Abstract
Mitogen-activated protein kinases (MAPKs) contribute to the establishment of plant disease resistance by regulating downstream signaling components, including transcription factors. In this study, we identified MAPK-interacting proteins, and among the newly discovered candidates was a Cys-2/His-2-type zinc finger protein named PtiZFP1. This putative transcription factor belongs to a family of transcriptional repressors that rely on an ERF-associated amphiphilic repression (EAR) motif for their repression activity. Amino acids located within this repression motif were also found to be essential for MAPK binding. Close examination of the primary protein sequence revealed a functional bipartite MAPK docking site that partially overlaps with the EAR motif. Transient expression assays in Arabidopsis (Arabidopsis thaliana) protoplasts suggest that MAPKs promote PtiZFP1 degradation through the 26S proteasome. Since features of the MAPK docking site are conserved among other EAR repressors, our study suggests a novel mode of defense mechanism regulation involving stress-responsive MAPKs and EAR repressors.
Plants have evolved molecular mechanisms to perceive and respond to a variety of biotic stresses (Dodds and Rathjen, 2010; Wu and Baldwin, 2010). Following the recognition of stress, the activation of complex signaling networks leads to a massive transcriptional reprogramming and de novo synthesis of defense-related proteins. Poplar (Populus spp.) trees, for instance, respond to herbivory or pathogen infection by reconfiguring their transcriptome, including the up-regulation of numerous defense-related genes (Ralph et al., 2006; Rinaldi et al., 2007; Major and Constabel, 2008; Azaiez et al., 2009; Duplessis et al., 2009; Philippe et al., 2010). Transcription factors (TFs) play a pivotal role in reprogramming the plant transcriptome, and several candidates have been identified as regulators of the plant defense response, including subgroups of zinc fingers, AP2-ERFs, bZIPs, WRKYs, and bHLHs (Eulgem, 2005), although few have been characterized for poplar. TFs are also responsible for the retrofeedback signal that terminates the elicited response (Eulgem, 2005; Kazan, 2006). This is crucial, as plant defense mechanisms are associated with high fitness costs; consequently, TFs must be tightly regulated. TFs can be regulated either at the transcriptional level or at the posttranslational level by protein modification such as phosphorylation. Phosphorylation by MAPK plays a crucial role in the signal transduction of stress and developmental cues in all eukaryotic cells. In yeast and metazoans, MAPK-mediated phosphorylation regulates the activity of several TFs, and large-scale analysis with protein chips also suggests that TFs represent primary targets for plant MAPKs (Popescu et al., 2009).
Based on homology, plant MAPKs are divided into four phylogenetic groups, A to D (MAPK Group, 2002), which are conserved across several plant species (Hamel et al., 2006; Nicole et al., 2006). These protein kinases are involved in signal transduction elicited after stress perception, along with the production of reactive oxygen species, Ca2+ influxes, and phytohormones like jasmonic acid (JA), salicylic acid (SA), and ethylene (ET; Asai et al., 2002; Mészáros et al., 2006). MAPK cascades usually relay a perceived signal through sequential phosphorylation of their tripartite kinase modules, where a MAPK kinase kinase (MAP3K) activates a MAPK kinase (MAP2K), which in turn activates a MAPK. MAPK further phosphorylates a variety of protein substrates, in turn affecting their biochemical properties. Phosphorylation, for instance, modulates TF function by altering their transcriptional activity, subcellular localization, or protein stability. Moreover, phosphorylation of different sites within the same protein often has different outcomes on protein function (Yoo et al., 2008). Thus, the outcome of phosphorylation, in addition to the protein target, determines how MAPKs affect signal transduction. In Arabidopsis (Arabidopsis thaliana), AtMPK3 and AtMPK6, for example, phosphorylate the bHLH AtSPCH, which is thus destabilized to alter stomatal development (Lampard et al., 2008). AtMPK3 and AtMPK6 also regulate TFs involved in ET signaling, where phosphorylation enhances the stability of AtEIN3 and the DNA-binding affinity of AtERF104 (Yoo et al., 2008; Bethke et al., 2009). Phosphorylation of the defense-related bZIP AtVIP1 by AtMPK3 was further shown to trigger its nuclear translocation (Djamei et al., 2007). Several studies have also shown that MAPK-mediated phosphorylation of TFs alters their transcriptional activity. For example, in tobacco (Nicotiana tabacum), NtWIPK (an AtMPK3 ortholog) phosphorylates NtWIF and NtSIPK (an AtMPK6 ortholog) phosphorylates NtWRKY1, both cases resulting in enhanced transcriptional activity (Yap et al., 2005; Menke et al., 2005). In loblolly pine (Pinus taeda), PtMAPK6 mediates the phosphorylation of R2R3-MYB PtMYB4, reducing its TF activity, which further prevents premature lignification during xylem development (Morse et al., 2009). These examples highlight the range of potential outcomes associated with TF phosphorylation and the diverse TFs and physiological processes regulated by plant MAPKs. The identification of new MAPK substrates and monitoring of their phosphorylation status are key issues to better understand how MAPKs fine-tune defense responses, especially with respect to how the phosphorylation of TFs shapes defense-related transcriptional reprogramming (Fiil et al., 2009).
In poplar, we previously identified TFs from various families that are differentially regulated in the poplar transcriptome in response to colonization by the biotrophic fungus Melampsora (Duplessis et al., 2009) or in response to herbivory (Ralph et al., 2006). This suggests that these TFs participate in regulation of the defense transcriptome; however, little is known about the posttranslational control of poplar TFs, and it is unclear how these TFs initiate the response. On the other hand, we have evidence of stress-activated MAPKs in poplar, which are prime candidates to modulate TF function. Poplar MAPKs were shown to be activated in response to chitosan, ozone treatment, and following infection by Melampsora foliar rust (Hamel et al., 2005; Boyle et al., 2010), yet no downstream substrate has been identified. Based on these data, and given the importance of AtMPK3 (and its orthologs) in both the priming (Beckers et al., 2009) and regulation of defense mechanisms against bacteria and fungi (Asai et al., 2002; Ren et al., 2008), a yeast two-hybrid screen was performed to find putative interactors of poplar PtiMPK3-1. This approach allowed the identification of a novel MAPK-interacting protein, PtiZFP1, a TF that belongs to the Cys-2/His-2-type zinc finger protein family and that is characterized by the presence of an ERF-associated amphiphilic repression (EAR) motif at the C terminus. The EAR motif was initially identified in class II ERF and Cys-2/His-2-type zinc finger TFs as a motif sufficient to confer transcriptional repression activity (Ohta et al., 2001). This repression domain has been identified in plant transcriptional regulators belonging to diverse families involved in plant development and stress response (Kagale et al., 2010; Kagale and Rozwadowski, 2010). Here, we demonstrate that PtiZFP1 interaction with MPK3- and MPK6-type MAPKs depends on a bipartite MAPK docking site (MDS) found in the functionally rich C terminus of PtiZFP1. Interestingly, MAPK interaction accelerates the turnover of PtiZFP1 in planta. Sequence comparison of EAR TFs also suggests that the MDS is conserved among other defense-related EAR repressors. Altogether, these results support a novel hypothesis in which MAPKs could serve as a molecular cue for the targeted degradation of EAR repressors and subsequent derepression of target genes.
RESULTS
Isolation of PtiZFP1: A MAPK-Interacting Partner
To address the question of MAPK function in stress signal transduction, we searched for proteins that interact with PtiMPK3-1. A yeast two-hybrid (Y2H) assay was used to screen a cDNA library generated from leaves of hybrid poplar NM6 (Populus nigra × Populus maximowiczii) infected with the foliar rust fungus Melampsora medusae f.sp. deltoidae (Mmd). Infection of NM6 poplar with Mmd rust results in host-specific partial resistance that still allows a certain extent of fungal growth. NM6 hybrids are thus considered susceptible, and the defense response (quantitative resistance) associated with this host-microbe interaction was described elsewhere (Azaiez et al., 2009; Boyle et al., 2010). Among the peptides selected for their potential interaction with PtiMPK3-1, we identified a truncated cDNA coding for the last 104 amino acids of a putative TF. Sequence analysis revealed one DNA-binding zinc finger domain as well as an EAR transcriptional repression motif [(L/F)DLN(L/F)XP, where X is any amino acid; Ohta et al., 2001]. The interaction was confirmed by cotransforming yeast cells with the candidate cDNA and with either PtiMPK3-1 or PtiMPK6-2, two MAPKs belonging to group A (Supplemental Fig. S1; Hamel et al., 2006). No interaction could be detected using empty vectors or the group C MAPK PtiMPK2, suggesting a specific interaction between the truncated TF and group A MAPKs.
