Abstract
Many eukaryotes have from one to three heat shock factors (Hsfs), but plants have more than 20 Hsfs, designated class A, B, and C. Class A Hsfs are activators of transcription, but details of the roles of individual Hsfs have not been fully characterized. We show here that Arabidopsis (Arabidopsis thaliana) HsfB1 and HsfB2b, members of class B, are transcriptional repressors and negatively regulate the expression of heat-inducible Hsfs (HsfA2, HsfA7a, HsfB1, and HsfB2b) and several heat shock protein genes. In hsfb1 hsfb2b double mutant plants, the expression of a large number of heat-inducible genes was enhanced in the non-heat condition (23°C) and the plants exhibited slightly higher heat tolerance at 42°C than the wild type, similar to Pro35S:HsfA2 plants. In addition, under extended heat stress conditions, expression of the heat-inducible Hsf genes remained consistently higher in hsfb1 hsfb2b than in the wild type. These data indicate that HsfB1 and HsfB2b suppress the general heat shock response under non-heat-stress conditions and in the attenuating period. On the other hand, HsfB1 and HsfB2b appear to be necessary for the expression of heat stress-inducible heat shock protein genes under heat stress conditions, which is necessary for acquired thermotolerance. We show that the heat stress response is finely regulated by activation and repression activities of Hsfs in Arabidopsis.
Acclimation to heat stress (HS) is critical for sessile plants, and complex systems have evolved that allow plants to respond to and survive HS. Heat shock factors (Hsfs) regulate the expression of genes for heat shock proteins (HSPs), which confer thermotolerance. Many eukaryotes have one, two, or three Hsfs, but plants have more than 20 Hsfs, which are divided into three classes (A, B, and C) based on peculiarities of their flexible linkers and oligomerization domain (HR-A/B; Nover et al., 2001; Baniwal et al., 2004). In Arabidopsis (Arabidopsis thaliana), class A Hsfs, with the exception of HsfA1d and HsfA9, include an AHA (for aromatic and large hydrophobic amino acid residues embedded in an acidic surrounding) motif that is involved in the activation activity of class A Hsfs (Nover et al., 2001), and HsfA1a, HsfA1b, HsfA1d, HsfA1e, and HsfA2 have been reported to act as positive regulators of the response to HS in plants (Busch et al., 2005; Nishizawa et al., 2006; Schramm et al., 2006; Charng et al., 2007; Liu et al., 2011; Nishizawa-Yokoi et al., 2011). HsfA1a, HsfA1b, and HsfA2 are localized in the cytoplasm under normal growth conditions and migrate to the nucleus in response to heat to activate HS-inducible genes (Kotak et al., 2004). Several HSPs have been shown to interfere with the nuclear localization of class A Hsfs by interacting with them (Lee and Schöffl, 1996; Port et al., 2004). Moreover, several plant Hsfs are inducible by HS, a characteristic limited to plant Hsfs (Busch et al., 2005; Schramm et al., 2008). The heat-inducible class A Hsfs, namely HsfA2, HsfA7a, and HsfA3, play an important role in thermotolerance (Nishizawa et al., 2006; Charng et al., 2007; Larkindale and Vierling, 2008; Schramm et al., 2008; Yoshida et al., 2008; Meiri and Breiman, 2009).
Class B Hsfs contain neither a nuclear export signal nor an activation domain, which are found in class A Hsfs (Nover et al., 2001; Kotak et al., 2004). HsfB1, HsfB2a, and HsfB2b are HS-inducible genes (Busch et al., 2005; Schramm et al., 2008). Tomato (Solanum lycopersicum) HsfB proteins were suggested to be coactivators of HsfAs (Bharti et al., 2004; Hahn et al., 2011). HsfB1 and HsfB2b were shown to regulate the expression of defensin genes, and their mutant lines exhibited higher disease resistance than in the wild type (Kumar et al., 2009), suggesting that HsfB1 and HsfB2b may negatively regulate the disease response.
The biological function of class B Hsfs in the HS response, however, remains to be clarified. We reported previously that HsfB1 and HsfB2b have repressive activities in transient expression assays and that the 11 amino acids (GEGLKLFGVWL) at the C terminus of HsfB1 act as a repression domain, designated the B3 repression domain (BRD; Ikeda and Ohme-Takagi, 2009). This sequence is conserved in HsfB1 of Arabidopsis, tomato, and soybean (Glycine max; Czarnecka-Verner et al., 2004). The repressive activity of HsfB1 was reported in tomato, Arabidopsis, and soybean (Czarnecka-Verner et al., 2000, 2004; Ikeda and Ohme-Takagi, 2009), but the functional role of the repressive activity of HsfB1 has not been identified.
In this report, we show that two members of class B Hsfs, HsfB1 and HsfB2b, are active repressors of the transcription of HS-inducible genes for Hsfs and HSPs. Moreover, we demonstrate that these class B heat shock transcriptional repressors are necessary for suppression of the general HS response under non-HS conditions and in the attenuating period and for acquired thermotolerance.
RESULTS
HsfB1 Is a Transcriptional Repressor
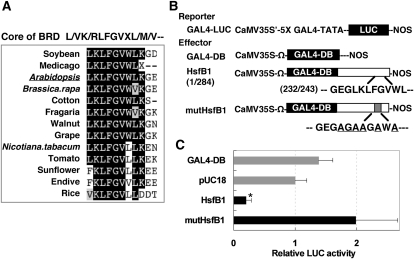
In Arabidopsis leaves, HsfB1 exhibited strong repressive activity in a yeast Gal4 DNA-binding domain (GAL4-DB) fusion transient expression assay (Ikeda and Ohme-Takagi, 2009). The L/VR/KLFGVXM/V/L sequence, which is the core of the BRD, is conserved in the orthologs of HsfB1 in a variety of plants (Fig. 1A; Czarnecka-Verner et al., 2004; Ikeda and Ohme-Takagi, 2009). When the BRD was mutated and six amino acid residues in LKLFGVWL were replaced by Ala and Gly residues (to yield AGAAGAWA in a mutant protein designated mutHsfB1; Fig. 1B), the repressive activity of HsfB1 was abolished in the transient expression assays (Fig. 1C). These results indicated that BRD is necessary for the repressive activity of HsfB1.
Figure 1.
Identification of the repression domain of HsfB1. A, Alignment of the core sequence of BRD of HsfB1 orthologs from 13 varieties of plants including Arabidopsis HsfB1. B, Schematic representation of the constructs used for transient expression analysis. Effector constructs encoded the gene for HsfB1 (white box) or for a mutant form of HsfB1 (white box with gray insert) fused to the yeast GAL4-DB (black box), as indicated. C, Relative LUC activities after cobombardment of Arabidopsis leaves with the effector and reporter constructs. The relative activity due to the pUC18 control vector was taken as 1. The asterisk indicates a P value below 0.05 between the pUC18 control and others.
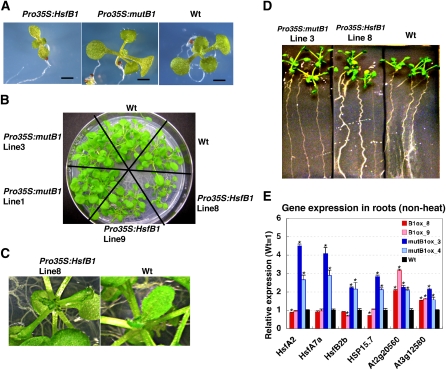
The transgenic seedlings that ectopically expressed HsfB1 (Pro35S:HsfB1) were much smaller than those of Pro35S:mutHsfB1 and wild-type plants, and Pro35S:HsfB1 plants had tiny rosettes with small, crinkly leaves and crooked roots (Fig. 2, C and D; Supplemental Fig. S1B). These morphological data suggest that repressive activity has an effect on the biological functions of HsfB1. It has been shown that the defective mutants for thermotolerance exhibit a short-hypocotyl phenotype (Hong and Vierling, 2000), and a quadruple mutant of HsfA1s, which is defective in thermotolerance, exhibits a tiny phenotype under normal growth conditions (Kwon et al., 2007; Liu et al., 2011). The tiny phenotype of Pro35S:HsfB1 plants might suggest the involvement of HsfB1 in the regulation of genes related to thermotolerance.
