Abstract
Nitric oxide (NO) is emerging as an important regulatory player in the Rhizobium-legume symbiosis, but its biological role in nodule functioning is still far from being understood. To unravel the signal transduction cascade and ultimately NO function, it is necessary to identify its molecular targets. This study provides evidence that glutamine synthetase (GS), a key enzyme for root nodule metabolism, is a molecular target of NO in root nodules of Medicago truncatula, being regulated by tyrosine (Tyr) nitration in relation to active nitrogen fixation. In vitro studies, using purified recombinant enzymes produced in Escherichia coli, demonstrated that the M. truncatula nodule GS isoenzyme (MtGS1a) is subjected to NO-mediated inactivation through Tyr nitration and identified Tyr-167 as the regulatory nitration site crucial for enzyme inactivation. Using a sandwich enzyme-linked immunosorbent assay, it is shown that GS is nitrated in planta and that its nitration status changes in relation to active nitrogen fixation. In ineffective nodules and in nodules fed with nitrate, two conditions in which nitrogen fixation is impaired and GS activity is reduced, a significant increase in nodule GS nitration levels was observed. Furthermore, treatment of root nodules with the NO donor sodium nitroprusside resulted in increased in vivo GS nitration accompanied by a reduction in GS activity. Our results support a role of NO in the regulation of nitrogen metabolism in root nodules and places GS as an important player in the process. We propose that the NO-mediated GS posttranslational inactivation is related to metabolite channeling to boost the nodule antioxidant defenses in response to NO.
Nitric oxide (NO) is widely recognized as an endogenous signaling molecule involved in various physiological processes, from stress responses to normal plant growth and development, including the legume-rhizobia symbiosis (Besson-Bard et al., 2007; Neil et al., 2008; Wilson et al., 2008). This symbiotic interaction involves complex signal-exchange mechanisms that culminate with the formation of newly differentiated organs called root nodules, which provide a niche for bacterial nitrogen fixation (Oldroyd et al., 2011). An increasing number of reports on the occurrence of NO during the legume-rhizobia symbiosis suggest an important, but yet unknown, signaling role of this molecule for the symbiosis (Wang and Ruby, 2011). The identification of the molecular elements involved in the nodule response to NO will be essential to understand its role and mechanisms of action. In functional nodules of Medicago truncatula, NO production has been located in the bacteroid-containing cells of the nodule fixation zone (Baudouin et al., 2006), where the ammonium generated by bacterial nitrogenase activity is released from the rhizobial bacteroids into the cytosol of the infected plant cells and assimilated into the organic pools by plant glutamine synthetase (GS; EC 6.3.1.2); thus, the enzyme can be considered a potential candidate to respond to NO.
GS is an extremely complex and highly regulated enzyme that, in addition to nitrogen fixation, is involved in a number of essential metabolic pathways in the plant. GS catalyzes the ATP-dependent condensation of ammonium with Glu to yield Gln, which is then used for the biosynthesis of essentially all nitrogenous compounds (Lea and Miflin, 2011). The ammonium for GS activity may be derived from primary sources (nitrogen fixation, nitrate assimilation, and ammonium uptake) or from secondary metabolism such as photorespiration and amino acid catabolism; therefore, the enzyme plays essential roles both in primary nitrogen assimilation and in nitrogen recycling (Lea and Miflin, 2011). Consistent with the diversity of metabolic roles, GS exists in the plant as a number of isoenzymes that are located both in the cytosol (GS1) and in the plastids (GS2) and are encoded by a small family of genes. Each of the GS genes shows distinct patterns of expression in different organs of the plant and appears to participate in different metabolic processes. In M. truncatula, the GS gene family consists of four expressed genes, MtGS1a, MtGS1b, MtGS2a, and MtGS2b (Stanford et al., 1993; Carvalho et al., 2000a, 2000b; Melo et al., 2003; Seabra et al., 2010). MtGS2a and MtGS2b encode precursors to plastid-located isoenzymes, with MtGS2b being a second plastid-located isoform unique to M. truncatula and closely related species and exclusively expressed in developing seeds (Seabra et al., 2010). The other three GS genes are expressed in root nodules, but MtGS1a is highly up regulated in the central infected cells, accounts for the production of over 90% of the total nodule GS activity, and encodes the isoenzyme responsible for the assimilation of the ammonia released by nitrogen fixation (Carvalho et al., 2000a).
The essentially of GS for plant life predicts the existence of several precise and tightly controlled mechanisms for the regulation of enzyme activity. The regulation of GS at the transcriptional level has been well studied, and it is generally accepted as the primary regulatory point, but a number of posttranslational regulatory controls were shown to be critical for the regulation of GS activity (Hoelzle et al., 1992; Temple et al., 1996, 1998; Ortega et al., 1999, 2001). In higher plants, GS activity is known to be modulated by oxidative turnover (Ortega et al., 1999; Palatnik et al., 1999; Ishida et al., 2002) and phosphorylation along with 14-3-3 interaction (Finnemann and Schjoerring, 2000; Man and Kaiser, 2001; Riedel et al., 2001; Lima et al., 2006a, 2006b). In Escherichia coli, the regulation of GS activity involves cyclic adenylylation and nitration of Tyr residues (Berlet et al., 1996). Tyr nitration has also been implicated in the regulation of GS in animals (Gorg et al., 2007), being conceivably a similar mechanism for the regulation of the plant enzyme. Protein Tyr nitration is a NO-mediated posttranslational modification that consists on the addition of a nitro (NO2) group to one of the equivalent ortho carbons of the aromatic ring of Tyr residues (Radi, 2004), which can alter the conformation and structure of proteins, the catalytic properties of enzymes, and their susceptibility to proteolysis (Abello et al., 2009). Protein Tyr nitration has been best studied in animal systems, and little is known regarding the functional effects of protein Tyr nitration in plants (Corpas et al., 2009), but recent proteomic studies revealed the existence of a high number of nitrated proteins in sunflower (Helianthus annuus; Chaki et al., 2009) and in Arabidopsis (Arabidopsis thaliana; Cecconi et al., 2009; Lozano-Juste et al., 2011), suggesting that Tyr nitration also represents an important NO-mediated regulatory mechanism in plants.
The formation of NO and its involvement in the legume-rhizobia symbiosis has been the subject of much research in the last few years, and it is believed that it plays an important, but yet unknown, signaling role in the symbiosis (Wang and Ruby, 2011). The model legume M. truncatula and its symbiotic partner Sinorhizobium meliloti provided the background for many of those studies. NO production has been located at the infection sites during the initial stages of nodule development of M. truncatula and seems to be required for the establishment of the symbiosis (Del Giudice et al., 2011), whereas in fully developed root nodules, it appears to be confined to the nodule fixation zone, suggesting an involvement of NO in root nodule metabolism (Baudouin et al., 2006; Horchani et al., 2011). Proposed sources of NO in root nodules include NO synthase-like proteins (Cueto et al., 1996; Baudouin et al., 2006; Leach et al., 2010) and nitrate reductase and electron transfer chains from both plants and bacteria (Kato et al., 2003; Mesa et al., 2004; Meakin et al., 2007; Gupta et al., 2011; Horchani et al., 2011).
NO appears to directly affect nodule metabolism by inhibiting nitrogenase activity. The involvement of NO in nitrogenase inactivation has been demonstrated in soybean (Glycine max) and Lotus after nitrate supply (Kanayama et al., 1990; Meakin et al., 2007; Kato et al., 2010). In Lotus japonicus, the artificial application of the NO donor sodium nitroprusside (SNP) decreased nitrogen fixation, whereas the application of a NO scavenger (2-4-carboxyphenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide) enhanced nitrogen fixation (Shimoda et al., 2009; Kato et al., 2010). In spite of the inhibitory effect of NO on nitrogenase, it appears that NO production is required for nodule development and functioning, and the plant antioxidant systems appear to be crucial to maintain nodule functioning (Pauly et al., 2006; Keyster et al., 2010; Leach et al., 2010; Sanchez et al., 2011). The levels of NO inside the nodule appear to be controlled by leghemoglobin, which is able to scavenge NO and in this way may protect nitrogenase from inactivation (Kanayama et al., 1990; Herold and Puppo, 2005; Meakin et al., 2007; Kato et al., 2010; Sanchez et al., 2011). Nitroso-leghemoglobin complexes have been detected in nodules of soybean and Lotus (Kanayama et al., 1990; Mathieu et al., 1998; Meakin et al., 2007; Sanchez et al., 2010). This NO-scavenging function has also been attributed to nonsymbiotic hemoglobins in L. japonicus, which are induced upon symbiotic infection and accumulate in nitrogen-fixing nodules (Shimoda et al., 2009).
There is still much controversy and uncertainty around the possible roles of NO in the legume-rhizobia symbiosis, and the identification of its molecular targets is a major asset to start dissecting its mechanisms of signaling and action. In this study, we provide evidence that GS is a molecular target of NO in root nodules of M. truncatula and that its activity is posttranslationally regulated by Tyr nitration in relation to active nitrogen fixation. A model is proposed to integrate the posttranslational regulation of GS by Tyr nitration within the context of root nodule metabolism.