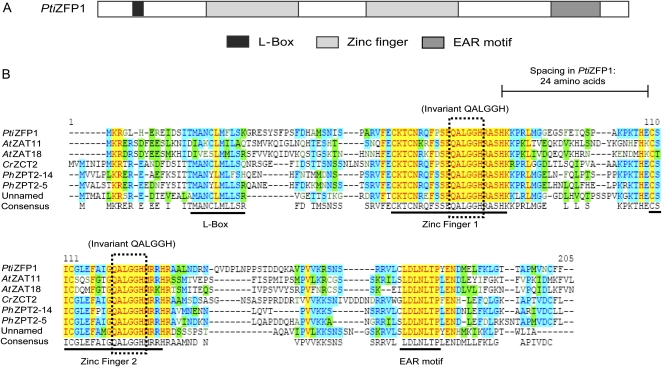
Search of the Populus trichocarpa genome identified a 540-bp gene model corresponding to the cDNA isolated using Y2H assay. The novel gene is located on chromosome X and displays no intron (Supplemental Fig. S2). The deduced amino acid sequence shares closest similarity to that of zinc finger proteins that possess two Cys-2/His-2 zinc finger domains (Fig. 1A) and was named P. trichocarpa Zinc Finger Protein1 (PtiZFP1; protein identifier 659738). Within the conserved sequence of each zinc finger domain (Cx2–4Cx3Fx5Lx2Hx3–5H), two Cys and two His residues tetrahedrally coordinate a zinc atom to produce a compact structure that interacts with the major groove of the DNA in a sequence-specific manner (Choo and Klug, 1997). Zinc fingers found in PtiZFP1 are separated by a 24-amino acid spacer, and both domains display a QALGGH motif that is the main DNA-contacting surface and is specific to plant zinc finger proteins (Fig. 1B; Kubo et al., 1998). Apart from the two zinc fingers, the primary sequence of this class of TF is overall poorly conserved, with only two other recognizable regions preserved across putative orthologs: an N-terminal L-box motif of unknown function and the C-terminal EAR motif (Fig. 1B).
Figure 1.
Structure and conservation of PtiZFP1. A, Schematic view of the PtiZFP1 protein and organization of its conserved motifs. B, Protein sequence alignment of homologous Cys-2/His-2-type zinc finger proteins. PtiZFP1 (protein identifier 659738) is derived from P. trichocarpa. CrZCT2 (CAF74934) is derived from C. roseus. PhZPT2-5 (BAA19110) and PhZPT2-14 (BAA21923) are derived from Petunia hybrida. AtZAT11 (NP_181279) and AtZAT18 (NP_190928) are derived from Arabidopsis. The unnamed product (ACB20696) is derived from Camellia sinensis.
With 176 members in Arabidopsis, Cys-2/His-2-type zinc finger proteins form one of the largest groups of transcriptional regulators in plants (Englbrecht et al., 2004). These proteins are further classified into different families, with the most studied members belonging to the C1 family (Ciftci-Yilmaz and Mittler, 2008). C1 members either possess one isolated or two to five separated zinc fingers, denoted by the acronym “i”; for example, PtiZFP1 belongs to subclass C1-2i. Phylogenetic analysis was conducted to identify the closest putative orthologs of PtiZFP1 (Supplemental Fig. S3). Among Arabidopsis candidates, AtZAT7, AtZAT10, and AtZAT12 are the most extensively characterized, as they have been associated with abiotic stress relief (Ciftci-Yilmaz and Mittler, 2008). Other members from this subclass are induced by biotic challenges, including AtZAT11 in response to Flg22 elicitor peptide (Zipfel et al., 2004). Several Cys-2/His-2-type zinc finger proteins have also been studied in Catharanthus roseus, where they were shown to repress the expression of genes involved in the biosynthesis of alkaloid metabolites (Pauw et al., 2004).
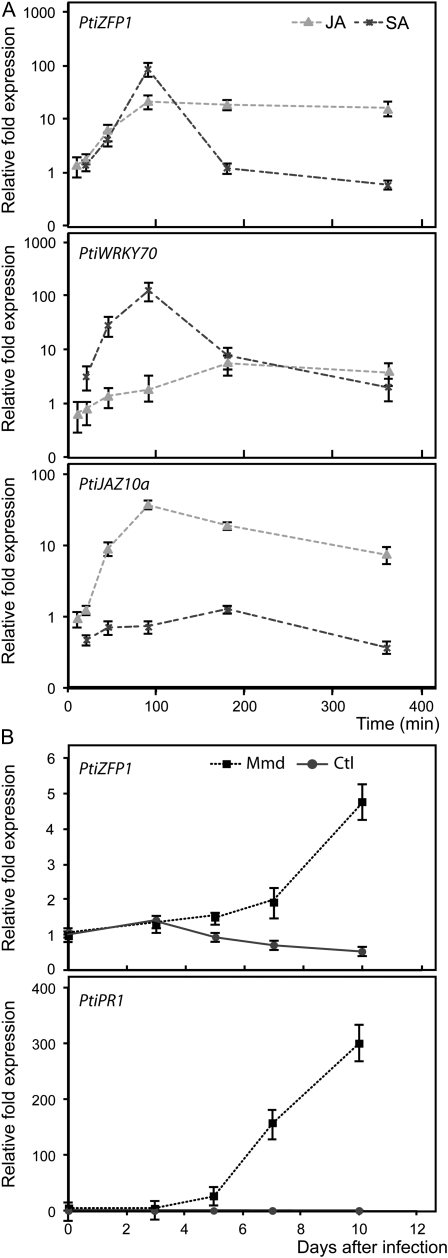
SA and JA are known to play major roles in the plant defense response against pathogens and pests, as they trigger a signal transduction network that leads to physiological adaptations to external stimuli (Bari and Jones, 2009). To explore the potential role of PtiZFP1 in biotrophic stress mechanisms, we analyzed PtiZFP1 induction in response to SA and JA. SA is thought to play a crucial role in disease resistance against biotrophic pathogens, while JA is more often associated with defense mechanisms against necrotrophic microbes and herbivorous insects (Bari and Jones, 2009). Poplar cell suspensions (hybrid H11-11; P. trichocarpa × Populus deltoides) treated with methyl jasmonate (MeJA) or SA were harvested after 10 to 350 min, and PtiZFP1 transcript accumulation was analyzed by quantitative real-time (RTq) PCR (Fig. 2A; Supplemental Tables S1 and S2). PtiZFP1 transcript shows a rapid accumulation in response to both SA and JA treatments, with respective 85- and 20-fold increases compared with untreated cells. The JA-responsive marker gene PtiJAZ10a (an ortholog of AtJAZ10) was used as a control for the MeJA treatment (Thines et al., 2007), and the TF WRKY70, which is known to be induced by the SA pathway (Li et al., 2004), was used as a marker gene for the SA treatment (Fig. 2A). These data suggest that PtiZFP1 is part of both SA and JA defense responses and, by correlation as a putative transcriptional repressor, participates in the regulation of the transcriptional responses downstream of these stress hormones. Moreover, PtiZFP1 expression is induced in poplar hybrid NM6 leaves in response to infection by Mmd (Fig. 2B). Mmd rust is known to elicit a strong defense response in this hybrid poplar (Azaiez et al., 2009; Boyle et al., 2010), as suggested here by the increase in PtiPR1 transcript in the infected leaves (Fig. 2B).
Figure 2.
Transcriptional control of PtiZFP1 following rust infection of leaves and hormone treatment of poplar cell suspensions. A, Transcript accumulation of PtiZFP1 following hormone treatment of poplar cell suspension H11-11 (P. trichocarpa × P. deltoides) with either SA or MeJA. Effective hormonal treatment was assessed by quantification of the JA-responsive marker gene PtiJAZ10a and the SA-responsive marker gene PtiWRKY70. Results are expressed in transcript fold increase compared with time zero and with untreated cells and normalized against reference genes. B, Transcript accumulation of PtiZFP1 and PtiPR1 following Mmd rust infection of leaves of poplar NM6 (P. nigra × P. maximowiczii). Results are expressed in transcript fold increase compared with time zero and normalized against reference genes.
Determination of the PtiZFP1-Interacting Domain
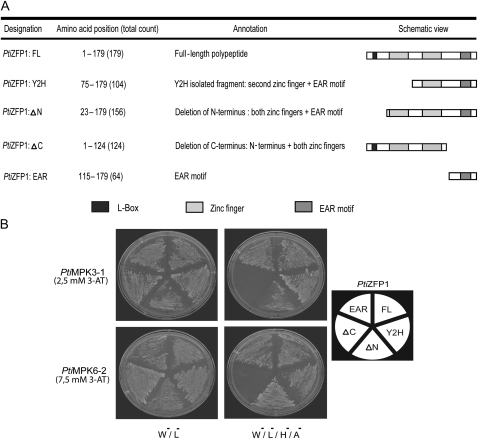
To determine the PtiZFP1 region that is specifically involved in MAPK interaction, Y2H assays were repeated with truncated versions of the TF (Fig. 3A). As shown in Figure 3B, full-length PtiZFP1 (PtiZFP1:FL) as well as both the initially isolated Y2H fragment (PtiZFP1:Y2H) and the N-terminal truncated version (PtiZFP1:ΔN) all interacted with PtiMPK3-1 and PtiMPK6-2. In contrast, C-terminal truncation of a 55-amino acid segment covering the EAR motif (PtiZFP1:ΔC) prevented protein-protein interaction. Finally, use of only the last 64 amino acids of PtiZFP1 (PtiZFP1:EAR) allowed MAPK interaction with similar strength compared with the FL, Y2H, and ΔN versions. Taken together, these results demonstrate that the C-terminal portion of PtiZFP1 is necessary and sufficient for interaction with MAPKs.
Figure 3.
Identification of PtiZFP1-interacting regions by Y2H assay. A, Schematic view of PtiZFP1 deletion mutants used for Y2H assay. B, To define the PtiZFP1 docking surface, directed Y2H experiments were conducted with PtiMPK3-1 or PtiMPK6-2. Petri dishes were incubated at 30°C for several days, and growing colonies were restreaked for presentation purposes.
