Abstract
Fruit ripening is a complex developmental process responsible for the transformation of the seed-containing organ into a tissue attractive to seed dispersers and agricultural consumers. The coordinated regulation of the different biochemical pathways necessary to achieve this change receives considerable research attention. The MADS-box transcription factor RIPENING INHIBITOR (RIN) is an essential regulator of tomato (Solanum lycopersicum) fruit ripening but the exact mechanism by which it influences the expression of ripening-related genes remains unclear. Using a chromatin immunoprecipitation approach, we provide evidence that RIN interacts with the promoters of genes involved in the major pathways associated with observed and well-studied ripening phenotypes and phenomena, including the transcriptional control network involved in overall ripening regulation, ethylene biosynthesis, ethylene perception, downstream ethylene response, cell wall metabolism, and carotenoid biosynthesis. Furthermore, in the cases of ethylene and carotenoid biosynthesis, RIN interacts with the promoters of genes encoding rate-limiting activities. We also show that RIN recruitment to target loci is dependent on a normally functioning allele at the ripening-specific transcription factor COLORLESS NONRIPENING gene locus, further clarifying the relationship between these two ripening regulators.
Ripening is a complex developmental process responsible for the transformation of the seed-bearing structure of fleshy fruit species into a palatable and nutritious tissue attractive to seed-dispersing organisms and agricultural consumers (Seymour, 1993). Although the exact physiological and chemical changes associated with fruit ripening differ among species, some general changes are characteristic of many. These include modification of tissue firmness and cell wall structure, changes in sugar/starch metabolism, alteration of composition and levels of secondary metabolites such as pigments, and increased susceptibility to pathogens (Seymour, 1993). These changes are the result of the coordinated activation of multiple genetic and biochemical pathways, the regulation of which has been a subject of research for more than 30 years (Seymour, 1993; Giovannoni, 2007). Critical transcription factors regulating these processes were identified only recently.
In fruits of climacteric species, including the fleshy fruit model species tomato (Solanum lycopersicum), a large amount of ethylene is produced at the onset of ripening. The presence of this gaseous hormone is essential for the initiation and completion of ripening in tomato and other climacteric species (Saltveit and Dilley, 1978; Hobson et al., 1984; Seymour, 1993; Dupille and Sisler, 1995). Ethylene biosynthesis, perception, and signaling pathways have been elucidated as a result of studies in both Arabidopsis (Arabidopsis thaliana) and tomato, and ethylene’s effect on gene expression during climacteric ripening are now well characterized (Wilkinson et al., 1997; Bleecker and Kende, 2000; Stepanova and Ecker, 2000; Klee, 2004). More recently, the cloning of genes underlying certain tomato nonripening mutants, such as ripening inhibitor (rin; Vrebalov et al., 2002), Colorless nonripening (Cnr; Manning et al., 2006), and nonripening (nor; Giovannoni, 2004), have provided new insights into mechanism of ripening competency acquisition. Ripening competency can be defined as the sum of developmentally controlled events occurring upstream of the ethylene-regulated ripening cascade that are required for ripening to proceed. The fruits of ripening competency acquisition mutants are unable to ripen and are repressed in phenotypes associated with climacteric ripening, remaining green and firm, and failing to produce the typical burst in ethylene. Lack of ethylene production is however not the only cause of their lack of ripening since exogenous application of ethylene fails to rescue their nonripening phenotypes (Giovannoni, 2007). The observation that ethylene-dependent transcription does occur after exogenous ethylene application however indicates that ethylene perception and signaling are at least partly functional in these mutants (Lincoln and Fischer, 1988). Together, these observations indicate that these mutations affect regulators of both ethylene- and nonethylene-mediated ripening pathways. The RIN, CNR, and NOR genes have been shown to encode transcriptional regulators and thus likely act to regulate the expression of other genes responsible for ripening phenotypes, including ethylene production (Vrebalov et al., 2002; Giovannoni, 2004; Manning et al., 2006). Other ripening transcriptional regulators have recently been identified via transcriptional profiling studies (Alba et al., 2005) and interaction with ethylene synthesis promoters (LeHB1; Lin et al., 2008). Functional studies demonstrated the critical roles of the TAGL1 transcription factor in both early fleshy fruit expansion and later ripening (Itkin et al., 2009; Vrebalov et al., 2009; Pan et al., 2010) and the role of an APETALA2 homolog (SlAP2a; Chung et al., 2010) in negative regulation of ethylene synthesis and ripening.
We and others have focused on the rin mutation and the RIN gene in part due to the wide use of Rin/rin hybrids for extending shelf life in commercial tomato production and especially due to its apparent conservation and ripening role in both climacteric and nonclimacteric species (Vrebalov et al., 2002). RIN is a member of the MADS-box family of transcription regulators, known to play essential roles in a variety of plant developmental processes including control of vegetative growth, flowering time control, and floral development (Ng and Yanofsky, 2001). The dramatic phenotypic effect of the rin mutation on virtually all ripening pathways supports its role as a master regulator of the ripening cascade. However, the exact mechanism by which RIN regulates the expression of genes involved in the different aspects of fruit ripening has only begun to be addressed. Ito et al. (2008) have shown that RIN can bind to CArG box primers in vitro and Fujisawa et al. (2011) confirmed RIN’s binding to ethylene synthesis and cell wall metabolism genes, the promoters of which contain CArG box sequences.
To gain a better understanding of the regulatory network underlying ripening competency acquisition, we have employed chromatin immunoprecipitation (ChIP) to validate numerous potential primary targets of RIN in a developmental time course through ripening, as well as in the context of both the rin and Cnr mutations. Here we show that RIN interacts with promoters of many genes belonging to all major ripening pathways including ethylene synthesis (Alexander and Grierson, 2002; Barry, 2007), ethylene perception (Klee and Tieman, 2002; Klee, 2004), cell wall metabolism (Marín-Rodríguez et al., 2002), carotenoid accumulation (Bramley, 2002), and regulation of additional ripening-related transcription factors (Giovannoni, 2007). We also demonstrate that RIN activity is dependent upon CNR and while it does not interact with all ethylene and carotenoid synthesis promoters it does interact with those coding for rate-limiting enzymes in both pathways. In short, we provide evidence that RIN is a master regulator of ripening that directly influences many ripening-associated processes in a developmental-specific pattern and via a mechanism that is dependent upon the presence of a functional CNR gene.
