Abstract
SHORT-ROOT (SHR) is a key regulator of root growth and development in Arabidopsis (Arabidopsis thaliana). Made in the stele, the SHR protein moves into an adjacent cell layer, where it specifies endodermal cell fate; it is also essential for apical meristem maintenance, ground tissue patterning, vascular differentiation, and lateral root formation. Much has been learned about the mechanism by which SHR controls radial patterning, but how it regulates other aspects of root morphogenesis is still unclear. To dissect the SHR developmental pathway, we have determined the genome-wide locations of SHR direct targets using a chromatin immunoprecipitation followed by microarray analysis method. K-means clustering analysis not only identified additional quiescent center-specific SHR targets but also revealed a direct role for SHR in gene regulation in the pericycle and xylem. Using cell type-specific markers, we showed that in shr, the phloem and the phloem-associated pericycle expanded, whereas the xylem and xylem-associated pericycle diminished. Interestingly, we found that cytokinin level was elevated in shr and that exogenous cytokinin conferred a shr-like vascular patterning phenotype in wild-type root. By chromatin immunoprecipitation-polymerase chain reaction and reverse transcription-polymerase chain reaction assays, we showed that SHR regulates cytokinin homeostasis by directly controlling the transcription of cytokinin oxidase 3, a cytokinin catabolism enzyme preferentially expressed in the stele. Finally, overexpression of a cytokinin oxidase in shr alleviated its vascular patterning defect. On the basis of these results, we suggest that one mechanism by which SHR controls vascular patterning is the regulation of cytokinin homeostasis.
As the primary site of water and inorganic nutrient uptake, the root is critically important for land plant growth and development. To meet the needs of a growing shoot, the root must also increase its surface area, which it does by continuous growth from the root apical meristem and by branching as a result of lateral or adventitious root formation. Because different cell types have distinct functions, each cell type must be precisely regulated in its fate determination and patterning.
The Arabidopsis (Arabidopsis thaliana) root has proven to be a very tractable system for dissection of the molecular basis of root development, mainly because it has a simple pattern of cell organization and stereotyped cell divisions that give rise to the various cell types (Fig. 1A; Benfey and Scheres, 2000). Along the longitudinal axis, three regions with distinct mitotic activity and morphology can be recognized: the apical meristem, which has small and rapidly dividing cells; the elongation zone, where mitotic activity has ceased and cells are elongating; and the maturation zone, where cells become differentiated, root hairs are produced, and at a later stage lateral roots emerge (Ishikawa and Evans, 1995). At the root tip are several layers of cells called the root cap, which has a dual role: sensing gravity and protecting the apical meristem. In longitudinal section, the epidermis, cortex, and endodermis are easily discernible as clearly defined cell files (Dolan et al., 1993). Adjacent to the endodermis is the pericycle, which also forms a well-delineated cell layer. It surrounds the central vascular tissue and is the site of lateral root formation (Dolan et al., 1993). In the center of the root meristem is a group of cells with very low mitotic activity called the quiescent center (QC). The QC cells do divide, though, producing cells that are mitotically active and are the immediate source of other cell types in the root (and therefore are called “initial cells”). The QC cells, along with the neighboring initial cells, form the stem-cell niche (Benfey and Scheres, 2000), which is indispensable for root growth. Because cell divisions in initial cells follow a seemingly predetermined pattern, the Arabidopsis root is particularly suitable for the study of cell fate specification (Benfey and Scheres, 2000). For example, the endodermis and cortex are derived from the same initial cell (the cortex and endodermis initial, CEI) through two consecutive asymmetric cell divisions (the first is transverse and the second is longitudinal) that are reiterated during the growth of the root meristem (Fig. 1A).
Figure 1.
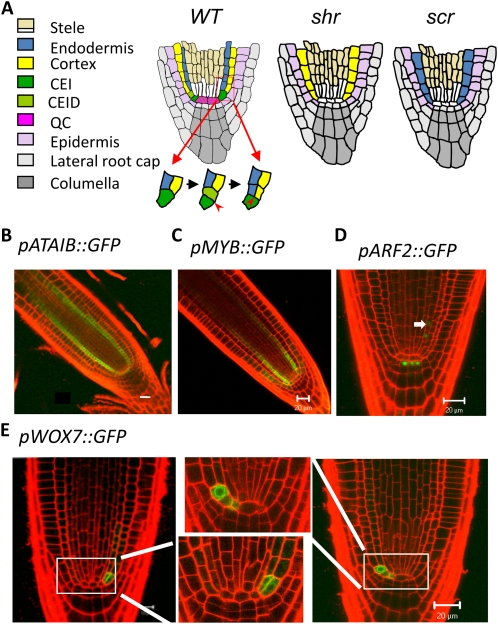
Expression patterns of novel SHR targets identified by ChIP-chip. A, Schematics of the radial patterns in the primary root of the wild type (WT), shr, and scr and the asymmetric division of the CEI that gives rise to the two layers of ground tissue. CEID, CEI daughter cell. B to E, Confocal microscopy images of transgenic lines expressing GFP under the control of the promoters of selected top-ranked SHR targets. Note the pARF2::GFP expression in the pericycle (arrow). Bars = 20 μm.
The Arabidopsis root also has a simple organization of vascular tissues: it has a single strand of xylem flanked by two separate domains of phloem (see Fig. 3A below; Parizot et al., 2008). The xylem can be distinguished from the phloem by the presence of large vessel element cells. On the basis of position and cell wall patterning, two types of xylem can be further distinguished: protoxylem, which is close to the pericycle and has annular wall thickening, and metaxylem, which is in the center of the stele and has net-like wall thickening. The pericycle was once regarded as a single cell type, but recent studies have shown that it consists of two populations of cells that are cytologically and functionally different: one associated with the xylem and the other with the phloem. The pericycle cells at the xylem pole have denser cytoplasm and are the sites of lateral root formation (Parizot et al., 2008). The two types of pericycle cells also respond differently to the phytohormones auxin and cytokinin. Auxin stimulates mitotic activity in the xylem-pole pericycle (Parizot et al., 2008), whereas cytokinin represses cell division in this domain (Li et al., 2006). The pericycle at the phloem pole does not appear to be affected by auxin (Parizot et al., 2008), but whether it responds to cytokinin is not clear.
Figure 3.