The C Terminus of PtiZFP1 Displays Amino Acids Reminiscent of a MDS
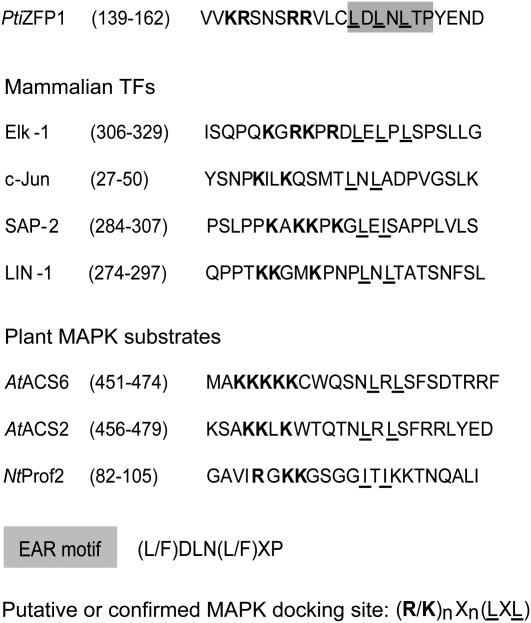
Since deletion of the PtiZFP1 C terminus prevents interaction with group A MAPKs, we took a closer look at the amino acids located within this protein segment in search of residues that could favor recognition by MAPKs. In animals and yeast, many MAPK-binding proteins interact with cognate MAPK through a specific surface-exposed site, the MDS (Sharrocks et al., 2000). This docking site is composed of several basic residues separated from an LxL motif by three to five amino acids [(R/K)nXn(LXL)]. The PtiZFP1 C terminus contains a potential MDS that partially overlaps the EAR motif, with both motifs sharing common LxL residues (Fig. 4). This potential MDS is highly similar to the MDS of mammalian TFs that interact with MAPKs (Fig. 4). Putative MDS are also present among distinct plant MAPK substrates, like the tobacco profilin NtProf2 (Limmongkon et al., 2004) and the Arabidopsis 1-aminocyclopropane-1-carboxylic acid synthases AtACS2/6 (Liu and Zhang, 2004). The functionality of plant MDS has never been formally assessed, yet these examples suggest that they could play an essential role in molecular recognition of certain plant MAPK substrates.
Figure 4.
Conservation of MDS. Sequence alignment of protein regions displaying functional or predicted MDS is shown. The consensus sequence of the EAR motif is highlighted in gray, and characteristics reminiscent of MDS are shown in boldface (basic amino acids) and underlined (bulky hydrophobic residues or the LXL motif).
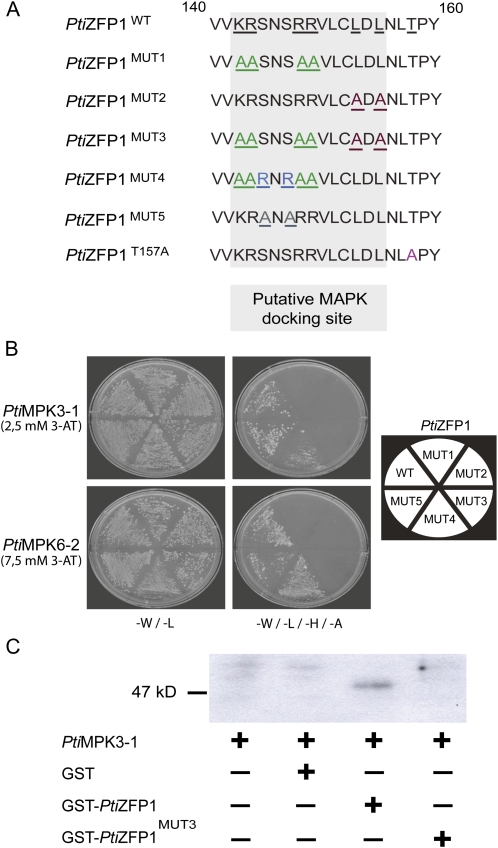
PtiZFP1 MDS Is Functional
To functionally characterize the putative MDS found in PtiZFP1, we used site-directed mutagenesis to generate variations within this region (Fig. 5A) and tested mutant proteins using Y2H assay. Mutations of either the basic amino acids (PtiZFP1MUT1) or the EAR motif core Leu residues (PtiZFP1MUT2) impaired interaction with MAPKs (Fig. 5B). Accordingly, a double mutant version (PtiZFP1MUT3) also failed to interact. Based on the PtiZFP1MUT1 construct, we also generated PtiZFP1MUT4, in which we reintroduced basic amino acids in front of the EAR motif (Fig. 5A). Interestingly, this mutant restored PtiZFP1MUT1 function, as interaction with MAPKs was observed (Fig. 5B). As a negative control, amino acids located outside the putative MDS were also converted to Ala (PtiZFP1MUT5), and this mutant had no impact on MAPK binding (Fig. 5B). Finally, in vitro glutathione S-transferase (GST) pull-down assays showed that recombinant PtiMPK3-1 can be affinity purified by GST-PtiZFP1 but not by GST or GST-PtiZFP1MUT3, attesting to the specificity of the interaction (Fig. 5C). Taken together, these approaches confirm the functionality of PtiZFP1 MDS.
Figure 5.
Functionality of PtiZFP1 MDS. A, Protein sequence alignment of the C-terminal region of PtiZFP1 mutants. For MUT1, MUT2, and MUT3, basic amino acid conversions are shown in green, whereas conversions of bulky hydrophobic residue are shown in red. MUT4 restores MUT1 activity, and amino acid changes are shown in blue. MUT5 was used as a negative control, and amino acid conversions are shown in gray. PtiZFP1T157A was also produced to study the involvement of phosphorylation in protein regulation. Amino acid conversion is shown in pink. B, To confirm the functionality of PtiZFP1 MDS, directed Y2H experiments were conducted using MAPKs and PtiZFP1 mutants. C, Pull-down assay between recombinant proteins confirms Y2H data. Pull-down assay of proteins with affinity matrix showed that PtiMPK3-1 interacted with GST-PtiZFP1 and failed to interact with GST-PtiZFP1MUT3. GST was used as a control, and PtiMPK3-1 was detected by immunoblotting with anti-PtiMPK3 antibody.
MAPKs and PtiZFP1 Colocalize and Interact in Planta
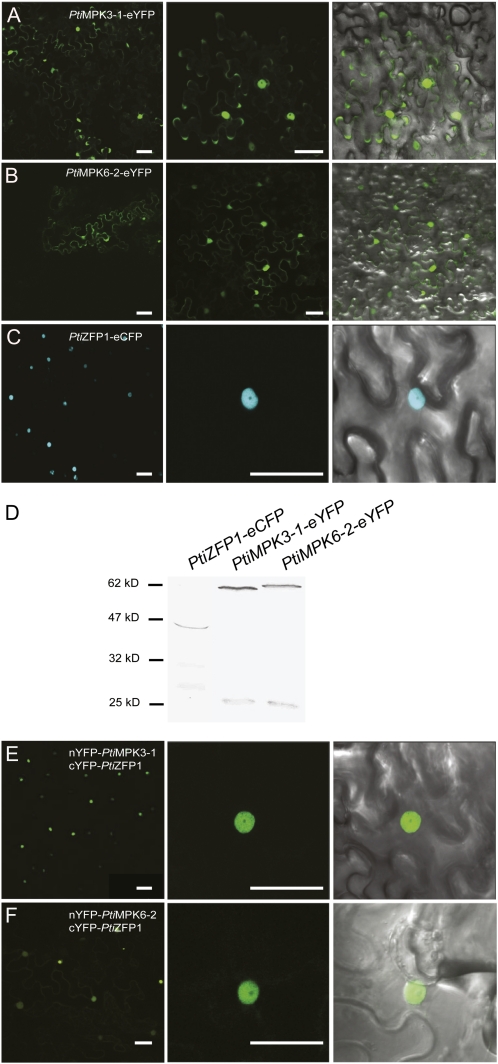
We further characterized PtiZFP1 and its MAPK-interacting partners by uncovering their respective subcellular localizations. PtiMPK3-1 and PtiMPK6-2 were each fused to the enhanced Yellow Fluorescent Protein (PtiMPK3-1-eYFP and PtiMPK6-2-eYFP), whereas PtiZFP1 was fused to the enhanced Cyan Fluorescent Protein (PtiZFP1-eCFP; Supplemental Fig. S4). Constructs were transiently expressed in Nicotiana benthamiana leaves using Agrobacterium tumefaciens, and fluorescence was monitored using confocal microscopy. Both PtiMPK3-1-eYFP and PtiMPK6-2-eYFP (Fig. 6, A and B) were found within the cytoplasm and nucleus of transformed cells. As for PtiZFP1-eCFP, fluorescence was solely detected in the nucleus (Fig. 6C), a result that is consistent with the predicted molecular function of PtiZFP1 as a TF. Despite the presence of some degradation products, western-blot analysis using anti-GFP antibodies confirmed the expression of full-length fusion proteins (Fig. 6D). As a whole, these results demonstrate that group A MAPKs and PtiZFP1 share nuclear localization, suggesting that they could interact in planta.
Figure 6.
PtiZFP1 and group A MAPKs colocalize and interact inside the nucleus. A to C, Subcellular localization of the chimeric proteins PtiMPK3-1-eYFP (A), PtiMPK6-2-eYFP (B), and PtiZFP1-eCFP (C) in N. benthamiana leaf epidermal cells. D, Anti-GFP western blot confirms the expression of fusion proteins used for localization experiments. E and F, BiFC experiments confirm the in planta interaction between nYFP-PtiMPK3-1 and cYFP-PtiZFP1 (E) and between nYFP-PtiMPK6-2 and cYFP-PtiZFP1 (F). Fluorescence and merged images are shown. Bars = 50 μm.
