Abstract
Tyrosine aminotransferase (TyrAT) catalyzes the transamination of l-Tyr and α-ketoglutarate, yielding 4-hydroxyphenylpyruvic acid and l-glutamate. The decarboxylation product of 4-hydroxyphenylpyruvic acid, 4-hydroxyphenylacetaldehyde, is a precursor to a large and diverse group of natural products known collectively as benzylisoquinoline alkaloids (BIAs). We have isolated and characterized a TyrAT cDNA from opium poppy (Papaver somniferum), which remains the only commercial source for several pharmaceutical BIAs, including codeine, morphine, and noscapine. TyrAT belongs to group I pyridoxal 5′-phosphate (PLP)-dependent enzymes wherein Schiff base formation occurs between PLP and a specific Lys residue. The amino acid sequence of TyrAT showed considerable homology to other putative plant TyrATs, although few of these have been functionally characterized. Purified, recombinant TyrAT displayed a molecular mass of approximately 46 kD and a substrate preference for l-Tyr and α-ketoglutarate, with apparent Km values of 1.82 and 0.35 mm, respectively. No specific requirement for PLP was detected in vitro. Liquid chromatography-tandem mass spectrometry confirmed the conversion of l-Tyr to 4-hydroxyphenylpyruvate. TyrAT gene transcripts were most abundant in roots and stems of mature opium poppy plants. Virus-induced gene silencing was used to evaluate the contribution of TyrAT to BIA metabolism in opium poppy. TyrAT transcript levels were reduced by at least 80% in silenced plants compared with controls and showed a moderate reduction in total alkaloid content. The modest correlation between transcript levels and BIA accumulation in opium poppy supports a role for TyrAT in the generation of alkaloid precursors, but it also suggests the occurrence of other sources for 4-hydroxyphenylacetaldehyde.
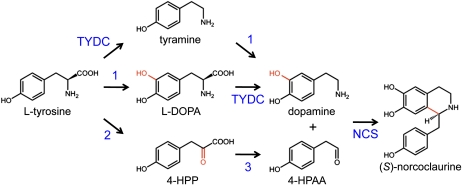
Although many downstream biosynthetic enzymes involved in the biosynthesis of natural products, including the narcotic analgesics codeine and morphine, the cough suppressant and potential anticancer agent noscapine, and the vasodilator papaverine, have been isolated from opium poppy (Papaver somniferum) and related plants, enzymes catalyzing the early steps of benzylisoquinoline alkaloid (BIA) biosynthesis are not well characterized. BIA biosynthesis has been purported to begin with a lattice of decarboxylations, meta-hydroxylations, and transaminations that convert l-Tyr to dopamine and 4-hydroxyphenylacetaldehyde (4-HPAA; Fig. 1; Desgagné-Penix and Facchini, 2011). 4-HPAA is produced by the decarboxylation of 4-hydroxyphenylpyruvate (4-HPP), which is the organic acid of l-Tyr derived through transamination. l-Dihydroxyphenylalanine (l-DOPA) and dopamine are derived via the corresponding 3-hydroxylation of l-Tyr and tyramine, whereas tyramine and dopamine result from the decarboxylation of Tyr and DOPA, respectively (Facchini and De Luca, 1994). Norcoclaurine synthase (NCS) catalyzes the stereoselective condensation of dopamine and 4-HPAA as the first committed step in BIA metabolism.
Figure 1.
Pathway leading to the formation of (S)-norcoclaurine, the central intermediate in the biosynthesis of BIAs in plants, from two molecules of l-Tyr. Dopamine is derived via decarboxylation and 3-hydroxylation of l-Tyr, although the prevailing reaction order is not known. Tyr/DOPA decarboxylase (TYDC) has been shown to accept l-Tyr and l-DOPA as substrates. However, the enzyme (1) responsible for the 3- hydroxylation of l-Tyr or tyramine has not been identified. 4-HPAA is suggested to result from the transamination of l-Tyr and the subsequent decarboxylation of 4-HPP. Tyr transaminase (2) and 4-HPP decarboxylase (3) activities have been reported in BIA-producing plants, but the corresponding enzymes have not been isolated. The condensation of dopamine and 4-HPPA is catalyzed by NCS.
Transamination reactions occur by a “ping-pong” mechanism whereby two half-reactions are required to complete one catalytic cycle and consequently transfer the α-amino group from an amino acid to an α-keto acid (Prabhu and Hudson, 2010). This reaction is readily reversible; thus, both amino acids and α-keto acids are substrates for transaminases. Pyridoxal-5′-phosphate (PLP) functions as a coenzyme forming a Schiff base with the amino acid, which is required for activity and involves a conserved Lys residue at the catalytic core of all aminotransferases (Hayashi, 1995). PLP-dependent aminotransferases are divided into four subgroups based on sequence identity. Subgroup I includes Asp aminotransferase, Ala aminotransferase, histidinol phosphate aminotransferase, and aromatic amino acid (including Tyr) aminotransferases. Subgroup II includes acetyl-Orn aminotransferase, Orn aminotransferase, and Lys aminotransferase. Subgroups III and IV include Ser aminotransferase, phospho-Ser aminotransferase, d-amino acid aminotransferases, and branched-chain amino acid aminotransferases (Hayashi, 1995).
Characterized aromatic amino acid aminotransferases have either specific or a broad range of amino acid and α-keto acid substrates, which are used as amino group donors and acceptors, respectively. Although Tyr, Phe, and Trp are primarily involved in protein synthesis, a vast array of secondary metabolites are also derived from these aromatic amino acids (Tzin and Galili, 2010). Although the biochemical and structural characterization of tyrosine aminotransferase (TyrAT) in mammals and fungi is well established (Blankenfeldt et al., 1999; Sobrado et al., 2003; Schneider et al., 2008; Mehere et al., 2010), considerably less is known about these enzymes in plants. TyrAT is regulated by coronatine, wounding, and methyl jasmonate (MeJA) and has been implicated as the initial enzyme in tocopherol biosynthesis in Arabidopsis (Arabidopsis thaliana) plants (Lopukhina et al., 2001; Holländer-Czytko et al., 2005) and Amaranthus caudatus and Chenopodium quinoa cell cultures (Antognoni et al., 2009). TyrAT activity was reported in rosmarinic acid-producing cell cultures of Anchusa officinalis and Coleus blumei and in MeJA-treated hairy root cultures of Salvia miltiorrhiza (De-Eknamkul and Ellis, 1987a; Xiao et al., 2009b). In plants, tocopherols and rosmarinic acid function as free radical scavengers and confer protection against a variety of biotic and abiotic environmental stress factors (Liu et al., 1992; Sattler et al., 2004; Xiao et al., 2009b). These natural products are also associated with potential benefits to human health.
In this study, we report the isolation and characterization of TyrAT involved in the generation of precursors required for the production of BIAs, such as morphine and codeine, in opium poppy. Although much work has been done to identify the genes responsible for downstream BIA metabolism, the enzymes involved in the supply of precursors are still poorly defined. Our work further demonstrates how the availability of high-throughout sequencing technologies, such as 454 pyrosequencing, and the emergence of functional genomics tools in opium poppy (Facchini and De Luca, 2008; Hagel and Facchini, 2010) provide new opportunities to characterize novel biosynthetic genes.
RESULTS
Identification of a TyrAT cDNA from Opium Poppy
A deep transcriptome database was generated by 454 GS-FLX Titanium pyrosequencing using a cDNA library prepared from opium poppy cell cultures (Desgagné-Penix et al., 2010) and several plant cultivars. The assembled and annotated database was initially screened for proteins related to PLP-dependent enzymes and sequences annotated as aminotransferases. Seven full-length cDNAs belonging to the PLP-dependent Asp aminotransferase superfamily (AAT-like proteins) were identified (Supplemental Fig. S1; Supplemental Table S1). One cDNA with substantial yet differential amino acid sequence identity to putative and functionally validated TyrATs was selected for further characterization. The cDNA contained a 1,257-bp open reading frame and encoded a predicted translation product of 418 amino acids with a molecular mass of 46.3 kD.
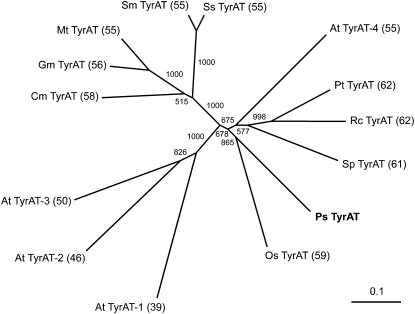
The predicted opium poppy TyrAT polypeptide contains a catalytic Lys residue found in all AAT-like proteins and 10 conserved domains that putatively bind a single PLP molecule as the enzymatic cofactor (Supplemental Fig. S2). The National Center for Biotechnology Information (NCBI) Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) structure prediction tool also suggests that the TyrAT candidate possesses several homodimer interfaces. The ClustalW2 program was used to compare the amino acid sequence of the predicted protein with known and putative TyrATs. The primary structures of all selected proteins were similar with respect to overall length and the positions of conserved domains (Supplemental Fig. S2). An unrooted neighbor-joining tree showing the phylogenetic relationships between the opium poppy TyrAT candidate and related plant enzymes is shown in Figure 2. Opium poppy TyrAT (PsTyrAT) showed the highest sequence identity (59%–62%) with Ricinus communis RcTyrAT (GenBank accession no. XP_002517869), Populus trichocarpa PtTyrAT (XP_002328046), Solanum pennellii SpTyrAT (ADZ24702), and Oryza sativa japonica group OsTyrAT (BAF95202). The PsTyrAT protein also exhibited considerable sequence identity (55%–56%) with S. miltiorrhiza SmTyrAT (ABC60050), Solenostemon scutellaridoides SsTyrAT (CAD30341), Medicago truncatula MtTyrAT (AAY85183), and Glycine max GmTyrAT (AAY21813). However, it is important to note that none of these purported TyrAT candidates has been demonstrated to accept Tyr as a substrate for transamination. In contrast, PsTyrAT showed relatively lower sequence identity with Arabidopsis AtTyrAT-1 (AAN15626), AtTyrAT-2 (NP_180058), and AtTyrAT-3 (AAG37062), which have been shown to function as TyrATs (Lopukhina et al., 2001; Holländer-Czytko et al., 2005). Recently, melon (Cucumis melo) CmTyrAT (ADC45389) and Arabidopsis AtTyrAT-4 (NM_124776) were characterized as TyrATs (Gonda et al., 2010; Prabhu and Hudson, 2010) and showed considerable sequence identity (55% and 58%, respectively) to PsTyrAT.
