Abstract
Medicago truncatula has been developed into a model legume. Its close relative alfalfa (Medicago sativa) is the most widely grown forage legume crop in the United States. By screening a large population of M. truncatula mutants tagged with the transposable element of tobacco (Nicotiana tabacum) cell type1 (Tnt1), we identified a mutant line (NF2089) that maintained green leaves and showed green anthers, central carpels, mature pods, and seeds during senescence. Genetic and molecular analyses revealed that the mutation was caused by Tnt1 insertion in a STAY-GREEN (MtSGR) gene. Transcript profiling analysis of the mutant showed that loss of the MtSGR function affected the expression of a large number of genes involved in different biological processes. Further analyses revealed that SGR is implicated in nodule development and senescence. MtSGR expression was detected across all nodule developmental zones and was higher in the senescence zone. The number of young nodules on the mutant roots was higher than in the wild type. Expression levels of several nodule senescence markers were reduced in the sgr mutant. Based on the MtSGR sequence, an alfalfa SGR gene (MsSGR) was cloned, and transgenic alfalfa lines were produced by RNA interference. Silencing of MsSGR led to the production of stay-green transgenic alfalfa. This beneficial trait offers the opportunity to produce premium alfalfa hay with a more greenish appearance. In addition, most of the transgenic alfalfa lines retained more than 50% of chlorophylls during senescence and had increased crude protein content. This study illustrates the effective use of knowledge gained from a model system for the genetic improvement of an important commercial crop.
Senescence of forage and turf species is of special interest because the market value of these widely grown plants is closely related to the visual appearance of their foliage. The most obvious phenomenon of plant senescence is leaf color change from green to yellow or red. Color change of leaves during senescence is caused by chlorophyll degradation, combined with carotenoid retention or anthocyanin accumulation (Park et al., 2007). Higher plants contain two types of chlorophylls, chlorophyll a (Chl a) and chlorophyll b (Chl b). The conversion of Chl b to Chl a is considered the first step of chlorophyll degradation (Hörtensteiner, 2009). Chl a is continuously converted by multistep pathways to linear colorless and nonfluorescent tetrapyrroles (Hörtensteiner, 2006).
stay-green (sgr) mutants with visible retention of chlorophyll have been identified from different plant species (Park et al., 2007; Ren et al., 2007; Alós et al., 2008; Sato et al., 2009). They have been classified into five types, A, B, C, D, and E, based on chlorophyll behavior during senescence (Thomas and Howarth, 2000). The most extensively investigated mutant is type C, in which chlorophylls are retained in senescent leaves while their photosynthetic competence decreases. This type of mutant was first found in a forage crop, meadow fescue (Festuca pratensis), and named senescence-induced degradation (Thomas and Stoddart, 1975; Thomas, 1987; Armstead et al., 2007). In recent years, type C stay-green-related genes have been identified in Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), pea (Pisum sativum), and other species and have been named SGR (Jiang et al., 2007; Park et al., 2007), NONYELLOWING (NYE1; Ren et al., 2007), GREEN-FLESH (GF), or CHLOROPHYLL RETAINER (Barry et al., 2008). Moreover, it has been shown that SGR cosegregates with the I locus controlling the green cotyledon trait in pea (Armstead et al., 2007; Sato et al., 2007), which was used as one of the seven traits by Gregor Mendel to establish his famous laws of genetic inheritance. SGR plays an important role in the dismantling of chlorophyll protein complexes during senescence (Park et al., 2007; Aubry et al., 2008). However, the exact function and mechanism of this family of SGR genes have not yet been elucidated.
Alfalfa (Medicago sativa) is widely cultivated in North America and is the third largest crop economically after corn (Zea mays) and soybean (Glycine max) in the United States (Bouton, 2007). Alfalfa is often referred to as “the queen of forage crops” because it is highly productive, drought tolerant, and provides high-quality forage over a long period of time. Most alfalfa is used for hay production. A key trait for the evaluation of hay value at the market is the color of alfalfa leaves. Bright green, instead of yellowish looking, is one of the physical characteristics of premium hay.
Alfalfa is an obligate outcrossing and tetraploid species (2n = 4x = 32); such features make genetic and genomic studies difficult and promoted the development of Medicago truncatula as a model legume. M. truncatula is a diploid, self-fertile species with a short life cycle and a small genome. Many tools and resources have been developed in M. truncatula, such as retrotransposon and fast-neutron mutant populations (Tadege et al., 2008), ecotype collections (Cook, 1999), EST and genespace sequencing information (Young et al., 2005; Young and Udvardi, 2009), and the Gene Expression Atlas (Benedito et al., 2008).
Like other legume species, alfalfa and M. truncatula have the ability to respond to nitrogen limitation by establishing symbiotic interactions with soil bacteria, collectively known as rhizobia, which are able to reduce atmospheric nitrogen (N2) to ammonium. Upon successful entry, rhizobia are hosted inside new organs, root nodules, where low oxygen levels allow the nitrogenase enzyme complex to carry out nitrogen fixation. When nodule cells senesce, nitrogen fixation ceases and the symbiotic relationship is lost. It has been demonstrated that nodule senescence is a complicated but regulated process, although the mechanism has not been elucidated. It is considered that crop yield and seed quality may benefit from delayed nodule senescence and the concomitant extended nitrogen fixation (Van de Velde et al., 2006).
In this study, we describe the identification and characterization of a sgr mutant from the M. truncatula population tagged with the transposable element of tobacco (Nicotiana tabacum) cell type1 (Tnt1). Genetic and molecular analyses revealed that the mutation was caused by Tnt1 insertion in the SGR gene of M. truncatula (MtSGR). Further analyses showed that SGR is involved not only in leaf senescence but also in nodule senescence. RNA interference (RNAi) was employed to silence the expression of the endogenous SGR gene (MsSGR) in alfalfa. The transgenic alfalfa plants retained much of their chlorophyll during senescence, stayed green during the natural drying process, and showed increased crude protein content.
RESULTS
Identification and Phenotypic Characterization of a M. truncatula sgr Mutant during Natural and Dark-Induced Senescence
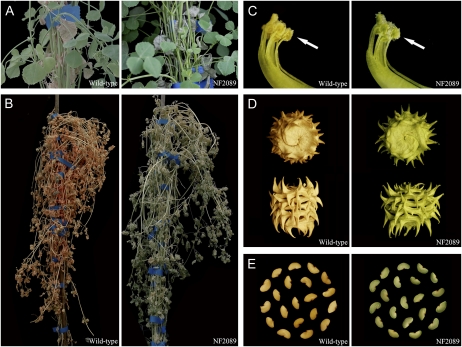
A mutant line, NF2089, caught our attention while screening large numbers (more than 10,000 lines) of the M. truncatula Tnt1 retrotransposon-tagged mutants. During natural senescence, the basal leaves of the wild type (ecotype R108) turned yellow first and the whole plant gradually became yellowish. However, the basal leaves of NF2089 did not show senescent yellow color in the same way as those of wild-type plants (Fig. 1A). Moreover, the whole plant remained green even after the leaves died (Fig. 1B). Interestingly, the stay-green phenomenon was evident not only in leaves but also in other organs, such as anthers and central carpels (Fig. 1C), mature pods (Fig. 1D), and seeds (Fig. 1E).
Figure 1.
Phenotypic characterization of the Tnt1-tagged M. truncatula mutant line NF2089. A, Natural senescence of leaves at the bottom of wild-type and NF2089 plants after 60 d of growth. B, Natural senescence of wild-type and NF2089 plants. C, Color change in mature anthers and central carpels of wild-type and NF2089 plants. D, Mature pods of wild-type and NF2089 plants. E, Mature seeds of wild-type and NF2089 plants.