RESULTS
In Vitro Inactivation of GS by NO
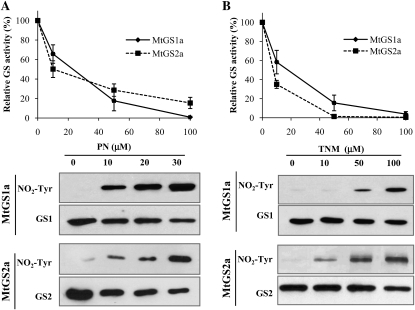
The major protein posttranslational modifications induced by NO are nitrosylation of Cys and nitration of Tyr residues (Radi, 2004). To investigate whether nodule GS could be affected by any of these modifications, we started by evaluating in vitro the effects of two reactive nitrogen species producers on the M. truncatula nodule isoenzyme MtGS1a and the plastid-located isoenzyme MtGS2a. The plant proteins were produced in E. coli with a His tag, purified by Ni-affinity chromatography, and used to evaluate GS activity following incubation with increasing concentrations of either peroxynitrite (PN) or tetranitromethane at pH 8 (TNM). GS activity was found to be inhibited by the two compounds in a dose-dependent manner for the two isoenzymes (Fig. 1), reaching total inactivation after incubation with 100 μm of either PN or TNM. To assess whether the inhibitory effect could be due to Tyr nitration, the treated enzymes were analyzed by western blotting using a specific anti-3-nitrotyrosine antibody. Immunoreactivity was found to increase with increasing PN or TNM concentration for both isoenzymes (Fig. 1).
Figure 1.
Effects of PN and TNM on GS activity and nitration. Purified MtGS1a or MtGS2a was incubated with PN (A) or TNM at pH 8 (B) at increasing concentrations, assayed for GS transferase activity, and immunodetected with an anti-3-nitrotyrosine specific antibody (NO2-Tyr). Loading controls were obtained by immunoblotting the membranes with anti-GS1 or anti-GS2 antibody (GS1 or GS2, respectively). GS activity was normalized to that found in the absence of NO donor and is represented as means ± sd of at least three independent experiments, assayed in triplicate. The average initial GS activities were 0.36 and 0.1 μmol min−1 μg−1 protein for MtGS1a and MtGS2a, respectively.
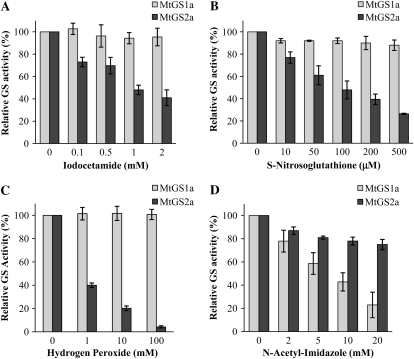
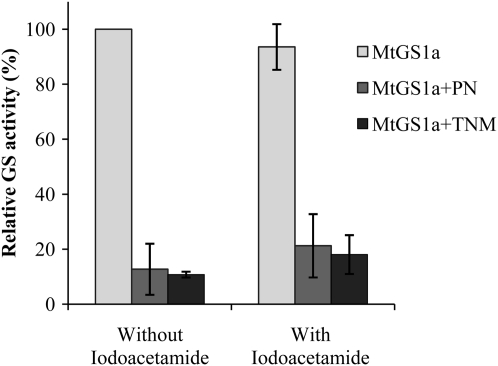
To further investigate whether the cause of the NO-induced GS inactivation is nitration or other oxidative processes triggered by the reactive nitrogen species, we tested the effect of a number of reagents known to selectively modify Tyr, Cys, or Met residues on the activity of each GS isoenzyme. The widely used Tyr O-acetylation agent N-acetyl-imidazole was used to specifically modify Tyr residues, the alkylating sulfhydryl reagent iodocetamide and the S-nitrosylation reagent S-nitrosoglutathione (S-GSNO) were used as Cys modification agents, and hydrogen peroxide was used as a potent Met and Cys oxidant. The activity of MtGS1a was not significantly affected by Cys alkylation (Fig. 2A), nitrosylation (Fig. 2B), or oxidation (Fig. 2C). Conversely, the acetylation of Tyr residues by N-acetyl-imidazole clearly induced MtGS1a inactivation (Fig. 2D). These results indicate the existence of Tyr residue(s) that, upon modification, lead to MtGS1a inactivation. Interestingly, this effect appears to be specific for the cytosolic isoenzyme MtGS1a, as treatment of the plastid-located MtGS2a isoenzyme with Cys-modifying reagents results in enzyme inactivation (Fig. 2, A–C), whereas N-acetyl-imidazole has a minor inhibitory effect on this isoenzyme (Fig. 2D). Additional evidence that MtGS1a inhibition is a result of Tyr modifications is given by the finding that Cys alkylation, induced by incubation of the enzyme with iodoacetamide, does not prevent MtGS1a from being inactivated by either PN or TNM (Fig. 3). Taken together, these results suggest that NO-dependent inactivation of MtGS1a is due to Tyr nitration, whereas MtGS2a activity is inhibited by thiol residue nitrosylation.
Figure 2.
Effects of modifications on Cys and Tyr residues on the activity of MtGS1a and MtGS2a. Purified MtGS1a or MtGS2a was treated with increasing concentrations of iodoacetamide (A), S-GSNO (B), hydrogen peroxide (C), or N-acetyl imidazole (D) and assayed for GS transferase activity. GS activity was normalized to that found in the absence of reagents and is represented as means ± sd of at least three independent experiments, assayed in triplicate. The average initial GS activity was 0.36 and 0.1 μmol min−1 μg−1 protein for MtGS1a and MtGS2a, respectively.
Figure 3.
Effects of iodoacetamide on MtGS1a inactivation induced by TNM and PN. Purified MtGS1a was treated with 500 μm iodoacetamide, followed by incubation with 50 μm PN or 50 μm TNM at pH 8, and assayed for GS activity. GS activity was normalized to that found in the absence of iodoacetamide and nitrating agent. GS activity is represented as means ± sd of at least three independent experiments, assayed in triplicate. The average initial GS activity was 0.36 μmol min−1 μg−1 protein.
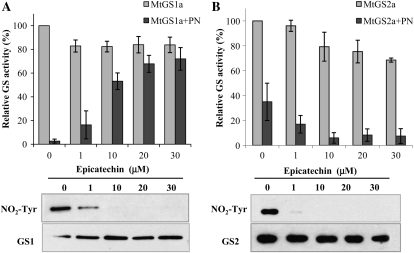
The flavonoid epicatechin has been shown to selectively prevent the nitration of Tyr residues (Schroeder et al., 2001); therefore, we examined whether this flavonoid could protect the enzymes from inactivation. Both MtGS1a and MtGS2a were incubated with increasing concentrations of epicatechin before treatment with 50 μm PN (Fig. 4). Epicatechin was able to protect MtGS1a from inactivation (Fig. 4A) but not MtGS2 (Fig. 4B). Furthermore, the incubation of both GS isoenzymes with epicatechin prior to PN treatment resulted in decreased immunoreactivity against nitrotyrosine, confirming that this flavonoid does indeed prevent Tyr nitration. These results substantiate that MtGS1a inactivation is a consequence of Tyr nitration mediated by reactive nitrogen species. As for MtGS2a, PN and TNM induced Tyr nitration, but this modification does not seem to be the cause of enzyme inactivation, as epicatechin was able to prevent nitration of the protein but not the inactivation induced by the reactive nitrogen species producers (Fig. 4B).
Figure 4.
Effects of epicatechin on GS inactivation and nitration. Purified MtGS1a (A) and MtGS2a (B) were incubated with different concentrations of epicatechin, followed by treatment with 50 μm PN, assayed for GS transferase activity, and immunodetected with anti-nitrotyrosine antibody (NO2-Tyr). Loading controls were obtained by immunoblotting the membranes with anti-GS1 or anti-GS2 antibody (GS1 or GS2, respectively). GS activity was normalized to that found in the absence of epicatechin and nitrating agent and is represented as means ± sd of at least three independent experiments, assayed in triplicate. The average initial GS activity was 0.36 and 0.1 μmol min−1 μg−1 protein for MtGS1a and MtGS2a, respectively.
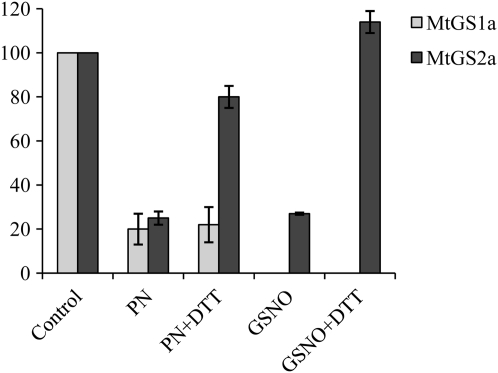
Tyr nitration is generally considered an irreversible process, whereas S-nitrosylation is rapidly reversible (Besson-Bard et al., 2007). In order to examine whether the NO-induced GS inactivation could be reversed, we incubated the NO-treated enzymes with dithiothreitol (DTT) and looked for activity recovery. Following incubation with DTT, the activity of MtGS2a could be totally recovered while MtGS1a inactivation could not be reversed (Fig. 5), further strengthening the idea that the MtGS1a NO-inhibitory effect is due to Tyr nitration whereas MtGS2 inactivation is likely to result from Cys nitrosylation.