Figure 2.
Involvement of the repression domain for the function of HsfB1. A and B, Phenotypes of 12-d-old seedlings (A) and 24-d-old rosettes (B) of Pro35S:HsfB1, Pro35S:mutHsfB1, and wild-type (Wt) plants grown on a agar plate at 23°C (non-heat condition). C, Magnified images from B showing small and crinkly leaves of Pro35S:HsfB1 plants. D, Phenotypes of roots of Pro35S:HsfB1, Pro35S:mutHsfB1, and wild-type plants grown on a vertical agar plate at 23°C for 18 d. E, Quantitative analysis by quantitative RT-PCR of the expression of HS-inducible genes in roots of two independent transgenic lines of Pro35S:HsfB1 (red and pink bars; B1ox), Pro35S:mutHsfB1 (blue and light blue bars; mutB1ox), and wild-type plants (black bars) at 23°C. RNAs were extracted from roots of more than 10 individual plants of each line. The level of expression in the wild-type roots was taken as 1. Error bars indicate the sd of the technical replicate (n = 3). Asterisks indicate P values below 0.05 between the wild type and others.
HsfB1 Represses the Transcription of Genes for HS-Inducible Hsfs and HSPs
To determine the role of the repressive activity of HsfB1 in the regulation of the HS response, the expression of transcripts of the HS-inducible Hsfs and several genes for HSPs was analyzed. Quantitative reverse transcription (RT)-PCR analysis using RNAs isolated from root tissues of 27-d-old plants grown on agar plates showed that the relative expression levels of HsfA2, HsfA7a, HsfB2b, and HSP15.7CI were higher in Pro35S:mutHsfB1 roots than in Pro35S:HsfB1 roots and in wild-type roots at the non-heat condition (23°C; Fig. 2E). The higher expression of the heat-inducible genes in Pro35S:mutHsfB1 plants is probably due to a dominant negative effect of mutHsfB1, which interfered with the repressive activity of endogenous HsfB1 by competing with it.
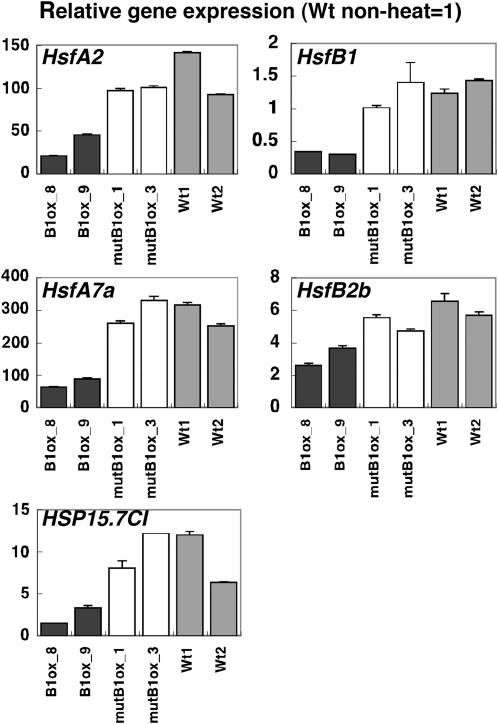
The repressive activity of HsfB1 to the heat-inducible genes was not observed when the Pro35S:HsfB1 plants were treated with HS (32°C, 34°C, and 37°C), probably due to the extremely high expression of HS-inducible genes (data not shown). However, under the moderate heat condition (28°C) at 45 min, induction of HsfA2, HsfA7a, HsfB2b, HSP15.7CI, and endogenous HsfB1 expression was suppressed or reduced in Pro35S:HsfB1 seedlings compared with Pro35S:mutHsfB1 and wild-type plants (Fig. 3). These results indicate that ectopically expressed HsfB1 acts as a repressor of the expression of these HS-inducible genes under moderate heat conditions (28°C).
Figure 3.
Effects of the repression domain of HsfB1 on the regulation of HS-inducible gene expression. Quantitative analysis by RT-PCR is shown for the expression of HS-inducible genes in seedlings of Pro35S:HsfB1 (black bars; B1ox), Pro35S:mutHsfB1 (white bars; mutB1ox), and wild-type (gray bars; Wt) plants treated with moderate heat (28°C) for 45 min. The expression level at the non-heat condition (23°C) in wild-type seedlings was taken as 1.
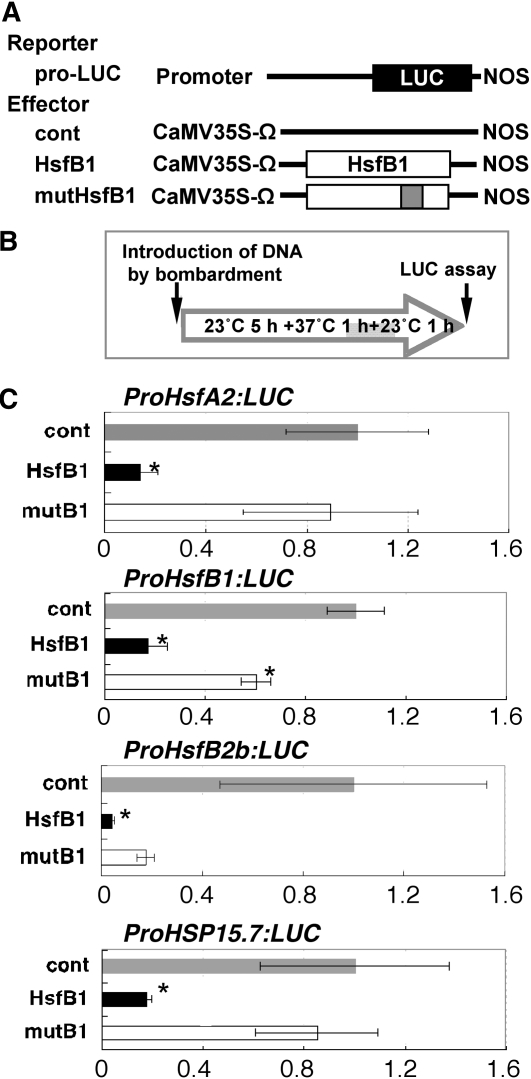
To evaluate the effect of HsfB1 for the promoter activity of HS-responsive genes, transient expression assays by bombardment of rosette leaves were performed using reporter genes in which the 5′ upstream regions of the translation initiation sites of HsfA2 (1,244 bp), HsfB1 (1,868 bp), HsfB2b (2,400 bp), and HSP15.7CI (1,462 bp) were placed upstream of the luciferase (LUC) reporter gene (Fig. 4A). The appropriate experimental condition for the transient assay was determined by analyzing the LUC activity of the ProHsfA2:LUC reporter under the three different conditions (Supplemental Fig. S2A). Increased activity of the ProHsfA2:LUC reporter gene was much higher with 1 h of postincubation at 23°C after treatment at 37°C for 1 h than without postincubation and heat treatment (Supplemental Fig. S2B); therefore, we used the condition of 1 h of postincubation at 23°C after the heat treatment in the transient assays (Fig. 4B). The activity of each reporter gene was up-regulated when treated with 37°C, but the activation by heat treatment was suppressed when the Pro35S:HsfB1 effector was coexpressed (Fig. 4C). Such suppression was not observed when Pro35S:mutHsfB1 was coexpressed. These results suggested that HsfA2, HsfB2b, and HSP15.7CI might be targets of HsfB1 and that HsfB1 might regulate its own expression.
Figure 4.
