Abstract
Germination represents a rapid transition from dormancy to a high level of metabolic activity. In-depth transcriptomic profiling at 10 time points in Arabidopsis (Arabidopsis thaliana), including fresh seed, ripened seed, during stratification, germination, and postgermination per se, revealed specific temporal expression patterns that to our knowledge have not previously been identified. Over 10,000 transcripts were differentially expressed during cold stratification, with subequal numbers up-regulated as down-regulated, revealing an active period in preparing seeds for germination, where transcription and RNA degradation both play important roles in regulating the molecular sequence of events. A previously unidentified transient expression pattern was observed for a group of genes, whereby a significant rise in expression was observed at the end of stratification and significantly lower expression was observed 6 h later. These genes were further defined as germination specific, as they were most highly expressed at this time in germination, in comparison with all developmental tissues in the AtGenExpress data set. Functional analysis of these genes using genetic inactivation revealed that they displayed a significant enrichment for embryo-defective or -arrested phenotype. This group was enriched in genes encoding mitochondrial and nuclear RNA-processing proteins, including more than 45% of all pentatricopeptide domain-containing proteins expressed during germination. The presence of mitochondrial DNA replication factors and RNA-processing functions in this germination-specific subset represents the earliest events in organelle biogenesis, preceding any changes associated with energy metabolism. Green fluorescent protein analysis also confirmed organellar localization for 65 proteins, largely showing germination-specific expression. These results suggest that mitochondrial biogenesis involves a two-step process to produce energetically active organelles: an initial phase at the end of stratification involving mitochondrial DNA synthesis and RNA processing, and a later phase for building the better-known energetic functions. This also suggests that signals with a mitochondrial origin and retrograde signals may be crucial for successful germination.
Seeds represent a crucial stage in the plant life cycle, as they are essential for the propagation of the species and allow dispersal to new locations. Seeds also allow plants to optimize survival strategy, as seeds display dormancy, which can be “broken” via a variety of environmental factors, thus allowing plants to optimize growth with reference to environmental conditions. As the seed is often the primary product utilized by humans, it is not surprising that seed germination is an intensively studied topic. There are a plethora of excellent articles and reviews characterizing seed dormancy and germination, with loss-of-function and gain-of-function mutants analyzed, across multiple species and utilizing many “omics” technologies (Gallardo et al., 2001; Fu et al., 2005; Nakabayashi et al., 2005; Cadman et al., 2006; Holdsworth et al., 2008a; Sreenivasulu et al., 2008; Howell et al., 2009). It has been observed that seed dormancy occurs in most plant species and provides seeds with a mechanism to survive extended periods of debilitating conditions prior to germination. In this way, germination can occur under favorable conditions, ensuring the greatest chance at seedling establishment (Baskin and Baskin, 2004). A range of factors have been identified in relation to dormancy initiation, maintenance, and alleviation, including temperature, moisture content, daylength, light quality, and mineral nutrition (Allen et al., 2007). These external triggers are perceived by the seed and elicit a series of signal transduction pathways, leading to the modulation of the phytohormones abscisic acid (ABA) and GA (Allen et al., 2007). Studies have shown that it is the antagonistic interaction between these two hormones that is at the core of dormancy maintenance (linked to ABA activity) and dormancy release and germination initiation (linked to GA activity) in plants (Allen et al., 2007; Holdsworth et al., 2008a; Sreenivasulu et al., 2008).
To dissect the molecular mechanisms that exist downstream of the hormonal signals regulating germination, transcriptomic and metabolomic profiling studies have been carried out in a range of species including Arabidopsis (Arabidopsis thaliana; Gallardo et al., 2001; Fu et al., 2005; Nakabayashi et al., 2005; Fait et al., 2006), barley (Hordeum vulgare; Sreenivasulu et al., 2008), and rice (Oryza sativa; Howell et al., 2009). These studies have revealed some common properties of transcriptomic changes that occur across these plant species. First, a large number of mRNA species (12,000–17,000) are present in the dry seeds or embryos; second, there appears to be a tightly regulated, temporally controlled transition through germination characterized by phasic changes in transcript abundance (Nakabayashi et al., 2005; Sreenivasulu et al., 2008; Howell et al., 2009). For Arabidopsis, transcriptomic changes were analyzed with a focus on profiling stored mRNAs in dry seed, to better differentiate the transition from late embryogenesis to seed germination (Nakabayashi et al., 2005). This profiling of dry seed (0 h) and seeds at 6, 12, and 24 h post imbibition under continuous light focused on ABA regulation and revealed transcriptomic differences between germination in the wild type and abi5 mutants (Nakabayashi et al., 2005). However, the maturation of freshly harvested seeds (prior to dark desiccation) and the breaking of dormancy over stratification were not examined. Likewise, in studies with rice and barley, germination of ripened seed, at 0, 1, 3, 12, and 24 h for rice (Howell et al., 2009) or at 0 and 24 h (and 48 and 72 h) for barley (Sreenivasulu et al., 2008), will not identify important steps from seed maturation (ripe seed) through desiccation and stratification. A recent systems-level approach, analyzing regulators of germination in flowering plants, utilized various publicly available transcriptome data sets and found that specific coordinated transcriptional regulation occurs separating the transition from dormancy to germination in flowering plants and that dormancy may have evolved by adjusting existing cellular phase transition and abiotic stress response-related genetic pathways (Bassel et al., 2011).
In Arabidopsis, dormancy is broken via the cold (4°C) imbibition of seeds in darkness. In most experiments involving the growth of Arabidopsis, seeds are typically sown and placed at 4°C in the dark for at least 48 h. This process is referred to as stratification, and studies examining the germination rate of stratified and nonstratified seeds alike reveal a significant increase in germination rates when seeds are subjected to stratification (Yamauchi et al., 2004; Dave et al., 2011). A role for GA- induced signaling during stratification has been shown using 8K Arabidopsis microarrays (Yamauchi et al., 2004). While the essential role for GA has been demonstrated for dormancy release, promoting embryonic expansion, inducing the mobilization of storage reserves, and mediating the weakening of tissues that envelope the embryo (Brady and McCourt, 2003; Yamauchi et al., 2004; Feurtado and Kermode, 2007; Holdsworth et al., 2008b), the detailed molecular networks and signaling that results in this increased germination rate and synchrony during and after stratification, are not yet elucidated. While the identities of some components responsive to these hormonal cues have been identified, such as the role of Della proteins in maintaining dormancy under ABA control (Tyler et al., 2004; Cao et al., 2006; Dohmann et al., 2010), a detailed molecular sequence of events, underpinning the transition from a dormant seed to the young seedling, is lacking;, especially with respect to temporal resolution.
Successful germination requires the mobilization of energy reserves to power germination until photosynthesis is established. Studies of mitochondrial biogenesis during germination in rice revealed that poorly differentiated mitochondria, lacking cristae and matrix structure, were present in dry seed and that the peak in transcript abundance for components encoding the machinery of oxidative phosphorylation was 24 h after imbibition (Howell et al., 2006). In maize, a similar study at the protein level showed that respiratory chain components did not peak until 48 h post imbibition (Logan and Leaver, 2000). However, other measures, such as analysis of cristae structure, increase in respiration, and ability to import proteins, reveal that mitochondrial biogenesis and activity are activated earlier than the peak in transcript abundance, encoding components involved in oxidative phosphorylation (Howell et al., 2006, 2007). In addition, protein import complexes were able to import protein just 30 min after imbibition (Howell et al., 2006, 2007). Complementing this, a transcriptomic study analyzing germination in rice revealed a surge in transcript abundance for genes encoding transport functions at 3 h after imbibition (Howell et al., 2009). This suggests that signals (and responses) affecting mitochondrial function are taking place earlier in germination.
To gain a greater temporal dissection of the processes occurring during germination in Arabidopsis, a detailed time course providing an expansive view of the process, before and after seed desiccation, over the course of stratification, to germination and postgermination, was analyzed. Analysis of this extensive time course enabled the identification of novel, stage-specific, and transient patterns of expression. The identification of these tight expression patterns provided the basis for determining the relationship between coexpression, colocalization, and function of encoded proteins. Functional analysis also revealed a link between function and the pattern of gene expression. GFP tagging was carried out to verify organellar localization for a large number of proteins, many of which were annotated as having “unknown functions.” This in-depth temporal analysis revealed a transient peak in expression for transcripts associated with ethylene metabolism, novel organelle proteins, and a role for RNA processing and mRNA decay at the earliest stages of germination in Arabidopsis. The greater dissection of these processes at the temporal level uncovers processes that have gone unnoticed and thus represent a mechanistic gap in our understanding of the transition from dormancy to germination.
RESULTS
Overview of Transcriptomic Changes: From Seed to Stratification and Germination
To gain a comprehensive insight into Arabidopsis germination, 10 time points were selected, including freshly harvested ecotype Columbia seeds (before desiccation, directly upon removal from the silique; H), seeds desiccated for 15 d in darkness (0 h), and seeds stratified at 4°C in the dark for 1 h (1 h S), 12 h (12 h S), and 48 h (48 h S). Stratified seeds were then transferred into continuous light and further collected at 1, 6, 12, 24, and 48 h into the light (1 h SL, 6 h SL, 12 h SL, 24 h SL, and 48 h SL, respectively). As germination is generally defined as concluding when a part of the embryo emerges from the testa (generally around 24 h after imbibition in the light), the final time point (48 h SL) is considered a postgermination time point. During the time course analyzed, 15,789 genes were found to be expressed at one or more time points, with more than 95% of these genes significantly up- or down regulated (following false discovery rate correction) during this crucial developmental stage, reflecting the extensive regulation occurring at the transcript level (Supplemental Table S1). While a previous study has examined the transcriptomic responses for less than 8,000 genes during stratification (Yamauchi et al., 2004), to date, there has been no global (22,000 genes) transcriptomic analysis carried out during stratification, which may reflect the general assumption that few significant processes, apart from an increase in water content, occur during stratification.
The inclusion of three time points during stratification revealed that greater than 10,000 genes are differentially expressed over the 48 h during stratification (S), with the greatest number of differentially expressed genes (DEGs) changing in transcript abundance between 12 h S and 48 h S (Fig. 1A; Supplemental Table S1). Notably, many of the changes observed were increases in transcript abundance, with a total of 7,517 unique transcripts increasing in transcript abundance over 48 h of stratification compared with 9,801 transcripts up-regulated during the first 48 h after transfer to light at 22°C (Fig. 1A). Thus, the observed differential expression is not only a reflection of the clearing out of stored transcripts upon imbibition but also the transcriptional up-regulation occurring as part of the germination process, which is supported by the observed functional categorization of the proteins encoded by these transcripts. The functional categories overrepresented in the earliest subsets of up-regulated DEGs during stratification included the oxidative pentose phosphate pathway and nitrogen and hormone metabolism (1 h S versus 12 h S; Fig. 1B). Upon closer examination, it is seen that genes encoding proteins involved in ethylene signaling are first induced in the early hours of stratification (1 h S versus 12 h S; Supplemental Table S2). This is in agreement with the role of ethylene, which has been shown to augment germination completion (Kecpczyński and Kecpczyńska, 1997; Beaudoin et al., 2000). The largest number of DEGs were observed between 12 h S and 48 h S, including an induction and overrepresentation of genes encoding nucleotide metabolism, RNA processing, and protein synthesis functions (12 h S versus 24 h S; Fig. 1B). Closer analysis of the subsets in the protein category (12 h S versus 48 h S; Fig. 1B) revealed an induction of genes encoding cytoplasmic and organellar ribosomal proteins (Supplemental Table S2). In contrast, protein modification and degradation functions were underrepresented in these subsets (12 h S versus 24 h S; Fig. 1B).
Figure 1.
Overview of transcriptomic changes during stratification and germination. A, The number of genes significantly (P < 0.05, PPDE > 0.96) differentially expressed during dark stratification at 4°C (in black boxes) and, following this, into continuous light. S, Stratification; SL, continuous light following stratification. The total number of genes up-regulated (red) or down-regulated (blue) in each comparison is shown above each bar. B, For each comparison, the significant fold change was analyzed using the PageMan tool (Usadel et al., 2006). Statistical analysis of overrepresented functional categories was carried out using the overrepresentation analysis Fisher method with Benjamini-Hochberg false discovery rate correction. For the larger functional categories, average z-scores are visualized. Functional categories that did not show significant changes were collapsed for display (detailed functional categories are shown in Supplemental Table S2). Statistical significance is represented by a false-color heat map (up, orange; down, green) where a z-score of 1.96 represents a P value of 0.05.