Figure 5.
Reversibility of the PN- and GSNO-induced inactivation of MtGS1a and MtGS2a by DTT. Purified MtGS1a or MtGS2a was treated with 50 μm PN, and MtGS2a was additionally treated with 500 μm GSNO, followed by incubation with 10 mm DTT, and assayed for GS activity. GS activity was normalized to that found in the absence of reagents and is represented as means ± sd of at least three independent experiments, assayed in triplicate. The average initial GS activity was 0.36 and 0.1 μmol min−1 μg−1 protein for MtGS1a and MtGS2a, respectively.
Identification of Tyr-167 as a Relevant Nitration Site
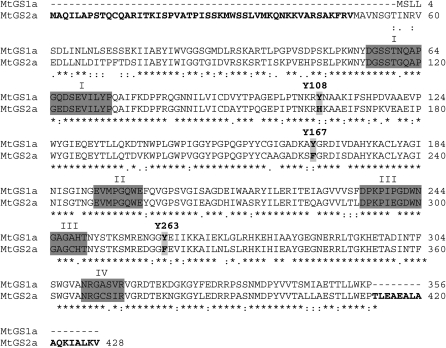
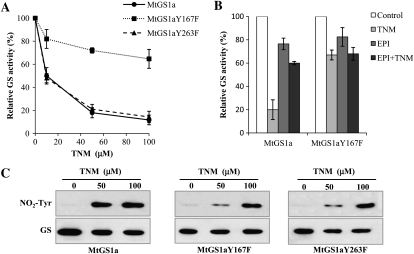
Once MtGS1a and MtGS2a appeared to be differentially regulated by NO-mediated modifications, either through Tyr nitration or Cys modification, respectively, we searched for Tyr residues present in MtGS1a but not in MtGS2a. An alignment of the amino acid sequences of the two proteins identified three Tyr residues on MtGS1a, Tyr-108, Tyr-167, and Tyr-263, that are substituted in MtGS2a (Fig. 6). From the analysis of the three-dimensional structure of MtGS1a (Seabra et al., 2009), it is predicted that only the residues Tyr-167 and Tyr-263 are placed within a local environment favorable to nitration (Abello et al., 2009); additionally, their modification is likely to affect GS activity. To investigate whether any of these residues could be a regulatory nitration site and account for the specific susceptibility of MtGS1a to inactivation by nitration, we mutated residues Tyr-167 and Tyr-263 to Phe by site-directed mutagenesis and tested the susceptibility of the mutated proteins to inactivation induced by TNM. The mutated recombinant proteins were found to be active, and their specific activities were similar to the nonmutated enzymes. However, the inhibitory effect of TNM was significantly reduced for the Tyr-167 mutated protein (MtGS1aY167F), whereas mutation of Tyr-263 did not appear to affect TNM-induced GS inhibition, with the protein showing a pattern of inhibition comparable with the nonmutated enzyme (Fig. 7A). These results demonstrate that Tyr-167 is a relevant site for the inhibition of MtGS1a activity by nitration. However, we cannot exclude the existence of additional Tyr residues important for the inactivation process or the contribution of other oxidative modifications induced by NO, as the mutated protein was still susceptible to inhibition by TNM, although to a much lower extent (20% inactivation). Pretreatment of the mutated enzyme with 20 μm epichatechin could not prevent this inactivation (Fig. 7B), suggesting that Tyr-167 is the only Tyr implicated in the regulation of MtGS1a activity by nitration and that the observed 20% reduction in GS activity results from oxidative modifications different from Tyr nitration. Western-blot analysis of the mutated proteins treated with TNM (Fig. 7C) showed a reduction in nitrotyrosine immunoreactivity of the two mutated enzymes (Tyr-167 and Tyr-263) relative to the nonmutated enzyme, indicating that the two residues are subjected to nitration, but only Tyr-167 appears to be critical for GS inactivation.
Figure 6.
Alignment of the amino acid sequences of MtGS1a and MtGS2a. The Tyr residues substituted in MtGS2a are shown in boldface, highlighted, and numbered. The GS conserved regions I to IV defined by Eisenberg et al. (1987) are highlighted in dark gray. The MtGS2a transit peptide and C-terminal extension characteristics of plastid-located GS are shown in boldface.
Figure 7.
Effects of in vitro nitration on the activity of MtGS1aY167F and MtGS1aY263F. A, Purified GS1a and mutated GS1aY167F and GS1aY263F were incubated with TNM, pH 8, at increasing concentrations and assayed for GS transferase activity. B, The effect of epicatechin on the inactivation of mutated GS1aY167F was assessed by incubation of the mutated and nonmutated enzymes with 20 μm epicatechin followed by treatment with 50 μm TNM, pH 8. GS activity was normalized to that found in the absence of TNM and epicatechin and is represented as means ± sd of at least three independent experiments, assayed in triplicate. C, Overall nitration of mutated and nonmutated TNM-treated enzymes was evaluated by western blots and immunodetected with anti-nitrotyrosine antibody (NO2-Tyr). Loading controls were obtained by immunoblotting the membranes with anti-GS antibody (GS). The average initial GS activity was 0.36 μmol min−1 μg−1 protein.
In Vivo GS Nitration in Effective versus Ineffective Root Nodules
To begin studying the physiological significance of GS Tyr nitration for root nodule functioning, we compared GS nitration in planta in situations where nitrogen fixation is impaired. For this purpose, a sandwich ELISA was developed as a tool to compare the amount of in vivo nitrated GS in nodules collected under different physiological conditions. This assay uses an anti-GS antibody as a capture antibody to immunoseparate GS from the other nodule proteins and a specific anti-3-nitrotyrosine antibody for detection and quantification of the relative amount of nitrated GS in the different nodule extracts. The relative quantification of GS nitration was based on standards using pure recombinant MtGS1a, which was previously subjected to in vitro nitration by incubation with 100 μm PN. Since the GS polypeptide content and GS activity change under different physiological conditions, the nodule protein extracts were also subjected to GS activity measurements and to immunoblot analysis using anti-GS antibodies. The cytosolic GS immunoreactive bands were quantified by densitometry and the values were plotted. The analyses were performed in at least four biological replicates, and for each biological sample GS nitration, GS quantity, and GS activity were compared.
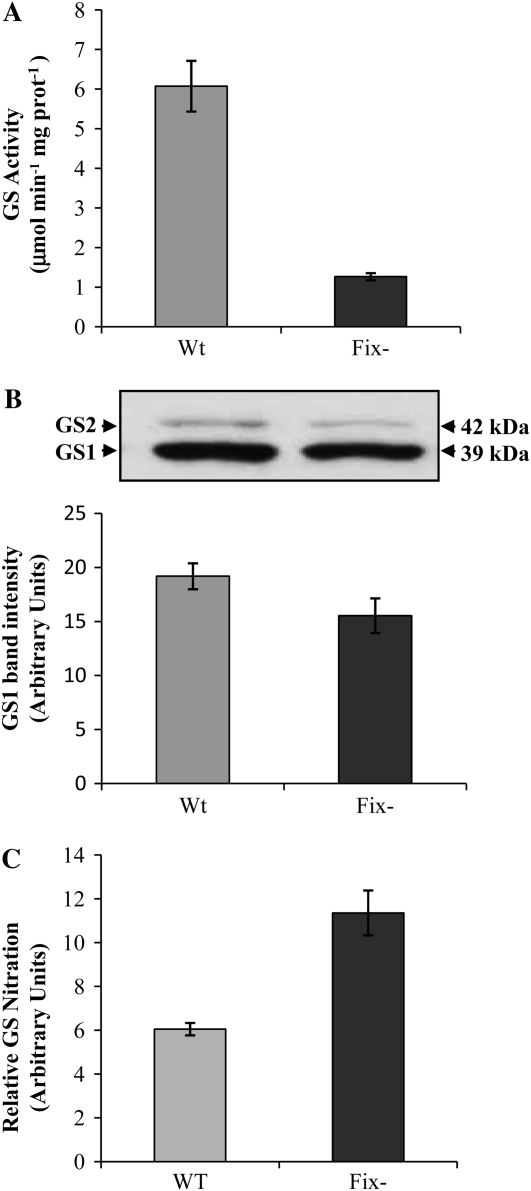
In ineffective root nodules formed by the Rhizobium mutant FixJ−, GS activity was significantly reduced (Fig. 8A), which is an expected result, since the Rhizobium mutant is unable to produce ammonium for GS assimilation, yet the GS polypeptide content was only slightly reduced (Fig. 8B). Interestingly, the amount of nitrated GS was found to be significantly higher in the mutant nodules when compared with wild-type nodules (Fig. 8C). Thus, the reduction in GS activity in ineffective root nodules can be related with an increase in in vivo GS nitration, strongly suggesting a direct relationship between GS nitration and nitrogen fixation.