RESULTS
Production of RIN-Specific Antibodies and Validation of the rin Mutation Predicted Chimeric Protein
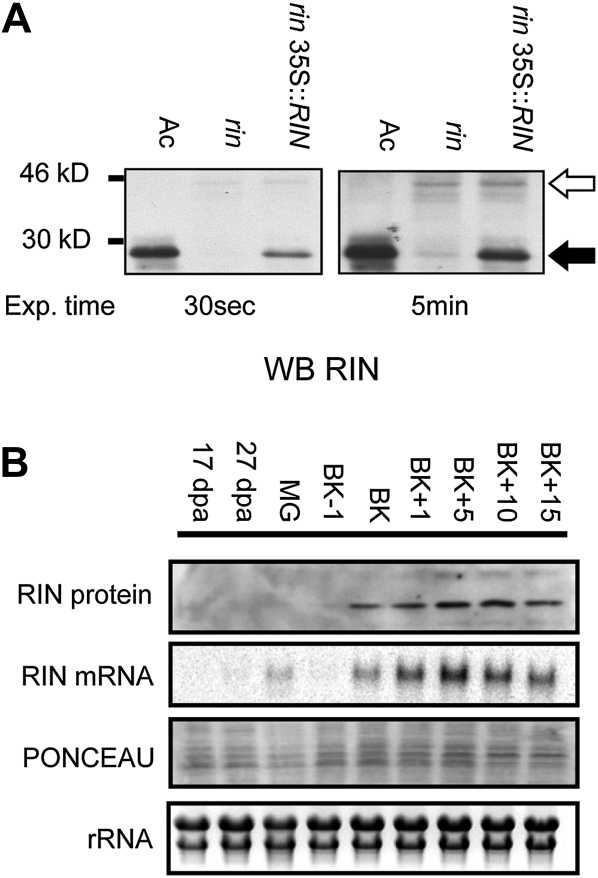
To study the endogenous function and regulation of the RIN gene, we developed polyclonal antibodies that can specifically detect the RIN protein. RIN shares a highly conserved N-terminal DNA-binding domain with other members of the MADS-box family, whereas the C-terminal portion, whose functions include protein-protein interactions and transcriptional activation, is more variable (Kaufmann et al., 2005). To obtain antibodies that would specifically recognize RIN and not other of the over 100 members of the tomato MADS family (www.solgenomics.net), we raised the antibodies against the C-terminal portion of RIN. A His-tagged recombinant protein encoding the RIN C-terminal portion was produced in Escherichia coli and purified on a His-binding column. The purified protein was used to raise polyclonal antibodies from rabbit. To test the specificity of the antibodies, protein extracts from tomato fruits of cultivar Ailsa Craig (Ac) and lines nearly isogenic for rin and rin plus a 35S::RIN transgene (Vrebalov et al., 2002) were isolated and used in a western-blot analysis. As shown in Figure 1A, RIN antibodies recognize a 28-kD band in the wild-type (Ac) fruit extract but not in the rin mutant extract. This size corresponds to the expected Mr of the RIN protein. The fact that this band is not detected in rin mutant protein extract but is present in the isogenic rin 35S::RIN line confirms that it corresponds to the RIN protein. A faint higher Mr band (45 kD) is detected in the rin mutant protein extract (Fig. 1A, 5-min exposure). As described previously (Vrebalov et al., 2002), the rin mutation is caused by a genomic deletion removing the 3′-untranslated region encoding exon of the RIN gene thus creating a translational fusion with the adjacent MACROCALYX (MC) MADS-box gene. Because the chimera still possess the majority of the RIN C-terminal region used to raise the RIN antibodies, said antibodies should detect the chimeric RIN-MC protein of the rin mutant. The 45-kD band observed in the rin pericarp protein extract (Fig. 1A, 5-min exposure) corresponds to the predicted size of the chimeric RIN-MC protein (Vrebalov et al., 2002).
Figure 1.
RIN gene expression and protein accumulation in tomato fruit. A, Western blot of BK + 2 d fruit from wild type (Ac), rin/rin mutant, and rin/rin 35S::RIN nearly isogenic genotypes using the RIN antibodies. Black arrow: RIN protein (28 kD), white arrow: RIN-MC chimera protein (45 kD) predicted by Vrebalov et al. (2002). B, Time course of RIN protein and mRNA accumulation during fruit development and ripening, as revealed by western blot (top) and northern blot (second from top), respectively. MG, Mature green stage; BK + X, BK + X day. Ponceau S staining and rRNA are used as loading controls for protein and mRNA gel-blot analyses, respectively.
RIN Expression and Protein Accumulation Are Tightly Correlated through Fruit Development
We next examined the behavior of RIN protein and mRNA accumulation during pericarp development and ripening using western- and northern-blot analyses. Figure 1B shows RIN protein and RNA accumulation at different stages of pericarp development and ripening. RIN protein and mRNA are absent during the initial preripening phase of pericarp development (0–35 DPA), but accumulate early during ripening. RIN protein is detected slightly before the breaker (BK) stage and its expression is maintained throughout ripening (up to 15 d after BK). Comparison of protein and RNA expression profiles indicates a tight correlation between the two. The simplest explanation for this observation is that the RIN protein is short lived in vivo and thus accumulation of the RIN protein and its corresponding effects are largely transcriptionally regulated, though we recognize that more intricate explanations could also account for this result.
Identification of Promoters That Associate with RIN
We suspect that RIN may act as master regulator of ripening by influencing the expression of numerous genes. Although its role in influencing ripening-associated traits, such as cell wall degradation, carotenoid accumulation, and ethylene production, is clearly supported by the strong phenotype of the rin mutant (Vrebalov et al., 2002), the mechanism and specific target genes by which it exerts its effect have only begun to be addressed. To better understand the mechanism by which RIN influences ripening, a ChIP strategy was employed (Wang et al., 2002). The above-described anti-RIN antibodies were used to isolate RIN-bound chromatin fragments from BK-stage tomato fruits. Quantitative PCR (qPCR) was used to measure an enrichment ratio at different loci. Association of RIN to a particular locus should result in an increase in the enrichment level following RIN immunoprecipitation compared to the level prior to immunoprecipitation (input) or following a nonspecific immunoprecipitation (using preimmune serum). The specificity of the immunoprecipitation was further assessed by performing the ChIP on rin fruits that we expect to have a nonfunctional RIN protein due to the rin mutation. To explore the depth of RIN activity, the ability of RIN to associate with important classes of ripening genes was examined. Candidate genes were selected among: (1) ripening-related transcription factors, (2) ethylene synthesis enzymes, (3) ethylene signal transduction components, (4) ethylene-responsive genes, (5) cell wall metabolism enzymes, and (6) carotenoid biosynthesis pathway enzymes.
Transcription Factors
In addition to the RIN gene itself that we assayed for in our ChIP analysis, other transcription factors have been shown to play important roles in fruit ripening. NOR is a NAC-domain transcription factor whose mutation leads to a nonripening phenotype similar to that observed in rin (Giovannoni, 2007). HB-1 is an HD-zip transcription factor that positively controls the expression of the ethylene-producing enzyme ACO1 during fruit development and ripening (Lin et al., 2008). CNR is a SBP-box transcription factor necessary for ripening (Manning et al., 2006). A mutation in this gene results in pleiotropic nonripening phenotypes, including a mealy and pale pericarp (Fraser et al., 2001; Orfila et al., 2002; Eriksson et al., 2004). TDR4 is another member of the MADS-box transcription family whose expression pattern suggests a possible role during tomato fruit ripening (Busi et al., 2003). A TDR4 ortholog from bilberry (Vaccinium myrtillus) was recently shown to influence ripening (Jaakola et al., 2010).
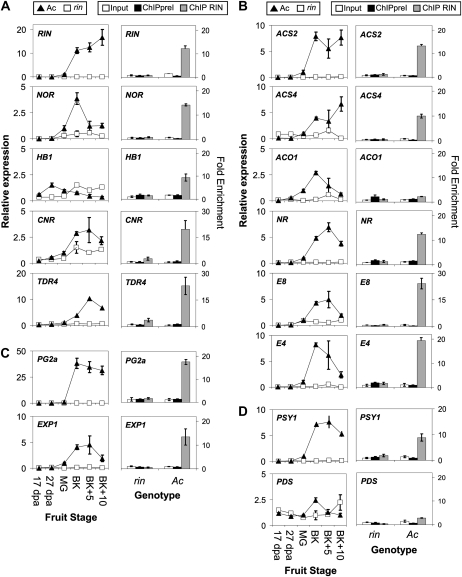
To examine the expression level of these candidate genes during the different stages of fruit development and ripening, quantitative reverse transcription (qRT)-PCR was first performed. Figure 2A shows the relative expression level of each gene in wild type and identically aged rin fruits. As expected, RIN, NOR, TDR4, and CNR transcript levels increased at the onset of ripening in wild-type fruit; these increases are not observed in the rin fruit, consistent with the hypothesis that RIN influences their expression as has been shown for at least RIN (Vrebalov et al., 2002) and CNR (Manning et al., 2006). The expression of HB1 remained constant during the early stage of fruit development and decreased during ripening, consistent with previously published data (Lin et al., 2008).