Effect of HsfB1 on the promoter activity of HS-responsive Hsf and HSP genes in a transient assay. A, Schematic representation of the constructs used in transient expression assays. The reporter gene contains the 5′ upstream region of a gene for Hsf and HSP, the firefly gene for luciferase (LUC; black box), and a nopaline synthase (NOS) terminator. Effector constructs for HsfB1 (white box) and a mutant form of HsfB1 (white box with gray insert) with the translation enhancer sequence derived from Tobacco mosaic virus were driven by the cauliflower mosaic virus (CaMV) 35S promoter. pUC18 vector was used as a control (cont). B, Sequence of manipulations of the heat treatment of Arabidopsis leaves after bombardment. The gray shading indicates incubation at 37°C. C, Relative LUC activities after cobombardment of Arabidopsis leaves with the Pro35S:HsfB1 (black bars; HsfB1) and Pro35S:mutHsfB1 (white bars; mutB1) effectors and ProHsfA2:LUC, ProHsfB1:LUC, ProHsfB2b:LUC, and ProHSP15.7CI:LUC reporter genes. After particle bombardment, the Arabidopsis leaves were incubated in the dark for 5 h at 23°C, 1 h at 37°C, and then 1 h at 23°C. The relative activity due to the pUC18 vector (gray bars; cont) was taken as 1. Asterisks indicate P values below 0.05 between the control and others.
HsfB1 and HsfB2b Repress the General HS Response under the Non-HS Condition and Hypocotyl Elongation
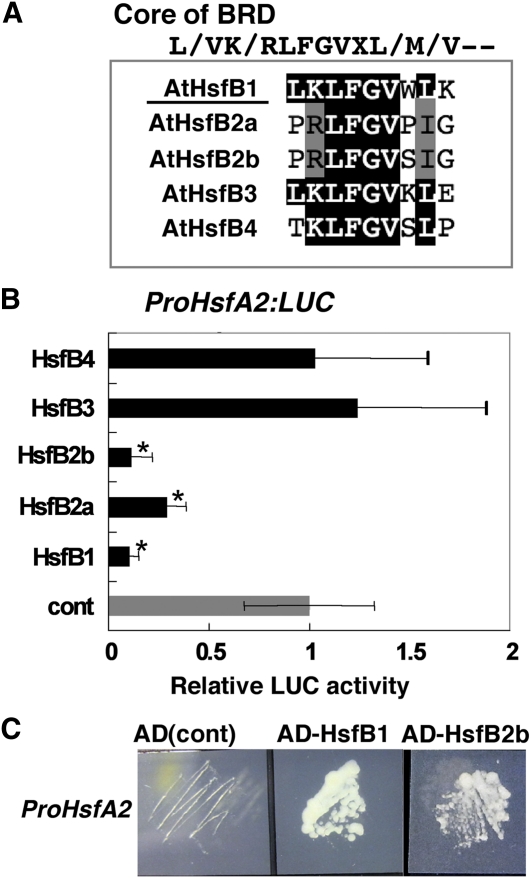
T-DNA disruption of HsfB1 (to yield the loss-of-function mutant hsfb1-2 [SALK_012292]; Supplemental Fig. S4A) did not affect the expression of any of the heat-inducible Hsf or HSP genes examined, with the exception of increased expression of HsfB2b, probably because of the presence of redundant factors to HsfB1 (Supplemental Fig. S3A). The five class B Hsfs in Arabidopsis contain the core sequence of BRD ([R/K]LFGV) and could act as repressors (Fig. 5A; Supplemental Fig. S3B; Ikeda and Ohme-Takagi, 2009). Transient expression analyses using the ProHsfA2:LUC reporter gene revealed that HsfB2b exhibited strong repressive activity and reduced the ProHsfA2:LUC reporter gene activity to less than 10% by coexpression, similar to HsfB1 (Fig. 5B). HsfB2a also repressed the activity of the reporter gene, but its repressive activity was weaker than that of HsfB1 and HsfB2b. On the contrary, neither HsfB3 nor HsfB4 repressed the expression of the ProHsfA2:LUC reporter gene (Fig. 5B). Expression of HsfB2a and HsfB2b was induced by heat, although the levels of expression were lower than that of HsfB1 (Supplemental Fig. S3C; Schramm et al., 2008). We confirmed the binding of HsfB1 and HsfB2b to the promoter of HsfA2 by yeast one-hybrid assay using 473-bp 5′ upstream regions from the translation initiation site of the HsfA2 (Fig. 5C).
Figure 5.
Exploring functional redundancy among class B Hsfs for the regulation of HsfA2 expression. A, An alignment of the core sequence of BRD of Arabidopsis class B Hsfs. B, Relative LUC activities after cobombardment of Arabidopsis leaves with plasmids encoding class B Hsf effectors and the ProHsfA2:LUC reporter gene. After particle bombardment, the Arabidopsis leaves were incubated in the dark for 5 h at 23°C, 1 h at 37°C, and then 1 h at 23°C. The relative activity due to the pUC18 vector (gray bar; cont) was taken as 1. Error bars indicate sd (n = 9). Asterisks indicate P values below 0.05 between the control and others. C, Interaction of HsfB1 and HsfB2b in the promoter region of HsfA2 analyzed in the yeast one-hybrid system. The transformed yeast cells were grown on medium containing 60 mm 3-amino-1,2,4-triazole. pDEST_GAD424 empty vector was used as a negative control that contains the activation domain only [AD(cont)]. [See online article for color version of this figure.]
Since HsfB2b appeared to be a functionally redundant gene of HsfB1, double knockout lines with mutations in both the HsfB1 and HsfB2b genes were prepared. From among the four resultant lines, the hsfb1-1 hsfb2b-1 null line rather than the hsfb1-2 hsfb2b-2 line was selected, because transcripts of the HsfB1 and HsfB2b genes were detected in hsfb1-2 hsfb2b-2 plants (Supplemental Fig. S4, A and B).
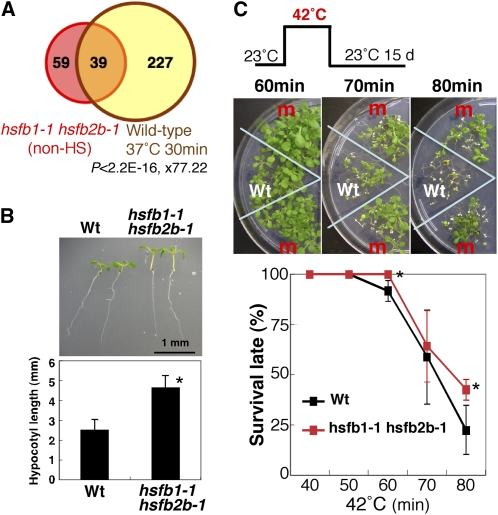
Microarray analysis revealed that genes related to the HS response were overrepresented among genes whose expression was enhanced in 7-d-old hsfb1-1 hsfb2b-1 seedlings in the non-HS condition (23°C; Fig. 6A; Table I; Supplemental Tables S1 and S2). Expression of almost all heat-inducible genes for Hsfs and small HSPs was enhanced in hsfb1-1 hsfb2b-1 plants (Table I; Supplemental Tables S3 and S4). By quantitative RT-PCR, we confirmed that the expression levels of HsfA2 and HsfA7a were more than 20-fold higher in hsfb1-1 hsfb2b-1 seedlings than in wild-type seedlings under the non-HS condition (Table I; Supplemental Fig. S4C). The elevated expression of these genes was also observed in hsfb1-2 hsfb2b-2 plants (Supplemental Fig. S4C). Because the expression of HsfA2 was not suppressed in the hsfb1-1 hsfb2b-1 double mutants, we considered that HsfB2b acts as a functionally redundant factor to HsfB1 and that HsfB2a may play a minor role in the regulation of the expression of HsfA2.