Figure 2.
Unrooted neighbor-joining tree showing the phylogenetic relationships between opium poppy TyrAT and related plant proteins. Numbers in the tree refer to the bootstrap values for each node over 1,000 iterations. Numbers in parentheses show the percentage amino acid identity of each protein compared with TyrAT from opium poppy. The annotations and GenBank accession numbers of each protein are as follows: PsTyrAT, opium poppy TyrAT (GU370929); OsTyrAT, O. sativa japonica group putative nicotianamine aminotransferase (BAF95202); SpTyrAT, S. pennellii putative TyrAT (ADZ24702); RcTyrAT, R. communis putative TyrAT (XP_002517869); PtTyrAT, P. trichocarpa aminotransferase family protein (XP_002328046); SsTyrAT, S. scutellaridoides putative TyrAT (CAD30341); SmTyrAT, S. miltiorrhiza putative TyrAT (ABC60050); MtTyrAT, M. truncatula putative TyrAT (AAY85183); GmTyrAT, G. max putative TyrAT (AAY21813); CmTyrAT, melon aromatic amino acid transaminase (ADC45389); AtTyrAT-1, Arabidopsis coronatine-regulated TyrAT (TAT1; AAN15626); AtTyrAT-2, Arabidopsis Tyr:2-oxoglutarate aminotransferase (TAT3; NP_180058); AtTyrAT-3, Arabidopsis rooty/superroot1 protein (AAG37062); AtTyrAT-4, Arabidopsis TyrAT (NM_124776).
Purification and Functional Characterization of Opium Poppy TyrAT
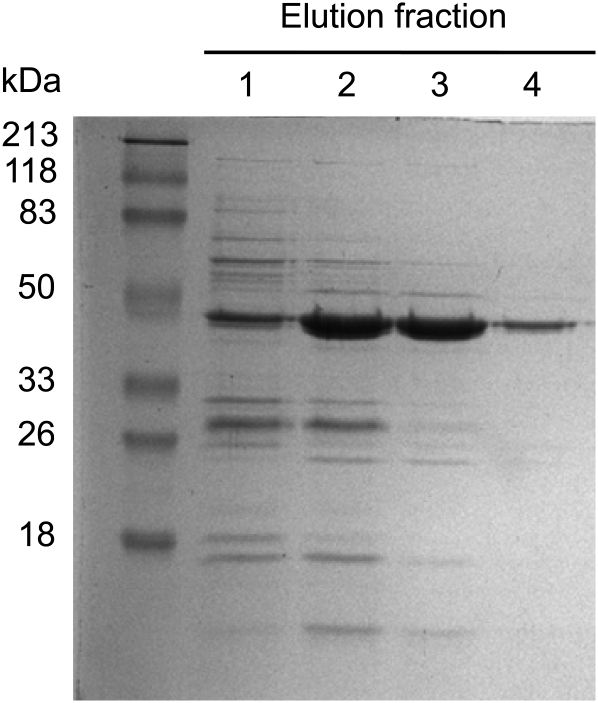
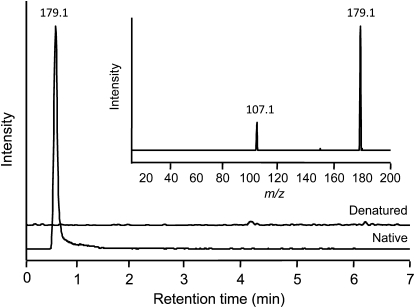
The full-length cDNA was cloned into the expression vector pQE30 with a translational fusion to an N-terminal His6 tag and expressed in Escherichia coli. Recombinant PsTyrAT exhibited a molecular mass of 46 kD and was isolated by cobalt-affinity chromatography to a high degree of purity (Fig. 3). To screen for the transamination of l-Tyr yielding 4-HPP, enzyme assays containing purified, recombinant PsTyrAT were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS; Fig. 4). To confirm TyrAT enzyme activity, the native enzyme was compared with heat-inactivated enzyme as a negative control. Reactions were monitored by LC-MS/MS in multiple-reaction monitoring (MRM) mode, and collision-induced dissociation (CID) mass spectra were generated for selected compounds. Using authentic standards, product ion spectra were used to determine compound-specific MRM transitions of 180.1 → 163.1 and 180.1 → 119.2 for l-Tyr and 179.1 → 157.1 and 179.1 → 107.1 for 4-HPP. The MRM transitions and the CID spectrum of the native TyrAT reaction product eluting at 0.54 min were identical to those of the authentic 4-HPP standard (Fig. 4). The CID mass spectrum of 4-HPP at mass-to-charge ratio (m/z) 179.1 contained a major fragment at m/z 107.1 [M-H]−. In contrast, the l-Tyr spectrum showed deprotonated molecular ions at m/z 163.1 and 119.2. Neither the m/z 179.1 precursor nor the m/z 107.1 fragment ions corresponding to 4-HPP were detected in reactions using heat-inactivated enzyme (Fig. 4). However, the m/z 179.8 [M-H]− precursor and the m/z 119.2 and 163.1 fragment ions corresponding to l-Tyr were present.
Figure 3.
Purification of His6-tagged, recombinant PsTyrAT from E. coli total soluble protein extracts by cobalt-affinity chromatography. Elutions were performed using increasing imidazole concentrations: 10 mm (lane 1), 30 mm (lane 2), 50 mm (lane 3), and 100 mm (lane 4). Lanes contained 20 μL of each fraction, which were separated by SDS-PAGE and visualized using Commassie Brilliant Blue G-250 stain.
Figure 4.
Extracted ion chromatograms at m/z 179.1 for enzyme assays using native and heat-inactivated PsTyrAT protein, and the corresponding CID spectrum for the compound eluting at the retention time of 0.54 min. Enzyme assays contained 2 μg of purified, recombinant PsTyrAT protein incubated with 0.1 mm PLP, 0.1 mm EDTA, 0.3 mm α-ketoglutarate, and 3 mm l-Tyr for 1 h at 30°C. Boiled PsTyrAT protein was used as the heat-inactivated control. Using the native enzyme, MRM in negative mode showed peaks corresponding to 4-HPP using the fragment ions at m/z 151.1 and 107.1 for the precursor ion at m/z 179.1. No peaks corresponding to 4-HPP were detected using the heat-inactivated enzyme. CID in the range of m/z 20 to 200 confirmed that the extracted ion spectra were derived from 4-HPP by the presence of a fragment ion at m/z 107.1 (inset).
TyrAT activity was also assayed using [14C-(U)]l-Tyr as the amino group donor, detecting the formation of [14C-(U)]4-HPP by thin-layer chromatography (TLC; Supplemental Fig. S3). The identity of the reaction product was determined by comparison of the RF value with that of an authentic [14C-(U)]4-HPP standard. The intensity of the reaction product was directly proportional to the amount of recombinant enzyme used in the assay up to 7 μg of purified protein (Supplemental Fig. S3B) and was directly proportional to the amount of [14C-(U)]l-Tyr up to at least 11 μmol (Supplemental Fig. S3C). [14C-(U)]4-HPP was not detected in assays using heat-inactivated enzyme. These data provided an empirical basis for the optimal amount of recombinant PsTyrAT and l-Tyr used in the standard enzyme assay.
Kinetic Parameters of Opium Poppy TyrAT
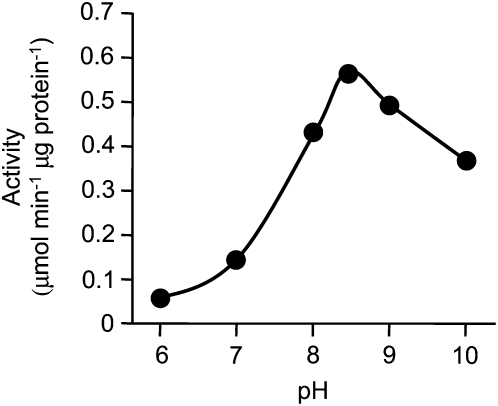
The effect of pH on TyrAT activity was determined between pH 6.0 and 10.0 in HEPES buffer. Optimal activity of PsTyrAT was measured at pH 8.5, which was about four times higher than the activity at pH 7.0 (Fig. 5). The substrate specificity of recombinant PsTyrAT was tested using the l-amino acids Tyr, Trp, and Phe and the α-keto acids α-ketoglutarate, pyruvate, and oxaloacetate as possible amino group donors and acceptors, respectively. Using α-ketoglutarate as the acceptor, l-Tyr was the preferred donor. l-Trp and l-Phe were also accepted as substrates with lower efficiency than l-Tyr (Supplemental Fig. S4). Using l-Tyr as the donor, α-ketoglutarate was the preferred acceptor, followed by pyruvate and oxaloacetate (Supplemental Fig. S4). Kinetic parameters were determined based on the Michaelis-Menten equation using a nonlinear least-squares approach. The apparent Km values for l-Tyr, l-Trp, and l-Phe at a saturating concentration (0.5 mm) of α-ketoglutarate were 1.82, 7.83, and 6.33 mm, respectively (Table I; Supplemental Fig. S5). Km values for α-ketoglutarate, pyruvate, and oxaloacetate at a saturating concentration (3 mm) of l-Tyr were 0.35, 2.45, and 56.13 mm, respectively (Table I; Supplemental Fig. S5). No substantial substrate or product inhibition was detected. The enzyme efficiencies (kcat/Km) for l-Tyr were 2.6- and 13-fold greater than those of l-Trp and l-Phe, respectively. The kcat/Km for α-ketoglutarate were 4.5- and 63-fold greater than those of pyruvate and oxaloacetate, respectively. Recombinant PsTyrAT activity did not increase in response to the addition of PLP to the reaction mixture. The purified enzyme was stable for several days in 100 mm HEPES buffer at −80°C, but its activity gradually decreased after longer term storage.