Once stratified, seeds were transferred into continuous light, and an induction and overrepresentation of genes encoding photosynthesis-related functions as well as lipid, hormone, and secondary metabolism functions were observed (Fig. 1). Interestingly, several RNA- and protein-related functions were seen to be significantly underrepresented in the subsets of DEGs up-regulated between 1 h SL and 48 h SL, possibly due to the earlier induction of these genes during stratification (Fig. 1B). As may be expected after transfer into light, an overrepresentation of lipid metabolism and developmental function-related genes, particularly storage proteins and late embryogenesis-abundant proteins, was observed in the down-regulated subsets of DEGs (1 h SL versus 6 h SL; Fig. 1; Supplemental Table S2). Closer examination of these data reveals an initial down-regulation of genes encoding lipid transfer functions between 0 h and 1 h SL, followed by a repeated overrepresentation of triacylglycerol synthesis functions in the genes down-regulated between 1 h SL and 48 h SL (Fig. 1; Supplemental Table S2). These changes complement the known breakdown of oil storage reserves that occurs during germination in oilseeds (Graham, 2008). Thus, there appear to be distinct processes occurring between stratification (0 h to 48 h S) and germination (48 h S to 48 h SL).
Correlation between Coexpression, Localization, and Function in Transcripts and Proteins
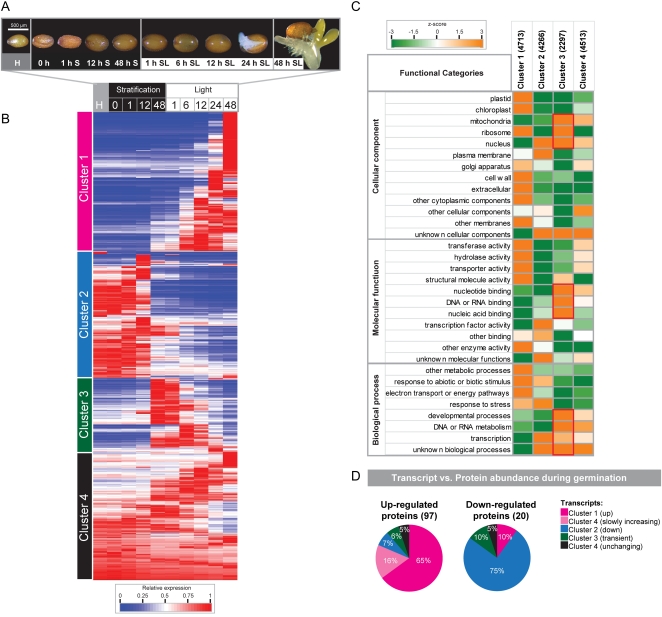
To visualize the expression profiles across stratification and germination (as defined in Fig. 2A), the normalized expression values for all 15,789 genes expressed over germination were made relative to the maximum expression over the time course and hierarchically clustered (see “Materials and Methods”), revealing four distinct clusters (Fig. 2B). Cluster 1 in Figure 2B represents approximately 30% of all genes expressed during germination and is characterized by low expression from dry seed to stratification and even up to 6 h SL, followed by significant up-regulation after 6 h SL (Fig. 2B). As expected, Gene Ontology (GO) overrepresentation analysis (see “Materials and Methods”) showed that this cluster was enriched in several GO categories, including genes encoding proteins targeted to the plastid, chloroplast, and ribosome (Fig. 2C). Genes encoding proteins with transferase, hydrolase, and transporter functions as well as structural molecular activity were also enriched in this cluster (cluster 1; Fig. 2C), corresponding with the increase in energy demand and significant morphological changes that are observed after 12 h SL, as the seed transcends to a seedling (Fig. 2A). The morphological changes observed during germination in this study (Fig. 2A) comply with previous observations during germination (Fu et al., 2005). Furthermore, this up-regulated expression pattern can be readily observed in a previous Arabidopsis germination study that analyzed global transcriptomic changes without stratification of seeds (cluster 1; Supplemental Fig. S1; Nakabayashi et al., 2005) and is also evidenced during germination in other species, such as rice and barley (Sreenivasulu et al., 2008; Howell et al., 2009). In contrast, cluster 2 represents the stored mRNAs present in dry seed that remain at a high level of expression up to 12 h S followed by a distinct down-regulation (Fig. 2C). Genes in this cluster represent 27% of the total genes expressed during germination and were enriched in nucleus-targeted proteins, including those showing transcription factor (TF) activity (cluster 2; Fig. 2C). Given that germination can still occur in the absence of transcription (Rajjou et al., 2004), the presence of these transcripts in dry seed, early in the time course, likely represents the crucial genes necessary for immediate response to imbibition (Fig. 2C). Again, the presence of these stored transcripts has also been observed in rice and barley (Sreenivasulu et al., 2008; Howell et al., 2009).
Figure 2.
Seed morphology, hierarchical clustering of expression profiles, and GO representation of genes during Arabidopsis germination. A, Light microscopy was used to examine the changes in morphology over the germination time course. B, All genes called present at a minimum of one time point were normalized to the highest level of expression over the time course of the study and hierarchically clustered using average linkage based on Euclidian distance. Four clusters were identified: cluster 1 (pink), transcripts that increased in abundance over the time period examined; cluster 2 (blue), transcripts highly expressed in dry seed to 12 h S that decreased in abundance over time; cluster 3 (green), transcripts with low or absent abundance in dry seeds that peaked between 48 h S and 6 h SL and then declined in abundance; cluster 4 (black), transcripts that displayed relatively stable levels of abundance throughout the time course. C, For each cluster, all genes were analyzed for representation in GO categories. Over/underrepresented functional categories were identified by z-score analysis, where statistical significance is represented by a false-color heat map (up, orange; down, green) where a z-score of 1.96 represents a P value of 0.05. D, Protein abundance profiles (from Fu et al., 2005) were compared with transcript abundance profiles during germination. The percentage of transcripts in each cluster that showed similar (up-regulated/down-regulated) protein abundance profiles during germination is indicated.
By analyzing this extensive time course, it was revealed that a group of genes (in cluster 3) are expressed at a very low level up to 12 h S, then dramatically increase in abundance between 48 h S and 6 h SL, before decreasing back to significantly lower levels by 48 h SL, the majority decreasing by 12 h SL (Fig. 2B). This transient expression pattern for a significant number of genes is not apparent during germination when seeds are not stratified (Supplemental Fig. S1; Nakabayashi et al., 2005). Examination of over/underrepresented GO categories for these transiently expressed genes revealed significant overrepresentation of genes encoding proteins targeted to the mitochondria, nucleus, and ribosomal proteins, corresponding with the observed overrepresentation of DNA, RNA-binding, and transcription functions (Fig. 2C, red boxes). The overrepresentation of TF functions in cluster 2 and DNA and RNA-binding functions in cluster 3 suggests a two-step regulation of genes encoding these regulatory factors, with the genes in cluster 2 likely encoding the regulatory proteins responsive to imbibition and required for the early stages of germination, while genes in cluster 3 are likely to encode the regulatory proteins required for normal germination progression and later plant development. In contrast, the genes in cluster 4 were enriched in proteins of unknown cellular localization and biological processes and slowly increased, decreased, or largely remained unchanging in abundance across the germination time course (Fig. 2, B and C). This set of genes may represent genes required for basic cellular functions, not highly responsive under germination conditions.
Previous studies have shown that while germination progresses to the point of radicle emergence in the absence of transcription, seedling establishment is prevented (Rajjou et al., 2004). Therefore, during this crucial developmental process, the translation of transcripts expressed during germination is clearly essential for further development. To identify any correlation between transcript and protein abundance, the transcript abundance profiles in this study were compared with previous studies examining protein abundance during germination (Gallardo et al., 2001, 2002; Fu et al., 2005; Chibani et al., 2006). Specifically, one study identified over 400 proteins expressed in dry seeds: seeds after 3 d of cold stratification and then 30, 48, 72, and 96 h into a 16/8-h light/dark cycle (Fu et al., 2005). This study categorized a large number of the protein abundance profiles as present = 1/absent = 0 for each time point (e.g. if a protein was present in dry seed and after stratification, but not following transfer into the light/dark cycle, it was categorized as 11000; Fu et al., 2005). In this way, present/absent profiles were observed for 117 unique proteins over this time course (Fu et al., 2005), for which parallel expression information is also available in our study (Supplemental Table S5). A number of the proteins identified by Fu et al. (2005) have also previously been identified in other germination studies, and these have been annotated in Supplemental Table S5 (Gallardo et al., 2001; Fu et al., 2005; Chibani et al., 2006). Using this present/absent (1/0) profiling of protein abundance, these proteins were matched to the corresponding transcript profiles from this study (Fig. 2D). Remarkably, it was seen that 81% of the up-regulated proteins over time showed comparable transcript expression profiles (16% slowly up in cluster 4 + 65% up in cluster 1; Fig. 2D; Supplemental Table S5). Similarly, of the 20 proteins highly expressed in seeds and not detected following stratification or transfer into the light/dark cycle, 15 proteins showed similar transcript expression profiles (i.e. for transcripts in cluster 2, highly expressed in freshly harvested and dry seed and decreasing in abundance over the germination time course; Fig. 2D). Interestingly, three genes encoding DNA/RNA-binding functions, Gly-rich protein 7 (At2g21660), a heterogeneous nuclear ribonucleoprotein (At4g14300), and a Isy1-like splicing domain-containing protein (At3g18790), were seen to have both a transient transcript expression profile (i.e. were in cluster 3 in this study) and also a transient protein abundance profile (Fu et al., 2005; Supplemental Table S5).
Identification of a Set of Germination-Specific Genes
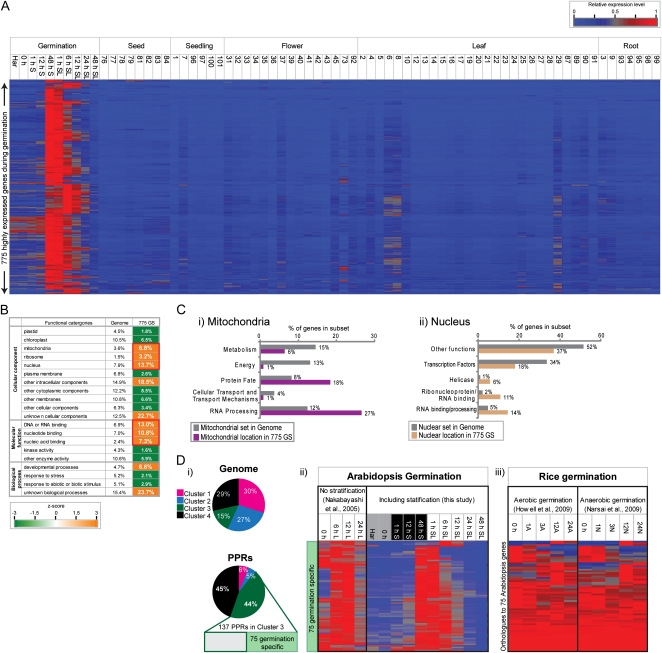
To further analyze the genes that showed transient expression during germination (cluster 3; Fig. 2B), publicly available microarrays carried out on a wide variety of tissues in the AtGenExpress developmental set (Supplemental Table S3; Schmid et al., 2005) were downloaded, normalized, and analyzed together with the 30 arrays in this study (see “Materials and Methods”). Expression levels of the 15,789 genes expressed during germination and development were visualized in the same row (and cluster) order as shown in Figure 2B and were hierarchically clustered by tissue samples (columns) to determine in what other tissues/organs these genes were also expressed (Supplemental Fig. S2). Examination of these revealed that the genes in clusters 1 and 4 (as shown in Fig. 2B) consist of genes that are highly expressed or unchanging in expression in most other developmental tissues as well as late germination (Supplemental Fig. S2). As expected, the genes in cluster 2 (Fig. 2B) that were highly expressed in dry seeds were also highly expressed across the microarrays analyzing developing seeds (Supplemental Fig. S2). In contrast, it was observed that the genes in cluster 3 were most highly expressed between 48 h S and 12 h SL, even in comparison with all other developmental tissues (Supplemental Fig. S2, blue box). To filter these genes further for primarily germination-specific (GS) expression, only those genes that had a relative expression level greater than 0.5 (relative to maximum expression level) between 1 h S and 24 h SL across all tissues were visualized (as these time points strictly represent germination). In this way, 775 unique genes were identified as showing the highest expression during germination (i.e. GS; Fig. 3A).