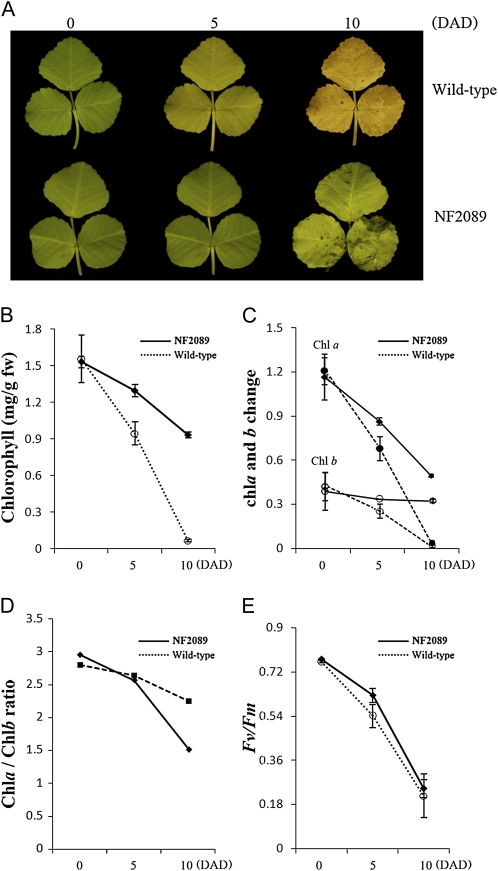
To determine if the NF2089 mutant exhibits a stable stay-green phenotype during dark-induced senescence, detached leaves of the mutant and wild type were placed in darkness for up to 10 d. The leaves of the mutant remained green on the 5th d after darkness and turned to light green after 10 d of dark treatment. In contrast, wild-type leaves became light green on the 5th d after darkness and turned yellow after 10 d of dark treatment (Fig. 2A).
Figure 2.
Stay-green phenotype and chlorophyll characterization of the NF2089 mutant during dark-induced senescence. A, Color change in detached leaves of wild-type and NF2089 plants during dark-induced senescence. B, Chlorophyll degradation during dark incubation. fw, Fresh weight. C, Changes in contents of Chl a and Chl b during senescence. Black circles, Chl a; white circles, Chl b. D, Chl a/b ratio of wild-type and NF2089 plants. E, Change in Fv/Fm values during dark incubation. Error bars indicate se (n = 3). Numbers (0, 5, and 10) indicate days after dark treatment (DAD).
To determine chlorophyll content and its possible impacts on leaf photosynthesis in the mutant, whole plants of the mutant and the wild type were transferred to a dark growth chamber. Much of the chlorophyll was retained in the mutant after dark treatment (Fig. 2B), with 43% of Chl a and 82% of Chl b retained after 10 d of dark treatment (Fig. 2C). The ratio of Chl a to Chl b in the mutant decreased at 10 d after dark treatment compared with that of the wild type (Fig. 2D). The maximal photochemical efficiency (Fv/Fm) of PSII, an important parameter of PSII activity, was also measured (Fig. 2E). The beginning Fv/Fm value before dark treatment was similar in both NF2089 and the wild type; while the value of the mutant was slightly higher than that of the wild-type after 5 d of dark treatment, the difference in Fv/Fm between the mutant and the wild type was not significant after 10 d of darkness. These results indicate that Fv/Fm in the mutant decreased in a similar way as the wild type during senescence, although the decrease in the mutant was slightly slower than that of the wild type.
Chloroplast Structure in Senescent Leaves
The ultrastructure of chloroplasts in both the wild type and the NF2089 mutant was examined using transmission electron microscopy. Under normal growth conditions, the structure of chloroplasts in the NF2089 mutant was similar to that in the wild type, both containing well-formed grana stacks along with distinct starch granules (Fig. 3, A, B, E, and F). After 10 d of dark treatment, clear differences between chloroplast structures of the wild type (Fig. 3, C and D) and the NF2089 mutant (Fig. 3, G and H) were observed. One difference was the large number of plastoglobules formed in the wild type, whereas only a few were observed in the mutant. Moreover, the structures of grana stacks became obscure in the wild type, while thick and wide grana stacks were still detected in chloroplasts of the NF2089 mutant. These results indicate that chloroplast decomposition in the NF2089 mutant was much slower than that in the wild type.
Figure 3.
Ultrastructure of chloroplasts in the wild type (A–D) and the NF2089 mutant (E–H). Chloroplast structures observed using transmission electron microscopy at 0 d (A, B, E, and F) and 10 d (C, D, G, and H) after dark treatment are shown. The insets show higher magnification images of chloroplast structures corresponding to the white squares in the main panels. G, Grana stack; P, plastoglobule; S, starch granule. Bars = 500 nm.
Molecular Cloning and Characterization of the SGR Gene in M. truncatula
To determine if the stay-green phenotype in NF2089 was caused by the mutation of a single gene, the mutant was crossed with the wild type and segregation analysis was performed in the progeny. The F1 plants did not show the stay-green phenotype when detached leaves were incubated in darkness. Segregation was observed in F1 seeds and F2 plants. The ratio of green versus yellow F1 seeds was close to 1:3 (148:457), and the ratio between stay-green F2 plants and non-stay-green F2 plants was approximately 1:3 (46:143; Supplemental Table S1). Furthermore, all the plants from the green seeds showed the stay-green phenotype after dark treatment, while the plants from yellow seeds did not show the phenotype. These results demonstrate that NF2089 is a recessive mutant and that the mutation was caused by the loss of function of a single gene. They also suggest that the stay-green phenotype in the leaves is associated with the green color of the seeds.
To determine which defective gene caused the stay-green phenotype in the mutant, thermal asymmetric interlaced PCR was performed to recover the flanking sequences of the Tnt1 retrotransposon in NF2089. In total, 13 retrotransposon insertions at different sites were recovered from the mutant. One of the insertions (NF2089-9) was found to segregate with the mutant phenotype, based on PCR genotyping (Supplemental Table S2). BLASTn analysis of the sequence flanking the Tnt1 insertion against the M. truncatula Gene Index database showed that NF2089-9 was located in a gene corresponding to TC126805. By reverse transcription (RT)-PCR and sequence analysis, the full-length coding sequence (792 bp) was obtained, which showed 87% identity with PsSGR (Sato et al., 2007), 76% identity with AtNYE1 (Ren et al., 2007), and 65% identity with rice SGR (Park et al., 2007). Sequence analysis of genomic DNA revealed that the gene, designated MtSGR, consists of four exons and three introns (Fig. 4A). PCR amplification of MtSGR from genomic DNA of the wild type and NF2089 confirmed that one 5.3-kb Tnt1 was inserted into this gene in NF2089 (Fig. 4B); therefore, the expression of MtSGR was abolished in the mutant (Fig. 4C). Analysis of transcription levels of MtSGR in wild-type plants showed that the expression of MtSGR was up-regulated during dark-induced senescence (Fig. 4D).
Figure 4.
Molecular cloning of the MtSGR gene. A, Diagram of the MtSGR gene (1,734 bp) structure showing the four exons (blocks), three introns (lines), and positions of Tnt1 insertions. B, PCR amplification of MtSGR from genomic DNA of the wild type (WT) and NF2089 showing the presence of a 5.3-kb Tnt1 insertion in the mutant. C, RT-PCR showing that transcription of MtSGR was interrupted in NF2089. D, Expression of MtSGR in leaves of the wild type. Numbers (0, 5, and 10) indicate days after dark treatment (DAD).
Two other Tnt1 mutants with insertions in MtSGR, NF6817 and NF8082, were obtained via a reverse genetics approach. The insertion sites in MtSGR of NF6817 and NF8082 were in the second intron and the promoter region, respectively (Fig. 4A). Homozygous NF6817 and NF8082 plants exhibited similar phenotypes as the wild type, indicating that the Tnt1 insertions of these lines did not impair MtSGR gene function.
