Abstract
Activities of 28 enzymes from central carbon metabolism were measured in pericarp tissue of ripe tomato fruits from field trials with an introgression line (IL) population generated by introgressing segments of the genome of the wild relative Solanum pennellii (LA0716) into the modern tomato cultivar Solanum lycopersicum M82. Enzyme activities were determined using a robotized platform in optimized conditions, where the activities largely reflect the level of the corresponding proteins. Two experiments were analyzed from years with markedly different climate conditions. A total of 27 quantitative trait loci were shared in both experiments. Most resulted in increased enzyme activity when a portion of the S. lycopersicum genome was substituted with the corresponding portion of the genome of S. pennellii. This reflects the change in activity between the two parental genotypes. The mode of inheritance was studied in a heterozygote IL population. A similar proportion of quantitative trait loci (approximately 30%) showed additive, recessive, and dominant modes of inheritance, with only 5% showing overdominance. Comparison with the location of putative genes for the corresponding proteins indicates a large role of trans-regulatory mechanisms. These results point to the genetic control of individual enzyme activities being under the control of a complex program that is dominated by a network of trans-acting genes.
The cultivated tomato (Solanum lycopersicum) is the second most consumed noncereal crop worldwide. Tomatoes are consumed raw, cooked, and in a variety of processed products. It is therefore important to understand the synthesis and storage of metabolites that affect nutritional or gustative qualities of tomato fruits. In addition, tomato is an important model for studies of fruit physiology and development and for quantitative genetics (Tanksley et al., 1995; Giovannoni, 2001; Zamir, 2001; Mueller et al., 2005; Lippman et al., 2007).
Modern tomato cultivars have limited genetic variability due to natural and artificial selection during domestication and evolution (Rick, 1976). Wild species are an especially rich source of desirable genetic diversity. Several inbred lines have been generated following the crossing of S. lycopersicum with wild relatives from the so-called “esculentum complex” (Knapp et al., 2004), including a set of 76 introgression lines (ILs) derived from a S. lycopersicum cv M82 × Solanum pennellii cross (Eshed and Zamir, 1994; Mueller et al., 2005). Each of these lines contains a small introgressed region of the S. pennellii genome, containing an estimated 200 to 1,000 genes (Kamenetzky et al., 2010) in a genetic background that otherwise derives from S. lycopersicum. The 76 ILs cover the entire S. pennellii genome. This IL population has been subjected to extensive agronomic, physiological, and molecular phenotyping (Lippman et al., 2007). Quantitative trait loci (QTLs) have been detected that affect morphology and yield (Semel et al., 2006), fruit coloration (Liu et al., 2003), metabolite levels (Causse et al., 2004; Fridman et al., 2004; Baxter et al., 2005; Schauer et al., 2006), volatile metabolites (Tieman et al., 2006), and antioxidants (Rousseaux et al., 2005). Tomato is a model for quantitative genetics (Tanksley et al., 1995) and one of the first examples of a crop plant that has benefited significantly from exotic germplasm introgression (Zamir, 2001; Lippman et al., 2007).
There have been numerous studies of natural variation on the levels of individual metabolites in tomato fruit (Causse et al., 1995, 2004; Eshed and Zamir, 1995; Fridman et al., 2000, 2002, 2004; Zamir, 2001; Lippman et al., 2007). Metabolite profiling (Fiehn et al., 2000; Sumner et al., 2003; von Roepenack-Lahaye et al., 2004; Kopka, 2006) has been used to detect hundreds of metabolite QTLs in inbred tomato populations (Schauer et al., 2006; Zanor et al., 2009; Do et al., 2010). Similar approaches have been applied to map metabolite QTLs in other species, like Arabidopsis (Arabidopsis thaliana; Kliebenstein et al., 2002, 2006; Keurentjes et al., 2008; Lisec et al., 2008; Rowe et al., 2008). Metabolite profiling has also been applied to analyze changes during tomato fruit development (Carrari et al., 2006) and to survey phenotypic diversity in wild relatives of tomato (Schauer et al., 2005). Networks obtained by combining transcript and metabolite profiles have been used to explore metabolic programs that underlie tomato fruit development (Carrari and Fernie, 2006) and to shortlist genes that may regulate fruit composition (Mounet et al., 2009).
Inbred lines can also be used to study the mode of inheritance of metabolic traits. For this, each IL is back-crossed to the cultivated parent S. lycopersicum M82, giving a set of heterozygote introgression lines (ILHs) that contain one copy of the S. pennellii introgressed section. Using gas chromatography-mass spectrometry (GC-MS)-based metabolite profiling, Schauer et al. (2008) investigated the mode of inheritance of 332 metabolite QTLs and found that over half of the metabolic QTLs (174 of 332) were dominantly inherited. Most of the remainder were additively (61 of 332) or recessively (80 of 332) inherited, with a negligible number displaying the characteristics of overdominant inheritance.
Metabolites are synthesized and degraded in reactions that are catalyzed by enzymes. Therefore, it can be expected that genetic diversity in enzyme activity will contribute to phenotypes that are linked to metabolic composition. One specific example is LIN5, which encodes a cell wall invertase and is a locus for a QTLs that positively affect tomato fruit sugar content and hence the important producer trait “solids” (Fridman et al., 2004; Schauer et al., 2006).
It might be anticipated that the genetic architecture that determines the levels of enzyme activities may be simpler than the networks that determine the level of a metabolite, at least in primary metabolism. Enzyme activities will depend on cis-variation, which influences the structure and properties of the protein, and cis- and trans-variation, which affect the rates of expression and degradation. Metabolite levels will additionally depend on complex interactions between many enzymes in metabolic networks (Sulpice et al., 2010).
There have been relatively few large-scale studies of the variation in enzyme activities. This is partly for technical reasons, because it is a challenge to perform high-throughput assays for a large number of enzymes. Available evidence indicates that enzyme activities exhibit considerable natural genetic variation. However, most previous studies have been restricted to only a small number of enzymes. In a maize (Zea mays) inbred population, a QTL for Suc phosphate synthase and invertase activity colocates with structural genes for these enzymes (Prioul et al., 1999; Causse et al., 2004; Thévenot et al., 2005). Sergeeva et al. (2006) found organ-specific variation in activity for invertase and other enzymes in Arabidopsis. In a study of six enzymes from primary metabolism and four enzymes from secondary metabolism in an Arabidopsis recombinant inbred line (RIL) population, Mitchell-Olds and Pedersen (1998) detected several enzyme activity QTLs that colocalized with structural genes and a trans-QTL (i.e. a QTL that does not colocate with a structural gene) for three glycolytic enzymes. The latter might represent a joint regulator of the three enzymes.
We have established a robot-based platform to determine the activities of over 30 enzymes from central metabolism in vitro in optimized conditions (Gibon et al., 2004; Steinhauser et al., 2010; Sulpice et al., 2010). Changes in measured activity in such conditions broadly reflect changes in the amount of protein for that enzyme (Piques et al., 2009). This platform was initially used to analyze changes in enzyme activity during physiological transitions (Gibon et al., 2004). It has also been used to profile 35 enzyme activities across 100 Arabidopsis accessions (Sulpice et al., 2010). The information about variation in enzyme activities was combined with information about biomass and metabolites to explore network structures in metabolism and the connectivity between metabolism and growth. This robotized platform has already been used to map enzyme activity QTLs in IL populations. In a study of 15 enzymes from central carbon and nitrogen metabolism in a Landsberg erecta × Cape verde islands Arabidopsis RIL population, a total of 15 enzyme activity QTLs were detected (Keurentjes et al., 2008). The majority was in trans to structural genes, but five QTLs colocated with structural genes for the corresponding enzyme, of which three also showed a strong correlation with transcript levels from the structural gene. This platform has also been used to map the genetic determinants of 10 enzymes from central carbon and nitrogen metabolism in maize using an intermated breeding population, which gives a much higher genetic resolution than conventional IL populations (Zhang et al., 2010). This study detected a total of 73 enzyme activity QTLs, which explained the majority of the genetic variance in each enzyme activity. The QTLs were almost all in trans to structural genes for the respective proteins.
We recently reported the optimization of this enzyme analysis platform for tomato fruit pericarp tissue and its use to investigate connectivity between enzyme activities during the development of S. lycopersicum and S. pennellii fruits (Steinhauser et al., 2010). We now report the mapping of QTLs for 28 enzymes in the S. lycopersicum M82 × S. pennellii IL population. We analyzed two independent field experiments from years with drastic differences in climate conditions. In total, 27 enzyme activity QTLs overlapped in the 2 years. In addition, the mode of inheritance was studied in a heterozygote IL population, and the locations of the QTLs were compared with the locations of putative structural genes in the tomato genome.
RESULTS
Enzyme Activity Assays and Choice of Biological Materials
Our initial investigations of enzyme activity changes during tomato fruit development revealed species-specific differences at late fruit development stages between S. lycopersicum M82 and S. pennellii (Steinhauser et al., 2010), which are the crossing parents for a widely used tomato IL population (Eshed and Zamir, 1994). Importantly, we documented that enzyme activities are stable in both genotypes during late development stages, providing a good basis for an analysis of fruit enzyme activity QTLs in the S. lycopersicum M82 × S. pennellii IL population.
Enzyme activities were determined in fruits harvested from two independent field experiments, run in 2003 and 2004. These two years were characterized by very different climate conditions: 2003 was warmer and drier than average, while 2004 was wet and cool (records of the Western Galilee Experimental Station, Akko, Israel). It was previously shown that the addition of the 2004 harvest strongly reduces the number of metabolite QTLs that are found in multiple years (Schauer et al., 2006, 2008). Therefore, we expected that comparison of these two experiments would allow us to identify robust enzyme activity QTLs with strong phenotypic effects.
A list of the analyzed enzymes, their abbreviations, and pathway assignments is provided in Supplemental Table S1; the number of samples analyzed per genotype and assay is given in Supplemental Data S1. The population analyzed within this study comprised 76 ILs (Mueller et al., 2005; Tomato IL map 6.5 and 6.9), an additional subline (IL-7-4-2), and the reference genotype S. lycopersicum M82, resulting in a total of 78 genotypes. We also investigated lines or sublines back-crossed to the reference genotype (i.e. ILHs; Semel et al., 2006).
Enzyme Activity Measurements and Data Analysis
For 2003, a total of 413 fruit samples (corresponding to 75 ILs, the additional subline, and the parental control M82) were analyzed for 28 enzyme activities (Supplemental Data S1). The line IL-3-3 was not included due to limited sample material. Out of 11,564 performed assays, 8,472 (73.3%) resulted in enzyme activity determinations, of which 279 (3.3%) values were identified as outliers and removed. The remaining 8,193 enzyme activity measurements allowed calculation of 1,723 (78.9% of the entire population: 78 × 28 = 2,184) mean-average values based on at least three replicates per genotype and assay (Fig. 1A; Supplemental Data S2). A large portion (35%) of the missing mean-average values were for ADP-Glc pyrophosphorylase (AGP; n = 57), glucokinase (GlcK; n = 56), and succinyl-coenzyme A ligase (SCS; n = 48), which had low activities in ripe fruits of the reference genotype M82 (Steinhauser et al., 2010). The total content of amino acids, proteins, Fru, Glc, and Suc (afterward metabolites) were also analyzed, resulting in 1,871 out of 2,065 (90.6%) determined metabolite data points and 379 (97.2% of the entire population: 78 × 5 = 390) considered mean-average values supported by at least three replicates per genotype and metabolite (Supplemental Data S2).