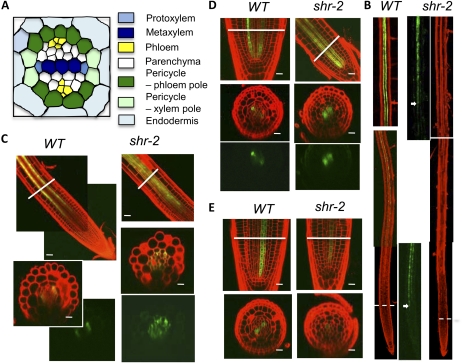
Confocal microscopy analysis of cell type-specific GFP markers in wild-type (WT) and shr-2 roots, showing the vascular patterning defect in shr. A, Diagram of the stele, showing the two types of pericycle as well as other cell types. B, Expression of J0121, a xylem-pole pericycle marker in the wild type and shr-2. The dashed lines mark the border between the apical meristem and the elongation zone. In the panels showing GFP only, the signal was enhanced to show more clearly the position where GFP was first detected, which is marked by the arrows. C, Expression of S17, a phloem-pole pericycle marker, in wild-type and shr-2 root. D, Expression of S32, an early phloem marker, in wild-type and shr-2 root. E, Expression of S4, an early xylem marker, in wild-type and shr-2 root. In C to E, the lines mark the positions where the cross-sections were made. Bars = 20 μm.
A pivotal role for auxin in vascular tissue differentiation, lateral root formation, and root apical meristem maintenance has been well established (Berleth et al., 2000; Bennett and Scheres 2010). Cytokinin, on the contrary, is known to inhibit root growth (Beemster and Baskin 2000; Dello Ioio et al., 2007) and lateral root formation (Laplaze et al., 2007). An important role for cytokinin in vascular tissue differentiation and patterning also begins to be appreciated. Mutation in cytokinin receptors, such as CYTOKININ RESPONSE1 (Mähönen et al., 2000), and loss of ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN6 (AHP6; Mähönen et al., 2006), an inhibitor of cytokinin signaling, cause the loss of phloem and a decrease in the total number of vascular cells.
Genetic studies in past decades have identified a number of proteins as important regulators of root morphogenesis, among them SHORT-ROOT (SHR; Helariutta et al., 2000), SCARECROW (SCR; Di Laurenzio et al., 1996), PLETHORA (Aida et al., 2004), BODENLOS (Hamann et al., 1999), and OBERON1 and OBERON2 (Thomas et al., 2009). Interestingly, many of these proteins are regulated by auxin, and some even function through the regulation of auxin distribution. PLETHORA, for example, is induced by auxin, but in turn, it activates the expression of some PIN-FORMED (PIN) family genes (Aida et al., 2004). PIN proteins are important players in auxin polar transport. The intimate interplay between PLETHORA and PIN proteins leads to the formation of auxin maxima in the root tip (Galinha et al., 2007), which is essential for maintaining the root apical meristem.
SHR and SCR play important roles not only in root apical meristem maintenance but also in radial patterning. In shr and scr, the QC cells are exhausted prematurely, causing a short-root phenotype. The asymmetric cell division in the CEI cells that generate the endodermis and cortex also fails to occur; consequently, only a single layer of ground tissue is produced (Di Laurenzio et al., 1996; Helariutta et al., 2000). Despite the similar developmental defects in shr and scr, SHR and SCR have different functions in ground tissue patterning. SCR appears to be required only for the asymmetric cell division (Di Laurenzio et al., 1996), whereas SHR controls both endodermis specification and asymmetric cell division (Helariutta et al., 2000). SHR and SCR also have different gene expression patterns: SCR is expressed in the endodermis, but SHR is expressed in the stele (Di Laurenzio et al., 1996; Helariutta et al., 2000). Made in the stele, the SHR protein moves into the adjacent cell layer (Nakajima et al., 2001), where it forms a heterodimer with SCR, which is already present at a low level, and the SHR-SCR complex activates the transcription of SCR through a positive feedback loop as well as other targets through feed-forward mechanisms (Cui et al., 2007).
Although SHR is expressed in the stele, its function in this tissue was not recognized until recently. Levesque et al. (2006) first noted that in shr the number of cell files in the stele was reduced. Subsequently, several studies showed that SHR has a role in promoting vascular tissue differentiation and lateral root formation (Carlsbecker et al., 2010; Gardiner et al., 2011; Yu et al., 2010). Carlsbecker et al. (2010) showed that the shr mutation causes the loss of protoxylem. This phenotype was also observed in scr (Carlsbecker et al., 2010). In further studies, they showed that at least two genes of the miR165/166 family, miR165A and miR166B, are direct targets of SHR and SCR and that these microRNAs move from their site of synthesis in the endodermis into the stele to repress homeodomain-leucine zipper (HD-ZIP III) transcription factors, thereby specifying protoxylem cell fate (Carlsbecker et al., 2010). How SHR regulates other cell types in the stele and lateral root formation is still unknown, however.
To dissect the SHR developmental pathway, we have recently identified a subset of its direct targets by chromatin immunoprecipitation followed by microarray analysis (ChIP-chip; Sozzani et al., 2010). We also performed ChIP-chip with SHR using another format of microarray. Combining the two sets of data, we determined the genome-wide location of direct SHR targets. Through clustering analysis and cell type GFP marker analysis, we found that shr causes alterations in the ratio between the xylem and phloem and the relative abundance of the two types of pericycle. Interestingly, we found that the response to cytokinin but not auxin was elevated in shr and that exogenous cytokinin mimicked the vascular patterning phenotype of shr. Consistent with this observation, we showed that cytokinin level was elevated in shr relative to that in the wild type. We further showed that SHR directly controls the expression of CYTOKININ OXIDASE3 (CKX3), a cytokinin catabolism enzyme that is preferentially expressed in the xylem, and that ectopic expression of a CKX gene in shr reversed its vascular pattern. These results together suggest that SHR controls vascular tissue patterning through the regulation of cytokinin homeostasis.
RESULTS
Genome-Wide Identification of SHR Direct Targets by ChIP-Chip
To dissect the SHR transcriptional network, we have developed a ChIP-chip method for genome-wide identification of direct targets by transcriptional regulators in Arabidopsis. Two types of microarray were used in the course of our study: a custom whole-genome microarray fabricated by means of the Nimblegen maskless lithography technology (Lee et al., 2007), and a custom promoter microarray made by Agilent (Gene Expression Omnibus accession no. GSE21338). The Agilent microarray has larger feature (probe spot) size than the Nimblegen and more probes corresponding to promoter sequences, which appear to produce higher signal-to-noise ratios and thus higher confidence in the data analysis. For the Agilent microarray, we also designed probes for microRNA genes (http://asrp.cgrb.oregonstate.edu/db/microRNAfamily.html), as microRNAs are a critical component of the transcriptional regulatory mechanism. Recently, we have used the data obtained with the Agilent microarray, coupled with a time-course study of the transcriptome in isolated ground tissue cells, in a study to identify SHR targets involved in ground tissue patterning (Sozzani et al., 2010). To obtain a global view of SHR target genes, we analyzed the ChIP-chip data independently of gene expression data. This alternative analysis was largely based on fold enrichment values, because they agree well with validation by ChIP-PCR (Supplemental Fig. S1; Cui et al., 2007). First, positive probes with greater than 2-fold enrichment in both biological ChIP-chip replicates were identified, and their corresponding genes were assigned. In this way, we identified 595 putative SHR direct targets (Supplemental Table S1). To select candidate genes with high confidence for further functional characterization, we next filtered this list by eliminating those that did not pass the same criterion (2-fold enrichment) from the Nimblegen data, producing a list of 200 genes (Supplemental Table S1). Finally, we ranked these genes on the basis of degree of enrichment as well as the number of positive probes for each promoter. Nine genes have been previously identified as SHR direct targets, including five transcriptional regulators (SCR, MAGPIE, JACKDAW, NUTCRACKER, SCARECROW-LIKE3; Levesque et al., 2006; Cui et al., 2007) and a microRNA gene (miR165/166; Carlsbecker et al., 2010). Remarkably, all of these transcriptional regulators except for SCR were among the 25 top-ranked SHR targets (Table I).