Figure 3.
Identification of GS gene expression. A, Publicly available microarrays carried out on wild-type tissues in the AtGenExpress developmental set (Schmid et al., 2005; E-AFMX-9) were normalized together with the arrays in this study. Hierarchical clustering of the relative expression levels are for 775 unique genes identified as showing the highest expression during germination, with expression levels less than 50% of these levels in all other tissues. B, Examination of over/underrepresented GO categories for these transiently expressed genes revealed significant overrepresentation of genes encoding proteins targeted to the mitochondria, nucleus, and ribosomes, corresponding with the observed overrepresentation of DNA- and RNA-binding functions. The overrepresentation of these functions, specifically during germination, implies that these genes are likely to encode the regulatory proteins required specifically for germination progression and later development. Over/underrepresented functional categories were identified by z-score analysis. Statistical significance is represented by a false-color heat map (up, orange; down, green) where a z-score of 1.96 represents a P value of 0.05. C, The distribution of genes into functional subcategories, categorized as follows: i, mitochondrial; ii, nuclear. D, i, The percentage of PPR-encoding genes in each cluster is displayed as a pie chart showing the percentage of genes in each cluster in the genome. ii, Expression levels during germination without (no stratification) and including stratification for the 75 PPR genes identified as possessing GS expression in this study. iii, Expression of the rice genes orthologous to the 75 Arabidopsis genes shown across germination under aerobic and anaerobic conditions in rice.
Intriguingly, analysis of over/underrepresented GO categories for these 775 GS genes revealed approximately double the expected percentage of genes in this set encoding proteins targeted to the mitochondria, nucleus, and ribosomes, corresponding with the observed overrepresentation of DNA- and RNA-binding functions (Fig. 3B). Genes encoding TFs represented only 53 of the 775 genes, which was not significantly greater/less than the expected percentage in the genome. This indicates that the genes encoding mitochondria- and nucleus-localized proteins were binding DNA or RNA but performing functions other than transcriptional regulation. To discover the nature of these other encoded protein functions, the genes in these mitochondrial and nuclear subsets from the 775 GS genes were viewed based on the subfunctional groups within these sets, revealing a significant enrichment of RNA-processing functions in both sets (mitochondrial and nuclear; Fig. 3C). Additionally, helicase and ribonucleoprotein/RNA-binding functions were also seen to be enriched in the nuclear set (Fig. 3Cii). For the mitochondrial set from the 775 GS genes, protein fate functions were observed to be enriched (18% versus 8% in the whole mitochondrial set), despite the majority of genes in the mitochondria encoding metabolism and energy functions (which were underrepresented in this set; Fig. 3Cii). Closer examination of the genes encoding RNA-processing functions revealed a significant (P < 0.001) overrepresentation of pentatricopeptide repeat domain (PPR)-containing genes in this GS subset, with nearly 10% (75 genes) of the 775 genes encoding a PPR domain-containing protein, while PPR domain-containing genes only make up less than 2% of all genes in the genome. To confirm that the observed PPR gene expression pattern was limited to the transiently expressed genes during germination, the percentage of PPR genes in each cluster was examined (Fig. 3Di). Interestingly, it was seen that 45% (137) of all PPRs were in cluster 3, showing transient expression during germination, and of these, 75 genes showed GS expression (Fig. 3D).
Despite the finding that this transient expression pattern cannot be readily identified when seeds are not stratified (Supplemental Fig. S1; Nakabayashi et al., 2005), when the 75 PPR genes showing GS expression were examined together with the microarrays from this study, it was observed that the transient expression pattern during germination was somewhat maintained (Fig. 3Dii); however, the temporal resolution was lost or not observed. This suggests that the stage-specific expression of these genes is a characteristic of germination, independent of whether seeds undergo stratification (Fig. 3Dii). To determine whether these transiently expressed genes also displayed transient expression during germination in other species, rice orthologs of the 775 Arabidopsis GS genes were visualized, showing their expression across germination and developmental tissues in rice (details of the arrays used are shown in Supplemental Table S4; Supplemental Fig. S3A). A total of 768 rice orthologs of the 775 Arabidopsis GS genes could be identified using InParanoid (version 7.0; Remm et al., 2001). Only 383 (approximately 50%) of the 768 rice orthologs displayed maximum expression during rice germination (Supplemental Fig. S3A, yellow box; Howell et al., 2009; Narsai et al., 2009); however, these did not display the transient pattern observed in Arabidopsis. Instead, the expression pattern observed for the rice orthologs was more similar to the patterns observed for clusters 1, 2, and 4 in Arabidopsis. As PPR genes display high levels of orthology between plant species (O’Toole et al., 2008), the subset of 68 rice PPR genes orthologous to the 75 GS genes in Arabidopsis (Fig. 3D, i and ii, green box) were isolated and the expression levels were hierarchically clustered (Fig. 3Diii). Expression of the 68 orthologous genes in rice revealed no transient expression pattern (Fig. 3Diii). In addition, visualization of these 68 rice PPR genes across germination and other developmental tissues (Supplemental Fig. S3B) further confirmed the divergence in the transcriptomic response of PPR gene expression between monocots and dicots, despite orthology.
Loss of Function of GS Genes Results in Embryo Lethality
A recent study defined 481 genes as seed essential, where a loss-of-function mutation in these genes was found to result in a seed-related phenotype, mostly embryo lethal. These genes are indicated in the SeedGenes database (Meinke et al., 2008). The list of SeedGenes characterized as showing a seed-related phenotype (e.g. seed lethal) was matched against the genes expressed during germination. Of the 481 genes in this database (referred to as “seed-genes”), expression of 422 genes could be detected during this germination time course. A significant enrichment of seed-genes was seen in cluster 1 (35% versus 30% in the genome) and cluster 3 (22% in cluster 3 versus 15% in the genome; Supplemental Fig. S4), with 35 seed-genes observed in the set of 775 GS genes (Fig. 3A). Interestingly, the rice orthologs for these 35 genes did not show a transient expression pattern during rice germination (Supplemental Fig. S4, B and C), suggesting that despite orthology, the controlled expression of these genes during germination may be specific to Arabidopsis germination.
Given the enrichment of seed-lethal genes in the 775 GS genes identified in Figure 3A, a search was carried out on the 775 to determine whether these genes encode crucial protein functions necessary for seed/seedling or even plant development. To do this, large-scale reverse genetic studies identifying phenotypes for knocked out/silenced genes were matched to the 775 GS genes. The studies/databases examined are outlined in Supplemental Table S6. In addition to the large-scale studies/databases, it was observed that 110 of the 775 GS genes encode proteins experimentally shown and/or predicted to be localized to the mitochondria (Fig. 2B); therefore, all genes encoding these proteins were individually searched for known phenotypes in previous publications. In this way, 114 genes were identified as having altered developmental phenotypes (Fig. 4A). To ensure that this process of searching for phenotypes did not have any particular bias, sets of 775 randomly selected genes were generated and examined for phenotypes exactly as carried out for the 775 GS set (i.e. all studies/databases in Supplemental Table S6, and individual searching for genes encoding mitochondrial proteins). It was seen that, overall, the 775 GS set consisted of significantly more genes with known phenotypes compared with the average number across the random gene sets (P < 0.01; 114 genes versus 79 genes expected). Moreover, the types of phenotypes also significantly differed, with more than three times the number of genes with seed-lethal/embryo-arrested phenotypes seen in the GS set (51 genes) compared with the 775 random gene sets (Fig. 4; Pagnussat et al., 2005; Meinke et al., 2008). Interestingly, closer examination of the 114 genes with known phenotypes also reveals an obvious enrichment of 40 genes encoding RNA-binding/processing functions, compared with only 16 in the random gene sets (Fig. 4). Examples of these genes encoding proteins involved in RNA-binding functions included RNA helicases (e.g. At5g08610), RNA-binding proteins (e.g. At4g32720 and At1g49400), various ribonucleases (e.g. At1g01040 and At2g17510), and proteins containing an RNA recognition motif (e.g. At4g24280; Supplemental Table S6). The identification of these functions as being most highly expressed during germination (Fig. 3A) combined with observations that the silencing/loss of function of a significant number of these genes results in seed-lethal/embryo-arrested phenotypes (Fig. 4; Supplemental Table S6) reveal the crucial requirement for the expression and function of RNA binding/processing during early germination and development in Arabidopsis.
Figure 4.
Functional analysis of proteins encoded by genes defined as GS. Of the 775 genes showing GS expression, 114 genes showed a published phenotype when mutated/silenced/knocked out. This number is significantly greater (P < 0.001) than the expected percentage of genes showing phenotypes in random sets of 775 genes. A, The types of phenotypes observed for the 114 genes with known phenotypes in the GS set. The number of genes displaying each phenotype is indicated (details can be seen in Supplemental Table S6). The significant overrepresentation of seed-lethal phenotypes is indicated with the red asterisk. Of the 114 genes, the number of those encoding RNA-binding/processing functions is also shown in the center of the pie chart. AA, Amino acids; FA, fatty acids; WT, wild type. B, From a list of 775 randomly selected genes, 79 genes (on average, across three independent, randomly selected sets) were found to have phenotypes. The number of genes and the distribution of phenotypes observed for these are shown. Of the 79 genes, the number of those encoding RNA-binding/processing functions is also shown in the center of the pie chart. Note that the pie chart in B is smaller than that of the 114 genes in A, as it is drawn to scale for the number of phenotypes observed.
Confirming the Organellar Location of Proteins with GS Expression
The transcriptomic results presented strongly suggest a clear link between localization and coexpression, with genes encoding proteins annotated as plastid/chloroplast localized being overrepresented in cluster 1 (Fig. 2B) and genes encoding mitochondria/nucleus-localized proteins being overrepresented in cluster 3 (Fig. 2B). Considering these correlations, it was hypothesized that a selection of genes, with hitherto unconfirmed protein localizations, could encode proteins localized to the mitochondria, plastids, and/or peroxisomes, based on their expression patterns and predicted localizations as annotated in the Arabidopsis SUBA localization database (Heazlewood et al., 2007). A range of 65 genes (Supplemental Table S7) largely showing GS expression (as in Fig. 3A) were analyzed by GFP targeting to determine protein localization. Fusion proteins were constructed and transiently transformed into Arabidopsis cell culture using biolistic transformation. Organelle targeting was verified using alternative oxidase-red fluorescent protein (AOX-RFP) as a mitochondrial control, targeted small subunit of Rubisco (SSU)-RFP as a plastid control, and RFP-SRL (S, Ser; R, Arg; L, Leu) as a control for peroxisomal targeting (see “Materials and Methods”). Protein accumulation was characterized as to the mitochondria, plastid, dual targeted to the mitochondria and the plastid, peroxisome, cytoplasm, endoplasmic reticulum, Golgi, or the nucleus. Examples of fluorescence micrographs for proteins targeted to the mitochondria, plastids, and peroxisomes are shown in Figure 4A, and the complete set of targeting results is shown in Supplemental Figure S5.
Most of the genes for which localization was determined exhibited low expression in dry seed and 48 h SL (i.e. they were highly expressed specifically during germination; genes with GS expression are indicated by carets in Fig. 5B). Analysis of the localization of these confirmed the predicted localization for most genes, with some exceptions, including a mitochondrial transcription termination factor (mTERF; At5g06810) and a PPR-containing protein, At4g21170, that were predicted to be mitochondrial but appear to be dual targeted to both mitochondria and plastids (Fig. 5B; Supplemental Fig. S5). It can be seen that several genes displayed in Figure 5B encode DNA/RNA-binding functions, including a mitochondrial intron maturase, a mTERF protein, many mitochondrial PPR-encoding proteins, as well as a plastid-targeted chaperone DNAJ protein, a plant homeodomain finger DNA-binding TF, and a tRNA methyltransferase (Fig. 5B). The transient expression of these genes encoding regulatory functions during germination and the confirmation of their mitochondrial and plastid localization reveal a specific step of transcriptional regulation that occurs for organellar proteins during this crucial stage of development. Further supporting the essential role of these genes during the earliest stages of germination, it was revealed that three of the 65 genes selected for localization studies encode proteins for which a loss of function results in a seed-lethal phenotype (denoted ED in Fig. 5B), as determined previously (Ding et al., 2006; Meinke et al., 2008; Hammani et al., 2011). Each of these genes, At1g79490 (Fig. 5A), At1g10270, At5g60960, and At5g39710, encoded proteins predicted to be mitochondrial, which was confirmed by GFP analysis (Fig. 5B; Supplemental Fig. S5). Notably, a number of recent studies have also confirmed the observed localization for four of these proteins, At5g46920 (Keren et al., 2009), At1g80270 (Doniwa et al., 2010), At5g60960 (Hammani et al., 2011), and At4g36040 (Chen et al., 2010).