The expression pattern of MtSGR in wild-type plants was analyzed by utilizing the M. truncatula Gene Expression Atlas (Benedito et al., 2008). The expression of MtSGR was detected in almost all the organs at various developmental stages, with higher levels in mature seeds and senescing nodules and relatively low levels in flowers, petioles, stems, pods, vegetative buds, leaves, roots, and young seeds (Supplemental Fig. S1).
Global Gene Expression Profiling of the NF2089 Mutant
To investigate if MtSGR affected downstream genes during dark-induced leaf senescence in M. truncatula, a microarray analysis was performed using the Affymetrix Medicago Genome Arrays. After 5 d of dark treatment, fully expanded leaves of both the NF2089 mutant and a corresponding control were used to isolate RNA for chip analysis. Compared with the control, 347 genes were repressed, and 1,251 genes were induced significantly by at least 2-fold in the NF2089 mutant (Supplemental Table S3). These data suggest that MtSGR probably mostly plays inhibitory roles in gene expression. The expression levels of 12 putative senescence-associated genes were changed, and all of them were up-regulated in NF2089 (Supplemental Table S4). Some of up-regulated genes are predicted to participate in the processes of cellular protein complex assembly, protein polymerization, and protein modification by small protein conjugation or removal (Supplemental Fig. S2), supporting the view that SGR is involved in the disassembly process of the light-harvesting chlorophyll-binding protein complexes (Park et al., 2007; Borovsky and Paran, 2008). On the other hand, the expression levels of M. truncatula homologs of chlorophyllase, pheide a oxygenase, and red chlorophyll catabolite reductase did not show any obvious changes in the mutant, indicating that the absence of MtSGR does not affect the expression of these genes in the chlorophyll degradation pathway.
Only 2.5% of up- or down-regulated genes (40 of 1,598) encode chloroplast-related proteins (Supplemental Table S5). This result implies that MtSGR may participate in broad biological processes during leaf senescence besides chlorophyll degradation. Gene Ontology (GO) analysis was performed using the GO Enrichment Analysis Software Toolkit (Zheng and Wang, 2008). The results showed that 168 GO classes and 49 GO classes were enriched, respectively, in up-regulated and down-regulated genes in the NF2089 mutant. The representations of probe sets in the enriched GO classes were examined with WEGO (Ye et al., 2006), and the number of genes up-regulated or down-regulated in each category was calculated (Supplemental Fig. S2). Different biological processes, cellular components, and molecular functions were found to be substantially affected in senescing leaves when MtSGR was absent (Supplemental Table S6). The involvement of multiple function categories suggests that MtSGR plays a broad role in plant development and senescence.
The Involvement of MtSGR in Nodule Senescence
The above-mentioned Gene Expression Atlas analysis revealed that MtSGR expression is relatively high in senescing nodules, second only to mature seeds (Supplemental Fig. S1). To test a possible association of MtSGR with nodule senescence, nodulated roots of the wild type and the NF2089 mutant were analyzed 4 weeks after inoculation with Sinorhizobium meliloti ABS7 strain. No obvious phenotypic differences between senescent nodules of the wild type and the NF2089 mutant were observed. Both nodule size and the extent of the senescence zone, as indicated by the greenish appearance at the proximal end of the nodule, were similar in the wild type and the mutant (Supplemental Fig. S3). Although the number of senescent nodules in the NF2089 mutant was similar to that in the wild type, at closer examination, we found a significantly higher number of young nodules on the mutant roots (P < 0.01; Fig. 5A). These young nodules were pink with no senescence zone (Supplemental Fig. S3), implying a potential correlation of MtSGR with the onset of nodule senescence. Furthermore, the phenotype of senescent nodules of the mutant was similar to that of the wild type, suggesting that other factors may also be involved in the process of nodule senescence.
Figure 5.
The involvement of MtSGR in nodule senescence of M. truncatula. A, Nodule number of the wild type and the NF2089 mutant. The two asterisks indicate a significance level corresponding to P < 0.01. B, Expression level of MtSGR in young nodules and senescing nodules of the wild type. C, Expression level of MtSGR in NO3-induced senescing wild-type nodules. dpi, Days after rhizobial inoculation. D, Different developmental zones of the wild-type nodule. E, Expression level of MtSGR in the different developmental zones of the wild-type nodule shown in D. F, Expression level of nodule senescence marker genes in senescing nodules of the wild type and the NF2089 mutant. Error bars indicate se (n = 20) in A and se (n = 3) in B, C, E, and F.
To study the relationship between MtSGR expression and nodule senescence, quantitative RT-PCR was used to analyze gene expression in nodules during natural senescence and NO3-induced senescence in M. truncatula. The relative expression level of MtSGR was measured in young nodules and naturally senescing nodules harvested 4 weeks postinoculation with rhizobia. MtSGR transcript levels in naturally senescing nodules was 2-fold higher than in young nodules (Fig. 5B). In addition, compared with naturally senescing nodules, the MtSGR transcript level was 8-fold higher in NO3-induced senescence (after 5 d of treatment; Fig. 5C).
MtSGR expression was also assessed in wild-type nodules along five functional zones corresponding to well-defined stages of rhizobial infection and nodule development. These zones are as follows: meristem; the invasion zone, where rhizobia are released from the tube-like structures called infection threads into the host cell cytoplasm and are surrounded by plant cell-derived membranes to form a new organelle, the symbiosome; interzone II-III, where rhizobia mature into rod-shaped, nitrogen-fixing bacteroids; the nitrogen fixation zone, where bacteroids actively reduce atmospheric nitrogen; and the senescence zone, where nitrogen fixation ceases and both bacteria and plant cells die (Fig. 5D). Quantitative RT-PCR analysis showed that MtSGR transcripts were present in all nodule zones, with the highest levels in the senescence zone (Fig. 5E).
To confirm that elevated levels of MtSGR expression in the senescence zone could link this gene function with degenerative processes of nodule senescence, we measured the expression of several nodule senescence markers in wild-type and NF2089 senescing nodules. These markers were selected on the basis of senescence-specific expression from a Medicago Affymetrix GeneChip data set obtained from spatially resolved nodule zones (C. Pislariu and M. Udvardi, unpublished data; Supplemental Fig. S3). Probe set target descriptions are listed in Supplemental Table S7. Quantitative RT-PCR analysis showed that seven out of nine markers were significantly down-regulated in the sgr mutant, while the expression of the other two markers was similar in mutant and wild-type senescent nodules (Fig. 5F). The most down-regulated are a phosphate transporter (Mtr.15323.1.S1_s_at) and a basic blue copper protein (Mtr.10725.1.S1_at), which may be involved in phosphate and copper salvage during senescence (Miller et al., 1999; Chapin and Jones, 2009). A putative ripening-related protein (Mtr.10361.1.S1_at) and a Cys proteinase (Mtr.43787.1.S1_at) have unchanged relative expression in NF2089 compared with the wild type (Fig. 5F).
Taken together, our findings suggest that MtSGR is possibly involved in certain nodule development and senescence pathways.
Isolation of an SGR Gene from Alfalfa
An SGR gene was cloned from alfalfa (designated MsSGR) based on the high sequence similarity between M. truncatula and alfalfa. Analysis of the deduced amino acid sequence revealed that the MsSGR protein contains 263 amino acids, with predicted pI of 8.716 and molecular mass of 30 kD. Predicted SGR protein sequences from several species were collected from GenBank and used for phylogenetic analysis (Supplemental Fig. S4A). The phylogenetic trees were rooted using Physcomitrella patens (moss) SGR and divided into two clades: one belonging to monocotyledonous species and the other belonging to dicotyledonous species. Phylogenetic analysis showed that MsSGR and MtSGR were closest to each other and were clustered close to PsSGR (Supplemental Fig. S4A). The MsSGR sequence showed 98% identity to MtSGR, 86% to PsSGR, 78% to GmSGR1, 67% to AtNYE1, and 64% to OsSGR. The SGR family members share a highly conserved central region but are divergent at N and C termini. The amino acid Arg-145 (Supplemental Fig. S4B, asterisk), at which site the Tnt1 was inserted into the mutant NF2089, is an invariant residue within the SGR family.