Figure 5.
Effect of pH on PsTyrAT activity. Assays were performed for 1 h at 30°C in HEPES buffer at the pH values indicated in the presence of 2 μg of purified enzyme, 3 mm l-Tyr, 0.1 mm PLP, 0.1 mm EDTA, and 0.5 mm α-ketoglutarate.
Table I. Kinetic parameters for TyrAT from opium poppy.
The data represent means of three independent measurements ± sd. The Km and Vmax values were calculated from the Michaelis-Menten equation using a least-squares method. The kcat value was calculated by dividing Vmax by Et (the number [pmol] of enzymes in each assay).
| Substrate | Cosubstrate (Concentration in Assay) | Km | Vmax | kcat | kcat/Km |
| mm | μmol min−1 mg−1 | s−1 | mm−1 s−1 | ||
| l-Tyr | α-Ketoglutarate (0.5 mm) | 1.82 ± 0.09 | 0.63 ± 0.02 | 0.24 ± 0.01 | 0.13 ± 0.07 |
| l-Phe | α-Ketoglutarate (0.5 mm) | 6.33 ± 1.09 | 0.19 ± 0.002 | 0.08 ± 0.001 | 0.01 ± 0.001 |
| l-Trp | α-Ketoglutarate (0.5 mm) | 7.83 ± 3.81 | 0.92 ± 0.02 | 0.36 ± 0.01 | 0.05 ± 0.002 |
| α-Ketoglutarate | l-Tyr (3 mm) | 0.35 ± 0.09 | 0.56 ± 0.02 | 0.22 ± 0.01 | 0.63 ± 0.39 |
| Pyruvate | l-Tyr (3 mm) | 2.45 ± 0.58 | 0.89 ± 0.09 | 0.34 ± 0.03 | 0.14 ± 0.06 |
| Oxaloacetate | l-Tyr (3 mm) | 56.13 ± 0.60 | 0.97 ± 0.05 | 0.37 ± 0.02 | 0.01 ± 0.03 |
Involvement of TyrAT in BIA Metabolism in Opium Poppy
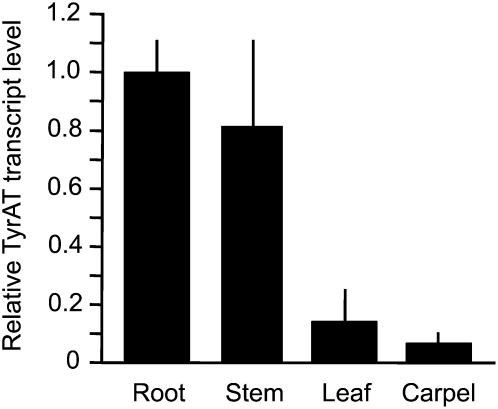
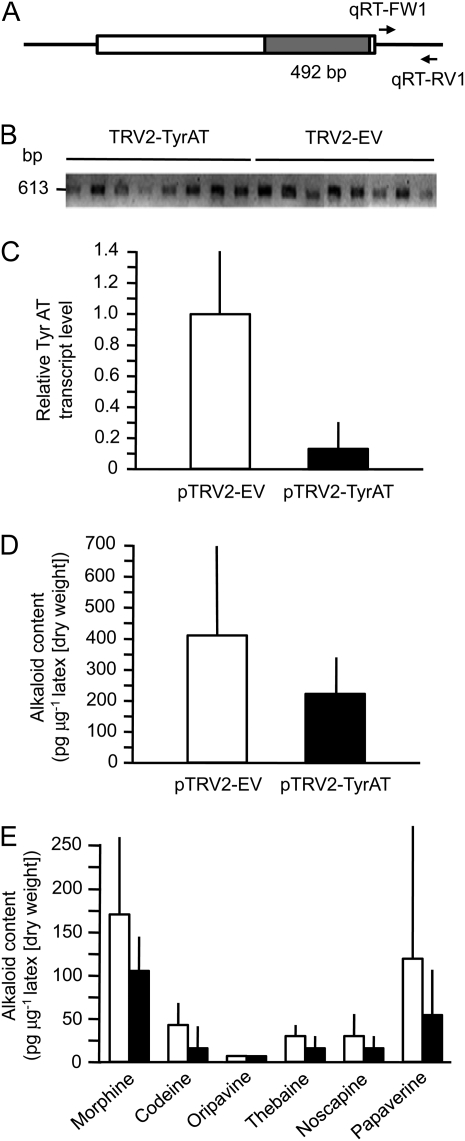
The highest PsTyrAT transcript levels were detected in the roots and stems of mature opium poppy plants, whereas lower transcript levels were detected in leaves and carpels (Fig. 6). The occurrence of relatively high transcript levels in opium poppy stems facilitated the analysis of stem tissue and latex using virus-induced gene silencing (VIGS) to examine the potential role of TyrAT in BIA metabolism. A 492-bp fragment of the PsTyrAT coding region was inserted into the pTRV2 vector used for tobacco rattle virus (TRV)-based VIGS (Fig. 7A). Agrobacterium tumefaciens containing pTRV1 and pTRV2-TyrAT or the pTRV2 empty vector (EV) was infiltrated into the apical meristems of 2-week-old opium poppy plants. The pTRV2 vector encodes the tobacco rattle viral coat protein, transcripts of which were detected in stem tissue approximately 10 weeks after infiltration to confirm the presence of the virus (Fig. 7B). Relative PsTyrAT transcript levels were reduced by at least 80% in plants infiltrated with A. tumefaciens harboring the pTRV2-TyrAT vector compared with EV controls (Fig. 7C). To test the effect of suppressing PsTyrAT transcript levels on the accumulation of BIAs, latex samples from plants infiltrated with pTRV2-TyrAT or pTRV2-EV and showing the occurrence of TRV2 coat protein transcripts were analyzed by HPLC. Levels of the six major BIAs in opium poppy were reduced in plants infiltrated with A. tumefaciens harboring the pTRV2-TyrAT vector compared with EV controls (Fig. 7D). The specific alkaloid content showed considerable variation in individual plants, which contributed to the relatively large sd in the mean values. The correlation between relative PsTyrAT transcript levels and alkaloid accumulation was not fully proportional, suggesting that other factors are involved in the supply of precursors for BIA metabolism
Figure 6.
Relative transcript abundance of PsTyrAT in different opium poppy organs. First-strand cDNAs were synthesized from total RNA and used as a template for RT-qPCR analysis. The transcript abundance of ubiquitin from opium poppy was used as an internal control, and relative values were normalized to the PsTyrAT transcript level in roots. Values represent means ± sd of triplicate experiments.
Figure 7.
Effect of reducing PsTyrAT transcript levels by VIGS in opium poppy plants. A, A 492-bp fragment of the PsTyrAT coding region was inserted into pTRV2 vector. Two-week-old poppy seedlings were coinfiltrated with pTRV1 and pTRV2-EV or with pTRV1 and pTRV2-TyrAT. After approximately 10 weeks, stem and latex samples were used to determine relative PsTyrAT transcript abundance and BIA levels, respectively. B, Ethidium bromide-stained agarose gel showing the detection of TRV2 coat protein transcripts in cDNAs synthesized from total stem RNA. C, Mean PsTyrAT transcript levels in plants infiltrated with pTRV2-TyrAT (black bar) and pTRV2-EV (white bar). D, Relative accumulation of total major BIAs in latex extracted from plants infiltrated with pTRV2-TyrAT (black bar) and pTRV2-EV (white bar). E, Relative accumulation of individual BIAs in latex extracted from plants infiltrated with pTRV-TyrAT (black bars) and pTRV2-EV (white bars). Values represent means ± sd of three technical replicates performed on each of three biological replicates for each of eight infiltrated plants.