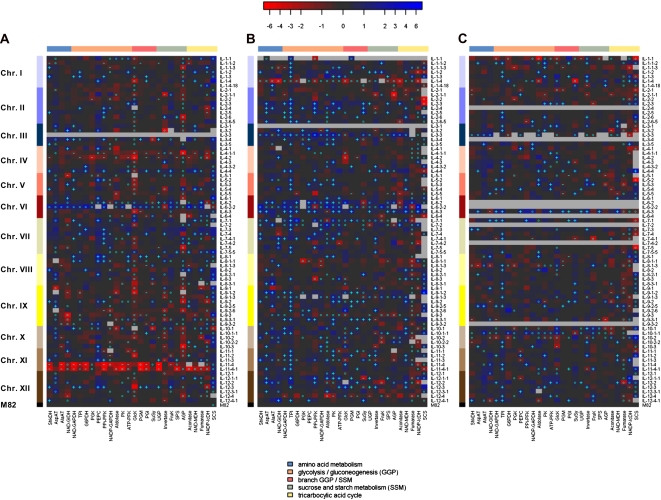
Figure 1.
Heat map visualization of log2-tranformed enzyme activity ratios of the S. pennellii IL population for independent field experiments performed in 2003 (A) and 2004 (B and C) for homozygote (A and B) and heterozygote (C) lines compared with the parental control M82. The ILs are represented by rows and sorted according to chromosomes depicted as color-coded row sidebars. The bottom row represents the parental control. The columns represent individual measured enzyme activities sorted according to their metabolic pathway assignment depicted as color-coded column sidebars. The effects are expressed as log2-transformed ratios based on mean average in each IL to the mean of the parental control in the respective field experiment. The strength of the effects is color coded according to the top color bar, with red colors reflecting decreasing (−, negative) and blue colors reflecting increasing (+, positive) effects. Missing values are represented by gray color. QTLs detected using t tests at a significance level of P < 0.05 are marked as follows: + = positive QTL; − = negative QTL. Heat map cells labeled with circles reflect data with fewer than three replicates; thus, a QTL assessment using a t test was not conducted for those combinations of IL and enzyme.
For the relatively cold year 2004, 583 samples corresponding to 75 ILs, the additional subline, and the parental control M82 were analyzed (Supplemental Data S1). Due to poor germination, line IL-3-1 was not included. Out of 16,324 individual assays performed, 9,911 (60.7%) resulted in enzyme activity determinations, of which 362 (3.7%) values were identified as outliers and removed. On the basis of the resulting 9,549 enzyme activity determinations, 1,714 (78.5% of 2,184) mean-average values based on at least three replicates per genotype and enzyme assay were available for further analyses (Fig. 1B; Supplemental Data S2). Many of the missing values were again for SCS (n = 73). For metabolite analyses, 2,736 out of 2,915 (93.9%) measurements provided values for metabolite pool sizes, resulting in 370 out of 390 (94.9%) mean-average values (Supplemental Data S2).
In addition, 636 samples derived from 69 ILHs were analyzed for the field experiment in 2004 (Supplemental Data S1). Out of 17,612 individual assays performed, 10,604 (60.2%) resulted in enzyme activity determinations, of which 475 (4.5%) outlying values were removed. The remaining 10,129 enzyme activity determinations allowed estimation of 1,670 (77.5% of 2,156 without M82) mean-average values (Fig. 1C; Supplemental Data S2). Similar to the ILs for the 2004 field experiment, SCS (n = 62) again comprises a large portion of the missing mean-average values. Metabolite analyses resulted in 3,008 out of 3,180 (94.6%) determined metabolite pool sizes, allowing estimation of 344 out of 345 (99.7%) mean-average values (Supplemental Data S2).
Heritability and Variation in Enzyme Activity
We first investigated what proportion of phenotypic variation is attributable to genetic variation among individuals. To do this, we estimated the broad-sense heritability (H2) as described by Semel et al. (2006). The resulting values were classified as follows (Schauer et al., 2008): H2 ≤ 20% is considered as low, 20% < H2 ≤ 40% is considered as intermediate, and H2 > 40% is considered as strong (Table I; Supplemental Table S2).
Table I. Heritability of enzyme activity traits in the S. pennellii IL population.
The coefficient of variation (CV) in percentage within and among the lines as well as the heritability (H2) for each enzyme activity trait are presented for the two independent field trials, 2003 and 2004. For each trait, the mean-average CV (ØCV) exhibits the average of CV values obtained for each IL; the CV among lines was computed using the mean enzyme activities among the lines. The columns Ø and CV show average heritability and the corresponding CV, respectively. The class (CL) column exhibits a grouping of the average H2 values as follows: ↑, high heritability; ↓, low (H2 ≤ 20); ↔, intermediate (20 < H2 ≤ 40); high (H2 > 40). Pearson’s (r) and Spearman’s (rs) correlation and the significance (* P < 0.05, ** P < 0.01) of enzyme activity levels in the ILs between the two field trials are also displayed.
| Enzymes | IL 2003 |
IL 2004 |
H2 |
Correlation |
|||||||
| ØCV (Within) | CV (Among) | H2 | ØCV (Within) | CV (Among) | H2 | Ø | CV | CL | r | rs | |
| ShkDH | 34 | 37 | 41 | 49 | 35 | 18 | 29.5 | 54 | ↔ | 0.42** | 0.27* |
| AspAT | 42 | 24 | 1 | 41 | 30 | 19 | 10 | 123 | ↓ | 0.17 | 0.24* |
| AlaAT | 37 | 31 | 30 | 59 | 42 | 13 | 21.4 | 58 | ↔ | 0.02 | 0.18 |
| NAD-GlDH | 55 | 48 | 31 | 65 | 47 | 17 | 24.1 | 41 | ↔ | 0.1 | 0.05 |
| NAD-GAPDH | 29 | 26 | 32 | 35 | 27 | 27 | 29.7 | 12 | ↔ | 0.5** | 0.41** |
| TPI | 40 | 30 | 22 | 41 | 27 | 14 | 17.9 | 32 | ↓ | 0.27* | 0.32** |
| G6PDH | 49 | 38 | 22 | 51 | 34 | 12 | 16.9 | 42 | ↓ | 0.04 | 0.14 |
| PGK | 60 | 30 | 0 | 46 | 24 | 3 | 1.4 | 141 | ↓ | 0.02 | −0.03 |
| PEPC | 60 | 46 | 16 | 55 | 32 | 7 | 11.4 | 51 | ↓ | 0.02 | 0.04 |
| PPi-PFK | 37 | 36 | 34 | 56 | 47 | 32 | 32.9 | 5 | ↔ | 0.31** | 0.42** |
| NADP-GAPDH | 46 | 26 | 0 | 52 | 35 | 10 | 5 | 141 | ↓ | 0.12 | 0.12 |
| Aldolase | 62 | 42 | 7 | 54 | 39 | 0 | 3.3 | 141 | ↓ | 0.03 | −0.08 |
| PK | 28 | 29 | 32 | 51 | 32 | 7 | 19.6 | 91 | ↓ | 0.19 | 0.18 |
| ATP-PFK | 44 | 33 | 18 | 47 | 36 | 21 | 19.5 | 10 | ↓ | 0.32** | 0.24* |
| GlcK | 71 | 44 | 10 | 60 | 40 | 12 | 11.1 | 8 | ↓ | 0.16 | 0.1 |
| PGM | 26 | 25 | 36 | 36 | 28 | 21 | 28.5 | 39 | ↔ | 0.01 | 0.15 |
| PGI | 28 | 24 | 30 | 34 | 21 | 13 | 21.4 | 56 | ↔ | 0.34** | 0.24* |
| SuSy | 56 | 34 | 0 | 56 | 40 | 17 | 8.3 | 141 | ↓ | 0.01 | −0.04 |
| UGP | 23 | 24 | 42 | 20 | 17 | 36 | 38.7 | 10 | ↔ | 0.32** | 0.41** |
| Invertase | 57 | 41 | 20 | 48 | 37 | 21 | 20.2 | 3 | ↔ | 0.29* | 0.34** |
| FruK | 55 | 33 | 2 | 60 | 37 | 4 | 2.8 | 51 | ↓ | 0.27* | 0.13 |
| SPS | 37 | 24 | 10 | 29 | 21 | 19 | 14.5 | 44 | ↓ | 0.25* | 0.3** |
| AGP | 71 | 72 | 13 | 62 | 40 | 0 | 6.6 | 141 | ↓ | 0.07 | 0.08 |
| Aconitase | 41 | 35 | 26 | 60 | 46 | 11 | 18.7 | 56 | ↓ | 0.05 | 0.09 |
| NAD-MDH | 44 | 35 | 18 | 44 | 37 | 25 | 21.9 | 23 | ↔ | 0.37** | 0.33** |
| Fumarase | 58 | 42 | 9 | 61 | 29 | 0 | 4.7 | 141 | ↓ | 0.15 | 0.06 |
| NADP-IcDH | 63 | 49 | 7 | 65 | 49 | 21 | 13.8 | 69 | ↓ | 0.11 | 0.14 |
| SCS | 71 | 58 | 3 | 67 | 68 | 0 | 1.7 | 141 | ↓ | 0 | 0.09 |
| Average | 47 | 36 | 18 | 50 | 36 | 14 | 16.3 | 67 | ↓ | 0.18 | 0.18 |
For the year 2003, two (7%) enzymatic traits, ShkDH and UGP, showed high heritability, 10 (36%) showed intermediate heritability, and 16 (57%) showed low heritability. Overall, considering all the enzymatic traits, the heritability in 2003 was low (H2 = 18%; Table I). For the year 2004, no enzymatic trait showed high heritability, eight (29%) showed intermediate heritability, and 20 (71%) showed low heritability. Again, considering all the enzymatic traits, the heritability in 2004 was low (H2 = 14%; Table I). Across both years, no enzymatic trait showed high heritability. UGP was at the upper limit of intermediate heritability, with H2 = 39%, another 10 (36%) enzymatic traits had an intermediate heritability, and 18 (64%) had a low heritability. When the enzymes were grouped according to their pathway assignment (Supplemental Table S1), each group showed a low average heritability except for amino acid metabolism, which had, on average, an intermediate heritability.
Metabolite traits for the 2003 field experiment showed, on average, an intermediate heritability (H2 = 21%), with three (60%) and two (40%) individual traits having intermediate and low heritability, respectively (Supplemental Table S2). For the 2004 field experiment, one (20%) and four (80%) metabolite traits showed intermediate and low heritability, with a low average heritability (H2 = 11%; Supplemental Table S2). Similarly, the heritability of metabolite traits across both field experiments was also on average low (H2 = 16%). No metabolite trait revealed strong heritability, three (60%) showed intermediate heritability, and two (40%) showed low heritability. Schauer et al. (2008) profiled a wider range of metabolites in the same biological material, including some covered in our study. In their analyses and based on the average heritability of the field experiments 2003 and 2004, nine (12%), 48 (64%), and 18 (24%) metabolite traits revealed strong, intermediate, and low heritability, respectively, with an average intermediate heritability of 28% (Schauer et al., 2008). We also inspected the heritability of metabolites that are substrates or products of the enzymes that were measured in our study. Overall, eight (50%) had an intermediate heritability, four (25%) had a high heritability, and four (25%) had a low heritability (Supplemental Table S3).
QTL Mapping
To identify introgressions that potentially harbor a QTL, t tests were first performed separately on the IL data sets of 2003 and 2004. This analysis was restricted to traits supported by three or more replicates in a given IL (n ≥ 3). Lines where the material was not available in one year were initially included in the analysis for the other year (Fig. 2). Thus, for the 2003 data set, fewer than three samples were available for IL-3-1 and IL-6-2-2, which prevented QTL detection in these lines. SCS activities could not be reliably determined in the parental line M82, which prevented QTL detection for SCS (Supplemental Data S1). For the 2004 data set, fewer than three samples were available for the lines IL-3-1, IL-1-1, IL-1-4, and IL-6-2-2 (Supplemental Data S1). The t test results were grouped according to the estimated probabilities (P < 0.01, P < 0.05, P ≥ 0.05, and not measured) and the direction of the observed phenotypic effect. The direction of change of a QTL (i.e. positive or negative, as illustrated by the sign) was defined as the level in the IL compared with the level in M82 and therefore indicates the impact of the S. pennellii introgression.