Figure 2.
Regulation of gene transcription by RIN. Left section, mRNA levels of genes in wild type (Ac) and rin mutant during tomato fruit development measured by qRT-PCR. MG, Mature green stage. Right section, ChIP of ripening-related promoters using RIN antibodies. BK-staged rin and Ac fruits were used for the assay. Input: enrichment before immunoprecipitation; ChIPpreI: enrichment following immunoprecipitation using preImmune serum; ChIP RIN: enrichment following immunoprecipitation using serum containing RIN antibodies. Gene categories: transcription factor (A), ethylene synthesis, perception and response genes (B), cell wall modifying genes (C), carotenoid biosynthesis genes (D).
Using the ChIP-qPCR procedure described above, we measured the ability of RIN protein to interact with the promoter region of each of these genes (Fig. 2A). Each promoter tested showed a clear enrichment following RIN immunoprecipitation in wild type but not rin fruit, indicating that RIN associates in vivo with NOR, CNR, TDR4, HB1, and its own promoter. This result suggests that RIN exerts it role in ripening in part via regulation of additional transcription factors known to be necessary for the ripening process.
Ethylene Components
Since RIN is expressed prior to the onset of climacteric ethylene synthesis and rin mutant fruit have a reduced ethylene production, RIN might be directly involved in regulating one or more components of the pathway. The increase in fruit ethylene production is largely driven by the biosynthetic genes ACS2, ACS4, and ACO1 (Barry, 2007). The ethylene receptor NEVER-RIPE (NR/LeETR3) has been shown to play a major role during fruit ripening since a dominant mutation leads to insensitivity to ethylene and inhibition of ripening (Klee, 2004). The E4 and E8 genes have long been known to be rapidly induced following ethylene induction and during normal fruit ripening (Lincoln et al., 1987). Figure 2B (left) shows that the rin mutation affects the normal mRNA accumulation of each of these genes. ChIP analyses (Fig. 2B) show that RIN associates with the ACS2 and ACS4 promoters but not with the ACO1 promoter. The RIN ChIP results for the promoter of ACS2, ACS4, and ACO1 are similar to those reported by others (Fujisawa et al., 2011). The promoter of the ethylene receptor NR showed significant enrichment following RIN immunoprecipitation as did the E8 and E4 promoters. Together these results indicate that not only are ethylene synthesis genes targets of RIN but so are ethylene signal transduction genes and downstream targets of the ethylene signaling pathway. We also note that while ACO1 is not directly targeted by RIN, the promoter of the previously described ACO1 regulator, HB1 (Lin et al., 2008), is a direct target of RIN (Fig. 2A).
Cell Wall Metabolism
Promoters of two well-characterized genes involved in ripening-related cell wall modification were tested for direct in vivo binding by RIN: Polygalacturonase2a (PG2a) and Expansin1 (Exp1). PG2a is an enzyme involved in pectin depolymerization and is highly up-regulated in close association with fruit ripening in tomato (Dellapenna et al., 1989). The promoter of the PG2a gene has been intensively studied and shown to possess both ethylene-dependent and independent cis-elements (Montgomery et al., 1993; Nicholass et al., 1995). Exp1 plays an important role in fruit softening during ripening although its exact mechanism of action is not known (Rose et al., 2000). qRT-PCR analyses confirm that both genes are up-regulated during wild-type fruit ripening, but fail to be in the rin mutant fruits (Fig. 2C). ChIP analyses (Fig. 2C) indicate that the RIN protein associates with the promoters of both PG2a and Exp1.
Carotenoid Metabolism
The PHYTOENE SYNTHASE1 (PSY1) and PHYTOENE DESATURASE (PDS) genes are involved in the production of carotenoids during fruit ripening (Cunningham and Gantt, 1998). PSY1 is the fruit-specific isoenzyme responsible for the initiation of the carotenoid biosynthetic cascade, combining two molecules of geranyl-geranyl diphosphate to form one molecule of phytoene (Bartley et al., 1992; Ray et al., 1992). PDS catalyzes the second step of the pathway by converting phytoene into ζ-carotene, a precursor of lycopene (Pecker et al., 1992). The PSY1 gene shows an increase in expression at the onset of ripening in wild type but not in rin mutant fruits (Fig. 2D). The expression of PDS remains fairly constant throughout fruit development although a slight but reproducible increase occurs at the BK stage. This suggests that PDS is not a major limiting enzyme in carotenoid biosynthesis during fruit ripening. Interestingly, only the promoter of the PSY1 gene was enriched following ChIP (Fig. 2D). In short, RIN interacts with the gene encoding the regulated rate-limiting step in fruit carotenoid biosynthesis, PSY1.
DNA-Binding Dynamics of RIN during Fruit Development and Ripening
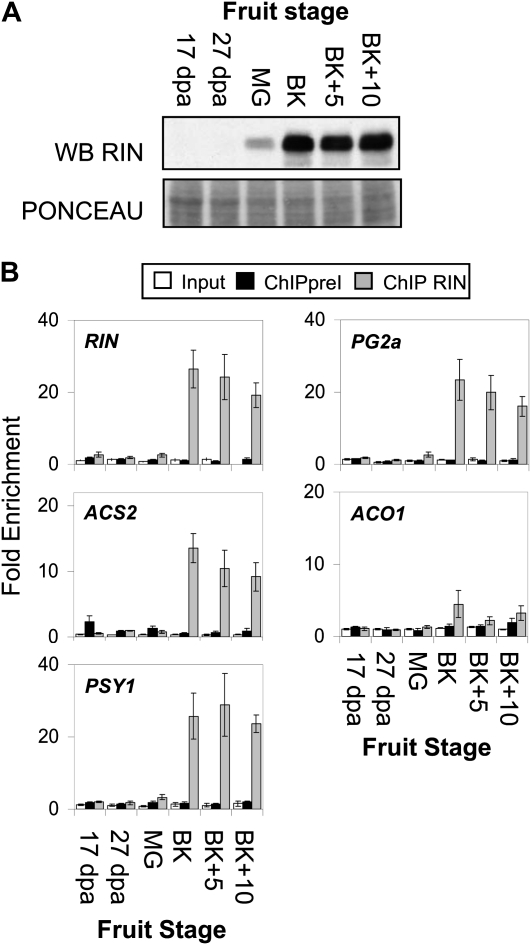
In light of previous results, two models can be proposed regarding the temporal action of RIN in controlling gene expression during fruit ripening. RIN could bind to and influence the expression of its target genes in early ripening (around BK stage and a few days before or after) but this binding might not be required to complete the ripening process. Alternatively, RIN might be required in a continuous fashion to both initiate and maintain the expression of ripening genes throughout the ripening process. Fujisawa et al. (2011) report active RIN binding in pink early ripening fruit to a number of ethylene synthesis and cell wall associated genes that could be consistent with either hypothesis. They also observed RIN binding to its own promoter at the ripe fruit stage, an observation that is consistent with the latter hypothesis. To better distinguish between these two hypotheses and to further validate the accuracy of our ChIP analyses, we performed the ChIP assay on a developmental series of both ripening wild type and identically aged rin fruits. Specifically, ChIP enrichment level of RIN, ACS2, PG2a, and PSY1 promoters was assayed in 17 DPA, 27 DPA, mature green, BK, 5-d post BK (BK + 5), and 10-d post BK (BK + 10) fruits. The nonbound target ACO1 promoter was also examined as a negative control. Western-blot analysis (Fig. 3A) shows the amount of RIN protein present in each of the stages used for the ChIP assay. As anticipated, the amount of RIN protein increases at the onset of ripening (mature green to BK) and remains high in the following stages. Figure 3B shows that a strong enrichment of all RIN targets is detected following RIN-ChIP on the BK, BK + 5 (red), and BK + 10 (red) stages when RIN protein is most prevalent. This result indicates that RIN binding occurs throughout ripening and better supports the hypothesis in which continuous RIN binding is required for RIN-mediated gene expression. This mechanism of action is also consistent with the observed RNA and protein expression profile of RIN (Fig. 1B). The tight correlation between RIN protein accumulation (Fig. 3A) and RIN ChIP promoter enrichment (Fig. 3B) also serves to validate the accuracy and specificity of the RIN ChIP analysis.