Figure 6.
hsfb1-1 hsfb2b-1 plants express the HS-inducible genes under the non-heat condition and exhibit the heat-treated phenotype. A, Venn diagram showing the overlap between genes up-regulated in hsfb1-1 hsfb2b-1 plants at the non-heat condition (23°C; more than 1.5-fold and q < 0.1; pink circle) and those up-regulated in 3-week-old seedlings of wild-type plants treated with 37°C for 30 min (yellow circle). The P value for Fisher’s exact test indicating statistical significance of the overlap and the odds ratio representing the number of overlapping genes/number of genes expected by chance are shown. B, Morphological analysis of hsfb1-1 hsfb2b-1 seedlings under the non-heat condition (23°C). Top panel, 6-d-old seedlings of wild-type (Wt) and hsfb1-1 hsfb2b-1 plants grown at 23°C; bottom panel, length of hypocotyls of 6-d-old seedlings of wild-type and hsfb1-1 hsfb2b-1 plants grown at 23°C. The asterisk indicates a P value below 0.05 between wild-type and hsfb1-1 hsfb2b-1 seedlings. C, Thermotolerance of 5-d-old seedlings of hsfb1-1 hsfb2b-1 plants treated with 42°C. The top panel shows, schematically, the conditions for exposure to HS. The middle panels show wild-type and hsfb1-1 hsfb2b-1 (m) seedlings after exposure to HS for the indicated times. The bottom panel shows survival rates of hsfb1-1 hsfb2b-1 (red line) and wild-type (black line) seedlings. Results are means ± sd (n ≥ 40). Asterisks indicate P values below 0.05 between wild-type and hsfb1-1 hsfb2b-1 seedlings.
Table I. Up-regulated genes among HS-related genes in hsfb1-1hsfb2b-1 plants compared with wild-type plants under the non-heat condition (23°C).
| Arabidopsis Genome Initiative No. | Annotation | Microarray |
Quantitative RT-PCR |
|||
| hsfb1-1 hsfb2b-1, Non Heat | 3-Week-Old Seedling Treated with 37°C for 30 min | hsfb1-1 hsfb2b-1, 32°C for 30 min | hsfb1-1 hsfb2b-1, Non Heat, Lot 1 | hsfb1-1 hsfb2b-1, Non Heat, Lot 2 | ||
| Hsfs | ||||||
| AT4G36990 | AtHSFB1, AT-HSF4, HSF4, HSFB1 | 0.09 | 18.48 | 0.01 | 0.00 | – |
| AT4G11660 | AtHSFB2b, HSFB2b | 0.14 | 15.65 | 0.01 | 0.00 | – |
| AT2G26150 | AtHSFA2 | 13.37 | 49.11 | 1 | 19.92 | 18.41 |
| AT3G51910 | AtHSFA7a | 2.63 | 22.1 | 1.09 | 36.09 | 32.62 |
| AT3G63350 | AtHSFA7b | 5.48 | 25.18 | 1.4 | – | – |
| AT5G62020 | AtHSFB2a | 2.3 | 13.27 | 0.99 | 3.04 | 2.09 |
| Small HSPs | ||||||
| AT1G30070 | SGS domain-containing protein | 1.91 | 31.75 | 0.87 | – | – |
| AT5G51440 | 23.5-kD mitochondrial small HSP | 1.78 | 70.4 | 0.81 | – | – |
| AT1G59860 | AtHSP17.6A-CI | 3.81 | 32.11 | 0.84 | – | – |
| AT1G07400 | AtHSP17.8-CI | 4.38 | 24.64 | 0.93 | – | – |
| AT5G37670 | AtHSP15.7-CI | 2 | 43.41 | 0.75 | 3.16 | 2.79 |
| AT1G54050 | AtHSP17.4-CIII | 1.6 | 66.62 | 0.77 | – | – |
| AT3G46230 | AtHSP17.4 | 2.4 | 23.67 | 0.66 | 2.04 | 4.07 |
| AT5G12020 | AtHSP17.6II | 2.49 | 36.79 | 0.76 | – | – |
| AT2G29500 | AtHSP17.6B-CI | 1.79 | 29.59 | 0.67 | – | – |
| AT5G12030 | AtHSP17.6A | 1.32 | 40.84 | 0.6 | – | – |
| AT4G10250 | AtHSP22.0 | 1.91 | 81.79 | 0.64 | – | – |
| AT1G52560 | 26.5-kD class I small HSP-like | 2.68 | 74.69 | 0.87 | 3.09 | 3 |
| AT5G59720 | AtHSP18.2 | 1.03 | 28.74 | 0.59 | 1.23 | 1.24 |
| AT4G25200 | AtHSP23.6-MITO | 2.51 | 43.13 | 0.71 | 3.08 | 3.75 |
| AT4G27670 | AtHSP21 | 4.83 | 31.14 | 0.59 | 1.53 | 4.29 |
| HSP70 | ||||||
| AT3G12580 | AtHSP70, ATP binding | 1.27 | 47.99 | 0.76 | 1.55 | 1.45 |
| AT5G49910 | CPHSC70-2 HEAT SHOCK PROTEIN70-2 | 1.24 | 2.09 | 0.91 | – | – |
| AT1G79920 | ATP binding | 1.38 | 4.22 | 0.97 | – | – |
| AT1G56410 | ERD2, ATP binding | 1.41 | 2.33 | 1.34 | – | – |
| HSP90 | ||||||
| AT5G52640 | AtHSP90.1, ATP binding | 1.19 | 27.73 | 0.8 | 1.71 | 1.4 |
| AT2G04030 | CR88, ATP binding | 1.17 | 2.23 | 0.94 | – | – |
| HSP90 | ||||||
| AT1G74310 | AtHSP101 | 1.07 | 37.72 | 0.62 | 1.60 | 1.79 |
| DNAJ | ||||||
| AT2G20560 | DNAJ heat shock family protein | 3.25 | 47.9 | 0.71 | 3.38 | 2.74 |
| AT3G14200 | DNAJ heat shock protein | 1.51 | 4.15 | 0.75 | – | – |
| AT3G13310 | DNAJ heat shock protein | 1.28 | 3.95 | 1.23 | – | – |
| At4G8480 | DNAJ heat shock protein | 1.24 | 3.56 | 0.79 | – | – |
| DREBs | ||||||
| AT5G05410 | DREB2A | 1.49 | 42.73 | 0.68 | 1.17 | – |
| AT3G11020 | DREB2B | 1.12 | 2.18 | 0.8 | – | – |
| AT1G75490 | DREB2D | 1.4 | 1.04 | 1.28 | – | – |
| AT1G12610 | DREB1F | 1.36 | 1.2 | 2.12 | – | – |
Quantitative RT-PCR showed that the expression levels of HsfB2a and genes for small HSPs, namely AtHSP23.6-MITO (At4g25200), AtHSP21 (At4g27670), AtHSP17.4 (At3g46230), HSP15.7CI (At5g37670), and DNAJ (At2g20560), were 3- to 4-fold higher in hsfb1-1 hsfb2b-1 plants than in wild-type plants at 23°C (Table I). The expression level of DNAJ (At2g20560) was also elevated in hsfb1-2 hsfb2b-1 plants (Kumar et al., 2009). Expression of the heat-inducible genes HSP101, AtHSP70 (At3g12580), AtHSP90.1 (At5g52640), DREB2A, AtHSP26.5 (At1g52560), and HSP18.2 (At5g59720) was also very slightly enhanced in hsfb1-1 hsfb2b-1 seedlings (Table I). In contrast, the expression of HsfA3 was not affected either in hsfb1-1 hsfb2b-1 or hsfb1-2 hsfb2b-2 plants, indicating that not all HS-responsive genes are regulated by HsfB1 and HsfB2b (Supplemental Fig. S4C). These results demonstrated that HsfB1 and HsfB2b act as repressors of HS-inducible genes for Hsfs and HSPs under a non-HS (23°C) temperature.