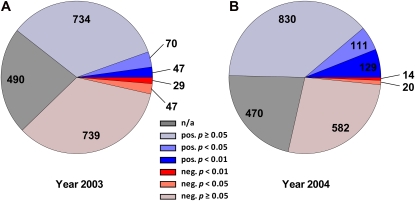
Figure 2.
Distribution of P values derived from t test analyses of enzyme activities observed in the homozygote ILs for the field trials 2003 (A) and 2004 (B). Blue sectors represent the number of positive traits (i.e. the introgression revealed higher values than the parental control M82). Red sectors reflect negative traits, where the observed value in the introgression is lower compared with the parental control. Gray sectors depict the portion of t tests that were not conducted, as fewer than three replicates were available with respect to genotype and enzyme activity. The observed P values are grouped accordingly as depicted: dark red/dark blue, significant portion in the range of 0 ≤ P < 0.01; red/blue, significant portion in the range of 0.01 ≤ P < 0.05; light red/light blue, not significant portion at P ≥ 0.05 ; n/a, P values were not calculated due to n < 3 samples available.
From a total of 1,666 mean-averaged (n ≥ 3) enzyme activities in the IL data set for 2003, 76 (4.6%) and 193 (11.6%) QTLs were identified at P < 0.01 and P < 0.05, respectively (Fig. 2; Supplemental Table S2). There were more positive than negative QTLs (Fig. 2). Of the significant QTLs at P < 0.05, 117 (60.6%) were positive and 76 (39.4%) were negative. At P < 0.01, 47 (61.8%) were positive and 29 (38.2%) were negative. For the 2004 data set, a total of 143 (8.5%; P < 0.01) and 274 (16.3%; P < 0.05) QTLs were identified out of 1,686 mean-averaged (n ≥ 3) enzyme activities (Fig. 2). Of the significant QTLs at P < 0.05, 240 (87.6%) were positive and 34 (12.4%) were negative. At P < 0.01, 129 (90.2%) were positive and 14 (9.8%) were negative (Fig. 2; Supplemental Table S2).
With respect to metabolites, out of 340 mean-averaged (n ≥ 3) values for the 2003 data set, 71 (20.9%) and 29 (8.5%) metabolite QTLs were detected at significance levels of P < 0.05 and P < 0.01. Of these, 45 (63.4%; P < 0.05) and 19 (65.5%; P < 0.01) were positive and 26 (36.6%; P < 0.05) and 10 (34.5%; P < 0.01) were negative (Supplemental Fig. S1; Supplemental Data S2). Accordingly, for the 2004 IL data set, out of 365 possible metabolite comparisons, 110 (30.1%) and 58 (15.9%) metabolite QTLs were detected at significance levels of P < 0.05 and P < 0.01, of which 103 (93.6%; P < 0.05) and 54 (93.1%; P < 0.01) were positive and only seven (6.4%; P < 0.05) and four (6.9%; P < 0.01) were negative, respectively (Supplemental Fig. S1; Supplemental Data S2).
QTLs Conserved across Both Years
As the climate in 2003 and 2004 was rather different, QTLs that are found in both experiments can be considered as robust candidates for further analysis. Due to limited sample material for some ILs and missing enzyme activity determinations (see above), the overlap analyses were only possible for 27 enzyme activity and five metabolite traits in 72 ILs (Supplemental Data S1). Furthermore, we only considered genotype-trait instances with three or more replicated measurements for both field experiments. This gave a combined data set of 1,406 (65.2%) out of 2,156 enzyme activity comparisons and 359 (93.3%) out of 385 metabolite comparisons (Supplemental Data S3).
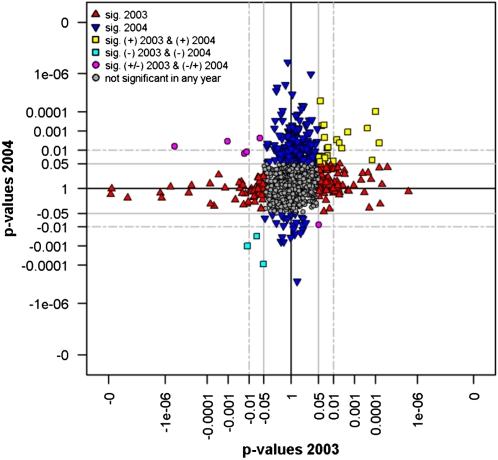
The overlap analysis revealed that 777 (55.3%) of the genotype-enzyme activity instances revealed consistent changes in the same direction in both years 2003 and 2004 (Fig. 3; Supplemental Data S3). Of these changes, 27 and eight were significant at P < 0.05 and P < 0.01, respectively, in both years (Table II). Of the positive enzyme activity QTLs, 24 were significant at P < 0.05 and seven at P < 0.01 in both years (Table II). Of the negative enzyme activity QTLs, three were significant at P < 0.05 and one at P < 0.01 (Table II; Supplemental Data S3). Interestingly, five significant traits (P < 0.05 in both years) were negative in 2003 and positive in 2004 and one significant trait (P < 0.05 in both years) was positive in 2003 and negative in 2004 (Fig. 3; Supplemental Data S3). While no negative overlapping metabolite QTL was identified for the two years, 31 (P < 0.05) and five (P < 0.01) positive overlapping QTLs were identified out of 223 (62.1%) genotype-metabolite instances with consistent phenotypic effects (Supplemental Fig. S2; Supplemental Data S3). Two metabolite QTLs (P < 0.05) displayed an opposite behavior among the two field experiments (negative in 2003 and positive in 2004; Supplemental Fig. S2; Supplemental Data S3).
Figure 3.
Scatterplot of the P value distribution derived from t test analyses of enzyme activities observed in the homozygote ILs for the field trials 2003 and 2004. The P values were computed separately by t tests, and traits were considered significant at P < 0.05. Observed positive (+, increasing effects) or negative (−, decreasing effects) traits compared with the parental control M82 are reflected by the signs on the P values. To aid visualization, P values were inverted and log10 transformed to separate significant from nonsignificant effects on enzyme activities. The significance levels of P < 0.05 and P < 0.01 are depicted as solid and dotted lines, respectively. Traits are represented by colored shapes as depicted in the key as follows: yellow squares, positive significant traits (P < 0.05) in both independent trials; cyan squares, negative significant traits (P < 0.05) in both trials; magenta circles, positive significant in one trial and negative significant in the other trial; blue triangles, significant traits in the 2004 trial; red triangles, significant traits in the 2003 trial; gray circles, no significant traits in either of the two independent field trials. Data with fewer than three replicates in any of the trials with respect to genotype and enzyme activity as well as ILs analyzed only in one trial were excluded.
Table II. Overview of enzyme activity QTLs identified, overlapping for both 2003 and 2004 harvests, on the basis of individual ILs.
Negative QTLs are colored red and positive QTLs are colored blue. Darker coloration reflects QTLs at P < 0.01, and lighter coloration reflects QTLs at P < 0.05 (in both years with P values derived from t test). Cells labeled with the letter S represent QTLs overlapping with structural genes. Only ILs considered to harbor a QTL or variables that resulted in traits identified are depicted here. Pathway abbreviations are as follows: AAM, amino acid metabolism; GGP, glycolysis/gluconeogenesis; SSM, Suc and starch metabolism; TCA, tricarboxylic acid cycle. All QTLs are defined with reference to ILs that were available for this study. The two QTLs for TPI in IL-1-2 and IL-1-3 and the two QTLs for PPi-PFK in IL-2-4 and IL-2-5 might each be due to a single QTL in the region of overlap between the adjacent ILs.
| Genotype | AAM |
GGP |
GGP/SSM |
SSM |
TCA: NAD-MDH | Total per Genotype | ||||||||
| ShkDH | NAD-GlDH | NAD-GAPDH | TPI | PEPC | PPi-PFK | ATP-PFK | PGM | PGI | UGP | FruK | Invertase | |||
| IL-1-2 | S | 2 | ||||||||||||
| IL-1-3 | 1 | |||||||||||||
| IL-2-4 | S | 2 | ||||||||||||
| IL-2-5 | S | 1 | ||||||||||||
| IL-3-2 | S | 1 | ||||||||||||
| IL-3-4 | 1 | |||||||||||||
| IL-4-1 | S | 1 | ||||||||||||
| IL-4-2 | 1 | |||||||||||||
| IL-4-4 | S | 3 | ||||||||||||
| IL-5-2 | 1 | |||||||||||||
| IL-5-4 | 1 | |||||||||||||
| IL-7-2 | 1 | |||||||||||||
| IL-7-4 | S | S | 2 | |||||||||||
| IL-7-4-1 | 1 | |||||||||||||
| IL-9-2 | 1 | |||||||||||||
| IL-10-1 | 1 | |||||||||||||
| IL-10-3 | S | 1 | ||||||||||||
| IL-11-3 | 2 | |||||||||||||
| IL-12-3 | 3 | |||||||||||||
| Total per enzyme | 2 | 1 | 4 | 5 | 1 | 4 | 1 | 1 | 1 | 2 | 1 | 2 | 2 | |
| Total per pathway | 3 | 15 | 2 | 5 | 2 | 27 | ||||||||
To further statistically evaluate the identified QTLs, two-way factorial ANOVA was performed on the combined data set. Overlapping QTLs were considered confirmed if they showed both a significant genotype effect at PG < 0.01 and a nonsignificant genotype-environment interaction (PGxE ≥ 0.01). While the majority of overlapping enzyme activity and metabolite QTLs were confirmed, one enzyme activity (NAD-MDH in IL-5-4) and four metabolite QTLs (all for total protein, in IL-4-4, IL-5-2, IL-8-1, and IL-12-3) need to be interpreted with caution, as they also displayed a significant genotype-environment interaction (Table III; Supplemental Data S3). Three further QTLs (FruK in IL-4-4, Glc in IL-5-1, and Suc in IL-8-3-1) revealed genotype effects that were above the chosen threshold of PG < 0.01 but were significant at PG < 0.05 (Table III; Supplemental Data S3).
Table III. ANOVA, t test, and mode of inheritance for 27 putative enzyme activity QTLs overlapping in both field experiments, 2003 and 2004.