Figure 3.
Time course of RIN-promoter interactions. A, Western-blot analysis using RIN antibodies of wild-type (Ac) fruit extract used for ChIP at different stages of development. B, ChIP of promoters at different stages of fruit development. Input: enrichment before immunoprecipitation; ChIPpreI: enrichment following immunoprecipitation using preImmune serum; ChIP RIN: enrichment following immunoprecipitation using serum containing RIN antibodies.
CNR Is Required for RIN-Binding Activity
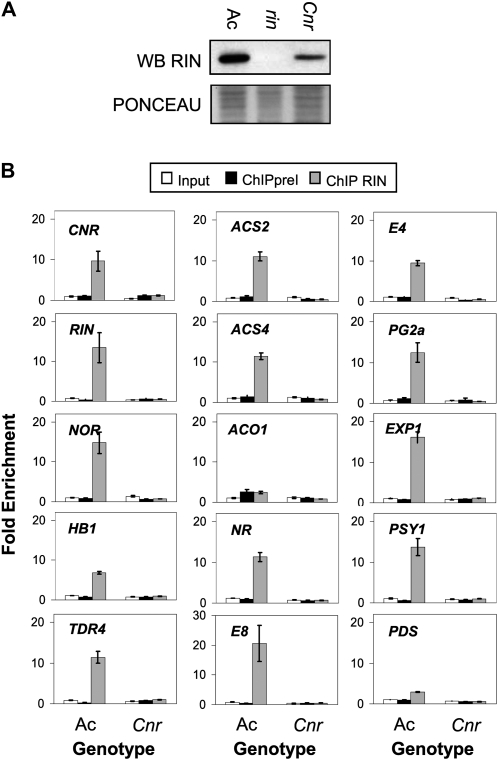
Like the rin mutation, the Cnr mutation has a broad inhibitory impact on ethylene- and nonethylene-mediated ripening activities (Fraser et al., 2001; Eriksson et al., 2004). The Cnr mutation is caused by changes in the methylation status of the CNR promoter (Manning et al., 2006). We showed that RIN protein associates with the CNR promoter and examination of the CNR promoter region reveals the presence of a CArG cis-element located 2-bp upstream of a cytosine whose methylation status is changed in the Cnr mutant. We were interested to see if the change in methylation level of the CNR promoter in the Cnr mutant impacts this binding. The ChIP assay was performed on Cnr fruits at the BK + 2 stage and enrichment levels of the CNR and other promoters were measured. Interestingly, no enrichment of the CNR promoter was observed in Cnr fruit following RIN ChIP (Fig. 4B) even though RIN protein is present (Fig. 4A). Other target loci should still be bound by RIN since the Cnr mutation affects only its own methylation status. Nevertheless, we found that RIN association with all tested target genes was lost in the Cnr mutant (Fig. 4B). This result suggests that RIN interaction with its target loci depends on CNR or a CNR-regulated gene product. Importantly, the lack of ChIP enrichment was not due to a lack of RIN protein accumulation in the Cnr mutant as western-blot analysis of Cnr mutant tissue clearly demonstrated the presence of RIN protein in the samples used for ChIP (Fig. 4A). The level of RIN protein accumulation in Cnr, although lower than in wild-type fruit, is likely sufficient to confer ripening activity in that a comparable level of RIN protein accumulation is observed in the 35S::RIN complementation lines that are able to ripen (Fig. 1A; Vrebalov et al., 2002). Eriksson et al. (2004) reported that PG2a, Exp, ACO1, E8, E4, and PSY1 expression are all down-regulated in the Cnr mutant consistent with the lack of RIN binding observed in Figure 4B.
Figure 4.
CNR requirement for RIN promoter-binding activity. A, Western-blot analysis using the RIN antibodies of proteins extracted from Ac, rin, and Cnr Bk + 2 fruits. B, Enrichment of ripening-related promoters following RIN ChIP in Ac and Cnr fruit at the BK + 2 stage. Input: enrichment before immunoprecipitation; ChIP preI: enrichment following immunoprecipitation using preImmune serum; ChIP RIN: enrichment following immunoprecipitation using serum containing RIN antibodies.
Two-Hybrid Screen Analysis Fails to Recover CNR But Does Capture Other Known Fruit Development and Ripening Regulators
To identify proteins that interact with RIN and thus indirectly ask whether the RIN and CNR proteins could directly interact with each other we performed a two-hybrid screen using reagents and procedures from Hybrigenics Inc. In summary, when the RIN protein was deployed as bait against a wild-type tomato fruit prey library, the primary proteins that interacted were almost all MADS-box proteins and included MPB7, TAGL1, TAG1, and TDR4 (Table I). These interacting proteins are known to be associated with fruit development or ripening (Pnueli et al., 1994; Lozano et al., 2009; Vrebalov et al., 2009). Importantly, CNR was not recovered as a significant RIN interacting protein (Table I), suggesting that CNR’s influence on RIN-binding activity may not be mediated by a direct interaction between the two proteins. We did not perform a two-hybrid assay with RIN and CNR as prey and bait to directly test this question.
Table I. Genes interacting with RIN (bait) in a two-hybrid screen against a red ripe tomato prey library.
PBS is computed score that represents the probability of an interaction to be nonspecific. A, Very high confidence in the interaction; B, high confidence in the interaction; D, moderate confidence in the interaction.
| Gene Name | Gene Family | PBS Score | No. of Interactions |
| A | 184 | ||
| B | 4 | ||
| LeMBP7 | MADS box | ||
| D | 3 | ||
| N/A | 2 | ||
| A | 117 | ||
| TDR4 | MADS box | B | 5 |
| N/A | 1 | ||
| TAGL1 | MADS box | A | 29 |
| A | 11 | ||
| TAG1 | MADS box | ||
| N/A | 1 | ||
| LeMBP21 | MADS box | D | 1 |
| TDR5 | MADS box | D | 2 |
| LAT61 | DUF | D | 1 |
DISCUSSION
The complex phenotype associated with the rin mutation suggests that RIN is acting as a master regulator of the ripening cascade by influencing numerous molecular pathways. Using a ChIP approach, we investigated the association of RIN with a subset of ripening-related loci and showed that RIN targets span the spectrum of previously described ripening pathways. In this regard RIN is a comprehensive ripening regulator explaining both the severe ripening inhibition of the mutation and its utility in coordinately slowing virtually all ripening processes in hybrid Rin/rin fruit predominant in current fresh market tomato production.
We used polyclonal antibodies directed against the C-terminal region of RIN to immunoprecipitate chromatin regions bound by the RIN protein in vivo. While the use of a polyclonal mixture could potentially result in nonspecific immunoprecipitation, we believe a series of controls support that our analysis reflects bona fide RIN binding. First, the RIN antibodies did not immunoprecipitate chromatin in the absence of a functional RIN protein as demonstrated by the lack of enrichment in rin mutant tissues (Figs. 2 and 3). Second, our western-blot analyses (Fig. 1A) indicate that the RIN antibodies are highly specific for the RIN protein as we detect a strong band of the predicted size in wild-type fruit, a lack of said band but the presence of a band corresponding to the larger predicted mutant protein in rin fruit, and a band of the anticipated size in transgenic rin fruit isogenic for a T-DNA sequence harboring a 35S:RIN construct (Fig. 1A). Furthermore, the ChIP analysis over a developmental time course shows that the RIN polyclonal antibodies mediate promoter enrichment in a manner in complete consistency with RIN gene expression (Fig. 1B) and protein accumulation (Fig. 3). Finally, our ChIP assay successfully identified several RIN target genes that have been independently reported to be bound by RIN (Ito et al., 2004; Fujisawa et al., 2011).