Figure 8.
Evaluation of GS nitration in effective and ineffective root nodules of M. truncatula. A, Total soluble proteins were extracted from root nodules formed by the wild type (Wt) and fixJ (Fix−) mutant strains of S. meliloti. GS activity was measured by the transferase reaction. B, The cytosolic GS (GS1) immunoreactive bands were quantified by densitometry and the values were plotted. A representative GS western blot of total soluble proteins (10 μg per lane) is shown. C, GS nitration was quantified by sandwich ELISA relative to GS1 polypeptide content. Values shown are means of four biological replicates ± sd, assayed in duplicate.
In Vivo GS Nitration in Roots and Root Nodules following Nitrate Application
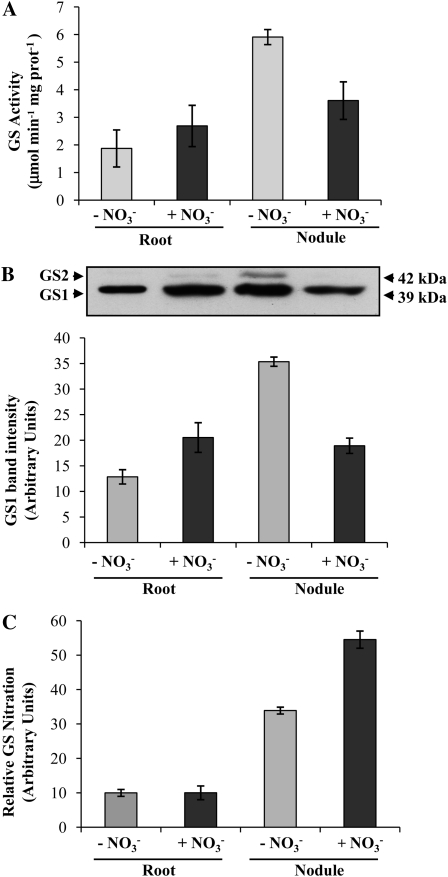
Nitrate is known to induce NO production inside the nodule, leading to the inhibition of nitrogenase activity (Kanayama et al., 1990; Meakin et al., 2007; Kato et al., 2010). To evaluate the effect of nitrate application on GS nitration in roots and nodules of M. truncatula, nodulated plants (19 d after inoculation) were fed with nitrate for 2 d, and the levels of GS nitration were compared in nitrate-treated and nontreated plants and both in roots and root nodules. Following nitrate application, GS activity was found to be significantly reduced in root nodules (Fig. 9A), and this reduction was accompanied by a decrease in GS polypeptide content (Fig. 9B). The amount of cytosolic GS polypeptides (39 kD) was 50% reduced, and the plastid-located polypeptides (42 kD) were reduced to undetectable levels. Interestingly, in spite of the observed reduction in GS abundance, the amount of nitrated GS was found to be significantly increased in the nitrate-treated nodules (Fig. 9C), further suggesting a relationship between GS nitration and active nitrogen fixation.
Figure 9.
Evaluation of GS nitration in roots and root nodules treated with nitrate. A, Total soluble proteins were extracted from roots and from 21-d-old root nodules collected from plants treated for 2 d with NH4NO3. GS activity was measured by the transferase reaction. B, The cytosolic GS (GS1) immunoreactive bands were quantified by densitometry and the values were plotted. A representative GS western blot of total soluble proteins (10 μg per lane) is shown. C, GS nitration was quantified by sandwich ELISA relative to GS1 polypeptide content. Values shown are means of three biological replicates ± sd, assayed in duplicate.
In roots, nitrate induced an increase in root GS activity (Fig. 9A) as well as in the amount of cytosolic and plastid-located polypeptides (Fig. 9B), reflecting the fact that the roots start to assimilate nitrate. In opposition to what was observed in the nodules, the amount of nitrated GS in the root, which is mainly composed of the isoenzyme MtGS1b (Carvalho et al., 2000b), does not seem to be affected by the nitrate treatment (Fig. 9C). In spite of the higher production of NO, which is likely to occur following nitrate feeding, the root enzyme does not appear to be regulated by nitration. This result supports a specific regulation of the nodule isoenzyme MtGS1a by Tyr nitration.
In Vivo GS Nitration in Root Nodules Treated with SNP
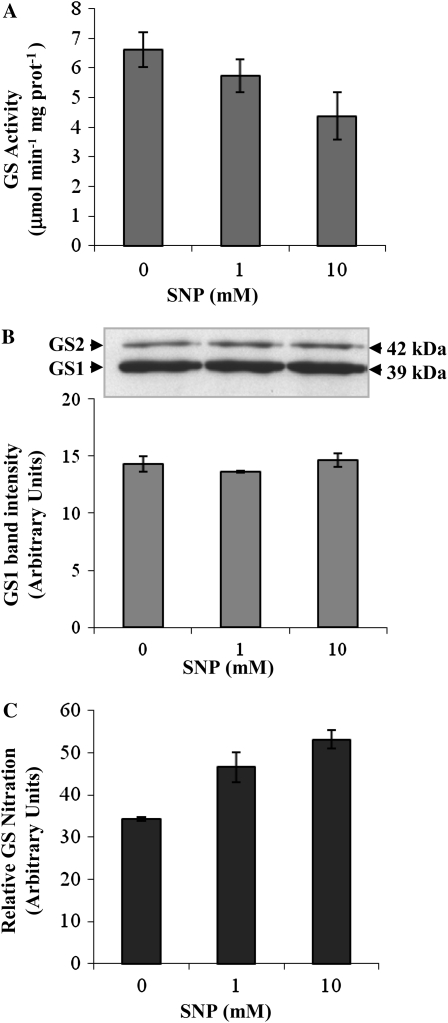
To further evaluate whether GS nitration is the cause of GS inactivation in vivo, we applied the widely used NO donor SNP to root nodules of M. truncatula and followed GS activity, GS polypeptide content, and GS nitration in nodules treated with 1 and 10 mm SNP. The GS polypeptide content was not affected by the SNP treatment (Fig. 10B), but GS activity was found to be reduced (Fig. 10A). The reduction in GS activity can be directly correlated with an increase in the levels of nitrated GS evaluated by ELISA (Fig. 10C).
Figure 10.
Evaluation of GS nitration in root nodules treated with SNP. A, Total soluble proteins were extracted from root nodules collected from plants treated with increasing concentrations of SNP. GS activity was measured by the transferase reaction. B, The cytosolic GS (GS1) immunoreactive bands were quantified by densitometry and the values were plotted. A representative GS western blot of total soluble proteins (10 μg per lane) is shown. C, GS nitration was quantified by sandwich ELISA relative to GS1 polypeptide content. Values shown are means of three biological replicates ± sd, assayed in duplicate.
DISCUSSION
There is increasing evidence for a role of NO in the legume-rhizobia symbiosis, but the nature of this role is still far from being understood. Elucidation of mechanisms of action of NO in root nodules will inevitably require the identification of its molecular targets. This study represents a first step in that direction, as it provides evidence that the key metabolic enzyme GS is a target of NO in root nodules of M. truncatula, being posttranslationally regulated by Tyr nitration in relation to active nitrogen fixation.
GS is abundantly present in root nodules and plays a pivotal role in the assimilation of the ammonium released by nitrogen fixation. We have previously shown that in M. truncatula, this function is fulfilled by the cytosolic isoenzyme MtGS1a, which is abundantly present in the infected cells of the nodule fixation zone (Carvalho et al., 2000a), coinciding with the major site of NO formation in this model species (Baudouin et al., 2006; Horchani et al., 2011). The enzyme is thus accessible to the oxidative effects induced by this reactive compound. NO can affect protein function and in this way may signal fundamental physiological processes, with S-nitrosylation of Cys residues and Tyr nitration being the major protein posttranslational modifications induced by NO (Besson-Bard et al., 2007). Therefore, we started by evaluating whether the M. truncatula nodule enzyme MtGS1a could be affected by either of these modifications. In vitro studies using purified recombinant proteins demonstrated that the nodule enzyme MtGS1a is subjected to Tyr nitration and that this modification provokes a loss of enzyme activity. To our knowledge, this is the first report on GS inactivation by Tyr nitration in plants, but previous studies reported an in vitro inhibition of the mammalian and bacterial GS activity by Tyr nitration (Berlet et al., 1996; Gorg et al., 2007). In Arabidopsis, the plastid-located GS (GS2) has been previously identified both as an S-nitrosylated protein (Lindermayr et al., 2005) and as a nitrated protein in more recent proteomic studies (Cecconi et al., 2009; Lozano-Juste et al., 2011), but the effect of either of these modifications on the activity of the enzyme was not assessed before. Our study demonstrates that the activity of the cytosolic enzyme is modulated by Tyr nitration and suggests that the plastid-located GS is susceptible to both nitration and S-nitrosylation, but the cause of GS2 inactivation appears to be S-nitrosylation. However, we cannot rule out the possibility that GS2 inactivation could result from other Cys oxidative processes triggered by reactive nitrogen species. The finding that two isoenzymes sharing a high degree of sequence homology and a remarkably conserved active site fold are differentially modified by NO strengthens the idea that the NO signaling effects are specific under different physiological contexts. In addition to a differential susceptibility of individual GS isoenzymes to NO, the differential localization of the isoenzymes in a specific plant tissue is likely to be implied in the NO-mediated regulation of GS activity.