The enzyme activity QTLs (aQTLs) are sorted according to genotype and enzyme activity trait analyzed. The ANOVA results depict the percentage variation for the factors genotype (G), environment (E), and genotype-environment interaction (GxE). An introgression was considered harboring a QTL if a significant genotype (PG < 0.01) and a nonsignificant genotype-environment interaction (PGxE ≥ 0.01) were observed. The results of t test analyses show the phenotypic responses as agronomical percentage values for the field experiments in 2003 [R(IL) 2003] and 2004 [R(IL) 2004]. QTLs were considered if significant changes at P < 0.05 in the same direction were observed for both field experiments. The responses of the respective heterozygote ILs [R(ILH) 2004] and associated P values from t test analysis compared with M82 are displayed. The mode of inheritance was computed and classified as described (Semel et al., 2006) as follows: R, recessive; A, additive; D, dominant; ODO, overdominant effects. The sign indicates the direction of the observed phenotypic effect of the S. pennellii allele introgressed into the S. lycopersicum M82 genome. All significant changes are depicted as follows in each cell: * P < 0.05, ** P < 0.01.
| Genotype | Enzyme | ANOVA: IL 2003/2004 |
t Test: IL 2003/2004 |
Inheritance (IL/ILH 2004) |
|||||||
| % (G) | % (E) | % (GxE) | QTL (P < 0.01) | R(IL) 2003 | R(IL) 2004 | QTL (P < 0.05) | R(ILH) 2004 | R(IL/H) 2004 | Mode (P < 0.05) | ||
| IL-1-2 | TPI | 32** | 4 | 0 | aQTL | 61* | 68** | aQTL | 35* | 0.129 | D+ |
| NAD-MDH | 21** | 20** | 7* | aQTL | 123* | 49* | aQTL | 10 | 0.147 | A+ | |
| IL-1-3 | TPI | 22** | 6 | 3 | aQTL | 94* | 58* | aQTL | 65** | 0.854 | D+ |
| IL-2-4 | TPI | 36** | 3 | 0 | aQTL | 52* | 76** | aQTL | |||
| PPi-PFK | 52** | 10** | 0 | aQTL | 99** | 164** | aQTL | ||||
| IL-2-5 | PPi-PFK | 46** | 14** | 1 | aQTL | 102** | 124** | aQTL | 33 | 0.012 | R+ |
| IL-3-2 | NAD-GlDH | 56** | 0 | 0 | aQTL | 204** | 142* | aQTL | 92 | 0.516 | A+ |
| IL-3-4 | NAD-GAPDH | 19** | 9* | 0 | aQTL | 53* | 38* | aQTL | |||
| IL-4-1 | PEPC | 36** | 0 | 1 | aQTL | 176** | 104** | aQTL | 109** | 0.942 | D+ |
| IL-4-2 | PGI | 29** | 1 | 1 | aQTL | −23* | −36** | aQTL | 15 | 0.044 | R− |
| IL-4-4 | NAD-GAPDH | 40** | 6* | 0 | aQTL | 89** | 80** | aQTL | 17 | 0.028 | R+ |
| UGP | 28** | 20** | 1 | aQTL | 44* | 36* | aQTL | 17 | 0.158 | A+ | |
| FruK | 14* | 17** | 1 | 136* | 94* | aQTL | 4 | 0.09 | A+ | ||
| IL-5-2 | PPi-PFK | 24** | 14** | 0 | aQTL | 48* | 96* | aQTL | 12 | 0.023 | R+ |
| IL-5-4 | NAD-MDH | 27** | 26** | 10** | 144** | 72** | aQTL | 39 | 0.262 | A+ | |
| IL-7-2 | TPI | 39** | 2 | 2 | aQTL | 55* | 105** | aQTL | |||
| IL-7-4 | PPi-PFK | 36** | 14** | 4 | aQTL | 127* | 101* | aQTL | −40 | 0.012 | R+ |
| ATP-PFK | 23** | 7 | 0 | aQTL | 63* | 60* | aQTL | 17 | 0.324 | A+ | |
| IL-7-4-1 | NAD-GAPDH | 20** | 12** | 1 | aQTL | 38* | 44** | aQTL | −36* | 0.030 | ODO−/R+ |
| IL-9-2 | ShkDH | 15** | 29** | 1 | aQTL | 52* | 53* | aQTL | −37 | 0.003 | R+ |
| IL-10-1 | NAD-GAPDH | 36** | 14** | 4 | aQTL | 42* | 75** | aQTL | 17 | ||
| IL-10-3 | Invertase | 31** | 1 | 1 | aQTL | −54* | −64** | aQTL | 34 | 0.009 | R− |
| IL-11-3 | ShkDH | 20** | 26** | 9* | aQTL | 73** | 142** | aQTL | 7 | 0.057 | A+ |
| Invertase | 26** | 4 | 0 | aQTL | −61** | −36** | aQTL | −19 | 0.551 | A− | |
| IL-12-3 | TPI | 26** | 3 | 0 | aQTL | 47* | 67* | aQTL | 52* | 0.716 | D+ |
| PGM | 17** | 40** | 1 | aQTL | 36* | 32** | aQTL | 3 | 0.099 | A+ | |
| UGP | 17** | 36** | 3 | aQTL | 33** | 18** | aQTL | −4 | 0.005 | R+ | |
It should be noted that our analysis attributes QTLs to the genomic regions denoted by ILs. In two cases, a pair of QTLs was detected in neighboring ILs, in which there was some overlap between the introgressed regions (TPI in IL-1-2 and IL-1-3; PPi-PFK in IL-2-4 and IL-2-5). The QTL pair may reflect a single QTL in the region of overlap. However, ILs that resolve the region of overlap would be needed to test this possibility. To avoid unsupported assumptions, the analyses in the following sections were performed using QTLs mapped to the ILs available for our study.
Mode of Inheritance
We next investigated the mode of inheritance of the enzyme activity QTL. This analysis was performed on the 2004 field trial, which included a set of ILHs. This hybrid population was obtained by crossing every individual IL to the reference parent M82 (for a detailed description, see Semel et al., 2006). We assembled a data set of the genotypes in which at least three sample replicates were available for both the IL and corresponding ILH, resulting in 66 homozygote and heterozygote introgressions (Supplemental Data S1). In an initial analysis, we compared the phenotypic effects of the corresponding IL and ILH lines with each other and the reference genotype M82 using t tests (Supplemental Data S2). If either the IL or the ILH was significantly different at P < 0.05 from M82, the introgression was considered to harbor a QTL. This analysis yielded 351 putative enzyme activity QTLs (24.6%; out of the 1,428 comparisons with n ≥ 3 for the IL and ILH populations) and 139 putative metabolite QTLs (42.2%; out of 329 valid comparisons; Supplemental Data S3). As only data from a single harvest were available for the ILH population, we also applied much more stringent filter criteria based on the ANOVA for the joint 2003/2004 IL data set (Supplemental Data S3; for details, see “Materials and Methods”). This yielded 92 enzyme activity QTLs and 57 metabolite QTLs.
To assess the mode of inheritance, we classified the identified QTLs according to Semel et al. (2006) in four broad categories: recessive, additive, dominant, and overdominant. The direction of the phenotypic effect of the S. pennellii allele compared with M82 is depicted by its sign (positive for increased, negative for decreased). Even though this approach was applied to the entire list of QTLs (Supplemental Data S3), we restricted our assessment to robust QTLs, filtered as described above (Supplemental Data S3). In this robust set, the vast majority (82 of 92) of the enzyme activity QTLs (Fig. 4) and all of the 57 metabolite QTLs revealed positive effects (Supplemental Fig. S3).
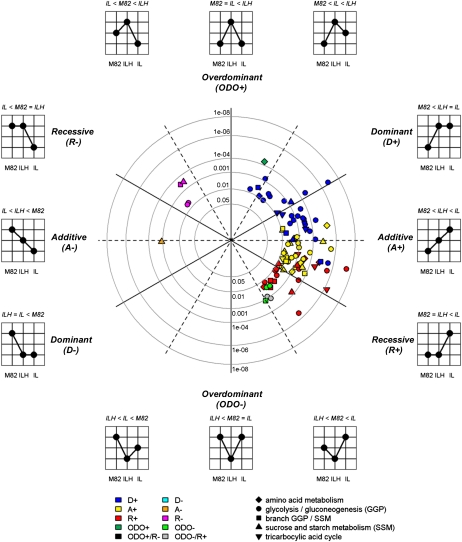
Figure 4.
Two-dimensional polar plot representation of the mode of inheritance and associated P values of detected enzyme activity QTLs estimated using the phenotypic effects in the ILs and ILHs and the parental control M82 based on the 2004 field trial. Traits positioned on the dashed black lines exhibit enzyme activity mean differences of one genotype that are exactly between the genotypes with low and high phenotypic effects. Traits exhibiting clear additive (A) or overdominant (ODO) effects are located on the horizontal and vertical lines, respectively. The distance to the center (radius) reflects the P value associated with a trait estimated using ANOVA of the corresponding homozygote IL data measured for both the 2003 and 2004 trials. The shape of the plotted traits corresponds to the metabolic pathway an enzyme is assigned, as depicted in the key. The color of each shape (see key) corresponds to the mode of inheritance of the trait classified using a decision tree, as suggested by Semel et al. (2006). Only those traits are visualized that were detected and evaluated using both t test analyses of the IL/ILH data from 2004 and ANOVA of IL data from 2003 and 2004 (see “Materials and Methods”; Supplemental Data S3).
Of the 92 robust enzyme activity QTLs, four revealed overdominant, 31 dominant, 29 additive, and 26 recessive inheritance. Two loci apparently harbored two QTLs, as the IL and ILH were significant but in opposite directions, namely one overdominant and one recessive (Fig. 4; Table IV). Thus, while a similar number of dominant, additive, and recessive QTLs were identified, the portion of overdominant enzyme activity QTLs was low. Of the 57 robust metabolite QTLs, 23 showed dominant, 21 additive, and 12 recessive effects, while one introgression apparently harbored two QTLs (Supplemental Data S3). For both enzyme activity and metabolite traits, the number of introgressions giving dominant or additive inheritance was similar to or larger than those conferring recessive inheritance. This was also the case when all QTLs identified for the joint data set were considered (Supplemental Data S3). Analysis by χ2 and Fisher’s exact test of the distribution of the individual mode of inheritance revealed no clear and significant differences at the levels of both positive and negative effects with respect to the pathway assignment of corresponding enzyme traits (Table IV; Fig. 4; Supplemental Fig. S3). Similarly, the distribution of the mode of inheritance of sets of enzymes that were assigned to different pathways was not significantly different (Table IV). Interestingly, the distribution of the mode of inheritance for enzyme activity QTLs mirrored the situation for metabolite QTLs identified in the same sample material using GC-MS-based metabolite profiling, as described by Schauer et al. (2008; see “Discussion”).
Table IV. Qualitative distribution of the mode of inheritance of enzyme activity QTLs grouped according to the metabolic pathway to which the measured enzymes are assigned.
Numbers in parentheses show the percentage of the mode of inheritance among all detected QTLs in each pathway group. Pathway abbreviations are as follows: AAM, amino acid metabolism; GGP, glycolysis/gluconeogenesis; SSM, Suc and starch metabolism; TCA, tricarboxylic acid cycle. The signs that precede the mode of inheritance indicate increasing (+) or decreasing (−) effects compared with the parental control M82. The columns P(χ2) and P(F) display the P values observed using χ2 and Fisher’s exact test, respectively, to evaluate the independence of the distribution among the pathway groups for each mode (row-wise assessment). The row P(χ2)|P(F) shows the P values obtained using χ2 and Fisher’s exact test to evaluate the independence of the distribution of the mode of inheritance for each pathway group (column-wise assessment, regardless of their change in direction [i.e. positive or negative]). n/a, Not measured.
| Mode | Total |
AAM (4 Traits) | GGP (10 Traits) | GGP|SSM (4 Traits) | SSM (5 Traits) | TCA (5 Traits) | P(χ2) | P(F) | |
| +Overdominant | 1 | 4 | 1 (10) | 0 | 0 | 0 | 0 | 0.08 | 0.22 |
| −Overdominant | 3 | 0 (0) | 2 (4) | 1 (8) | 0 | 0 | 0.69 | 0.76 | |
| +Dominant | 31 | 31 | 1 (10) | 21 (45) | 3 (25) | 2 (15) | 4 (40) | 0.11 | 0.12 |
| −Dominant | 0 | 0 | 0 | 0 | 0 | 0 | n/a | n/a | |
| +Additive | 28 | 29 | 3 (30) | 12 (26) | 4 (33) | 6 (46) | 3 (30) | 0.72 | 0.7 |
| −Additive | 1 | 0 | 0 | 0 | 1 (8) | 0 | 0.19 | 0.49 | |
| +Recessive | 22 | 26 | 5 (50) | 8 (17) | 3 (25) | 3 (23) | 3 (30) | 0.27 | 0.27 |
| −Recessive | 4 | 0 | 2 (4) | 1 (8) | 1 (8) | 0 | 0.79 | 0.79 | |
| ODO+/R− | 0 | 2 | 0 | 0 | 0 | 0 | 0 | n/a | n/a |
| ODO−/R+ | 2 | 0 | 2 (4) | 0 | 0 | 0 | 0 | 0 | |
| P(χ2)|P(F) | 0.31|0.22 | 0.09|0.07 | 0.87|0.73 | 0.3|0.35 | 0.93|1 | ||||
| Total | 92 | 10 | 47 | 12 | 13 | 10 | |||
Mapping of Structural Genes
We next asked how often enzyme activity QTLs colocate with a structural gene that putatively encodes the enzyme function. To do this, we identified protein sequences from the iTAG2.3 tomato genome release. The results were aligned against the S. lycopersicum and S. pennellii genomes and finally mapped using EXPN1992 (Mueller et al., 2005). Due to the incomplete physical marker information, it was not possible to map all the resulting sequences precisely to the physical map. Loci that map within 100 to 1,000 kb of the introgressed segment and thus might be in the locus are depicted, and corresponding results are summarized in Supplemental Data S4. A structural gene that putatively encodes the corresponding enzyme activity could be mapped to the corresponding introgression locus for nine (33%) of the 27 robust QTLs (Table II). For the remaining 18 (67%) QTLs, no corresponding structural gene mapped to the IL locus or its close proximity (Supplemental Data S4). There would be eight (32%) cis-QTLs and 17 (68%) trans-QTLs if the QTL pairs for TPI in IL-1-2 and IL-1-3 and for PPi-PFK in IL-2-4 and IL-2-5 were due to single QTLs in the region of overlap between the neighboring ILs.