Table I. Top-ranked direct SHR targets identified by ChIP-chip.
AGI, Arabidopsis Genome Initiative identification number; FC, fold change.
| Rank | AGI | No. of Probes | FC | Gene Annotation |
| 1 | AT1G03840a | 14 | 3.06 | MAGPIE (MGP) |
| 2 | AT1G50420a | 9 | 4.18 | SCARECROW-LIKE3 (SCL3) |
| 3 | AT2G46510b | 7 | 3.65 | ATAIB, bHLH transcription factor |
| 4 | AT1G68670b | 7 | 2.98 | MYB family transcription factor |
| 5 | AT5G62000b | 5 | 3.96 | ARF2, transcription factor B3 family protein |
| 6 | AT1G24120 | 4 | 3.65 | ARG-like protein |
| 7 | AT2G28550 | 4 | 3.2 | TOE1, AP2 domain-containing transcription factor |
| 8 | AT4G01720 | 4 | 3.07 | WRKY47 |
| 9 | AT4G25560 | 4 | 2.95 | MYB18 |
| 10 | AT3G02140 | 4 | 2.86 | TMAC2, TWO OR MORE ABRES-CONTAINING GENE2 |
| 11 | AT3G02150 | 4 | 2.86 | TCP transcription factor |
| 12 | AT5G44160a | 4 | 2.73 | NUTCRACKER (NUC) |
| 13 | AT2G03730 | 3 | 4.33 | ACT domain-containing protein (ACR5) |
| 14 | AT5G03150a | 3 | 3.33 | JACKDAW (JKD) |
| 15 | AT1G13260 | 3 | 3.18 | DNA-binding protein RAV1 (RAV1) |
| 16 | AT3G04570 | 3 | 2.94 | DNA-binding protein-related |
| 17 | AT3G50060 | 3 | 2.84 | MYB77 |
| 18 | AT3G61890 | 3 | 2.65 | AtHB12 |
| 19 | AT3G61897a | 3 | 2.65 | miR166B (targeting At1g52150 [HD-ZIP]) |
| 20 | AT2G36080 | 3 | 2.65 | RAV1/2 like DNA-binding protein |
| 21 | AT2G18160 | 3 | 2.55 | GBF5/bZIP2 transcription factor |
| 22 | AT5G44180 | 3 | 2.33 | Homeobox transcription factor |
| 23 | AT3G07360 | 2 | 3.61 | Armadillo/β-catein repeat family factor |
| 24 | AT5G05760 | 2 | 3.13 | SYP31 (syntaxin 31)/SED5 |
| 25 | AT5G05770b | 2 | 3.13 | WUSCHEL-RELATED HOMEOBOX7 (WOX7) |
| . . . | ||||
| 154 | AT3G54220a | 1 | 2.26 | SCR |
Previously identified SHR target.
Novel SHR target confirmed in this study.
To assess how well our analysis methods performed, we confirmed SHR binding to the promoters of some novel top-ranked SHR targets by ChIP-PCR using primer pairs that tile the entire promoter region. As shown in Supplemental Figure S2, SHR bound to the promoters of all four genes examined. As a first step toward characterizing their function, we next examined their in vivo expression patterns by examining transgenic plants carrying promoter-GFP fusion constructs. Remarkably, all these genes showed a cell type-specific expression pattern within the SHR domain (Fig. 1). Of particular interest is WUSCHEL-RELATED HOMEOBOX7 (WOX7; Fig. 1E), which is expressed in the cortex/endodermis initial, in its daughter cell, or in the first cortex or endodermis cell, and therefore may play a role in controlling the asymmetric cell division that patterns the ground tissue. When these GFP reporter constructs were introduced into the shr mutant background (by crossing), their expression was dramatically reduced or no longer detectable (Supplemental Fig. S3). These results validated our analysis, suggesting that it is a straightforward but reliable approach for the identification of transcription factor targets and the assignment of priorities for further functional characterization.
Clustering Analysis Revealed a Direct Role for SHR in Vascular Patterning
Protein function is dictated by the protein’s location of expression. To get a clue to the function of other SHR targets in cell fate specification, we examined their expression pattern according to RootMap, which was produced by profiling of the transcriptome in every cell type as well as at different developmental stages in the Arabidopsis root (Birnbaum et al., 2003; Brady et al., 2007). On the basis of the gene expression data, we then performed K-means clustering analysis of the 200 putative SHR targets. SHR targets were first divided into two clusters, because SHR has distinct subcellular localization in the endodermis and stele: it is present in both the cytoplasm and the nucleus in the stele but is exclusively nuclear localized in the endodermis. The first cluster of genes (46 genes; Supplemental Table S2) had a lower level of expression in the stele (Fig. 2A), suggesting transcriptional repression by SHR. In supporting this notion, we confirmed that IAA16, which appears to have lowest expression in the xylem (Supplemental Fig. S4A), was a direct target of SHR (Supplemental Fig. S4B) and that its transcript level was higher in the shr mutant (Supplemental Fig. S4C).
Figure 2.