We found that seedlings of hsfb1-1 hsfb2b-1 plants had longer hypocotyls than those of the wild type under the non-heat condition, although their roots and adult plants were similar to those of the wild type (Fig. 6B; Supplemental Fig. S5, A and B). A relationship between thermotolerance and altered hypocotyl elongation has been reported. High temperature (greater than 37°C) is known to inhibit the elongation of hypocotyls, and the decreased thermotolerance mutant, hot2, exhibits a short-hypocotyl phenotype (Hong and Vierling, 2000). On the other hand, elongation of hypocotyls was induced under the moderate heat condition (27°C–29°C; Gray et al., 1998; Koini et al., 2009; Kumar and Wigge, 2010). Our microarray analyses revealed that the expression of heat-inducible genes was enhanced in hsfb1-1 hsfb2b-1 plants under the non-HS condition (Table I). This suggests that hsfb1-1 hsfb2b-1 plants are constitutively in a state similar to a moderate HS and that this may be the reason why seedlings of hsfb1-1 hsfb2b-1 plants had longer hypocotyls.
HsfB1 and HsfB2b Suppress the Expression of HsfA2 and HsfA7a under HS Conditions and Are Involved in Thermotolerance
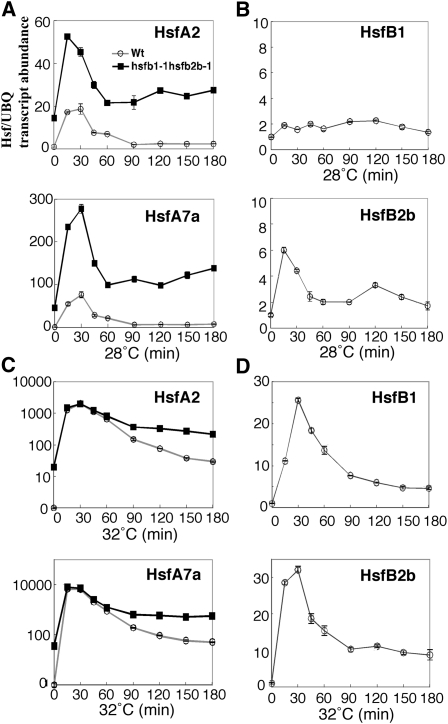
Under the moderate heat condition (28°C), the expression levels of HsfA2 and HsfA7a in 7-d-old seedlings of hsfb1-1 hsfb2b-1 plants were also much higher than in wild-type seedlings (Fig. 7A). These findings suggest that HsfB1 and HsfB2b act as repressors of the expression of HsfA2 and HsfA7a under the moderate heat condition. Actually, HsfB1 was expressed under normal conditions, especially in root, although the expression of HsfB1 was not up-regulated at 28°C, whereas the expression of HsfB2b was induced at 28°C (Fig. 7B; Busch et al., 2005; Schramm et al., 2008).
Figure 7.
The relative expression of HsfA2, HsfA7a, HsfB1, and HsfB2b in hsfb1-1 hsfb2b-1 and wild-type (Wt) seedlings under the moderate heat condition (28°C) and in the extended HS period of the HS response (32°C). A and C, Relative expression of HsfA2 (top graph) and HsfA7a (bottom graph) in 5-d-old seedlings of hsfb1-1 hsfb2b-1 (black squares) and the wild type (white circles) during incubation for 3 h at 28°C (A) and 32°C (C). B and D, Relative expression of HsfB1 (top graph) and HsfB2b (bottom graph) in 5-d-old seedlings of the wild type during incubation for 3 h at 28°C (B) and 32°C (D). The level of expression under the normal growth condition (23°C) of wild-type seedlings was taken as 1. The y axis indicates expression relative to an internal control and uses a linear scale in A, B, and D and a log10 scale in C. UBQ, Ubiquitin1.
The expression of HsfA2 and HsfA7a in hsfb1-1 hsfb2b-1 seedlings was induced at 32°C to a similar extent to that in the wild type within 30 min (Fig. 7C). However, under continuous HS at 32°C, the expression levels of these genes decreased rapidly after 30 min in wild-type seedlings, while those in hsfb1-1 hsfb2b-1 seedlings remained consistently higher than in the wild type (Fig. 7C). Since the expression of HsfB1 and HsfB2b was induced at 32°C (Fig. 7D), HsfB1 and HsfB2b might play a role in attenuation of the strongly induced expression of HsfA2 and HsfA7a during the extended HS response.
Because seedlings of hsfb1-1 hsfb2b-1 plants overexpressed HS-responsive genes at the normal growth temperature (23°C), we examined the thermotolerance of hsfb1-1 hsfb2b-1 plants. Five-day-old seedlings of hsfb1-1 hsfb2b-1 plants were treated with 42°C for 40 to 80 min and then grown at 23°C for 15 d. Seedlings of hsfb1-1 hsfb2b-1 plants exhibited higher thermotolerance than those of the wild type when treated with 42°C for 80 min (Fig. 6C). This enhanced thermotolerance of hsfb1-1 hsfb2b-1 plants might have been due to higher levels of expression of HsfA2 and HSP (Table I), whose ectopic expression has been shown to induce stress tolerance (Nishizawa et al., 2006).
HsfB1 and HsfB2b Are Necessary for the Acquired Thermotolerance
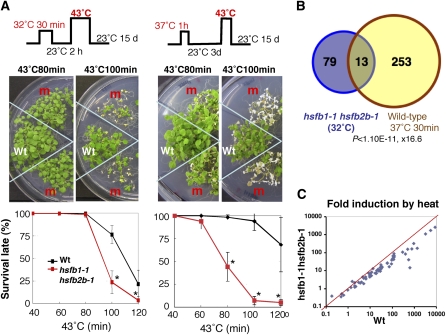
It has been reported that prior heat treatment enhances the subsequent thermotolerance in plants, which is called acquired thermotolerance, and that HsfA2 is necessary for the acquired thermotolerance (Hong and Vierling, 2000; Charng et al., 2007). To analyze whether HsfB1 and HsfB2b, which affect the expression of HsfA2, are involved in acquired thermotolerance, we exposed seedlings to 43°C for 40 to 120 min after a 2-h recovery at 23°C following a conditioning treatment at 32°C for 30 min (Fig. 8A, top left panel) and then grew them at 23°C for 15 d. The results showed that the relative number of hsfb1-1 hsfb2b-1 seedlings that survived HS at 43°C for 100 min was 50% lower than that of wild-type seedlings (Fig. 8A, left panels). In addition, this phenomenon was similarly observed when plants were exposed to 43°C after 3 d of recovery at 23°C following a conditioning treatment at 37°C for 1 h (Fig. 8A, right panels). These results indicate that HsfB1 and HsfB2b are required for the acquired thermotolerance in Arabidopsis.
Figure 8.
Roles of HsfB1 and HsfB2b in acquired thermotolerance. A, Thermotolerance of hsfb1-1 hsfb2b-1 plants at 43°C after 2 h of recovery at 23°C following a conditioning treatment at 32°C for 30 min (left panel) and at 43°C after 3 d of recovery at 23°C following a conditioning treatment at 37°C for 1 h (right panel). The top panel shows, schematically, the conditions for exposure to HS. The middle panels show wild-type (Wt) and hsfb1-1 hsfb2b-1 (m) seedlings after exposure to HS for the indicated times and then grown at 23°C for 15 d. The bottom panel shows survival rates of hsfb1-1 hsfb2b-1 (red lines) and wild-type (black lines) seedlings. Results are means ± sd (n ≥ 40). Asterisks indicate P values below 0.05 between wild-type and hsfb1-1 hsfb2b-1 seedlings. B, Venn diagram showing the overlap between genes down-regulated in hsfb1-1 hsfb2b-1 plants at 32°C for 30 min (more than 1.5-fold and q < 0.1; purple circle) and those up-regulated in 3-week-old seedlings of wild-type plants treated with 37°C for 30 min (yellow circle). The P value and odds ratio for this overlap are shown. C, The extent of heat induction of HS-inducible genes in wild-type (x axis) and hsfb1-1 hsfb2b-1 (y axis) plants. Each point represents one gene. The red line indicates equal induction of expression between wild-type and hsfb1-1 hsfb2b-1 plants.