A QTL for NAD-MDH in IL-1-2 colocates with a structural gene for this enzyme revealed by our mapping analysis. QTLs for PPi-PFK were detected in IL-2-4 and in the neighboring, and partly overlapping, IL-2-5. Causse et al. (2004) mapped Pfpb (encoding PPi-PFK) to this genomic region. Our analyses confirm the presence of this gene in the close vicinity of this locus (100–1,000 kb from the marker), locating it in the region of overlap of IL-2-4 with IL-2-5. A QTL for NAD-GlDH in IL-3-2 colocates with a gene encoding Glu dehydrogenase revealed by our mapping. IL-4-1 contains a QTL for PEPC. Causse et al. (2004) mapped LePPC3 (encoding PEPC) to bin 4-B, a region in IL-4-1 that does not overlap with any other IL, and this mapping is confirmed by our analyses. IL-4-4 contains QTLs for NAD-GAPDH, UGP, and FruK. Interestingly, a significant increase in the expression of the transcripts for fructokinase 2 and NAD-GAPDH in IL-4-4 was reported by Baxter et al. (2005). Causse et al. (2004) mapped Fk(1) (for fructokinase-like gene) to this locus. Our colocalization analyses could not confirm this but did identify a NAD-GAPDH-encoding gene within this locus. We mapped potential fructokinase candidates to chromosomes 2, 3, 5, 6, 9, 10, and 11. It is possible, however, that an as yet unsequenced fructokinase gene is located in the genomic region corresponding to IL-4-4. IL-7-4 contains QTLs for ATP-PFK and PPi-PFK. Our mapping analyses indicate the presence of structural genes for both of these enzymes in this genomic region. IL-10-3 contains a QTL for invertase, and our mapping analyses indicate the presence of five potential structural genes for invertase in this genomic region. While a QTL for TPI in IL-1-3 putatively colocated with a structural gene encoding the chloroplastic triose-P isomerase reported by Causse et al. (2004), our analyses using the available whole-genome information indicates that while this gene is localized on chromosome 1, it does not map to IL-1-3. IL-3-4 contains a QTL for NAD-GAPDH. Our analyses indicate that a gene encoding for GAPDH is located close to this locus but is not colocated. Similarly, IL-10-1 contains a QTL for NAD-GAPDH. While our analysis of genome sequence data revealed a gene encoding for GAPDH in close proximity, it is not exactly at this locus.
Summarizing, colocation of an enzyme activity QTL and a structural gene was found in nine instances: PPi-PFK for IL-2-4/IL-2-5 and IL-7-4, NAD-GlDH for IL-3.2, PEPC for IL-4-1, NAD-GAPDH for IL-4-4, ATP-PFK for IL-7-4, NAD-MDH for IL-1-2, and invertase for IL-10-3. In some other cases, a gene is located in the vicinity but probably not in the introgressed region (NAD-GAPDH for IL-3-4 and IL10-1, TPI for IL-1-3).
Given the large size of the introgressed S. pennellii segments, some of the identified colocations are likely to be fortuitous. Using a strategy similar to Lisec et al. (2008), we estimated that the most likely number for spurious colocations is about four (Supplemental Fig. S4), and even seven colocations could well be expected by chance. Nevertheless, the observed number of colocations (nine) is significantly more than expected by chance (0.01 < P < 0.001).
We also assessed whether the colocated QTLs contained any notable differences between the sequences of the structural genes in the two parent species. Sequences were aligned to identify single nucleotide polymorphisms (SNPs) that cause a change in the protein sequence, potential splice site changes, and insertions/deletions. While we found several SNPs in potentially colocated genes, we found a similar frequency for structural genes that are not colocated with a QTL (Supplemental Data S5). There was no excess of potentially important SNPs in colocating genes; indeed, the most likely candidates for potentially deleterious changes, such as a SNP in a splice acceptor site, were found in genes that did not colocate with a QTL. However, these observations have to be considered preliminary due to the provisional status of the S. pennellii genome and because our analyses involve a comparison with cv Heinz rather than cv M82.
We asked whether the mode of inheritance was different for QTLs that were clearly in trans to known structural genes and QTLs that may colocate with a structural gene. For the cis-QTLs, one showed a positive dominant, three a positive additive, three a positive recessive, and one a negative/recessive inheritance mode (Supplemental Data S3). The mode of inheritance for trans-QTLs included three positive dominant, six additive (five positive, one negative), four recessive (three positive, one negative), and one overdominant/recessive (Supplemental Data S3). Due to missing data for the ILH lines, the mode of inheritance could not be determined for one cis-QTL and four trans-QTLs (Supplemental Data S3). Statistical analysis by Fisher’s exact test of the distribution of the individual mode of inheritance with respect to cis- and trans-enzyme activity QTLs revealed no significant differences in the mode of inheritance (data not shown).
Colocation of Enzyme Activity QTLs and Metabolite QTLs
Schauer et al. (2006) determined metabolite QTLs in the same material as we used for measurements of enzyme activities. We compared the two data sets to identify colocations of enzyme activity QTLs and QTLs for metabolites that are in the same pathways or directly downstream of the pathways in which the enzyme operates. This is summarized in Supplemental Data S4. Based on this analysis, we identified seven enzyme activity QTLs that colocate with metabolite QTLs associated with the same pathways or closely connected pathways (Supplemental Data S4).
One example is summarized in Figure 5, where Schauer et al. (2006) have already identified IL-4-4 as a “pathway QTL” based on its showing changes in several metabolites, including Suc, Glc, Fru, Fru-6-P, glycerate-3-phosphate, citrate, isocitrate, Asp, Glu, and succinate. Metabolite analysis using enzyme assays conducted in this study revealed two further metabolite QTLs (overlapping for both the 2003 and 2004 field experiments) for total amino acids and total protein. The protein QTLs showed a significant genotype-environment interaction (Supplemental Data S3). IL-4-4 also contained enzyme activity QTLs for fructokinase and UGP, which are involved in sugar metabolism, and the glycolytic enzyme NAD-GAPDH (Fig. 5; Supplemental Data S3).
Figure 5.
QTLs for S. pennellii IL-4-4 mapped onto a simplified scheme of the glycolysis pathway including associated reactions. Positive metabolic QTLs are depicted with a blue background and positive enzyme activity QTLs by blue arrows. Dark blue QTLs reflect significant differences at P < 0.01, and light blue QTLs reflect significant differences at P < 0.05. White boxes indicate metabolic traits with no significant differences; gray boxes depict nonmeasured metabolic traits (Schauer et al., 2006). EC numbers in italics indicate nonmeasured enzyme activity QTLs.
Further putative colocations of enzyme activity and metabolite QTLs were found for PPi-PFK and TPI enzyme activity with Glc and Asp in IL-2-4, for NAD-GAPDH with Fru-6-P, Ala, and total amino acids in IL-3-4, for AspAT and NAD-MDH with Glc-6-P and glycerate-3-phosphate (data for one year only) in IL-7-1, and TPI activity and glycerate-3-phosphate in IL-7-2 (Supplemental Data S3 and S4).
DISCUSSION
Johannsen (1911) and East (1910) established in the early 20th century that quantitative variation results from the combination of multiple segregating genes and environmental factors. However, unraveling the number of loci responsible for a particular trait is still challenging. Breeders focus on the identification of a small number of loci with large effects, such as the major fruit weight QTL fw2.2 (Alpert and Tanksley, 1996) or genotypes showing an increased Brix index (Fridman et al., 2002, 2004).
To identify robust enzyme activity quantitative traits that are not strongly dependent on environmental factors, this study used two separate field experiments with the S. lycopersicum × S. pennellii tomato IL population (Eshed and Zamir, 1994) that were performed in years with very different climate conditions. Fruit material was harvested when more than 80% of the ILs displayed fully ripe fruits. We showed in a previous study that enzyme activities are relatively stable in the later stages of fruit development in S. lycopersicum and S. pennellii (Steinhauser et al., 2010).
Heritability of Enzyme Activity Traits
The heritability of a trait is the proportion of phenotypic variation attributable to additive genetic effects. Our analysis of enzyme activity traits within the tomato IL population revealed that a majority of enzyme traits showed intermediate to low heritability (Table I). Interestingly, a recently published metabolite profiling study on the same sample material (Schauer et al., 2008) revealed that metabolite traits, especially when limited to the products or substrates of enzymes analyzed in this study (Supplemental Table S3), displayed largely intermediate heritability. Consequently, enzymes showed lower heritability than metabolites, even though we might expect them to be closer to the gene expression when considering the flow of genetic information from the genotype to the phenotype.
Keurentjes and coworkers (2008) analyzed enzyme activity and metabolite traits on seedling samples of an Arabidopsis RIL population. Out of 18 analyzed enzyme traits, five showed low, seven showed intermediate, and six showed strong heritability. In comparison, from the 11 metabolite traits, none displayed low, three displayed intermediate, and eight displayed strong heritability. Thus, as in our study, metabolite traits in general revealed a higher heritability than enzyme activity traits. One possible explanation might be that there is more inherent variability in enzyme activities than in metabolites. It could be argued that levels of metabolites will be highly regulated, while enzyme activities can show more variation, because changes in one enzyme can be compensated by changes in other enzymes. Furthermore, many of the metabolites investigated in these studies are products, which may accumulate with time. An alternative explanation would be that there is more technical variation in measurements of enzyme activities than metabolite levels. However, Zhang et al. (2010) described the heritability of 10 enzyme activity traits in expanding leaves of the maize intermated B73 × Mo17 mapping population grown in a greenhouse, measured using the same analytic platform as the one used in this study. One enzyme showed intermediate heritability and the other nine all displayed strong heritability. Even though metabolite traits were not investigated by Zhang et al. (2010), their study illustrates that the heritability of enzyme traits can be quite high under some conditions. Zhang et al. (2010) used greenhouse-grown material, whereas we used material from field experiments. Furthermore, in Zhang et al. (2010) and Keurentjes et al. (2008), each sample was pooled from several leaves or rosettes, whereas our analysis is based on material from a single tomato fruit.
Identification of Environmentally Robust Enzyme Activity QTLs
It was previously published that mutations with major effects occur most often in domesticated or artificially disturbed populations (Lande, 1983). This supports the use of inbred populations between a nondomesticated and a domesticated population (Eshed and Zamir, 1994) to identify traits with strong and repeatable phenotypes; however, we still do not know whether the number of loci responsible for most genetic variation is small or large (Barton and Turelli, 1989).