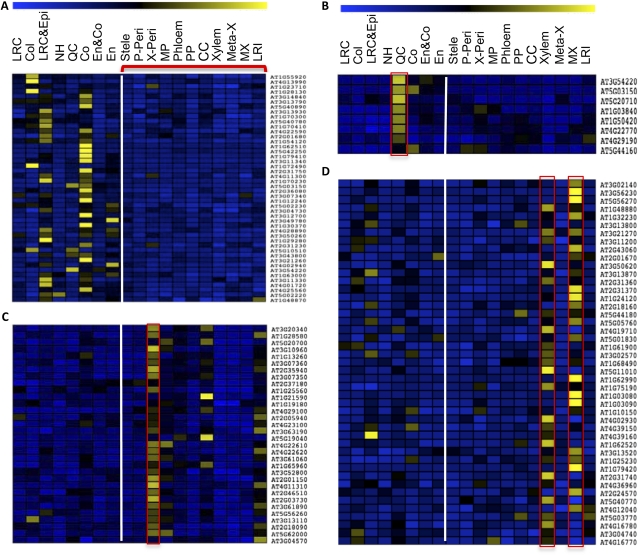
K-means clustering analysis of SHR direct targets. A, Gene cluster with a low level of expression in the stele. B, The QC subcluster. C and D, Gene clusters with preferential expression in the pericycle and xylem, respectively. Yellow and blue indicate high and low levels of gene expression, respectively. The cell types from left to right are lateral root cap (LRC), columella (Col), lateral root cap and epidermis (LRC&Epi), non-root hair cells (NH), QC, cortex (Co), endodermis and cortex (En&Co), endodermis (En), stele, phloem-associated pericycle (P-Peri), xylem-associated pericycle (X-Peri), pericycle in the maturation zone (MP), phloem, protophloem (PP), companion cell (CC), xylem, metaxylem (Meta-X), xylem in the maturation zone (MX), and lateral root initial (LRI). The vertical lines mark the boundary between the stele and outer cell types.
Interestingly, the majority of putative SHR targets (the second cluster) were preferentially expressed in the stele, suggesting that SHR also acts as an activator there. Further clustering of the second cluster resulted in three subclusters with prominent expression in the QC, xylem, and pericycle (Fig. 2). The QC subcluster has eight genes (Fig. 2B; Supplemental Table S2), including SCL3, MGP, and NUC as well as SCR and JKD, which were initially clustered into the stele-low cluster but were added to the QC subcluster because of their prominent expression pattern in the QC (Fig. 2A). Three novel SHR targets were also present in this subcluster, including two putative transcription factors (At4g29190, a zinc finger-containing transcription factor, and At4g22770, an AT-Hook transcription factor) and At5g20710, a β-galactosidase. Their QC-specific expression pattern suggests that these genes play a role in stem-cell renewal or physiology. The pericycle and xylem subclusters include 32 and 45 genes, respectively (Supplemental Table S2).
The preferential expression of SHR direct targets in the xylem and pericycle suggests a direct role for SHR in the specification of these cell types. A role for SHR in xylem and phloem differentiation has been identified (Carlsbecker et al., 2010; Gardiner et al., 2011; Yu et al., 2010), but whether SHR has a role in pericycle specification is unclear, so we examined the effect of the shr mutation on pericycle cell fate using the pericycle markers J0121 (Laplaze et al., 2005) and S17 (Lee et al., 2006). In wild-type root, J0121 was first detected in the early elongation zone, but in shr, it was not detected until late into the maturation zone (Fig. 3B). In contrast, the S17 expression domain increased dramatically in shr (Fig. 3C). Because S17 and J0121 are markers for the pericycle associated with the phloem and xylem, respectively, we reasoned that the balance between the phloem and xylem might also be altered by the shr mutation. Using S32, a phloem marker, and S4, a xylem marker (Lee et al., 2006), we indeed found that the phloem domain was enlarged, whereas the xylem domain was reduced, in the shr mutant (Fig. 3, D and E). These observations suggest that SHR controls the balance between phloem and xylem as well as the relative abundance of the two types of pericycle associated with them.
Exogenous Cytokinin Confers a shr-Like Vascular Phenotype
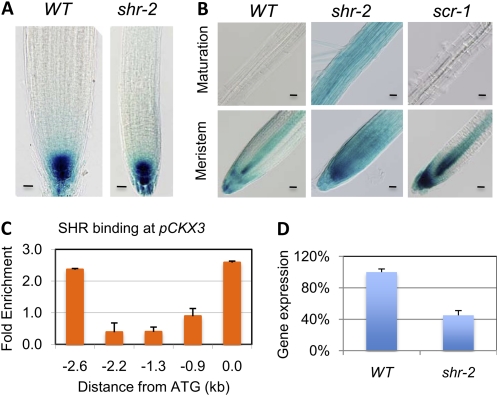
A recent publication reported that shr has an elevated level of auxin (Lucas et al., 2011). Because auxin is known to play a key role in vascular differentiation, it seems most likely to control vascular patterning through auxin signaling. To test this hypothesis, we examined the effect of exogenously applied 1-naphthaleneacetic acid (NAA), a synthetic auxin, on S17, J0121, S32, and S4 expression in a wild-type background. Surprisingly, we observed no apparent change in the expression pattern of these markers (Supplemental Fig. S5), except for J0121, the expression domain of which was enlarged at the sites of lateral root initiation (Fig. 4D, arrows), as reported previously (Parizot et al., 2008). Therefore, we next determined auxin response in shr using the DR5::GUS reporter construct but found that its expression pattern and intensity were not altered by the shr mutation (Fig. 5A). These results argue against a role for SHR in regulating vascular patterning through auxin homeostasis or signaling.
Figure 4.
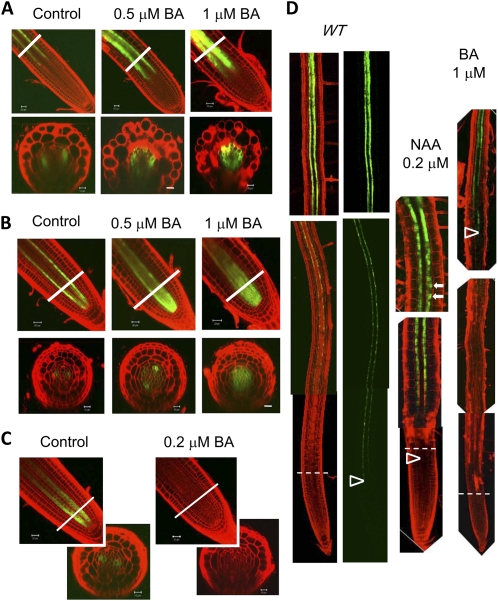
Exogenous cytokinin confers a shr-like phenotype in root vascular patterning. A, Expression of the phloem-pole pericycle marker, S17, in response to BA. B, Response of the early phloem marker S32 to BA. C, Response of the early xylem marker S4 to BA. The lines in A to C mark the positions of cross-sectioning. D, Response of the xylem-pole pericycle marker, J0121, to NAA or BA. The dashed lines indicate the border between the root apical meristem and the elongation zone. The arrowheads mark the positions where GFP was first detected; the arrows mark the locations of lateral root primordia. WT, Wild type. Bars = 20 μm.
Figure 5.