To approach the molecular events and the roles of class B Hsfs under HS conditions, we performed microarray analysis using RNA from seedlings that had been incubated at 32°C for 30 min. In contrast to the results obtained under normal conditions (23°C), under HS conditions at 32°C, the group of genes related to the HS response was clearly overrepresented among genes whose expression was suppressed in hsfb1-1 hsfb2b-1 seedlings as compared with wild-type seedlings (Fig. 8B; Supplemental Tables S1 and S5). The level of expression of the genes for HSP101, 10 small HSPs, AtHSP70, HSP90, and DNAJ was lower in hsfb1-1 hsfb2b-1 plants than in wild-type plants under the 32°C HS condition (Table I; Supplemental Tables S4 and S5). These data indicate that the extent of induction of HS-inducible genes (for definition, see “Materials and Methods”) at 32°C was weaker in hsfb1-1 hsfb2b-1 plants than in wild-type plants (0.55-fold on average; Fig. 8C). These results suggest that HsfB1 and HsfB2b are necessary for the expression of HS-inducible genes during the pretreatment at 32°C for acquired thermotolerance.
DISCUSSION
Class B Hsfs Repress the Expression of HS-Responsive Hsf and HSP Genes under Non-Heat Conditions and during the Attenuation Period
This study demonstrated that the Arabidopsis class B Hsfs, HsfB1 and HsfB2b themselves, are transcriptional repressors and that the BRD (Czarnecka-Verner et al., 2004; Ikeda and Ohme-Takagi, 2009), which is conserved in orthologs of HsfB1 in a variety of plant species, is necessary for the HsfB1-mediated repression of the expression of downstream genes (Figs. 1–4). BRD is conserved in all five members of Arabidopsis class B Hsfs (Fig. 5A). However, neither HsfB3 nor HsfB4 has repressive activity to the promoter of HsfA2, which was shown to be a direct target of HsfB1 in this study (Fig. 5B). In addition, HsfB2b, but not HsfB2a, appears to be a functionally redundant factor to HsfB1, because the expression of HsfA2 and HsfA7a was highly induced in hsfb1-1 hsfb2b-1 plants under the non-heat condition (Table I; Fig. 7; Supplemental Fig. S4C). HsfB2a may regulate the expression of other HS-inducible genes than those regulated by HsfB1 and HsfB2b.
Under non-HS conditions, HsfB1 and HsfB2b are localized in the nucleus (Scharf et al., 1998; Heerklotz et al., 2001; Kotak et al., 2004) and directly repress expression of the HS-inducible genes for Hsfs (HsfA2, HsfA7a, HsfB1, and HsfB2b) and some HSPs (Figs. 3 and 4). The class A HsfA2 was reported as a key regulator for HS-inducible genes in plants (Nishizawa et al., 2006; Schramm et al., 2006; Charng et al., 2007). Our microarray analyses revealed that the expression profile of hsfb1-1 hsfb2b-1 plants under normal conditions is similar to that of the wild type treated with moderate heat at 28°C. These data suggest that hsfb1-1 hsfb2b-1 plants under non-HS conditions are under similar conditions as the wild type suffering moderate HS. HsfB1 and HsfB2b appeared to suppress the expression of numerous HS-inducible genes, including their nondirect targets (Table I; Fig. 6; Supplemental Tables S1–S4).
Repressive activities of HsfBs during the attenuation period of the HS response were also confirmed (Fig. 7C). These results indicate that HsfB1 and HsfB2b repress the general HS response under non-HS conditions and attenuation period via direct repression of the expression of the HS-inducible HsfA2. Our analyses of microarray data revealed that the groups of genes that are repressed by HsfB1 and HsfB2b show considerable overlap with the groups of genes whose expression is enhanced by HsfA1a, HsfA1b, HsfA2, or HsfA3 (Supplemental Table S1; Nishizawa et al., 2006; Yoshida et al., 2008). In addition, HsfA7a, HsfB1, and HSP15.7CI, which were shown to be direct targets of HsfB1 in this study, are reported to be targets of HsfA1s and HsfA2 (Lohmann et al., 2004; Busch et al., 2005; Nishizawa et al., 2006; Liu et al., 2011). These results indicate that class B HsfB1 and HsfB2b repressors and class A HsfA1a, HsfA1b, HsfA2, and HsfA3 activators appear to regulate similar groups of genes.
HsfB1 and HsfB2b Are Required for Acquired Thermotolerance
Although hsfb1-1 hsfb2b-1 plants exhibited higher thermotolerance, they exhibited lower acquired thermotolerance than the wild type (Figs. 6C and 8A). It seems plausible that hsfb1-1 hsfb2b-1 plants might be unable to accumulate sufficient amounts of the HSPs that are necessary for resistance to severe HS during the pretreatment. Microarray analyses revealed that the levels of expression of various HS-inducible genes for HSPs, including the HSP101 gene, which has been shown to be necessary for acquired thermotolerance, were lower in the double mutant than in wild-type plants under HS conditions (32°C, 30 min; Table I; Supplemental Tables S1, S5, and S6; Hong and Vierling, 2000; Queitsch et al., 2000). We showed that the group of genes that are regulated by HsfA1a and HsfA1b were significantly overrepresented among genes down-regulated in hsfb1-1 hsb2b-1 plants at 32°C (Supplemental Tables S1, S5, and S6). HsfA1s are activators of genes for HSPs and are involved in the early response to HS (Lohmann et al., 2004; Busch et al., 2005; Nishizawa-Yokoi et al., 2011). In addition, a quadruple mutant of HsfA1s has been shown to have dramatically decreased the capacity of the acquired thermotolerance (Liu et al., 2011). The lower acquired thermotolerance in hsfb1-1 hsfb2b-1 plants might be due to the impaired activities of HsfA1s. HsfB1 and HsfB2b may promote the activity of HsfA1s under HS conditions, probably by repressing the expression of genes for HSPs that interfere with the nuclear migration of HsfA1s.
Why Do Plants Have So Many Different Types of Hsfs?
In many eukaryotes, a single type of Hsf controls the HS response. It activates the expression of HSP genes, of which the products negatively regulate Hsf activity by feedback regulation. In contrast, plants have more than 20 Hsfs with different functions. HsfA1a and HsfA1b of Arabidopsis, which show similar characteristics to the Hsfs of other eukaryotes, regulate the early response to HS (Kotak et al., 2004; Lohmann et al., 2004; Busch et al., 2005). HsfA2 is rapidly induced by HS and regulates the extended response to HS by enhancing and maintaining the early HS response (Charng et al., 2007; Meiri and Breiman, 2009). Accordingly, plants acquired class B Hsfs, which act as repressors of the expression of the HS-inducible Hsfs.