In our analysis, 467 enzymatic QTLs were detected in two experiments in differing conditions (193 in 2004 and 274 in 2003). These years were characterized by very different environmental conditions. To identify robust QTLs, we searched for QTLs that were shared in both years. A robust set of 27 QTLs were shared in both years (24 positive and three negative). Of these, 15 QTLs are for enzymes from the glycolysis/gluconeogenesis pathway. Three glycolysis/gluconeogenesis pathway enzymes have four or five QTLs: NAD-GAPDH (four), PPi-PFK (four), and TPI (five). This contrasts with enzymes from other pathways, which have zero or one to two QTLs.
In both of the individual years, there was a large excess of positive (i.e. the trait was changed in the direction of S. pennellii) over negative (i.e. the trait was changed in the direction of M82) QTLs. Also, of the 27 robust QTLs shared in both years, 24 were positive and only three were negative. It was previously reported for morphological traits that alleles that derive from the S. pennellii parent tend to affect the trait in the direction of the S. pennellii value (Semel et al., 2006). This trend was partly confirmed for metabolic traits. However, it is not a universal rule, as several amino acids increased in the ILs but were lower in S. pennellii than in M82 (Schauer et al., 2006, 2008). The behavior of enzyme activities at the ripe stage of development in S. lycopersicum and S. pennellii was described by Steinhauser et al. (2010). While in the glycolysis and tricarboxylic acid pathway, enzyme activities are generally higher in S. pennellii than in S. lycopersicum M82, in other pathways, the opposite behavior is seen. For the population described in this work, most of the enzyme activity QTLs were in the glycolysis and tricarboxylic acid pathway and showed increased activity in the ILs compared with M82, mirroring the changes in S. pennellii.
Mode of Inheritance of Enzyme Activity Traits Associated with Primary Metabolism
Recently, there has been increasing interest in the mode of inheritance of traits, with the objective of improving breeding qualities. Biomass traits sometimes show hybrid vigor or heterosis in the F1 when cultivated and/or wild species are crossed (rice [Oryza sativa], Li et al., 2008; Arabidopsis, Meyer et al., 2004; maize, Springer and Stupar, 2007). Such enhancement compared with the behavior of the parents is considered to be an outcome orchestrated by partial-to-complete dominance, overdominance, and epistasis (Lippman and Zamir, 2007; Li et al., 2008). While epistasis is difficult to assign (Rieseberg et al., 1999), the mode of inheritance of a trait can be more precisely studied. For this purpose, ILs can be used as a tool to reveal the mode of inheritance of specific traits. Thus, comparison of the recipient genotype, with pairs of ILs and ILHs, allows the determination of the mode of inheritance of a trait for each introgressed segment and, in particular, reveals if a heterotic response can be generated by the dominance or overdominance of one or more independent QTLs, either alone or in association with possible epistatic effects with the remainder of the genome.
In general, the mode of inheritance of the enzyme activities is fairly well distributed between dominant (34%), additive (32%), and recessive (28%) modes, with only a minority (4%) showing overdominance (Table IV). This resembles the inheritance of those metabolic traits measured by Schauer et al. (2008) that are closest to the enzymatic traits measured (subcategories organic acids, sugars, sugar alcohols, and phosphates), where 50% showed dominance, 19% were additive, 26% were recessive, and 9% showed overdominance.
It might have been expected that enzymes would show a simpler genetic determination than metabolites. As outlined in the introduction, whereas enzyme activities will depend on cis-variation, which influences protein structure and properties, and cis- and trans-variation, which affects the rate of expression and degradation of the protein, metabolite levels will also depend on complex interactions between many enzymes in metabolic networks (Sulpice et al., 2010). On the other hand, it could be argued that levels of metabolites will be more highly regulated, because changes in the activity or expression of one enzyme can be compensated by changes in the expression or activity of other enzymes. The similar modes of inheritance indicate that the genetic architecture affecting enzyme activities and metabolites is not that different in complexity. This may be in part because the majority of the QTLs that determine enzyme activities act in trans and, thus, via a regulatory network that may already be highly complex (see below). Furthermore (see above), for both enzyme activities and metabolites, an introgressed genomic segment has a similar qualitative effect on the trait to that seen between the recipient (S. lycopersicum M82) and the donor (S. pennellii) genotype.
Colocation of Enzyme Activity and Metabolite QTLs with a Structural Gene
This is the most extensive enzyme activity QTL study to date in terms of the number of enzymes covered (n = 28). However, three other substantial studies are also available. Mitchell-Olds and Pedersen (1998) analyzed 10 enzyme activities in an Arabidopsis RIL population, Keurentjes et al. (2008) measured 15 enzyme activities in a population of 160 Arabidopsis RILs, and Zhang et al. (2010) measured 10 enzyme activities in 94 intermated maize RILs.
Based on these studies, a picture is emerging about the role of cis- and trans-regulation in the genetic determination of the activities of enzymes in central metabolism. In Mitchell-Olds and Pedersen (1998), while three activity QTLs colocalized close to putative structural genes, at least three trans-QTLs were detected. In Keurentjes et al. (2008), the majority of the 15 detected enzyme activity QTLs were in trans to structural genes; only five QTLs colocated with structural genes for the corresponding enzyme. Of these, three showed a strong correlation with transcript levels from the structural gene, and two of these were particularly strong QTLs. In the study of Zhang et al. (2010) with an intermated maize RIL population, the vast majority (70 of 73) of the QTLs were in trans to structural genes for the corresponding enzymes. In our study, the majority (18 of 27) of the robust enzyme activity QTLs are in trans to putative structural genes. Two enzyme activity QTLs colocated with an expression QTL reported in a separate study by Baxter et al. (2005), and of these, one was for a gene in trans to structural genes, while one might be in cis.
The picture emerging from these studies is that most of the major genetic determinants of enzyme activity act in trans, with cis-regulation only occurring in a minority of cases. Two important technical issues will affect the validity of this conclusion. First, depending on the extent of replication, the precision of the technical analysis, and the power of the genetic population, the detected QTLs represent only a small number of the total number of loci that genetically determine the activity of the enzyme in question. In this sense, the study of Zhang et al. (2010) is the most exhaustive study to date, as the intermated mapping population allowed very fine-mapping and the detection of multiple QTLs per enzyme, accounting for over half of the total genetic variance for the activity of most of the enzymes. Zhang et al. (2010) actually detected the lowest contribution of cis-regulation. In the other studies, none or only one to four QTLs were detected per enzyme. Nevertheless, these are likely to be the major QTLs, and therefore instructive for learning about the relative importance of cis- and trans-regulation. Second, as introgressions contain many genes, fortuitous colocation cannot be excluded. In their study with an intermated RIL population, in which loci were defined to genomic regions containing 20 to 30 genes, Zhang et al. (2010) estimated that 50% of the colocations of QTLs might still be fortuitous. The estimated proportion of spurious colocations was even higher in the S. lycopersicum × S. pennellii tomato IL population. This is probably because in this population, as in other conventional IL populations, the size of the introgressed genomic sectors, and hence the number of genes per designed genomic sector, is 10 or more times higher. Thus, it is likely that the frequency of cis-QTLs is still overestimated in our and most other published studies. The emerging picture is that enzyme activities are largely regulated in trans-QTLs, with a small contribution from cis-QTLs, whose importance will vary from case to case.
In this study, as in Keurentjes et al. (2008) and Zhang et al. (2010), enzyme activities were measured using optimized assays with saturating substrate concentrations. We have shown elsewhere that there is good agreement between the enzyme activities measured in this way and protein abundance as estimated by MS (Piques et al., 2009). The likely causes for trans-regulation are the effects of transcription factors or differential enzyme degradation (e.g. through proteasome and ubiquitination pathways). The comparatively rare cis-effects detected using this enzyme activity platform are likely to be nonsynonymous polymorphisms that modify transcription, translation, the enzymatic properties of the protein, or the stability of the mRNA or protein. Since the assay contains saturating concentrations of substrates, cis- or trans-effects are unlikely to be caused by allosteric regulation of the enzyme activity (changes in binding properties or in genes affecting the levels of allosteric inhibitors, respectively). It is also unlikely that changes in enzyme activity due to phosphorylation or redox regulation are retained, because the extraction and assay were not performed in the presence of protein phosphatase inhibitors or conditions that would maintain the in vivo redox form of the enzyme (Hendriks et al., 2003).
Finally, some enzyme activity QTLs in tomato may be part of a “network QTL,” where several elements of a metabolic network are affected by expression QTLs, enzyme activity QTLs, or metabolite QTLs. For example, the genomic region denominated by IL-4-4 contains three positive enzyme activity QTLs for the enzymes in the glycolysis/gluconeogenesis pathway (NAD-GAPDH, UGP, and FruK). For two of these enzymes, Baxter et al. (2005) reported increased levels of the corresponding transcripts (NAD-GAPDH, fructokinase). Furthermore, Schauer et al. (2006) described positive metabolite QTLs at this same locus for Fru, Suc, Glc, Fru-6-P, glycerate-3-phosphate, citrate, isocitrate, Asp, Glu, and succinate, including metabolites that are direct substrates or products of NAD-GAPDH, UGP, and FruK or metabolites farther upstream or downstream in the glycolysis/gluconeogenesis pathway.
In summary, we have mapped 27 robust enzyme activity QTLs for enzymes from central carbon metabolism in an IL population generated from the S. lycopersicum M82 × S. pennellii IL population. In all cases, the change in enzyme activity in the IL qualitatively resembles the change in activity seen between S. pennellii and M82. In the vast majority of cases, the mode of inheritance is dominant, additive, or recessive, with only a very low frequency of overdominance, resembling the mode of inheritance of metabolites in the same population. Furthermore, in the majority of cases, the QTLs are in trans to structural genes for that enzyme. Together with studies in Arabidopsis and maize, these results point to individual enzyme activities being under the genetic control of a complex program that is dominated by a network of trans-acting genes.
MATERIALS AND METHODS
Materials
Inorganic compounds were purchased from Merck and organic compounds from Sigma, except ethanol (Merck) and NAD+, NADH, NADP+, NADPH, and phosphoenolpyruvate (Roche). Enzymes were purchased from Roche except phosphoglycerokinase and glycerokinase (Sigma-Aldrich). UMP-kinase, a clone provided by Octavian Barzu (Institut Pasteur), was overexpressed and purified as described by Serina et al. (1995).
Plant Growth and Materials
Enzyme activity measurements were performed on fruit pericarp tissue from a field-grown tomato (Solanum lycopersicum) ‘M82’ × Solanum pennellii IL population comprising 76 genotypes (Eshed and Zamir, 1994; Mueller et al., 2005; Tomato IL map 6.5 and 6.9). Line IL-7-4-2, harboring a genomic subsegment of IL-7-4, was additionally included. For QTL analysis, fruit samples of homozygote ILs and the reference genotype M82 were obtained from two independent field experiments in 2003 and 2004 (Semel et al., 2006). Samples of ILHs, the lines of the respective IL back-crossed to the S. lycopersicum M82 parent, were additionally received for the field experiment in 2004.
For both experiments, plants were first propagated under greenhouse conditions for 35 to 40 d and then field grown in a completely randomized design at the Western Galilee Experimental Station in Akko, Israel, under the conditions described (Schauer et al., 2006; Semel et al., 2006). Because of poor germination behavior (IL-3-1, ILH-2-4, ILH-3-4, ILH-6-2, ILH-6-2-2, ILH-6-4, ILH-7-2, and ILH-7-4-2) and limited sample material (IL-3.3 and ILH-9-3-2), 10 ILs could not be included in some or all analyses. Morphological, reproductive, and metabolite traits have been described previously for this tomato IL population (Schauer et al., 2006, 2008; Semel et al., 2006).
All tomato fruits were harvested when 80% to 100% of the tomatoes revealed red coloration (Eshed and Zamir, 1995). Fruits were cut with a scalpel blade into two parts, peeled, and separated from the placental tissue. All analyses were performed on the fleshy part of the tomato, the skinned pericarp, which is the major edible part of a tomato fruit. The sample material used in this study is precisely the same as used to analyze metabolite QTLs (Schauer et al., 2006, 2008).