Tyr nitration is considered a selective process, and typically, only one or two of the Tyr residues present in a protein become preferentially nitrated, depending on the structural environment (Abello et al., 2009). The two M. truncatula GS enzymes MtGS1a and MtGS2a are very similar in their amino acid sequences, so the fact that only MtGS1a is regulated by Tyr nitration is in accordance with the selectivity of the Tyr nitration process and prompted us to search for specific Tyr residues on MtGS1a not present in MtGS2a. This search identified Tyr-108, Tyr-167, and Tyr-263. Previously characterized factors determining the selectivity of Tyr nitration in proteins include the exposure of the aromatic ring at the surface of the protein and the location of the Tyr residue on a loop and in the neighborhood of a negatively charged residue (Souza et al., 1999; Ischiropoulos 2003; Chaki et al., 2009). For the identification of the Tyr residue(s) potentially nitrated in MtGS1a, we analyzed the structural environment within the three-dimensional structure of the protein and identified Tyr-167 to be favorably placed for nitration (Seabra et al., 2009). The residue is located in a solvent-accessible loop, close to the enzyme active site and in close proximity to a basic residue (Lys-137). Tyr-167 establishes a hydrogen bond with Lys-137, which may be important to maintain a correct conformation of the active site and is likely to interfere with the catalytic activity of the enzyme. Experimental evidence that Tyr-167 is a regulatory nitration site is given by the fact that a mutation of this residue to Phe results in a significantly reduced NO-mediated inhibitory effect. These results strongly suggest that Tyr-167 is the only Tyr implicated in the modulation of MtGS1a activity by nitration, but Tyr-263 constitutes an additional nitration target without apparent functional significance, as mutation of this residue to Phe does not affect the susceptibility of the enzyme to NO, in spite of its reduced nitrotyrosine immunoreactivity in response to NO treatment. The nitration sites have been appointed for some plant proteins in a recent proteomic approach, but the functional significance of nitration at a particular residue was not demonstrated (Lozano-Juste et al., 2011). Here, by site-directed mutagenesis, we present unequivocal evidence that Tyr-167 is the regulatory nitration site responsible for enzyme inactivation. To our knowledge, this is the first report that unambiguously identifies a nitration site in a plant enzyme and determines its functional significance.
Contrary to S-nitrosylation, Tyr nitration is generally considered an irreversible process; however, it has been reported that the inhibition of mammalian GS by Tyr nitration could be reversed by a putative denitrase existing in spleen extracts from lipopolysaccharide-treated mouse (Gorg et al., 2007). We tested whether the NO-induced inactivation of MtGS1a could be reversed by incubation of the in vitro nitrated protein with protein extracts from different organs of the plant and even from spleen extracts from lipopolysaccharide-treated mouse (data not shown). All our attempts failed to recover MtGS1a activity; thus, the inhibitory effect of NO on MtGS1a activity does not appear to be reversible. Conversely, the NO-mediated inactivation of MtGS2a could be readily reversed by incubation with reducing agents, further suggesting that the plastid-located enzyme is inactivated by S-nitrosylation.
Due to the interest in the physiological significance of GS nitration for root nodule functioning, we investigated whether GS was nitrated in vivo in root nodules and whether the levels of GS nitration could be related to lower levels of GS activity in vivo. Central to these studies was the development of a sandwich ELISA to quantify the amount of in vivo nitrated GS in root nodules under different physiological conditions. Using this method, the levels of in vivo GS nitration were compared in two situations where nitrogen fixation is impaired, GS activity is known to change, and NO is expected to be produced, namely, in nodules fed with nitrate and in effective versus ineffective nodules. Interestingly, a direct relationship could be established between reduced nodule GS activity, increased GS nitration, and reduced nitrogen fixation activity, strongly suggesting that GS is posttranslationally inactivated by NO-mediated nitration in response to lower nitrogen fixation rates. Further evidence for the effect of in vivo GS nitration on the reduction of GS activity is given by the finding that nodules treated with the NO donor SNP show reduced GS activity, which can be directly correlated with increased GS nitration. Since the GS polypeptide content was not altered by SNP treatment, the reduction in GS activity can be directly attributed to the increased GS nitration levels. Given that the ammonium released by nitrogen fixation is assimilated in the cytosol of the infected cells by GS, it seems reasonable that the enzyme activity is modulated in response to the cell requirements to shut down ammonia assimilation, if it is not being produced.
It is noteworthy that the NO-induced inactivation of GS by nitration appears to be somehow specific in the root nodules. Following nitrate application, the root enzyme behaved in an opposite manner in relation to the nodule enzyme. Root GS activity increased following nitrate treatment, a process accompanied by de novo synthesis of the enzyme. However, in spite of the elevated GS polypeptide content, there were no detectable changes in the amount of nitrated GS in the root, further suggesting a specific regulation of the nodule enzyme by NO. As mentioned above, in M. truncatula roots, GS is mainly composed of a different cytosolic isoenzyme, MtGS1b, that is mainly located in the root cortex, whereas MtGS1a is confined to the root vascular tissues (Carvalho et al., 2000b). It is likely that both the formation of NO at the sites of expression of each individual isoenzyme and the differential susceptibility of the two isoenzymes to NO account for the differential regulation of GS in roots and root nodules.
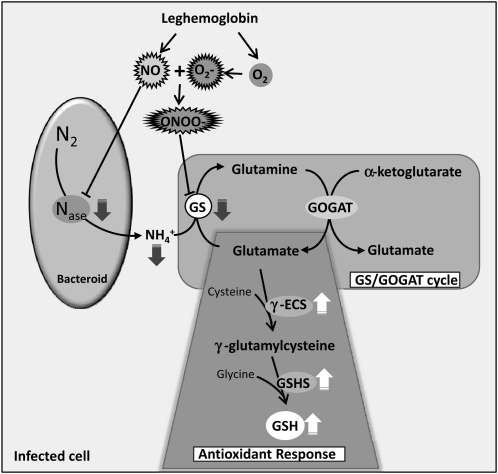
Altogether, the results presented in this work indicate that GS is posttranslationally regulated by Tyr nitration in root nodules of M. truncatula in relation to nitrogen fixation. A model for the regulation of the nodule enzyme is proposed in Figure 11. According to this model, the inhibition of GS activity by Tyr nitration would be directly related to NO-induced nitrogenase inhibition. In view of the fact that elevated levels of NO in root nodules lead to decreased production of ammonium for GS assimilation, the enzyme would be shut down by posttranslational inactivation through Tyr nitration in response to the signal NO, the same signal that shuts down nitrogenase. On the other hand, it is proposed that GS inhibition is involved in the nodule antioxidant response to NO. Glu, a substrate for GS activity, is also the precursor for the synthesis of glutathione (GSH), which is known to be highly abundant in root nodules of several plant species and to play a major role in the antioxidant defense, participating in the ascorbate/GSH cycle (Matamoros et al., 1999, 2003; Becana et al., 2010). Upon NO-mediated GS inhibition, Glu could be channeled for the synthesis of GSH, contributing to neutralize the deleterious effects of reactive nitrogen species. Consistent with this model, the synthesis of the two enzymes involved in GSH production from Glu, γ-glutamyl-Cys synthetase and glutathione synthetase, was found to be up-regulated by NO in root nodules (Matamoros et al., 2003; Innocenti et al., 2007). The proposed model also predicts that leghemoglobin and possibly nonsymbiotic hemoglobins are important players in the process by regulating the levels of both oxygen and NO, controlling in this way the formation of PN (ONOO−). PN is probably the main nitrating agent in vivo and is formed rapidly in the reaction of the superoxide anion (O2−) with NO (Abello et al., 2009, Arasimowicz-Jelonek and Floryszak-Wieczorek, 2011). It is documented that leghemoglobin has the capacity to scavenge NO, forming nitrosylleghemoglobin complexes and contributing in this way to modulate NO bioactivity and protecting nitrogenase from inactivation (Kanayama et al., 1990; Herold and Puppo, 2005; Meakin et al., 2007; Sanchez et al., 2011). According to the proposed model, GS would be involved in the NO signaling response in root nodules by contributing to the antioxidant defenses, and like this the NO signaling events would meet the nodule metabolic pathways to provide an adaptive response to the inhibition of symbiotic nitrogen fixation by reactive nitrogen species.
Figure 11.
Proposed model to integrate the posttranslational regulation of GS by Tyr nitration within the context of root nodule metabolism. Enzymes are as follows: nitrogenase (Nase), GS, Glu synthase (GOGAT), γ-glutamyl-Cys synthetase (γ-ECS), and glutathione synthetase (GSHS). Dark gray arrows indicate down-regulation by NO, and white arrows indicate up-regulation by NO.