Suppression of MsSGR Expression in Alfalfa by RNAi
To suppress the activity of the endogenous MsSGR in alfalfa, an MsSGR-RNAi vector was constructed and introduced into alfalfa plants by Agrobacterium tumefaciens-mediated transformation. Twenty transgenic lines were identified through PCR analysis (Fig. 6A). Quantitative RT-PCR analysis revealed that five transgenic lines, SGRi-10, SGRi-17, SGRi-21, SGRi-29, and SGRi-39, had SGR transcript levels reduced by more than 60% when compared with the empty vector control line (Fig. 6B). These five transgenic alfalfa lines were used for further analyses.
Figure 6.
Molecular characterization of alfalfa MsSGR-RNAi transgenic lines. A, PCR analysis of regenerated alfalfa plants together with the positive control (pANDA35HK-SGR plasmid), negative control (Wild-type), and empty vector control (CTRL). The sizes of DNA fragments are 699 bp for nptII, 375 bp for hph, and 633 bp for gus linker. B, Quantitative RT-PCR analysis of MsSGR gene expression in transgenic lines. All values were normalized using the empty vector control. Error bars indicate se (n = 3). C, Southern-blot analysis of KpnI-digested genomic DNA from leaves of the wild type and MsSGR-RNAi transgenic lines. The DNA was probed with 699 bp of gus linker fragment from the pANDA35HK vector.
Southern-blot hybridization analysis confirmed that the transgene was stably integrated in the alfalfa genome and that the regenerated positive lines were truly independent transformants. Both single-copy and multiple-copy integrations of the transgene were observed in the transgenic lines. Transgenic line SGRi-21 had single-copy integration; SGRi-29 contained two copies of the transgene; and SGRi-10, SGRi-17, and SGRi-39 had at least three copies of the transgene (Fig. 6C).
Leaf Senescence of the MsSGR-RNAi Transgenic Alfalfa Plants
The effects of MsSGR down-regulation on transgenic alfalfa were analyzed by incubating detached leaves in darkness to induce senescence. The transgenic lines exhibited a stable nonyellowing phenotype during a continuous 20-d dark treatment. Although the detached leaves from the transgenics turned light green on the 5th d of dark incubation, the green color remained until the end of the 20-d treatment (Fig. 7A). In contrast, the empty vector control and wild-type leaves began to turn yellow by the 5th d, and the yellow color spread gradually to full leaves by the 10th d of dark treatment.
Figure 7.
Dark incubation of alfalfa MsSGR-RNAi transgenic lines. A, Detached leaves of the wild type, empty vector control (CTRL), and MsSGR-RNAi transgenic lines in the dark. B, Living plants of the wild type and a transgenic line (SGRi-39) in the dark. Numbers (0, 5, 10, 15, and 20) indicate days after dark treatment (DAD).
To determine the effects of MsSGR suppression at the whole plant level, live transgenic plants were placed in a dark growth chamber. During natural senescence of alfalfa plants, the lower leaves usually senesce earlier than do the upper leaves. A similar phenomenon was observed in dark-induced senescence in alfalfa. On the 5th d of dark treatment, basal leaves of the control plants began to turn yellow, while the leaves of all the transgenic lines remained green. On the 10th d of dark treatment, whole plants of the controls became yellowish, with some basal leaves turning red, while the transgenic lines kept the stay-green phenotype, although some of the leaves at the top became wrinkled. After 15 and 20 d of dark treatment, almost all the leaves in the control became yellow and red, while the leaves of the transgenic lines remained a similar green color to that of the 10th d of dark treatment (Fig. 7B).
Transmission electron microscopy analysis revealed that chloroplast outline, shape, and content in the transgenic alfalfa lines showed similar temporal changes as did the M. truncatula NF2089 mutant during dark-induced senescence (Supplemental Fig. S5).
To determine if delayed leaf senescence was associated with the repression of MsSGR expression, the transcript level of MsSGR was measured in leaves of the wild type and the SGRi-39 line under dark treatment (Supplemental Fig. S6). MsSGR was up-regulated dramatically in leaves of the wild type, especially after 5 and 10 d in darkness. The expression level of MsSGR in SGRi-39 varied slightly and was consistently lower than that of the control at different time points of the dark treatment.
Under natural growth conditions, as plants grow, the old leaves lose function and drop off the plant. A color difference was observed in these fallen leaves, with control leaves yellow while transgenic leaves remained green (Supplemental Fig. S7).
Chlorophyll Content and Physiological Changes in Transgenic Alfalfa during Dark Incubation
Chlorophyll content, Chl a/b ratio, and PSII functionality (measured as Fv/Fm) were measured during dark treatment of living transgenic alfalfa plants. No difference was observed in chlorophyll loss after 5 d of dark treatment (Fig. 8A). Significant changes were observed, however, after 10 d of dark treatment. By the 20th d of dark treatment, only 5% of chlorophylls remained in the control leaves, while more than 50% of chlorophylls remained in most of the MsSGR-RNAi lines (50.6% in SGRi-10, 52.8% in SGRi-21, 55.6% in SGRi-29, and 60.2% in SGRi-39). Among the transgenic lines, SGRi-39 retained the highest level of chlorophylls (60.2%), which was double the amount of chlorophylls retained in SGRi-17 (30.0%).
Figure 8.
Measurement of chlorophyll content and Fv/Fm values in control and independent MsSGR-RNAi transgenic alfalfa lines during dark incubation. A, Change of chlorophyll content during dark incubation. fw, Fresh weight. B, Chl a/b ratio of the same samples as in A. C, Fv/Fm values. CTRL, Control plants. Numbers (0, 5, 10, 15, and 20) indicate days after dark treatment (DAD). Error bars indicate se (n = 3).
No difference in Chl a/b ratio between transgenic and control plants was observed after 0 and 5 d of dark treatment. However, after 10 and 20 d of darkness, the RNAi lines showed large decreases in Chl a/b ratios (Fig. 8B). As shown in Figure 8C, Fv/Fm values decreased similarly during the course of senescence in the transgenic lines and the control.
Improvement of Forage Quality of MsSGR-RNAi Transgenic Alfalfa
When harvesting alfalfa for hay production, the fresh-cut plants are usually dried and baled in the field. To evaluate the potential application of MsSGR-RNAi alfalfa, we mimicked the harvest and drying process using these transgenic materials. The transgenics were greener than the control after 5 d of drying. This difference in visual quality of the transgenics and the control became more obvious after 10 d of drying, with transgenics showing a more attractive greenish color (Fig. 9).
Figure 9.
Phenotypic appearance of control and MsSGR-RNAi transgenic alfalfa under natural drying conditions. A, Control plants. B, SGRi-39 transgenic line. 0d, 5d, 10d, 15d, and 20d indicate days after cutting of the materials.
Forage nutritive quality analysis revealed that most of the transgenic lines (except SGRi-17) had increased crude protein content (Fig. 10). Compared with the control, the level of increase in crude protein content of SGRi-29, -21, -10, and -39 varied from 2.3% to 5.5%. Other nutritive quality traits, including in vitro true dry matter digestibility, total digestible nutrients, acid detergent fiber content, neutral detergent fiber content, magnesium concentration, and phosphorus concentration, did not show consistent changes between the transgenic lines and the control (Supplemental Fig. S8).