To confirm the in planta interaction between PtiZFP1 and MAPKs, we conducted bimolecular fluorescence complementation (BiFC) assays. PtiMPK3-1 and PtiMPK6-2 were each fused to the N-terminal portion of the YFP (nYFP-PtiMPK3-1 and nYFP-PtiMPK6-2), whereas PtiZFP1 was fused to the C-terminal portion of the YFP (cYFP-PtiZFP1; Supplemental Fig. S4). nYFP-fused MAPKs were separately expressed in tobacco epidermal cells along with cYFP-PtiZFP1. In both cases, a fluorescent signal was detected by microscopy, suggesting that both MAPKs interacted with PtiZFP1 in planta. As expected from subcellular localization data, fluorescence was exclusively detected in the nucleus (Fig. 6, E and F). No fluorescent signal was visible when only one of the fusion proteins was expressed with the complementary portion of YFP (data not shown).
Specific MAPKs Reduce PtiZFP1 Protein Accumulation in Arabidopsis Protoplasts
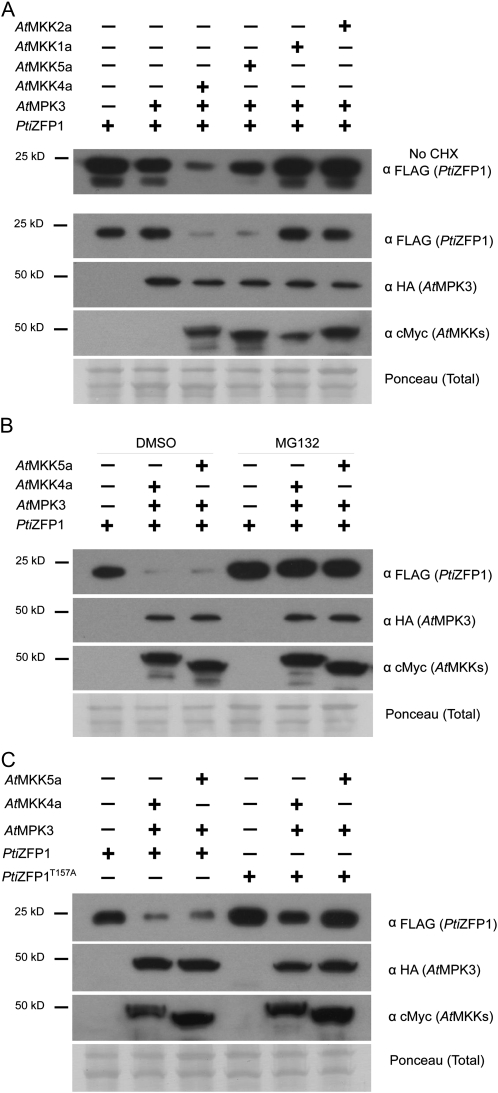
To assess the role of the interaction between group A MAPKs and PtiZFP1, we focused on PtiZFP1 protein stability following its coexpression with MAPKs in Arabidopsis mesophyll protoplasts. FLAG-tagged PtiZFP1 was expressed alone or with hemagglutinin (HA)-tagged AtMPK3/6. These Arabidopsis MAPKs are the putative orthologs of PtiMPK3-1/3-2 and PtiMPK6-1/6-2, respectively (Hamel et al., 2006). To mimic stress signaling and obtain insights into signal specificity, additional transfections were conducted with constitutively activated forms of cMyc-tagged AtMKKs. Expression of AtMKK4a/5a was used to activate AtMPK3/6, while AtMKK1a/2a were used as a negative control. PtiZFP1 protein level was then evaluated by western blotting. PtiZFP1 accumulation was similar when expressed alone or in combination with AtMPK3 or AtMPK3 and AtMKK1a/2a (Fig. 7A, top panel). In contrast, cotransfection with AtMPK3 and AtMKK4a/5a notably reduced PtiZFP1 protein level (Fig. 7A, top panel). PtiZFP1 accumulation was also reduced when expressed with AtMPK6 and AtMKK4a/5a, whereas little or no change was observed when expressed with AtMPK6 and AtMKK1a/2a (Supplemental Fig. S5). As a poplar TF was used in combination with Arabidopsis MAPKs, we performed a coimmunoprecipitation experiment to confirm interaction between heterologous proteins. Western-blot analysis indicates that PtiZFP1 can be detected following immunoprecipitation of HA-tagged AtMPK3 (Supplemental Fig. S6A) or AtMPK6 (Supplemental Fig. S6B), confirming physical interaction of coexpressed proteins in protoplasts. We also monitored the effect of MAPK activation on AtZAT11 and AtZAT18, the closest homologs of PtiZFP1 in Arabidopsis (Supplemental Fig. S3). As shown in Supplemental Figure S7, accumulation of both TFs was compromised when coexpressed with AtMPK3 and AtMKK4a or AtMPK6 and AtMKK4a. Taken together, these results suggest that following the activation of group A MAPKs, similar regulation is exerted on homologous zinc finger proteins from various plant species.
Figure 7.
AtMPK3 and its upstream MAP2Ks promote PtiZFP1 degradation through the 26S proteasome. A, Western-blot analysis depicting PtiZFP1 protein level following coexpression with various protein kinases. Samples were harvested 5.5 h after transfection. The experiment depicted in the top panel (α FLAG [PtiZFP1]) was conducted without CHX. CHX was added 1.5 h before sample collection in all other experiments. Expression of AtMPK3 and MAP2Ks is also shown. Ponceau staining confirms equal loading among various protein samples. B, MAPK-promoted degradation of PtiZFP1 through the 26S proteasome. MG132 was used to inhibit 26S proteasome activity and was added 1.5 h before sample collection. Dimethyl sulfoxide was used as a control. C, Thr-157 partially controls PtiZFP1 protein fate.
Following the overexpression and activation of group A MAPKs in protoplasts, compromised accumulation of PtiZFP1 could simply be associated with a general inhibition of translation or with a global increase in cellular proteolytic activity. To rule out these possibilities, the assay was repeated with unrelated TFs AtWRKY22 and AtERF99. In both cases, TF protein levels were not reduced when expressed in combination with AtMPK3 and AtMKK4a/5a (Supplemental Fig. S8), suggesting that MAPK signaling components did not affect overall rates of protein synthesis and turnover. Moreover, PtiZFP1 accumulation was unaltered when expressed with AtMPK4 or AtMPK4 and AtMKK1a/2a (Supplemental Fig. S9), which are MAPK signaling components also known for their involvement in stress responses (Gao et al., 2008; Qiu et al., 2008). Overall, these experiments suggest that PtiZFP1 protein level is modulated by specific MAPKs, which previously need to be activated by relevant upstream MAP2Ks.
In the protoplast assay, transfected constructs continuously produce proteins, which could overrun the downstream cellular machinery regulating PtiZFP1 accumulation. This perhaps explains why a significant quantity of intact PtiZFP1 proteins is still detected following coexpression with relevant MAPKs and MAP2Ks. To address this potential issue, constructs were expressed for 4 h before protein synthesis was inhibited by addition of the translation inhibitor cycloheximide (CHX). Samples were collected 90 min later, and PtiZFP1 protein level was again evaluated. In the presence of CHX, accumulation of PtiZFP1 was clearly impaired when coexpressed with AtMPK3 and AtMKK4a/5a (Fig. 7A, bottom panels). This confirms previous observations made without CHX and reinforces the idea of group A MAPKs having a posttranslational effect on PtiZFP1 protein accumulation.
MAPKs Promote PtiZFP1 Degradation through the 26S Proteasome
Lack of PtiZFP1 protein accumulation following coexpression with specific MAPKs suggested that the EAR repressor is targeted for proteolytic cleavage. To confirm this hypothesis and obtain insights into downstream players, the 26S proteasome machinery was inhibited with MG132. Protoplasts were transfected with PtiZFP1 alone or in combination with AtMPK3 and AtMKK4a/5a in the presence of CHX and of either MG132 or dimethyl sulfoxide as a control. When expressed alone, PtiZFP1 accumulated to a much greater extent when protoplasts were treated with MG132 (Fig. 7B), which suggests a role for the 26S proteasome in PtiZFP1 basal protein turnover. Moreover, MAPK-promoted degradation of PtiZFP1 was completely compromised in the presence of MG132 (Fig. 7B), demonstrating that upon activation by appropriate MAP2Ks, group A MAPKs promote PtiZFP1 degradation via the 26S proteasome.
Thr-157 Partially Controls PtiZFP1 Protein Fate
We next wanted to assess the involvement of phosphorylation in the promotion of PtiZFP1 degradation. This TF contains three SP or TP sites (Ser-42, Ser-85, and Thr-157; see asterisks in Supplemental Fig. S2), which are the usual specificity determinants for phosphorylation by Pro-directed Ser/Thr kinases (Pearson et al., 2001). As only Thr-157 is strictly conserved among closely related homologs (Fig. 1B), we mutated this residue to a nonphosphorylable Ala (PtiZFP1T157A; Fig. 5A). Wild-type or mutant versions of PtiZFP1 were then transfected into protoplasts in the presence or absence of MAPK signaling components. When expressed alone, PtiZFP1T157A accumulated to a greater extent than the wild-type protein (Fig. 7C), showing that Thr-157 mutation slows down basal protein turnover. Furthermore, PtiZFP1T157A was more stable than PtiZFP1 following coexpression with AtMPK3 and AtMKK4a/5a (Fig. 7C). This suggests that Thr-157 is phosphorylated by MAPKs and that this posttranslational modification participates in the promotion of PtiZFP1 degradation. The stabilization effect of the T157A mutation, however, remains limited, as PtiZFP1T157A level still decreases upon coexpression with particular protein kinases (e.g. AtMPK3/AtMKK4a).