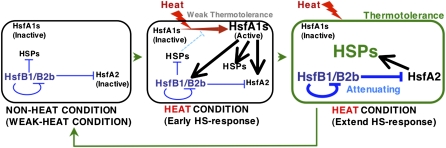
Plants might have evolved complex systems to respond to and survive HS by obtaining Hsfs that play a role in an extended HS response (Fig. 9). Under normal growth conditions, the HsfA1 activators are localized in the cytoplasm and are inactive (Kotak et al., 2004), while the HsfB1 and HsfB2b repressors, which localize and form a homomeric oligomer in the nucleus (Scharf et al., 1998; Heerklotz et al., 2001; Kotak et al., 2004; Li et al., 2010), suppress the general HS response by repressing the expression of HS-inducible genes for Hsfs and HSPs (Figs. 3–6 and Fig. 9, left panel). When the plants are exposed to heat, HsfA1a and HsfA1b rapidly migrate to the nucleus and strongly activate HS-inducible genes, with their activation activity overcoming the repressive activity of HsfBs (Fig. 9, middle panel). In tomato, it has been reported that HsfAs and HsfBs form a complex to activate HS-inducible genes (Bharti et al., 2004; Hahn et al., 2011). The HSPs that are induced by HsfA1s in the early HS response may confer thermotolerance to the plant cell. The resultant activation of HS-inducible class A Hsfs (HsfA2, HsfA7a, and HsfA3) may enhance and maintain the HS response and promote the production of HSPs necessary for thermotolerance (Fig. 9, right panel; Nishizawa et al., 2006; Schramm et al., 2006, 2008; Charng et al., 2007; Larkindale and Vierling, 2008; Yoshida et al., 2008; Meiri and Breiman, 2009).
Figure 9.
Model of the HS response in plants. The left panel shows possible activities of Hsfs under non-HS and moderate HS conditions. HsfB1 and HsfB2b repress the expression of HS-inducible Hsfs and HSPs; the HsfA1s are inactive under non-HS conditions. The middle panel shows the activity of Hsfs in the early stage of the HS condition. HsfA1s are activated rapidly in response to HS to overcome the repressive activities of HsfBs. The expression of genes for Hsfs (HsfA2, HsfA7a, HsfB1, and HsfB2b) and HSPs is strongly induced by HsfA1s. The right panel shows the activity of Hsfs in an extended HS response. Under this condition, the plant cells acquire thermotolerance by the activities of HSPs, which have been produced in the early stage of the HS response. Several HSPs suppress the activity of class A Hsfs by interacting with them to interfere with their trimer formation and nuclear import. On the other hand, the HS response is enhanced and maintained by HS-induced HsfAs (e.g. HsfA2). At the same time, the HS-induced HsfBs (HsfB1 and HsfB2b), the expression of which is induced by HsfA1s in the early stage of the HS response, repress the expression of heat-inducible HsfA2 to attenuate the HS response. The font size indicates the quantity of active proteins. Black and blue arrows indicate positive (black) and negative (blue) regulation of the transcription of each gene. Red and light blue arrows indicate activation (red) and inactivation (light blue) of HsfA1 proteins.
In parallel with the drastic induction of HS-responsive genes in response to HS, plants appear to operate an attenuation system. Several HSPs suppress the activity of some class A Hsfs by interacting with them to interfere with their trimer formation and nuclear import (Lee and Schöffl, 1996; Port et al., 2004; Hahn et al., 2011). In addition to the inactivation of class A Hsfs by HSPs, the HS-inducible HsfB1 and HsfB2b by HsfA1s (Busch et al., 2005) directly repress the expression of HS-inducible Hsfs to attenuate the HS response (Fig. 9, right panel).
Plants are sessile organisms, and thermotolerance is critical to their survival. This study demonstrated that the responses of Arabidopsis to HS are regulated by the finely tuned activation and repression activities of Hsfs. In contrast to many eukaryotes, which control the HS response using simple feedback regulation between Hsfs and HSPs, the HS-responsive system in plants is composed of two activation steps for early (HsfA1s) and extended (HsfA2) responses and two attenuating steps for suppression of the activation activity of class A Hsfs by HSPs and repression of the expression of Hsf genes by class B Hsfs. Thus, plants have evolved a unique HS-response system using Hsfs and HSPs to acquire the ability to survive HS.
MATERIALS AND METHODS
Construction of Plasmids
The coding regions and the 5′ upstream regions of genes used in this study were amplified from a cDNA library or from the genomic DNA of Arabidopsis (Arabidopsis thaliana) with appropriate primers (Supplemental Table S7). The mutations in HsfB1 were introduced by the use of appropriate mutagenic primers. Construction of the effector and reporter plasmids for the transient expression assay was described previously (Hiratsu et al., 2002). Effector plasmids were constructed by fusion of the yeast Gal4-DB coding region to the coding sequence of each gene, in frame, under the control of the cauliflower mosaic virus 35S promoter (−800 to +8). The reporter gene 35S-Gal4-TATA-LUC-NOS was described previously (Hiratsu et al., 2002).
Growth and Transformation of Plants
Arabidopsis ecotype Columbia was used in all experiments. Plants used for transient assay and for transformation were grown in soil at 23°C (normal growth temperature) with a photoperiod of 10 h/14 h and 16 h/8 h of light/dark, respectively. Transformation of Arabidopsis was performed using the floral dip method (Clough and Bent, 1998). Plants used for RNA isolation and analysis of thermotolerance were grown on agar plates at 23°C with a photoperiod of 16 h/8 h of light/dark.
Heat Treatment
Five-day-old seedlings grown on horizontal agar plates, which contained 30 mL of solid Gamborg’s B5 medium, at 23°C under 60 μmol m−2 s−1 light were submerged in a water bath at 28°C, 32°C, 42°C, and 43°C for appropriate times.
Transient Expression Assays
Transient expression assays were performed using Arabidopsis rosette leaves of 2-month-old plants, as described previously (Hiratsu et al., 2004). For heat treatment, Arabidopsis leaves were incubated at 37°C under air conditions for 1 h in the dark after particle bombardment.
Isolation of RNA and Analysis of RNA Expression
Total RNA was isolated with the RNeasy Plant Mini Kit (Qiagen) from seedlings or roots of more than 10 individual plants in every case. Quantitative RT-PCR was performed as described previously (Mitsuda et al., 2005) with appropriate primers (Supplemental Table S7). Relative levels of transcripts were calculated by an absolute quantification method, with transcription of Ubiquitin1 as an internal control. More than three replicates were included in each experiment. Results are presented as means ± sd The absence of an error bar indicates that the bar falls within the symbol.
Yeast One-Hybrid Assays
The yeast one-hybrid assays were performed as described previously (Mitsuda et al., 2010). For bait construction, a 473-bp upstream region from the translation initiation site of HsfA2 was amplified from the genomic DNA of Arabidopsis with appropriate primers (Supplemental Table S7) and cloned into pLacZi and pHisi vectors. For the construction of prey, cDNAs with the stop codon of HsfB1 and HsfB2b were amplified by a pair of primers with attB1/B2 sites and cloned into pDONR207 vector (Life Technologies) by BP cloning. Cloned cDNAs were transferred into pDEST_GAD424 vector by LR clonase.
Microarray Analysis
The microarray experiments were performed using the Agilent Arabidopsis 3 (44k) microarray (Agilent Technologies) according to the manufacturer’s instructions. Four biological replicates were tested with a two-color method in which wild-type RNA was labeled by Cy3 in two replicates and by Cy5 in the other replicates. Spot signal values were calculated with Feature Extraction version 9.1 software (Agilent). The quality control (QC) value was defined as 1 when a spot passed the “FeatNonUnifOL” filter and as 2 when the spot further passed the “FeatPopnOL” filter. The detection value was defined as 1 when a spot passed the “IsPosAndSignif” filter and as 2 when the spot further passed the “IsWellAboveBG” filter. All signal values were divided by the median value among spots with QC = 2 to enable comparison with other microarray data. Spot-to-gene conversion was accomplished based on a table provided by The Arabidopsis Information Resource (TAIR; ftp://ftp.arabidopsis.org/home/tair/Microarrays/Agilent/agilent_array_elements-2008-9-17.txt). The average values were used for the genes corresponding to two or more probes. Genes with an average QC value of less than 1.5 in the “target” sample or the “control” sample were excluded from the following analyses. Only genes with an average detection value of 1.5 or greater in the target sample were analyzed when selecting up-regulated genes, while only genes with an average detection value greater than 1.5 in the control sample were analyzed when selecting down-regulated genes. The P value for each gene was calculated using Welch’s t test. To control type I family-wise error, we calculated the q value from the P value using QVALUE software using the default settings (Storey and Tibshirani, 2003) and selected up- or down-regulated genes with q < 0.1 in addition to fold-change filters (greater than 1.5-fold or less than 0.67-fold). Fisher’s exact test was performed using R (http://www.r-project.org/). We defined the HS-inducible genes as those annotated as “response to heat” by Gene Ontology (GO:0009408), up-regulated by 37°C for 30-min heat treatment (Sakuma et al., 2006), up-regulated in the shoot by 30-min heat treatment (Kilian et al., 2007), up-regulated in cultured cells by 30-min heat treatment (Kilian et al., 2007), up-regulated in HsfA3ox plants (Yoshida et al., 2008), up-regulated in HsfA2ox plants (Nishizawa et al., 2006), or potentially up-regulated by HsfA1a/1b (Busch et al., 2005).