Figure 10.
Crude protein content of control (CTRL) and MsSGR-RNAi transgenic alfalfa lines. Five-week-old plants were collected for analysis. Error bars indicate se (n = 3). Columns labeled with the same letters are not significantly different at P = 0.05.
DISCUSSION
Both M. truncatula Mutant NF2089 and MsSGR-RNAi Alfalfa Show Nonfunctional Type C Stay-Green Phenotype during Senescence
A stay-green mutant, NF2089, was identified through screening a M. truncatula mutant collection generated by Tnt1 retrotransposon insertional mutagenesis. The stay-green phenotype is caused by a Tnt1 insertion in the MtSGR gene, which is induced by senescence and responsible for chlorophyll breakdown. During both natural and dark-induced senescence, the sgr mutant showed the stay-green phenotype not only in leaves, stems, mature pods, and seeds, as reported in other species (Sato et al., 2007; Aubry et al., 2008), but also in anthers and central carpels, which has not been reported before. In addition, we isolated MsSGR from alfalfa and produced transgenic plants with an MsSGR-RNAi construct. The transgenic alfalfa with down-regulated expression of MsSGR showed the same stay-green phenotype.
In both the sgr M. truncatula mutant and MsSGR-RNAi transgenic alfalfa, a large portion of Chl a and Chl b was retained after senescence, suggesting that the SGR gene plays an important role in degradation mechanisms of both Chl a and Chl b in the two species. Although much of the chlorophylls were retained after senescence, the Fv/Fm values of the mutant and the transgenics decreased in a similar manner, as did those of the controls. These results indicate that the sgr mutant and MsSGR-RNAi transgenic alfalfa showed the nonfunctional type C stay-green phenotype during senescence.
SGR Is Implicated in Nodule Development and Senescence
In indeterminate nodules such as those of M. truncatula, where an active meristem is permanently maintained, functional zonation along the nodule is easily distinguishable. Adjacent to the meristem is the invasion zone, where rhizobia are released from infection threads into the host cell cytoplasm and are entrapped by plant-derived membranes in symbiosomes. Proximal to the invasion zone is interzone II-III of maturation and the onset of nitrogen fixation. The nitrogen fixation zone constitutes the bulk of the nodule and consists of large host cells packed with functional bacteroids alternating with uninfected plant cells. The characteristic pink color of this zone is given by leghemoglobin, a high-affinity oxygen carrier that helps to establish the low oxygen levels required for nitrogenase activity. At a certain stage during nodule development, nitrogen fixation ceases and senescence is triggered in the proximal part of the nodule adjacent to the root (Van de Velde et al., 2006). Nodule senescence is a complex process that has not yet been clarified. In our study, the expression of MtSGR was found to be low in most organs but was induced during seed maturation and nodule senescence. Because SGR is associated with seed color, its increased expression during seed maturation is not surprising. What was unexpected was the increase in MtSGR expression during nodule senescence. Nodule senescence was initiated after NO3 treatment, and the expression level of MtSGR was induced 8-fold in M. truncatula as a result. Increased expression of SGR in M. truncatula was also associated with natural senescence in older nodules.
We also found that a selection of nodule senescence markers displayed differential expression patterns in the sgr mutant compared with wild-type senescing nodules. This finding points to a possible role of MtSGR in certain steps of nodule senescence. The fact that MtSGR regulates leaf senescence and also appears to be involved in nodule senescence is not completely surprising. Previously, cDNA amplification fragment length polymorphism analysis identified 545 transcripts that were differentially expressed during M. truncatula leaf senescence (De Michele et al., 2009) and 508 transcripts that were regulated during M. truncatula nodule senescence (Van de Velde et al., 2006). While the overlap between these two processes in terms of the gene families involved was found to be relatively low, only 7% of the leaf data set, these findings indicate the existence of conserved mechanisms governing both leaf and nodule senescence (De Michele et al., 2009).
In addition to being up-regulated during nodule senescence, MtSGR is also expressed in all nodule zones in the wild type, suggesting that SGR may play a broader role during nodule development and is not restricted to senescence.
MtSGR Plays a Broad Role in the Plant Senescence Process
It is known that leaf senescence is accompanied by the degradation of proteins, lipids, and nucleic acids (Buchanan-Wollaston et al., 2003). Our microarray experiments revealed nearly 1,600 genes that were either down- or up-regulated in the sgr mutant. Based on the GO analysis, we observed that the MtSGR gene directly or indirectly affects the expression of a large portion of genes involved in different metabolic processes and biosynthetic processes. These data support that MtSGR plays multiple roles in the senescing process and may explain the multiple effects on other organs, including anthers, seeds, and seed pods, in the mutant. Transcript profiling was used to characterize a stay-green mutant (nan) of Citrus sinensis. Detailed analysis showed that the nan mutation is distinct from sgr (Alós et al., 2008). The global profiling analysis of the mtsgr mutant provides valuable data for further research in elucidating functional mechanisms of SGR.
To date, the mechanism of SGR function remains unknown (Hörtensteiner, 2009). Transcript profiling revealed that only a small portion (2.5%) of the genes with altered expression are associated with chloroplast-related proteins, indicating that the role of SGR may extend beyond chlorophyll degradation. Since no obvious change was observed in genes involved in the chlorophyll degradation pathway, it is unlikely that SGR directly affects a specific component of the chlorophyll catabolism pathway. Furthermore, the number of repressed genes was much smaller than the number of up-regulated genes in the mutant, indicating that SGR may play a preferential role in gene repression. Furthermore, MtSGR is involved in nodule development and senescence, indicating that MtSGR may be related to nitrogen remobilization. In Arabidopsis and rice, SGR has been shown to localize to the chloroplast (Park et al., 2007; Ren et al., 2007). Therefore, it is possible that the changes in transcript profiling of the sgr mutant are the consequences of secondary effects. A clearer picture can be drawn by comparing transcript profiling data of different species after such information becomes available in other plants.
Down-Regulation of MsSGR Improves the Forage Quality of Alfalfa
Cultivated alfalfa or lucerne is the most important forage legume in the United States, with approximately 9.6 million ha in production and an estimated value for alfalfa hay alone of US$8.1 billion (Bouton, 2007). Most of the alfalfa grown in the United States is used for hay production. Color is an important characteristic of hay. The ideal color is one that most closely resembles the bright green color of an immature legume crop in situ. Senescence causes leaf yellowing and reduces the market value of hay. Because alfalfa is a tetraploid species with a high degree of self-incompatibility, recessive mutations can be easily masked after crossing. To date, there has been no report of successful conventional breeding of alfalfa with delayed leaf senescence. In this study, silencing the MsSGR gene using the RNAi approach led to the production of stay-green transgenic alfalfa. This beneficial trait offers the opportunity to produce premium alfalfa hay with a more greenish appearance.
Another important aspect of forage quality is the actual nutritive value of hay. Routine forage analyses usually include levels of crude protein, dry matter digestibility, acid detergent fiber, neutral detergent fiber, total digestible nutrients, as well as mineral contents (Collins and Fritz, 2003). Leaf senescence negatively affects the nutritional quality of forage crops. Chlorophyll degradation and the progressive loss of protein in the chloroplasts happen during senescence and chloroplast degeneration (Lim et al., 2007). It has been shown that the leaf stay-green feature of the rice sgr mutant is associated with a failure in the destabilization of the light-harvesting chlorophyll-binding protein complexes (Park et al., 2007). Thus, delayed chlorophyll breakdown during leaf senescence may benefit the conservation of leaf protein. This hypothesis was supported by the data of nutritive quality analysis in MsSGR-RNAi transgenic alfalfa. Most of the transgenic lines showed a high level of chlorophyll retention and a simultaneous significant increase in crude protein content. Furthermore, the analysis of chloroplast ultrastructure also supported that the chloroplast breakdown was delayed in senescent leaves of transgenic alfalfa. Because the alfalfa plants used for forage quality analysis were not inoculated with rhizobia and grown in soil with full nutrition, the increase in crude protein content in transgenic alfalfa is not related to the nitrogen-fixing nodules. These data suggest that the crude protein content is positively associated with chlorophyll retention in transgenic alfalfa and that the increased chlorophyll stability has more positive impact beyond the visual phenotype. On the other hand, down-regulation of MsSGR did not have any obviously negative impact on plant phenotype and other quality traits, such as in vitro dry matter digestibility and total digestible nutrients. These results further support the potential usefulness of the transgenic materials for alfalfa improvement.