Figure 5.
GFP analysis confirms the localization of proteins encoded by genes expressed during germination. A subset of 65 genes predicted to encode organelle-targeted proteins were analyzed to determine subcellular localization. Only those genes most highly expressed during germination (i.e. showing low expression in dry seed and postgermination [48 h SL]) were selected for analysis. A, Examples of fluorescence images for proteins targeted to the mitochondria, plastids, and peroxisomes. The images for every protein identified as targeting to an organelle can be found in Supplemental Figure S5. Fusion proteins containing GFP and the first 100 amino acids (AA) of the protein of interest (or the last 100 amino acids in the case of the peroxisomal predicted proteins) were constructed and transiently transformed into cell culture using biolistic transformation. Organelle targeting was verified using AOX-RFP as a mitochondrial control, SSU-RFP as a plastid control, and RFP-SRL as a control for peroxisomal targeting. B, A heat map showing expression levels during germination for the 65 genes analyzed by GFP targeting. The Arabidopsis Gene Identifier for each gene, brief gene symbol/description, and localization determined by GFP are shown next to the heat map. Predicted localizations are indicated in parentheses (M, mitochondria; Pl, plastids; Per, peroxisomes) next to the experimentally determined localization. Note that genes that were defined as most highly expressed during germination in Figure 3a are indicated by carets. Known phenotypes when the gene is knocked out/silenced are as follows: defective in flower (F), stress (St), embryo (ED), growth (G), and seedling (SL). Reference for phenotypes are as follows: 1 Boavida et al. (2009); 2 Kwak et al. (2011); 3 Ding et al. (2006); 4 Keren et al. (2009); 5 Hilson et al. (2004); 6 Hammani et al. (2011); 7 Meinke et al. (2008); 8 Kuromori et al. (2006); 9 Chen et al. (2010); 10 Saiga et al. (2008); 11 Osakabe et al. (2005). AGI numbers in gray indicate those annotated as involved in RNA binding/processing.
Furthermore, including the two seed-lethal genes annotated as “unknown function,” it can be seen that localization was determined for 13 other genes of unknown function (Fig. 5B). These analyses reveal clues about the possible functions of these genes, inferred from their organellar localization and GS expression, forming the basis of further functional studies of these genes (Fig. 5B). In addition, it was observed that two small auxin-responsive RNA-like-encoding genes were predicted to encode mitochondria-targeted proteins; however, the localization of these was determined to be nuclear/cytoplasmic (Fig. 5B). The transient expression of these was particularly interesting, as it is known that these genes are specifically regulated at the level of mRNA decay, allowing tight control of mRNA levels (Newman et al., 1993). This subset of tightly controlled, transiently expressed nucleus-, mitochondria-, and/or plastid-targeted genes (Fig. 5B) may represent crucial control factors responsible for the normal regulation of gene expression during germination.
Factors Affecting Transcript Abundance during Arabidopsis Germination
It is known that, upon imbibition during germination, stored mRNAs are degraded as in vivo transcription begins, and this pattern of decrease in abundance of stored transcripts appears to be conserved independently of whether seeds are stratified or not (cluster 2; Fig. 6Aii). Similarly, the up-regulation of specific transcripts over the germination time course is also conserved (cluster 1; Fig. 6Ai). Although these expression patterns are relatively conserved during germination with/without stratification, the temporal development sequence is different due to different experimental designs. Two distinct phases of RNA degradation were evidenced (i.e. clusters 2 and 3). A comparison of these genes with the mRNA half-lives of approximately 13,000 genes that have been previously determined in Arabidopsis (Narsai et al., 2007) allows insight into the regulatory processes that may affect transcript stability or degradation. Given that transcripts encoding core cellular functions, such as those involved in energy (e.g. photosynthesis), have relatively long mRNA half-lives (Narsai et al., 2007), it was not surprising to observe a significant (P < 0.05) enrichment of transcripts with longer half-lives in cluster 1 (Fig. 6B). Similarly, given the sharp decrease that occurs after the transient expression seen in cluster 3, it was also expected to find that this cluster was enriched in transcripts with relatively short half-lives (less than 6 h; Fig. 6B). In contrast, it was surprising to see that there was no enrichment of transcripts with shorter half-lives in cluster 2 (Fig. 6B), given that this group is characterized by a significant decrease in transcript abundance (cluster 2; Fig. 6, A and B). Although this decrease is seen to occur much slower for stratified seeds (this study), it does appear that when seeds are imbibed under continuous light, with no stratification (Nakabayashi et al., 2005), these stored transcripts decrease to about 50% of their dry seed levels within 6 h, indicating a relatively rapid rate of decrease in abundance (cluster 1; Fig. 6A). These findings indicate that the rate of degradation is controlled in a developmental stage-specific manner (Fig. 6, A and B).
Figure 6.
Analysis of putative regulatory factors affecting transcript abundance. A, Parallel comparison showing the average profile of genes in clusters 1 to 4 (i–iv) for transcripts expressed during germination without stratification (no stratification) and including stratification (this study). B, The mRNA half-lives of all genes expressed during germination are shown as a pie chart indicating the percentage of genes that had mRNA half-lives of differing lengths (i.e. 0 h to over 24 h). For each cluster (1–4; i–iv), the distribution of genes into the different mRNA half-life increments is indicated, with red/blue font indicating an over/underrepresentation of genes with mRNA half-lives for that increment. C, Expression profiles of all the genes encoding TFs in Arabidopsis (Riaño-Pachón et al., 2007; 1,299 expressed during germination) were used to determine whether there was an overall over/underrepresentation of TFs in each cluster. i, The percentage of TFs in each cluster is compared with the percentage of genes in that cluster in the genome. ii, The representation of specific TF families within each cluster is compared with the percentage present in the genome to show if specific families were over/underrepresented, which is indicated by the red or blue font, respectively (P < 0.05). TF families that were also overrepresented and showed the same expression pattern in rice germination are indicated in black boxes (Perrin et al., 2004; Pagnussat et al., 2005; Nakagawa and Sakurai, 2006; Lister et al., 2007; Meinke et al., 2008; Boavida et al., 2009; Yu et al., 2009).
The other factor controlling transcript abundance is transcription; therefore, without directly measuring transcription, focus was shifted to how the transcripts encoding these regulatory factors respond during germination. Thus, the Arabidopsis TF database was queried (Riaño-Pachón et al., 2007), resulting in the observation that cluster 2 was significantly enriched in genes encoding TFs (35% of all TFs are in cluster 2; Fig. 6Ci). This is particularly interesting as it further supports the important role of mRNA stability during germination, given that genes encoding TFs are known to have short mRNA half-lives (Narsai et al., 2007) and yet are seen to remain stably abundant up to 12 h S (Fig. 6A). A closer look at the families of TFs in each cluster reveals specific over/underrepresentation patterns, including the significant overrepresentation of the MYB, LOB, Orphans, AUX/IAA, and bHLH families in cluster 1 (Fig. 6Cii). Interestingly, the AUX/IAA and bHLH families were also seen to be overrepresented in the group of genes showing the same up-regulation pattern over germination in rice (Fig. 6Cii, black boxes; Howell et al., 2009). Several studies have examined the role of AUX/IAA proteins and have revealed that these proteins have repressor functions crucial for normal development (Ulmasov et al., 1997; Tiwari et al., 2004). Given their conserved expression pattern, it is possible that their role in regulating germination and development is conserved across monocots and dicots. Similarly, cluster 2 was significantly enriched in the HB, Trihelix, and HSF families, with the latter also seen to be overrepresented in the parallel cluster during rice germination (Fig. 6Cii; Howell et al., 2009). Previous studies have implicated a role for HSFs for normal development in plants and other eukaryotic species (Kotak et al., 2007a), with one study even showing a build up of both transcript and protein abundance for HSFs over the course of seed development (Kotak et al., 2007b). Therefore, it was not surprising to see the conserved expression pattern of these genes across rice and Arabidopsis (Fig. 6Cii).
Although cluster 3 was not enriched in TFs, it was seen that of the TFs present, there was an enrichment of C3H, HB, C2C2 GATA, and the mTERFs, with 15 of the 23 mTERFs expressed during germination present in cluster 3 (Fig. 6Cii). It has been shown that HB TFs have a role in cell differentiation and growth; thus, their overrepresentation in cluster 3 may reflect an increase in demand for TFs controlling these vital processes during germination (Kappen, 2000). A previous study has shown the crucial role these factors play, not only in mitochondrial transcription termination but also in the initiation of transcription and the control of mitochondrial DNA replication (Roberti et al., 2009). The role for CH3 was also characterized in Arabidopsis embryos and was seen to be expressed from globular to late cotyledon stages (Li and Thomas, 1998). Thus, it was not surprising to see the overrepresentation of these in cluster 3 (Fig. 6Cii).
To analyze putative cis-elements that may be involved in regulating transcript abundance during germination, genes in each cluster and the 775 GS set identified in Figure 3 were examined for overrepresented putative 6-mers in the 1-kb upstream region of the transcriptional start site. The 775 GS set was specifically chosen because the abundance of these transcripts increases and decreases in a relatively short time period, and thus they are likely to be coregulated at the transcriptional level, accounting for the observed increase and possibly actively degraded to produce the decrease observed. Overall, the 1-kb upstream promoter elements displayed significant enrichment of 6-mers, with some elements found in up to 49% of the genes that showed GS expression (Table I). All promoter elements were matched against known cis-element-binding sites within the AGRIS database (Davuluri et al., 2003; Palaniswamy et al., 2006; Yilmaz et al., 2011) and studies that have characterized specific binding sites (Kosugi et al., 1995; Schöffl et al., 1998). Any elements showing a significant overrepresentation (P < 0.05) for genes in clusters 1 to 4 and the GS subset are shown in Table I. Promoter analysis of stored mRNAs was carried out previously, and a significant overrepresentation of ABRE-binding sites was observed for these (Nakabayashi et al., 2005), a feature confirmed in this study, with ABRE elements also observed in genes of cluster 2 (Fig. 6A; Table I). Interestingly, there was an overwhelmingly large number of known motifs seen in the GS subset, suggesting that numerous factors may be involved in the controlled expression pattern observed for these genes. An example of these included an overrepresentation of Telobox and Site II elements for the genes in cluster 3 and the GS subset (Table I), complying with the role of these genes in the control of genes encoding organellar proteins during development and the circadian regulation of genes encoding mitochondrial proteins (Giraud et al., 2010). Additionally, it can be seen that genes encoding HSF TFs were enriched in cluster 2 (Fig. 6C). It is possible that these HSFs have a role in the control of genes in the GS subset, as several heat shock element (HSE)-binding site motifs were seen to be enriched in the GS subset (Table I), implying that the stored HSF encoding transcripts in dry seed (cluster 2) may be translated upon imbibition and have a downstream role in the control of GS transcript abundance.
Table I. Known motifs that were found to be significantly overrepresented in genes expressed over germination.