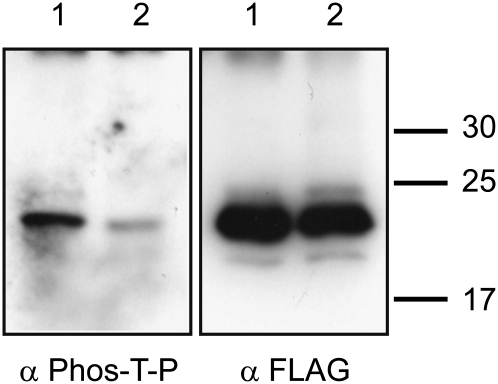
To confirm the phosphorylation of PtiZFP1 and mutant PtiZFP1T157A by group A MAPKs, we created recombinant proteins with the aim of performing in vitro kinase assays. PtiMPK3-1 and PtiMKK5a were successfully expressed and purified from Escherichia coli. On the other hand, both PtiZFP1 and PtiZFP1T157A were completely insoluble when expressed in bacteria, abrogating enzymatic assays with native proteins. PtiZFP1 and PtiZFP1T157A were thus produced in baculovirus expression systems, but purified protein extracts likely contained a contaminant kinase from insect cells, which masked the visualization of recombinant MAPK enzymatic activity during our assays. Using an antibody specific to phosphorylated Thr-Pro residues (also known to cross-react with phosphorylated Ser-Pro residues), we show that PtiZFP1 and to a much lesser extent the mutant PtiZFP1T157A are both phosphorylated proteins (Fig. 8), thus suggesting that PtiZFP1 has multiple phosphorylation sites including Thr-157. We have also verified by directed Y2H assay that the T157A mutation does not impair interaction with PtiMPK3-1 (Supplemental Fig. S10). Based on the canonical nature of the MDS and MAPK substrate specificity determinants (Sharrocks et al., 2000; Bardwell, 2006), a protein is likely to be phosphorylated, albeit with varying degrees of affinity, by kinases from different species. Although we were unable to directly demonstrate the phosphorylation of recombinant PtiZFP1 by group A MAPKs, these experiments suggest the functionality of PtiZFP1 MDS and the accessibility to kinase(s) of the closely positioned phosphorylation site.
Figure 8.
PtiZFP1 is a phosphorylated protein. Recombinant proteins FLAG-PtiZFP1 (lane 1) and FLAG-PtiZFP1T157A (lane 2) were separated by SDS-PAGE and immunodetected with anti-phospho-Thr-Pro antibody (α Phos-T-P). Protein loads were monitored by anti-FLAG antibody (α FLAG).
DISCUSSION
It is well established that plant MAPK cascades play key functions in defense responses (Pitzschke et al., 2009). MAPK signaling is mediated at the transcriptional, translational, and posttranslational levels (Beckers et al., 2009) and was for instance shown to promote the synthesis of camalexin, an antimicrobial compound (Ren et al., 2008). MAPKs also participate in signal transduction leading to transcriptional reprogramming of defense-related genes (Fiil et al., 2009). MAPKs are rapidly and transiently activated in poplar in response to various stress conditions (Hamel et al., 2005). In poplar-rust interactions, enhanced expression of PtiMPK3-1 as well as sustained activation of 44- and 47-kD MAPKs illustrate the crucial role of MAPK signaling pathways in resistance to pathogen attacks (Boyle et al., 2010). Yet, downstream targets of poplar MAPKs remain unidentified, a situation that precludes a connection between signaling cascade and defense-related outputs. To discover novel substrates and obtain insights into MAPK functions during biotic stress responses, we identified MAPK-interacting proteins via Y2H screening. The use of PtiMPK3-1 as bait allowed us to identify several potential interacting partners, including the novel TF PtiZFP1. Consistent with interactions observed in yeast, PtiZFP1 and PtiMPK3-1/6-2 are directed to the plant cell nucleus, and in planta interaction was confirmed by BiFC. PtiZFP1 contains two zinc finger DNA-binding domains and an EAR transcriptional repression motif. This TF belongs to the Cys-2/His-2-type zinc finger protein family that was previously reported to regulate defense responses against biotic and abiotic challenges (Ciftci-Yilmaz and Mittler, 2008). A Cys-2/His-2-type zinc finger protein was previously identified in a poplar hybrid (Populus tremula × Populus alba; Martin et al., 2009). This gene, named PtaZFP2, was shown to be up-regulated in response to mechanical stress and wounding as well as cold and salt treatments (Martin et al., 2009). Here, RTqPCR analysis shows that PtiZFP1 transcripts increase in poplar cells elicited with JA or SA as well as in poplar hybrid NM6 leaves infected with Mmd rust (partial resistance interaction). These induction profiles are consistent with our hypothesis that PtiZFP1 is an important component of the gene response associated with stress conditions.
Mutations within the EAR motif render PtiZFP1 incapable of interacting with PtiMPK3-1 and PtiMPK6-2. Using directed Y2H assays, we were able to identify two conserved Leu residues that are located within the EAR motif core sequence and that are critical for MAPK binding. These Leu residues, however, are insufficient by themselves, as the interaction with MAPKs also requires Lys and Arg residues located before the EAR motif. When combined, these amino acids create a functional bipartite MDS that partially overlaps with the transcriptional EAR repression motif. To our knowledge, the clear functionality of MDS had not been assessed in plants. However, mammalian MAPKs use their common docking (CD) domain to interact with MDS of various proteins, including activating MAP2Ks, inactivating protein phosphatases, and downstream substrates (Tanoue et al., 2000). Consensus sequences of the CD domain (Supplemental Fig. S4) comprise adjacent acidic residues (D and E), which interact with basic amino acids of the MDS (K and R) as well as bulky hydrophobic amino acids (L, F, W, H, and Y) that bind their MDS counterpart (LXL motif; Tanoue et al., 2000). In plants, group A and B MAPKs contain evolutionarily conserved CD domains similar to the ones found in mammalian JNK-, p38-, and ERK-type MAPKs (MAPK Group, 2002; Tanoue and Nishida, 2002). In contrast, group C MAPKs possess a modified CD domain, whereas evolutionarily divergent group D MAPKs have no recognizable CD domain (MAPK Group, 2002). Considering these evolutionary traits, PtiZFP1, which displays a canonical MDS, is more likely to interact with group A and B MAPKs. Our data confirm MDS-dependent binding of PtiZFP1 to group A MAPKs PtiMPK3-1 and PtiMPK6-2. This provides preliminary evidence of the molecular recognition between MDS and CD domains in plants.
The results presented here also show that group A MAPKs promote PtiZFP1 degradation through the 26S proteasome. Interestingly, coexpression with MAPKs alone does not dramatically affect PtiZFP1 stability, as accelerated degradation also requires the expression of constitutively active MAP2Ks. This suggests that upstream signaling is necessary for increased PtiZFP1 breakdown. Our results also suggest that phosphorylation of Thr-157 is involved in controlling PtiZFP1 protein fate. Antibody raised against phosphorylated Thr-Pro residues confirmed that Thr-157 is a phosphorylation site, and mutation of this residue to a nonphosphorylable Ala allows greater accumulation of the mutant protein following expression under basal conditions in protoplasts. PtiZFP1T157A was also more stable than its wild-type counterpart when coexpressed with activated MAPKs. Although it was not possible to directly demonstrate the phosphorylation of PtiZFP1 by MPK3- or MPK6-type MAPKs, these data, in combination with previous demonstrations of the MAPK-PtiZFP1 interaction, are consistent with the hypothesis that MAPKs phosphorylate PtiZFP1, leading to its enhanced degradation. The fact that the T157A mutation only results in partial stabilization of PtiZFP1, however, suggests that protection against proteolytic cleavage is limited and that additional mechanisms and/or phosphorylation sites may be involved in the control of PtiZFP1 turnover. Another possibility that we cannot rule out is that PtiZFP1 may be constitutively phosphorylated by an unknown kinase and that protein interaction with activated MAPKs from group A is sufficient to trigger accelerated degradation of this TF. It has been proposed that AtERF7, an EAR repressor, is phosphorylated in an abscisic acid-dependent manner by the protein kinase PKS3 (Song et al., 2005). The authors, however, have not addressed the question of the impact of this posttranslational modification on AtERF7 stability. Five phosphorylated EAR repressors have also been identified in an Arabidopsis phosphoproteome database, although the protein kinases involved in the EAR repressor phosphorylation and the impact of the phosphorylation itself on protein function have not been characterized (Heazlewood et al., 2008, in Kagale et al., 2010).