SHR regulates cytokinin homeostasis. A, DR5::GUS activity in wild-type (WT) and shr-2 roots. Bars = 20 μm. B, GUS staining of wild-type, shr-2, and scr-1 roots containing the pARR5::GUS reporter construct, showing the elevated response to cytokinin in shr-2 root. Bars = 20 μm. C, ChIP-PCR assay showing SHR binding to the CKX3 promoter. D, Quantitative RT-PCR assay of CKX3 transcript in wild-type and shr-2 roots. The wild type was taken as 100%.
Because cytokinin also plays an important role in vascular tissue differentiation, we next checked the effect of exogenous cytokinin on the same set of cell markers. Strikingly, on medium containing 6-benzylaminopurine (BA), a synthetic cytokinin, S17 and S32 marker lines expanded (Fig. 4, A and B), whereas J0121 and S4 expression domains were reduced (Fig. 4, C and D). The effect of cytokinin on vascular differentiation depended on concentration: an apparent effect was observed when the cytokinin concentration was 0.5 μm and above, except for S4, which lost expression at a BA concentration as low as 0.2 μm (Fig. 4C). At a concentration of 1 μm BA, S17 and S32 were seen nearly throughout the stele (Fig. 4, A and B), whereas S4 and J0121 became barely detectable (Fig. 4, C and D). These results suggest that the vascular patterning defect in shr is due to an elevated level of cytokinin response.
SHR Controls Cytokinin Homeostasis
To determine whether the cytokinin response is altered in shr, we first compared the expression of pARR5::GUS, a cytokinin response marker (D’Agostino et al., 2000), in wild-type and shr roots. In wild-type root, GUS activity was visible in the root cap and stele but not detected in the maturation zone. In contrast, in shr, strong GUS activity was seen in all cell types in the root, including root hairs, and all development stages (Fig. 5B). The elevated response of ARR5 in shr is unlikely to be a secondary effect from the shr mutation, because only a slight increase in pARR5::GUS activity was observed in scr (Fig. 5B), which has developmental defects similar to those in shr. These results suggest that SHR regulates cytokinin homeostasis or signaling independently of SCR. To distinguish between these possibilities, we next measured cytokinin content in shr and wild-type plants. As shown in Table II, the concentrations of several types of cytokinin were significantly higher in both root and shoot of shr, consistent with the expression of SHR in both organs (Helariutta et al., 2000; Dhondt et al., 2010; Gardiner et al., 2011). This result, along with the observations that exogenous cytokinin causes shr-like vascular patterning, strongly suggests that high cytokinin concentration is a major cause of the vascular patterning defect in shr.
Table II. Cytokinin content in the root and shoot of shr-2 and wild-type seedlings.
The values for cytokinin content are means ± sd from triplicate measurements. Units are pmol g−1 fresh weight. tZ, Transzeatin; tZR, tZ riboside; tZRPs, tZR mono-, di-, and triphosphates; cZ, cis-zeatin; cZR, cZ riboside; cZRPs, cZR mono-, di-, and triphosphates; iP, N6-(Δ2-isopentenyl)adenine; iPR, iP riboside; iPRPs, iPR mono-, di-, and triphosphates; tZ7G, tZ-7-glucoside; tZ9G, tZ-9-glucoside; tZOG, tZ-O-glucoside; tZROG, tZR-O-glucoside; cZROG, cZR-O-glucoside; DZ9G, dihydrozeatin riboside; iP7G, iP-7-glucoside; iP9G, iP-9-glucoside; N.D., not detected.
| Cytokinin | Wild-Type Root | shr-2 Root | Wild-Type Shoot | shr-2 Shoot |
| tZ | 0.23 ± 0.02 | 0.98 ± 0.39a | 0.33 ± 0.16 | N.D. |
| tZR | 1.22 ± 0.12 | 8.78 ± 3.22b | 1.67 ± 0.73 | 3.60 ± 0.23 |
| tZRPs | 32.22 ± 11.39 | 22.01 ± 4.53 | 53.19 ± 5.10 | 9.21 ± 6.45b |
| cZ | 2.40 ± 0.17 | 1.89 ± 0.49a | 0.61 ± 0.04 | N.D. |
| cZR | 14.30 ± 2.44 | 30.97 ± 10.38b | 8.66 ± 1.69 | 44.01 ± 10.00b |
| cZRPs | 5.52 ± 0.85 | 25.81 ± 6.53b | 11.68 ± 4.90 | 3.09 ± 1.62 |
| iP | 0.42 ± 0.08 | 0.53 ± 0.12 | 0.56 ± 0.14 | 1.12 ± 0.004 |
| iPR | 5.67 ± 0.66 | 7.70 ± 0.79b | 22.10 ± 7.94 | 22.49 ± 3.19 |
| iPRPs | 0.36 ± 0.06 | 3.70 ± 0.74b | 11.53 ± 1.23 | 2.27 ± 2.01b |
| tZ7G | 1.78 ± 0.20 | 6.08 ± 1.19b | 10.63 ± 1.68 | 10.09 ± 2.55 |
| tZ9G | 1.93 ± 0.15 | 7.84 ± 1.58b | 12.36 ± 1.81 | 11.16 ± 3.64 |
| tZOG | 1.16 ± 0.13 | 4.88 ± 1.01b | 3.11 ± 0.97 | 5.21 ± 0.27 |
| tZROG | 0.04 ± 0.004 | 0.25 ± 0.04b | 0.33 ± 0.06 | 0.44 ± 0.14 |
| cZROG | 0.03 ± 0.002 | 0.10 ± 0.02a | 0.81 ± 0.28 | 1.66 ± 0.52 |
| DZ9G | 0.01 ± 0.002 | 0.06 ± 0.01b | 0.05 ± 0.01 | 0.07 ± 001 |
| iP7G | 10.13 ± 2.1 | 12.13 ± 0.10 | 37.88 ± 11.52 | 35.99 ± 4.77 |
| iP9G | 1.02 ± 0.1 | 1.36 ± 0.09b | 3.26 ± 0.93 | 2.66 ± 0.07 |
0.05 ≤ P ≤ 0.10 (from t test).
P ≤ 0.05 (from t test).
SHR could regulate cytokinin homeostasis directly or indirectly. To determine which, we searched the list of SHR targets for genes that are involved in cytokinin biosynthesis or catabolism. Among the top-ranked SHR targets identified by the Nimblegen microarray, we found CKX3, which encodes a cytokinin oxidase that is responsible for cytokinin inactivation (Werner et al., 2003). Another putative SHR target that is involved in cytokinin homeostasis is IPT5, which encodes an adenylate isopentenyltransferase that plays a positive role in cytokinin biosynthesis. We could not use the Agilent data to compare the ranking of CKX3 and IPT5, because CKX3 was not represented on that micorarray. According to the Nimblegen data, however, CKX3 was ranked much higher than IPT5 (nos. 76 and 1,437, respectively; Supplemental Table S1). Therefore, we decided to focus on CKX3 for further study. We first confirmed SHR binding to the CKX3 promoter by ChIP-PCR (Fig. 5C). Reverse transcription (RT)-PCR assay showed that the CKX3 transcript level was significantly reduced in shr root (Fig. 5D). According to RootMap (Brady et al., 2007), CKX3 is expressed preferentially in the xylem (Supplemental Fig. S6). These results suggest that SHR controls vascular patterning by maintaining a low level of cytokinin in the xylem through CKX3.