In summary, we identified stay-green mutants in the model legume M. truncatula, characterized the SGR gene, and successfully applied the knowledge to alfalfa improvement. We found that SGR not only played an important role in leaf senescence but was also involved in nodule senescence. Transcript profiling revealed that large numbers of genes were either up-regulated or down-regulated in the mutant. Significant improvement in forage quality was achieved in the transgenic alfalfa lines down-regulated with MsSGR. One expected outcome from the study of the model plant Medicago truncatula is that the information obtained would benefit alfalfa improvement (Choi et al., 2004; Zhu et al., 2005). This study illustrates the effective use of a model system for the genetic improvement of an important commercial crop.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Generation of the Medicago truncatula Tnt1 insertional mutant population was described previously by Tadege et al. (2008). M. truncatula ecotype R108 was used as the wild type. Mutant and wild-type seeds were scarified with concentrated sulfuric acid and treated at 4°C for 5 d on filter paper. Small plantlets were transferred to Metro-Mix 830 soil mix and grown in the greenhouse or growth chamber under the following conditions: 24°C day/22°C night temperature, 16-h/8-h photoperiod, 70% relative humidity, and 150 μmol m−2 s−1 light intensity.
An alfalfa (Medicago sativa) genotype, Regen SY-4D, was used for Agrobacterium tumefaciens-mediated transformation to produce transgenic plants. Both transgenic and wild-type alfalfa plants were vegetatively propagated using shoot cuttings. All plants were grown in a greenhouse at 24°C/22°C with a 16-h/8-h photoperiod and relative humidity of 70%.
Both detached leaves and whole plants were used for dark-induced senescence experiments. For the senescence treatment with detached leaves, fully expanded leaves were excised from M. truncatula and alfalfa plants and then incubated on sterilized wet filter paper at 25°C in the dark for up to 20 d. For the senescence treatment with living plants, 7-week-old M. truncatula and alfalfa plants were placed in a growth chamber without light. Chlorophyll content was measured every 5 d.
To mimic the harvesting and drying process of alfalfa hay, aboveground parts of 5-week-old transgenic and control alfalfa lines were cut off and put outside in a well-ventilated area; the average temperature was 27°C during the drying period.
Rhizobium Inoculation and Bacterial Strains
Seeds of M. truncatula wild type (R108 and A17) and Tnt1 insertion mutant NF2089 were germinated at room temperature in the dark for 24 h. Seedlings were planted in a mixture of turface and vermiculite (2:1, v/v) and transferred to a growth chamber set to the following conditions: 16-h/8-h photoperiod, 22°C temperature, 40% humidity, and 200 μmol m−2 s−1 light intensity. Fertilization was carried out with half-strength B&D nutrient solution (Broughton and Dilworth, 1971) containing 0.5 mm KNO3 (low nitrogen). Five-day-old seedlings were inoculated with Sinorhizobium meliloti wild-type strain ABS7. Nodulation phenotype and natural nodule senescence were evaluated at 28 d after inoculation. Induced nodule senescence was assessed at 33 d after inoculation (5 d after application of 2 mm NH4NO3).
The following nodule zones, meristem, invasion zone, interzone II-III, nitrogen fixation, and senescence zones, were dissected out from fresh, senescing wild-type A17 nodules, immediately frozen in liquid nitrogen, and stored at −80°C before carrying out RNA extractions.
Screening of a M. truncatula Stay-Green Mutant and Cloning of MtSGR
A mutant line, NF2089, was identified from a Tnt1 insertional population (more than 10,000 lines) of M. truncatula based on the segregation of leaf and seed color, green versus yellow. Tnt1 flanking sequences of mutant NF2089 were recovered using the thermal asymmetric interlaced PCR method (Liu et al., 2005; Zhou et al., 2011). The PCR products were purified and cloned into pGEM-T Easy vector (Promega) for sequencing. The flanking sequences were BLASTed against the M. truncatula genome sequence at the National Center for Biotechnology Information (Supplemental Table S2). Based on the flanking sequences recovered at the ninth insertion, the sequence of MtSGR was obtained from the M. truncatula Gene Index (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=medicago). Genomic sequences and the coding sequences of MtSGR were obtained through PCR and RT-PCR amplification using primers Mtsgr-F and Mtsgr-R (Supplemental Table S8). The amplifications were performed using Ex Taq polymerase (TaKaRa). The Tnt1 insertion site in the M. truncatula genome was confirmed using primers Mtsgr-F and Mtsgr-R coupled with Tnt1 border primers Tnt1-F and Tnt1-R (Supplemental Table S8). Homozygous or heterozygous status of MtSGR in M. truncatula plants was checked using the primer pair Mtsgr-F and Mtsgr-R. Primers Mtsgr-2F and Mtsgr-2R were coupled with Tnt1 border primers for reverse screening of other mtsgr mutants. Two other Tnt1 mutants, NF8082 and NF6817, were identified.
Phylogeny Analysis, Protein Alignment, and Expression Pattern of MtSGR
SGR amino acid sequences from various plant species were directly downloaded from GenBank (http://www.ncbi.nlm.nih.gov) or retrieved via ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). All sequences were subsequently aligned using the MAFFT program and the G-INS-i strategy (http://align.bmr.kyushu-u.ac.jp/mafft/software/). The resulting alignment was edited with GeneDoc software version 2.6.02 (http://www.nrbsc.org/gfx/genedoc/; Nicholas et al., 1997). A phylogenetic tree was built using the neighbor-joining method with a pairwise deletion option and 1,000 replicate analyses by employing the MEGA 4.0 software (Kumar et al., 2008). In addition, the phylogeny of these plant SGRs was reevaluated using the maximum likelihood method with the JTT model and 100 replicates (Guindon and Gascuel, 2003). Both trees showed complete agreement to each other, and bootstrap support values were printed on each branch.
SGR sequences of moss were obtained using the tBLASTn program against http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html, with AtNYE1 protein sequence as query. Protein sequences of moss were then predicted by Genescan at http://genes.mit.edu/GENSCAN.html. The resulting protein sequences were compared with other plant SGRs by protein alignment analysis and were further annotated manually to fix errors in accordance with the GT-AG rule. Consequently, these moss protein sequences were included in the phylogenetic analysis.
The coding sequence of MtSGR was used for the analysis of expression pattern based on the M. truncatula Gene Expression Atlas through the BLAST function (http://bioinfo.noble.org/gene-atlas/v2).
Quantification of Chlorophyll and Photochemical Efficiency
First fully expanded leaves (0.1 g) were immediately frozen and ground in liquid N2 quickly. Chlorophyll was extracted with 3 mL of 80% acetone containing 1 μm KOH (Schelbert et al., 2009). After centrifugation (10,000g, 2 min), the supernatant was quantified using a spectrophotometer (Arnon, 1949).