The specific hexamer, name of the known motif, reference (Ref.) showing the source of the motif (1 = AGRIS, 2 = Schöffl et al. [1998], 3 = Kosugi et al. [1995]), and occurrence in all genes expressed during germination in each cluster and the GS subset are shown (boldface data indicate significant overrepresentation). Note that motifs with asterisks indicate that these were also identified as overrepresented in dry seed by Nakabayashi et al. (2005).
| Hexamer | Motif Name | Ref. | Genome (15,789) | Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 | GS |
| CACGTG* | ABRE, CBF2, G-box motif | 1 | 2,927 (19%) | 886 (19%) | 978 (23%) | 375 (16%) | 688 (15%) | 98 (13%) |
| CGTGTC | ABRE-like binding site motif | 1 | 1,799 (11%) | 508 (11%) | 636 (15%) | 239 (10%) | 416 (9%) | 79 (10%) |
| ACGTGT | ABRE-like binding site motif, Z box | 1 | 2,888 (18%) | 850 (18%) | 967 (23%) | 351 (15%) | 720 (16%) | 102 (13%) |
| ACACGT* | ACE promoter motif | 1 | 2,916 (18%) | 854 (18%) | 973 (23%) | 365 (16%) | 724 (16%) | 99 (13%) |
| GCCGAC | CBF1 BS in cor15a | 1 | 799 (5%) | 222 (5%) | 282 (7%) | 100 (4%) | 195 (4%) | 29 (4%) |
| CCACGT* | CBF2 binding site motif, GBF | 1 | 2,240 (14%) | 701 (15%) | 729 (17%) | 276 (12%) | 534 (12%) | 96 (12%) |
| TGCCGA | DRE-like promoter motif | 1 | 781 (5%) | 221 (5%) | 266 (6%) | 103 (4%) | 191 (4%) | 35 (5%) |
| TTCCCG | E2F binding site motif | 1 | 1,342 (9%) | 347 (7%) | 363 (9%) | 235 (10%) | 397 (9%) | 77 (10%) |
| CAAGGG | EIL1 BS in ERF1 | 1 | 1,246 (8%) | 338 (7%) | 345 (8%) | 221 (10%) | 342 (8%) | 90 (12%) |
| GCCGCC | ERE promoter motif, GCC box | 1 | 748 (5%) | 181 (4%) | 201 (5%) | 137 (6%) | 229 (5%) | 44 (6%) |
| CTAGGG | LFY BS in AP3 | 1 | 992 (6%) | 274 (6%) | 243 (6%) | 184 (8%) | 291 (6%) | 66 (9%) |
| CGGGCC | SORLIP2 | 1 | 381 (2%) | 123 (3%) | 89 (2%) | 70 (3%) | 99 (2%) | 27 (3%) |
| GGGCCG | SORLIP2 | 1 | 715 (5%) | 194 (4%) | 194 (5%) | 127 (6%) | 200 (4%) | 56 (7%) |
| GGGCCT | SORLIP2 | 1 | 1,793 (11%) | 522 (11%) | 447 (10%) | 324 (14%) | 500 (11%) | 112 (14%) |
| AAACCC | TELO-box promoter motif | 1 | 4,979 (32%) | 1,442 (31%) | 1,242 (29%) | 925 (40%) | 1,370 (30%) | 377 (49%) |
| AACCCT | TELO-box promoter motif | 1 | 3,412 (22%) | 939 (20%) | 793 (19%) | 716 (31%) | 964 (21%) | 313 (40%) |
| ACCCTA | TELO-box promoter motif | 1 | 2,570 (16%) | 727 (15%) | 606 (14%) | 547 (24%) | 690 (15%) | 239 (31%) |
| CCCTAA | TELO-box promoter motif | 1 | 2,726 (17%) | 739 (16%) | 684 (16%) | 542 (24%) | 761 (17%) | 226 (29%) |
| AGCCGA | DRE-like promoter motif | 1 | 1,140 (7%) | 319 (7%) | 345 (8%) | 168 (7%) | 308 (7%) | 68 (9%) |
| GCCGAA | DRE-like promoter motif | 1 | 1,073 (7%) | 306 (6%) | 311 (7%) | 162 (7%) | 294 (7%) | 68 (9%) |
| CGACAT | DRE promoter motif | 1 | 1,949 (12%) | 579 (12%) | 523 (12%) | 330 (14%) | 517 (11%) | 115 (15%) |
| TCCCGC | E2F binding site motif | 1 | 750 (5%) | 231 (5%) | 189 (4%) | 119 (5%) | 211 (5%) | 47 (6%) |
| TCAAGG | EIL in ERF1 | 1 | 2,401 (15%) | 675 (14%) | 626 (15%) | 380 (17%) | 720 (16%) | 149 (19%) |
| AGGGGG | EIL in ERF1 | 1 | 507 (3%) | 145 (3%) | 147 (3%) | 88 (4%) | 127 (3%) | 30 (4%) |
| AGAGCC | ERE promoter motif | 1 | 1,726 (11%) | 509 (11%) | 455 (11%) | 275 (12%) | 487 (11%) | 102 (13%) |
| GATTGC | GATA-like | 1 | 2,087 (13%) | 597 (13%) | 514 (12%) | 341 (15%) | 635 (14%) | 126 (16%) |
| GCGATT | GATA-like | 1 | 1,476 (9%) | 385 (8%) | 396 (9%) | 235 (10%) | 460 (10%) | 90 (12%) |
| GGATAC | GATA | 1 | 1,624 (10%) | 464 (10%) | 424 (10%) | 244 (11%) | 492 (11%) | 99 (13%) |
| AGAACG | HSE binding site motif | 1 | 1,856 (12%) | 533 (11%) | 519 (12%) | 278 (12%) | 526 (12%) | 110 (14%) |
| AGCTTC | HSE binding site motif | 1 | 3,672 (23%) | 1,001 (21%) | 953 (22%) | 622 (27%) | 1,096 (24%) | 224 (29%) |
| CGTTCT | HSE binding site motif | 1 | 1,691 (11%) | 473 (10%) | 456 (11%) | 258 (11%) | 504 (11%) | 102 (13%) |
| GCTTCT | HSE binding site motif | 1 | 3,817 (24%) | 1,054 (22%) | 1,015 (24%) | 618 (27%) | 1,130 (25%) | 239 (31%) |
| CAGAAC | HSE binding site motif | 2 | 2,531 (16%) | 734 (16%) | 658 (15%) | 405 (18%) | 734 (16%) | 154 (20%) |
| CCAGAA | HSE binding site motif | 2 | 2,781 (18%) | 773 (16%) | 766 (18%) | 472 (21%) | 770 (17%) | 170 (22%) |
| TTCCGG | HSE binding site motif | 2 | 1,581 (10%) | 394 (8%) | 446 (10%) | 265 (12%) | 476 (11%) | 98 (13%) |
| TTCGCG | HSE binding site motif | 2 | 634 (4%) | 165 (4%) | 173 (4%) | 106 (5%) | 190 (4%) | 41 (5%) |
| CGTCGT | JASE2 motif in OPR2 | 1 | 1,811 (11%) | 512 (11%) | 497 (12%) | 298 (13%) | 504 (11%) | 107 (14%) |
| TAAACC | LFY BS in AP3 | 1 | 5,756 (36%) | 1,648 (35%) | 1,526 (36%) | 952 (41%) | 1,630 (36%) | 374 (48%) |
| ATCGAC | Nonamer promoter motif | 1 | 1,817 (12%) | 515 (11%) | 479 (11%) | 285 (12%) | 538 (12%) | 117 (15%) |
| TCGACG | Nonamer promoter motif | 2 | 1,326 (8%) | 380 (8%) | 354 (8%) | 207 (9%) | 385 (9%) | 80 (10%) |
| CGGATC | Octamer promoter motif | 1 | 1,503 (10%) | 401 (9%) | 400 (9%) | 254 (11%) | 448 (10%) | 91 (12%) |
| CAACAG | RAV1-A binding site motif | 1 | 2,763 (18%) | 775 (16%) | 764 (18%) | 415 (18%) | 809 (18%) | 170 (22%) |
| TGGGCT | Site II | 3 | 2,769 (18%) | 805 (17%) | 697 (16%) | 475 (21%) | 792 (18%) | 186 (24%) |
| TGGGCC | Site II | 3 | 2,922 (19%) | 838 (18%) | 749 (18%) | 500 (22%) | 835 (19%) | 183 (24%) |
| AGGGCC | SORLIP2 | 1 | 562 (4%) | 174 (4%) | 151 (4%) | 93 (4%) | 144 (3%) | 34 (4%) |
| GGGCCA | SORLIP2 | 1 | 1,197 (8%) | 382 (8%) | 287 (7%) | 187 (8%) | 341 (8%) | 72 (9%) |
| TGGGCC | SORLIP2 | 1 | 2,922 (19%) | 838 (18%) | 749 (18%) | 500 (22%) | 835 (19%) | 183 (24%) |
| CTCAAG | SORLIP3 | 1 | 3,105 (20%) | 901 (19%) | 819 (19%) | 489 (21%) | 896 (20%) | 184 (24%) |
DISCUSSION
Profiling analysis has been carried out at different levels during germination in Arabidopsis, from transcriptomic (Nakabayashi et al., 2005) to proteomic (Gallardo et al., 2001; Rajjou et al., 2004; Fu et al., 2005; Chibani et al., 2006), to metabolite (Fait et al., 2006) analysis. These analyses have contributed to the understanding of the various processes that occur during germination. This study set out to observe if a greater temporal dissection of germination in Arabidopsis would reveal additional time- or stage-specific molecular processes that enable the transition from dormancy to active metabolism and also to gain a deeper understanding of the role of stratification in the process of germination. While the above studies have uncovered many novel aspects of germination and given insights into various regulatory processes, the results in this study revealed specific features of germination that, to our knowledge, have previously gone undetected, specifically, the number of changes in transcript abundance during 48 h of stratification almost equaled that observed during 48 h of germination in continuous light; the identification of a specific set of genes that appear to be predominantly expressed during the earliest stages of germination on the transition from stratification to germination, which are enriched in functions required for germination; and lastly, the environmental effects on the rate of RNA degradation during germination.
Stratification-Specific Regulation during Germination
The process of germination is strongly affected by environmental cues, which can significantly affect the rate and success of germination. Exposing seeds to cold temperatures to assist breaking dormancy, or seed stratification, is an established practice that is routinely utilized to maximize germination potential (Russell et al., 2000; Yamauchi et al., 2004; Dave et al., 2011). However, the molecular mechanisms underpinning the role of seed stratification have not been explored in depth, with only one previous study examining the effect of stratification on the transcriptome, utilizing 8K DNA microarrays and focusing on the role of GA during stratification (Yamauchi et al., 2004). Thus, our study, utilizing the 22K Arabidopsis genome microarrays, represents the most in-depth transcriptomic analysis during stratification to date. The first overall effects of stratification are evidenced in the number of DEGs, where there was in fact a greater rate of induction of genes upon exposure to light (cluster 1; Fig. 5A) and a significantly slower rate of decrease in abundance for stored mRNAs (cluster 2; Fig. 5A) for seeds that have been stratified versus not stratified, supporting the positive effect of stratification on germination rate. Also, comparing the DEGs in this study (including stratification) with germination without stratification (Nakabayashi et al., 2005) reveals that the up-regulation of transcripts encoding protein synthesis and hormone metabolism functions occurs between 12 h S and 48 h S in this study, revealing, to our knowledge for the first time, that the up-regulation of these genes occurs almost exclusively during stratification in the germination process (Fig. 1; Supplemental Fig. S6). It has been shown that although germination can succeed when transcription is inhibited, seedling establishment is prevented (Rajjou et al., 2004), revealing a crucial role for transcription postgermination. Taken together with the finding that germination is prevented when translation is inhibited (Rajjou et al., 2004), it is likely that these genes encoding protein synthesis functions represent the next wave of translational machinery required for germination progression.
In addition to protein synthesis functions, hormone metabolism was also seen to be affected during stratification (Fig. 1; Supplemental Fig. S6). Specifically, a significant up-regulation of genes encoding Ethylene Response Factor family members (ERFs; e.g. ERF1 [At3g23240], ERF2 [At5g47220], and ERF5 [At5g47230]) was observed within the first 12 h of stratification (Fig. 1; Supplemental Table S2). Notably, this early up-regulation of ERFs cannot be distinguished from genes highly expressed in the dry seed or other hormone-responsive genes when seeds are not stratified (Supplemental Figs. S6, A versus B, and S7; Nakabayashi et al., 2005). It has been shown that ethylene counteracts the inhibitory actions of ABA (Kecpczyński and Kecpczyńska, 1997; Beaudoin et al., 2000; Linkies et al., 2009) and promotes endosperm cap weakening and endosperm rupture in Lepidium sativum and Arabidopsis (Linkies et al., 2009). Thus, the rapid induction of these genes during stratification, as revealed in this study, not only confirms these roles for ethylene but reveals that this regulation is possibly one of the earliest contributing factors to the greater germination rates and synchrony generally observed after stratification. Taken together, the findings in this study provided novel insight into why stratification can lead to greater germination rates (i.e. hormonal signaling is activated at this stage before the other changes occur that drive germination and growth). In contrast, when seeds are not stratified, these changes occur simultaneously and thus are not as efficient at ensuring successful germination. This demonstrates that the specific temporal sequence of events that occur during germination is important for developmental progression.