DISCUSSION
A full-length cDNA encoding TyrAT was isolated based on its annotation in a deep transcript library generated by 454 pyrosequencing of opium poppy (Desgagné-Penix et al., 2010), and the predicted amino acid sequence was used to reanalyze the transcriptome databases through a BLASTx search. Six full-length cDNAs were revealed (Supplemental Fig. S1), five of which were annotated as Ala aminotransferase or hypothetical protein and displayed only 16% to 22% amino acid identity compared with PsTyrAT (Supplemental Table S1). The other predicted protein (cl.10988) showed 68% amino acid identity with PsTyrAT but was not selected for further analysis because corresponding gene transcripts were not detected in elicitor-treated cell cultures (Desgagné-Penix et al., 2010). In contrast, PsTyrAT transcripts were detected in the 454 databases of elicitor-treated cell cultures and stems, which are both capable of BIA biosynthesis. TyrAT converts l-Tyr to 4-HPP via the pyridoxal phosphate-dependent transamination reaction. The proposed early steps in the formation of the central BIA intermediate (S)-norcoclaurine were based on the incorporation of radiolabeled precursors (Fig. 1; Holland et al., 1979; Schumacher et al., 1983; Rueffer and Zenk, 1987). Only [14C]l-Tyr was equally incorporated into both the “upper” isoquinoline and the “lower” benzylic portions of the BIA backbone. In contrast, l-DOPA, dopamine, and tyramine were predominantly incorporated into the isoquinoline moiety. Enzyme activities corresponding to the purported transamination, decarboxylation, and hydroxylation reactions have been reported in crude protein extracts (Rueffer and Zenk, 1987; Hara et al., 1994). However, only cDNAs encoding Tyr/DOPA decarboxylase (EC 4.1.1.25), which converts l-Tyr to tyramine and l-DOPA to dopamine, have been isolated from BIA-producing plants (Facchini and De Luca, 1994). Through the isolation and characterization of a cDNA encoding TyrAT from opium poppy, we provide biochemical and physiological support for the involvement of 4-HPP as an intermediate in the formation of the 4-HPAA precursor used in BIA biosynthesis, in agreement with the proposed pathway (Fig. 1). Previously, TyrAT cDNAs implicated in the biosynthesis of tocopherols or in fruit ripening have been characterized in Arabidopsis and melon, respectively (Lopukhina et al., 2001; Holländer-Czytko et al., 2005; Gonda et al., 2010; Prabhu and Hudson, 2010).
l-TyrATs are well characterized in mammals, E. coli, and Trypanosoma cruzi. Several cDNAs encoding TyrAT have been isolated and characterized from rat, mouse, and human (Andersson and Pispa, 1982; Shinomiya et al., 1984; Grange et al., 1985; Müller et al., 1985), and the crystal structures have been solved (Protein Data Bank code 3dyd; Blankenfeldt et al., 1999; Ko et al., 1999; Mehere et al., 2010). Several amino acids have been implicated in catalyzing the transamination reaction based on the structural features of TyrAT from various organisms. Highly conserved catalytic Lys and Asp residues interact with the pyridine nitrogen of PLP (Mehere et al., 2010), which is covalently bound to the ϵ-amino group of the Lys residue via a Schiff base linkage (Hayashi, 1995). Once the amino acid substrate interacts with the active site, a new Schiff base is generated. Opium poppy TyrAT shares approximately 30% amino acid identity with TyrATs from rat, mouse, and T. cruzi but less than 10% amino acid identity with TyrAT from E. coli. In opium poppy TyrAT, Lys-251 and Asp-222 are assumed to serve in the same capacity as Lys-280 and Asp-247 from mouse TyrAT in the binding of PLP (Supplemental Fig. S2). Arg-417 is one of the residues responsible for the interaction of Tyr with mouse and rat TyrAT (Sobrado et al., 2003; Mehere et al., 2010), and Arg-390 of PsTyrAT might perform the same function. As reported for other investigations of plant aminotransferases, the in vitro activity of PsTyrAT was not affected by the addition of exogenous PLP. Transamination activity occurred without the addition of PLP in crude protein extracts of bushbean (Phaseolus vulgaris; Forest and Wightman, 1972), tomato (Solanum lycopersicum; Gibson et al., 1972), mung bean (Vigna radiata; Truelsen, 1972), peanut (Arachis hypogaea; Mazelis and Fowden, 1969), wheat (Triticum aestivum; Cruickshank and Isherwood, 1958), and cauliflower (Brassica oleracea; Ellis and Davies, 1961). Plant aminotransferases appear to function as holoenzymes composed of an apoenzyme tightly bound to the coenzyme moiety, whereas the apoenzyme and coenzyme components of mammalian aminotransferases can be separated (Forest and Wightman, 1972; Wightman and Forest, 1978).
In rat, TyrAT activity is regulated by glucocorticoids, insulin, and glucagon (Rettenmeier et al., 1990). TyrAT deficiency leads to type II tyrosinemia in humans, which is associated with microcephaly, tremor, ataxia, language deficits, and convulsions (Bein and Goldsmith, 1977; Cavelier-Balloy et al., 1985). Fungal TyrATs have been implicated in the biosynthesis of atromentin in Tapinella panuoides (Schneider et al., 2008). Unlike mammalian and microbial enzymes, the physiological functions of plant TyrATs are not well understood. Among the plant enzymes included in the phylogenetic analysis (Fig. 2), only TyrATs from Arabidopsis and melon have been isolated and characterized (Lopukhina et al., 2001; Holländer-Czytko et al., 2005; Gonda et al., 2010; Prabhu and Hudson, 2010). ArAT from melon (FJ896816; designated here as CmTyrAT) displayed aromatic and branched-chain amino acid transaminase activities in flesh and rind tissues during melon fruit ripening (Gonda et al., 2010). The TAT1 (COR13; At4g23600) gene and six related sequences in the Arabidopsis genome, which encode class I aminotransferases, were identified by differential display analysis in Arabidopsis plants treated with the phytotoxin coronatine (Lopukhina et al., 2001). Transcript levels for TAT1 (designated here as AtTyrAT-1) increased in response to MeJA or methyl 12-oxophytodienoic acid treatment and to wounding. The deduced amino acid sequence of TAT1 showed 35% identity to human and rat TyrATs and shares extensive sequence similarity with nicotianamine aminotransferase, involved in the biosynthesis of mugineic acid family phytosiderphores (Takahashi et al., 1999). However, the TyrAT activity of TAT1 remains controversial. In one study, the TAT1 gene was suggested to encode a PLP-dependent cystine lyase (Cys-lyase) and not TyrAT, since recombinant TAT1 protein showed higher Cys-lyase than TyrAT activity. Moreover, TAT1 shares substantial (79%) amino acid identity with the Brassica oleracea BOCL3 gene product, which exhibits Cys-lyase activity (Jones et al., 2003). In support of this suggestion, human kynerenine aminotransferase I/Gln transaminase K (EC 2.6.1.64) was proposed to play a dual function in catalyzing the transamination of several amino acids and also showed Cys S-conjugate β-lyase activity (EC 4.4.1.13; Cooper, 2004). Ala and Asp aminotransferase from procine heart was also able to cleave Cys conjugates (Adcock et al., 1996). Aminotransferases might generally possess multifunctional potential in complex metabolic networks.
The TAT3 gene (At2g24850; designated AtTyrAT-2 here) was shown to encode the enzyme catalyzing the first step in tocopherol biosynthesis and was induced by MeJA and methyl 12-oxophytodienoic acid, wounding, high light intensity, UV light, and the herbicide oxyfluorfen (Sandorf and Holländer-Czytko, 2002). The rooty/superroot1 gene (At2g20610; designated AtTyrAT-3 here) is a locus on Arabidopsis chromosome 2 encoding a protein suggested to have TyrAT activity (Gopalraj et al., 1996). However, the major role of the rooty/superroot1 gene product is apparently either Trp aminotransferase (EC 2.6.1.27) or Cys-lyase, implying a role in modulating indole-3-acetic acid levels in Trp-derived specialized metabolism such as indole glucosinolate biosynthesis (Nonhebel et al., 1993; Gopalraj et al., 1996; Jones et al., 2003). Recently, another Arabidopsis TyrAT gene (At5g36160; designated here as AtTyrAT-4) was reported as an aminotransferase capable of interconverting l-Tyr and 4-HPP as well as and l-Phe and phenylpyruvate (Prabhu and Hudson, 2010). The transcript abundance of TyrAT in crude protein extracts of S. miltiorrhiza hairy root cultures increased along with Phe ammonia lyase, cinnamic acid 4-hydroxylase, 4-hydroxyphenylpyruvate reductase (HPPR), and 4-hydroxyphenylpyruvate dioxygenase (HPPD) transcript levels in response to MeJA treatment (Xiao et al., 2009b). TyrAT with a high substrate specificity for Tyr and broad specificity toward amino group acceptors was purified from rosmarinic acid-producing cell cultures of A. officinalis and C. blumei (De-Eknamkul and Ellis, 1987b), and an increase in TyrAT activity in response to MeJA treatment was accompanied by higher α-tocopherol levels in A. caudatus cell cultures (Antognoni et al., 2009). Overall, TyrATs are regulated by stress factors via a complex signaling network and are associated with multiple metabolic and other physiological processes. A role for TyrAT in BIA metabolism was proposed based on the detection of enzyme activity in cell cultures (Rueffer and Zenk, 1987). The isolation and functional characterization of a TyrAT cDNA in opium poppy plants validates the contribution of the enzyme in the provision of 4-HPAA for the formation of (S)-norcoclaurine.
The relatively high pH optimum of pH 8.5 (Fig. 5) for PsTyrAT is consistent with the reported maximal activity at pH 8.2 for TyrATs from other plants (De-Eknamkul and Ellis, 1987a; Prabhu and Hudson, 2010). Opium poppy TyrAT also showed a pronounced preference for l-Tyr over other aromatic amino acid substrates but relatively broad specificity toward the amino group acceptors α-ketoglutarate, pyruvate, and oxaloacetate (Supplemental Fig. S4). The apparent Km and Vmax for l-Tyr (using α-ketoglutarate as the amino group acceptor) of 1.82 mm and 0.63 μmol min−1 mg−1, respectively, are within the range of values reported for other plant enzymes with TyrAT activity. Three purified TyrAT isoforms from A. officinalis cell cultures displayed Km values for l-Tyr between 0.45 and 20 mm (De-Eknamkul and Ellis, 1987a). One isoform showed a relatively strict specificity toward l-Tyr/α-ketoglutarate, whereas another exhibited higher specificity for l-aspartate/α-ketoglutarate compared with l-Tyr/α-ketoglutarate. In Arabidopsis, an apparent Km of 0.19 mm and Vmax of 5 μmol min−1 mg−1 were reported for a recently isolated enzyme with TyrAT activity (Prabhu and Hudson, 2010). In contrast, mouse TyrAT displayed a Km of 1.8 mm for l-Tyr, 11.4 mm for Phe, and 4.9 mm for Glu (Mehere et al., 2010). T. cruzi TyrAT showed a broader substrate specificity that extended to l-Ala (Nowicki et al., 2001). Interestingly, the affinity of PsTyrAT for l-Tyr is lower than that of one reported enzyme from Arabidopsis, similar to that of mouse TyrAT, and higher than that of TyrAT isoforms from A. officinalis cell cultures. TyrATs from mammals (Andersson and Pispa, 1982; Mehere et al., 2010) and T. cruzi (Blankenfeldt et al., 1999) have been shown to function as homodimers, whereas TyrATs from Arabidopsis (Lopukhina et al., 2001) and A. officinalis (De-Eknamkul and Ellis, 1987a) were reported as a homodimer and a homotetramer, respectively. Accordingly, the kcat of PsTyrAT was calculated assuming that the enzyme functions as a homodimer. The catalytic efficiencies (kcat/Km) toward l-Phe and l-Trp were much lower than that of l-Tyr (with α-ketoglutarate), in support of a predominant role in the interconversion of l-Tyr and α-ketoglutarate (Table I).