The live plants were placed in a dark growth chamber. For M. truncatula, the duration of darkness was 0, 5, and 10 d. For alfalfa, the duration of darkness was 0, 5, 10, 15, and 20 d. Fv/Fm was measured at the different time points using the LI-COR 6400 photosynthesis system (Li-Cor) with a CO2 concentration of 380 μmol mol−1 and a flow rate of 400 μmol s−1 (Jiang et al., 2007). For measurement of Fv/Fm at 0 d, the plants were adapted in darkness for 30 min.
Chloroplast Observation by Transmission Electron Microscopy
Leaf tissues (5 mm × 5 mm) were fixed in 3% (v/v) glutaraldehyde (Electron Microscopy Sciences) in 1× phosphate-buffered saline buffer (pH 7.0) for 12 h at 4°C. The samples were then washed with 1× phosphate-buffered saline and postfixed in buffered 1% (v/v) osmium tetroxide (Electron Microscopy Sciences) for 2 h at 0°C. Then, all specimens were washed, dehydrated in a series of ethanol, and embedded in LR White resin (London Resin Co.). The resin was polymerized at 55°C for 3 d. Ultrathin sections (0.1 μm thick) were cut with a diamond knife on an MT-X ultramicrotome (Boeckeler Instruments) and put onto formvar-carbon coated copper grids. They were stained with saturated uranyl acetate for 20 min followed by staining with Sato’s lead for 2 min. The specimens were observed with a transmission electron microscope operated at 80 kV (JEOL 2000FX; JEOL).
Construction of pANDA35HK-SGR Vector and Alfalfa Transformation
A 543-bp fragment of SGR was PCR-amplified from alfalfa using primers MsSGR-F and MsSGR-R (Supplemental Table S8). The fragment was inserted into pENTR/D-TOPO cloning vector (Invitrogen) and transferred into the pANDA35HK vector by attL × attR recombination reactions (Invitrogen).The final binary vector, pANDA35HK-SGR, was transferred into Agrobacterium strain EHA105 using the freezing/heat-shock method.
Transgenic alfalfa plants were obtained by Agrobacterium-mediated transformation (Austin et al., 1995). Alfalfa lines transformed with the original pANDA35HK vector were used as an empty vector control.
RNA Extraction and Quantitative RT-PCR Analysis
Total RNA was extracted from leaves or nodules of alfalfa or M. truncatula using Trizol Reagent (Invitrogen) followed by chloroform extraction, isopropanol precipitation, and quantification with a NanoDrop spectrophotometer (ND-1000). After treatment with Turbo DNase I (Ambion), 1 μg of RNA was reverse transcribed with SuperScriptIII (Invitrogen). The cDNA was diluted 1:20 and subsequently used as a template for quantitative RT-PCR. The 10-μL reaction mix included 2 μL of primers (0.5 μm each primer), 5 μL of Power Sybr (Applied Biosystems), 2 μL of diluted cDNA, and 1 μL of water. The primers used for quantitative RT-PCR are listed in Supplemental Table S8. Quantitative RT-PCR data were analyzed with SDS 2.2.1 software (Applied Biosystems). PCR efficiency (E) was estimated using LinRegPCR software (Ramakers et al., 2003), and the transcript levels were determined by relative quantification (Pfaffl, 2001) using the M. truncatula Actin gene (TC107326) as a reference.
Microarray Analysis
Plants of the NF2089 mutant and the corresponding control (wild-type-like sibling from the same line) were kept in darkness for 5 d, then RNA was isolated from the fully expanded leaves of both the NF2089 mutant and the control. RNA was extracted from triplicate biological replicates of the above samples, and 10 μg of purified RNA samples, in total, was used for microarray analysis. Probe labeling, hybridization, and scanning for microarray analysis were conducted according to the manufacturer’s instructions (Affymetrix; http://www.affymetrix.com). Functional analysis of differentially expressed genes from microarray data was performed using the GOEAST program, which by default adjusts the raw P values into a false discovery rate using the Benjamini-Yekutieli method (Zheng and Wang, 2008; http://omicslab.genetics.ac.cn/GOEAST). The enriched GO annotation results were then classified using WEGO (Ye et al., 2006).
Southern-Blot Analysis
Genomic DNA was extracted from each transgenic line and the wild type using the cetyl-trimethyl-ammonium bromide method and then treated with RNase (Qiagen). A 50-ng aliquot of plasmid DNA and 15 μg of purified DNA of each line were digested with restriction endonuclease KpnI, electrophoresed on 0.8% agarose gels, and transferred to positively charged nylon membranes (Roche) by alkaline capillary blotting. The hybridization probe (gus) was labeled with digoxigenin by PCR, and hybridization was carried out using the DIG Luminescent Detection kit (Roche).
Forage Analysis of Transgenic Alfalfa Lines
Transgenic and control alfalfa were grown in the soil with full nutrition. The 5-week-old plants were harvested and dried immediately. After being dried in natural conditions for 20 d to mimic the harvesting and drying process of alfalfa hay, the samples were dried completely at 50°C for 72 h and ground through a Thomas-Wiley Laboratory Mill (Lehman Scientific) with a 1-mm sieve. Near-infrared reflectance spectroscopy (NIRS) was performed using a Foss NIRS 6500 monochromator with a scanning range of 1,100 to 2,500 nm (Foss NIR Systems). Each sample was scanned eight times, and the average spectra were used for calibration. Mathematical and statistical treatments of all spectra were performed with WinISI III calibration development software (Foss NIR Systems). The existing commercial NIRS prediction equations (07AHY50) developed by the NIRS Forage and Feed Testing Consortium were employed to calculate quality characteristics of alfalfa. The precision of NIRS has been assessed by regression analysis of the predicted values and actual determined values. All data were analyzed using the SAS GLM procedure (SAS Institute). Statistical significance was determined by Student’s t test according to lsd. P < 0.05 was considered to be statistically significant.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Medicago truncatula SGR, HQ849484; alfalfa (Medicago sativa) SGR, HQ849485; Arabidopsis thaliana NYE1, At4g22920; Arabidopsis NYE2, At4g11910; soybean (Glycine max) SGR1, AY850141; soybean SGR2, AY850142; pea (Pisum sativum) SGR, AB303331; tobacco (Nicotiana tabacum) SGR, ABY19382; pepper (Capsicum annuum) SGR/CL, EU414631; tomato (Solanum lycopersicum) SGR/GF, EU414632; rice (Oryza sativa) SGR, AY850134; sorghum (Sorghum bicolor) SGR, AY850140; corn (Zea mays) SGR1, AAW82956; corn SGR2, NP_001105771; moss (Physcomitrella patens) SGR1, EDQ70701; and moss SGR2, EDQ62217.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression pattern of MtSGR in the wild type.
Supplemental Figure S2. Statistically enriched GO terms in biological processes in the NF2089 mutant.
Supplemental Figure S3. Nodule development and the expression pattern of nodule senescence marker genes.
Supplemental Figure S4. Phylogenetic analysis of SGRs among diversified plant species.
Supplemental Figure S5. Structures of chloroplasts from the control and the transgenic line SGRi-39.
Supplemental Figure S6. Transcript levels of MsSGR in transgenic alfalfa during leaf senescence.
Supplemental Figure S7. Naturally fallen leaves of the wild type, empty vector control, and transgenic alfalfa lines.
Supplemental Figure S8. Evaluation of the nutritive quality of transgenic alfalfa lines.
Supplemental Table S1. Genetic segregation analysis of the NF2089 mutant.
Supplemental Table S2. BLASTn analysis of Tnt1 flanking sequences retrieved from the mutant NF2089.
Supplemental Table S3. Microarray analysis of the NF2089 mutant.
Supplemental Table S4. Expression levels of changed genes in the NF2089 mutant microarray encoding putative SAG.
Supplemental Table S5. Up- and down-regulated genes in the NF2089 mutant microarray encoding chloroplast-related proteins.