Reduction of Cytokinin Reverses the Vascular Patterning Defect in shr
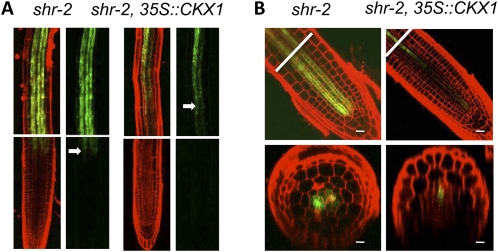
To determine whether cytokinin homeostasis and vascular patterning are causally related in shr, we introduced the 35S::CKX1 transgene into the shr background by crossing and then examined the expression pattern of the phloem and phloem-associated markers S32 and S17. The 35S::CKX1 gene was used instead of CKX3 because CKX1, CKX2, and CKX3 are functionally equivalent (Werner et al., 2003) and the 35S::CKX1 gene has been demonstrated to be an efficient tool for reducing cytokinin levels in plants (Werner et al., 2003). By this approach, we expected to alleviate the vascular patterning defect in shr. As shown in Figure 6, S17 and S32 expression was dramatically reduced in shr. In shr root, S17 was visible at the border between the meristem and the elongation zone, and it became strong in the late elongation zone and the maturation zone (Fig. 6A). In contrast, in shr root with the 35S::CKX1 transgene, S17 was no longer expressed in the elongation zone and only weakly expressed in the maturation zone (Fig. 6A). Similar changes occurred to S32. In shr root, S32 was expressed not only early (close to the first phloem initial cell) but also in more cell files (up to six; Fig. 6B), but in shr root with 35S::CKX1, S32 was barely detectable in the meristem zone, and in the elongation zone it was restricted to fewer cells (one or two) and a single phloem pole (Fig. 6B). These results lend strong support to the conclusion that SHR controls vascular patterning through its effect on cytokinin homeostasis.
Figure 6.
CKX1 overexpression in shr resulted in decreases in the size of the phloem and phloem-associated pericycle. A, Expression of the phloem-pole pericycle marker, S17, in shr-2 or shr-2 containing the 35S::CKX1 transgene. The arrows mark the position where GFP was first detected. The top panels show the maturation zone. B, Expression of the phloem marker, S32, in shr-2 or shr-2 containing the 35S::CKX1 transgene. The lines mark the position where the cross-sectioning was made. Bars = 20 μm.
DISCUSSION
SHR is a key regulator of root growth and development. Initially identified as a factor essential for root apical meristem maintenance and ground tissue patterning (Helariutta et al., 2000), SHR was subsequently found to also play an important role in vascular tissue differentiation and lateral root formation (Gardiner et al., 2011; Yu et al., 2010). Although studies in past decades have shed significant light on the mechanism by which SHR regulates ground tissue patterning (Nakajima et al., 2001; Gallagher et al., 2004; Levesque et al., 2006; Cui et al., 2007; Gallagher and Benfey, 2009; Sozzani et al., 2010), how SHR controls other aspects of root morphogenesis is still unclear.
In the study reported here, we identified direct SHR targets at the genome scale using a ChIP-chip method. This systems approach not only revealed additional SHR targets that are preferentially expressed in the QC or CEI and thus potentially play a role in stem cell renewal and ground tissue patterning but also identified a direct role for SHR in regulating gene expression in the pericycle and xylem. Using cell type-specific markers, we found that, in shr root, phloem and phloem-associated pericycle were enlarged, whereas xylem and xylem-associated pericycle were reduced. Interestingly, this vascular patterning phenotype was observed when wild-type root was treated with cytokinin but not auxin, suggesting that an elevated level of cytokinin response is a cause of the vascular patterning defect in shr. In support of this hypothesis, we showed that SHR directly controls the cytokinin oxidase gene CKX3 in the stele and that shr had a substantially higher level of cytokinin. We further showed that reduction in cytokinin content in shr by CKX1 overexpression reversed its vascular patterning defect. Based on these results, we suggest that SHR controls root vascular patterning through the regulation of cytokinin homeostasis.
Our finding concerning the role of SHR and cytokinin in vascular patterning is consistent with other reports that cytokinin promotes phloem formation and represses metaxylem differentiation (Mähönen et al., 2000, 2006; Bishopp et al., 2011). In addition, we showed that cytokinin also determines the relative abundance of xylem- or phloem-associated pericycle: while promoting pericycle formation at the phloem pole, cytokinin inhibits pericycle cell identity at the xylem pole. Our study thus reveals a broader role for cytokinin in vascular patterning. Our demonstration that the two groups of pericycle cells also respond to auxin differently lends support to the proposition that the pericycle consists of two cell types. More importantly, it provides a mechanistic explanation for the lateral root defect in shr, as an elevated level of cytokinin would reduce the size of xylem-associated pericycle, the site of lateral root formation, and consequently compromise the ability of shr to produce lateral roots.
Besides cytokinin, auxin also plays a pivotal role in vascular tissue differentiation. Recent studies have suggested that auxin and cytokinin together control root vascular patterning through an interactive feedback loop (Bishopp et al., 2011). According to this model, cytokinin delivered to the root through phloem induces PIN7 expression in phloem initial cells and AHP6 expression in the initial cells for protoxylem and xylem-associated pericycle. Because PIN7 removes auxin from the phloem initial cells, it will effectively generate a zone with relatively high cytokinin that will promote phloem specification. In contrast, cytokinin signaling is blocked by AHP6, which thereby produces a zone without PIN7 expression, which will ultimately differentiate into protoxylem. Because of the funneling role of PIN7, auxin will not accumulate in the phloem, and its effect will be restricted to the AHP6 expression domain, even in the presence of exogenous auxin. This feedback model, therefore, also offers a plausible explanation for our observation that exogenous auxin had no obvious effect on vascular patterning and that no difference in DR5::GUS expression was noted between shr and wild-type roots despite a higher auxin level in shr (Lucas et al., 2011). Although the expression domain of J0121, a xylem-associated pericycle marker, was expanded in response to auxin, the enlargement was due to increased mitotic activity rather than its conversion into other cell types.