A high Km for Tyr suggests that the cellular pools of this aromatic amino acid are comparatively abundant. Although its absolute levels in opium poppy have not been determined, Tyr appears to be generally abundant in plants. The concentration of Tyr in the phloem sap of barley leaves was reported as high as 1.6 ± 1.3 mm (Winter et al., 1992). In the phloem sap of Papaver dubium, Tyr levels were higher than other amino acids (Wilkinson et al., 2001). Similarly, Tyr was also more abundant than other amino acids in elicitor-treated opium poppy cell cultures (Zulak et al., 2008).
The highest transcript levels for most biosynthetic genes involved in BIA metabolism occur in the stems and roots of opium poppy (Bird et al., 2003; Samanani et al., 2006). In particular, transcripts encoding Tyr/DOPA decarboxylase and NCS, the other known enzymes involved in the early step of the BIA pathway, were abundant in stems and roots and were found only at low levels in leaves and developing seed capsules (Facchini and De Luca, 1995; Lee and Facchini, 2010). The distribution of PsTyrAT transcripts is consistent with a preeminent role of stems and roots in the biosynthesis of BIAs in opium poppy (Fig. 6). NCS (Lee and Facchini, 2010) and other BIA biosynthetic enzymes are also specifically localized or at least most abundant in sieve elements of the phloem (Bird et al., 2003; Samanani et al., 2006). The occurrence of TyrAT in sieve elements would facilitate access to l-Tyr translocated in the phloem to ensure adequate precursor availability for BIA biosynthesis. The localization of aminotransferase isoforms to different subcellular compartments has been reported previously (Wightman and Forest, 1978).
The application of VIGS as an effective method to specifically silence targeted genes in opium poppy allows direct investigation of the physiological roles of putative biosynthetic enzymes (Hagel and Facchini, 2010; Lee and Facchini, 2010). The VIGS mechanism is based on cosuppression of a transgene and an endogenous gene through the formation of double-stranded RNA (Robertson, 2004). Close homologs (greater than 90% nucleotide sequence identity) might also be affected, but none of the TyrAT homologs displayed identities in this range (Supplemental Table S1). Transcripts corresponding to cl.10988 were unaffected by the VIGS-mediated reduction in TyrAT transcript levels (Supplemental Fig. S6), in support of the specificity of silencing. VIGS facilitated a significant (P <0.01) suppression of PsTyrAT transcript levels in opium poppy stems to less than 20% of that found in control plants (Fig. 7C). The combined mean abundance of the six major BIAs (i.e. morphine, codeine, oripavine, thebaine, noscapine, and papaverine) was also significantly reduced by almost 50% in plants that showed a reduction in PsTyrAT transcript levels compared with controls using one-tailed (P < 0.07) and two-tailed (P < 0.14) t test analyses (Fig. 7D). The statistical analysis showed a balanced effect among TyrAT-VIGS plants. The alkaloid levels in several control plants were significantly higher than those in TyrAT-VIGS plants, whereas most TyrAT-VIGS plants contained lower to similar alkaloid levels compared with controls. A reduction in the accumulation of several individual BIAs was also detected in plants with suppressed PsTyrAT transcript levels (Fig. 7E), but the natural variation in the ratio of some pathway end products and intermediates reduced the statistical confidence when metabolites were considered separately. The correlation between transcript levels and total alkaloid accumulation supports a major physiological role for TyrAT in the generation of precursors for BIA metabolism. The modest relationship between transcript and total alkaloid levels was potentially influenced by several factors. The major reason might be that plant aminotransferases generally exhibit broad substrate specificity, which suggests that other gene products can utilize l-Tyr as an amino donor and contribute to the cellular pool of 4-HPP. Purified plant aromatic aminotransferases possess properties similar to most animal and microbial aminotransferases. Several reports have suggested that plant Trp and Asp aminotransferases have the same substrate multispecificity (Bonner and Jensen, 1985) as mammalian and microbial Asp aminotransferases (Mavrides and Orr, 1975). Plant Asp aminotransferases have been shown to transaminate five l-amino acids, Asp, Glu, Phe, Tyr, and Trp, using α-ketoglutarate or oxaloacetate as the amino group acceptor (Forest and Wightman, 1972). Moreover, Trp aminotransferase was able to catalyze the transamination of other aromatic amino acids, including l-Tyr (Truelsen, 1972; Noguchi and Hayashi, 1980; McQueen-Mason and Hamilton, 1989; Koshiba et al., 1993). The contribution of TyrAT homologs (Supplemental Fig. S1; Supplemental Table S1) to maintaining balance in the cellular pools of l-Tyr and 4-HPP cannot be ruled out.
The complex network of aromatic amino acid metabolism provides substrates leading to the biosynthesis of numerous specialized metabolites with diverse physiological functions. In the formation of rosmarinic acid, l-Phe and l-Tyr are converted to 4-coumaroyl-CoA and 4-hydroxyphenylacetate, respectively. The synthesis of tocopherols, tocotrienols, and plastoquinones shares the same aromatic precursor, homogentisic acid, which is synthesized from 4-HPP by HPPD. Interestingly, the content of 4-HPP was stable after treatment of S. miltiorrhiza hairy root cultures with MeJA, despite an increase in the transcript levels of HPPD and HPPR, which catalyzes the conversion of 4-HPP to 4-hydroxylphenylacetic acid (Xiao et al., 2009a). In this case, 4-HPP was suggested not only as an amino group acceptor but also as a cosubstrate for HPPD and HPPR. Additional metabolic pressure on the cellular pool of 4-HPP could account, in part, for the modest correlation between TyrAT transcript levels and BIA accumulation in opium poppy.
Negative feedback mechanisms have been implicated in the regulation of metabolic pathways involving TyrAT. For example, TyrAT activity was inhibited by α-aminooxyacetic acid or α-aminooxy-β-phenylpropionic acid, which are also inhibitors of Phe ammonia lyase and PLP-dependent enzymes in general (De-Eknamkul and Ellis, 1987b). Furthermore, TyrAT activity was also inhibited by 3-(3,4-dihydroxyphenyl)lactic acid, a Tyr metabolite and an intermediate in rosmarinic acid biosynthesis (De-Eknamkul and Ellis, 1987a). Clearly, further investigation is required to better understand the regulation of l-Tyr catabolism in the context of BIA metabolism. Nevertheless, the biochemical characterization of a recombinant TyrAT in vitro coupled with the physiological evaluation of function in the plant supports the role of 4-HPP as an intermediate in the formation of BIA precursors.
MATERIALS AND METHODS
Chemicals
l-Tyr and 4-HPP were purchased from Sigma-Aldrich (www.sigmaaldrich.com). [14C-(U)]l-Tyr (74 kBq; specific activity of 450 mCi mmol−1) was purchased from American Radiolabeled Chemicals (www.arc-inc.com). Benzylisoquinoline alkaloid standards were obtained or synthesized as described previously (Hagel and Facchini, 2010). [14C-(U)]4-HPP was synthesized from [14C-(U)]l-Tyr by modifying methods described previously (Rueffer and Zenk, 1987; Barta and Böger, 1996). Briefly, a 20-μL portion of [14C-(U)]l-Tyr was diluted with phosphate buffer (0.1 m, pH 6.5, 130 μL). Then, 5,000 units of bovine liver catalase (activity of 2,000–5,000 units mg−1; Sigma-Aldrich) and 5 mg of crude l-amino acid oxidase from Crotalus adamanteus (Sigma-Aldrich) were added. The mixture was incubated in an open vial with shaking at room temperature for 80 min and then was loaded onto a column containing 500 μL of Dowex 50W X8 (Sigma-Aldrich) resin equilibrated with 1.0 m hydrochloric acid. The radioactive product was eluted with 2 mL of 0.1 m hydrochloric acid and subsequently purified on a silica gel 60 F254 TLC plate (EMD Chemicals; www.emdchemicals.com) using ethyl acetate:methanol (4:1, v/v) as the mobile phase.
Isolation and Cloning of Opium Poppy TyrAT
A full-length cDNA encoding PsTyrAT from opium poppy (Papaver somniferum) was identified by screening an in-house 454 pyrosequencing database (Desgagné-Penix et al., 2010) for unigenes annotated as PLP-dependent aminotransferases. BLASTx analysis of the NCBI database (http://www.ncbi.nlm.nih.gov/BLAST/) was performed to identify TyrAT orthologs. A codon-optimized synthetic gene encoding the PsTyrAT enzyme was constructed to improve recombinant protein production in Escherichia coli (GenScript; www.genscript.com). The synthetic gene was amplified by PCR using forward (5′-GAGCTCATGGAAAAAGGCGGCAAAA-3′) and reverse (5′-AAGCTTTTACTGCTGTTTAGCGTGAC-3′) primers containing SacI and HindIII restriction sites, respectively. The amplicon was digested with SacI and HindIII and cloned into the corresponding sites of pQE-30 vectors (Qiagen; www.qiagen.com).