All data were deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE31888.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotype of Pro35S:HsfB1 and Pro35S:mutHsfB1 plants.
Supplemental Figure S2. Conditions of heat treatment and transient expression assays.
Supplemental Figure S3. Analysis of functional redundancy among class B Hsfs.
Supplemental Figure S4. Analysis of T-DNA-tagged lines for hsfB1 and hsfB2b.
Supplemental Figure S5. Morphological analysis of hsfb1-1 hsfb2b-1 double knockout plants.
Supplemental Table S1. Groups of genes that overrepresented significantly among genes whose expression was enhanced in hsfb1-1 hsfb2b-1 plants.
Supplemental Table S2. Average changes (-fold) in expression levels of genes enhanced in hsfb1-1 hsfb2b-1 plants at 23°C from seven experiments.
Supplemental Table S3. Average changes (-fold) in expression levels of genes for Hsfs from seven experiments.
Supplemental Table S4. Average changes (-fold) in expression levels of genes for small HSPs from seven experiments.
Supplemental Table S5. Average changes (-fold) in levels of expression of genes whose expression was suppressed in hsfb1-1 hsfb2b-1 plants treated with 32°C from seven experiments.
Supplemental Table S6. Average changes (-fold) in levels of expression of HS-inducible genes, which are enhanced or suppressed in normal growth temperature or HS condition (32°C) of hsfb1-1 hsfb2b-1 plants, respectively, from seven experiments.
Supplemental Table S7. Oligonucleotides used in this study.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for seeds of SALK_104713, SALK_012292, SALK_047291, and SALK_045982 mutant plants, Ms. Yoko Ooi and Ms. Yuko Takiguchi for skilled technical assistance, and Ms. Sumiko Takahashi, Ms. Yoshimi Sugimoto, and Ms. Manami Watanabe (National Institute of Advanced Industrial Science and Technology) for cultivation of plants.
References
- Baniwal SK, Bharti K, Chan KY, Fauth M, Ganguli A, Kotak S, Mishra SK, Nover L, Port M, Scharf KD, et al. (2004) Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J Biosci 29: 471–487 [DOI] [PubMed] [Google Scholar]
- Bharti K, Von Koskull-Döring P, Bharti S, Kumar P, Tintschl-Körbitzer A, Treuter E, Nover L. (2004) Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 16: 1521–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W, Wunderlich M, Schöffl F. (2005) Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J 41: 1–14 [DOI] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT. (2007) A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol 143: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Czarnecka-Verner E, Pan S, Salem T, Gurley WB. (2004) Plant class B HSFs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Mol Biol 56: 57–75 [DOI] [PubMed] [Google Scholar]
- Czarnecka-Verner E, Yuan CX, Scharf KD, Englich G, Gurley WB. (2000) Plants contain a novel multi-member class of heat shock factors without transcriptional activator potential. Plant Mol Biol 43: 459–471 [DOI] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M. (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn A, Bublak D, Schleiff E, Scharf KD. (2011) Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 23: 741–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerklotz D, Döring P, Bonzelius F, Winkelhaus S, Nover L. (2001) The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Mol Cell Biol 21: 1759–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Mitsuda N, Matsui K, Ohme-Takagi M. (2004) Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem Biophys Res Commun 321: 172–178 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Ohta M, Matsui K, Ohme-Takagi M. (2002) The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett 514: 351–354 [DOI] [PubMed] [Google Scholar]
- Hong SW, Vierling E. (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA 97: 4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Ohme-Takagi M. (2009) A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol 50: 970–975 [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA. (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Kotak S, Port M, Ganguli A, Bicker F, von Koskull-Döring P. (2004) Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J 39: 98–112 [DOI] [PubMed] [Google Scholar]
- Kumar M, Busch W, Birke H, Kemmerling B, Nürnberger T, Schöffl F. (2009) Heat shock factors HsfB1 and HsfB2b are involved in the regulation of Pdf1.2 expression and pathogen resistance in Arabidopsis. Mol Plant 2: 152–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA. (2010) H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Kwon Y, Kim SH, Jung MS, Kim MS, Oh JE, Ju HW, Kim KI, Vierling E, Lee H, Hong SW. (2007) Arabidopsis hot2 encodes an endochitinase-like protein that is essential for tolerance to heat, salt and drought stresses. Plant J 49: 184–193 [DOI] [PubMed] [Google Scholar]
- Larkindale J, Vierling E. (2008) Core genome responses involved in acclimation to high temperature. Plant Physiol 146: 748–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Schöffl F. (1996) An Hsp70 antisense gene affects the expression of HSP70/HSC70, the regulation of HSF, and the acquisition of thermotolerance in transgenic Arabidopsis thaliana. Mol Gen Genet 252: 11–19 [DOI] [PubMed] [Google Scholar]
- Li M, Doll J, Weckermann K, Oecking C, Berendzen KW, Schöffl F. (2010) Detection of in vivo interactions between Arabidopsis class A-HSFs, using a novel BiFC fragment, and identification of novel class B-HSF interacting proteins. Eur J Cell Biol 89: 126–132 [DOI] [PubMed] [Google Scholar]
- Liu HC, Liao HT, Charng YY. (2011) The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ 34: 738–751 [DOI] [PubMed] [Google Scholar]
- Lohmann C, Eggers-Schumacher G, Wunderlich M, Schöffl F. (2004) Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol Genet Genomics 271: 11–21 [DOI] [PubMed] [Google Scholar]
- Meiri D, Breiman A. (2009) Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J 59: 387–399 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Ikeda M, Tanaka S, Takiguchi Y, Kondou Y, Yoshizumi T, Fujita M, Shinozaki K, Matsui M, Ohme-Takagi M. (2010) Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors. Plant Cell Physiol 51: 2145–2151 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17: 2993–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. (2006) Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J 48: 535–547 [DOI] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A, Nosaka R, Hayashi H, Tainaka H, Maruta T, Tamoi M, Ikeda M, Ohme-Takagi M, Yoshimura K, Yabuta Y, et al. (2011) HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiol 52: 933–945 [DOI] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD. (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port M, Tripp J, Zielinski D, Weber C, Heerklotz D, Winkelhaus S, Bublak D, Scharf KD. (2004) Role of Hsp17.4-CII as coregulator and cytoplasmic retention factor of tomato heat stress transcription factor HsfA2. Plant Physiol 135: 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S. (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. (2006) Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA 103: 18822–18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Heider H, Höhfeld I, Lyck R, Schmidt E, Nover L. (1998) The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol Cell Biol 18: 2240–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm F, Ganguli A, Kiehlmann E, Englich G, Walch D, von Koskull-Döring P. (2006) The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol Biol 60: 759–772 [DOI] [PubMed] [Google Scholar]
- Schramm F, Larkindale J, Kiehlmann E, Ganguli A, Englich G, Vierling E, von Koskull-Döring P. (2008) A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J 53: 264–274 [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Sakuma Y, Todaka D, Maruyama K, Qin F, Mizoi J, Kidokoro S, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K. (2008) Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem Biophys Res Commun 368: 515–521 [DOI] [PubMed] [Google Scholar]