Enzyme and Metabolite Assays
For enzyme assays, sample extraction, handling, and enzyme activity determination were performed exactly as described by Steinhauser et al. (2010). Additionally, metabolites were extracted using ethanol from frozen and ground sample tissue (Geigenberger et al., 1999). The soluble sugars Suc, Glc, and Fru were measured in ethanolic extracts as described (Geigenberger and Stitt, 2000) with volumes adapted to microplate format using a microplate spectrophotometer. Total amino acid content was determined in ethanolic extracts by the fluorescamine method (Bantan-Polak et al., 2001). Total protein content was assayed from the remaining pellet of ethanolic extracts with the Bio-Rad Bradford reagent (Bio-Rad Laboratories) according to the manufacturer’s instructions.
Graphical Visualization and Heat Maps
All graphs were created using Sigma Plot 10 (Systat Software) or the R statistical framework (version 2.9.1; R Development Core Team, 2009). For heat map visualization, enzyme activity effects are expressed as log2-transformed ratios based on the mean average in each tomato IL to the mean of the reference genotype S. lycopersicum M82. The mean-average enzyme activity patterns of the heterozygote and homozygote ILs and their respective reference genotype M82 were visualized as a two-dimensional polar coordinate plot according to Swanson-Wagner et al. (2006).
Statistical Analyses
All statistical analyses were implemented and performed, if not otherwise stated, according to Sokal and Rohlf (1995) with R (version 2.9.1), or available R functions were used. To identify robust effects on enzyme activities, one-dimensional outliers were detected using a box-plot approach performed with standard parameters as implemented in R. Data points that lie beyond the extremes of the whiskers (lower whisker Q0.25 − 1.5 × interquartile range [IQR] and upper whisker Q0.75 + 1.5 × IQR, with Q0.25 and Q0.75 as the continuous sample quantiles for 0.25 and 0.75 probability, respectively, and the IQR given as the difference of both quantiles, IQR = Q0.75 − Q0.75) were labeled as outliers. In the case of more than six replicates for a combination of genotype and enzyme/metabolite, identified outliers were completely removed (multiple outlier removal); if six or fewer replicates were available, the most deviant data point from median was removed (single outlier removal). For all further statistical analyses, only combinations of genotype and enzyme/metabolite were considered, with at least three replicates per year.
The homogeneity of variance across groups was analyzed with Levene’s test on the basis of the absolute deviations from the group medians. The two-sample t test for the difference in mean was performed two-sided with equal or unequal variance estimated by Levene’s test. Two-way factorial ANOVA was performed with genotype, environment, and genotype-environment interaction as factors. The percentage of variation due to each factor was computed as described (Prudent et al., 2009). The coefficient of variation was calculated as the ratio of the sd to the mean and expressed as a percentage. χ2 and Fisher’s exact test were performed to test for the independence of count data.
Trait Analyses
Heritability was calculated for each trait and year using one-way factorial ANOVA, and genetic variation was expressed as a percentage from total variation (genetic × environment) by H2 = σ2G/σ2G+E × 100. Additionally, the parametric Pearson’s product moment (r) and nonparametric Spearman’s rank order correlation (rs) were computed for each trait and year across the ILs.
QTL Mapping
QTL mapping for each independent field experiment was performed using a t test by comparing the measurements of each IL with those of the reference genotype S. lycopersicum M82 for each enzyme/metabolite and year separately. The P values were grouped in (1) nonsignificant differences (P ≥ 0.05), where the introgression contains no QTL, and (2) significant differences (P < 0.05 and P < 0.01), where the introgression was considered harboring a QTL in the respective year. Additionally, each group was further subdivided regarding the direction of change in (1) negative QTLs, where the mean-average value of the IL is lower than in M82 (ØIL < ØM82), and (2) positive QTLs, where the mean-average value of the IL is equal to or higher compared with M82 (ØIL ≥ ØM82).
To identify overlapping QTLs in both field experiments, the t test-derived P values were filtered according (1) P < 0.05 and (2) P < 0.01 in conjunction with a consistent change in direction (i.e. in both years, ØIL < ØM82 or ØIL > ØM82) for both years, respectively. Thus, introgressions that reveal significant differences of at least P < 0.05 in both years together with observed mean-average changes in the same direction (i.e. both lower and higher) were considered as harboring an environment-independent QTL. Additionally, a two-way factorial ANOVA as described above was performed to evaluate identified overlapping QTLs. Overlapping QTLs revealing a nonsignificant (PG ≥ 0.01) genotype effect or a significant genotype-environment interaction (PGxE < 0.01) were marked as weak or questionable QTLs, respectively.
As sample material of ILs and ILHs were available for the field experiment in 2004, a refined QTL mapping within this year was conducted. For this, each IL and corresponding ILH were compared by t test among each other and with the reference genotype S. lycopersicum M82. If either of them, the IL or the ILH, was significantly different at P < 0.05 from M82, the introgression was evaluated for harboring a QTL. For stringent downstream analysis (e.g. mode of inheritance), the list of identified QTLs in 2004 was filtered: only those QTLs were further considered that revealed a significant genotype (PG < 0.01) and nonsignificant genotype-environment interaction (PGxE ≥ 0.01) effect, estimated using ANOVA on the basis of the IL measurements in 2003 and 2004, as well as those that displayed mean-average changes in the same direction for both years.
Mode of Inheritance Analysis
The phenotypic effect of a QTL was considered to be the effect of the significant line, either IL or ILH, and was expressed as a percentage of the reference genotype M82 and by using the sign to indicate positive (+, increasing) and negative (−, decreasing) QTLs (i.e. the direction of the observed effect). In the case of significant effects within the same direction (+/+ or −/−) for both the IL and the respective ILH, the higher value was considered to be the QTL phenotypic effect. A QTL was considered to harbor two QTLs, namely one increasing and another decreasing, if both the IL and the respective ILH revealed significant effects but in opposite directions (+/− or −/+).
The mode of inheritance for a QTL under investigation was classified according to the decision tree proposed by Semel et al. (2006) as implemented in R and defined for the S. pennellii allele. Briefly, a QTL was considered as recessive if the ILH was significantly different from the IL but not from M82, as additive if the ILH differed from both parents or did not differ from either of them, and as dominant if the ILH differed from M82 but not from the IL. If the ILH phenotype was significantly higher or lower than either parent, the QTL was considered to be positively or negatively overdominant, respectively.
Gene Mapping
For each IL where an enzyme QTL was identified, we first mapped the genetic markers that defined this IL to the physical map. To this end, marker sequences were extracted from the SOL Genomics Network Web site (Mueller et al., 2005) and aligned to the S. lycopersicum genome (release 2.31) using BLASTN. In a next step, protein sequences were extracted from Swiss-Prot and The Arabidopsis Information Resource databases for the given enzyme function. These sequences were aligned against the S. lycopersicum genome to identify potential coding sequences using BLAST and exonerate software (Slater and Birney, 2005). In addition, the iTAG2 annotation from the SOL Genomics Network Web site (Mueller et al., 2005) was queried using enzyme name keywords as well as Swiss-Prot identifiers to extract corresponding S. lycopersicum gene models. The corresponding candidate genes for a given enzyme QTL are provided as Supplemental Data S4.
Colocation Simulation
To assess the number of colocations that might occur by chance, a strategy similar to that of Lisec et al. (2008) was adopted. All considered enzymes and their locations on the different ILs were first determined based on the tomato genome release (iTAG2.3; http://solgenomics.net/organism/solanum_lycopersicum/genome). If multiple isoforms (e.g. tandem duplications) were located on the same IL, they were counted as one enzyme for the sake of the simulation, as a separation would require a better resolution for the QTL locations. This gave a total of 27 QTLs for 13 different enzymes. A total of 27 QTLs (i.e. ILs) for the 13 different enzymes were randomly drawn from the pool of 72 ILs that were studied in both years. The sampling was performed without replacement for each enzyme. The whole procedure was repeated 100,000 times, and this empirical colocation distribution was compared with the observed nine colocations.
Assessment of Gene Families
The iTAG2.3 annotation was searched to find candidate genes for all 13 enzymes within the QTL. The candidate genes were extracted from the S. lycopersicum genome release version 2.41 and aligned to the respective version of the predraft S. pennellii genome. Using the predicted cDNA of iTAG2.3 as a guide to splice sites, the reading frames were detected using gmap (Wu and Watanabe, 2005). SNPs causing amino acid changes in the protein as well as insertions/deletions and splice junction changes were recorded after a manual assessment.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Distribution of P values derived from t test analyses of metabolites observed in the homozygote ILs for the field trials 2003 and 2004.
Supplemental Figure S2. Scatterplot of the P value distribution derived from t test analyses of metabolites observed in the homozygote ILs for the field trials 2003 and 2004.
Supplemental Figure S3. Two-dimensional polar plot representation of the mode of inheritance and associated P values of detected metabolite QTLs, estimated using the phenotypic effects in the ILs and ILHs and the parental control M82.
Supplemental Figure S4. Estimation of the frequency of spurious colocations of enzyme activity QTLs and structural genes for that enzyme.
Supplemental Table S1. Overview of the optimized enzyme assays, their EC numbers, the abbreviations used in this work, and the pathways they belong to.
Supplemental Table S2. Trait heritability of metabolite traits in the S lycopersicum M82 × S. pennellii IL population.
Supplemental Table S3. Trait heritability of selected metabolite traits in the S lycopersicum M82 × S. pennellii IL population analyzed using GC-MS-based metabolite profiling by Schauer et al. (2008).
Supplemental Data S1. Overview of processed samples per genotype and mean-average values per genotype and assay.
Supplemental Data S2. Mean-average values and t test statistics of maximal enzyme activities and metabolite pool sizes, determined on tomato fruit pericarp tissue harvested at the ripe stage of fruit development, of S. pennellii ILs compared with the reference genotype S. lycopersicum M82.
Supplemental Data S3. Overview of QTL mapping and mode of inheritance statistics for maximal enzyme activities and metabolite pool sizes, determined on tomato fruit pericarp tissue harvested at the ripe stage of fruit development, for the S. lycopersicum × S. pennellii IL population.
Supplemental Data S4. Colocalization analyses of enzyme activity, metabolite, and expression QTLs with structural genes.
Supplemental Data S5. Analysis of polymorphisms in structural genes that potentially colocate with enzyme activity QTLs and of small gene families where no colocated QTLs were found.
Acknowledgments
We thank the International Tomato Genome Sequencing Consortium for making tomato genome sequence data available.