Phosphorylation has been reported, in some cases, to increase protein stability (Liu and Zhang, 2004) and, in others, to promote protein degradation. In fact, an increasing number of studies support the idea that phosphorylation may target plant proteins for degradation via the proteasome pathway (Vierstra, 2009). For example, in phytochrome signaling, which controls plant photomorphogenesis, different light conditions induce the phosphorylation of bHLH phytochrome-interacting factors prior to their proteasome-mediated degradation (Al-Sady et al., 2006; Castillon et al., 2007; Lorrain et al., 2008). In ET signaling, phosphorylation on two sites of the Arabidopsis Ethylene Insensitive3 (AtEIN3) TF has opposite functions. Without ET, phosphorylation of Thr-592 by an unknown kinase promotes degradation of the TF, while ET stabilizes AtEIN3 through MAPK-mediated phosphorylation of Thr-174 (Yoo et al., 2008). Recently, the transcriptional coactivator AtNPR1 (for Arabidopsis nonexpressor of pathogenesis related 1), which is essential for SA-mediated plant immunity, was also found to be phosphorylated following SA elicitation and subsequently degraded via the proteasome pathway (Spoel et al., 2009). The case of PtiZFP1 is thus a novel example where protein kinases control the fate of downstream substrates.
An increasing body of evidence shows that proteolytic cleavage of transcriptional repressors is a general mechanism used by plants to enhance gene induction (Huq, 2006; Vierstra, 2009). This mechanism is emerging, for instance, as a common theme of plant hormone signaling (Santner and Estelle, 2009). In auxin, GA, and jasmonate signaling, for example, expression of hormone-responsive genes depends on the proteasome-mediated degradation of specific transcriptional repressors; AUX/IAA for auxin, DELLA for GA, and JAZs for jasmonate (Gray et al., 2001; Dill et al., 2004; Chini et al., 2007; Thines et al., 2007). In general, by binding and inhibiting specific transcriptional activators, these repressors maintain a low level of responsive gene expression (Kim et al., 1997). Increasing the concentration of hormones leads to ubiquitin-proteasome-mediated degradation of the repressors and thus to derepression of target genes. Several of the transcriptional repressors are themselves up-regulated in response to hormone signaling, ensuring a negative feedback to constrain the response.
Interestingly, the EAR motif is required for the repression activity of many plant transcriptional regulators (Ohta et al., 2001). Genome-wide analysis suggests that the EAR motif is conserved across all plant species and is involved in developmental and stress-related processes (Kagale et al., 2010). The exact nature of the molecular mechanism enabling this transcriptional repression activity remains unclear, but recent reports suggest that the EAR motif is engaged in protein-protein interaction. For example, AtIAA12/BDL, a member of the EAR transcriptional repressors AUX/IAA, was shown to utilize its EAR motif to interact with the corepressor protein TPL (Szemenyei et al., 2008). TPL is thought to be involved in chromatin remodeling (Long et al., 2006), and AtIAA12/BDL repression activity depends on TPL complex formation. The authors thus hypothesized that binding of corepressors such as TPL could explain the EAR-dependent repression mechanism (Szemenyei et al., 2008). In the JA signaling pathway, transcriptional repression is mediated by JAZ repressor proteins that associate with both bHLH transcriptional activators MYC and an EAR protein named NINJA (Pauwels et al., 2010). The authors have further demonstrated that NINJA recruits a TPL corepressor to the complex, again supporting the hypothesis of an EAR-dependent association with a corepressor (Pauwels et al., 2010). Our data demonstrate that the C-terminal region of PtiZFP1 is responsible for MAPK interaction; the EAR motif is located within this region, again stressing its importance in intermolecular binding. As described in the previous examples, PtiZFP1 could also use its EAR motif to interact with other signaling components, so it can repress the expression of target genes. Following binding and most likely phosphorylation, MAPKs may create steric hindrance and/or conformational changes that interfere with repressor complex formation, affecting PtiZFP1 function and stability.
The crucial role played by MAPKs in PtiZFP1 stability and the overlap between PtiZFP1 MDS and its EAR motif prompted us to extend our search for conserved features of MDS among other EAR repressors. Interestingly, most defense-related Cys-2/His-2-type zinc finger proteins as well as all class II AP2/ERFs of the B1a subfamily (McGrath et al., 2005) contain basic amino acid enrichment followed by the EAR motif core sequence (Fig. 9). The presence of a MDS at the C terminus is thus conserved among various EAR repressors belonging to two TF families. The functionality of these putative docking motifs will need to be empirically assayed, but considering our data for PtiZFP1, these sites could also act as genuine MAPK-binding surfaces. Degradation of transcriptional repressors is an efficient means for the release of target gene repression and could thus be a common regulatory mechanism for other EAR repressors. MAPK regulation could serve as a molecular cue for targeted degradation and link the activation of MAPK signal transduction to an effective gene up-regulation. The facts that many EAR repressors possess putative MDS and that MAPKs form a multigene family suggest the existence of a regulatory network between these key signaling components. Further studies on the relations existing between stress-responsive MAPKs and other EAR repressors will surely help to better define the mechanisms involved in the derepression of target genes during the establishment of plant defense responses.
Figure 9.
Conservation of MDS in EAR repressors. Protein sequence alignments of the C-terminal region of various EAR repressors. The consensus sequence of the EAR motif is highlighted in gray, and characteristics reminiscent of MDS are shown in boldface (basic amino acids) and underlined (bulky hydrophobic residues or the LXL motif). Species acronyms are as follows: At, Arabidopsis thaliana; Ph, Petunia hybrida; Nb, Nicotiana benthamiana; Nt, Nicotiana tabacum; Os, Oryza sativa; Pti, Populus trichocarpa.
MATERIALS AND METHODS
Plant Material and Fungal Inoculation Procedure
In vitro-grown plantlets of hybrid poplar NM6 (Populus nigra × Populus maximowiczii) were transferred to Promix soil. Plants were allowed to develop under controlled greenhouse conditions (22°C/19°C day/night, 16-h day) for 2 weeks. Thereafter, young trees were placed in a growth chamber (Conviron Environmental Growth Chambers), where conditions were as follows: 26°C/22°C day/night, 16-h day, 60% relative humidity, light intensity of 100 μmol m−2 s−1. Plants were grown until plastochron index 16 stage was reached (Larson and Isebrands, 1971) and fertilized once every 2 weeks alternatively with 10/52/10 (0.5 g L−1) plus 15.5/0/0 (calcium, 19%; 0.5 g L−1) or with 20/20/20 (1 g L−1). Poplar rust infections were conducted as described previously using 6-cm2 leaf sections (Boyle et al., 2010). Nicotiana benthamiana seeds were spread on Agro-Mix soil and placed in a growth chamber, where conditions were as follows: 22°C/22°C day/night, 16-h day, 100% relative humidity, light intensity of 100 μmol m−2 s−1. Once germinated, relative humidity was lowered to 60% and young plants were kept growing for 5 weeks with fertilization every fourth day: 20/20/20 (1 g L−1). Arabidopsis (Arabidopsis thaliana) Columbia ecotype seeds were spread on Agro-Mix soil and placed in a growth chamber, where conditions were as follows: 22°C/18°C day/night, 13-h day, 100% relative humidity, light intensity of 75 μmol m−2 s−1. After 1 week, humidity was lowered to 60%, and seedlings were grown for 3 weeks in the same conditions.
Hormonal Treatment of Poplar Cell Suspension
Poplar cell suspension H11-11 (hybrid Populus trichocarpa × Populus deltoides) was treated with MeJA (100 μm), SA (100 μm), or left untreated. Cells were harvested by filtration after 10, 20, 45, 90, 180, and 360 min, frozen in liquid nitrogen, and RNA was extracted with the Plant Mini-RNA extraction kit (Qiagen) followed by a DNase treatment. RNA quality was verified by an Agilent 2100 Bioanalyzer (Agilent Technologies), and cDNA was reverse transcribed using SuperScript II (Invitrogen) prior to RTqPCR analysis.
RTqPCR Analysis
Poplar gene expression analysis by RTqPCR was conducted using the Stratagene Mx3000P system (Agilent). Each PCR had a 10 μL volume and contained 0.6 μm forward and reverse primers, 5 ng of cDNA as template, and 1× master mix solution (QuantiTect SYBR Green mix; Qiagen). The sequence of each primer used for RTqPCR analysis is given in Supplemental Table S3. Using the SYBR Green amplification mode from Stratagene’s software, PCR cycling conditions were as follows: 15 min of incubation at 95°C, followed by 40 amplification cycles (94°C for 15 s, 64°C for 30 s, and 65°C for 90 s). Reactions containing no DNA template were conducted as a negative control. Fluorescence readings were taken at the end of each cycle, and the specificity of amplification as well as the absence of primer dimers were confirmed with a melting curve analysis at the end of each reaction. Fluorescence and cycle threshold (Ct) values were exported and analyzed in Microsoft Excel. The relative number of transcripts (1/2Ct) was then averaged for technical RTqPCR duplicates and used for subsequent normalization.
To correct for technical variations in RNA extraction, total RNA quantification, reverse transcription, and RTqPCR (e.g. inhibitors of PCR) as well as biological variation, expression data were normalized against the geometric mean of four reference genes, Eukaryotic Translation Initiation Factor4A-2, Actin, Cell Division Control Protein2, and Ubiquitin (for primer sequence, see Supplemental Table S3). Stability of reference genes was evaluated using geNORM VBA applet for Microsoft Excel (http://medgen.ugent.be/~jvdesomp/genorm; Vandesompele et al., 2002). For cell suspension experiments, fold change is expressed as treatment relative to the control calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001; Bustin et al., 2009) For the poplar leaf-Mmd experiment, fold change was calculated relative to time zero. sd related to the biological variation within one line was calculated in accordance with error propagation rules.