RIN Regulation of Other Transcription Factors
Recent efforts have identified a number of transcription factors from multiple gene families (MADS-box, NAC, HB-zip, APETALA2, SBP) that are necessary for broad fruit ripening effects. We show here that RIN interacts with the promoters of several of these transcription factors influencing the major ripening processes and phenotypes of tomato, demonstrating the central role played by RIN in this regulatory process. The observed binding of RIN to the NOR promoter is interesting given the similar and strong ripening inhibition conferred by both mutations, suggesting interaction between the RIN and NOR genes. Regulation of a NAC domain transcription factor by a MADS-box protein was previously reported by Sablowski and Meyerowitz (1998) who showed that the floral identity dimer AP3/PI directly regulates the NAC domain protein, NAP. Our result suggests that the binding of RIN to the NOR could be required to promote NOR expression. Our result also suggests that a positive feedback loop is involved in RIN regulation, since an enrichment of the RIN promoter was observed. Several other examples of MADS regulatory feedback loops have been described (Schwarz-Sommer et al., 1992; Tröbner et al., 1992; Hill et al., 1998). Arabidopsis AGL15 was shown to have a direct inhibitory effect on its own expression in embryonic tissue (Heck et al., 1995). Tilly et al. (1998) showed that the expression of AP3, a B-class floral identity MADS-box gene, is directly influenced by the binding of the AP3-PI dimer to one or more CArG boxes present in its promoter. PI gene expression is similarly controlled by the AP3-PI dimer but does not possess a CArG box motif in its promoter (Chen et al., 2000). This latter observation could indicate that the AP3-PI dimer is recruited to the promoter of PI through another DNA-binding protein, or could result from indirect regulation. Our results suggests that RIN may also operate in such a regulatory loop and further interaction with ethylene synthesis gene and regulatory promoters (see below) may explain the autocatalytic production of ethylene occurring during climacteric ripening (Barry et al., 2000). Another interesting observation is the binding of RIN to the CNR promoter. The SBP family is known to be involved in the regulation of the SQUAMOSA family of MADS-box proteins (Klein et al., 1996); however, little is known regarding their own regulation at the transcriptional level. Our results suggest that RIN, a MADS-box protein, may be involved in CNR regulation.
RIN and Ethylene Regulation
A hallmark of climacteric fruit ripening is the production of elevated ethylene at the onset of ripening. rin fruits are unable to produce climacteric ethylene, hence our interest in testing the binding of RIN to ethylene-producing genes. Interestingly, both the ACS2 and ACS4 gene promoters show enrichment following RIN ChIP, indicating that RIN associates with their respective promoters. ACS2 and ACS4 have previously been reported to be bound by RIN by Ito et al. (2008) and Fujisawa et al. (2011), respectively. The ACO1 promoter was not significantly bound by RIN in our assay. A regulatory link might however be postulated between RIN and ACO1 expression since RIN binds to the promoter of HB1. HB1 has been shown to interact with the promoter of ACO1 and be necessary for its expression during fruit ripening (Lin et al., 2008). Our results suggest that RIN binding to the promoter of HB1 could indirectly influence ACO1 expression.
Other loci of components of the ethylene-signaling cascade also associate with RIN, including the ethylene receptor NR. Ito et al. (2008) and Fujisawa et al. (2011) also tested interaction between RIN and the NR promoter but failed to detect a significant enrichment following ChIP. This discrepancy might be due to a difference in the promoter region tested. The ChIP primers used by Ito et al. (2008) and Fujisawa et al. (2011) fall inside a large intron (2.35 kb) located between the 5′-untranslated region coding exon and the ATG coding exon of the NR gene. We selected our ChIP primers in the 3-kb region located upstream of the first exon of the gene. Together these results suggest that RIN association with the NR gene is mediated by its interaction with the genomic region located upstream of the first exon of the gene and not by interaction with putative CArG boxes in the intron located upstream of the ATG. Interestingly, the promoter of the NR gene possesses two putative CArG box elements. Further studies are needed to determine if these motifs are involved in RIN recruitment to the promoter.
CNR-Dependent RIN-Binding Activity
We showed that RIN-binding activity was greatly diminished in the Cnr mutant background (Fig. 4). This observation suggests that RIN may not engage in direct DNA binding with its target promoter but might be recruited to them through the DNA-binding ability of other members of a complex. No direct interaction between RIN and CNR proteins could be detected by two-hybrid assay, however we cannot exclude the possibility that CNR and RIN are part of the same protein complex in vivo and that the absence of CNR from this complex prevents its recruitment to target loci. Alternatively, CNR could be required for the expression of a cofactor needed for proper DNA-binding activity of the RIN-containing complex. CNR is a member of the SBP family of transcription factors (Klein et al., 1996) known to regulate the transcription of certain MADS-box genes. One possible candidate for bridging CNR and RIN activity is the MADS-box gene TDR4. As reported here, TDR4 protein is able to interact directly with RIN in a two-hybrid assay and TDR4 gene expression is highly diminished in the Cnr mutant (Eriksson et al., 2004). Although a direct interaction between CNR and the TDR4 promoter has yet to be reported, it is interesting to note that TDR4 belongs to the SQUAMOSA class of MADS-box gene, a class of gene known to be regulated by SBP-box proteins, of which CNR is a member. Other potential partners of RIN were uncovered by a two-hybrid screen for all available interacting partners in maturing fruit (Table I), including the recently described MADS-box gene TAGL1 (Itkin et al., 2009; Vrebalov et al., 2009). The expression of this gene is independent from RIN and its suppression by RNAi produces a ripening phenotype similar to that of RIN-silenced lines (Vrebalov et al., 2002, 2009). Furthermore, Itkin et al. (2009) showed that TAGL1 interacts directly with the ACS2 promoter, a gene also bound by RIN (Ito et al., 2008).
RIN-Binding Motif
Previous studies have reported the ability of RIN to bind to CArG motif in vitro and that a number of promoters that are bound by RIN appear to be enriched in CArG sites (Ito et al., 2004; Fujisawa et al., 2011). We analyzed the promoter region (3-kb upstream of the transcription start site) of the genes used in this study for the presence of putative CArG boxes defined as the consensus sequence C(C/T)(A/T)6(A/G)G. All but two promoters possesses one or more CArG boxes (data not shown). The two promoters for which no CArG box could be detected are the CNR and the EXP1 promoters, both of which associate with RIN in our ChIP assay. Relaxing the parameter of the search to allow one mismatch in the CArG box consensus sequence allows the identification of two additional motifs in EXP1 but not CNR promoters. The promoters of ACO1 and PDS also possess CArG boxes sequences but fail to associate with RIN in vivo. Based on these results, it is not clear whether RIN association to its target promoter is solely mediated by CArG boxes or if other cis-elements are required for specificity of recruitment. A search for other common motifs using the MEME software (Multiple Em for Motif Elicitation, http://meme.nbcr.net/meme/intro.html) did not reveal any motif significantly overrepresented in the ChIPed promoters. More than one motif could be involved in the recruitment of the RIN complex, a mechanism that has previously been described for other transcription factors with a large number of target genes (Chakravarthy et al., 2003; Kaufmann et al., 2009). Although this work has focused on the ability of RIN to bind to the 5′ promoter region of ripening genes, it would be of interest in future studies to determine if RIN could also be recruited to other regions of the gene (coding, 3′ region) especially in the cases of genes such as PDS where RIN upstream binding has not been detected.