Identification of a Novel Transient Expression Pattern during Germination
Upon examination of expression profiles during germination, it was revealed that approximately 14% of genes show transient expression, mostly peaking in abundance between 48 h S and 6 h SL (cluster 3; Fig. 2B). Extensive comparisons of these genes across other developmental tissues then revealed that a subset of these genes are specifically expressed during germination (GS set of 775 genes; Fig. 3A). Given that this transient expression pattern is not observed for a significant number of genes during germination without stratification (Supplemental Fig. S1), this suggests that during stratification, there is a time- and temperature-specific regulation that occurs that allows these genes to peak in expression before the observed synchronous decrease in abundance occurs. Thus, this expression pattern for these genes cannot be identified in unstratified seeds, as they are grouped with genes in cluster 2, which decline in abundance during germination. Examination of the genes in this GS set reveals an enrichment of genes encoding RNA-binding functions, nuclear and mitochondrial proteins, particularly those encoding PPR proteins (Fig. 3, B and C). This study revealed a tightly controlled stage of regulation for PPR-encoding genes over the course of germination (Fig. 3D). PPR proteins are defined by a repeating 35-amino acid motif, predicted to form an α-helix and to be targeted to mitochondria and plastids (Schmitz-Linneweber and Small, 2008). To date, PPR proteins have been shown to have roles in RNA splicing, cleavage, editing, stability, and translation (Schmitz-Linneweber and Small, 2008). A recent study has even shown that AtPPR2 binds to 23S rRNA and plays a role in mitotic division and cell proliferation during embryogenesis (Lu et al., 2011). Another study showed that a point mutation in the PPR domain of a chloroplast PPR protein delayed chloroplast development (Cao et al., 2011). In addition to this, many of the genes identified as showing embryo-lethal phenotypes under loss-of-function conditions (Meinke et al., 2008) encode PPRs, and a significant number of these showed this transient expression pattern in this study, suggesting a finely controlled, stage-specific requirement for RNA processing during germination. Overall, a significant proportion of the genes defined as displaying a GS profile in this study were observed to display altered phenotypes associated with embryo development or male or female gametophyte development (Fig. 4). This shows that the functions of many of the proteins, largely with RNA-binding functions, encoded by these genes are essential for germination, thus giving insight into some of the earliest processes that occur during germination.
The role of RNA-binding proteins has been examined in a number of studies, from those specifically focused on plastid RNA-binding proteins (Wang et al., 2006) to analysis of families of proteins such as the Gly-rich RNA-binding proteins (Kim et al., 2007; Kwak et al., 2011). Specifically, transcriptomic analysis revealed that two genes, cp29A and cp29B, were highly expressed in germinating seedlings and therefore were selected for further analysis (Wang et al., 2006). Interestingly, a correlation between the transcript and protein abundance for these genes was observed, and it was observed that new isoforms of these proteins were generated following posttranslational modification of these proteins during seedling development (Wang et al., 2006). Similarly, studies examining Gly-rich RNA-binding proteins have also indicated a crucial role for these proteins during Arabidopsis germination and seedling development (Kim et al., 2007) as well as in the regulation of gene expression at the posttranscriptional level during abiotic stress (Schmidt et al., 2010). A recent study analyzing the protein structure of GRP4 and GRP7 during cold acclimation revealed the crucial role of specific sequence domains necessary for the correct RNA chaperone activity of these proteins (Kwak et al., 2011). Interestingly, GFP localization in this study has revealed, to our knowledge for the first time, that GRP4 is localized to the mitochondria (Fig. 5). Also, notably, comparison of the protein abundance data during germination and the transcriptomic analysis from this study has also revealed that GRP7 is transiently expressed both at the transcript (this study) and protein (Fu et al., 2005) levels during germination and seedling establishment. Collectively, these findings suggest a finely controlled but crucial role of RNA-binding GRPs during germination.
A recent study used publicly available microarray data to examine the phase transitions from dormancy to germination and generated a condition-dependent network model of transcriptional interactions in Arabidopsis called SeedNet (Bassel et al., 2011; http://vseed.nottingham.ac.uk). This network comprises 8,261 nodes and demonstrates two major regions of interaction enriched in transcripts identified by significance analysis microarrays: region 1, which is associated with nongermination; region 2, which represents a transition between nongermination and germination; and region 3, which is associated with germination (Bassel et al., 2011). Closer examination reveals that region 2 is significantly enriched in transcripts encoding RNA metabolism functions and represents a unique cluster of interactions that bridge these two regions, suggesting a mediating role for these genes in the transition from nongerminating to germinating states (Bassel et al., 2011). Interestingly, when the 775 transcripts comprising the GS set (identified in Fig. 3A) from this study were queried in SeedNet, the majority localized to region 2 and region 3, with only a small proportion being identified in region 1. The large number of GS genes seen in region 2, together with the association of these genes with RNA metabolism as well and the enrichment of RNA-binding/processing functions, suggest that the transient up-regulation of these genes represents a crucial regulatory switch necessary for germination progression.
In addition to RNA-processing and PPR protein-encoding genes, closer examination of the genes in the GS set (775 genes; Fig. 3A) rapidly reveals other genes encoding mitochondrial proteins that also show this tight, transient expression pattern. GFP localization for a selection of the proteins encoded by these genes (Fig. 4) confirmed their mitochondrial localization and supported the coexpression and colocalization pattern observed for many of these genes. The crucial role of mitochondria during germination has been examined before, and it has been suggested that the biogenesis of new mitochondria is more important in oilseeds (Morohashi et al., 1981; Weitbrecht et al., 2011). Notably, this burst in the expression of genes encoding mitochondrial proteins does not represent the building of the bioenergetic functions required to power germination but rather a specific phase of DNA replication, RNA synthesis, and processing that occurs during germination. The genes encoding mitochondrial proteins that were most highly expressed (transiently) at this time included a significant percentage of genes encoding mTERFs (15 of the 23 genes encoding mTERFs were expressed during germination; Fig. 5C) and PPR proteins (137 of the 309 genes encoding PPRs were expressed during germination; Fig. 3Di). It has been shown that mTERFs not only function in mitochondrial transcription termination but also in the initiation of transcription and the control of mitochondrial DNA replication (Roberti et al., 2009). Taken together, these findings reveal a possible pinpoint in time during germination when mitochondrial biogenesis is activated. This activation of mitochondrial DNA replication and transcription may play a crucial role in signaling the later expression of other bioenergetics components required for germination. It has been previously reported that a dual-targeted protein that plays a role in translation in plastids and mitochondria is required for organelle retrograde signaling (Pesaresi et al., 2006). Thus, these findings during germination suggest the possibility that mitochondrial retrograde signaling may play a crucial role in the process of germination.
Interestingly, the transcriptomic analysis of rice germination also identified a subset of transiently expressed genes (Howell et al., 2009); however, this occurred later in germination, and the functions of genes in these sets differed. While the transiently expressed genes during rice germination were enriched in genes encoding TFs (Howell et al., 2009), genes transiently expressed in Arabidopsis germination were enriched in RNA-processing functions (Fig. 3A). However, it is possible that the controlled expression of these RNA-processing functions also occurs in rice but is missed due to rice seed maturity, as rice seeds do not undergo stratification; thus, dormancy is broken in an alternative manner. Another possibility is that these differences represent divergences between monocots and dicots or starch seeds and oilseeds. However, more intensive profiling studies in rice or other cereals, from fresh seed, during ripening and during dormancy alleviation may identify a similar process.
Crucial Role of mRNA Decay during Germination
One of the first processes seen to occur upon imbibition is the clearing out of stored transcripts (cluster 2; Fig. 6A). For these, it was seen that even transcripts with mRNA half-lives longer than 6 h decrease to less than 50% of their starting abundance in under 6 h upon imbibition in optimal light and temperature conditions (no stratification; cluster 2; Fig. 6, Aii and Bii), while transcripts in this cluster decrease in abundance at a much slower rate during stratification (this study; cluster 2; Fig. 6A). Finding the genes in cluster 2 (i.e. the stored mRNAs) to be enriched in TFs was somewhat surprising, given that transcripts encoding TFs have been shown to have short half-lives, allowing these mRNAs to be rapidly degraded (Narsai et al., 2007). Thus, these findings suggest that the stored seed transcripts are somehow highly stabilized in the dry seed and that controlled degradation of these specific transcripts occurs upon imbibition, even beginning during stratification. The importance of the role of mRNA decay during germination has been displayed in studies that have shown that normal germination and development suffer when mRNA degradation is affected (Delseny et al., 1977; Nishimura et al., 2005; Hirayama and Shinozaki, 2007). Given that the expression profiles compared for stratified versus nonstratified seeds revealed differences in the rate of decrease for transcripts in cluster 2, this indicates that the germination “clock” is highly controlled, with gene expression being tightly regulated and controlled by environmental conditions. A perfect example of this controlled regulation during germination is for genes in cluster 3 (Fig. 2B), which show a strong induction followed by an equivalently strong reduction of these genes, suggesting that tight regulation occurs, most likely both at the transcriptional and degradation levels. A previous study has shown a role for controlled/active mRNA decay in yeast in response to anaerobic conditions (Dagsgaard et al., 2001). Thus, the tightly controlled regulation of transcript abundance observed in clusters 2 and 3, as well as the differences in the rates of decrease in abundance under stratified or unstratified conditions, present an argument suggesting a possible role for active mRNA decay that occurs in response to the imbibition or light during germination.
CONCLUSION
The greater temporal dissection of germination combined with functional analysis reveals molecular mechanisms occurring during germination that have previously gone undetected. Identification of these processes provides a molecular explanation for the greater rates of germination that occur during germination after stratification and also provide greater resolution of the processes that occur during germination. If such processes also occur in monocots, it may provide new targets to prevent precocious germination in cereals.
MATERIALS AND METHODS
Arabidopsis Tissue Collection and Microarrays
To analyze a range of time points before and during Arabidopsis (Arabidopsis thaliana ecotype Columbia) germination, 10 time points were analyzed including freshly harvested seeds (H; which were collected from a single batch of wild-type plants that were exactly the same age) and then the seeds following 15 d of ripening (0 h) and after 1 h of stratification (1 h S), 12 h of stratification (12 h S), and 48 h of stratification (48 h S); plates were then transferred to continuous light and collected 1 h into the light (1 h SL), 6 h into the light (6 h SL), 12 h into the light (12 h SL), 24 h into the light (24 h SL), and 48 h into the light (48 h SL). Eighty milligrams of wild-type Arabidopsis seeds was plated onto Murashige and Skoog medium (containing 3% Suc) for each individual time point collection in the time course. Collections were repeated three times for three biological replicates.
RNA Isolation and Microarray and Differential Expression Analyses
For all samples collected, the Ambion Plant RNA isolation aid and RNAqeous RNA isolation kit were used for effective isolation of RNA. A total of 400 ng of total RNA was used as the starting amount of RNA for the ATH1 Arabidopsis genome expression array. Using the IVT Express kit (Affymetrix), microarrays were carried out according to the manufacturers’ instructions. Before beginning the microarray experiments as well as during the course of the microarrays, the Agilent Bioanalyzer was used to ensure high-quality starting RNA, generated amplified RNA, and effective fragmentation prior to hybridization to the microarray. All raw intensity CEL files were imported into Avadis 4.3 (Strand Genomics), and standard MAS5.0 normalization was first carried out to determine present/absent/marginal calls for each probe set. All probe sets that encoded hybridization controls, bacterial genes, and more than one single gene were excluded, leaving a global Arabidopsis expression set consisting of 15,789 probe sets. Probe sets that were called present in two or more replicates were considered to be expressed and were then used for further analysis. GC-robust multiarray average normalization was carried out for all 30 microarrays, and the resulting normalized intensities were used as the input for the differential expression analysis. This was carried out using the Cyber-T method, which implements a Bayesian method (Baldi and Long, 2001) for the determination of probe sets showing significant changes in transcript abundance. The posterior probability of differential expression (PPDE) method within Cyber-T was used for false discovery rate calculation (Choe et al., 2005). All input criteria were set according to Cyber-T recommendations applicable for each experimental set. A probe set was defined as significantly changing at P < 0.05, with a PPDE of greater than 0.96 (false discovery rate). These cutoffs and this Bayesian method of differential expression have been verified and been used in previous microarray studies (Narsai et al., 2010). In this way, step-wise differential expression analysis was carried out. All original microarray data files have been deposited to the Gene Expression Omnibus at the National Center for Biotechnology Information under accession number GSE30223.