In conclusion, this study identifies GS as a target of NO in root nodules of M. truncatula and shows that the enzyme is in vivo posttranslationally regulated by Tyr nitration in relation to nitrogen fixation. These findings are significant, as the identification of the molecular targets of NO is determinant for the assembly of the signal transduction cascade that will ultimately help to unravel NO function. This study also highlighted subtle differences between the regulatory effects of NO on different GS isoenzymes and under different physiological contexts, corroborating the idea that NO signaling is specific in determined physiological backgrounds. This specificity is probably governed by the compartmentalization and dynamics of NO production as well as the differential regulation of individual GS isoenzymes by NO. Particularly interesting is the clear relationship that was observed between reduced nitrogen fixation and the inactivation of nodule GS by Tyr nitration. This NO-mediated posttranslational inactivation, which is possibly related to metabolite channeling to the cell antioxidant defenses, provides a link between NO signaling and nitrogen metabolism in root nodules.
MATERIALS AND METHODS
Materials
PN was prepared as described by Radi et al. (1991). PN concentration was determined spectrophotometrically at 302 nm using ε302 of 1,670 m−1 cm−1. TNM, S-GSNO, and SNP were obtained from Sigma; epicatechin and N-acetyl-imidazole were from Fluka; and iodocetamide was from Bio-Rad.
Plant Material and Growth Conditions
Plants of Medicago truncatula ‘Jemalong J5’ were grown in aeroponic conditions under 16-h-light (22°C)/8-h-dark (19°C) cycles and under a light intensity of 150 to 200 μmol m−2 s−1 in the growth medium supplemented with ammonium nitrate as described by Lullien et al. (1987). For nodule induction, the growth medium was replaced with fresh medium lacking a nitrogen source 3 d before inoculation with either the wild-type Sinorhizobium meliloti effective wild-type strain Rm1021 pXLGD4 RCR 2011(GMI 151) or the ineffective fixJ S. meliloti mutant (GMI 347). Nodules were harvested at 21 d after infection. For the experiment with nitrate, nodulated plants (19 d after inoculation) were aeroponically grown for 2 d in a growth medium containing 10 mm NH4NO3 before harvesting. For the treatment with SNP, mature nodules of M. truncatula were infiltrated with a solution containing 1 or 10 mm SNP in phosphate buffer, pH 7.0, under vacuum for 10 min and incubated at room temperature for 3 h. All plant material was immediately frozen in liquid nitrogen and stored at –80°C.
Protein Extraction from Plant Tissues
Roots or root nodules of M. truncatula were homogenized at 4°C with a mortar and pestle with 2 volumes of extraction buffer (phosphate-buffered saline [PBS], pH 7.4, containing 0.05% Triton X-100 and 1 mm phenylmethylsulfonyl fluoride), and the homogenates were centrifuged at 13,000g for 20 min at 4°C.
Expression and Purification of Recombinant GS in Escherichia coli
The M. truncatula GS isoenzymes MtGS1a and MtGS2a were produced in E. coli BL21-CodonPlus (DE3)-RP cells harboring previously described gene constructs pET28a-MtGS1a and pET28a-MtGS2a encoding N-terminally His-tagged fusion proteins (Lima et al., 2006b).
Expression was induced with isopropylthio-β-galactoside (final concentration, 1 mm) at midexponential growth (optical density at 600 nm = 0.5) and proceeded for 3 to 5 h at 37°C. The cells were harvested by centrifugation at 2,800g, resuspended in 0.02 m potassium phosphate, pH 7.4, 0.01 m magnesium sulfate, 0.005 m Glu, 0.5 m NaCl, and 0.02 m imidazole (buffer A), disrupted by sonication, and centrifuged (60 min, 36,700g, 4°C) to remove cell debris. The crude protein extract was filtered through a 5-μm low-protein-binding filter and loaded onto a 5-mL nickel Sepharose column (GE Healthcare) equilibrated with buffer A. Elution of the bound fusion protein was achieved with buffer A supplemented with 0.23 m imidazole. MtGS1a-containing fractions were pooled and dialyzed against 0.02 m potassium phosphate, pH 7.4, 0.005 m Glu, and 0.01 m magnesium sulfate and further purified by size-exclusion chromatography on a Sephacryl S-300 16/60 column (GE Healthcare) equilibrated in the same buffer. The fractions containing purified MtGS1a were pooled and concentrated on a centrifugal concentration device with a 10-kD molecular weight cutoff membrane.
Site-Directed Mutagenesis
The Quick-Change Site-Directed Mutagenesis Kit (Agilent/Stratagene) was used to mutate Tyr-167 and Tyr-263 to Phe in M. truncatula MtGS1a protein. Mutations were generated by PCR amplification using the pET28a-MtGS1a plasmid as template and the appropriate primers containing the desired mutations. The sense sequences of the primers used were as follows: Y167F, 5′-GCTGATAAAGCCTTTGGTCGTGACATTGTCGACGCACATTAC-3′, and Y363F, 5′-GGAGGCTTTGAAATCATCAAGAAGGCCATTGAAAAACTTGGATTG-3′, where the underlined bases represent the nucleotide changes to mutate Tyr (codon TAT) to Phe (codon TTT) and to create a SalI restriction site (boldface) or eliminate the HindIII restriction site, respectively, to facilitate the initial screening for mutations. The PCR-amplified products were transformed into XL1 Blue competent cells, and the plasmid DNA was extracted and sequenced to confirm production of the desired mutations.
Determination of GS Activity
GS activity was determined using the transferase assay (Cullimore and Sims, 1980). One unit of transferase activity is equivalent to 1 μmol min−1 γ-glutamyl hydroxamate produced at 30°C. Activity data were expressed as means ± sd of at least three independent experiments, with triplicate technical assays for each experiment. The specific activity of the E. coli-produced purified GS isoenzymes is 144 μmol min−1 mg−1 protein for MtGS1a and 40 μmol min−1 mg−1 protein for MtGS2.
In Vitro GS Nitration
Purified GS (2.5 μg) was diluted in 0.02 m potassium phosphate buffer and incubated with either PN or TNM at concentrations of 0 to 100 μm in a final volume of 100 μL. Treatment with PN (diluted in 0.001 n NaOH) was performed for 10 min at room temperature, pH 7.4. Control assays were performed by incubating the enzyme with 50 μm decomposed PN; for this purpose, PN was added to the buffer 10 min before addition of the enzyme. The addition of PN to the buffer system did not lead to a pH change. Treatments with TNM (diluted in ethanol) were performed for 1 h at room temperature, pH 8. Control assays were performed by adding equivalent volumes of ethanol. To evaluate the protective effect of epicatechin on GS nitration, the purified enzymes were incubated for 30 min at 30°C with epicatechin at concentrations of 0 to 30 μm, prior to the addition of the nitrating agents. GS activity was assessed in the absence of nitrating reagents to evaluate the effect of epicatechin on enzyme activity. Aliquots were taken for the determination of enzyme activity and in some cases for western-blot analysis.
Reaction of GS with S-GSNO, Hydrogen Peroxide, Iodoacetamide, and N-Acetyl-Imidazole
We assessed the effects of several chemical reagents known to modify Cys (S-GSNO, hydrogen peroxide, and iodoacetamide) and Tyr residues (N-acetyl-imidazole). To this end, 2.5 μg of purified GS, diluted in 0.02 m potassium phosphate buffer (pH 7.4), was incubated with various concentrations of each chemical reagent for 30 min at 30°C. GS activity was assayed as described above. To assess the contribution of thiol oxidation to the PN- and TNM-induced GS inactivation, purified His-tagged MtGS1a was treated with 500 μm iodoacetamide for 30 min at 30°C, prior to incubation with 50 μm of each of the NO donor reagents.
Protein Determination, SDS-PAGE, and Western-Blot Analysis
Soluble protein concentration was measured by the Coomassie dye-binding assay (Bio-Rad) using bovine serum albumin as a standard. Soluble protein extracts were separated by 12.5% (w/v) SDS-PAGE in nonreducing conditions (using Laemmli’s sample buffer without β-mercaptoethanol) and electroblotted onto nitrocellulose membranes (Schleicher & Schuell) using a Criterion Blotter from Bio-Rad.
Immunodetection of nitrotyrosine was performed using a biotinylated monoclonal antibody against 3-nitrotyrosine (Cayman Chemicals) diluted 1:3,000, and detection of the polypeptides was performed using NeutrAvidin-HRP-conjugate (Invitrogen) diluted 1:5,000. GS immunodetection was carried out using rabbit serum antibody raised against a GS isoform from Phaseolus vulgaris (kindly provided by Dr. Julie Cullimore from INRA Toulouse; Cullimore and Miflin, 1984) and a secondary goat anti-rabbit peroxidase-conjugated antibody (Vector Laboratories). The immunocomplexes were detected by chemiluminescence, using the enhanced chemiluminescence (GE Healthcare, Lifesciences) detection system, with a photographic film. Representative blots of at least three independent experiments are shown in each of the figures and in Supplemental Figure S1. GS protein quantification was performed using the Molecular Imager GS800 calibrated densitometer (Bio-Rad) and Quantity 1-D Analysis software (Bio-Rad).