Supplemental Table S6. GOEAST analysis of the differentially expressed genes in the NF2089 mutant microarray.
Supplemental Table S7. Nodule senescence markers.
Supplemental Table S8. Primers used in this study.
Acknowledgments
We thank Xiaofei Cheng, Kuihua Zhang, and Jiangqi Wen for assistance with screening the Tnt1 mutants; Yuanhong Han for assistance with operating LI-COR 6400; Crystal Marris for assistance with plant growth; Ivone Torres-Jerez for quantitative RT-PCR; Dennis Walker for assistance with NIRS analysis; and Ko Shimamoto for providing the pANDA35HK vector.
References
- Alós E, Roca M, Iglesias DJ, Mínguez-Mosquera MI, Damasceno CM, Thannhauser TW, Rose JK, Talón M, Cercós M. (2008) An evaluation of the basis and consequences of a stay-green mutation in the navel negra citrus mutant using transcriptomic and proteomic profiling and metabolite analysis. Plant Physiol 147: 1300–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstead I, Donnison I, Aubry S, Harper J, Hörtensteiner S, James C, Mani J, Moffet M, Ougham H, Roberts L, et al. (2007) Cross-species identification of Mendel’s I locus. Science 315: 73. [DOI] [PubMed] [Google Scholar]
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry S, Mani J, Hörtensteiner S. (2008) Stay-green protein, defective in Mendel’s green cotyledon mutant, acts independent and upstream of pheophorbide a oxygenase in the chlorophyll catabolic pathway. Plant Mol Biol 67: 243–256 [DOI] [PubMed] [Google Scholar]
- Austin S, Bingham ET, Mathews DE, Shahan MN, Will J, Burgess RR. (1995) Production and field performance of transgenic alfalfa (Medicago sativa L.) expressing alpha-amylase and manganese-dependent lignin peroxidase. Euphytica 85: 381–393 [Google Scholar]
- Barry CS, McQuinn RP, Chung MY, Besuden A, Giovannoni JJ. (2008) Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol 147: 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Borovsky Y, Paran I. (2008) Chlorophyll breakdown during pepper fruit ripening in the chlorophyll retainer mutation is impaired at the homolog of the senescence-inducible stay-green gene. Theor Appl Genet 117: 235–240 [DOI] [PubMed] [Google Scholar]
- Bouton J. (2007) The economic benefits of forage improvement in the United States. Euphytica 154: 263–270 [Google Scholar]
- Broughton WJ, Dilworth MJ. (1971) Control of leghaemoglobin synthesis in snake beans. Biochem J 125: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D. (2003) The molecular analysis of leaf senescence: a genomics approach. Plant Biotechnol J 1: 3–22 [DOI] [PubMed] [Google Scholar]
- Chapin LJ, Jones ML. (2009) Ethylene regulates phosphorus remobilization and expression of a phosphate transporter (PhPT1) during petunia corolla senescence. J Exp Bot 60: 2179–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HK, Mun JH, Kim DJ, Zhu H, Baek JM, Mudge J, Roe B, Ellis N, Doyle J, Kiss GB, et al. (2004) Estimating genome conservation between crop and model legume species. Proc Natl Acad Sci USA 101: 15289–15294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M, Fritz J. (2003) Forage quality. Barnes RF, Nelson CJ, Collins M, Moore KJ, , Forages: An Introduction to Grassland Agriculture, Ed 6. Iowa State University Press, Ames, pp 363–390 [Google Scholar]
- Cook DR. (1999) Medicago truncatula: a model in the making! Curr Opin Plant Biol 2: 301–304 [DOI] [PubMed] [Google Scholar]
- De Michele R, Formentin E, Todesco M, Toppo S, Carimi F, Zottini M, Barizza E, Ferrarini A, Delledonne M, Fontana P, et al. (2009) Transcriptome analysis of Medicago truncatula leaf senescence: similarities and differences in metabolic and transcriptional regulations as compared with Arabidopsis, nodule senescence and nitric oxide signalling. New Phytol 181: 563–575 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S. (2006) Chlorophyll degradation during senescence. Annu Rev Plant Biol 57: 55–77 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S. (2009) Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci 14: 155–162 [DOI] [PubMed] [Google Scholar]
- Jiang H, Li M, Liang N, Yan H, Wei Y, Xu X, Liu J, Xu Z, Chen F, Wu G. (2007) Molecular cloning and function analysis of the stay green gene in rice. Plant J 52: 197–209 [DOI] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9: 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Liu YG, Chen Y, Zhang Q. (2005) Amplification of genomic sequences flanking T-DNA insertions by thermal asymmetric interlaced polymerase chain reaction. Methods Mol Biol 286: 341–348 [DOI] [PubMed] [Google Scholar]
- Miller JD, Arteca RN, Pell EJ. (1999) Senescence-associated gene expression during ozone-induced leaf senescence in Arabidopsis. Plant Physiol 120: 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas K, Nicholas H, Deerfield D. (1997) GeneDoc: analysis and visualization of genetic variation. European Molecular Biology Network News 4: 1–4 [Google Scholar]
- Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, Lee SK, Jeong SW, Seo HS, Koh HJ, et al. (2007) The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19: 1649–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Ren G, An K, Liao Y, Zhou X, Cao Y, Zhao H, Ge X, Kuai B. (2007) Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol 144: 1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Morita R, Katsuma S, Nishimura M, Tanaka A, Kusaba M. (2009) Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J 57: 120–131 [DOI] [PubMed] [Google Scholar]
- Sato Y, Morita R, Nishimura M, Yamaguchi H, Kusaba M. (2007) Mendel’s green cotyledon gene encodes a positive regulator of the chlorophyll-degrading pathway. Proc Natl Acad Sci USA 104: 14169–14174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelbert S, Aubry S, Burla B, Agne B, Kessler F, Krupinska K, Hörtensteiner S. (2009) Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21: 767–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al. (2008) Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Thomas H. (1987) Sid: a Mendelian locus controlling thylakoid membrane disassembly in senescing leaves of Festuca pratensis. Theor Appl Genet 73: 551–555 [DOI] [PubMed] [Google Scholar]
- Thomas H, Howarth CJ. (2000) Five ways to stay green. J Exp Bot 51: 329–337 [DOI] [PubMed] [Google Scholar]
- Thomas H, Stoddart JL. (1975) Separation of chlorophyll degradation from other senescence processes in leaves of a mutant genotype of meadow fescue (Festuca pratensis L.). Plant Physiol 56: 438–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde W, Guerra JC, De Keyser A, De Rycke R, Rombauts S, Maunoury N, Mergaert P, Kondorosi E, Holsters M, Goormachtig S. (2006) Aging in legume symbiosis: a molecular view on nodule senescence in Medicago truncatula. Plant Physiol 141: 711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, et al. (2006) WEGO: a Web tool for plotting GO annotations. Nucleic Acids Res 34: W293–W297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Cannon SB, Sato S, Kim D, Cook DR, Town CD, Roe BA, Tabata S. (2005) Sequencing the genespaces of Medicago truncatula and Lotus japonicus. Plant Physiol 137: 1174–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Udvardi M. (2009) Translating Medicago truncatula genomics to crop legumes. Curr Opin Plant Biol 12: 193–201 [DOI] [PubMed] [Google Scholar]
- Zheng Q, Wang XJ. (2008) GOEAST: a Web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res 36: W358–W363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Han L, Hou C, Metelli A, Qi L, Tadege M, Mysore KS, Wang Z-Y. (2011) Developmental analysis of a Medicago truncatula smooth leaf margin1 mutant reveals context-dependent effects on compound leaf development. Plant Cell 23: 2106–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Choi HK, Cook DR, Shoemaker RC. (2005) Bridging model and crop legumes through comparative genomics. Plant Physiol 137: 1189–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]