Heterologous Expression and Purification of Recombinant TyrAT
The pQE-TyrAT plasmid encoding a translational fusion between PsTyrAT and an N-terminal His6 purification tag was expressed in E. coli strain SG13009. Transformed bacteria were grown at 37°C to an optical density at 600 nm of 0.4 and were then induced with 0.3 mm isopropyl β-thiogalactopyranoside at room temperature for 4 h. The bacteria were collected by centrifugation at 13,000g for 10 min, resuspended in 200 mm Tris, 100 mm KCl, and 10% (w/v) glycerol, pH 7.5, and lysed by sonication (five times, 10 s each). Debris was collected by centrifugation at 13,000g for 10 min, and the supernatant was used for affinity purification of the recombinant protein over Talon His-Tag Purification Resin (Clontech; www.clontech.com). Proteins eluting between 10 and 100 μm imidazole were analyzed by SDS-PAGE on a 12% (w/v) acrylamide gel and visualized using Coomassie Brilliant Blue R-250 stain. Protein concentrations were determined using the Bradford assay (Bio-Rad Laboratories; www.bio-rad.com).
Enzyme Assays and Recombinant Protein Characterization
Purified, recombinant TyrAT protein was desalted using a PD-10 column (GE Healthcare; www.gehealthcare.com) equilibrated with 100 mm HEPES buffer, pH 8.0. For LC-MS/MS analysis, the reaction mixture contained 2 μg of purified PsTyrAT protein, 0.1 mm PLP, 0.1 mm EDTA, 0.3 mm α-ketoglutarate, and 3 mm l-Tyr in HEPES buffer at pH 8.2, to a total volume of 100 μL. As a control, purified TyrAT protein was denatured by boiling for 10 min. For TLC analysis, enzyme assays consisted of 0.1 mm PLP, 0.1 mm EDTA, 0.3 mm α-ketoglutarate, [14C-(U)]l-Tyr, and purified, recombinant TyrAT protein in 100 mm HEPES buffer, pH 8.0, in a total volume of 100 μL. Different concentrations of [14C-(U)]l-Tyr and amounts of recombinant protein were tested to optimize the enzyme assay. Reactions were incubated for 1 h at 30°C and terminated by adding 50 μL of 1.0 n HCl. Products were extracted with 1.0 mL of ethyl acetate, which was subsequently evaporated under reduced pressure. The residue was dissolved in 10 μL of methanol, and samples were applied to a silica gel 60 F254 TLC plate (EMD Chemicals). Compounds were separated using a mobile phase of ethyl acetate:methanol (4:1, v/v). The reaction product was identified based on its RF value compared with that of authentic [14C-(U)]4-HPP and quantified relative to [14C-(U)]l-Tyr.
Enzyme kinetics were obtained by monitoring the absorbance of various transamination reaction products as described previously (Collier and Kohlhaw, 1972; De-Eknamkul and Ellis, 1987a); 4-HPP at 331 nm (ɛ331 = 19,500 m−1 cm−1), phenylpyruvate at 320 nm (ɛ331 = 17,500 m−1 cm−1), and indole-3-pyruvate at 328 nm (ɛ331 = 10,000 m−1 cm−1), corresponding to the substrates l-Tyr, l-Phe, and l-Trp. The standard assay contained the indicated concentrations of an aromatic amino acid and an organic acid, 2 μg of purified recombinant TyrAT protein, 0.1 mm PLP, 0.1 mm EDTA, and 100 mm HEPES buffer, pH 8.2, in a total volume of 250 μL. The following substrate concentrations were used: 0.1 to 30 mm l-Tyr, 0.1 to 40 mm l-Phe, 0.1 to 50 mm l-Trp, 0.05 to 30 mm α-ketoglutarate, 0.05 to 150 mm pyruvate, and 0.05 to 300 mm oxaloacetate. Reactions were incubated for 1 h at 30°C and terminated by adding 70 μL of 2.0 n NaOH. Vmax and Km values were calculated according to a nonlinear regression of the Michaelis-Menten equation, where V = (VmaxS)/(Km + S) (Hernández and Ruiz, 1998). The kcat value is defined as Vmax/Et, where Et is the total enzyme concentration.
VIGS
A 492-bp fragment of the PsTyrAT coding region was amplified by PCR to introduce EcoRI and SacI restriction sites using forward (5′-GAATTCGATAGTGCCTGGTTTACGAC-3′) and reverse (5′-GAGCTCCTGTTGTTTGGCGTGCCTAC-3′) primers. The amplicon was cloned into the corresponding restriction sites of pTRV2 vector to produce the pTRV2-TyrAT construct. Agrobacterium tumefaciens strain GV3101 harboring pTRV1 and pTRV2-EV or pTRV1 and pTRV2-TyrAT was cultured at 28°C in 300 mL of Luria-Bertani medium containing 10 mm MES, 20 μm acetosyringone, and 50 μg mL−1 kanamycin. Bacteria were pelleted at 3,000g for 15 min and resuspended in infiltration buffer (10 mm MES, 200 μm acetosyringone, and 10 mm MgCl2) to an optical density at 600 nm of 2.5. Two-week-old opium poppy (cv Bea’s Choice) seedlings were infiltrated using a 1-mL syringe with a 1:1 (v/v) mixture of A. tumefaciens cultures harboring pTRV1 and either pTRV2-TyrAT or pTRV2-EV. Infiltrated plants were analyzed at maturity (i.e. the emergence of flower buds). Stem sections were excised below the flower bud, young stem tissue was flash frozen in liquid N2, for reverse transcription-quantitative (RT-q)PCR analysis, and 10 μL of exuding latex was collected for HPLC analysis.
RT-qPCR
Plant tissue was ground under liquid nitrogen and extracted in 0.8 m guanidinium thiocyanate, 0.4 m ammonium thiocyanate, 0.1 m sodium acetate, pH 5.0, 5% (v/v) glycerol, and 38% (v/v) Tris-buffered phenol. Subsequently, 200 μL of CHCl3 was added and the mixture was emulsified. Samples were centrifuged, and 400 μL of the aqueous phase was precipitated with 500 μL of isopropanol. After centrifugation, the supernatant was discarded and the pellet was washed with 70% (v/v) ethanol. The RNA was reduced to dryness and resuspended in 30 μL of sterile water. First-strand cDNA was synthesized from 100 to 400 ng of total RNA using SuperScript II reverse transcriptase (Invitrogen; www.invitrogen.com), an oligo(dT)20VN primer (2.5 μm), RT buffer (5× first-strand buffer), deoxyribonucleotide triphosphates (0.5 mm each), and dithiothreitol (5 mm) in a total reaction volume of 20 μL. The occurrence of cDNAs corresponding to TRV2 coat protein was determined by PCR as described previously (Rotenberg et al., 2006).
RT-qPCR was performed using SYBR Green detection on triplicate technical and biological assays for each of eight plants shown to contain TRV2 coat protein transcripts, and the plants were infiltrated with A. tumefaciens harboring pTRV2-EV or pTRV2-TyrAT. Reactions were performed in a total volume of 10 μL and contained 5 μL of SYBR Green PCR mix (Applied Biosystems; www.appliedbiosystems.com), 1 μL of cDNA, and 5 μm PCR primers. PCR conditions were 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of denaturation at 95°C for 15 s each and annealing/extension at 72°C for 60 s each. Primers used for the relative quantification of transcripts were qRT-FW1 (5′-GTGAAAACAACACATAAATTC-3′) and qRT-RV1 (5′-CCAACTGCTACGATTGAGCAC-3′) for PsTyrAT and qRT-FW2 (5′-CAACAGTAGCAATAATCCGC-3′) and qRT-RV2 (5′-ACTAGCAGATACTTGCAACA-3′) for cl.10988. Threshold cycle (Ct) values of PsTyrAT and cl.10988 were normalized against the Ct of ubiquitin from opium poppy (GenBank accession no. JN402989), which served as the reference transcript. Primers used for the quantification of ubiquitin transcripts were qRT-FW3 (5′-TACCCTCCATTTGGTGCTTC-3′) and qRT-RV3 (5′-CCTCTGCTGATCTGGAGGAA-3′). Fluorescent signal intensities were recorded and analyzed on an Applied Biosystems 7300 Real-Time PCR System and SDS software. Dissociation curves for each amplicon were generated to confirm the presence of a single amplification product. The relative gene expression of PsTyrAT was compared in plants infiltrated with pTRV1/pTRV2-EV and pTRV1/pTRV2-TyrAT using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Gene Expression Analysis
Total RNA was isolated from opium poppy organs, and cDNAs were synthesized as described above. The cDNA samples served as templates for RT-qPCR analysis to determine the relative abundance of PsTyrAT transcripts in each organ using the primers and conditions described for the analysis of plants subjected to VIGS. PCR products were purified and sequenced to verify the identity of amplified cDNAs.
HPLC
Latex samples were reduced to dryness to determine dry weight and subsequently resuspended in methanol at a concentration of 30 mg mL−1. Ten microliters of each extract was diluted to a total volume of 100 μL with solvent A (98% [v/v] water:2% [v/v] acetonitrile:0.02% [v/v] phosphoric acid). HPLC was performed using a System Gold HPLC device (Beckman-Coulter; www.beckmancoulter.com) equipped with a LiChrospher 60 RP Select B column (146 × 4.1 mm, 5 μm; Merck; www.merck.com) and a mobile phase consisting of solvent A (2% [v/v] acetonitrile and 98% [v/v] water) and solvent B (98% [v/v] acetonitrile and 2% [v/v] water), each containing 0.02% (v/v) phosphoric acid. The column was equilibrated in solvent A, and alkaloids were eluted at a flow rate of 1.5 mL min−1 using the following gradient: 0 to 1 min, to 10% (v/v) solvent B; 1 to 50 min, to 100% (v/v) solvent B; 50 to 53 min, to 2% (v/v) solvent B; 53 to 60 min, hold at 2% (v/v) solvent B. Dextromethorphan was used as an internal standard. Peaks corresponding to morphine, codeine, thebaine, noscapine, papaverine, and dextromethorphan were monitored at 210 nm and identified on the basis of retention times and UV spectra compared with those of authentic standards. BIA levels were expressed as pg alkaloid μg−1 dry weight of latex based on standard quantification curves determined using authentic compounds.