Quantification of Nitrated GS by ELISA
A microtiter plate (F8 Maxisorp Nunc-Immuno module; Nunc) was coated with the anti-GlnA antibody (Cullimore and Miflin, 1984), diluted in PBS (50 μL well−1), and incubated overnight at 4°C. After two washings with a washing buffer (PBS, pH 7.4, and 0.5% Tween 20), the plate was blocked with a blocking buffer (PBS, pH 7.4, and 3% bovine serum albumin; 100 μL well−1) for 2 h at room temperature. The blocking buffer was removed, the plant extracts were diluted in extraction buffer to 10 μg in 50 μL, and the calibration standard solution was added to the plate (50 μL well−1) and incubated for 4 h at room temperature. All experiments were performed in duplicate. After five washings, a diluted biotin-labeled monoclonal antibody against 3-nitrotyrosine (100 μL well−1) was added to the plate and incubated for 2 h at room temperature. After washing, a NeutrAvidin-HRP-conjugate (Invitrogen) was added to the plate and incubated for 1h at room temperature.
The plates were washed four times with wash buffer before adding 100 μL well−1 p-nitrophenyl phosphate substrate (Sigma Fast; Sigma-Aldrich). The plates were incubated in the dark for 30 min at room temperature before stopping the reaction by the addition of 100 μL well−1 0.25 m H2SO4. The absorbance was measured at 405 nm using a microplate reader (μQuant Microplat Spec; Biotek). Standard curves were established by serial dilutions (1–5 ng) of in vitro nitrated GS per well. For this purpose, the purified enzyme was preincubated with 100 μm PN for 15 min at room temperature. Since it is not possible to ensure that GS is nitrated to completion in the in vitro nitration assays of the standards used for the quantification, GS nitration values are presented as arbitrary units in relation to the relative amount of cytosolic GS polypeptides quantified by densitometry.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers Y10267 and AY225150 for MtGS1a and MtGS2a, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Western-blot analysis of crude protein extracts from root nodules.
Acknowledgments
We gratefully acknowledge Julie Cullimore (Laboratoire des Interaction Plantes-Microrganismes) for helpful suggestions and discussions and Pedro Pereira (Instituto de Biologia Molecular e Celular da Universidade do Porto) and João Morais Cabral (Instituto de Biologia Molecular e Celular da Universidade do Porto) for invaluable help on the analysis and interpretation of the structural features related to GS Tyr nitration.
References
- Abello N, Kerstjens HAM, Postma DS, Bischoff R. (2009) Protein tyrosine: selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J Proteome Res 8: 3222–3238 [DOI] [PubMed] [Google Scholar]
- Arasimowicz-Jelonek M, Floryszak-Wieczorek J. (2011) Understanding the fate of peroxynitrite in plant cells: from physiology to pathophysiology. Phytochemistry 72: 681–688 [DOI] [PubMed] [Google Scholar]
- Baudouin E, Pieuchot L, Engler G, Pauly N, Puppo A. (2006) Nitric oxide is formed in Medicago truncatula-Sinorhizobium meliloti functional nodules. Mol Plant Microbe Interact 19: 970–975 [DOI] [PubMed] [Google Scholar]
- Becana M, Matamoros MA, Udvardi M, Dalton DA. (2010) Recent insights into antioxidant defenses of legume root nodules. New Phytol 188: 960–976 [DOI] [PubMed] [Google Scholar]
- Berlet BS, Friguet B, Yim MB, Chock PB, Stadtman ER. (1996) Peroxynitrite-mediated nitration of tyrosine residue in Escherichia coli glutamine synthetase mimics adenylylation: relevance of signal transduction. Proc Natl Acad Sci USA 96: 7809–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson-Bard A, Pugin A, Wendehenne D. (2007) New insights into nitric oxide signaling in plants. Annu Rev Plant Biol 59: 21–39 [DOI] [PubMed] [Google Scholar]
- Carvalho H, Lescure N, de Billy F, Chabaud M, Lima L, Salema R, Cullimore J. (2000a) Cellular expression and regulation of the Medicago truncatula cytosolic glutamine synthetase genes in root nodules. Plant Mol Biol 42: 741–756 [DOI] [PubMed] [Google Scholar]
- Carvalho H, Lima L, Salema R, Cullimore J. (2000b) Differential expression of the two cytosolic glutamine synthetase genes in various organs of Medicago truncatula. Plant Sci 159: 301–312 [DOI] [PubMed] [Google Scholar]
- Cecconi D, Orzetti S, Vandelle E, Rinalducci S, Zolla L, Delledone M. (2009) Protein nitration during defense response in Arabidopsis thaliana. Electrophoresis 30: 2460–2468 [DOI] [PubMed] [Google Scholar]
- Chaki M, Valderrama R, Fernandez-Ocana AM, Carreras A, López-Jaramillo J, Luque F, Palma JM, Pedrejas JR, Begara-Morales JC, Sánchez-Calvo B, et al. (2009) Protein targets of tyrosine nitration in sunflower (Helianthus annuus L.) hypocotyls. J Exp Bot 60: 4221–4234 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Chaki M, Leterrier M, Barroso JB. (2009) Protein tyrosine nitration: a new challenge in plants. Plant Signal Behav 4: 920–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cueto M, Hernández-Pereira O, Martin R, Bentura ML, Rodrigo J, Lamas S, Golvano MP. (1996) Presence of nitric oxide synthetase activity in roots and nodules of Lupinus alba. FEBS Lett 398: 159–164 [DOI] [PubMed] [Google Scholar]
- Cullimore JV, Miflin BJ. (1984) Immunological studies on glutamine synthetase using antisera raised to the two plant forms of the enzyme from Phaseolus vulgaris root nodules. J Exp Bot 35: 581–587 [Google Scholar]
- Cullimore JV, Sims AP. (1980) An association between photorespiration and protein catabolism: studies in Chlamydomonas. Planta 150: 392–396 [DOI] [PubMed] [Google Scholar]
- Del Giudice J, Cam Y, Damiani I, Fung-Chat F, Meilhoc E, Bruand C, Brouquisse R, Puppo A, Boscari A. (2011) Nitric oxide is required for an optimal establishment of the Medicago truncatula-Sinorhizobium meliloti symbiosis. New Phytol 191: 405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Almassy RJ, Janson CA, Chapman MS, Suh SW, Cascio D, Smith WW. (1987) Some evolutionary relationships of the primary biological catalysts glutamine synthetase and Rubisco. Cold Spring Harb Symp Quant Biol 52: 483–490 [DOI] [PubMed] [Google Scholar]
- Finnemann J, Schjoerring JK. (2000) Post-translational regulation of cytosolic glutamine synthetase by reversible phosphorylation and 14-3-3 protein interaction. Plant J 24: 171–181 [DOI] [PubMed] [Google Scholar]
- Gorg B, Qvartskhava N, Voss P, Grune T, Haussinger D, Schliess F. (2007) Reversible inhibition of mammalian glutamine synthetase by tyrosine nitration. FEBS Lett 581: 84–90 [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Fernie AR, Kaiser WM, van Dongen JT. (2011) On the origins of nitric oxide. Trends Plant Sci 16: 160–168 [DOI] [PubMed] [Google Scholar]
- Herold S, Puppo A. (2005) Oxyleghemoglobin scavengers nitrogen monoxide and peroxynitrite: a possible role in functioning nodules. J Biol Inorg Chem 10: 935–945 [DOI] [PubMed] [Google Scholar]
- Hoelzle I, Finner JJ, McMullen MD, Streeter JG. (1992) Induction of glutamine synthetase activity in nonnodulated roots of Glycine max, Phaseolus vulgaris and Pisum sativum. Plant Physiol 100: 525–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horchani F, Prévot M, Boscari A, Evangelisti E, Meilhoc E, Bruand C, Raymond P, Boncompagni E, Ascgi-Smiti S, Puppo A, et al. (2011) Both plant and bacterial nitrate reductase contribute to nitric oxide production in Medicago truncatula nitrogen-fixing nodules. Plant Physiol 155: 1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti G, Pucciariello C, Le Gleuher M, Hopkins J, de Stefano M, Delledonne M, Puppo A, Baudouin E, Frendo P. (2007) Glutathione synthesis is regulated by nitric oxide in Medicago truncatula roots. Planta 225: 1597–1602 [DOI] [PubMed] [Google Scholar]
- Ischiropoulos H. (2003) Biological selectivity and functional aspects of protein tyrosine nitration. Biochem Biophys Res Commun 305: 776–783 [DOI] [PubMed] [Google Scholar]
- Ishida H, Anzawa D, Kokubun N, Marino A, Mae T. (2002) Direct evidence for non-enzymatic fragmentation of chloroplastic glutamine synthetase by reactive oxygen species. Plant Cell Environ 2: 625–631 [Google Scholar]
- Kanayama Y, Watanabe I, Yamamoto Y. (1990) Inhibition of nitrogen fixation in soybean plants supplied with nitrate. I. Nitrite accumulation and formation of nitrosylleghemoglobin in nodules. Plant Cell Physiol 31: 341–346 [Google Scholar]