Hierarchical Clustering and z-Score Analyses of GO Categories
To view the profiles of expression changes over the time course, all GC-robust multiarray average studies were made relative to the maximum intensity over the time course. These data were then hierarchically clustered using average linkage (based on Euclidean distance), and four cluster profiles were drawn using Partek Genomics Suite (version 6.5). To determine if there was a statistically significant overrepresentation or underrepresentation of a particular subcategory of GO (i.e. GO cellular component, GO molecular function, and GO biological process), a z-score analysis was carried out to compare the two proportions (subset versus genome), where π refers to the mean and n is the number of genes in the subset.
A cumulative standard normal table was used to match the z-score, and based on this, P values were determined.
PageMan Analysis
To analyze the functional representation of the genes differentially expressed during the time course, differential PageMan analyses were carried out using the unique set of probe sets representing the DEGs (Usadel et al., 2006). Fisher’s test for overrepresentation analysis (1.0 cutoff; Benjamini-Hochberg [Benjamini and Hochberg, 1995] false discovery rate correction) was carried out in PageMan to determine statistically significant over/underrepresentation of genes classified into specific bins.
Publicly Available Arabidopsis and Rice Microarrays
To compile the publicly available Affymetrix Arabidopsis and rice (Oryza sativa) microarrays, all experiments containing CEL files were downloaded from the Gene Expression Omnibus within the National Centre for Biotechnology Information database or from the MIAME ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/). The Gene Expression Omnibus Series or Experiment numbers for the respective studies are shown in Supplemental Table S3. The Arabidopsis AtGenExpress developmental data set was downloaded as CEL files (E-AFMX-9). These CEL files in addition to the 30 CEL files from this study were imported and quantile normalized together to enable comparability across these arrays using Partek Genomics Suite version 6.5. To carry out parallel analysis for developmental conditions in rice, 41 developmental tissues for rice (Supplemental Table S4) were also downloaded and analyzed in the same manner as described for the Arabidopsis developmental set.
Analysis of Orthologs
The InParanoid: Eukaryotic Ortholog Groups database (version 7.0; Remm et al., 2001) was utilized to analyze all orthologs between Arabidopsis and rice. The orthologous group file containing the rice versus Arabidopsis set was downloaded for the whole-genome comparison. This generated information for orthologs is identified by Arabidopsis Genome Initiative (AGI) numbers for Arabidopsis and The Institute for Genomic Research identifiers for rice.
Analysis of Phenotypes for 775 GS Genes and Random Gene-Sets
For this analysis, only the list of 21,192 Affymetrix probe sets that matched to individual AGI numbers was isolated and used. From those, three sets of 775 randomly selected gene-sets were generated using Partek Genomic Suite version 6.5. These sets were analyzed and scrutinized for phenotypes in exactly the same manner as was carried out for the 775 GS set identified in Figure 3A. First, large-scale forward genetics studies were mined and matched to any genes in these sets; in addition, all genes encoding proteins localized to the mitochondria were individually searched for publications indicating phenotypes, and these were collated (numbers for each set are shown in Supplemental Table S6B). Full references for all phenotypes identified in the 775 GS set are shown in Supplemental Table S6C).
Construction of GFP Fusion Proteins to Confirm Organelle Targeting
Given that the expression profiles showed correlation between coexpression and colocalization (e.g. the genes showing a transient expression pattern [cluster 3] were enriched in genes encoding mitochondrial proteins, while cluster 1 was enriched in genes encoding plastid proteins; Fig. 2C), a selection of 65 genes, mostly encoding proteins predicted to be mitochondrial, were selected for GFP localization analysis. For each gene, the region of the protein encoding the targeting signals was cloned in frame with GFP by Gateway cloning (Invitrogen), as described previously (Carrie et al., 2009). For proteins predicted to target to the mitochondria or plastid, the first 100 amino acids were fused to the N terminus of GFP. For proteins predicted to target to the peroxisome, the last 100 amino acids were fused to the C terminus of GFP. Three RFP fusion proteins were utilized as controls for subcellular localization. To control for mitochondrial targeting, the 42-amino acid mitochondrial targeting signal of AOX was fused to RFP (Murcha et al., 2007). To control for plastid targeting, the full-length cDNA of the plastid-targeted SSU was fused to RFP (Murcha et al., 2007). To control for peroxisomal targeting, the peroxisomal targeting signal 1 was fused to the C terminus of RFP, which has been used effectively previously (Carrie et al., 2007).
Confirmation of Organelle Targeting of the GFP Fusion Proteins
Biolistic transformation of the GFP constructs of interest, with their associated RFP controls, was carried out on Arabidopsis cell suspensions as reported previously (Carrie et al., 2007). Approximately 5 μg of both the GFP fusion protein and the RFP control was coprecipitated onto gold particles and transiently transformed using the biolistic PDS-1000/He system (Bio-Rad; http://bio-rad.com/). Gold particles were bombarded into 2 mL of Arabidopsis cell suspension spread on filter paper placed on osmoticum plates, followed by incubation at 22°C for 24 h in the dark. Visualization of the fluorescent proteins was carried out using an Olympus BX61 fluorescence microscope (http://www.olympusmicro.com) with excitation wavelengths of 460/480 nm for GFP and 535/555 nm for RFP; emission wavelengths were measured at 495 to 540 nm for GFP and 570 to 625 nm for RFP. Micrographs were captured and processed using cell-imaging software as described previously (Carrie et al., 2007). Localizations as determined by GFP analysis were as follows: At5g60730, mitochondrial; At5g40660, mitochondrial; At1g14620, mitochondrial; At4g14790, mitochondrial; At3g23830, mitochondrial; At1g10270, mitochondrial; At5g39960, mitochondrial; At5g46920, mitochondrial; At1g67620, mitochondrial; At1g61990, mitochondrial; At1g09680, mitochondrial; At1g20300, mitochondrial; At1g80270, mitochondrial; At2g15630, mitochondrial; At2g40240, mitochondrial; At4g02820, mitochondrial; At5g60960, mitochondrial; At5g61370, mitochondrial; At3g15140, mitochondrial; At3g19440, mitochondrial; At5g54580, mitochondrial; At4g29540, mitochondrial; At1g71850, mitochondrial; At2g39120, mitochondrial; At1g08220, mitochondrial; At1g28395, mitochondrial; At1g79490, mitochondrial; At5g39710, mitochondrial; At5g47455, mitochondrial; At5g03455, mitochondrial; At2g16570, plastid; At1g30270, plastid; At4g36040, plastid; At2g27510, plastid; At1g09940, plastid; At4g08790, plastid; At3g07780, plastid; At1g78050, plastid; At5g66950, plastid; At3g56330, plastid; At1g51080, plastid; At1g52720, plastid; At3g19680, plastid; At3g24506, plastid; At3g56360, plastid; At5g06810, mitochondrial/plastid; At4g21170, mitochondrial/plastid; At1g06270, plastid membrane/endoplasmic reticulum; At3g60680, peroxisome/cytoskeleton; At1g69270, peroxisome; At4g14680, nucleus; At2g44510, nucleus; At2g45210, nucleus; At1g02870, nucleus; At1g07220, nucleus; At1g18680, cytoplasmic; At1g64185, cytoplasmic; At5g51770, cytoplasmic; At5g50760, cytoplasmic; At5g65450, cytoplasmic; At1g31940, cytoplasmic; At1g74680, endoplasmic reticulum; At3g21140, endoplasmic reticulum; At1g04900, endoplasmic reticulum; At4g00750, Golgi.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Gene expression profiles during germination without stratification (data from Nakabayashi et al., 2005) were also hierarchically clustered in parallel.
Supplemental Figure S2. Gene expression across Arabidopsis germination and development.
Supplemental Figure S3. Expression of rice genes orthologous to 775 Arabidopsis GS genes across rice germination and development.
Supplemental Figure S4. Expression profiles of all 422 genes with known seed phenotypes (described in the SeedGenes database; Meinke et al., 2008).
Supplemental Figure S5. GFP fluorescence images for the 65 proteins targeted to the mitochondria, plastids, and/or peroxisomes.
Supplemental Figure S6. PageMan output for DEGs during Arabidopsis germination without stratification (A), original microarray data from Nakabayashi et al. (2005), and including stratification (B), as in this study.
Supplemental Figure S7. Gene expression profiles during germination with stratification (this study) and without stratification (data from Nakabayashi et al., 2005) are visualized in parallel for genes encoding proteins involved in ethylene-related functions (based on PageMan bins).
Supplemental Table S1. Differential expression analysis in dry seeds and stratified and germinating seedlings.
Supplemental Table S2. PageMan output for DEGs during Arabidopsis germination.
Supplemental Table S3. Details of the developmental tissue set (Schmid et al., 2005; E-AFMX-9) AtGenExpress (ATGE) accessions, number of biological replications, sample details, genotype, tissue, age, photoperiod, growth, and substrate.
Supplemental Table S4. Details of the developmental tissue microarrays used for rice.
Supplemental Table S5. Comparison of protein abundance data from Fu et al. (2005) with the parallel transcript abundance profiles in this germination study.
Supplemental Table S6. Details of the phenotypes shown in Figure 4, names of database resources used, and full references for these genes.
Supplemental Table S7. A selection of genes (more than 75% from cluster 3) with predicted organellar localizations were analyzed by GFP tagging to confirm protein localization.