Significance of RIN Binding
Our ChIP results suggest that RIN binds to a wide variety and large number of ripening-associated genes spanning all major classes of ripening pathways. Recent work by others have provided data concerning the number of binding sites of transcription factors on a genomic scale using ChIP-sequencing technology, and have shown that transcription factors generally bind to a very large number of target genes. For example, the Arabidopsis SEP3 protein (a close homolog of the RIN protein) was shown to bind to about 4,000 sites throughout the genome (Kaufmann et al., 2009). Interestingly, only a fraction of the bound regions contained a typical CArG box consensus motif, while many other motifs associated with other known transcription factors were also enriched. Similarly, Zheng et al. (2009) showed that the MADS-box protein AGL15 was bound to 2,000 sites, only 64% of which had a clearly defined CArG cis-element. The HY5 protein involved in regulating light signal transduction was also shown to bind to more than 3,000 sites (Lee et al., 2007). Considering these studies, the ability of RIN to bind to numerous ripening-associated genes is not unexpected, though the meaning of such global binding remains unclear. An interesting question raised by such a high number of bound target genes is its relation with gene expression. It is becoming clear that binding of a transcription factor to a specific promoter does not immediately activate transcription (Wyrick and Young, 2002). Instead, it is thought that some transcription factors may bind to their targets yet will not affect transcription until other conditions are met (e.g. interaction with other transcription factors). In line with this model, the expression of only a small subset of AGL15-bound targets are influenced by the presence of AGL15 (Zheng et al., 2009). Similarly, the expression of only 6% of HY5-bound targets are affected in a hy5 mutant (Lee et al., 2007). Previous studies and qRT-PCR analyses presented here (Fig. 2) clearly demonstrate that RIN is required for the regulation of most of the genes that we have identified by ChIP as target of RIN. However, it is unclear whether RIN binding alone is sufficient to affect expression. Indeed a few genes do not show a clear correlation between RIN binding and changes in gene expression. The HB1 promoter is bound by RIN in our ChIP assay but its expression decreases during normal ripening and is elevated in post-BK rin mutant fruits. This observation may indicate that alternative regulators operate in the absence of RIN. We also note that ACO1 doesn’t seem to be bound by RIN even if its expression increases during ripening. These discrepancies highlight the fact that RIN binding does not necessarily result in a change in gene expression and that other cofactors might be required for efficient expression of RIN-bound targets (for example, RIN itself presumably responds to additional regulators beyond the product of its own transcription/translation as evidenced in Fig. 4 where RIN binding is absent in the Cnr mutant but RIN protein still accumulates to a reduced level). RIN is a member of the SEP3 clade of MADS-box proteins that are known to be involved in the bridging of other MADS-box proteins to form higher-order complexes involved in regulating flower development (ABCE model; Robles and Pelaz, 2005; Immink et al., 2009). RIN activity could similarly be influenced by the formation of transcriptional complexes through protein-protein interactions with other transcriptional regulators such as those recovered in the two-hybrid results presented here (Table I).
Our analysis expands the work of Ito et al. (2008) and Fujisawa et al. (2011) to demonstrate that RIN interacts not just with cell wall metabolism and ethylene synthesis promoters but also ethylene signaling, ethylene response, and carotenoid synthesis genes in addition to promoters of other ripening-related transcription factors. In the specific cases of ethylene and carotenoid biosynthesis RIN interacts with the promoters of the rate-limiting steps in each pathway (ACS and PSY, respectively). Furthermore, demonstration of CNR necessity for RIN promoter binding yet absent evidence for direct RIN-CNR interaction provides an initial foundational insight on which to expand our understanding of transcriptional ripening control.
MATERIALS AND METHODS
Plants
Wild type and rin (rin/rin) mutant tomato (Solanum lycopersicum ‘Ac’) were grown under normal greenhouse condition until maturity. Fruits were staged based on the number of days from anthesis to BK stage as defined by the detection of orange color at the base of wild-type fruits. Age of the nonripening cultivars rin and Cnr were calculated based on DPA using wild type as a reference (hence BK stage in rin correspond to fruits having the same age as wild-type BK fruits).
Constructs
The construct pET-RIN-KC was obtained by PCR amplification of pET-RIN using primers RIN KC-F (5′-TATAGGTACCGGTGAGGATTTGGGACAATTG-3′) and pET28a R (5′-TATAGGTACCCATTTGCTGTCCACCAGTC-3′), digesting with KpnI and DpnI, and ligating. pET-RIN was obtain by PCR, amplifying full-length RIN cDNA using primers RIN F (5′-TTTTGGATCCGAATTCATGGGTAGAGGGA-AAGTAG-3′) and RIN R (5′-TTTTCTCGAGTCAAAGCATCCATCCAGGTA-CAAC-3′), digesting with EcoRI and XhoI restriction enzymes, and cloning into pET28a vector (Novagen) previously digested with the same enzymes.
Recombinant Protein Purification and Antibodies Production
Escherichia coli BL21 star (DE3; 2TInvitrogen2T) cells containing pETRIN-KC were grown in Luria-Bertani broth at 30°C overnight. The next day a fresh Luria-Bertani broth culture was inoculated with the overnight culture diluted to 0.1 ODR600R and grown for 3 h at 30°C. Isopropylthio-β-galactoside was then added to a final concentration of 1 mm and the culture incubated for another 3 h at 30°C. The cells were pelleted by centrifugation and resuspended in TALON equilibration buffer 1× pH 8 (Clontech) containing 2 mg/mL of lysozyme. Cells were lysed by sonication on ice using a Branson 450 sonicator (settings: power 4.5, duty 50%). The lysate was centrifuged at 14,000g for 10 min at 4°C and the supernatant discarded. The pellet was solubilized in TALON equilibration buffer 1× pH 8 containing 6 m guanidine. Purification of the HIS-tagged RIN-KC protein was performed according to the TALON His batch/gravity-flow column purification protocol (Clontech). One milligram of purified His-RIN-KC protein, quantified using the bicinchoninic acid assay (Pierce), was sent to Covance Research Products for injection into rabbits to raise antibodies.
Protein Extraction from Fruit
Tomato (cv Ac) wild type and rin fruits at different stages of development were frozen and ground in liquid nitrogen. Proteins were isolated from ground tissue using the protocol described by Wang et al. (2006). Protein was quantified using the bicinchoninic acid assay (Pierce).
Western Blot
Twenty-five-microgram aliquots of protein extracts were separated in SDS-PAGE gels and transferred to a nitrocellulose membrane using standard procedures. Immunoblotting analyses were performed using the rabbit polyclonal RIN antibodies at a 1:1,000 dilution and 1:100,000 anti-rabbit HRP secondary antibodies (Sigma).