LC-MS/MS
Enzyme assays contained 2 μg of native or heat-inactivated PsTyrAT protein, 3 mm l-Tyr, 1 mm PLP, 0.1 mm EDTA, and 0.3 mm α-ketoglutarate in a total volume of 100 μL of HEPES buffer, pH 8.2. Reactions were incubated for 1 h at 30°C and subsequently diluted with 250 μL of acetonitrile:water (55:45, v/v). l-Tyr and 4-HPP standards were prepared by diluting the pure compound dissolved in water with acetonitrile:water (55:45, v/v) to yield a final concentration of 500 nm. Enzyme assays were analyzed by LC-MS/MS using a 6400 Triple Quadrupole electrospray ionization-MS/MS apparatus (Agilent Technologies; www.agilent.com). The Zorbax SB-C18 2.1-mm × 50-mm column containing 1.8-μm particles was run at 45°C. The mobile phase was set for elution with gradients from 5% to 95% (v/v) acetonitrile in water and at a flow rate 0.5 mL min−1 for 5 min. The mobile phase returned to 5% (v/v) acetonitrile with a 3-min reequilibration period. Fragment voltages of 90 and 60 V were used for the analyses of l-Tyr and 4-HPP, respectively. In negative electrospray ionization mode, the voltage was 4,000 kV, the gas flow was 10 L min−1, nebulizing pressure was 30 ψ, and the gas temperature was 350°C. Collision energy was set at 0 eV and/or −10 eV, which showed the most specific and intense fragment ion for each compound. In full-scan mode, nine [M-H]− ions for l-Tyr (i.e. m/z 180.1, 163.1, 136.8, 119.2, 107.2, 105.5, 93.1, 74.3, and 71.8) and four [M-H]− ions for 4-HPP (i.e. m/z 179.1, 151.1, 114.7, and 107.1) were detected.
Phylogenetic Analysis
BLASTx searches of the NCBI (http://www.ncbi.nlm.nih.gov/BLAST) and in-house opium poppy nucleotide sequence databases were used to identify orthologs of PsTyrAT. The multiple sequence alignments and neighbor-joining phylogenetic tree were generated using ClustralW2 (http://www.ebi.ac.uk). The bootstrap analysis was performed using TREECON (Van de Peer and De Wachter, 1994). Differences in the transcript and alkaloid levels of pTRV2-TyrAT and TRV2-EV plants were analyzed by one- or two-tailed Student’s t test and regression analysis.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers GU370929 (PsTyrAT), JN402988 (synthetic PsTyrAT), JN402989 (ubiquitin), JN542549 (cl.10988), JN542550 (cl.1232), JN542551 (cl.1670), JN542552 (cl.8533), JN542553 (cl.615), and JN542554 (cl.5707).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of six homologs isolated from opium poppy transcriptome databases and encoding putative PLP-dependent aminotransferases.
Supplemental Figure S2. Amino acid sequence alignment of TyrAT (PsTyrAT) from opium poppy with other plant TyrATs.
Supplemental Figure S3. Detection of TyrAT enzyme activity by thin-layer chromatography.
Supplemental Figure S4. Relative substrate specificity of TyrAT from opium poppy.
Supplemental Figure S5. Steady-state enzyme kinetics of purified recombinant PsTyrAT with various substrates at different concentrations.
Supplemental Figure S6. Mean cl.10988 transcript levels in plants infiltrated with pTRV2-TyrAT (black bar) and pTRV2-EV (white bar).
Supplemental Table S1. Sequence similarity of six homologs isolated from opium poppy transcriptome databases and encoding putative PLP-dependent aminotransferases compared with PsTyrAT.
Acknowledgments
We thank Dr. Shaobo Wu for constructing the pQE30-TyrAT and pTRV2-TyrAT vectors and Scott Farrow for assistance with the LC-MS/MS analysis.
References
- Adcock HJ, Gaskin PJ, Shaw PN, Teesdale-Spittle PH, Buckberry LD. (1996) Novel sources of mammalian C-S lyase activity. J Pharm Pharmacol 48: 150–153 [DOI] [PubMed] [Google Scholar]
- Andersson SM, Pispa JP. (1982) Purification and properties of human liver tyrosine aminotransferase. Clin Chim Acta 125: 117–123 [DOI] [PubMed] [Google Scholar]
- Antognoni F, Faudale M, Poli F, Biondi S. (2009) Methyl jasmonate differentially affects tocopherol content and tyrosine amino transferase activity in cultured cells of Amaranthus caudatus and Chenopodium quinoa. Plant Biol (Stuttg) 11: 161–169 [DOI] [PubMed] [Google Scholar]
- Barta IC, Böger P. (1996) Purification and characterization of 4-hydroxyphenylpyruvate dioxygenase from maize. Pestic Sci 48: 109–116 [Google Scholar]
- Bein NN, Goldsmith HS. (1977) Recurrent massive haemorrhage from benign hepatic tumours secondary to oral contraceptives. Br J Surg 64: 433–435 [DOI] [PubMed] [Google Scholar]
- Bird DA, Franceschi VR, Facchini PJ. (2003) A tale of three cell types: alkaloid biosynthesis is localized to sieve elements in opium poppy. Plant Cell 15: 2626–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenfeldt W, Nowicki C, Montemartini-Kalisz M, Kalisz HM, Hecht HJ. (1999) Crystal structure of Trypanosoma cruzi tyrosine aminotransferase: substrate specificity is influenced by cofactor binding mode. Protein Sci 8: 2406–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner CA, Jensen RA. (1985) Novel features of prephenate aminotransferase from cell cultures of Nicotiana silvestris. Arch Biochem Biophys 238: 237–246 [DOI] [PubMed] [Google Scholar]
- Cavelier-Balloy B, Venencie PY, Lemonnier V, Verola O, Servant JM, Puissant A, Civatte J. (1985) [Histiocytoid hemangioma of the scalp]. Ann Dermatol Venereol 112: 965–972 [PubMed] [Google Scholar]
- Collier RH, Kohlhaw G. (1972) Nonidentity of the aspartate and the aromatic aminotransferase components of transaminase A in Escherichia coli. J Bacteriol 112: 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AJ. (2004) The role of glutamine transaminase K (GTK) in sulfur and α-keto acid metabolism in the brain, and in the possible bioactivation of neurotoxicants. Neurochem Int 44: 557–577 [DOI] [PubMed] [Google Scholar]
- Cruickshank DH, Isherwood FA. (1958) Glutamic-alanine and glutamic-aspartic transaminases of wheat germ. Biochem J 69: 189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-Eknamkul W, Ellis BE. (1987a) Purification and characterization of tyrosine aminotransferase activities from Anchusa officinalis cell cultures. Arch Biochem Biophys 257: 430–438 [DOI] [PubMed] [Google Scholar]
- De-Eknamkul W, Ellis BE. (1987b) Tyrosine aminotransferase: the entrypoint enzyme of tyrosine-derived pathway in rosmarinic acid biosynthesis. Phytochemistry 26: 1941–1946 [Google Scholar]
- Desgagné-Penix I, Facchini P. (2011) Benzylisoquinoline alkaloid biosynthesis. Ashihara H, Crozier A, Komamine A, , Plant Metabolism and Biotechnology. John Wiley & Sons, Chichester, UK, pp 241–261 [Google Scholar]
- Desgagné-Penix I, Khan MF, Schriemer DC, Cram D, Nowak J, Facchini PJ. (2010) Integration of deep transcriptome and proteome analyses reveals the components of alkaloid metabolism in opium poppy cell cultures. BMC Plant Biol 10: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ, Davies DD. (1961) Glutamic-oxaloacetic transaminase of cauliflower. 1. Purification and specificity. Biochem J 78: 615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini PJ, De Luca V. (1994) Differential and tissue-specific expression of a gene family for tyrosine/dopa decarboxylase in opium poppy. J Biol Chem 269: 26684–26690 [PubMed] [Google Scholar]
- Facchini PJ, De Luca V. (1995) Phloem-specific expression of tyrosine/dopa decarboxylase genes and the biosynthesis of isoquinoline alkaloids in opium poppy. Plant Cell 7: 1811–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini PJ, De Luca V. (2008) Opium poppy and Madagascar periwinkle: model non-model systems to investigate alkaloid biosynthesis in plants. Plant J 54: 763–784 [DOI] [PubMed] [Google Scholar]
- Forest JC, Wightman F. (1972) Amino acid metabolism in plants. 3. Purification and some properties of a multispecific aminotransferase isolated from bushbean seedlings (Phaseolus vulgaris L.). Can J Biochem 50: 813–829 [DOI] [PubMed] [Google Scholar]
- Gibson RA, Barrett G, Wightman F. (1972) Biosynthesis and metabolism of indole-3yl-acetic acid. III. Partial purification and properties of tryptamine-forming L-tryptophan decarboxylase from tomato shoots. J Exp Bot 23: 775–786 [Google Scholar]
- Gonda I, Bar E, Portnoy V, Lev S, Burger J, Schaffer AA, Tadmor Y, Gepstein S, Giovannoni JJ, Katzir N, et al. (2010) Branched-chain and aromatic amino acid catabolism into aroma volatiles in Cucumis melo L. fruit. J Exp Bot 61: 1111–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalraj M, Tseng TS, Olszewski N. (1996) The rooty gene of Arabidopsis encodes a protein with highest similarity to aminotransferases. Plant Physiol 111: S-114 [Google Scholar]
- Grange T, Guénet C, Dietrich JB, Chasserot S, Fromont M, Befort N, Jami J, Beck G, Pictet R. (1985) Complete complementary DNA of rat tyrosine aminotransferase messenger RNA: deduction of the primary structure of the enzyme. J Mol Biol 184: 347–350 [DOI] [PubMed] [Google Scholar]