References
- Allen PS, Benech-Arnold RL, Batlla D, Bradford KJ. (2007) Modeling of seed dormancy. Bradford KJ, Nonogaki H, , Seed Development, Dormancy and Germination, Vol 27 Blackwell Publishing, Oxford, doi/10.1002/9780470988848.ch4 [Google Scholar]
- Baldi P, Long AD. (2001) A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17: 509–519 [DOI] [PubMed] [Google Scholar]
- Baskin CC, Baskin JM. (2004) A classification system for seed dormancy. Seed Sci Res 14: 1–16 [Google Scholar]
- Bassel GW, Lan H, Glaab E, Gibbs DJ, Gerjets T, Krasnogor N, Bonner AJ, Holdsworth MJ, Provart NJ. (2011) Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc Natl Acad Sci USA 108: 9709–9714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin N, Serizet C, Gosti F, Giraudat J. (2000) Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12: 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Boavida LC, Shuai B, Yu HJ, Pagnussat GC, Sundaresan V, McCormick S. (2009) A collection of Ds insertional mutants associated with defects in male gametophyte development and function in Arabidopsis thaliana. Genetics 181: 1369–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, McCourt P. (2003) Hormone cross-talk in seed dormancy. Plant Growth Regul 22: 25–31 [Google Scholar]
- Cadman CS, Toorop PE, Hilhorst HW, Finch-Savage WE. (2006) Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J 46: 805–822 [DOI] [PubMed] [Google Scholar]
- Cao D, Cheng H, Wu W, Soo HM, Peng J. (2006) Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol 142: 509–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao ZL, Yu QB, Sun Y, Lu Y, Cui YL, Yang ZN. (2011) A point mutation in the pentatricopeptide repeat motif of the AtECB2 protein causes delayed chloroplast development. J Integr Plant Biol 53: 258–269 [DOI] [PubMed] [Google Scholar]
- Carrie C, Kuhn K, Murcha MW, Duncan O, Small ID, O’Toole N, Whelan J. (2009) Approaches to defining dual-targeted proteins in Arabidopsis. Plant J 57: 1128–1139 [DOI] [PubMed] [Google Scholar]
- Carrie C, Murcha MW, Millar AH, Smith SM, Whelan J. (2007) Nine 3-ketoacyl-CoA thiolases (KATs) and acetoacetyl-CoA thiolases (ACATs) encoded by five genes in Arabidopsis thaliana are targeted either to peroxisomes or cytosol but not to mitochondria. Plant Mol Biol 63: 97–108 [DOI] [PubMed] [Google Scholar]
- Chen KM, Holmström M, Raksajit W, Suorsa M, Piippo M, Aro EM. (2010) Small chloroplast-targeted DnaJ proteins are involved in optimization of photosynthetic reactions in Arabidopsis thaliana. BMC Plant Biol 10: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibani K, Ali-Rachedi S, Job C, Job D, Jullien M, Grappin P. (2006) Proteomic analysis of seed dormancy in Arabidopsis. Plant Physiol 142: 1493–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe SE, Boutros M, Michelson AM, Church GM, Halfon MS. (2005) Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control dataset. Genome Biol 6: R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagsgaard C, Taylor LE, O’Brien KM, Poyton RO. (2001) Effects of anoxia and the mitochondrion on expression of aerobic nuclear COX genes in yeast: evidence for a signaling pathway from the mitochondrial genome to the nucleus. J Biol Chem 276: 7593–7601 [DOI] [PubMed] [Google Scholar]
- Dave A, Hernández ML, He Z, Andriotis VM, Vaistij FE, Larson TR, Graham IA. (2011) 12-Oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell 23: 583–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, Grotewold E. (2003) AGRIS: Arabidopsis Gene Regulatory Information Server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delseny M, Aspart L, Guitton Y. (1977) Effect of the protein synthesis inhibitor cycloheximide on RNA synthesis in radish seedlings. Biochimie 59: 51–57 [DOI] [PubMed] [Google Scholar]
- Ding YH, Liu NY, Tang ZS, Liu J, Yang WC. (2006) Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell 18: 815–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmann EM, Nill C, Schwechheimer C. (2010) DELLA proteins restrain germination and elongation growth in Arabidopsis thaliana COP9 signalosome mutants. Eur J Cell Biol 89: 163–168 [DOI] [PubMed] [Google Scholar]
- Doniwa Y, Ueda M, Ueta M, Wada A, Kadowaki K, Tsutsumi N. (2010) The involvement of a PPR protein of the P subfamily in partial RNA editing of an Arabidopsis mitochondrial transcript. Gene 454: 39–46 [DOI] [PubMed] [Google Scholar]
- Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochniak E, Fernie AR, Galili G. (2006) Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol 142: 839–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feurtado JA, Kermode AR. (2007) A merging of paths: abscisic acid and hormonal cross-talk in the control of seed dormancy maintenance and alleviation. Bradford KJ, Nonogaki H, , Seed Development, Dormancy and Germination, Vol 27 Blackwell Publishing, Oxford, doi/10.1002/9780470988848.ch8 [Google Scholar]
- Fu Q, Wang BC, Jin X, Li HB, Han P, Wei KH, Zhang XM, Zhu YX. (2005) Proteomic analysis and extensive protein identification from dry, germinating Arabidopsis seeds and young seedlings. J Biochem Mol Biol 38: 650–660 [DOI] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D. (2001) Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol 126: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D. (2002) Proteomics of Arabidopsis seed germination: a comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol 129: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Ng S, Carrie C, Duncan O, Low J, Pong Lee C, Van Aken O, Millar AH, Murcha MW, Whelan J. (2010) TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22: 3921–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham IA. (2008) Seed storage oil mobilization. Annu Rev Plant Biol 59: 115–142 [DOI] [PubMed] [Google Scholar]
- Hammani K, Gobert A, Hleibieh K, Choulier L, Small I, Giegé P. (2011) An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. Plant Cell 23: 730–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. (2007) SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res 35: D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, et al. (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res 14: 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. (2007) Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci 12: 343–351 [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJ. (2008a) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Finch-Savage WE, Grappin P, Job D. (2008b) Post-genomics dissection of seed dormancy and germination. Trends Plant Sci 13: 7–13 [DOI] [PubMed] [Google Scholar]
- Howell KA, Cheng K, Murcha MW, Jenkin LE, Millar AH, Whelan J. (2007) Oxygen initiation of respiration and mitochondrial biogenesis in rice. J Biol Chem 282: 15619–15631 [DOI] [PubMed] [Google Scholar]
- Howell KA, Millar AH, Whelan J. (2006) Ordered assembly of mitochondria during rice germination begins with pro-mitochondrial structures rich in components of the protein import apparatus. Plant Mol Biol 60: 201–223 [DOI] [PubMed] [Google Scholar]
- Howell KA, Narsai R, Carroll A, Ivanova A, Lohse M, Usadel B, Millar AH, Whelan J. (2009) Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol 149: 961–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappen C. (2000) Analysis of a complete homeobox gene repertoire: implications for the evolution of diversity. Proc Natl Acad Sci USA 97: 4481–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kecpczyński J, Kecpczyńska E. (1997) Ethylene in seed dormancy and germination. Physiol Plant 101: 720–726 [Google Scholar]
- Keren I, Bezawork-Geleta A, Kolton M, Maayan I, Belausov E, Levy M, Mett A, Gidoni D, Shaya F, Ostersetzer-Biran O. (2009) AtnMat2, a nuclear-encoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA 15: 2299–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YO, Pan S, Jung CH, Kang H. (2007) A zinc finger-containing glycine-rich RNA-binding protein, atRZ-1a, has a negative impact on seed germination and seedling growth of Arabidopsis thaliana under salt or drought stress conditions. Plant Cell Physiol 48: 1170–1181 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Suzuka I, Ohashi Y. (1995) Two of three promoter elements identified in a rice gene for proliferating cell nuclear antigen are essential for meristematic tissue-specific expression. Plant J 7: 877–886 [DOI] [PubMed] [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD. (2007a) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10: 310–316 [DOI] [PubMed] [Google Scholar]
- Kotak S, Vierling E, Bäumlein H, von Koskull-Döring P. (2007b) A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19: 182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Wada T, Kamiya A, Yuguchi M, Yokouchi T, Imura Y, Takabe H, Sakurai T, Akiyama K, Hirayama T, et al. (2006) A trial of phenome analysis using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant J 47: 640–651 [DOI] [PubMed] [Google Scholar]
- Kwak KJ, Park SJ, Han JH, Kim MK, Oh SH, Han YS, Kang H. (2011) Structural determinants crucial to the RNA chaperone activity of glycine-rich RNA-binding proteins 4 and 7 in Arabidopsis thaliana during the cold adaptation process. J Exp Bot 62: 4003–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Thomas TL. (1998) PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell 10: 383–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkies A, Müller K, Morris K, Turecková V, Wenk M, Cadman CS, Corbineau F, Strnad M, Lynn JR, Finch-Savage WE, et al. (2009) Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 21: 3803–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Carrie C, Duncan O, Ho LH, Howell KA, Murcha MW, Whelan J. (2007) Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell 19: 3739–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan DC, Leaver CJ. (2000) Mitochondria-targeted GFP highlights the heterogeneity of mitochondrial shape, size and movement within living plant cells. J Exp Bot 51: 865–871 [PubMed] [Google Scholar]
- Lu Y, Li C, Wang H, Chen H, Berg H, Xia Y. (2011) AtPPR2, an Arabidopsis pentatricopeptide repeat protein, binds to plastid 23S rRNA and plays an important role in the first mitotic division during gametogenesis and in cell proliferation during embryogenesis. Plant J 67: 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D, Muralla R, Sweeney C, Dickerman A. (2008) Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci 13: 483–491 [DOI] [PubMed] [Google Scholar]
- Morohashi Y, Bewley JD, Yeung EC. (1981) Biogenesis of mitochondria in imbibed peanut cotyledons. II. Development of light and heavy mitochondria. Plant Physiol 68: 318–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcha MW, Elhafez D, Lister R, Tonti-Filippini J, Baumgartner M, Philippar K, Carrie C, Mokranjac D, Soll J, Whelan J. (2007) Characterization of the preprotein and amino acid transporter gene family in Arabidopsis. Plant Physiol 143: 199–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Nakagawa N, Sakurai N. (2006) A mutation in At-nMat1a, which encodes a nuclear gene having high similarity to group II intron maturase, causes impaired splicing of mitochondrial NAD4 transcript and altered carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol 47: 772–783 [DOI] [PubMed] [Google Scholar]
- Narsai R, Castleden I, Whelan J. (2010) Common and distinct organ and stress responsive transcriptomic patterns in Oryza sativa and Arabidopsis thaliana. BMC Plant Biol 10: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Carroll A, Ivanova A, Millar AH, Whelan J. (2009) Defining core metabolic and transcriptomic responses to oxygen availability in rice embryos and young seedlings. Plant Physiol 151: 306–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Millar AH, O’Toole N, Small I, Whelan J. (2007) Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19: 3418–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman TC, Ohme-Takagi M, Taylor CB, Green PJ. (1993) DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell 5: 701–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Kitahata N, Seki M, Narusaka Y, Narusaka M, Kuromori T, Asami T, Shinozaki K, Hirayama T. (2005) Analysis of ABA hypersensitive germination2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant J 44: 972–984 [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Maruyama K, Seki M, Satou M, Shinozaki K, Yamaguchi-Shinozaki K. (2005) Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17: 1105–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, Sugita M, Small I. (2008) On the expansion of the pentatricopeptide repeat gene family in plants. Mol Biol Evol 25: 1120–1128 [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V. (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Palaniswamy SK, James S, Sun H, Lamb RS, Davuluri RV, Grotewold E. (2006) AGRIS and AtRegNet: a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol 140: 818–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin R, Meyer EH, Zaepfel M, Kim YJ, Mache R, Grienenberger JM, Gualberto JM, Gagliardi D. (2004) Two exoribonucleases act sequentially to process mature 3′-ends of atp9 mRNAs in Arabidopsis mitochondria. J Biol Chem 279: 25440–25446 [DOI] [PubMed] [Google Scholar]
- Pesaresi P, Masiero S, Eubel H, Braun HP, Bhushan S, Glaser E, Salamini F, Leister D. (2006) Nuclear photosynthetic gene expression is synergistically modulated by rates of protein synthesis in chloroplasts and mitochondria. Plant Cell 18: 970–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D. (2004) The effect of α-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol 134: 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M, Storm CE, Sonnhammer EL. (2001) Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol 314: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Riaño-Pachón DM, Ruzicic S, Dreyer I, Mueller-Roeber B. (2007) PlnTFDB: an integrative plant transcription factor database. BMC Bioinformatics 8: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberti M, Polosa PL, Bruni F, Manzari C, Deceglie S, Gadaleta MN, Cantatore P. (2009) The MTERF family proteins: mitochondrial transcription regulators and beyond. Biochim Biophys Acta 1787: 303–311 [DOI] [PubMed] [Google Scholar]
- Russell L, Larner V, Kurup S, Bougourd S, Holdsworth M. (2000) The Arabidopsis COMATOSE locus regulates germination potential. Development 127: 3759–3767 [DOI] [PubMed] [Google Scholar]
- Saiga S, Furumizu C, Yokoyama R, Kurata T, Sato S, Kato T, Tabata S, Suzuki M, Komeda Y. (2008) The Arabidopsis OBERON1 and OBERON2 genes encode plant homeodomain finger proteins and are required for apical meristem maintenance. Development 135: 1751–1759 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schmidt F, Marnef A, Cheung MK, Wilson I, Hancock J, Staiger D, Ladomery M. (2010) A proteomic analysis of oligo(dT)-bound mRNP containing oxidative stress-induced Arabidopsis thaliana RNA-binding proteins ATGRP7 and ATGRP8. Mol Biol Rep 37: 839–845 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I. (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Schöffl F, Prändl R, Reindl A. (1998) Regulation of the heat-shock response. Plant Physiol 117: 1135–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N, Usadel B, Winter A, Radchuk V, Scholz U, Stein N, Weschke W, Strickert M, Close TJ, Stitt M, et al. (2008) Barley grain maturation and germination: metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiol 146: 1738–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun TP. (2004) Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BC, Wang HX, Feng JX, Meng DZ, Qu LJ, Zhu YX. (2006) Post-translational modifications, but not transcriptional regulation, of major chloroplast RNA-binding proteins are related to Arabidopsis seedling development. Proteomics 6: 2555–2563 [DOI] [PubMed] [Google Scholar]
- Weitbrecht K, Muller K, Leubner-Metzger G. (2011) First off the mark: early seed germination. J Exp Bot 62: 3289–3309 [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S. (2004) Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16: 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, Mejia-Guerra MK, Kurz K, Liang X, Welch L, Grotewold E. (2011) AGRIS: the Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res 39: D1118–D1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu QB, Jiang Y, Chong K, Yang ZN. (2009) AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J 59: 1011–1023 [DOI] [PubMed] [Google Scholar]