- Pottosin II, Schönknecht G. (1996) Ion channel permeable for divalent and monovalent cations in native spinach thylakoid membranes. J Membr Biol 152: 223–233 [DOI] [PubMed] [Google Scholar]
- Rengasamy A, Selvam R, Gnanam A. (1981) Isolation and properties of an acid phosphatase from thylakoid membranes of Sorghum vulgare. Arch Biochem Biophys 209: 230–236 [DOI] [PubMed] [Google Scholar]
- Rombauts S, Déhais P, Van Montagu M, Rouzé P. (1999) PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res 27: 295–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M, Petersson UA, Haas BJ, Funk C, Schröder WP, Kieselbach T. (2002) Proteome map of the chloroplast lumen of Arabidopsis thaliana. J Biol Chem 277: 8354–8365 [DOI] [PubMed] [Google Scholar]
- Sirpiö S, Allahverdiyeva Y, Suorsa M, Paakkarinen V, Vainonen J, Battchikova N, Aro EM. (2007) TLP18.3, a novel thylakoid lumen protein regulating photosystem II repair cycle. Biochem J 406: 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroebel D, Choquet Y, Popot JL, Picot D. (2003) An atypical haem in the cytochrome b(6)f complex. Nature 426: 413–418 [DOI] [PubMed] [Google Scholar]
- Sun G, Bailey D, Jones MW, Markwell J. (1989) Chloroplast thylakoid protein phosphatase is a membrane surface-associated activity. Plant Physiol 89: 238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta A, Wrzaczek M, Scharte J, Tikkanen M, Konert G, Rahikainen M, Holmström M, Hiltunen HM, Rips S, Sipari N, et al. (2011) Regulatory subunit B′γ of protein phosphatase 2A prevents unnecessary defense reactions under low light in Arabidopsis. Plant Physiol 156: 1464–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umena Y, Kawakami K, Shen JR, Kamiya N. (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473: 55–60 [DOI] [PubMed] [Google Scholar]
- Vener AV, Harms A, Sussman MR, Vierstra RD. (2001) Mass spectrometric resolution of reversible protein phosphorylation in photosynthetic membranes of Arabidopsis thaliana. J Biol Chem 276: 6959–6966 [DOI] [PubMed] [Google Scholar]
- Vener AV, Rokka A, Fulgosi H, Andersson B, Herrmann RG. (1999) A cyclophilin-regulated PP2A-like protein phosphatase in thylakoid membranes of plant chloroplasts. Biochemistry 38: 14955–14965 [DOI] [PubMed] [Google Scholar]
- Weiner MP, Costa GL, Schoettlin W, Cline J, Mathur E, Bauer JC. (1994) Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene 151: 119–123 [DOI] [PubMed] [Google Scholar]
- Zouni A, Witt HT, Kern J, Fromme P, Krauss N, Saenger W, Orth P. (2001) Crystal structure of photosystem II from Synechococcus elongatus at 3.8 Å resolution. Nature 409: 739–743 [DOI] [PubMed] [Google Scholar]