The current model does not, however, explain the observation that, in shr or in the presence of exogenous cytokinin, phloem and phloem-associated pericycle markers expanded their expression domain into the xylem and xylem-associated pericycle. According to this model, AHP6 as a cytokinin-inducible gene should be expressed regardless of the concentration of cytokinin, and as a consequence, a cytokinin-insensitive zone should be maintained. This is clearly not the case, because the phloem and phloem-associated cell markers S32 and S17 could expand their expression into the whole stele when the cytokinin level was sufficiently high. Our data showing that SHR directly regulates CKX3 expression could add an important piece to this model. Because CKX3 is preferentially expressed in the xylem (including the initial cells), it would generate a domain of low cytokinin and a PIN7 domain with a normal level of cytokinin. The resulting asymmetric pattern of cytokinin distribution is opposite to that of auxin, and the two contrasting gradients could act synergistically in vascular differentiation. The asymmetric pattern of cytokinin distribution would be disrupted when SHR is absent or cytokinin is applied exogenously, causing the loss of the xylem domain.
Despite our observation that exogenous auxin did not affect vascular patterning, we do not exclude the possibility that SHR controls some aspects of vascular patterning through auxin signaling. This is because among SHR targets are a number of genes that are apparently involved in auxin signaling, such as IAA16, ARF2 (Okushima et al., 2005), and MYB77 (Shin et al., 2007), which all appear to be expressed preferentially in the stele (Okushima et al., 2005; Shin et al., 2007). These auxin-signaling components could be involved in vascular tissue patterning or meristem renewal and warrant further investigation.
In summary, we have uncovered an important role for SHR in cell patterning in the Arabidopsis root vasculature. Several lines of evidence support the conclusion that SHR controls vascular patterning by regulating cytokinin homeostasis. By reducing the cytokinin concentration in the xylem initial cells, SHR facilitates an asymmetric pattern of cytokinin distribution, thereby promoting the formation of the two major cell types in the stele: xylem and phloem. By promoting the specification of the xylem-associated pericycle, SHR also plays an important role in lateral root formation. Unlike that in ground tissue patterning, the role of SHR in root vascular patterning appears to be independent of SCR.
MATERIALS AND METHODS
Plant Growth and Treatments
The Columbia ecotype of Arabidopsis (Arabidopsis thaliana) was used throughout this study. Seedling growth conditions and chemical treatments were as described previously (Cui and Benfey, 2009). The synthetic auxin NAA and the cytokinin BA were purchased from Caisson Laboratories (catalog nos. N001-25gm and B001-5gm, respectively).
ChIP-Chip Experiment and Data Analysis
To enrich DNA bound by SHR, we performed ChIP experiments with the transgenic line pSHR::SHR-GFP/shr-2, using an anti-GFP antibody, as described previously (Cui et al., 2007). As a control, an aliquot of the same chromatin preparation was subjected to the ChIP procedure but with the antibody replaced by bovine serum albumin. The efficiency of each ChIP was evaluated by real-time PCR assay to determine the degree of enrichment (between the ChIP and mock samples) for known SHR targets such as SCL3 and MGP, and only the best ChIP samples were used for subsequent ChIP-chip experiments. Both ChIP and mock DNA samples were amplified by a random-primed genome amplification method (Bozdech et al., 2003). The DNA was then modified by aminoallyl-dUTP with a mixture of aminoallyl-dUTP (Sigma) and dTTP (a ratio of 1:5) in the last round of PCR amplification, and the ChIP and mock samples were labeled with Cy3 or Cy5 dye (GE Life Sciences), respectively, or by Cy5 and Cy3 in a dye-swap experiment, by means of an amino-allyl dye-coupling method (Bozdech et al., 2003). After thorough mixing, both the ChIP and mock DNA samples were hybridized to the same microarray slide by a standard microarray hybridization protocol. Both the Agilent microarray and Nimblegen microarrays were scanned at 480 nm (for Cy3) and 635 nm (for Cy5) at a resolution of 5 μm pixel−1 with Agilent scanner G2565BA and Axon Genepix scanner 4000B, respectively. Posthybridization image processing was conducted with the Agilent Feature Extraction software for the Agilent microarray or custom software for the Nimblegen microarray.
Computational analysis of the ChIP-chip data was conducted as follows. First, for each type of microarray, the gene that a probe corresponded to was assigned mainly according to two criteria: (1) if it was located between two genes that have the same orientation (on either the Watson or Crick strand), it was assigned to the gene that lies downstream; (2) if it was located in the intergenic region between two genes whose sense strand runs outward, it was assigned to both genes. Second, for each slide, the fold enrichment for all probes was calculated as the ratio between the signal intensity for the ChIP and the corresponding mock samples, and probes with greater than 2-fold enrichment were identified. Third, putative SHR targets were identified as those that had at least one probe that met the 2-fold-enrichment threshold among all biological replicates (duplicates for the Agilent microarray and triplicates plus a dye swap for the Nimblegen array). Finally, putative SHR targets identified by both microarray experiments were ranked according to fold enrichment and the number of positive probes.
Molecular Cloning and Analysis of GFP Reporter Constructs
The pARF2::GFP line has been described previously (Schruff et al., 2006). Gateway technology (Invitrogen) was used for cloning the GFP transcriptional fusion constructs. For all the genes examined in our study, including ATAIB (At2g46510), MYB (At1g68670), and WOX7 (At5g05770), we took the 3-kb sequence upstream of the translational start site as the promoter, because SHR-binding sites were located within this region by our ChIP-PCR assay. The sequence was amplified with the high-fidelity Phusion DNA polymerase (New England Biolabs) and cloned into the pDONOR-P4-P1R vector. The primers used for cloning these promoter sequences are listed in Supplemental Table S3. The promoter clones were first confirmed by sequencing and then fused to GFP with an endoplasmic reticulum localization signal, which had been cloned into the pDONR221 vector by means of the Multi-Site Gateway system. After confirmation by restriction analysis, the final constructs were transformed into wild-type Arabidopsis (Columbia ecotype) by the Agrobacterium tumefaciens-mediated method. Transgenic lines showing GFP signal were selected by light microscopy.
To examine the expression of GFP and GUS marker lines, we crossed them with shr and analyzed seedlings from the F2 segregating population or F3 generation. For confocal microscopy, seedling roots were stained with propidium iodide (10 μg mL−1), dissolved in hydrogen peroxide, and images were taken with a Zeiss LSM510 confocal microscope.
Other Methods
ChIP-PCR and RT-PCR assays were performed as described previously (Cui et al., 2007), with a StepOnePlus real-time PCR system (Applied Biosystems) and with 18S DNA or RNA as a control for normalization. For those genes whose upstream sequence was less than 3 kb, the whole intergenic region was analyzed; for others, the 3-kb region upstream of the translational start site was analyzed. Primers used for these assays are listed in Supplemental Table S3.