Preparation of Protein Extracts and Western Blots
Protein extraction and western-blot analysis were conducted as described previously (Hamel et al., 2005). Antibody dilutions were as follows: anti-FLAG-horseradish peroxidase (HRP; 1:2,000), anti-HA-HRP (1:2,000), anti-cMYC (1:1,500), anti-GFP (1:5,000), and mouse or rabbit HRP-conjugated secondary antibody (1:10,000 or 1:20,000).
Recombinant Protein Expression and Purification
PtiMKK5a was cloned in frame with GST in pGEX-6P-3 expression vector (GE Healthcare), expressed in Escherichia coli, and purified with reduced glutathione Sepharose 4B (GE Healthcare) as specified by the supplier. PtiMPK3-1 was cloned in the pET29b expression vector, expressed in E. coli, and purified by nickel chromatography using His-Bind Resin (Novagen). PtiZFP1 and PtiZFP1T157A were purified from a baculovirus expression system (Molecular Throughput).
Site-Directed Mutagenesis
Site-directed mutagenesis experiments were conducted using the QuikChange Site-Directed Mutagenesis kit (Stratagene) following the manufacturer’s instructions. The sequences of primers used for mutagenesis experiments are given in Supplemental Table S3.
Y2H Experiments
Y2H experiments were performed using the MATCHMAKER Gal4 Two-Hybrid System 3 (BD Biosciences/Clontech). PtiMPK3-1 and PtiMPK6-2 were cloned in the pGBKT7 vector. Constructs were transformed in yeast strain AH109 and tested for autoactivation of the reporter genes. Both MAPKs were found to weakly activate reporter gene His3 (data not shown). To prevent false-positive detection, various concentrations of 3-amino-1,2,4-triazole were tested. Following optimization, Y2H media were supplemented with appropriate inhibitor concentrations (Supplemental Figs. S1, S3B, S5B, and S10).
To discover new interacting partners, a cDNA library was produced using BD MATCHMAKER Library Construction and Screening Kits (BD Biosciences/Clontech). Leaves from hybrid poplar NM6 were infected with Mmd, and tissues were collected at 5 and 10 d post infection. Harvested tissues were pooled, ground into powder using liquid nitrogen, and frozen at −80°C until use. Total RNA was extracted as described elsewhere (Chang et al., 1993). From 1 mg of total RNA, the mRNA fraction was isolated using the PolyATtract mRNA Isolation System (Promega). From this enrichment, 350 ng of mRNA was used as a template for PCR. Screening was accomplished by mating yeast strains Y187 (containing PtiMPK3-1) and AH109 (containing clones from the library). A total of 2.4 × 107 clones were screened for interaction, and the identification of interacting candidates was done by scoring prototrophy for His and adenine on selection medium (W−/L−/H−/A−/X-α-GAL). Positive colonies were liquid cultured, and plasmids were isolated using the ChargeSwitch Plasmid Yeast Mini Kit (Invitrogen). PCR specifically amplified DNA inserts of the prey vectors. Amplicons were purified and directly sequenced using the dideoxy nucleotide termination method with an ABI 373 Stretch XL sequencer (Applied Biosystems). Other experiments using MAPKs and truncated/mutated versions of PtiZFP1 were conducted by cotransforming yeast strain AH109. For all Y2H experiments, petri dishes containing low-stringency selection medium (W−/L−) were used to assess vector transformation and yeast cell viability.
Subcellular Localization and BiFC
MAPKs and PtiZFP1 were PCR amplified to integrate Gateway recombination sites. Using the Gateway Technology with Clonase II system (Invitrogen), entry clones were generated using vector pDONR221. For subcellular localization, MAPKs and PtiZFP1 were recombined in binary vectors pH7YWG2,0 (eYFP) and pH7CWG2,0 (eCFP) respectively (Karimi et al., 2005). For BiFC, MAPKs and PtiZFP1 were recombined in Gateway-compatible vector pE3136 (pSAT4-DEST-nEYFP-C1; amino acids 1–174 of eYFP:nYFP) and pE3130 (pSAT5-DEST-cEYFP-C1; amino acids 175–end of eYFP:cYFP), respectively. Expression cassettes were linearized and cloned in the binary vector pCambia 2300 (CAMBIA).
For transient protein expression in N. benthamiana, binary vectors were transformed by heat shock in Agrobacterium tumefaciens (AGL1 strain). Freshly transformed colonies were liquid cultured at 28°C in yeast extract peptone medium supplemented with appropriate antibiotics and 50 μm acetosyringone. Culture medium was removed after centrifugation, and cells were resuspended in Murashige and Skoog plant growth medium (Murashige and Skoog, 1962) supplemented with 10 mm MgCl2 and 100 μm acetosyringone. Bacteria were infiltrated in leaves from 5-week-old N. benthamiana plants along with Agrobacterium cells carrying the P19 silencing suppressors (Voinnet et al., 2003). Microscopic observations of infiltrated plants were done after 3 d. To validate the specificity of BiFC experiments, interaction between PtiZFP1 and PNek1 was tested. PNek1 is a poplar protein kinase that localizes to the plant cell cytoplasm and nucleus (Cloutier et al., 2005). For this pair of fusion proteins, no fluorescence signal could be detected (data not shown).
Fluorescence Microscopy
Sections from infiltrated N. benthamiana leaves were cut out using a clean scalpel and placed upside down on a microscope slide. Microscopy images were obtained using an IX81 confocal scanning microscope accompanied by the FV1000 package (Olympus). eCFP was excited using a 458-nm argon laser, and emission was measured from 465 to 505 nm. eYFP was excited using a 514-nm wavelength, and emission was measured from 525 to 600 nm. Selected scan speed was 40 μs per pixel, and a 512 × 512 resolution was used. Acquired images were processed using version 1.7a of the FV10 ASW software (Olympus).
Mesophyll Protoplast Transient Expression Assays
Arabidopsis protoplasts were isolated and transfected by a polyethylene glycol method as described previously (Yoo et al., 2007). Briefly, 0.1 mL of protoplast suspension (200,000 cells mL−1) was transfected with a mixture of 20 μg of DNA. All samples contained 8 μg of PtiZFP1 DNA along with 12 μg of unspecific DNA, 6 μg of MAPK DNA, and 6 μg of unspecific DNA or 6 μg of MAPK DNA and 6 μg of MAP2K DNA. Transfected protoplasts were incubated at 25°C for 5.5 h before harvesting. For inhibitor analysis, expression was performed for 4 h before the addition of 10 μm CHX. Inhibition of the 26S proteasome was conducted using 10 μm MG132. Protoplast incubation was then continued for 1.5 h before protoplasts were collected by brief centrifugation. Samples were frozen on dried ice and conserved at −80°C until use. Cells were disrupted using Laemmli loading buffer and heating at 95°C for a few minutes. Extracted proteins were separated by SDS-PAGE and analyzed by western blotting.
Gene Model Numbers and Nomenclature
For identification purposes and despite the use of hybrids NM6 and H11-11, the “Pti” acronym (Kumar et al., 2009) was attributed to each studied gene following mining of the P. trichocarpa genome (http://www.phytozome.net/poplar). Reported gene model numbers correspond to version 2.2 of the genome assembly (Supplemental Table S4). Sequences from hybrid NM6 (P. nigra × P. maximowiczii) may display polymorphism and should thus be preceded by the “Pnm” acronym. Following sequencing and mining of ESTs databases, polymorphisms were considered for optimal primer design in RTqPCR.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number JN700523.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Interaction of two MAPKs with a truncated TF.
Supplemental Figure S2. The PtiZFP1 gene and corresponding protein.
Supplemental Figure S3. Phylogenetic relationships between C1-2i Cys-2/His-2-type zinc finger proteins.
Supplemental Figure S4. Organization of the fusion proteins used for subcellular localization and BiFC.
Supplemental Figure S5. AtMPK6 and its upstream MAP2Ks have an effect on PtiZFP1 protein accumulation.
Supplemental Figure S6. PtiZFP1 physically interacts with AtMPK3 and AtMPK6.
Supplemental Figure S7. Group A MAPKs compromise the accumulation of Arabidopsis zinc finger proteins.
Supplemental Figure S8. Activation of group A MAPKs does not prevent the accumulation of all TFs.
Supplemental Figure S9. AtMPK4 and its upstream MAP2Ks have no effect on PtiZFP1 protein accumulation.
Supplemental Figure S10. Interaction of PtiMPK3-1 and PtiZFP1 or PtiZFP1T157A.
Supplemental Table S1. RTqPCR data following hormonal treatment of H11-11 suspension cells.
Supplemental Table S2. RTqPCR data following pathogen infection of NM6 leaves.
Supplemental Table S3. Sequences of primers used in this study.
Supplemental Table S4. Gene model numbers of the studied genes.
Acknowledgments
We are obliged to Dr. Lan-Ying Lee and Dr. Stanton Gelvin (Department of Biological Sciences, Purdue University) for their sharing of the BiFC vectors. We also thank Dr. Kamal Bouarab (Département de Biologie, Université de Sherbrooke) for the P19 binary vector. Finally, we express our gratitude to Carl St-Pierre, Dr. Anne Loranger, and Dr. Normand Marceau (Centre de Recherche de l’Hôtel-Dieu de Québec), who provided valuable technical assistance for microscopy experiments. We also thank Pamela Cheers (Laurentian Forestry Centre) for editorial work.