- Kato K, Kanahama K, Kanayama Y. (2010) Involvement of nitric oxide in the inhibition of nitrogenase activity by nitrate in Lotus root nodules. J Plant Physiol 167: 238–241 [DOI] [PubMed] [Google Scholar]
- Kato K, Okamura Y, Kanahama K, Kanayama Y. (2003) Nitrate-independent expression of plant nitrate reductase in Lotus japonicus root nodules. J Exp Bot 388: 1685–1690 [DOI] [PubMed] [Google Scholar]
- Keyster M, Klein A, Ludidi N. (2010) Endogenous NO levels regulate nodule functioning: potential role of cGMP in nodule functioning? Plant Signal Behav 5: 1679–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea P, Miflin B. (2011) Nitrogen assimilation and its relevance for crop improvement. Foyer C, Zang H, , Nitrogen Metabolism in the Post-Genomic Era. Plant Annual Reviews, Vol 42. Wiley-Blackwell, London, pp 1–40 [Google Scholar]
- Leach J, Keyster M, Du Plessis M, Ludidi N. (2010) Nitric oxide synthetase activity is required for development of functional nodules in soybean. J Plant Physiol 167: 1584–1591 [DOI] [PubMed] [Google Scholar]
- Lima L, Seabra A, Melo P, Cullimore J, Carvalho H. (2006a) Phosphorylation and subsequent interaction with 14-3-3 protein regulates plastid glutamine synthetase in Medicago truncatula. Planta 223: 558–567 [DOI] [PubMed] [Google Scholar]
- Lima L, Seabra A, Melo P, Cullimore J, Carvalho H. (2006b) Post-translational regulation of cytosolic glutamine synthetase of Medicago truncatula. J Exp Bot 57: 2751–2761 [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J. (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol 137: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Juste J, Colom-Moreno R, León J. (2011) In vivo protein tyrosine nitration in Arabidopsis thaliana. J Exp Bot 62: 3501–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lullien V, Barker DG, da Lajudie P, Huguet T. (1987) Plant gene expression in effective and ineffective root nodules of alfalfa (Medicago sativa). Plant Mol Biol 9: 469–478 [DOI] [PubMed] [Google Scholar]
- Man H-M, Kaiser WM. (2001) Increased glutamine synthetase activity and changes in amino acid pools in leaves treated with 5-aminoimidazole-4-carboxyamide ribonucleoside (AICAR). Physiol Plant 111: 291–296 [DOI] [PubMed] [Google Scholar]
- Matamoros MA, Dalton DA, Ramos J, Clemente MR, Rubio MC, Becana M. (2003) Biochemistry and molecular biology of antioxidants in the rhizobia-legume symbiosis. Plant Physiol 133: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamoros MA, Moran JF, Iturbe-Ormaetxe I, Rubio MC, Becana M. (1999) Glutathione and homoglutathione synthesis in legume root nodules. Plant Physiol 121: 879–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu C, Moreau S, Frendo P, Puppo A, Davies MJ. (1998) Direct detection of radicals in intact soybean nodules: presence of nitric oxide-leghemoglobin complexes. Free Radic Biol Med 24: 1242–1249 [DOI] [PubMed] [Google Scholar]
- Meakin GE, Bueno E, Jepson B, Bedmar EJ, Richardson DJ, Delgado MJ. (2007) The contribution of bacteroidal nitrate and nitrite reduction to the formation of nitrosylleghaemoglobin complexes in soybean root nodules. Microbiology 153: 411–419 [DOI] [PubMed] [Google Scholar]
- Melo PM, Lima LM, Santos IM, Carvalhjo HG, Cullimore JV. (2003) Expression of plastid-located glutamine synthetase of Medicago truncatula: accumulation of the precursor in root nodules reveals an in vivo control at the level of protein import into plastids. Plant Physiol 132: 390–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa S, de Dios Alché J, Bedmar EJ, Delgado MJ. (2004) Expression of nir, nor and nos desnitrifiction genes from Bradyrhizobium japonicum in soybean root nodules. Physiol Plant 120: 205–211 [DOI] [PubMed] [Google Scholar]
- Neil S, Bright J, Desikan R, Hancock J, Harrison J, Wilson I. (2008) Nitric oxide evolution and perception. J Exp Bot 59: 25–35 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Murray JD, Poole PS, Downie JA. (August 11, 2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet http://dx.doi.org/10.1146/annurev-genet-110410-132549 [DOI] [PubMed] [Google Scholar]
- Ortega JL, Roche D, Sengupta-Gopalan C. (1999) Oxidative turnover of soybean root glutamine synthetase: in vitro and in vivo studies. Plant Physiol 119: 1483–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega JL, Temple SJ, Sengupta-Gopalan C. (2001) Constitutive overexpression of cytosolic glutamine synthetase (GS1) gene in transgenic alfalfa demonstrates that GS1 may be regulated at the level of RNA stability and protein turnover. Plant Physiol 126: 109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Carrilo N, Valle EM. (1999) The role of photosynthetic electron transport in the oxidative degradation of chloroplastic glutamine synthetase. Plant Physiol 121: 471–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly N, Pucciariello C, Mandon K, Innocenti G, Jamet A, Baudouin E, Hérouart D, Frendo P, Puppo A. (2006) Reactive oxygen and nitrogen species and glutathione: key players in the legume-Rhizobium symbiosis. J Exp Bot 57: 1769–1776 [DOI] [PubMed] [Google Scholar]
- Radi R. (2004) Nitric oxide, oxidants, and protein tyrosine nitration. Proc Natl Acad Sci USA 23: 4003–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radi R, Beckman JS, Bush KM, Freeman BA. (1991) Peroxynitrite oxidation of sulfhydryls: the cytotoxic potential of superoxide and nitric oxide. J Biol Chem 266: 4244–4250 [PubMed] [Google Scholar]
- Riedel J, Tischner R, Mack G. (2001) The chloroplastic glutamine synthetase (GS-2) of tobacco is phosphorylated and associated with 14-3-3 proteins inside the chloroplast. Planta 213: 396–401 [DOI] [PubMed] [Google Scholar]
- Sanchez C, Cabrera JJ, Gates AJ, Bedmar EJ, Richarrdson DJ, Delgado MJ. (2011) Nitric oxide detoxification in the rhizobia-legume symbiosis. Biochem Soc Trans 39: 184–188 [DOI] [PubMed] [Google Scholar]
- Sanchez C, Gates AJ, Meakin GE, Uchiumi T, Girard L, Richarrdson DJ, Bedmar EJ, Delgado MJ. (2010) Production of nitric oxide and nitrosylleghemoglobin complexes in soybean nodules in response to flooding. Mol Plant Microbe Interact 23: 702–711 [DOI] [PubMed] [Google Scholar]
- Schroeder P, Klotz L-O, Buchczyk DP, Sadik CD, Schewe T, Sies H. (2001) Epicatechin selectively prevents nitration but not oxidation reactions of peroxynitrite. Biochem Biophys Res Commun 285: 782–787 [DOI] [PubMed] [Google Scholar]
- Seabra AR, Carvalho H, Pereira PJB. (2009) Crystallization and preliminary crystallographic characterization of glutamine synthetase from Medicago truncatula. Acta Crystallogr Sect F Struct Biol Cryst Commun 65: 1309–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabra AR, Vieira CP, Cullimore JV, Carvalho HG. (2010) Medicago truncatula contains a second gene encoding a plastid located glutamine synthetase exclusively expressed in developing seeds. BMC Plant Biol 10: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda Y, Shimoda-Sasakura F, Kucho K, Kanamori M, Suzuki A, Abe M, Higashi S, Uchhiumi T. (2009) Overexpression of class 1 plant hemoglobin genes enhances symbiotic nitrogen fixation activity between Mesorhizobium loti and Lotus japonicus. Plant J 57: 254–263 [DOI] [PubMed] [Google Scholar]
- Souza JM, Daikhin E, Yudkoff M, Raman CS, Ischiropoulos H. (1999) Factors determining the selectivity of protein tyrosine nitration. Arch Biochem Biophys 371: 169–178 [DOI] [PubMed] [Google Scholar]
- Stanford AC, Larsen K, Barker DG, Cullimore JV. (1993) Differential expression within the glutamine synthetase gene family of the model legume Medicago truncatula. Plant Physiol 103: 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple SJ, Bagga S, Sengupta-Gopalan C. (1998) Down-regulation of specific members of the glutamine synthetase gene family by antisense RNA technology. Plant Mol Biol 37: 535–547 [DOI] [PubMed] [Google Scholar]
- Temple SJ, Heard J, Kunjibettu S, Roche D, Sengupta-Gopalan C. (1996) Total glutamine synthetase activity during soybean nodule development is controlled at the level of transcription and holoprotein turnover. Plant Physiol 112: 1723–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Ruby EG. (2011) The roles of NO in microbial symbioses. Cell Microbiol 13: 518–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I, Neill SJ, Hancock JT. (2008) Nitric oxide synthesis and signaling in plants. Plant Cell Environ 31: 622–631 [DOI] [PubMed] [Google Scholar]