For cytokinin measurement, shr-2 and wild-type seeds were sown on a nylon mesh placed on Murashige and Skoog medium, and root and shoot were collected 10 d after germination. For each sample, about 100 mg fresh tissue was lyophilized for cytokinin extraction. An HPLC-mass spectrometry-based method was used for cytokinin quantification, as described previously (Kojima et al., 2009).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Profile of SHR binding to known target genes as defined by ChIP-chip data.
Supplemental Figure S2. Confirmation of top-ranked SHR targets by ChIP-PCR.
Supplemental Figure S3. Selected top-ranked SHR targets have reduced expression in shr-2 roots.
Supplemental Figure S4. IAA16 is a direct SHR target that is repressed by SHR in the stele.
Supplemental Figure S5. Expression pattern of cell-type-specific markers in the absence (control) or presence of NAA.
Supplemental Figure S6. CKX3 is preferentially expressed in the xylem.
Supplemental Table S1. Complete list of SHR target genes identified with the Agilent microarry, the Nimblegen microarray, or both.
Supplemental Table S2. SHR target genes in different clusters shown in Figure 2.
Supplemental Table S3. Primers used in this study.
Acknowledgments
We thank Dr. George Bates (Florida State University) for critical reading and Dr. Anne B. Thistle (Florida State University) for careful editing of the manuscript. We also thank Dr. Joseph Kieber (University of North Carolina, Chapel Hill), Dr. Thomas Schmulling (Institute of Biology/Applied Genetics, Berlin), and Dr. Rod J. Scott (University of Bath, UK) for providing the pARR5::GUS transgenic line, the 35S::CKX1 transgenic line, and the pARF2::GFP line, respectively.
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Beemster GT, Baskin TI. (2000) Stunted plant 1 mediates effects of cytokinin, but not of auxin, on cell division and expansion in the root of Arabidopsis. Plant Physiol 124: 1718–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Scheres B. (2000) Root development. Curr Biol 10: R813–R815 [DOI] [PubMed] [Google Scholar]
- Bennett T, Scheres B. (2010) Root development: two meristems for the price of one? Curr Top Dev Biol 91: 67–102 [DOI] [PubMed] [Google Scholar]
- Berleth T, Mattsson J, Hardtke CS. (2000) Vascular continuity and auxin signals. Trends Plant Sci 5: 387–393 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Bishopp A, Help H, El-Showk S, Weijers D, Scheres B, Friml J, Benková E, Mähönen AP, Helariutta Y. (2011) A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr Biol 21: 917–926 [DOI] [PubMed] [Google Scholar]
- Bozdech Z, Zhu J, Joachimiak MP, Cohen FE, Pulliam B, DeRisi JL. (2003) Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol 4: R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, et al. (2010) Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Benfey PN. (2009) Interplay between SCARECROW, GA and LIKE HETEROCHROMATIN PROTEIN 1 in ground tissue patterning in the Arabidopsis root. Plant J 58: 1016–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. (2007) An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425 [DOI] [PubMed] [Google Scholar]
- D’Agostino IB, Deruère J, Kieber JJ. (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17: 678–682 [DOI] [PubMed] [Google Scholar]
- Dhondt S, Coppens F, De Winter F, Swarup K, Merks RM, Inzé D, Bennett MJ, Beemster GT. (2010) SHORT-ROOT and SCARECROW regulate leaf growth in Arabidopsis by stimulating S-phase progression of the cell cycle. Plant Physiol 154: 1183–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. (1993) Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Gallagher KL, Benfey PN. (2009) Both the conserved GRAS domain and nuclear localization are required for SHORT-ROOT movement. Plant J 57: 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher KL, Paquette AJ, Nakajima K, Benfey PN. (2004) Mechanisms regulating SHORT-ROOT intercellular movement. Curr Biol 14: 1847–1851 [DOI] [PubMed] [Google Scholar]
- Gardiner J, Donner TJ, Scarpella E. (2011) Simultaneous activation of SHR and ATHB8 expression defines switch to preprocambial cell state in Arabidopsis leaf development. Dev Dyn 240: 261–270 [DOI] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jürgens G. (1999) The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126: 1387–1395 [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Evans ML. (1995) Specialized zones of development in roots. Plant Physiol 109: 725–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, Maes L, Vanneste S, Swarup R, Weijers D, Calvo V, Parizot B, Herrera-Rodriguez MB, et al. (2007) Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19: 3889–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Parizot B, Baker A, Ricaud L, Martinière A, Auguy F, Franche C, Nussaume L, Bogusz D, Haseloff J. (2005) GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J Exp Bot 56: 2433–2442 [DOI] [PubMed] [Google Scholar]
- Kojima M, Kamada-Nobusada T, Komatsu H, Takei K, Kuroha T, Mizutani M, Ashikari M, Ueguchi-Tanaka M, Matsuoka M, Suzuki K, et al. (2009) Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol 50: 1201–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN. (2006) Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA 103: 6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU. (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mo X, Shou H, Wu P. (2006) Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiol 47: 1112–1123 [DOI] [PubMed] [Google Scholar]
- Lucas M, Swarup R, Paponov IA, Swarup K, Casimiro I, Lake D, Peret B, Zappala S, Mairhofer S, Whitworth M, et al. (2011) SHORT-ROOT regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol 155: 384–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Törmäkangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y. (2006) Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98 [DOI] [PubMed] [Google Scholar]
- Mähönen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y. (2000) A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev 14: 2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN. (2001) Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Mitina I, Quach HL, Theologis A. (2005) AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J 43: 29–46 [DOI] [PubMed] [Google Scholar]
- Parizot B, Laplaze L, Ricaud L, Boucheron-Dubuisson E, Bayle V, Bonke M, De Smet I, Poethig SR, Helariutta Y, Haseloff J, et al. (2008) Diarch symmetry of the vascular bundle in Arabidopsis root encompasses the pericycle and is reflected in distich lateral root initiation. Plant Physiol 146: 140–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ. (2006) The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133: 251–261 [DOI] [PubMed] [Google Scholar]
- Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP. (2007) The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19: 2440–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JA, Benfey PN. (2010) Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CL, Schmidt D, Bayer EM, Dreos R, Maule AJ. (2009) Arabidopsis plant homeodomain finger proteins operate downstream of auxin accumulation in specifying the vasculature and primary root meristem. Plant J 59: 426–436 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T. (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu NI, Lee SA, Lee MH, Heo JO, Chang KS, Lim J. (2010) Characterization of SHORT-ROOT function in the Arabidopsis root vascular system. Mol Cells 30: 113–119 [DOI] [PubMed] [Google Scholar]