References
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH. (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Azaiez A, Boyle B, Levée V, Séguin A. (2009) Transcriptome profiling in hybrid poplar following interactions with Melampsora rust fungi. Mol Plant Microbe Interact 22: 190–200 [DOI] [PubMed] [Google Scholar]
- Bardwell L. (2006) Mechanisms of MAPK signalling specificity. Biochem Soc Trans 34: 837–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Jones JD. (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Beckers GJM, Jaskiewicz M, Liu Y, Underwood WR, He SY, Zhang S, Conrath U. (2009) Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21: 944–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G, Unthan T, Uhrig JF, Pöschl Y, Gust AA, Scheel D, Lee J. (2009) Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc Natl Acad Sci USA 106: 8067–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle B, Levée V, Hamel LP, Nicole MC, Séguin A. (2010) Molecular and histochemical characterisation of two distinct poplar Melampsora leaf rust pathosystems. Plant Biol (Stuttg) 12: 364–376 [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622 [DOI] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E. (2007) Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci 12: 514–521 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Choo Y, Klug A. (1997) Physical basis of a protein-DNA recognition code. Curr Opin Struct Biol 7: 117–125 [DOI] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S, Mittler R. (2008) The zinc finger network of plants. Cell Mol Life Sci 65: 1150–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier M, Vigneault F, Lachance D, Séguin A. (2005) Characterization of a poplar NIMA-related kinase PNek1 and its potential role in meristematic activity. FEBS Lett 579: 4659–4665 [DOI] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP. (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H. (2007) Trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signaling. Science 318: 453–456 [DOI] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Duplessis S, Major I, Martin F, Séguin A. (2009) Poplar and pathogen interactions: insights from Populus genome-wide analyses of resistance and defense gene families and gene expression profiling. Crit Rev Plant Sci 28: 309–334 [Google Scholar]
- Englbrecht CC, Schoof H, Böhm S. (2004) Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T. (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci 10: 71–78 [DOI] [PubMed] [Google Scholar]
- Fiil BK, Petersen K, Petersen M, Mundy J. (2009) Gene regulation by MAP kinase cascades. Curr Opin Plant Biol 12: 615–621 [DOI] [PubMed] [Google Scholar]
- Gao M, Liu J, Bi D, Zhang Z, Cheng F, Chen S, Zhang Y. (2008) MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res 18: 1190–1198 [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Hamel LP, Miles GP, Samuel MA, Ellis BE, Séguin A, Beaudoin N. (2005) Activation of stress-responsive mitogen-activated protein kinase pathways in hybrid poplar (Populus trichocarpa x Populus deltoides). Tree Physiol 25: 277–288 [DOI] [PubMed] [Google Scholar]
- Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, Ehlting J, Beaudoin N, Barbazuk B, Klessig D, Lee J, et al. (2006) Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci 11: 192–198 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Durek P, Hummel J, Selbig J, Weckwerth W, Walther D, Schulze WX. (2008) PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res 36: D1015–D1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E. (2006) Degradation of negative regulators: a common theme in hormone and light signaling networks? Trends Plant Sci 11: 4–7 [DOI] [PubMed] [Google Scholar]
- Kagale S, Links MG, Rozwadowski K. (2010) Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol 152: 1109–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Rozwadowski K. (2010) Small yet effective: the ethylene responsive element binding factor-associated amphiphilic repression (EAR) motif. Plant Signal Behav 5: 691–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P. (2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Kazan K. (2006) Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci 11: 109–112 [DOI] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A. (1997) Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA 94: 11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo K, Sakamoto A, Kobayashi A, Rybka Z, Kanno Y, Nakagawa H, Takatsuji H. (1998) Cys2/His2 zinc-finger protein family of petunia: evolution and general mechanism of target-sequence recognition. Nucleic Acids Res 26: 608–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Thammannagowda S, Bulone V, Chiang V, Han K-H, Joshi CP, Mansfield SD, Mellerowicz E, Sundberg B, Teeri T, et al. (2009) An update on the nomenclature for the cellulose synthase genes in Populus. Trends Plant Sci 14: 248–254 [DOI] [PubMed] [Google Scholar]
- Lampard GR, Macalister CA, Bergmann DC. (2008) Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Larson PR, Isebrands JG. (1971) The plastochron index as applied to developmental studies of cottonwood. Can J For Res 1: 1–11 [Google Scholar]
- Li J, Brader G, Palva ET. (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limmongkon A, Giuliani C, Valenta R, Mittermann I, Heberle-Bors E, Wilson C. (2004) MAP kinase phosphorylation of plant profilin. Biochem Biophys Res Commun 324: 382–386 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S. (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM. (2006) TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523 [DOI] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Major IT, Constabel CP. (2008) Functional analysis of the Kunitz trypsin inhibitor family in poplar reveals biochemical diversity and multiplicity in defense against herbivores. Plant Physiol 146: 888–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAPK Group (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Martin L, Leblanc-Fournier N, Azri W, Lenne C, Henry C, Coutand C, Julien J-L. (2009) Characterization and expression analysis under bending and other abiotic factors of PtaZFP2, a poplar gene encoding a Cys2/His2 zinc finger protein. Tree Physiol 29: 125–136 [DOI] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K. (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke FL, Kang HG, Chen Z, Park JM, Kumar D, Klessig DF. (2005) Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR-like cell death in tobacco. Mol Plant Microbe Interact 18: 1027–1034 [DOI] [PubMed] [Google Scholar]
- Mészáros T, Helfer A, Hatzimasoura E, Magyar Z, Serazetdinova L, Rios G, Bardóczy V, Teige M, Koncz C, Peck S, et al. (2006) The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J 48: 485–498 [DOI] [PubMed] [Google Scholar]
- Morse AM, Whetten RW, Dubos C, Campbell MM. (2009) Post-translational modification of an R2R3-MYB transcription factor by a MAP kinase during xylem development. New Phytol 183: 1001–1013 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol 15: 473–497 [Google Scholar]
- Nicole MC, Hamel LP, Morency MJ, Beaudoin N, Ellis BE, Séguin A. (2006) MAP-ping genomic organization and organ-specific expression profiles of poplar MAP kinases and MAP kinase kinases. BMC Genomics 7: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauw B, Hilliou FA, Martin VS, Chatel G, de Wolf CJ, Champion A, Pré M, van Duijn B, Kijne JW, van der Fits L, et al. (2004) Zinc finger proteins act as transcriptional repressors of alkaloid biosynthesis genes in Catharanthus roseus. J Biol Chem 279: 52940–52948 [DOI] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, Chico JM, Bossche RV, Sewell J, Gil E, et al. (2010) NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. (2001) Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev 22: 153–183 [DOI] [PubMed] [Google Scholar]
- Philippe RN, Ralph SG, Mansfield SD, Bohlmann J. (2010) Transcriptome profiles of hybrid poplar (Populus trichocarpa × deltoides) reveal rapid changes in undamaged, systemic sink leaves after simulated feeding by forest tent caterpillar (Malacosoma disstria). New Phytol 188: 787–802 [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Schikora A, Hirt H. (2009) MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Gerstein M, Snyder M, Dinesh-Kumar SP. (2009) MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev 23: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Zhou L, Yun BW, Nielsen HB, Fiil BK, Petersen K, Mackinlay J, Loake GJ, Mundy J, Morris PC. (2008) Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol 148: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph S, Oddy C, Cooper D, Yueh H, Jancsik S, Kolosova N, Philippe RN, Aeschliman D, White R, Huber D, et al. (2006) Genomics of hybrid poplar (Populus trichocarpa x deltoides) interacting with forest tent caterpillars (Malacosoma disstria): normalized and full-length cDNA libraries, expressed sequence tags, and a cDNA microarray for the study of insect-induced defences in poplar. Mol Ecol 15: 1275–1297 [DOI] [PubMed] [Google Scholar]
- Ren D, Liu Y, Yang KY, Han L, Mao G, Glazebrook J, Zhang S. (2008) A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 105: 5638–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi C, Kohler A, Frey P, Duchaussoy F, Ningre N, Couloux A, Wincker P, Le Thiec D, Fluch S, Martin F, et al. (2007) Transcript profiling of poplar leaves upon infection with compatible and incompatible strains of the foliar rust Melampsora larici-populina. Plant Physiol 144: 347–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Estelle M. (2009) Recent advances and emerging trends in plant hormone signalling. Nature 459: 1071–1078 [DOI] [PubMed] [Google Scholar]
- Sharrocks AD, Yang S-H, Galanis A. (2000) Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem Sci 25: 448–453 [DOI] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK. (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17: 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X. (2009) Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Tanoue T, Adachi M, Moriguchi T, Nishida E. (2000) A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol 2: 110–116 [DOI] [PubMed] [Google Scholar]
- Tanoue T, Nishida E. (2002) Docking interactions in the mitogen-activated protein kinase cascades. Pharmacol Ther 93: 193–202 [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034.1–research0034.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wu J, Baldwin IT. (2010) New insights into plant responses to the attack from insect herbivores. Annu Rev Genet 44: 1–24 [DOI] [PubMed] [Google Scholar]
- Yap YK, Kodama Y, Waller F, Chung KM, Ueda H, Nakamura K, Oldsen M, Yoda H, Yamaguchi Y, Sano H. (2005) Activation of a novel transcription factor through phosphorylation by WIPK, a wound-induced mitogen-activated protein kinase in tobacco plants. Plant Physiol 139: 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J. (2008) Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]