- Hagel JM, Facchini PJ. (2010) Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nat Chem Biol 6: 273–275 [DOI] [PubMed] [Google Scholar]
- Hara M, Tanaka S, Tabata M. (1994) Induction of a specific methyltransferase activity regulating berberine biosynthesis by cytokinin in Thalictrum minus cell cultures. Phytochemistry 36: 327–332 [Google Scholar]
- Hayashi H. (1995) Pyridoxal enzymes: mechanistic diversity and uniformity. J Biochem 118: 463–473 [DOI] [PubMed] [Google Scholar]
- Hernández A, Ruiz MT. (1998) An EXCEL template for calculation of enzyme kinetic parameters by non-linear regression. Bioinformatics 14: 227–228 [DOI] [PubMed] [Google Scholar]
- Holland HL, Jeffs PW, Capps TM, McLean DB. (1979) The biosynthesis of protoberberine and related isoquinoline alkaloids. Can J Chem 57: 1588–1597 [Google Scholar]
- Holländer-Czytko H, Grabowski J, Sandorf I, Weckermann K, Weiler EW. (2005) Tocopherol content and activities of tyrosine aminotransferase and cystine lyase in Arabidopsis under stress conditions. J Plant Physiol 162: 767–770 [DOI] [PubMed] [Google Scholar]
- Jones PR, Manabe T, Awazuhara M, Saito K. (2003) A new member of plant CS-lyses: a cystine lyase from Arabidopsis thaliana. J Biol Chem 278: 10291–10296 [DOI] [PubMed] [Google Scholar]
- Ko TP, Wu SP, Yang WZ, Tsai H, Yuan HS. (1999) Crystallization and preliminary crystallographic analysis of the Escherichia coli tyrosine aminotransferase. Acta Crystallogr D Biol Crystallogr 55: 1474–1477 [DOI] [PubMed] [Google Scholar]
- Koshiba T, Mito N, Miyakado M. (1993) L- and D-tryptophan aminotransferases from maize coleoptiles. J Plant Res 106: 25–29 [Google Scholar]
- Lee EJ, Facchini PJ. (2010) Norcoclaurine synthase is a member of the pathogenesis-related 10/Bet v1 protein family. Plant Cell 22: 3489–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GT, Zhang TM, Wang BE, Wang YW. (1992) Protective action of seven natural phenolic compounds against peroxidative damage to biomembranes. Biochem Pharmacol 43: 147–152 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lopukhina A, Dettenberg M, Weiler EW, Holländer-Czytko H. (2001) Cloning and characterization of a coronatine-regulated tyrosine aminotransferase from Arabidopsis. Plant Physiol 126: 1678–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrides C, Orr W. (1975) Multispecific aspartate and aromatic amino acid aminotransferases in Escherichia coli. J Biol Chem 250: 4128–4133 [PubMed] [Google Scholar]
- Mazelis M, Fowden L. (1969) Conversion of ornithine into proline by enzymes from germinating peanut cotyledons. Phytochemistry 8: 801–809 [Google Scholar]
- McQueen-Mason SJ, Hamilton RH. (1989) The biosynthesis of indole-3-acetic acid from D-tryptophan in Alaska pea plastids. Plant Cell Physiol 30: 999–1005 [Google Scholar]
- Mehere P, Han Q, Lemkul JA, Vavricka CJ, Robinson H, Bevan DR, Li J. (2010) Tyrosine aminotransferase: biochemical and structural properties and molecular dynamics simulations. Protein Cell 1: 1023–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller G, Scherer G, Zentgraf H, Ruppert S, Herrmann B, Lehrach H, Schütz G. (1985) Isolation, characterization and chromosomal mapping of the mouse tyrosine aminotransferase gene. J Mol Biol 184: 367–373 [DOI] [PubMed] [Google Scholar]
- Noguchi T, Hayashi S. (1980) Peroxisomal localization and properties of tryptophan aminotransferase in plant leaves. J Biol Chem 255: 2267–2269 [PubMed] [Google Scholar]
- Nonhebel HM, Cooney TP, Simpson R. (1993) The route, control and compartmentation of auxin synthesis. Aust J Plant Physiol 20: 527–539 [Google Scholar]
- Nowicki C, Hunter GR, Montemartini-Kalisz M, Blankenfeldt W, Hecht H, Kalisz HM. (2001) Recombinant tyrosine aminotransferase from Trypanosoma cruzi: structural characterization and site directed mutagenesis of a broad substrate specificity enzyme. Biochim Biophys Acta 1546: 268–281 [DOI] [PubMed] [Google Scholar]
- Prabhu PR, Hudson AO. (2010) Identification and partial characterization of L-tyrosine aminotransferase (TAT) from Arabidopsis thaliana. Biochem Res Int 2010: 549572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettenmeier R, Natt E, Zentgraf H, Scherer G. (1990) Isolation and characterization of the human tyrosine aminotransferase gene. Nucleic Acids Res 18: 3853–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D. (2004) VIGS vectors for gene silencing: many targets, many tools. Annu Rev Plant Biol 55: 495–519 [DOI] [PubMed] [Google Scholar]
- Rotenberg D, Thompson TS, German TL, Willis DK. (2006) Methods for effective real-time RT-PCR analysis of virus-induced gene silencing. J Virol Methods 138: 49–59 [DOI] [PubMed] [Google Scholar]
- Rueffer M, Zenk MH. (1987) Distant precursors of benzylisoquinoline alkaloids and their enzymatic formation. Z Naturforsch 42c: 319–332 [Google Scholar]
- Samanani N, Alcantara J, Bourgault R, Zulak KG, Facchini PJ. (2006) The role of phloem sieve elements and laticifers in the biosynthesis and accumulation of alkaloids in opium poppy. Plant J 47: 547–563 [DOI] [PubMed] [Google Scholar]
- Sandorf I, Holländer-Czytko H. (2002) Jasmonate is involved in the induction of tyrosine aminotransferase and tocopherol biosynthesis in Arabidopsis thaliana. Planta 216: 173–179 [DOI] [PubMed] [Google Scholar]
- Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D. (2004) Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 16: 1419–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider P, Bouhired S, Hoffmeister D. (2008) Characterization of the atromentin biosynthesis genes and enzymes in the homobasidiomycete Tapinella panuoides. Fungal Genet Biol 45: 1487–1496 [DOI] [PubMed] [Google Scholar]
- Schumacher HM, Rüffer M, Nagakura N, Zenk MH. (1983) Partial purification and properties of (S)-norlaudanosoline synthase from Eschscholtzia tenuifolia cell cultures. Planta Med 48: 212–220 [DOI] [PubMed] [Google Scholar]
- Shinomiya T, Scherer G, Schmid W, Zentgraf H, Schütz G. (1984) Isolation and characterization of the rat tyrosine aminotransferase gene. Proc Natl Acad Sci USA 81: 1346–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrado VR, Montemartini-Kalisz M, Kalisz HM, De La Fuente MC, Hecht HJ, Nowicki C. (2003) Involvement of conserved asparagine and arginine residues from the N-terminal region in the catalytic mechanism of rat liver and Trypanosoma cruzi tyrosine aminotransferases. Protein Sci 12: 1039–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Yamaguchi H, Nakanishi H, Shioiri T, Nishizawa NK, Mori S. (1999) Cloning two genes for nicotianamine aminotransferase, a critical enzyme in iron acquisition (strategy II) in graminaceous plants. Plant Physiol 121: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truelsen TA. (1972) Indole-3-pyruvic acid as an intermediate in the conversion of tryptophan to indole-3-acetic acid. I. Some characteristics of tryptophan transaminase form mung bean seedlings. Physiol Plant 26: 289–295 [Google Scholar]
- Tzin V, Galili G. (2010) New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol Plant 3: 956–972 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, De Wachter R. (1994) TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci 10: 569–570 [DOI] [PubMed] [Google Scholar]
- Wightman F, Forest JC. (1978) Properties of plant aminotransferases. Phytochemistry 17: 1455–1471 [Google Scholar]
- Wilkinson TL, Adams D, Minto LB, Douglas AE. (2001) The impact of host plant on the abundance and function of symbiotic bacteria in an aphid. J Exp Biol 204: 3027–3038 [DOI] [PubMed] [Google Scholar]
- Winter H, Lohaus G, Heldt HW. (1992) Phloem transport of amino acids in relation to their cytosolic levels in barley leaves. Plant Physiol 99: 996–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Di P, Chen J, Liu Y, Chen W, Zhang L. (2009a) Characterization and expression profiling of 4-hydroxyphenylpyruvate dioxygenase gene (Smhppd) from Salvia miltiorrhiza hairy root cultures. Mol Biol Rep 36: 2019–2029 [DOI] [PubMed] [Google Scholar]
- Xiao Y, Gao S, Di P, Chen J, Chen W, Zhang L. (2009b) Methyl jasmonate dramatically enhances the accumulation of phenolic acids in Salvia miltiorrhiza hairy root cultures. Physiol Plant 137: 1–9 [DOI] [PubMed] [Google Scholar]
- Zulak KG, Weljie AM, Vogel HJ, Facchini PJ. (2008) Quantitative 1H NMR metabolomics reveals extensive metabolic reprogramming of primary and secondary metabolism in elicitor-treated opium poppy cell cultures. BMC Plant Biol 8: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]