References
- Alpert KB, Tanksley SD. (1996) High-resolution mapping and isolation of a yeast artificial chromosome contig containing fw2.2: a major fruit weight quantitative trait locus in tomato. Proc Natl Acad Sci USA 93: 15503–15507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantan-Polak T, Kassai M, Grant KB. (2001) A comparison of fluorescamine and naphthalene-2,3-dicarboxaldehyde fluorogenic reagents for microplate-based detection of amino acids. Anal Biochem 297: 128–136 [DOI] [PubMed] [Google Scholar]
- Barton NH, Turelli M. (1989) Evolutionary quantitative genetics: how little do we know? Annu Rev Genet 23: 337–370 [DOI] [PubMed] [Google Scholar]
- Baxter CJ, Sabar M, Quick WP, Sweetlove LJ. (2005) Comparison of changes in fruit gene expression in tomato introgression lines provides evidence of genome-wide transcriptional changes and reveals links to mapped QTLs and described traits. J Exp Bot 56: 1591–1604 [DOI] [PubMed] [Google Scholar]
- Carrari F, Baxter C, Usadel B, Urbanczyk-Wochniak E, Zanor MI, Nunes-Nesi A, Nikiforova V, Centero D, Ratzka A, Pauly M, et al. (2006) Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol 142: 1380–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Fernie AR. (2006) Metabolic regulation underlying tomato fruit development. J Exp Bot 57: 1883–1897 [DOI] [PubMed] [Google Scholar]
- Causse M, Duffe P, Gomez MC, Buret M, Damidaux R, Zamir D, Gur A, Chevalier C, Lemaire-Chamley M, Rothan C. (2004) A genetic map of candidate genes and QTLs involved in tomato fruit size and composition. J Exp Bot 55: 1671–1685 [DOI] [PubMed] [Google Scholar]
- Causse M, Rocher J, Henry A, Charcosset A, Prioul J, de Vienne D. (1995) Genetic dissection of the relationship between carbon metabolism and early growth in maize, with emphasis on key-enzyme loci. Mol Breed 1: 259–272 [Google Scholar]
- Do PT, Prudent M, Sulpice R, Causse M, Fernie AR. (2010) The influence of fruit load on the tomato pericarp metabolome in a Solanum chmielewskii introgression line population. Plant Physiol 154: 1128–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- East EM. (1910) A Mendelian interpretation of variation that is apparently continuous. Am Nat 44: 65–82 [Google Scholar]
- Eshed Y, Zamir D. (1994) Introgressions from Lycopersicon pennellii can improve the soluble solids yield of tomato hybrids. Theor Appl Genet 88: 891–897 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Zamir D. (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L. (2000) Metabolite profiling for plant functional genomics. Nat Biotechnol 18: 1157–1161 [DOI] [PubMed] [Google Scholar]
- Fridman E, Carrari F, Liu YS, Fernie AR, Zamir D. (2004) Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Fridman E, Liu YS, Carmel-Goren L, Gur A, Shoresh M, Pleban T, Eshed Y, Zamir D. (2002) Two tightly linked QTLs modify tomato sugar content via different physiological pathways. Mol Genet Genomics 266: 821–826 [DOI] [PubMed] [Google Scholar]
- Fridman E, Pleban T, Zamir D. (2000) A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc Natl Acad Sci USA 97: 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P, Muller-Rober B, Stitt M. (1999) Contribution of adenosine 5′-diphosphoglucose pyrophosphorylase to the control of starch synthesis is decreased by water stress in growing potato tubers. Planta 209: 338–345 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Stitt M. (2000) Diurnal changes in sucrose, nucleotides, starch synthesis and AGPS transcript in growing potato tubers that are suppressed by decreased expression of sucrose phosphate synthase. Plant J 23: 795–806 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Höhne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M. (2004) A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J. (2001) Molecular biology of fruit maturation and ripening. Annu Rev Plant Physiol Plant Mol Biol 52: 725–749 [DOI] [PubMed] [Google Scholar]
- Hendriks JH, Kolbe A, Gibon Y, Stitt M, Geigenberger P. (2003) ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannsen W. (1911) The genotype conception of heredity. Am Nat 45: 129–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenetzky L, Asís R, Bassi S, de Godoy F, Bermúdez L, Fernie AR, Van Sluys MA, Vrebalov J, Giovannoni JJ, Rossi M, et al. (2010) Genomic analysis of wild tomato introgressions determining metabolism- and yield-associated traits. Plant Physiol 152: 1772–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJ, Sulpice R, Gibon Y, Steinhauser MC, Fu J, Koornneef M, Stitt M, Vreugdenhil D. (2008) Integrative analyses of genetic variation in enzyme activities of primary carbohydrate metabolism reveal distinct modes of regulation in Arabidopsis thaliana. Genome Biol 9: R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D, Pedersen D, Barker B, Mitchell-Olds T. (2002) Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidopsis thaliana. Genetics 161: 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, West MA, van Leeuwen H, Loudet O, Doerge RW, St Clair DA. (2006) Identification of QTLs controlling gene expression networks defined a priori. BMC Bioinformatics 7: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S, Bohs L, Nee M, Spooner DM. (2004) Solanaceae: a model for linking genomics with biodiversity. Comp Funct Genomics 5: 285–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J. (2006) Current challenges and developments in GC-MS based metabolite profiling technology. J Biotechnol 124: 312–322 [DOI] [PubMed] [Google Scholar]
- Lande R. (1983) The response to selection on major and minor mutations affecting a metrical trait. Heredity 50: 47–65 [Google Scholar]
- Li L, Lu K, Chen Z, Mu T, Hu Z, Li X. (2008) Dominance, overdominance and epistasis condition the heterosis in two heterotic rice hybrids. Genetics 180: 1725–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman ZB, Semel Y, Zamir D. (2007) An integrated view of quantitative trait variation using tomato interspecific introgression lines. Curr Opin Genet Dev 17: 545–552 [DOI] [PubMed] [Google Scholar]
- Lippman ZB, Zamir D. (2007) Heterosis: revisiting the magic. Trends Genet 23: 60–66 [DOI] [PubMed] [Google Scholar]
- Lisec J, Meyer RC, Steinfath M, Redestig H, Becher M, Witucka-Wall H, Fiehn O, Törjék O, Selbig J, Altmann T, et al. (2008) Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J 53: 960–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YS, Gur A, Ronen G, Causse M, Damidaux R, Buret M, Hirschberg J, Zamir D. (2003) There is more to tomato fruit colour than candidate carotenoid genes. Plant Biotechnol J 1: 195–207 [DOI] [PubMed] [Google Scholar]
- Meyer RC, Törjék O, Becher M, Altmann T. (2004) Heterosis of biomass production in Arabidopsis: establishment during early development. Plant Physiol 134: 1813–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T, Pedersen D. (1998) The molecular basis of quantitative genetic variation in central and secondary metabolism in Arabidopsis. Genetics 149: 739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounet F, Moing A, Garcia V, Petit J, Maucourt M, Deborde C, Bernillon S, Le Gall G, Colquhoun I, Defernez M, et al. (2009) Gene and metabolite regulatory network analysis of early developing fruit tissues highlights new candidate genes for the control of tomato fruit composition and development. Plant Physiol 149: 1505–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, Lin C, Wright MH, Ahrens R, Wang Y, et al. (2005) The SOL Genomics Network: a comparative resource for Solanaceae biology and beyond. Plant Physiol 138: 1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piques M, Schulze WX, Höhne M, Usadel B, Gibon Y, Rohwer J, Stitt M. (2009) Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol Syst Biol 5: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioul JL, Pelleschi S, Sene M, Thevenot C, Causse M, de Vienne D, Leonardi A. (1999) From QTLs for enzyme activity to candidate genes in maize. J Exp Bot 50: 1281–1288 [Google Scholar]
- Prudent M, Causse M, Génard M, Tripodi P, Grandillo S, Bertin N. (2009) Genetic and physiological analysis of tomato fruit weight and composition: influence of carbon availability on QTL detection. J Exp Bot 60: 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2009) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Rick CM. (1976) Natural variability in wild species of Lycopersicon and its bearing on tomato breeding. Genetica Agraria 30: 249–259 [Google Scholar]
- Rieseberg LH, Archer MA, Wayne RK. (1999) Transgressive segregation, adaptation and speciation. Heredity 83: 363–372 [DOI] [PubMed] [Google Scholar]
- Rousseaux MC, Jones CM, Adams D, Chetelat R, Bennett A, Powell A. (2005) QTL analysis of fruit antioxidants in tomato using Lycopersicon pennellii introgression lines. Theor Appl Genet 111: 1396–1408 [DOI] [PubMed] [Google Scholar]
- Rowe HC, Hansen BG, Halkier BA, Kliebenstein DJ. (2008) Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell 20: 1199–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Balbo I, Steinfath M, Repsilber D, Selbig J, Pleban T, Zamir D, Fernie AR. (2008) Mode of inheritance of primary metabolic traits in tomato. Plant Cell 20: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, et al. (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Schauer N, Zamir D, Fernie AR. (2005) Metabolic profiling of leaves and fruit of wild species tomato: a survey of the Solanum lycopersicum complex. J Exp Bot 56: 297–307 [DOI] [PubMed] [Google Scholar]
- Semel Y, Nissenbaum J, Menda N, Zinder M, Krieger U, Issman N, Pleban T, Lippman Z, Gur A, Zamir D. (2006) Overdominant quantitative trait loci for yield and fitness in tomato. Proc Natl Acad Sci USA 103: 12981–12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva LI, Keurentjes JJ, Bentsink L, Vonk J, van der Plas LH, Koornneef M, Vreugdenhil D. (2006) Vacuolar invertase regulates elongation of Arabidopsis thaliana roots as revealed by QTL and mutant analysis. Proc Natl Acad Sci USA 103: 2994–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serina L, Blondin C, Krin E, Sismeiro O, Danchin A, Sakamoto H, Gilles AM, Bârzu O. (1995) Escherichia coli UMP-kinase, a member of the aspartokinase family, is a hexamer regulated by guanine nucleotides and UTP. Biochemistry 34: 5066–5074 [DOI] [PubMed] [Google Scholar]
- Slater GS, Birney E. (2005) Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. (1995) Biometry: The Principles and Practice of Statistics in Biological Research, Ed 3. W.H. Freeman, New York [Google Scholar]
- Springer NM, Stupar RM. (2007) Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. Plant Cell 19: 2391–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauser MC, Steinhauser D, Koehl K, Carrari F, Gibon Y, Fernie AR, Stitt M. (2010) Enzyme activity profiles during fruit development in tomato cultivars and Solanum pennellii. Plant Physiol 153: 80–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R, Trenkamp S, Steinfath M, Usadel B, Gibon Y, Witucka-Wall H, Pyl ET, Tschoep H, Steinhauser MC, Guenther M, et al. (2010) Network analysis of enzyme activities and metabolite levels and their relationship to biomass in a large panel of Arabidopsis accessions. Plant Cell 22: 2872–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW, Mendes P, Dixon RA. (2003) Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry 62: 817–836 [DOI] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Jia Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS. (2006) All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc Natl Acad Sci USA 103: 6805–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, Ganal MW, Martin GB. (1995) Chromosome landing: a paradigm for map-based gene cloning in plants with large genomes. Trends Genet 11: 63–68 [DOI] [PubMed] [Google Scholar]
- Thévenot C, Simond-Côte E, Reyss A, Manicacci D, Trouverie J, Le Guilloux M, Ginhoux V, Sidicina F, Prioul JL. (2005) QTLs for enzyme activities and soluble carbohydrates involved in starch accumulation during grain filling in maize. J Exp Bot 56: 945–958 [DOI] [PubMed] [Google Scholar]
- Tieman D, Taylor M, Schauer N, Fernie AR, Hanson AD, Klee HJ. (2006) Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc Natl Acad Sci USA 103: 8287–8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Roepenack-Lahaye E, Degenkolb T, Zerjeski M, Franz M, Roth U, Wessjohann L, Schmidt J, Scheel D, Clemens S. (2004) Profiling of Arabidopsis secondary metabolites by capillary liquid chromatography coupled to electrospray ionization quadrupole time-of-flight mass spectrometry. Plant Physiol 134: 548–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK. (2005) GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875 [DOI] [PubMed] [Google Scholar]
- Zamir D. (2001) Improving plant breeding with exotic genetic libraries. Nat Rev Genet 2: 983–989 [DOI] [PubMed] [Google Scholar]
- Zanor MI, Rambla JL, Chaïb J, Steppa A, Medina A, Granell A, Fernie AR, Causse M. (2009) Metabolic characterization of loci affecting sensory attributes in tomato allows an assessment of the influence of the levels of primary metabolites and volatile organic contents. J Exp Bot 60: 2139–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Gibon Y, Gur A, Chen C, Lepak N, Höhne M, Zhang Z, Kroon D, Tschoep H, Stitt M, et al. (2010) Fine quantitative trait loci mapping of carbon and nitrogen metabolism enzyme activities and seedling biomass in the maize IBM mapping population. Plant Physiol 154: 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]