ChIP
ChIP was adapted from Fiil et al. (2008, CSH protocol), Manzara et al. (1991), and Nelson et al. (2006). For chromatin cross-linking, tomato fruit pericarp tissue was diced and place in a 50-mL Falcon tube filled with MC buffer (10 mm KHPOR4R pH 7, 50 mm NaCl, 0.1 m Suc, 1% formaldehyde). Vacuum was applied for 10 min at a time for four times (5-min rest between vacuum applications) and one additional time in presence of 0.125 m Gly to stop the cross-linking. Tissue was then rinsed with water, frozen in liquid nitrogen, and ground with mortar and pestle. For each ChIP, 3 g of the cross-linked tissue was resuspended for 30 min at 4°C in 45 mL of buffer 1 (0.4 m Suc, 10 mm Tris pH 8, 5 mm β-mercapto-ethanol [BME], 1× plant protease inhibitors [Sigma]), filtered through two layers of Miracloth (Calbiochem), and centrifuged for 20 min at 3,000g. Pellets were resuspended in 1 mL of buffer 2 (0.25 m Suc, 10 mm Tris pH 8, 10 mm MgClR2R, 1% Triton X-100, 5 mm BME, 1× plant protease inhibitor [Sigma]), transferred to a 1-mL eppendorf, and centrifuged for 10 min at 12,000g. Pellets were resuspended in 300 μL of buffer 3 (1.7 m Suc, 10 mm Tris pH 8, 2 mm MgClR2R, 0.15% Triton X-100, 5 mm BME, 1× plant protease inhibitors), carefully layered on top of 1.5-mL of buffer 3 (in a 2-mL tube), and centrifuged for 60 min at 16,000g. The resulting pellet was resuspended in 1 mL of freezing buffer (100 mm NaCl, 50 mm HEPES pH 7.6, 25% glycerol, 1 mm EDTA, 5 mm BME, protease inhibitor), frozen in liquid nitrogen, and thawed on ice. A total of 250 μL of lysing buffer (2.5 m NaCl, 50 mm HEPES pH 7.6, 1 mm EDTA, 5 mm BME) was added to the resupsended pellet and incubated for 30 min at 4°C. The solution was then sonicated on ice (Branson Sonifier 450) for 10 s at a time (duty 15%, power 3) for a total of 40 s (30 s on ice between sonication), centrifuged 10 min at 16,000g, and the chromatin-containing supernatant was transferred to a 2-mL tube. Supernatant was diluted 2-fold with 50 mm Tris-Cl pH 8, 1 mm EDTA, 0.1% Triton X-100, and incubated for 1 h at 4°C on a rotating wheel with 25 μL of preimmune serum and 40 μL of blocked protein A sepharose beads (GE Healthcare), blocked with 10 μg/mL of bovine serum albumin and 10 μg/mL of salmon sperm DNA. The mixture was centrifuged for 10 min at 16,000g and the supernatant separate in three tubes for INPUT, preimmune, and RIN ChIP treatments. ChIP was performed on the preimmune and RIN ChIP samples by incubation with 25 μL of blocked proteinA sepharose and 3 μL of rabbit preimmune serum or RIN antibodies serum, respectively, for 16 h at 4°C on a rotating wheel. Samples were centrifuged 2 min at 2,000g and the supernatant discarded. The sepharose beads were then washed five times with washing buffer (150 mm NaCl, 50 mm Tris pH 8, 1 mm EDTA, 1% Triton X-100) by rotation for 10 min at room temperature followed by 2-min centrifugation at 2,000g. After the last wash was removed, 100 μL of 10% Chelex resin solution (BioRad) was added to the beads and the INPUT sample and boiled for 10 min. After cooling at room temperature, samples were incubated for 45 min at 55°C with 20 μg/μL of proteinase K. Samples were then boiled for 10 min and centrifuged for 10 min at 16,000g (4°C) and the supernatant recovered. An additional 100 μL of water was added to each tube, vortexed, centrifuged, and the supernatant was then pooled with the previous supernatant solution. Samples were further purified using a Qiagen PCR purification column (Qiagen) following the manufacturer’s instructions, except that the final elution was 100 μL.
qPCR
qPCR was performed using SYBR green on an AB 7900 real-time PCR platform following the manufacturer’s instructions. Briefly, 2 μL of gDNA sample obtained as described above was mixed with 5 μL of SYBR Green and 300 nm of promoter-specific primer. Relative-fold enrichments are calculated by dividing the amount of gene-specific amplification by 18S amplification. Primers used in the qPCR reaction are listed in Supplemental Table S1. At least three biological replicates of each data point were used to calculate statistical significance of the enrichment. Error bars represent the se calculated from those samples.
qRT-PCR
RNA was extracted from fruit tissue at different stages of development (days after anthesis) using the plant RNA kit (Invitrogen) and following the manufacturer’s instructions. RNA was then digested with RQ1 DNase (Promega), and further purified on Qiagen RNA purification columns. qRT-PCR was performed using SYBR Green on the AB7900 using 18S as the internal control. Gene-specific primers used in the qRT-PCR assay are listed in Supplemental Table S1. ses were calculated based on a minimum of three biological replicates.
Two-Hybrid
The yeast-two-hybrid assay was performed by Hybrigenics Inc (www.hybrigenics.com) using the RIN protein (amino acid 1 to amino acid 185) as bait to screen a cDNA prey library derived from wild-type (cv Ac) fruits of stages from immature through red ripe (BK + 10). Calculation of predicted biological score (PBS) has been described previously (Rain et al., 2001; Formstecher et al., 2005).
Supplemental Data
Supplemental Table S1. Primers used for gene expression and ChIP assay.
References
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ. (2005) Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17: 2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander L, Grierson D. (2002) Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J Exp Bot 53: 2039–2055 [DOI] [PubMed] [Google Scholar]
- Barry CGJ. (2007) Ethylene and fruit ripening. J Plant Growth Regul 26: 143–159 [Google Scholar]
- Barry CS, Llop-Tous MI, Grierson D. (2000) The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol 123: 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartley GE, Viitanen PV, Bacot KO, Scolnik PA. (1992) A tomato gene expressed during fruit ripening encodes an enzyme of the carotenoid biosynthesis pathway. J Biol Chem 267: 5036–5039 [PubMed] [Google Scholar]
- Bleecker AB, Kende H. (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16: 1–18 [DOI] [PubMed] [Google Scholar]
- Bramley PM. (2002) Regulation of carotenoid formation during tomato fruit ripening and development. J Exp Bot 53: 2107–2113 [DOI] [PubMed] [Google Scholar]
- Busi MV, Bustamante C, D’Angelo C, Hidalgo-Cuevas M, Boggio SB, Valle EM, Zabaleta E. (2003) MADS-box genes expressed during tomato seed and fruit development. Plant Mol Biol 52: 801–815 [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Tuori RP, D’Ascenzo MD, Fobert PR, Despres C, Martin GB. (2003) The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 15: 3033–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Riechmann JL, Jia D, Meyerowitz E. (2000) Minimal regions in the Arabidopsis PISTILLATA promoter responsive to the APETALA3/PISTILLATA feedback control do not contain a CArG box. Sex Plant Reprod 13: 85–94 [Google Scholar]
- Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J. (2010) A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 64: 936–947 [DOI] [PubMed] [Google Scholar]
- Cunningham FX, Gantt E. (1998) Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 49: 557–583 [DOI] [PubMed] [Google Scholar]
- Dellapenna D, Lincoln JE, Fischer RL, Bennett AB. (1989) Transcriptional analysis of polygalacturonase and other ripening associated genes in rutgers, rin, nor, and Nr tomato fruit. Plant Physiol 90: 1372–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupille E, Sisler E. (1995) Effect of an ethylene receptor antagonist on plant material. Ait-Oubahou A, El-Otmani M, , Postharvest Physiology, Pathology and Technologies for Horticultural Commodities: Recent Advances. Institut Agronomique & Veterinaire Hassan II, Agadir, Morocco, pp 294–301 [Google Scholar]
- Eriksson EM, Bovy A, Manning K, Harrison L, Andrews J, De Silva J, Tucker GA, Seymour GB. (2004) Effect of the colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiol 136: 4184–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiil BK, Qiu JL, Petersen K, Petersen K, Petersen M, Mundy J. (2008) Coimmunoprecipitation (co-IP) of Nuclear Proteins and Chromatin Immunoprecipitation (ChIP) from Arabidopsis. CSH Protoc 2008: pdb.prot5049 [DOI] [PubMed] [Google Scholar]
- Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, et al. (2005) Protein interaction mapping: a Drosophila case study. Genome Res 15: 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Bramley P, Seymour GB. (2001) Effect of the Cnr mutation on carotenoid formation during tomato fruit ripening. Phytochemistry 58: 75–79 [DOI] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Ito Y. (2011) Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biol 11: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. (2004) Genetic regulation of fruit development and ripening. Plant Cell (Suppl) 16: S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. (2007) Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol 10: 283–289 [DOI] [PubMed] [Google Scholar]
- Heck GR, Perry SE, Nichols KW, Fernandez DE. (1995) AGL15, a MADS domain protein expressed in developing embryos. Plant Cell 7: 1271–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TA, Day CD, Zondlo SC, Thackeray AG, Irish VF. (1998) Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development 125: 1711–1721 [DOI] [PubMed] [Google Scholar]
- Hobson GE, Nichols R, Davies JN, Atkey PT. (1984) The inhibition of tomato fruit ripening by silver. J Plant Physiol 116: 21–29 [DOI] [PubMed] [Google Scholar]
- Immink RGH, Tonaco IAN, de Folter S, Shchennikova A, van Dijk ADJ, Busscher-Lange J, Borst JW, Angenent GC. (2009) SEPALLATA3: the ‘glue’ for MADS box transcription factor complex formation. Genome Biol 10: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A. (2009) The TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J 60: 1081–1095 [DOI] [PubMed] [Google Scholar]
- Ito T, Wellmer F, Yu H, Das P, Ito N, Alves-Ferreira M, Riechmann JL, Meyerowitz EM. (2004) The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430: 356–360 [DOI] [PubMed] [Google Scholar]
- Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, Ito H, Inakuma T, Hiroi S, Kasumi T. (2008) DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J 55: 212–223 [DOI] [PubMed] [Google Scholar]
- Jaakola L, Poole M, Jones MO, Kämäräinen-Karppinen T, Koskimäki JJ, Hohtola A, Häggman H, Fraser PD, Manning K, King GJ, et al. (2010) A SQUAMOSA MADS-box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Melzer R, Theissen G. (2005) MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347: 183–198 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC. (2009) Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee H, Tieman D. (2002) The tomato ethylene receptor gene family: form and function. Physiol Plant 115: 336–341 [DOI] [PubMed] [Google Scholar]
- Klee HJ. (2004) Ethylene signal transduction: moving beyond Arabidopsis. Plant Physiol 135: 660–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Saedler H, Huijser P. (1996) A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol Gen Genet 250: 7–16 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D. (2008) A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J 55: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln JE, Cordes S, Read E, Fischer RL. (1987) Regulation of gene expression by ethylene during Lycopersicon esculentum (tomato) fruit development. Proc Natl Acad Sci USA 84: 2793–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln JE, Fischer RL. (1988) Regulation of gene expression by ethylene in wild-type and rin tomato (Lycopersicon esculentum) fruit. Plant Physiol 88: 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano R, Giménez E, Cara B, Capel J, Angosto T. (2009) Genetic analysis of reproductive development in tomato. Int J Dev Biol 53: 1635–1648 [DOI] [PubMed] [Google Scholar]
- Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Manzara T, Carrasco P, Gruissem W. (1991) Developmental and organ-specific changes in promoter DNA-protein interactions in the tomato rbcS gene family. Plant Cell 3: 1305–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Rodríguez MC, Orchard J, Seymour GB. (2002) Pectate lyases, cell wall degradation and fruit softening. J Exp Bot 53: 2115–2119 [DOI] [PubMed] [Google Scholar]
- Montgomery J, Pollard V, Deikman J, Fischer RL. (1993) Positive and negative regulatory regions control the spatial distribution of polygalacturonase transcription in tomato fruit pericarp. Plant Cell 5: 1049–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Sova P, Bomsztyk K. (2006) Fast chromatin immunoprecipitation assay. Nucleic Acids Res 34: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Yanofsky MF. (2001) Function and evolution of the plant MADS-box gene family. Nat Rev Genet 2: 186–195 [DOI] [PubMed] [Google Scholar]
- Nicholass FJ, Smith CJ, Schuch W, Bird CR, Grierson D. (1995) High levels of ripening-specific reporter gene expression directed by tomato fruit polygalacturonase gene-flanking regions. Plant Mol Biol 28: 423–435 [DOI] [PubMed] [Google Scholar]
- Orfila C, Huisman MM, Willats WG, van Alebeek GJ, Schols HA, Seymour GB, Knox JP. (2002) Altered cell wall disassembly during ripening of Cnr tomato fruit: implications for cell adhesion and fruit softening. Planta 215: 440–447 [DOI] [PubMed] [Google Scholar]
- Pan IL, McQuinn R, Giovannoni JJ, Irish VF. (2010) Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J Exp Bot 61: 1795–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecker I, Chamovitz D, Linden H, Sandmann G, Hirschberg J. (1992) A single polypeptide catalyzing the conversion of phytoene to zeta-carotene is transcriptionally regulated during tomato fruit ripening. Proc Natl Acad Sci USA 89: 4962–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Hareven D, Rounsley SD, Yanofsky MF, Lifschitz E. (1994) Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell 6: 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, Lenzen G, Petel F, Wojcik J, Schächter V, et al. (2001) The protein-protein interaction map of Helicobacter pylori. Nature 409: 211–215 [DOI] [PubMed] [Google Scholar]
- Ray J, Moureau P, Bird C, Bird A, Grierson D, Maunders M, Truesdale M, Bramley P, Schuch W. (1992) Cloning and characterization of a gene involved in phytoene synthesis from tomato. Plant Mol Biol 19: 401–404 [DOI] [PubMed] [Google Scholar]
- Robles P, Pelaz S. (2005) Flower and fruit development in Arabidopsis thaliana. Int J Dev Biol 49: 633–643 [DOI] [PubMed] [Google Scholar]
- Rose JK, Cosgrove DJ, Albersheim P, Darvill AG, Bennett AB. (2000) Detection of expansin proteins and activity during tomato fruit ontogeny. Plant Physiol 123: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski RW, Meyerowitz EM. (1998) A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92: 93–103 [DOI] [PubMed] [Google Scholar]
- Saltveit ME, Dilley DR. (1978) Rapidly induced wound ethylene from excised segments of etiolated Pisum sativum L., cv. Alaska: III. Induction and transmission of the response. Plant Physiol 62: 710–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lönnig WE, Saedler H, Sommer H. (1992) Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J 11: 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour GB. (1993) Biochemistry of Fruit Ripening. Chapman & Hall, London, pp 1–454 [Google Scholar]
- Stepanova AN, Ecker JR. (2000) Ethylene signaling: from mutants to molecules. Curr Opin Plant Biol 3: 353–360 [DOI] [PubMed] [Google Scholar]
- Tilly JJ, Allen DW, Jack T. (1998) The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125: 1647–1657 [DOI] [PubMed] [Google Scholar]
- Tröbner W, Ramirez L, Motte P, Hue I, Huijser P, Lönnig WE, Saedler H, Sommer H, Schwarz-Sommer Z. (1992) GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J 11: 4693–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJ, McQuinn R, Chung M, Poole M, Rose J, Seymour G, Grandillo S, Giovannoni J, et al. (2009) Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21: 3041–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wang H, Tang W, Zhu C, Perry SE. (2002) A chromatin immunoprecipitation (ChIP) approach to isolate genes regulated by AGL15, a MADS domain protein that preferentially accumulates in embryos. Plant J 32: 831–843 [DOI] [PubMed] [Google Scholar]
- Wang W, Vignani R, Scali M, Cresti M. (2006) A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 27: 2782–2786 [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Clark DG, Bleecker AB, Chang C, Meyerowitz EM, Klee HJ. (1997) A dominant mutant receptor from Arabidopsis confers ethylene insensitivity in heterologous plants. Nat Biotechnol 15: 444–447 [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Young RA. (2002) Deciphering gene expression regulatory networks. Curr Opin Genet Dev 12: 130–136 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE. (2009) Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]